Abstract
The eukaryotic cell cycle is typically divided into distinct phases with cytokinesis immediately following mitosis. To ensure proper cell division, each phase is tightly coordinated via feedback controls named checkpoints. During its asexual replication cycle, the malaria parasite Plasmodium falciparum undergoes multiple asynchronous rounds of mitosis with segregation of uncondensed chromosomes followed by nuclear division with intact nuclear envelope. The multi-nucleated schizont is then subjected to a single round of cytokinesis that produces dozens of daughter cells called merozoites. To date, no cell cycle checkpoints have been identified that regulate the Plasmodium spp. mode of division. Here, we identify the Plasmodium homologue of the Mini-Chromosome Maintenance Complex Binding Protein (PfMCMBP), which co-purified with the Mini-Chromosome Maintenance (MCM) complex, a replicative helicase required for genomic DNA replication. By conditionally depleting PfMCMBP, we disrupt nuclear morphology and parasite proliferation without causing a block in DNA replication. By immunofluorescence microscopy, we show that PfMCMBP depletion promotes the formation of mitotic spindle microtubules with extensions to more than one DNA focus and abnormal centrin distribution. Strikingly, PfMCMBP-deficient parasites complete cytokinesis and form aneuploid merozoites with variable cellular and nuclear sizes. Our study demonstrates that the parasite lacks a robust checkpoint response to prevent cytokinesis following aberrant karyokinesis.
1 |. INTRODUCTION
The eradication of human malaria would save more than 400,000 lives annually and prevent approximately 200 million cases of the disease each year (World Health Organization, 2018). Malaria is caused by unicellular protozoan parasites of the Plasmodium genus with P. falciparum being the most virulent of the five Plasmodium parasites that readily infect humans. Despite major progress to decrease the burden of malaria, emerging resistance of the parasite to current medications and the lack of an effective vaccine remain obstacles to eradication. Thus, the identification of novel targets for new therapeutics to block parasite replication is crucial. Clinical malaria results from the proliferation of the parasite in human red blood cells, where it replicates via schizogony, a division mode that can be separated into a replicative phase and a budding phase (Francia & Striepen, 2014). During the replicative phase, the parasite undergoes multiple asynchronous rounds of mitosis with segregation of uncondensed chromosomes followed by nuclear division with an intact nuclear envelope (White & Suvorova, 2018). The budding phase occurs when the multi-nucleated cell is subjected to a semi-synchronous round of karyokinesis and segmentation of dozens of daughter cells known as merozoites (Rudlaff, Kraemer, Marshman, & Dvorin, 2020; Rudlaff, Kraemer, Streva, & Dvorin, 2019). The progression and regulation of these two phases in Plasmodium are poorly understood. Given the peculiarity of cell division in Plasmodium parasites, understanding the molecular mechanisms that drive and regulate these processes in the parasite could unveil novel targets for the treatment of malaria.
The typical eukaryotic cell cycle is divided into multiple phases during which the cell grows (interphase), replicates its DNA (S phase), and divides its nucleus with DNA segregation and mitosis (M phase) - usually followed immediately by cytokinesis. To ensure proper division of a cell, each phase is tightly coordinated via a system of feedback controls named checkpoints (reviewed in [Morgan, 2007]). The first decision point for a cell to commit to divide happens at the interval between mitosis and initiation of DNA replication referred to as the G1 checkpoint. An intra-S-phase checkpoint monitors for damaged DNA present during duplication (Iyer & Rhind, 2017). After DNA replication and prior to initiating mitosis, the G2 checkpoint prevents cell cycle progression in presence of damaged DNA or incomplete DNA replication. Finally, the M checkpoint, also known as the spindle assembly checkpoint (SAC), monitors for proper spindle attachment prior to anaphase (Lara-Gonzalez, Westhorpe, & Taylor, 2012). To date, none of the expected cell cycle checkpoints have been identified that regulate Plasmodium spp. proliferation in the asexual blood stage (Robbins, Absalon, Streva, & Dvorin, 2017).
In Plasmodium spp., the microtubule organising center (MTOC) that nucleates spindle microtubules and drives mitotic spindle formation is embedded in the nuclear envelope, which remains intact throughout the cell cycle (Arnot, Ronander, & Bengtsson, 2011; Gerald, Mahajan, & Kumar, 2011). During each round of DNA replication, each sister chromatid of the uncondensed chromosomes must attach to one of the duplicated MTOCs to be accurately segregated. In many eukaryotic cells, the SAC is the surveillance mechanism that ensures amphitelic attachment of kinetochores to spindle microtubules prior to cell cycle progression into anaphase (Foley & Kapoor, 2013). Remarkably, whether a SAC ensures proper DNA segregation in Plasmodium spp. has not been fully established. Previous studies have used small molecule inhibitors of microtubules, DNA polymerase, and cyclin-dependent kinases and monitored DNA replication in P. falciparum (Naughton & Bell, 2007; Usanga, O’Brien, & Luzzato, 1986). Parasites treated with inhibitors of microtubule polymerisation (vinblastine, vincristine, or colcemid) arrest as trophozoites, likely prior to first nuclear division, demonstrating that microtubules are critical for nuclear division in P. falciparum (Usanga et al., 1986). In a separate study, parasites treated with vinblastine were observed to arrest development but continue with DNA replication, suggesting the absence of a conventional G1 checkpoint (Naughton & Bell, 2007). Parasites treated with the cyclin-dependent kinase inhibitor roscovitine had impaired DNA replication and nuclear division (Naughton & Bell, 2007). While these studies confirm the presence of some similarities between model eukaryotes and P. falciparum, many regulators of the cell cycle in the parasite have not been found experimentally or bioinformatically.
The Mini-Chromosome Maintenance (MCM) complex is a replicative helicase made of six subunits (MCM2-7) that forms a hexameric ring. Highly conserved among eukaryotes, the MCM complex is required to form and elongate the replication fork (Bochman & Schwacha, 2009). While no MCM complex proteins have been functionally characterised in Plasmodium, the MCM protein family has been bioinformatically identified (Patterson et al., 2006). In P. falciparum, the molecular mechanism of DNA replication licensing, which ensures a single complete round of DNA replication prior to nuclear division, remains poorly understood. The human MCM Complex Binding Protein (MCMBP) of the MCM complex was first identified by tandem affinity purification (Sakwe, Nguyen, Athanasopoulos, Shire, & Frappier, 2007). Interestingly, recent works showed that MCMBP depletion or overexpression in plants (Takahashi et al., 2010), yeast (Ding & Forsburg, 2011), human cells (Jagannathan, Sakwe, Nguyen, & Frappier, 2012), and protozoans such as Trypanosoma brucei (Kim, 2019; Kim, Park, Gunzl, & Cross, 2013) results in common phenotypic traits, including defective sister chromatid cohesion, abnormal nuclear morphology, and the appearance of zoid cells with an accumulation of multinucleated cells with poly-lobed nuclei, which led to the activation of the G2 checkpoint. Here we identify the P. falciparum MCMBP ortholog (PfMCMBP/PF3D7_1412100) and show that by association with MCM proteins, it is likely a component of the replisome in Plasmodium. By utilising a conditional knockdown of PfMCMBP, we demonstrate its essential role during the asexual replication of P. falciparum. Following depletion of PfMCMBP, schizont-stage parasites displayed an abnormal nuclear morphology and MTOC-like ectopic foci with extended spindle microtubules that attached to chromosomes in spatially separated clusters of DNA. Despite the uneven segregation of chromosomes, PfMCMBP-depleted schizonts segmented and formed aneuploid merozoites with varied nuclear and cellular body size. Thus, our study demonstrates that P. falciparum parasites lack a robust cell cycle checkpoint to prevent cytokinesis after asymmetric karyokinesis has produced nuclei with aberrant karyotype.
2 |. RESULTS
2.1 |. PfMCMBP homologue is associated with the MCM protein complex
Through a BLAST search of the P. falciparum genome with the human MCMBP sequence (Aurrecoechea et al., 2009; Takahashi et al., 2010), we identified PF3D7_1412100 as the Plasmodium falciparum MCMBP homologue. Phylogenetic analysis of MCMBPs from diverse eukaryotic organisms largely recapitulates established phylogenetic trees (Burki, 2014), suggesting that horizontal gene transfer has not occurred (Figures S1 and S2). Interestingly, the budding yeast S. cerevisiae does not code for MCMBP (Santosa, Martha, Hirose, & Tanaka, 2013). In silico analysis of the primary PfMCMBP sequence, together with orthologous proteins from other model organisms, revealed four regions with similarity to the MCM replisome factor domain (IPR019140) (Figure 1a) (Aurrecoechea et al., 2009; Mitchell et al., 2019). This domain has alternating regions of conservation and low complexity — and has been identified as a common feature of other MCMBPs (Mitchell et al., 2019; Takahashi et al., 2010). The four identified regions within PfMCMBP likely represent a single discontinuous domain because the full domain is approximately 600 amino acids (Mitchell et al., 2019).
