Abstract
BACKGROUND:
The ability of a genetic risk score to predict risk in established cardiovascular disease and identify individuals who derive greater benefit from PCSK9 (proprotein convertase subtilisin/kexin type 9) inhibition has not been established.
METHODS:
We studied 14 298 patients with atherosclerotic cardiovascular disease from the FOURIER trial (Further Cardiovascular Outcomes Researh With PCSK9 Inhibition in Subjects With Elevated Risk). A 27–single-nucleotide polymorphism genetic risk score defined low (quintile 1), intermediate (quintiles 2–4), and high (quintile 5) genetic risk. Patients were also categorized by major atherosclerotic risk factors including diabetes mellitus, hypertension, low-density lipoprotein cholesterol ≥100 mg/dl, and smoking; multiple (≥2) risk factors was considered high clinical risk. Outcomes consisted of major coronary events (coronary heart death, myocardial infarction, or coronary revascularization) and major vascular events (major coronary events and stroke). Median follow-up was 2.3 years.
RESULTS:
After we adjusted for clinical factors, the genetic risk score was associated with risk for both major vascular events (Ptrend=0.005) and major coronary events (Ptrend<0.0001). Individuals with intermediate and high genetic risk scores had 1.23- and 1.65-fold increased hazard for major coronary events, respectively. Elevated genetic risk was additive to major atherosclerotic risk factors and identified patients more likely to benefit from evolocumab. There was no benefit for major vascular events in patients without multiple clinical risk factors or high genetic risk (hazard ratio [HR], 1.02; absolute risk reduction [ARR], −0.2%, P=0.86). In contrast, there was a 13% relative risk reduction (HR, 0.87 [0.75–0.998], P=0.047) and a 1.4% ARR in patients with multiple clinical risk factors but without high genetic risk and a 31% relative risk reduction (HR, 0.69 [0.55–0.86], P=0.0012), and 4.0% ARR in patients with high genetic risk, irrespective of clinical risk (Ptrend for HR=0.017, ARR Ptrend=0.004). Patients with high genetic risk who received evolocumab had event rates similar to patients with a low burden of both genetic and clinical risk.
CONCLUSION:
Patients without multiple clinical risk factors or high genetic risk had a low event rate and did not appear to derive benefit from evolocumab over 2.3 years. Conversely, patients with multiple clinical risk factors but without high genetic risk had intermediate risk and intermediate risk reduction. Patients with high genetic risk, regardless of clinical risk, had a high event rate and derived the greatest relative and absolute benefit from evolocumab, which mitigated this risk.
Keywords: cardiovascular disease; evolocumab; genetics; PCSK9 protein, human; risk factors
Despite significant progress in treating patients with established atherosclerotic cardiovascular disease, recurrent cardiovascular events are common and lead to substantial morbidity.1 Although traditional clinical risk factors are essential to patient risk stratification, as much as 30% to 60% of the variation in risk in an individual may be explained by genetic factors.2,3 Population-based, genome-wide association studies have identified many single-nucleotide polymorphisms (SNPs) that have been associated with increased risk of incident coronary artery disease. Genetic risk scores (GRS) provide an opportunity to improve individual risk stratification.4
We have previously shown that a GRS using 27 loci individually associated with myocardial infarction could identify individuals with and without coronary artery disease who were more likely to have coronary events and benefit from statin therapy.5 More recently, PCSK9 (proprotein convertase subtilisin/kexin type 9) inhibitors have emerged as an effective new class of low-density lipoprotein cholesterol (LDL-C)–lowering drugs that lower LDL-C by approximately 60% and reduce the risk of major vascular events.6,7 The 2018 cholesterol guidelines now recommend PCSK9 inhibitors for patients with atherosclerotic disease deemed to be very high clinical risk after initiating statins and ezetimibe. Whether genetic risk can add to clinical risk assessment and help determine who will derive the greatest benefit from additional lipid lowering therapy with PCSK9 inhibition is unclear.
