Abstract
Summary
Fine-mapping human leukocyte antigen (HLA) genes involved in disease susceptibility to individual alleles or amino acid residues has been challenging. Using information regarding HLA alleles obtained from HLA typing, HLA imputation or HLA inference, our software expands the alleles to amino acid sequences using the most recent IMGT/HLA database and prepares a dataset suitable for fine-mapping analysis. Our software also provides useful functionalities, such as various association tests, visualization tools and nomenclature conversion.
Availability and implementation
1 Introduction
Human leukocyte antigen (HLA) genes encode the major histocompatibility complex (MHC) proteins, which control the immune responses and affect one’s susceptibility to a diverse set of diseases, including autoimmune and neuropsychiatric diseases. HLA fine-mapping is a technique that identifies the allele or amino acid residue in the HLA genes causing the disease. Recently, HLA fine-mapping has grown in popularity owing to the development of HLA imputation and inference technologies. These technologies allowed researchers to collect information on HLA alleles from numerous samples without performing expensive HLA typing analyses (Dilthey et al., 2011, 2016; Jia et al., 2013; Xie et al., 2017; Zheng et al., 2014).
Unfortunately, performing a systematic HLA fine-mapping analysis remains challenging for many researchers. To investigate which amino acid residue or DNA base position is contributing to the disease, each HLA allele needs to be expanded to the amino acid and DNA sequences from the IMGT/HLA database and be processed into a format that facilitates various downstream analyses. It is important to use the most recent database containing the updated sequence and information on the allele names for the analysis because the IMGT/HLA database is dynamically expanding and evolving over time as information on new alleles is incorporated. For example, the number of HLA allele entries in the database has increased by more than 2-fold over the past 5 years. Moreover, due to the complex history of the HLA naming convention, researchers may need to convert HLA allele names to the updated nomenclature. Often, specific statistical tests are required to account for the multi-allelic nature of the amino acid residues at a single amino acid position.
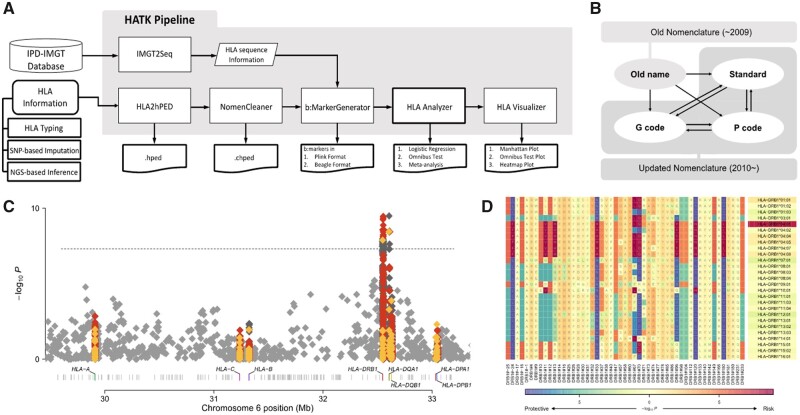
In this article, we introduce a novel software package, HLA Analysis Tool Kit (HATK), which helps in overcoming these obstacles (Fig. 1A). Our software takes the HLA alleles of samples as input, for which the output of various imputation or inference technologies can be directly fed. HLA allele names are automatically checked and corrected to conform to the present naming standards. Next, our software generates binary markers for HLA alleles, amino acid residues, and intragenic single-nucleotide polymorphisms (SNPs), which can be conveniently used for the fine-mapping of association signals. These markers are created based on the sequence information in the IMGT/HLA database, in which a specific version may be selected. Our streamlined pipeline performs fine-mapping of the association signals using various association test methods and provides an effective method of visualizing the results.
Fig. 1.
(A) Overall diagram of the HATK pipeline. (B) Nomenclature conversions provided by NomenCleaner. (C) MHC Manhattan plot and (D) MHC heatmap plot examples. The shading under an amino acid residue shows the amino acid association, whereas the shading under an allele shows the allele association. Both plots were generated using rheumatoid arthritis case/control data from the Wellcome Trust Case Control Consortium (Burton et al., 2007). Red: amino acid, yellow: HLA allele, dark gray: intragenic SNP markers, light gray: intergenic SNP markers in the Manhattan plot
2 Methods
2.1 Automatic HLA file conversion
The HLA2HPED module converts the output from various HLA imputation or inference technologies (Dilthey et al., 2011; Jia et al., 2013; Zheng et al., 2014) to HPED (HLA PED), which is a file format that was introduced to handle HLA allele information in a unified manner. It is similar to the PED file format that was defined in PLINK (Purcell et al., 2007), but the genomic alleles are replaced by the alleles of the eight classical HLA genes (A, B, C, DPA1, DPB1, DQA1, DQB1 and DRB1).
2.2 Obtaining the HLA sequence and nomenclature
The IMGT2Seq module preprocesses the raw database files that users can download from the IMGT/HLA database (Robinson et al., 2016). Specifically, IMGT2Seq generates the following: (i) the amino acid and DNA sequence dictionaries of the eight HLA genes and (ii) the table of Old/Standard/G-group/P-group names for HLA alleles based on the HLA nomenclature of the given database files. Users can either use the most recent version or a specific version of the database. The processed files are used for b:MarkerGenerator and NomenCleaner modules in the following steps.