FIGURE 1.
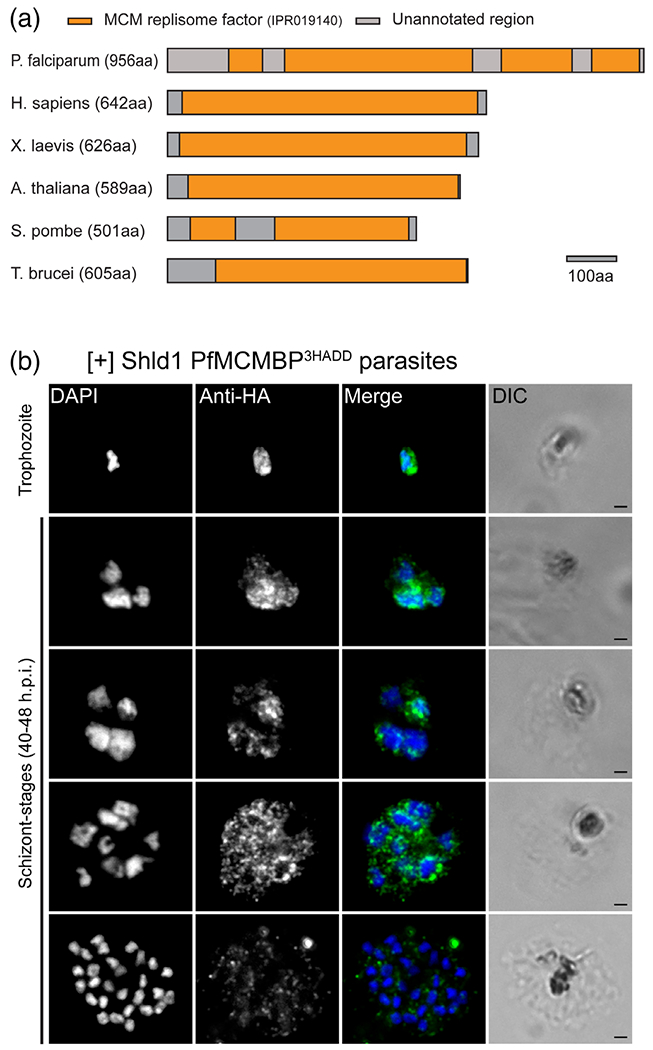
Identification of the Plasmodium homologue of the Mini-Chromosome Maintenance Complex Binding Protein (PfMCMBP) homologue and expression profile during asexual life cycle. (A) A diagram of the PfMCMBP highlights the position of the multiple Mini-Chromosome Maintenance replisome factor domains in PfMCMBP and among other species where MCMBP orthologue has characterised. Scale bar at 100 amino acids (B) Representative pictures from [+] Shld1 PfMCMBP3HADDparasites after PFA-fixation and probing with a rat anti-HA antibody (1:50) and goat anti-rat antisera conjugated to Alexa 488 (1:1000). Images were obtained on a Nikon E800 microscope using the 100X objective. Scale bar is 1 μm
To investigate the role of PfMCMBP during the asexual life cycle of P. falciparum parasites, we fused three copies of the hemagglutinin epitope (HA) tag with a destabilisation domain (DD) to the carboxy-terminus of the endogenous PfMCMBP in the 3D7 strain by homologous recombination, generating the 3D7-PfMCMBP3HADD transgenic parasite line (Figure S3). In the presence of the small molecule Shield-1 (Shld1), PfMCMBP3HADD protein is stabilised, while in absence of Shld1, the protein is rapidly degraded (Banaszynski, Chen, Maynard-Smith, Ooi, & Wandless, 2006; Dvorin et al., 2010). This system allowed us to inducibly regulate PfMCMBP level in the parasite. We assessed the localization of the protein throughout the parasite asexual replication cycle by immunofluorescence microscopy (Figure 1b). Through the intraerythrocytic development cycle, we observed a strong fluorescent signal with multiple cytoplasmic foci that also overlapped with the 4’,6’-diamidino-2-phenylindole (DAPI)-stained region of the nuclei from trophozoite-stage (~ 24–32 hr post invasion [h.p.i.]) to unsegmented schizont-stage (40–46 h.p.i.) (Figure 1b and Figure S4). Interestingly, in fully segmented schizonts, PfMCMBP fluorescent signal was low and equivalent to background (Figure S5). To confirm the timing of PfMCMBP signal loss, we monitored the localization of PfMCMBP and PfMORN1, a component of the basal complex (Ferguson et al., 2008; Rudlaff et al., 2019), simultaneously. Prior to segmentation, when PfMORN1 is not yet visible by immunofluorescence, we observed a strong fluorescent signal for PfMCMBP in the cytoplasm as well as bright foci near the nuclei (Figure S6). As segmentation progressed, PfMORN1 is visualised as small rings at the apical end of the parasites, which maintain high PfMCMBP expression. As segmentation progressed further, the PfMORN1 rings enlarged and PfMCMBP fluorescent signal decreased. Finally, once PfMORN1 had reached the basal end of the parasite, marking the end of segmentation, PfMCMBP was no longer detectable (Figure S6). In addition, no fluorescent signal was detected in ring stage (Figure S4). Altogether, PfMCMBP is present during the replicative phase of schizogony and is undetectable during the budding phase (i.e., during segmentation).
To identify potential PfMCMBP-interacting proteins, we performed co-immunoprecipitation in the PfMCMBP3HADD transgenic line followed by unbiased mass spectrometry. Biologic duplicates of co-immunoprecipitations were performed on soluble (cytosolic) and insoluble (nuclear-enriched) protein fractions. In parallel, the wild-type 3D7 parental line was used as a control for nonspecific interactions. In Table 1, we list the proteins present in both sets of the PfMCMBP pulldown with more than one unique peptide in at least one set and either exclusively found in the PfMCMBP co-immunoprecipitation or enriched by >30-fold in both runs. In Table 1, we ranked hit proteins based on their unique peptide counts and Figure S7 provides the representation of the average fractional coverage and number of unique peptides of identified proteins. The complete list of identified proteins is provided in Table S1. We note that five of the six subunits that compose the MCM complex (Bochman & Schwacha, 2009), were pulled down as the top five hits in the soluble protein fraction. In contrast, only three MCM subunit proteins were significantly pulled down in the insoluble protein fraction with PfMCM7 and five the strongest hits in both fractions. Two structural maintenance of chromosome proteins, SMC2 and SMC4, as well as a DNA-directed RNA polymerase II subunit putative protein (RBP2) and ADP-dependent DNA helicase (RecQ) were found associated with PfMCMBP in the insoluble fraction.
TABLE 1.
Proteins uniquely present in PfMCMBP pulldown
| [Gene ID] | Unique peptide #1 | Unique peptide #2 | [Product description] |
|---|---|---|---|
| Insoluble protein fraction | |||
| PF3D7_1211700 | 45 | 30 | DNA replication licensing factor MCM5, putative |
| PF3D7_1412100 | 36 | 24 | MCMBP |
| PF3D7_0705400 | 44 | 16 | DNA replication licensing factor MCM7 |
| PF3D7_0215700 | 8 | 7 | DNA-directed RNA polymerase II subunit RPB2, putative |
| PF3D7_1464600 | 1 | 6 | Serine/threonine protein phosphatase UIS2, putative |
| PF3D7_1318400 | 11 | 5 | Structural maintenance of chromosomes protein 2, putative |
| PF3D7_0825200 | 2 | 3 | Translation initiation factor IF-3 |
| PF3D7_0310300 | 9 | 3 | Phosphoglycerate mutase, putative |
| PF3D7_0937800 | 1 | 2 | PfEMP1 |
| PF3D7_0603400 | 4 | 2 | Trophozoite exported protein 1 |
| PF3D7_0624600 | 2 | 2 | SNF2 helicase, putative |
| PF3D7_0509100 | 10 | 2 | Structural maintenance of chromosomes protein 4, putative |
| PF3D7_1355100 | 7 | 1 | DNA replication licensing factor MCM6 |
| PF3D7_1443400 | 3 | 1 | WD repeat-containing protein |
| PF3D7_1429900 | 2 | 1 | ADP-dependent DNA helicase RecQ |
| Soluble protein fraction | |||
| PF3D7_1211700 | 37 | 63 | DNA replication licensing factor MCM5, putative |
| PF3D7_0705400 | 42 | 56 | DNA replication licensing factor MCM7 |
| PF3D7_1317100 | 23 | 51 | DNA replication licensing factor MCM4 |
| PF3D7_1412100 | 39 | 47 | MCMBP |
| PF3D7_1355100 | 8 | 20 | DNA replication licensing factor MCM6 |
| PF3D7_0527000 | 7 | 11 | DNA replication licensing factor MCM3, putative |
| PF3D7_0310300 | 4 | 3 | Phosphoglycerate mutase, putative |
| PF3D7_1355900 | 1 | 3 | RWD domain-containing protein, putative |
| PF3D7_1107200 | 2 | 2 | Conserved protein, unknown function |
| PF3D7_1408700 | 1 | 2 | Conserved protein, unknown function |
2.2 |. PfMCMBP knockdown impaired schizogony without causing a stage-specific arrest
Upon Shld1 removal from a synchronous culture of early ring stage PfMCMBP3HADD parasites, we quantified a 69% ± 9% depletion of PfMCMBP levels in early schizont-stage (40–44 h.p.i.) cultures (Figure 2a). To determine the requirement of PfMCMBP for asexual parasite replication, we monitored the growth of PfMCMBP3HADD parasites in the presence or absence of Shld1, representing PfMCMBP-sufficient and PfMCMBP-deficient status, respectively, over two complete asexual replication cycles. As a control, we monitored replication of the wild-type 3D7 parental line with and without Shld1. Upon the removal of Shld1 in PfMCMBP3HADD parasites, we observed a 74% ± 1.9% reduction in growth within the first cycle of replication and a 94% ± 0.95% reduction in growth within the second cycle of replication (Figure 2b), demonstrating that PfMCMBP is important for asexual replication. We note that the replication block is not 100%, and there are a few successful reinvasions (~25% compared to the plus Shld1 condition after the first cycle). This incomplete arrest may be because the knockdown was insufficient for complete arrest or because some daughter merozoites, by chance, have correct or nearly correct DNA content/physical geometry (discussed more below) and result in an invasive daughter parasite.