METHODS
Study Population
We performed a nested cohort study of 14 298 unrelated European-ancestry patients enrolled in the FOURIER trial (Further Cardiovascular Outcomes Researh With PCSK9 Inhibition in Subjects With Elevated Risk; 7163 in the evolocumab arm and 7135 in the placebo arm; Figure I in the online-only Data Supplement). There were no clinically important differences between the overall trial participants and the individuals in the genetic subset (Table I in the online-only Data Supplement). The FOURIER trial was a multinational, randomized, double-blind, placebo-controlled trial of the efficacy of evolocumab in patients with clinically evident atherosclerotic cardiovascular disease.8 The key inclusion criteria for the trial were an age between 40 and 85 years, LDL-C ≥70 mg/dl or non–high-density lipoprotein cholesterol ≥100 mg/dl, and a history of either myocardial infarction, nonhemorrhagic stroke, or symptomatic peripheral artery disease. All patients included in this study consented for genetic analyses at the time of trial enrollment, had genotyped data that passed quality control, and were of European ancestry. The study was approved by the local institutional review committee. The data, analytical methods, and study materials will not be made universally available to other researchers for purposes of reproducing the results or replicating the procedure. However, we encourage parties interested in collaboration and data sharing to contact the corresponding author directly for further discussions.
Genetic Risk Scores
We set out to validate whether the 27-SNP genetic risk score (GRS-27) could risk-stratify patients with established atherosclerotic cardiovascular disease and predict benefit from evolocumab therapy. The 27-SNP GRS was calculated using the genotype dosage for each allele, multiplied by its weight (Table II in the online-only Data Supplement), and then summed across all variants. The most updated weight was used for each SNP based on a recent large-scale meta-analysis of coronary artery disease genome-wide association studies.9 Each patient received a raw score standardized per 1 SD (continuous) and a percentile score relative to the study population. All scoring was done using PLINK v2.0 (www.cog-genomics.org/plink/2.0/).10 A similar analysis was done to explore the predictive value of a polygenic risk score with 6 334 602 SNPs (PRS-6M).11
Clinical End Points
On the basis of prior work,5,12 we examined 2 outcomes: major coronary events, which reflect the clinical events used to originally discover the genetic variants; and major vascular events, which is the traditional outcome for lipid-lowering therapy. Specifically, major coronary events were defined as coronary heart death, myocardial infarction, and coronary revascularization. Major vascular events included all major coronary events plus stroke. Patients in the genetic cohort were followed for a median of 2.3 years. All end points were formally adjudicated by a blinded clinical-events committee during the trial.
Genotyping and Imputation
Samples were genotyped on the Infinium Global Screening Array chip and called with the Illumina AutoCall algorithm. Preimputation quality control removed variants with a call rate <98% and with a Hardy–Weinberg equilibrium test in European ancestry samples of P<1E-6, and removal of duplicated variants. Twenty samples were removed because of sex discrepancy. Samples with call rate <98% and heterozygosity rate deviating more than 3 SDs from the mean were excluded. PLINK v2.0 was used for preimputation quality control.10 The Will Rayner preimputation checker script v4.2.7 was applied (https://www.well.ox.ac.uk/~wrayner/tools/). A total of 16 469 samples and 509 657 variants were submitted for imputation. Imputation was conducted using Minimac413 on the Michigan Imputation server14 to the TOPMed Freeze5 reference panel (National Heart, Lung, and Blood Institute’s Trans-Omics for Precision Medicine [TOPMed] program).15 Eagle v2.4 was used for phasing.16,17 Postimputation quality control on variants included imputation quality filter of Rsq >0.3. Cryptic relatedness was calculated through identity by descent, and a pi-hat threshold of 0.2 was used to identify unrelated samples. European ancestry individuals were identified using the ADMIXTURE tool,18 using the 1000 Genomes phase 3 v5 reference population19 and cutoff >0.8.