2.3 HLA nomenclature cleaning
The NomenCleaner module is a versatile tool for converting HLA allele names to the most up-to-date naming conventions (Fig. 1B). First, the NomenCleaner can convert old names, which had been used until 2009, to updated names (e.g. A*9202 to A*02:102). Second, it can convert standard names to the appropriate P-group or G-group name and vice versa. If multiple solutions exist (e.g. A*24:02P can correspond with A*24:02:01:01, A*24:02:01:02L, …), our module maps to the solution that appears first in the database (A*24:02:01:01) and reports the possible candidates in the log. Thirdly, it can clean the names in the nonstandard convention without colons to the standard convention, since the numbers in each field are often ambiguous without colons. For example, it is unclear whether the allele DPB1*101101 corresponds with DBP1*10:11:01, DPB1*101:101 or DPB1*1011:01. NomenCleaner searches the entire IMGT database and determines that DPB1*1011:01 is the only valid solution. This module is embedded in our preprocessing step for input cleaning in HATK, but it can also be used as a standalone program.
2.4 b:Marker generation
For facilitating fine-mapping, the b:MarkerGenerator module generates binary markers that represent genetic variations in the HLA genes. Since HLA genes are highly polymorphic, one HLA gene can have multiple alleles, multiple residues may be present at one amino acid position, and one exonic SNP can have multiple SNP alleles. Similar to the function of SNP2HLA (Jia et al., 2013), b:MarkerGenerator generates binary markers (b:markers) that represent the presence/absence of each allele or amino acid residue. We designed an extended marker name format; for example, AA_DRB1_11_32552129_exon2_V represents the presence of valine at the 11th amino acid position of HLA-DRB1, of which the center of the codon is located at chr6:32552129 in the second exon. The b:marker data are generated in the PLINK format for which the genomic coordinate can be selected (hg18, hg19 or GRCh38).
2.5 HLA analyzer
HLA analyzer is a wrapper script that runs PLINK and in-house R scripts for performing the association analyses of HLA genes using the b:marker data. It can perform logistic regression analyses with covariates, stepwise conditional analyses, omnibus tests for each amino acid position and meta-analyses of multiple datasets.
2.6 HLA visualizer
HLA visualizer is a wrapper script that runs in-house R scripts for visualizing the association results of HLA. The Manhattan plot presents the P-values of all b:markers within the MHC region (Fig. 1C). The amino acid Manhattan plot shows the omnibus test P-values for the amino acid positions within each HLA gene. The MHC heatmap plot shows the P-values for the HLA alleles and amino acid residues in a matrix form (Fig. 1D).
Funding
B.H. and W.C. were supported by the National Research Foundation of Korea (NRF) (grant number 2019R1A2C2002608) funded by the Korean Government, Ministry of Science and ICT. S.R. was supported by funding from NIH/NIAID (U19 AI111224-01). This work was supported by the Creative-Pioneering Researchers Program funded by Seoul National University (SNU).
Conflict of Interest: B.H. is the CTO of Genealogy Inc.
Contributor Information
Wanson Choi, Department of Biomedical Sciences, BK21 Plus Biomedical Science Project, Seoul National University College of Medicine, Seoul 03080, Republic of Korea.
Yang Luo, Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115, USA; Program in Medical and Population Genetics, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA; Center for Data Sciences, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115, USA; Division of Rheumatology, Inflammation, and Immunity, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115, USA; Department of Biomedical Informatics, Harvard Medical School, Boston, MA 02115, USA.
Soumya Raychaudhuri, Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115, USA; Program in Medical and Population Genetics, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA; Center for Data Sciences, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115, USA; Division of Rheumatology, Inflammation, and Immunity, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA 02115, USA; Department of Biomedical Informatics, Harvard Medical School, Boston, MA 02115, USA; Centre for Genetics and Genomics Versus Arthritis, Manchester Academic Health Science Centre, University of Manchester, Manchester M13 9PL, UK.
Buhm Han, Department of Biomedical Sciences, BK21 Plus Biomedical Science Project, Seoul National University College of Medicine, Seoul 03080, Republic of Korea.
References
- Burton P.R. et al. (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature, 447, 661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilthey A.T. et al. (2011) HLA*IMP—an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics, 27, 968–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilthey A.T. et al. (2016) High-accuracy HLA type inference from whole-genome sequencing data using population reference graphs. PLoS Comput. Biol., 12, e1005151–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X. et al. (2013) Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One, 8, e64683–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. et al. (2016) The IPD-IMGT/HLA database—new developments in reporting HLA variation. Hum. Immunol., 77, 233–237. [DOI] [PubMed] [Google Scholar]
- Xie C. et al. (2017) Fast and accurate HLA typing from short-read next-generation sequence data with xHLA. Proc. Natl. Acad. Sci. USA, 114, 8059–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X. et al. (2014) HIBAG—HLA genotype imputation with attribute bagging. Pharmacogenomics J., 14, 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]