FIGURE 2.
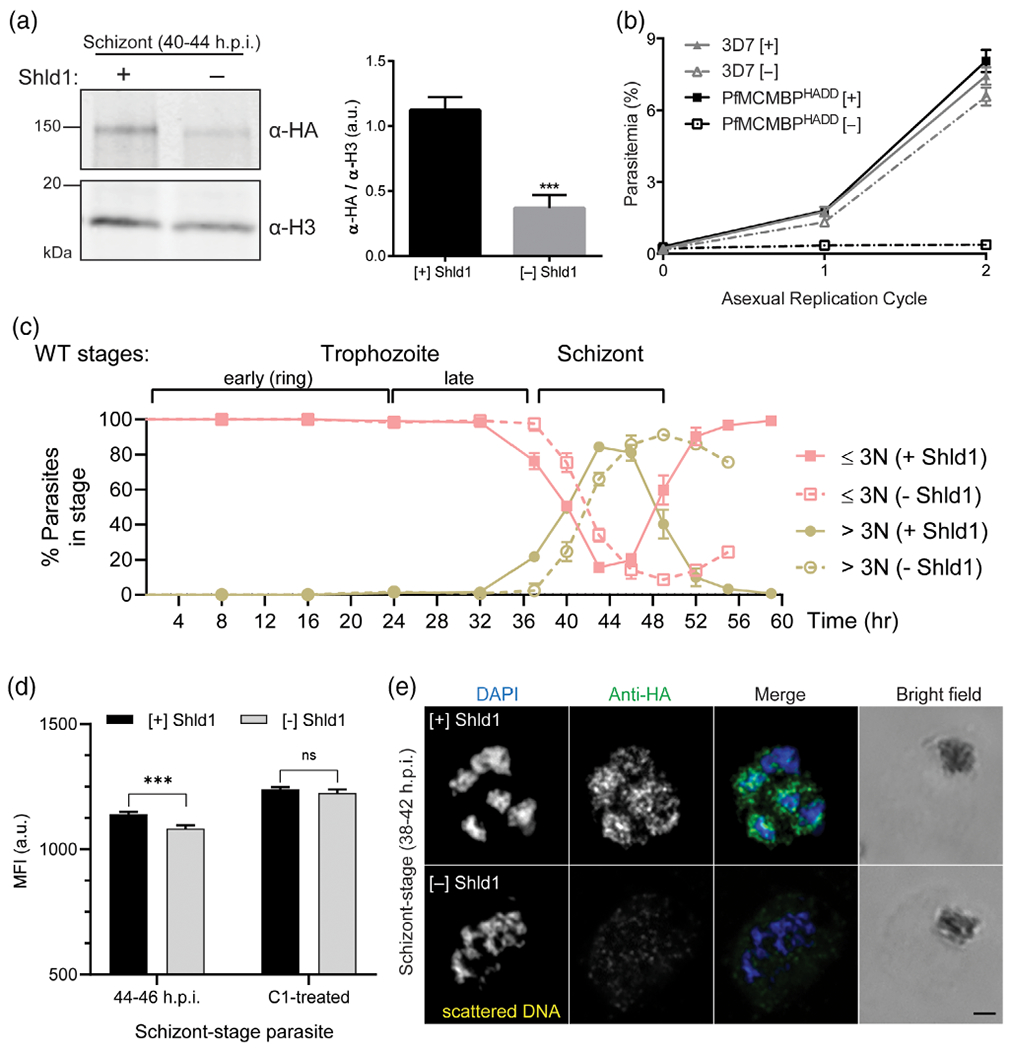
Following Plasmodium homologue of the Mini-Chromosome Maintenance Complex Binding Protein (PfMCMBP) knockdown parasite growth was prolonged three hours and parasite proliferation failed. (A) Immunoblot of protein lysates from PfMCMBP3HADDschizont stage parasites (36-48 h.p.i.) cultured in [+]/[−] 250 nM Shld1 and probed with antibodies against the HA epitope or Histone H3 (loading control). Quantification of immunoblot was performed by volumetric measurement of fluorescence intensity with the LiCor Odyssey system (n = 3, mean ±SD). (B) Replication curves of PfMCMBP3HADDparasites cultured [+]/[−] Shld1 reveal the essentiality of PfMCMBP for parasite growth (n = 3, mean with ±SDerror bars). (C) PfMCMBP3HADDparasites were tightly synchronised and grown for 1.2 cycles with eight-hour time points for the first 32 hrs, then 3-hr time points until the egress of the minus Shld1 condition was complete. At each time point parasite-stage was assessed by counting 100 infected-RBCs for each biological replicate using Hemacolor stain of thin smears (n = 3 biological replicates, mean with ±SDerror bars). (D) SYBR Green I staining demonstrated no significant difference in total DNA content following RNAse A treatment (n = 3 biological replicates) on C1-treated schizont parasites (arrested prior to parasitophorous vacuole rupture). (E) Representative pictures from [+]/[−] Shld1 PfMCMBP3HADDparasites at schizont-stage (38-42 h.p.i) after PFA-fixation and probing with rat anti-HA (1:50) and goat anti-rat conjugated with Alexa 488 (1:1000). Images were taken with Nikon E800 using the 100× objective. Scale bar is 1 μm
The growth defect in PfMCMBP-deficient parasites may be due to disruption of DNA replication, errant nuclear division, or potentially both. To evaluate if the observed growth defect occurred at a specific stage of asexual development, we monitored the progression of a ring-stage synchronised culture for 59 hr by light microscopy (Figure 2c). Samples were taken every 8 hr for the first 32 hr, then every 3 hr from 37 h.p.i. for the remaining 22 hr of the experiment. At each time point, we measured the proportion of parasites with less than three nuclei (ring and trophozoite stage) or equal to or greater than three nuclei (schizont stage). For the first 32 hr, we observed no significant differences in parasite stage between the [+] and [−] Shld1 conditions. At 37 h.p.i, we observed 21.7% ± 1.2 of parasites with ≥3 nuclei in the [+] Shld1 condition and 2.3% ± 4.0 in the [−] Shld1 condition. However, at the next time point, PfMCMBP-deficient parasites had progressed into the schizont-stage, indicating a three-hour delay. Because PfMCMBP interacts with the MCM complex, we hypothesized that DNA replication was likely affected, causing the delayed transition to the schizont stage in MCMBP-deficient parasites. To test this hypothesis, we measured the DNA content by quantifying the mean fluorescence intensity (MFI) of SYBR Green I stained-parasites using flow cytometry, under [+] and [−] Shld1 conditions (Figure 2d and Figure S8). At 37 h.p.i. we measured no significant difference in total DNA content between parasites grown in [+] or [−] Shld1 conditions. To monitor the effect of PfMCMBP depletion on DNA replication throughout the remainder of schizogony, we collected samples every 3 hr from 37 to 46 h.p.i. (Figure S8). While parasites grown in the presence of Shld1 had a 39% increase in DNA content (measured by MFI) by 40 h.p.i, the PfMCMBP-deficient parasites gained only 9%. However, in the next three-hour interval, from 40 until 43 h.p.i., we observed a similar rate of DNA content increase with 35% increase in parasites grown in absence of Shld1 and 31% in presence of Shld1. From 43 to 46 h.p.i., parasites in [+] Shdl1 condition had only 19% increase in MFI compared to the 43 h.p.i time point, while parasites in [−] Shld1 condition showed the highest rate with 40% increase in MFI of parasites between the 43 and 46 h.p.i. time point. At the end of the time course, [−] Shld1 condition parasites had 11% less DNA compared to the [+] Shld1 condition. Next, we tested if DNA replication could catch up to the [+] Shld1 condition if the PfMCMBP-deficient parasites were prevented from egressing. These experiments tested if DNA replication was defective or merely delayed in PfMCMBP-deficient parasites. When parasites were treated with Compound 1 (4-[2-(4-fluorophenyl)-5-(1-methylpiperidine-4-yl)-1H-pyrrol-3-yl] pyridine or C1), a reversible protein kinase G (PfPKG) inhibitor, they accumulated as late-stage, segmented schizonts prior to exoneme release (Collins et al., 2013; Gurnett et al., 2002; Taylor et al., 2010). As previously observed, we measured a significant difference of MFI at 44–46 h.p.i. between parasites grown in [−] Shld1 compared to [+] Shld1 condition. However, following an additional 5 hr C1 treatment, we observed no significant difference in DNA content between parasites cultured with or without Shld1. From these data, we conclude that total DNA replication was equivalent, albeit with a moderate delay (Figure 2d). Next, we monitored nuclear division by immunofluorescence microscopy by probing mid-schizont-stage parasites (36–42 h.p.i.) with anti-HA to recognise PfMCMBP and counterstained the nucleus with DAPI. In the presence of Shld1, mid-schizont stage parasites revealed uncondensed DNA contained in multiple round nuclear compartments. The largest fraction of PfMCMBP fluorescent signal was present in the cytoplasm and a small fraction overlapped with the DAPI staining (Figure 2e upper panel). In contrast, when parasites were cultured in absence of Shld1, PfMCMBP depletion caused a total loss of PfMCMBP fluorescent signal and, more strikingly, an abnormal nuclear morphology. The chromatin appeared condensed, scattered, and bridged (Figure 2e lower panel). It is important to note that nuclear morphology remained unaffected during ring-stage and until late trophozoite-stage (at 28 h.p.i.) when parasites contained one nucleus (Figure S9). Together, these results demonstrate that knockdown of PfMCMBP disrupted nuclear morphology and caused a delay to, but not inhibition of, DNA replication.