Statistical Analyses
Patients were stratified into quintiles on the basis of their GRS. In keeping with prior work,5 genetic risk categories were then defined as low (quintile 1), intermediate (quintiles 2–4), and high (quintile 5). Continuous variables within each genetic risk group are presented as median and interquartile range and compared using the Wilcoxon rank-sum test. Categorical variables are compared using chi-square test. Cox proportional hazards regression was used to calculate unadjusted and adjusted hazard ratios (HRs) across genetic risk categories, using the low-genetic-risk category as a reference. Adjusted analysis included age, sex, hypertension, diabetes mellitus, smoking, estimated glomerular filtration rate, and ancestry (using the first 5 principal components). All analyses were performed for both major vascular events and major coronary events. For comparison, we also categorized patients by burden of major atherosclerotic clinical risk factors including diabetes, hypertension, baseline LDL-C ≥100 mg/dl, and smoking;20,21 the presence of multiple (≥2) risk factors was considered high clinical risk.
For the GRS-27, we calculated the absolute and relative risk reductions with evolocumab therapy across each genetic risk category for major vascular events and major coronary events. Absolute risk reduction (ARR) was calculated using the difference in proportion of events between placebo arm and treatment arm. Significance testing for gene-treatment interaction was performed for absolute and relative risk reductions across genetic risk categories. The likelihood ratio test was performed to evaluate the goodness of fit of the model based on the GRS-27 versus the model based on both GRS-27 and traditional major clinical risk factors. Treatment interactions were again tested across genetic and clinical risk groups. All P values used a threshold for significance of <0.05. SAS software version 9.4 (SAS Institute Inc.) and R version 3.5.4 (R Core Team, 2019) were used for statistical and quality control analyses.
RESULTS
Patients were an average of 63 years old, 76% male, and had a median follow-up time of 2.3 years (interquartile range, 1.9–2.6). Of these patients, 29% were smokers, 33% had diabetes mellitus, 81% had hypertension, and the median baseline LDL-C was 92 mg/dl. The majority of patients had a history of myocardial infarction (82%), whereas 18% had ischemic stroke, and 15% had peripheral artery disease. In total, 1235 patients had a major vascular event, 1074 of which were major coronary events.
Genetic Risk Score
The GRS was normally distributed across the study cohort (Figure II in the online-only Data Supplement). Baseline characteristics by genetic risk category based on the GRS-27 are presented in the Table. Patients in the high-genetic-risk category were less likely to have traditional clinical risk factors. They were younger, more likely to be female, and less likely to smoke or have diabetes mellitus. A higher proportion of patients in the high-genetic-risk category had a myocardial infarction as their qualifying atherosclerotic event, resulting in higher proportions of stroke in the low-genetic-risk group. The median LDL-C in the patients with high genetic risk was 94 mg/dl, compared with 92 mg/dl in the patients with intermediate risk, and 91 mg/dl in the patients with low genetic risk.
Table.
Baseline Characteristics, by Genetic Risk Category Based on the GRS-27
| Variable | Low (N=2859) | Intermediate (N=8580) | High (N=2859) | P Value |
|---|---|---|---|---|
| Demographics | ||||
| Age, y | 63.4 (8.7) | 62.9 (8.9) | 62.2 (8.7) | <0.0001 |
| Male | 2194 (77) | 6591 (77) | 2121 (74) | 0.01 |
| White | 2852 (99.8) | 8566 (99.8) | 2851 (99.7) | 0.42 |
| Weight, kg | 87.3 (16.3) | 87.4 (16.4) | 86.6 (16.6) | 0.04 |
| Medical history | ||||
| Myocardial infarction | 2230 (78.0) | 7026 (81.9) | 2432 (85.1) | <0.0001 |
| Ischemic stroke | 599 (21.0) | 1498 (17.5) | 408 (14.3) | <0.0001 |
| Peripheral artery disease | 388 (13.6) | 1286 (15.0) | 428 (15.0) | 0.16 |
| Hypertension | 2309 (80.8) | 6908 (80.5) | 2278 (79.7) | 0.55 |
| Diabetes mellitus | 1002 (35.0) | 2817 (32.8) | 872 (30.5) | 0.001 |
| Current cigarette use | 853 (29.8) | 2551 (29.7) | 767 (26.8) | 0.009 |
| Lipid measures, mg/dl | ||||
| LDL cholesterol | 91.0 (79.0–108.5) | 92.0 (79.5–108.5) | 94.0 (82.0–110.0) | <0.0001 |
| Total cholesterol | 168.5 (152.0–189.5) | 169.0 (152.5–190.0) | 170.0 (153.0–191.0) | 0.13 |
| HDL cholesterol | 45.0 (38.0–53.5) | 44.5 (37.5–53.5) | 45.0 (38.0–53.5) | 0.68 |
| Triglycerides | 137.5 (101.0–184.5) | 135.0 (102.0–184.5) | 131.0 (100.0–179.5) | 0.02 |
Values indicate n (%) or median (interquartile range), unless otherwise indicated. GRS indicates genetic risk score; HDL, high-density lipoprotein; and LDL, low-density lipoprotein.
