Abstract
Summary
Ribbon is an alignment visualization tool that shows how alignments are positioned within both the reference and read contexts, giving an intuitive view that enables a better understanding of structural variants and the read evidence supporting them. Ribbon was born out of a need to curate complex structural variant calls and determine whether each was well supported by long-read evidence, and it uses the same intuitive visualization method to shed light on contig alignments from genome-to-genome comparisons.
Availability and implementation
Ribbon is freely available online at http://genomeribbon.com/ and is open-source at https://github.com/marianattestad/ribbon.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Visualization has played an important role in the genomic revolution, enabling scientists to investigate variants, expression patterns, evolutionary changes and a number of other relationships (Kent et al., 2002; Krzywinski et al., 2009; Robinson et al., 2011). Long-standing genome browsers have taken a very useful reference-based perspective, which is largely sufficient for representing short-read alignments. However, advances in long-read sequencing (Sedlazeck et al., 2018) have shed light on examples where visualizing the portions of reads that maps to different parts of the reference is crucial. When a read is over 10 kb long, it can cover large compound structural variations on several loci, but that context is lost using IGV (Robinson et al., 2011) or other reference-centric viewers without adding a read-based perspective and showing all relevant reference loci. In recent years, other visualization tools have tackled some challenging aspects of validating structural variant calls. In particular, SV-plaudit (Belyeu et al., 2018) and SVCurator (Chapman et al., 2019) have enabled crowd-sourced curation of structural variants, and svviz (Spies et al., 2015) has used realignment against a putative structural variant allele to better evaluate the evidence. Even with these very useful approaches, more complex patterns of structural variation remain hard to represent. A more flexible approach can be helpful by showing the full picture of alignments from both the reference and read perspective, allowing researchers to explore and communicate about complex variation in a more intuitive way. The true power of long reads is hidden when the tools we use are built for short reads. We have addressed this problem by creating Ribbon (http://genomeribbon.com) an interactive online visualization tool that displays alignments along both reference and query sequences, along with variant calls, genes and other genomic features.
2 Materials and methods
Ribbon can load alignments from SAM (Li et al., 2009) and BAM files with long, short or paired-end reads. While the main strengths of Ribbon are more apparent with long sequences, short and paired-ends reads are also supported and can benefit from the more structurally focused visualization Ribbon provides (Supplementary Fig. S9). BAM files can be read from either a local file on the user's computer or from a remote server by entering the file's URL (Miller et al., 2014). To support parsing local BAM files, we compiled the command-line utility SAMtools (Li et al., 2009) from C to WebAssembly (Haas et al., 2017), which allows us to process BAM files efficiently inside a web browser.
In addition to long-read alignment, Ribbon also has features for the related field of whole-genome alignment visualization. First, we added support in Ribbon for visualizing genome-to-genome alignments, such as for comparing genomes of two related species or comparing a new genome assembly to an existing reference. This is done by supporting a simple tab-delimited, human-readable coordinate file format that can be created by the whole genome aligner MUMmer (Kurtz et al., 2004; Robinson et al., 2011) or by scripting from another file format. Second, we adopted a version of the dot plot visualization as an optional alternative to the classic ribbon visualization, since dot plots are often used in the genome assembly/comparison field. The ribbon and dot plot visualizations show the same coordinates in different ways, but most users have found the ribbon visualization more intuitive, while the dot plot remains a common visualization method in the field of whole-genome alignment.
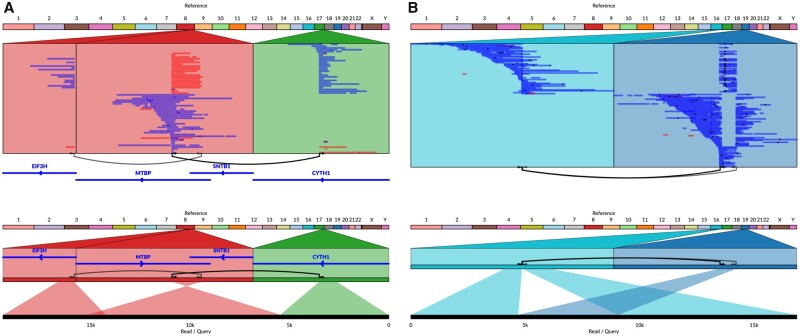
Finally, in addition to alignments with SAM, BAM and general coordinate files, Ribbon can also show variants in VCF or BEDPE format, and genes or any other features in BED format. The user can select a variant to jump to that locus (or both loci for two-breakpoint variants) in the genome, and all the reads with alignments in the given region(s) will show up, including all their alignments to other places in the genome. This is particularly powerful because it pulls other relevant regions into focus; for example, in Figure 1A, the left-most locus is included automatically because it contains alignments of reads also found at the other two loci.
Fig. 1.