2.3 |. PfMCMBP depletion promotes the formation of MTOCs with extended spindle microtubules that are attached to chromosomes in spatially separated clusters of DNA
To evaluate mitosis in the PfMCMBP-deficient parasites, we monitored MTOC formation/duplication and spindle microtubule formation in early schizont-stage parasites (36–42 h.p.i.), where the number of nuclei did not exceed four to five based on DAPI staining to facilitate the microscopy analysis. For this analysis, the spindle microtubules were stained with anti-alpha-tubulin antibodies, the centrosomes were stained with an antibody against Chlamydomonas reinhardtii centrin that recognises PfCentrin-3 (Mahajan et al., 2008), and nuclei were counterstained with DAPI. Assembly of a bipolar spindles, defined as microtubules that are attached to two spatially separated MTOCs, requires duplication and segregation of the MTOCs. Three different z-slices from a single representative [+] Shld1 parasite are shown in Figure 3a, rows i-iii. Each of the three nuclei within this parasite displayed one of the expected patterns: short microtubules in non-dividing nuclei (Figure 3a, panel i), short microtubules following MTOC duplication (Figure 3a, panel ii, yellow asterisk), or bipolar spindle microtubules in mitotic nuclei (Figure 3a, panel iii, yellow arrow). In contrast, PfMCMBP-deficient parasites revealed extended spindle microtubules (Figure 3a, panels iv–vi), likely corresponding to the scattered nuclear morphology. In the absence of Shld1, we performed the same staining with parasites that were obtained 3 hrs later, to accommodate the previously observed delay in DNA replication following PfMCMBP depletion. Three different [−] Shld1 parasites are shown in Figure 3, rows iv–vi. Given the abnormal nuclear morphology with scattered DNA (and the absence of a reliable nuclear envelope marker), determination of number of nuclei per schizont was challenging, and the three staining patterns described for the [+] Shld1 parasite above were largely absent. We estimated four and five nuclei in the [−] Shld1 parasites shown in rows iv and v, respectively. In both cells, microtubule staining revealed multiple sets of extended spindle microtubules that were interconnected to each other and each spindle emerged from a bright centrosomal focus. Additionally, we detected multiple dim ectopic centrin staining, co-localising with several spindle microtubules arising from the brighter centrosomal foci. In the third [−]Shld1 parasite (row vi), we observed greater distance between the foci of nuclear staining, and within each we detected multiple extended microtubules rising from a brighter centrin focus. Additional representative images from late trophozoite-stage (with two nuclei) to early schizont-stage (with an estimated number of nuclei between 3 and 5) revealing the presence of multiple extended bipolar microtubules in PfMCMBP-deficient parasites are presented in Figure S10.
FIGURE 3.
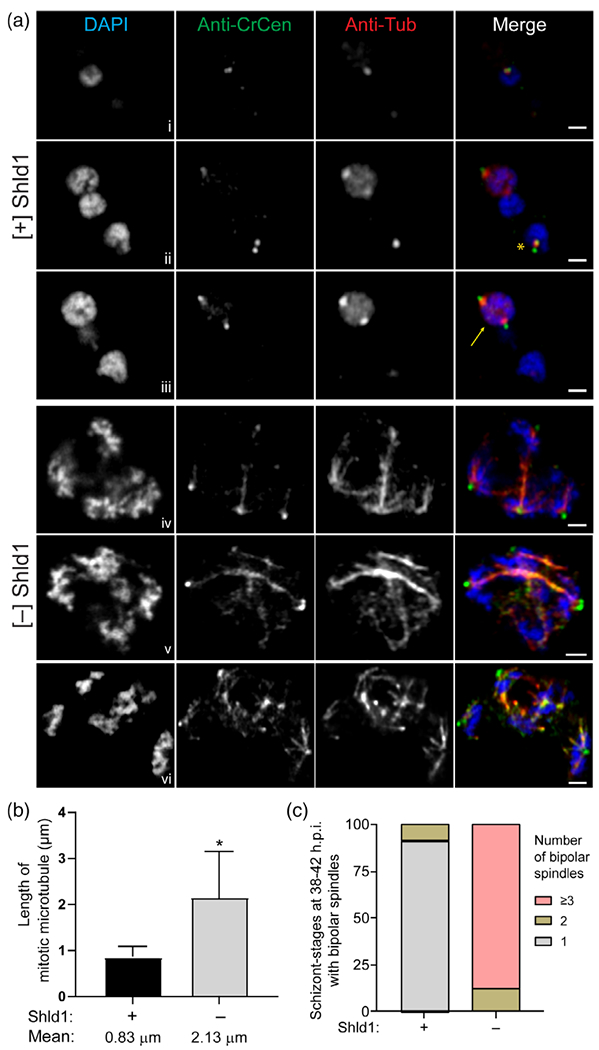
Plasmodiumhomologue of the Mini-Chromosome Maintenance Complex Binding Protein (PfMCMBP) depletion causes abnormal nuclear morphology with additional MTOC ectopic foci and formation of extended mitotic spindle microtubules. (A) Representative pictures from [+]/[−] Shld1 PfMCMBP3HADDparasites (40-44 h.p. i.) after PFA-fixation and probing with mouse anti-CrCen1 (1:500, green) and mouse anti-αTubulin (1:200, red) followed by isotype-specific goat anti-mouse antibodies. Three different z-slices from a single representative [+] Shld1 parasite are shown in rows i-iii. Panels iv-vi represent three different PfMCMBP-deficient parasites cultured [−] Shld1. Images were taken with LSM880 Airyscan using the 63× Objective. Yellow arrows indicate bipolar spindle and yellow asterisk indicate short microtubule between duplicated MTOC in WT condition. Scale bar at 1 μm. (B) The length of mitotic microtubules was obtained using ImageJ software and (C) the count of bipolar spindles per cell was done using the Nikon E800 microscope. Schizonts with three or four nuclei and at least one bipolar microtubule were selected for analysis. Two biological replicates with 50 schizonts per replicate were counted. Values that were significantly different (p < .0001) by unpaired t-test are indicated by asterisk (n = 2, mean with ±SD error bars)
We quantified the length of mitotic microtubules and the percentage of schizonts with one, two, or greater than three bipolar spindles. In the presence of Shld1, we measured a mean length of 0.83 ± 0.24 μm for spindle microtubules (Figure 3b) and found 91% of parasites with one bipolar spindle, and only 9% with two (Figure 3c). In contrast, parasites grown in the absence of Shld1 presented extended microtubules with a mean length of 2.13 ± 1.01 μm and 88% of parasites had three or more bipolar spindles simultaneously. Together, these results demonstrate that PfMCMBP depletion impacted MTOC integrity and promoted the formation of extended mitotic spindle microtubules and increased numbers of bipolar microtubules.