In the placebo arm, 774 patients had a major vascular event, 673 of which were major coronary events. The Kaplan–Meier event rates at 2.5 years in the low-, intermediate-, and high-genetic-risk categories were 10.1%, 11.3%, and 13.8%, respectively, for major vascular events, and 8.0%, 9.7%, and 13.2%, respectively, for major coronary events (Figure 1). After adjusting for clinical factors, the GRS-27 was significantly and independently associated with risk for both major vascular events (Ptrend for HR=0.005) and major coronary events (Ptrend for HR<0.0001). Individuals with a high genetic risk had a 1.65-fold increased hazard for major coronary events (HRadj, 1.65 [95% CI, 1.30, 2.10]), whereas individuals with intermediate genetic risk had a 1.23-fold increased hazard compared with those with low genetic risk (HRadj, 1.23 [95% CI, 0.99, 1.52]). A similar gradient was present for major vascular events, with a 1.37-fold increased hazard in the high-genetic-risk category (HRadj, 1.37 [95% CI, 1.10, 1.71]) and a 1.14-fold increased hazard in the intermediate-genetic-risk category (HRadj, 1.14 [95% CI, 0.94, 1.38]). The hazard ratio per 1 SD increase in GRS was 1.17 (95% CI, 1.08, 1.26; P=0.0001) and 1.10 (95% CI, 1.03, 1.18; P=0.005) for major coronary events and major vascular events, respectively. The continuous relationship between GRS-27 and log hazard ratio of major coronary events is presented in Figure III in the online-only Data Supplement. Data on additional end points are included in Figure IV in the online-only Data Supplement.
Figure 1. Kaplan–Meier rates for major vascular events and major coronary events in the placebo arm, by genetic risk category based on the GRS-27.
(A) Major vascular events and (B) major coronary events. GRS indicates genetic risk score; HR, hazard ratio; Int, intermediate; KM, Kaplan–Meier; and No., number.
The PRS-6M had a comparable distribution of baseline characteristics (Table III in the online-only Data Supplement) and a qualitatively similar risk stratification in the placebo arm (Table IVA and IVB in the online-only Data Supplement). Specifically, patients with high genetic risk had a 1.55-fold increased hazard for major coronary events (HRadj, 1.55 [95% CI, 1.21, 1.98]) and individuals with intermediate genetic risk had a 1.26-fold increased hazard compared with those with low genetic risk (HRadj, 1.26 [95% CI, 1.02, 1.56]). For major vascular events, there was a 1.31-fold increased hazard in the high-genetic-risk group (HRadj, 1.31 [95% CI, 1.05, 1.64]) and a 1.11-fold hazard in the intermediate-genetic-risk group (HRadj, 1.11 [95% CI, 0.91, 1.34]). The hazard ratio per 1 SD increase in PRS-6M was 1.16 (1.07, 1.25; P=0.0002) and 1.10 (1.02, 1.18; P=0.013), for major coronary events and major vascular events, respectively. The continuous relationship between PRS-6M and log hazard ratio of major coronary events is presented in Figure III in the online-only Data Supplement.