Ribbon shows all alignments from the perspective of both the reference and the read or query. Here, long reads from SMRT sequencing show evidence for complex variants in the SK-BR-3 breast cancer genome. (A) The CYTH1-EIF3H gene fusion takes place through a series of two events that are captured within some of the long reads. (B) Long reads from SMRT sequencing show evidence of a full interchromosomal translocation where a sequence is homozygous deleted from chromosome 19 and homozygous inserted into chromosome 16
3 Results
By showing a synchronized read and reference perspective, Ribbon shows patterns in alignments of many reads across multiple chromosomes, while allowing detailed inspection of individual reads (Supplementary Note S1). For example, here we show a gene fusion in the SK-BR-3 breast cancer cell line linking the genes CYTH1 and EIF3H revealed by PacBio long-read sequencing (Nattestad et al., 2018). While it has been found in the transcriptome previously (Chen et al., 2013; Edgren et al., 2011; Kim and Salzberg, 2011), genome sequencing did not identify a direct chromosomal fusion between these two genes. Using long-read sequencing, Ribbon shows that there are indeed reads that span from one gene to the other, going through not one but two variants, for the first time showing the genomic link between these two genes (Nattestad et al., 2018) (Fig. 1A). More gene fusions of this cancer cell line are investigated with Ribbon in Supplementary Note S2. Figure 1B shows another complex event in this sample made simple in Ribbon: the translocation of a 4.4-kb sequence deleted from chr19 and inserted into chr16.
With the support for genome-genome alignments, Ribbon can also be used to test assembly algorithms or inspect the similarity between species. Supplementary Note S4 shows a comparison of gorilla (Gordon et al., 2016) and human genomes using Ribbon, highlighting major structural differences.
4 Discussion
Ribbon enables understanding of complex variants, and it may also help in the detection of sequencing and sample preparation issues, testing of aligners and variant-callers and rapid curation of structural variant candidates (Supplementary Note S3). Ribbon is a powerful and versatile visualization tool for investigating complex structural differences between any two genomes, using intuitive methods that take advantage of the rich context of long reads and contigs.
Funding
This work was supported by the National Science Foundation (NSF) [DBI-1350041] and the National Human Genome Research Institute (NHGRI) [R01-HG006677].
Conflict of Interest: During the initial work, M.N. was a contractor and C.-S.C. was an employee and stockholder of Pacific Biosciences, a company commercializing DNA sequencing technologies. M.N. is currently an employee and stockholder of Google. C.-S.C. is currently an employee and stockholder of DNAnexus. R.A. is an employee and stockholder of Invitae. The other author declared no conflict of interest.
Supplementary Material
Contributor Information
Maria Nattestad, Simons Center for Quantitative Biology, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 11724, USA.
Robert Aboukhalil, Invitae, San Francisco, CA 94103, USA.
Chen-Shan Chin, DNAnexus, Mountain View, CA 94040, USA.
Michael C Schatz, Simons Center for Quantitative Biology, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 11724, USA; Department of Computer Science, Johns Hopkins University, Baltimore, MD 21218, USA; Department of Biology, Johns Hopkins University, Baltimore, MD 21218, USA.
References
- Belyeu J.R. et al. (2018) SV-plaudit: a cloud-based framework for manually curating thousands of structural variants. GigaScience, 7, giy064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman L.M. et al. (2019) SVCurator: a crowdsourcing app to visualize evidence of structural variants for the human genome. BioRxiv, doi: 10.1101/581264. [Google Scholar]
- Chen K. et al. (2013) BreakTrans: uncovering the genomic architecture of gene fusions. Genome Biol., 14, R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgren H. et al. (2011) Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol., 12, R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. et al. (2016) Long-read sequence assembly of the gorilla genome. Science, 352, aae0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas A. et al. (2017) Bringing the web up to speed with WebAssembly. ACM SIGPLAN Notices, 52, 185–200. [Google Scholar]
- Kent W.J. et al. (2002) The human genome browser at UCSC. Genome Res., 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Salzberg S.L. (2011) TopHat-fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol., 12, R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M. et al. (2009) Circos: an information aesthetic for comparative genomics. Genome Res., 19, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S. et al. (2004) Versatile and open software for comparing large genomes. Genome Biol., 5, R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. ; 1000 Genome Project Data Processing Subgroup. (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C.A. et al. (2014) Bam.iobio: a web-based, real-time, sequence alignment file inspector. Nat. Methods, 11, 1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nattestad M. et al. (2018) Complex rearrangements and oncogene amplifications revealed by long-read DNA and RNA sequencing of a breast cancer cell line. Genome Res., 28, 1126–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J.T. et al. (2011) Integrative genomics viewer. Nat. Biotechnol., 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlazeck F.J. et al. (2018) Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat. Rev. Genet., 19, 329–346. [DOI] [PubMed] [Google Scholar]
- Spies N. et al. (2015) Svviz: a read viewer for validating structural variants. Bioinformatics, 31, 3994–3996. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.