2.4 |. Formation of aneuploid merozoites fails to prevent completion of cytokinesis
To ensure proper cell division, higher eukaryotic cells tightly coordinate each mitotic step via a system of feedback controls named checkpoints (Morgan, 2007). As noted above, these checkpoints have not been demonstrated during the asexual replication of P. falciparum. Upon PfMCMBP depletion, we demonstrated that despite severe disruption of the nuclear morphology, DNA replication continued without causing a stage-specific arrest (Figure 2(D)). This result suggested that the P. falciparum asexual replication cycle may lack a robust mechanism to halt progression of the cell cycle, allowing PfMCMBP-deficient parasites to complete cytokinesis and form aneuploid merozoites. To test this hypothesis, we evaluated 52 h.p.i. schizonts cultivated with and without Shld1 that had been treated with E64, a cysteine protease inhibitor that prevents rupture of the RBC plasma membrane while still allowing rupture of the parasitophorous vacuole membrane and the development of viable merozoites (Hale et al., 2017). E64-treated schizonts were stained with anti-PfGAP45 (an inner membrane complex-associated protein [Jones, Kitson, & Rayner, 2006]), and SYBR Green I as a marker of the merozoite membrane and nuclear DNA, respectively. Strikingly, at this late fully segmented stage, the nuclear morphology in the PfMCMBP-deficient parasites resembled the [+] Shld1 condition, and we no longer observed scattered DNA (Figure 4a). This result showed that nuclear condensation and karyokinesis at the end of schizogony occurred in both the PfMCMBP-deficient and PfMCMBP-sufficient states. In addition, we compared [+] and [−] Shld1 E64-treated schizonts by immunofluorescence assay (IFA) using a primary antibody against the rhoptry neck marker, PfRON4 (Figure S11a) co-stained with anti-PfGAP45. In both conditions, merozoites contained a strong fluorescent staining indicating that the lack of PfMCMBP does not affect the biogenesis of apical organelles. This result was confirm using transmission electron microscopy (Figure S11b). Despite the similar smooth nuclear perimeters, we encountered merozoites with varied cellular and nuclear sizes as well as merozoites lacking DNA, known as zoids (Figure 4a). To quantify this observation, we measured the cell body and nuclear area using the PfGAP45 and SYBR Green I staining of more than 300 merozoites for each [+] and [−] Shld1 cultures. In the presence of Shld1, all merozoites clustered within 1.36 to 2.34 μm2 for the body size and 0.36 to 0.96 μm2 for the nucleus area (Figure 4b). In contrast, the cell body size of PfMCMBP-deficient merozoites ranged from 0.13 to 2.94 μm2 compared to [+] Shld1 and with a nucleus area from 0.08 to 2.94 μm2 (Figure 4b). In addition to this increased range of cell body and nucleus areas, 17.6% of the PfMCMBP-deficient merozoites lacked DNA staining and were classified as zoid. Furthermore, we assessed the ploidy of the same merozoites by measuring the integrated fluorescence of SYBR Green I for each nucleus. The total DNA content of PfMCMBP-deficient merozoites ranged from 0.02 to 3.72, normalised to the average of the integrated fluorescence of [+] Shld1 nuclei (assumed to be 1 N). As expected, in [+] Shld1 parasites, the DNA content of each nuclei narrowly clustered around 1. Here, we report that PfMCMBP depletion results in the formation of aneuploid merozoites with varied cellular and nuclear size, thus demonstrating that P. falciparum parasites lack a robust mechanism to prevent progression of the cell cycle through cytokinesis despite have aberrant nuclear karyotype.
FIGURE 4.
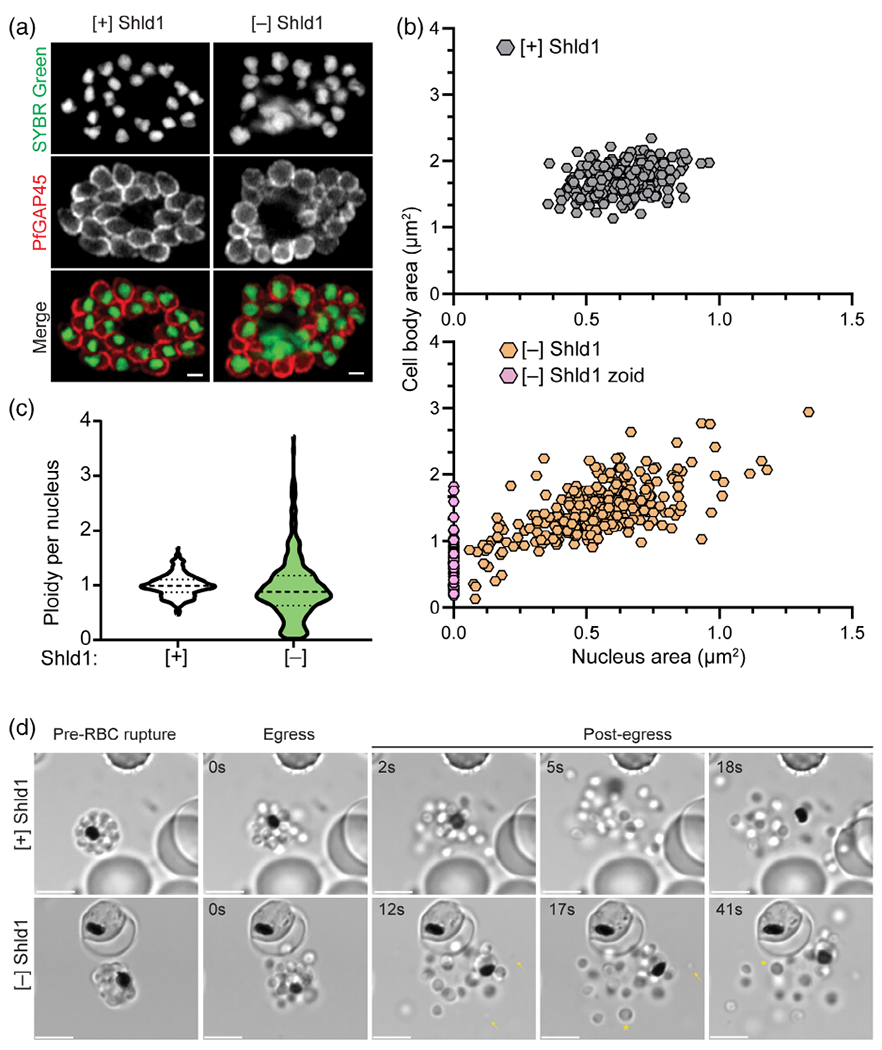
Plasmodium homologue of the Mini-Chromosome Maintenance Complex Binding Protein (PfMCMBP)-deficient parasites complete budding and form merozoites with varied cell body size and ploidy. (A) Representative pictures from [+]/[−] Shld1 PfMCMBP3HADDparasites treated with E64, stained with SYBR Green I and probed with rabbit anti-PfGAP45. Images were taken with LSM880 Airyscan using the 60× objective and the scale bar is at 1 μm. (B) The measurement of cytoplasm and nucleus area were obtained using Image J software. Three biological replicates with a total of 284 and 392 merozoites were, respectively, measured for [+] and [−] Shld1 conditions. (C) The ploidy per nucleus was obtained using Image J software and calculating the corrected total cell fluorescence (CTCF) of the SYBR Green I fluorescence intensity calculated using the following the formula: CTCF = Integrated Density - (Area of selected cell × Mean fluorescence of background readings). Three biological replicates and a total of 306 and 361 merozoites were respectively counted for [+] and [−] Shld1 conditions. (D) Representative still images from time lapse microscopy of [+]/[−] Shld1 PfMCMBP3HADDparasites during egress. Scale bar at 5 μm
2.5 |. PfMCMBP-deficient schizonts egressed and released largely non-invasive merozoites
To demonstrate that PfMCMBP-deficient schizonts undergo physiological egress, we performed time-lapse microscopy of parasites treated with C1 (Figure 4d). We added C1 to 44–46 h.p.i. schizonts grown in [+] or [−] Shld1 condition, and, after an additional 6 hr, we washed cells with C1-free media and used time-lapse microscopy to monitor egress. In both conditions, parasites egressed rapidly (Videos S1 and S2). Remarkably, the time-lapse videos confirmed that PfMCMBP depletion does not cause a stage specific arrest and results in the release of mutant daughter parasites of various sizes (Figure 4d arrows and asterisks mark small and large merozoites, respectively). When visualised by time-lapse microscopy, these mutant merozoites initiate contact and glide on the surface of the RBC but fail to invade (Video S3). Although not visualised in these movies, we note that we observe a small number of newly reinvaded rings following the first cycle of PfMCMBP depletion. However, the majority of egressed merozoites do not successfully reinvade. Together, these results demonstrate that despite the severe impairment of parasite cytokinesis in PfMCMBP-depleted schizonts, newly formed merozoites egress but most fail to successfully complete invasion.