GRS and Clinical Benefit of Evolocumab Versus Placebo
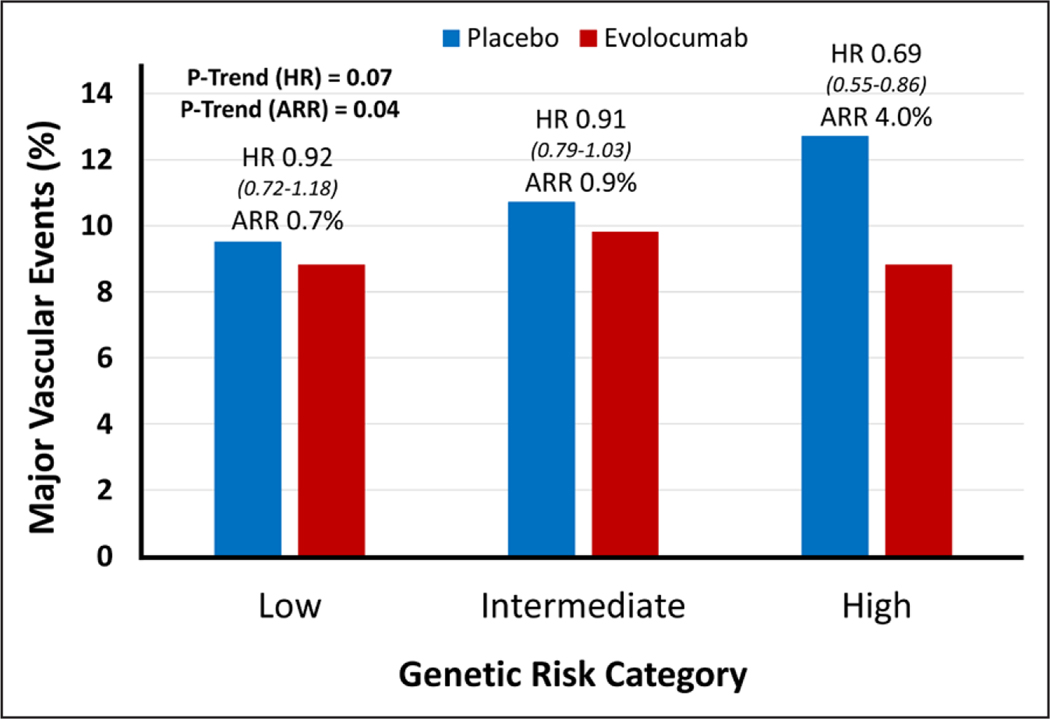
Overall, in both arms, 1446 patients had a major vascular event, 1269 of which were major coronary events. When assessing the benefit of evolocumab therapy by genetic risk categories alone, there was an increased treatment effect in patients with higher genetic risk (Figure 2 and Figure V in the online-only Data Supplement). The hazard ratios (95% CI) for major vascular events in the low-, intermediate-, and high-genetic-risk categories were 0.92 (0.72–1.18), 0.91 (0.79–1.03), and 0.69 (0.55–0.86), respectively (Ptrend for HR=0.07). There was a significant gradient in the overall ARR in major vascular events across the same categories was 0.7%, 0.9%, and 4.0%, respectively (Ptrend=0.04). Treatment with evolocumab completely mitigated the increased risk in the high-genetic-risk category, lowering their event rate to that of the low-genetic-risk category (proportion of patients with events is 8.8% in both groups after treatment).
Figure 2. Relative and absolute risk reduction of major vascular events with evolocumab, by genetic risk category.
ARR indicates absolute risk reduction, and HR, hazard ratio.
Genetics and Clinical Risk
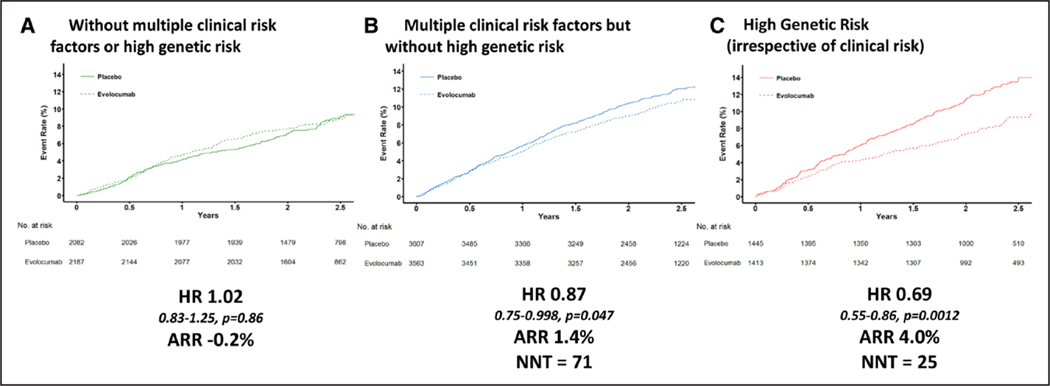
The GRS was independently associated with major coronary or vascular events even after adjustment for major atherosclerotic clinical risk factors (for major coronary events, likelihood ratio test ; for major vascular events, likelihood ratio test ). The combination of both genetic and major clinical risk factors helped define a wide gradient of relative (Ptrend for HR=0.017) and absolute (Ptrend=0.004) clinical benefit from PCSK9 inhibition (Figure 3 and Figure VI in the online-only Data Supplement). Specifically, in patients without multiple clinical risk factors or high genetic risk, no benefit was observed over a median of 2.3 years (HR, 1.02, ARR, –0.2%, P=0.86). In contrast, there was a 13% relative risk reduction (HR, 0.87 [95% CI, 0.75–0.998], P=0.047) and 1.4% ARR in major vascular events in patients with multiple clinical risk factors but without high genetic risk, and a 31% relative risk reduction (HR, 0.69 [95% CI, 0.55–0.86], P=0.0012) and 4.0% ARR in patients with high genetic risk (irrespective of major clinical risk factors). There was no significant difference for the ARR across clinical risk factor burden in the high-genetic-risk category for either major vascular events or major coronary events (Table V in the online-only Data Supplement). The hazard ratio with evolocumab as a function of genetic risk modeled as a continuous variable and further subsetted by clinical risk is shown in Figure VII in the online-only Data Supplement. The treatment benefit started to take effect at approximately 9 months in patients with multiple clinical risk factors but without high genetic risk and at approximately 3 months in patients with high genetic risk. Patients with high genetic risk who received evolocumab had event rates similar to patients with a low burden of both genetic and clinical risk factors (2.5-year Kaplan–Meier rate of 9.3% and 9.1%, respectively).
Figure 3. Treatment effect for major vascular events of evolocumab therapy, by genetic and clinical risk category.
(A) Patients without multiple clinical risk factors or high genetic risk, (B) patients with multiple clinical risk factors but without high genetic risk, and (C) patients with high genetic risk (irrespective of clinical risk). ARR indicates absolute risk reduction; HR, hazard ratio; NNT, number needed to treat; and No., number.
DISCUSSION
This study highlights 3 important observations when considering genetic risk in clinical practice. The first is the validation of a 27-SNP GRS for identifying patients in a secondary prevention population who are at elevated risk of cardiovascular events. Second, genetic risk is independent of traditional clinical risk factors that we currently use to assess cardiovascular risk. Third, a 27-SNP GRS combined with traditional major atherosclerotic clinical risk factors identifies a gradient of risk and benefit. Patients without multiple clinical risk factors or high genetic risk did not appear to derive benefit from evolocumab over 2.3 years. In contrast, patients with multiple clinical risk factors but without high genetic risk had an intermediate risk reduction, and patients with high genetic risk derived the greatest relative and absolute benefit from evolocumab therapy.
Building on our prior work,5 we demonstrate that the GRS-27 provides a significant gradient of risk stratification in a secondary prevention population. This risk gradient is most significant for coronary events, which is expected as these SNPs were originally identified in genome-wide association studies for incident CAD. Specifically, patients at high genetic risk showed evidence of increased risk as early as 3 months into the study and were ultimately at a 65% increased risk of coronary events. Likewise, PRS-6M identified a subset of patients who were at a 55% increased risk of coronary events.