3 |. DISCUSSION
We identified PF3D7_1412100 as the P. falciparum homologue of the MCM Binding Protein. Here, we sought to assess the impact of PfMCMBP-deficiency on asexual replication of Plasmodium falciparum. During the replicative phase of schizogony, PfMCMBP is present in both the cytoplasmic and nuclear compartments. As the budding phase (i.e., segmentation) progresses, however, we observed loss of PfMCMBP expression. This expression profile suggests a role during asexual mitosis. In agreement with a central role of PfMCMBP during DNA replication, PfMCMBP coimmunoprecipitates with the PfMCM complex (for which it was named) indicating that PfMCMBP is likely a component of the Plasmodium DNA replication licensing complex. In addition, the specific presence of core subunit proteins of the parasite RNA Polymerase II in our pull-downs compared to controls suggests that PfMCMBP may contribute to the control of gene expression. In human cells, subunits of the MCM complex control gene expression through interaction with Histone H3, the transcription factor STAT1, and RNA Pol II (Holland, Gauthier, Bell-Rogers, & Yankulov, 2002; Snyder, He, & Zhang, 2005). Furthermore, in the insoluble fraction PfMCMBP coimmunoprecipitated with SMC2 and SMC4 proteins. In eukaryotic cells, SMC2 and SMC4 are components of the condensin complex and are required for chromosome architecture and segregation (Hirano, 2016). SMC2 and SMC4 are essential for Plasmodium spp, however their functions are not completely understood (Pandey et al., 2020). In other systems, down-regulation or overexpression of an MCMBP homologue in plants (Takahashi et al., 2010), yeast (Ding & Forsburg, 2011), human cancer cell lines (Quimbaya et al., 2014), and protozoans such as Trypanosoma brucei (Kim et al., 2013), result in some common phenotypic traits, including abnormal nuclear morphology, the appearance of zoid cells, and accumulation of multinucleated cells with poly-lobed nuclei. PfMCMBP is required for blood-stage parasite growth, and PfMCMBP-deficient parasites share phenotypic traits with other eukaryotic MCMBP mutant models. Using fluorescence microscopy, the most striking common phenotypic appearance of parasites with scattered, elongated, and branched DNA with a nuclear compartment that no longer forms a rounded structure (at least during early and mid-schizogony). The coimmunoprecipitation of PfMCMBP with PfSMC2/4 raises the possibility that PfMCMBP depletion impedes the interaction between the condensin complexes and nuclear DNA, causing impairment of nuclear morphology. Future studies of condensin complex components will be necessary to determine the mechanism by which PfMCMBP and PfSMC2/4 regulate the formation and folding of chromatin during blood-stage mitosis in Plasmodium.
In many eukaryotic organisms, molecular checkpoints ensure even replication and separation of nuclear DNA, but these checkpoints have not yet been identified for asexual replication of Plasmodium spp. Our previous study of PfCyclin1 unveiled an unexpected requirement of PfCyclin1 for licensing segmentation but not for entering and completing mitosis (Robbins et al., 2017), providing evidence that the molecular control of and/or checkpoints within the asexual development cycle in Plasmodium are different from metazoan cells. Our current functional genetic study of PfMCMBP revealed that PfMCMBP depletion promotes the formation of extended mitotic spindle microtubules without causing a stage-specific arrest. PfMCMBP-deficiency does cause an approximately 3 hr delay in DNA replication. However, in total, our data suggest the absence of a cell cycle checkpoint to prevent progression in the presence of abnormal karyokinesis. It is possible that in the absence of PfMCMBP, DNA replication is no longer coordinated with karyokinesis, resulting in the emergence of interconnected and extended spindle microtubules. Nevertheless (and unexpectedly), once parasites reached the stage of segmentation, depolymerization of those abnormal microtubules is likely to have occurred as well as formation of individual nuclei. Despite the abnormal nuclear morphology and impaired DNA segregation, PfMCMBP-deficient parasites enter and complete cytokinesis, forming aberrant daughter cells with variably sized cell bodies and nuclei. Consistent with the observation that PfMCMBP level decreased as segmentation progresses, we demonstrated that cytokinesis occurs by a PfMCMBP-independent pathway. It is likely that once DNA replication is complete, PfMCMBP is no longer involved in the regulation of DNA compaction.
Previous studies from our group have demonstrated normal egress after severe impairment of cytokinesis, suggesting that the egress pathway is independent of the success (or failure) of the cell cycle (Absalon, Robbins, & Dvorin, 2016; Rudlaff et al., 2019). Likewise, despite the severe impact of PfMCMBP depletion on DNA segregation and merozoite segmentation, mutant parasites were still able to egress – but the majority of merozoites remained non-invasive.
In summary, our findings show that the PfMCMBP homologue is required for asexual replication, and its deficiency leads to uneven nuclear division and abnormal karyotype. Taken together, these findings unveil the lack of a robust checkpoint to prevent cytokinesis following abnormal karyokinesis and highlight the independence of the segmentation and egress pathways from nuclear division.
4 |. EXPERIMENTAL PROCEDURES
4.1 |. Plasmid construction
To generate a PfMCMBP targeting plasmid for homologous recombination, the 3′end of Pf3D7_1412100 gene locus was PCR amplified from 3D7 genomic DNA with oJDD1646 (5′-taggcggccgcGACGTAATCGAAATTATTGGAA-3′)/oJDD1647 (5′-gatctcgagTATATATATTTTGTTAAG- 3′) and cloned into pSAB01 (Absalon et al., 2016) cut with NotI/XhoI to obtain pSAB60.
4.1.1 |. Plasmodium falciparum culture and transfection
The 3D7 Plasmodium falciparum strain, obtained from the Walter & Eliza Hall Institute (Australia), and descendant transgenic parasites were maintained in vitro in human red blood cells in RPMI-1640 supplemented with 50 mg/L hypoxanthine, 25 mM HEPES, and 0.5% Albumax II as previously described (Trager & Jensen, 1976). To generate the 3D7-PfMCMBP3HADD strain, sorbitol-synchronised ring-stage parasites at 1%–2% parasitemia were transfected with ~100 μg of plasmid DNA using electroporation. Upon transfection, parasites were cultured with 250 nM Shield-1 and stable single crossover parasites were selected by cycling on and off WR99210 as previously described (Absalon et al., 2016). Individual transgenic clones were obtained by limiting dilution in presence of Shld1 only, then placed back on WR99210 selection during clonal expansion.
4.2 |. Southern and immuno (western) blot analysis
Genomic DNAs from 3D7 and 3D7-PfMCMBP3HADD strains were collected with QIAamp Blood Mini Kit and digested with BamHI, NotI, PacI, and XhoI restriction enzymes. Digested DNAs were resolved on 0.7% agarose gel, transferred overnight to GeneScreen Plus membrane, and hybridised with a radiolabeled probe specific against PMCMBP (NotI/XhoI fragment from pSAB60). For immunoblot assays, sorbitol-synchronised ring-stage parasites at 0.5% parasitemia were cultured in presence or absence of 250 nM Shld1. At the schizont-stage (40–46 h.p.i.), proteins were extracted using 0.2% saponin and pellets were resuspended in Laemmli sample buffer. Protein samples were loaded on 4%–20% mini-Protean TGX gels (Bio-Rad) and transferred to nitrocellulose membranes using the Tran-blot Turbo transfer system. Blocking was performed with Odyssey blocking buffer (LiCor) diluted 1:5 in PBS. Mouse anti-HA (clone 2–2.2.14, Pierce) and mouse anti-H3 (ab1791, Abcam) were diluted into 3% BSA-PBS at 1:1000 and 1:2000, respectively. Secondary antibodies were directly labelled with near-infrared dyes (LiCor; 800CW donkey anti-mouse or 680LT donkey anti-mouse) were diluted into TBST buffer at 1:10,000. ImageStudio Software was utilised to visualise and quantify immunoblot data on a LiCor Odyssey CLx imager.