For years, clinical risk factors and scoring systems have been used to estimate a patient’s risk of cardiovascular events.22,23 There is now the ability to consider a patient’s genetic risk, but whether this approach offered additional value in secondary prevention remained uncertain. One hypothesis was that genetic risk would no longer be relevant after decades of exposure to traditional risk factors and establishment of overt atherosclerotic disease. However, we demonstrate that genetics remain an important predictor of risk, independent of and beyond traditional major clinical risk factors. In fact, genetic risk further stratifies patients with both low and high clinical risk factor burden, such that the presence of both is worse than either one alone, and the presence of neither carries the lowest risk of all. Other studies have supported the additive value of clinical and genetic risk stratification for cardiovascular disease.24
As the list of available cardiovascular drugs grows, strategies that match patients with the best individualized therapies will be necessary. In this study, we demonstrate that genetics may be one such strategy. We found that patients with the highest genetic risk for coronary events also derived the greater benefit from evolocumab therapy. Specifically, patients with high genetic risk achieved a 31% relative risk reduction and 4% ARR, approximately double the benefit seen in the overall trial. In addition, the benefit of therapy with evolocumab began to emerge very early in treatment, at approximately 3 months, which likely reflects that high genetic risk imparts a substantial atherosclerotic burden that is potentially modifiable by powerful lipid lowering therapy. Of note, the elevated genetic risk seen in these patients was completely mitigated by treatment with evolocumab, neutralizing their risk to the level of patients with low genetic risk.
Only a minority of the SNPs are found near genes that are known to be a part of a lipid pathway. Other SNPs are near genes implicated in inflammation, vascular tone or stability, but the mechanisms through which these polymorphisms contribute to increased atherosclerosis has not yet been established.25 Regardless, imaging studies have demonstrated that patients with higher genetic risk using a similar risk score had a higher burden of atherosclerosis.26 These data, in turn, are concordant with prior clinical observations that patients with a greater clinical burden of atherosclerosis enjoy a larger and more rapid benefit with PCSK9 inhibition.27 In the case of genetics, the predisposition to atherosclerosis would be lifelong.
In addition to tailoring aggressive LDL-C–lowering therapy for patients, genetics could be used to run more efficient clinical trials. Specifically, incorporating genetic risk as an enrichment criterion would lead to higher event rates, greater absolute and relative risk reduction, and the ability to enroll fewer patients for a shorter period of time to demonstrate efficacy at a lower cost, albeit with the results applying to a more focused population.
The observation that more SNPs does not necessarily translate to better risk prediction may be counterintuitive but should be taken in context. The genome-wide polygenic risk score, like others, was developed based on patients with a primary event of coronary artery disease versus controls free of coronary artery disease. This is a much different comparison than a secondary prevention population in which one is trying to determine which patients with atherosclerotic disease will have recurrent events. This more subtle difference between patients in a secondary prevention population may have led to an attenuated risk prediction with the polygenic risk score. In addition, the strength of the polygenic risk score is at the extremes of the population, where a small number of patients will have many nominally weighted SNPs that add up to an important increase in risk. When focusing on the top 20% of a cohort, the polygenic score may not offer any greater precision in risk estimation in this population.
Limitations
This was a subgroup analysis of a clinical trial population and therefore the results may not be generalizable to all populations. Specifically, this study focused on patients of European ancestry because this is where the majority of genome-wide association study data is derived. Furthermore, genotyping was only performed in consenting patients. We chose to do the bulk of the analysis treating genetic risk as a categorical variable. This is more clinically interpretable but leads to a loss of power, so continuous data was also presented. In addition, patients were divided into categories based on percentile relative to the study population, not a healthy reference population. This likely resulted in patients with higher genetic risk being forced into lower risk categories than would otherwise be expected. This could have weakened the signal for event prediction. Reference values for GRSs are not yet established but are an important step for future research and necessary for implementation into clinical practice.