4.3 |. Immunoprecipitation assays
For co-immunoprecipitation studies, experimental and control parasites lines (250 mL with 3% parasitemia at 4% haematocrit) were sorbitol-synchronised and collected at the schizont-stage (36–46 h.p.i.). At collection, erythrocytes were lysed in 0.05% saponin in PBS with protease inhibitors (SigmaFast Protease Inhibitor Cocktail, Sigma). Resulting parasite pellets were lysed in Cell Lysis Buffer (CLB: 20 mM HEPES, 10 mM KCl, 1 mM EDTA, 1 mM EGTA, 0.65% NP-40, pH 7.9, EDTA-free proteinase inhibitors [SigmaFast tablets]) for 30 min on ice and spun for 3 min at 2000 g at 4°C. Supernatants were kept as the soluble protein fraction, and pellets were washed three times with CLB buffer, resuspended in 0.5% Tx-100 in PBS for 20 min on ice, sonicated three times at 25% amplitude for 30 s every 2 min, and spun for 3 min at 2000 g at 4°C to collect the supernatant as the insoluble protein fraction. Each protein fraction was applied to 25 μL of magnetic anti-HA beads (Pierce #88836) and incubated for 60 min at room temperature. Beads were washed four times with TBST, resuspended in 50 μL of 5 mM ammonium bicarbonate and submitted for on-bead digestion followed by peptide identification by LC-MS/MS.
4.4 |. Flow cytometry analysis of parasite replication
PfMCMBP3HADD and parental parasite lines were synchronised at the schizont stage by density centrifugation using 60% Percoll, incubated for 2–3 hr at 37°C with fresh red blood cells, and then new ring-stage parasites were isolated with 5% (w/v) sorbitol treatment. Ring-stage parasite cultures were diluted to 0.25% parasitemia at 2% haematocrit in triplicate and incubated at 37°C in presence or absence of Shld1. For each time point, 100 μl of culture were collected and stained with SYBR green I (Life Technologies) as previously described (Absalon et al., 2016). For each time point, 100,000 events were acquired using a BD FACSCalibur platform. For the mean of fluorescence intensity analysis, parasites were fixed in PBS with 2% PFA and 0.2% glutaraldehyde for 1 hr at 4°C, permeabilized for 10 min with 0.3% Triton X-100 at room temperature, incubated with 0.5 mg/mL RNAse-PBS for 1 hr at 37° C, and stained with SYBR green I as above.
4.5 |. Time-lapse microscopy
PfMCMBP3HADD parasites were tightly synchronised by density centrifugation with 60% Percoll, allowed to reinvade for 3 hr, and new rings were isolated with 5% (w/v) sorbitol then allowed to mature in the presence or absence of Shld1. Magnetic-activated cell sorting columns were used to purify 44–46 h.p.i. schizont-stage parasites. The purified schizonts were incubated in fresh media with 2.5 uM compound 1, (4-[2-(4fluorophenyl)-5-(1-methylpiperidine-4-yl)-1H-pyrrol-3-yl]pyridine [C1]) at 37°C. At 48 h.p.i., these [+]/[−] Shld1 schizonts were washed with C1-free media, applied to dishes coated with 0.5 mg/mL concanavalin A solution, and allowed to adhere for 10 min at 37°C. For egress visualisation, brightfield images were recorded every 1–5 s for 30 min with a Nikon Ti Eclispe microscope equipped with a 37°C chamber using a 60X objective. Time-lapse videos were processed using NIS Elements software.
4.6 |. Immunofluorescence assays
For all IFAs, thin dried blood smears were made on a glass slide (Azer scientific), fixed for 10 min with 4% PFA-PBS, washed three times with PBS, permeabilized for 10 min with a fresh dilution of 0.1%-Triton X-100-PBS, washed three times with PBS, and blocked with 3%BSA-PBS for 1 hr at room temperature. Slides were incubated with primary antibodies diluted in blocking solution overnight at 4°C. The following morning, slides were washed three times with PBS, incubated with diluted secondary antibodies in blocking solution for 45 min at room temperature (anti-rat Alexa 488, anti-mouse IgG1 Alexa 595, anti-mouse IgG2a Alexa 488, anti-mouse Alexa 594 and anti-rabbit Alexa 594 [Life Technologies] all used at 1:1000), washed three times with PBS, and mounted with a DAPI-mounting media (VectaShield) and 1.5 mm coverslip (Corning #2940–224). Images were captured either on a Nikon E800 using a 100X objective or on an ECHO Revolve using a 100X objective or on a Zeiss LSM880 with Airyscan mode microscope using a 60X objective and processed using ZEN 2012 Black software (Zeiss). Primary antibodies were diluted as follows: rat anti-HA (1:50, clone 3F10, Sigma), mouse anti-Cen3 (1:500, clone 20H5, EMD Millipore), mouse anti-alpha Tubulin (1:200, clone B-E-1–2, Sigma), and rabbit anti-PfGAP45 (1:5000, gift from Julian Rayner, University of Cambridge). For electron microscopy, samples were prepared as previously described (Dvorin et al., 2010). The length of mitotic microtubules and the ploidy per nucleus were measured using Image J software. The Corrected Total Cell Fluorescence (CTCF) of the SYBR Green fluorescence intensity was calculating using the formula CTCF = Integrated Density - (area of selected cell × mean fluorescence of background readings). Based on the assumption that the mean ploidy of the [+]Shld1 merozoites was i. each individual CTCF measurement was normalised by the mean CTCF of all merozoites from the [+]Shld1 condition.
Supplementary Material
ACKNOWLEDGEMENTS
We thank D.S. Roos (University of Pennsylvania) for insightful idea regarding ploidy measurement and M. Duraisingh for guidance and sharing of resources. We thank Rebecca Clements, Rachel Rudlaff, and Benjamin Scott Liffner for critical reading of the manuscript. We thank M. Makler (Flow, Inc.), A. Cowman, J. Thompson, K. Wycherley, R. Anders (The Walter & Elizabeth Hall Medical Research Institute), R. Coppel (Monash University), D. Richard (Université Laval), J. Rayner (University of Cambridge), A. Holder (The Francis Crick Institute), S. Prigge (Johns Hopkins University), and MR4 via contribution from J. Adams for provision of antibodies. We thank R. Tomaino (Taplin Mass Spectrometry Facility), M. Ericsson and L. Trakimas (Electron Microscopy Facility), A. Hill and C. Innocent (Cellular Imaging Core), and D. Richardson (Harvard Center for Biological Imaging) for core training and management. This work was supported by NIH R01 AI102907 (J.D.D.) and R01 AI145941 (J.D.D.).
Funding information
National Institutes of Health, Grant/Award Numbers: R01AI102907, R01AI145941
Footnotes
CONFLICT OF INTEREST
The authors have no conflicts of interest to declare.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available on request from the corresponding author.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
REFERENCES
- Absalon S, Robbins JA, & Dvorin JD (2016). An essential malaria protein defines the architecture of blood-stage and transmission-stage parasites. Nature Communications, 7, 11449. 10.1038/ncomms11449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnot DE, Ronander E, & Bengtsson DC (2011). The progression of the intra-erythrocytic cell cycle of plasmodium falciparum and the role of the centriolar plaques in asynchronous mitotic division during schizogony. International Journal for Parasitology, 41(1), 71–80. 10.1016/j.ijpara.2010.07.012 [DOI] [PubMed] [Google Scholar]
- Aurrecoechea C, Brestelli J, Brunk BP, Dommer J, Fischer S, Gajria B, … Wang H (2009). PlasmoDB: A functional genomic database for malaria parasites. Nucleic Acids Research, 37(Database issue), D539–543. doi: 10.1093/nar/gkn814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski LA, Chen LC, Maynard-Smith LA, Ooi AG, & Wandless TJ (2006). A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell, 126(5), 995–1004. 10.1016/j.cell.2006.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochman ML, & Schwacha A (2009). The mcm complex: Unwinding the mechanism of a replicative helicase. Microbiology and Molecular Biology Reviews, 73(4), 652–683. 10.1128/mmbr.00019-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burki F (2014). The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harbor Perspectives in Biology, 6(5), a016147. 10.1101/cshperspect.a016147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CR, Hackett F, Strath M, Penzo M, Withers-Martinez C, Baker DA, & Blackman MJ (2013). Malaria parasite cGMP-dependent protein kinase regulates blood stage Merozoite secretory organelle discharge and egress. PLoS Pathogens, 9(5), e1003344. 10.1371/journal.ppat.1003344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, & Forsburg SL (2011). Schizosaccharomyces pombe minichromosome maintenance-binding protein (MCM-BP) antagonizes MCM helicase. The Journal of Biological Chemistry, 286(38), 32918–32930. 10.1074/jbc.M111.282541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorin JD, Martyn DC, Patel SD, Grimley JS, Collins CR, Hopp CS Duraisingh MT. (2010). A plant-like kinase in Plasmodium falciparum regulates parasite egress from erythrocytes. Science, 328(5980), 910–912. doi: 10.1126/science.1188191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson DJP, Sahoo N, Pinches RA, Bumstead JM, Tomley FM, & Gubbels M-J (2008). MORN1 has a conserved role in asexual and sexual development across the Apicomplexa. Eukaryotic Cell, 7(4), 698–711. 10.1128/ec.00021-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley EA, & Kapoor TM (2013). Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nature Reviews. Molecular Cell Biology, 14(1), 25–37. 10.1038/nrm3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia ME, & Striepen B (2014). Cell division in apicomplexan parasites. Nature Reviews. Microbiology, 12(2), 125–136. 10.1038/nrmicro3184 [DOI] [PubMed] [Google Scholar]
- Gerald N, Mahajan B, & Kumar S (2011). Mitosis in the human malaria parasite Plasmodium falciparum. Eukaryotic Cell, 10(4), 474–482. 10.1128/ec.00314-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurnett AM, Liberator PA, Dulski PM, Salowe SP, Donald RGK, Anderson JW Schmatz DM. (2002). Purification and molecular characterization of cGMP-dependent protein kinase from apicomplexan parasites. A novel chemotherapeutic target. Journal of Biological Chemistry, 277(18), 15913–15922. 10.1074/jbc.m108393200 [DOI] [PubMed] [Google Scholar]
- Hale VL, Watermeyer JM, Hackett F, Vizcay-Barrena G, van Ooij C, Thomas JA, … Saibil HR. (2017). Parasitophorous vacuole poration precedes its rupture and rapid host erythrocyte cytoskeleton collapse in Plasmodium falciparum egress. Proceedings of the National Academy of Sciences of the United States of America, 114(13), 3439–3444. 10.1073/pnas.1619441114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T (2016). Condensin-based chromosome organization from bacteria to vertebrates. Cell, 164(5), 847–857. 10.1016/j.cell.2016.01.033 [DOI] [PubMed] [Google Scholar]
- Holland L, Gauthier L, Bell-Rogers P, & Yankulov K (2002). Distinct parts of minichromosome maintenance protein 2 associate with histone H3/H4 and RNA polymerase II holoenzyme. European Journal of Biochemistry, 269(21), 5192–5202. 10.1046/j.1432-1033.2002.03224.x [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, Serra F, & Bork P (2016). ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Molecular Biology and Evolution, 33(6), 1635–1638. 10.1093/molbev/msw046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer DR, & Rhind N (2017). The intra-S checkpoint responses to DNA damage. Genes (Basel), 8(74), 1–25. 10.3390/genes8020074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan M, Sakwe AM, Nguyen T, & Frappier L (2012). The MCM-associated protein MCM-BP is important for human nuclear morphology. Journal of Cell Science, 125(Pt 1), 133–143. 10.1242/jcs.089938 [DOI] [PubMed] [Google Scholar]
- Jones ML, Kitson EL, & Rayner JC (2006). Plasmodium falciparum erythrocyte invasion: A conserved myosin associated complex. Molecular and Biochemical Parasitology, 147(1), 74–84. 10.1016/j.molbiopara.2006.01.009 [DOI] [PubMed] [Google Scholar]
- Kim HS (2019). Genome-wide function of MCM-BP in Trypanosoma brucei DNA replication and transcription. Nucleic Acids Research, 47(2), 634–647. 10.1093/nar/gky1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Park SH, Gunzl A, & Cross GA (2013). MCM-BP is required for repression of life-cycle specific genes transcribed by RNA polymerase I in the mammalian infectious form of Trypanosoma brucei. PLoS One, 8(2), e57001. 10.1371/journal.pone.0057001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara-Gonzalez P, Westhorpe FG, & Taylor SS (2012). The spindle assembly checkpoint. Current Biology, 22(22), R966–R980. 10.1016/j.cub.2012.10.006 [DOI] [PubMed] [Google Scholar]
- Mahajan B, Selvapandiyan A, Gerald NJ, Majam V, Zheng H, Wickramarachchi T, … Kumar S (2008). Centrins, cell cycle regulation proteins in human malaria parasite Plasmodium falciparum. The Journal of Biological Chemistry, 283(46), 31871–31883. 10.1074/jbc.M800028200 [DOI] [PubMed] [Google Scholar]
- Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A Finn RD. (2019). InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Research, 47 (D1), D351–D360. doi: 10.1093/nar/gky1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DO (2007). The cell cycle: Principles of control, London: New Science Press. [Google Scholar]
- Naughton JA, & Bell A (2007). Studies on cell-cycle synchronization in the asexual erythrocytic stages of Plasmodium falciparum. Parasitology, 134(Pt 3), 331–337. 10.1017/S0031182006001466 [DOI] [PubMed] [Google Scholar]
- Pandey R, Abel S, Boucher M, Wall RJ, Zeeshan M, Rea E, … Tewari R. (2020). Plasmodium Condensin Core subunits SMC2/SMC4 mediate atypical mitosis and are essential for parasite proliferation and transmission. Cell Reports, 30(6), 1883–1897 e1886. 10.1016/j.celrep.2020.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson S, Robert C, Whittle C, Chakrabarti R, Doerig C, & Chakrabarti D (2006). Pre-replication complex organization in the atypical DNA replication cycle of Plasmodium falciparum: Characterization of the mini-chromosome maintenance (MCM) complex formation. Molecular and Biochemical Parasitology, 145(1), 50–59. 10.1016/j.molbiopara.2005.09.006 [DOI] [PubMed] [Google Scholar]
- Quimbaya M, Raspe E, Denecker G, De Craene B, Roelandt R, Declercq W, … Berx G. (2014). Deregulation of the replisome factor MCMBP prompts oncogenesis in colorectal carcinomas through chromosomal instability. Neoplasia, 16(9), 694–709. 10.1016/j.neo.2014.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins JA, Absalon S, Streva VA, & Dvorin JD (2017). The malaria parasite cyclin H homolog PfCyc1 is required for efficient cytokinesis in blood-stage Plasmodium falciparum. MBio, 8(3), e00605–17. 10.1128/mBio.00605-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudlaff RM, Kraemer S, Marshman J, & Dvorin JD (2020). Three-dimensional ultrastructure of Plasmodium falciparum throughout cytokinesis. PLoS Pathogens, 16(6), e1008587. 10.1371/journal.ppat.1008587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudlaff RM, Kraemer S, Streva VA, & Dvorin JD (2019). An essential contractile ring protein controls cell division in Plasmodium falciparum. Nature Communications, 10(1), 2181. 10.1038/s41467-019-10214-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakwe AM, Nguyen T, Athanasopoulos V, Shire K, & Frappier L (2007). Identification and characterization of a novel component of the human minichromosome maintenance complex. Molecular and Cellular Biology, 27(8), 3044–3055. 10.1128/MCB.02384-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santosa V, Martha S, Hirose N, & Tanaka K (2013). The fission yeast minichromosome maintenance (MCM)-binding protein (MCM-BP), Mcb1, regulates MCM function during prereplicative complex formation in DNA replication. The Journal of Biological Chemistry, 288(10), 6864–6880. 10.1074/jbc.M112.432393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M, He W, & Zhang JJ (2005). The DNA replication factor MCM5 is essential for Stat1-mediated transcriptional activation. Proceedings of the National Academy of Sciences of the United States of America, 102(41), 14539–14544. 10.1073/pnas.0507479102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Quimbaya M, Schubert V, Lammens T, Vandepoele K, Schubert I, … De Veylder L (2010). The MCM-binding protein ETG1 aids sister chromatid cohesion required for postreplicative homologous recombination repair. PLoS Genetics, 6(1), e1000817. 10.1371/journal.pgen.1000817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor HM, McRobert L, Grainger M, Sicard A, Dluzewski AR,Hopp CS, … Baker DA. (2010). The malaria parasite cyclic GMP-dependent protein kinase plays a central role in blood-stage schizogony. Eukaryotic Cell, 9(1), 37–45. 10.1128/ec.00186-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trager W, & Jensen J (1976). Human malaria parasites in continuous culture. Science, 193(4254), 673–675. 10.1126/science.781840 [DOI] [PubMed] [Google Scholar]
- Usanga EA, O’Brien E, & Luzzato L (1986). Mitotic inhibitors arrest the growth of Plasmodium falciparum. FEBS Letters, 209(1), 23–27. 10.1016/0014-5793(86)81077-8 [DOI] [PubMed] [Google Scholar]
- White MW, & Suvorova ES (2018). Apicomplexa cell cycles: Something old, borrowed, lost, and new. Trends in Parasitology, 34(9), 759–771. 10.1016/j.pt.2018.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. (2018). World malaria report 2018 (pp. 1–210). Geneva, Switzerland: World Health Organization. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