CONCLUSION
In established cardiovascular disease, a GRS identified subsets of patients who had significantly increased risk of recurrent events beyond clinical risk factors and identified a gradient of benefit from evolocumab. Patients without multiple clinical risk factors or high genetic risk had a low event rate and did not appear to derive benefit from evolocumab over 2.3 years in this study. This does not preclude a benefit in these patients if treated for a longer period of time. At the other extreme, patients with high genetic risk had a high event rate and over a relatively short time frame derived large relative and absolute benefit from evolocumab therapy, which mitigated this risk.
Supplementary Material
Clinical Perspective.
What Is New?
This is the first study to demonstrate an interaction between a genetic risk score and treatment benefit from a PCSK9 (proprotein convertase subtilisin/ kexin type 9) inhibitor.
Patients in the top 20% of genetic risk received a 2-fold greater benefit from evolocumab than the overall trial population.
What Are the Clinical Implications?
This 27–single-nucleotide polymorphism genetic risk score can be used to personalize therapy, identifying patients with higher risk in whom PCSK9 inhibition should be strongly considered.
Treatment with evolocumab in patients with high genetic risk completely mitigates the increased genetic risk, lowering their event rate to that of the patients with low genetic risk.
Acknowledgments
Dr Marston contributed to the study design, literature search, statistical analysis, data interpretation, figures, and drafting of the article. Drs Kamanu, Nordio, and Gurmu and C. Roselli contributed to the data preparation, study design, and statistical analysis. Drs Sever, Pedersen, Keech, Wang, Pineda, and Giugliano contributed to the data interpretation and critical review of the article. Drs Lubitz, Ellinor, Sabatine, and Ruff contributed to the study design, statistical analysis, data interpretation, figures, and critical review of the article. Drs Sabatine and Ruff are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and accuracy of the data analysis.
Sources of Funding
The present study was funded by Amgen.
Disclosures
Dr Marston reports a significant research grant from the US National Institutes of Health’s National Research Service Award. Drs Kamanu, Nordio, Gurmu, and Ruff are members of the TIMI Study Group, which has received institutional research grant support through Brigham and Women’s from Abbott, Amgen, Aralez, AstraZeneca, Bayer HealthCare Pharmaceuticals, Inc., BRAHMS, Daiichi-Sankyo, Eisai, GlaxoSmithKline, Intarcia, Janssen, MedImmune, Merck, Novartis, Pfizer, Poxel, Quark Pharmaceuticals, Roche, Takeda, The Medicines Company, and Zora Biosciences. Dr Sever reports research grants and honoraria for speaker’s bureau from Amgen and Pfizer. Dr Pedersen reports modest speaker honoraria from as well as consults for Sanofi and Amgen. Dr Keech reports grants and personal fees from Abbott, personal fees from Amgen, personal fees from AstraZeneca, grants and personal fees from Mylan, personal fees from Pfizer, grants from Sanofi, grants from Novartis, and personal fees from Bayer, outside the submitted work. Dr Wang reports salary support from Amgen during the conduct of the study and salary support from Amgen, outside the submitted work. Dr Pineda reports salary support from Amgen during the conduct of the study. Dr Giugliano reports grants from Merck during the conduct of the study; personal fees from Akcea; grants and personal fees from Amarin; personal fees from the American College of Cardiology; grants and personal fees from Amgen; personal fees from Angel Med; personal fees from Beckman-Coulter; personal fees from Boeringer-Ingelheim; personal fees from Bristol-Myers Squibb; personal fees from CVS Caremark; grants and personal fees from Daiichi Sankyo; personal fees from GlaxoSmithKline; personal fees from Janssen; personal fees from Lexicon; grants and personal fees from Merck; personal fees from Portola; personal fees from Pfizer; personal fees from St Jude; and personal fees from Stealth Peptide, outside the submitted work; and an institutional research grant to the TIMI Study Group at the Brigham and Women’s Hospital for research he is not directly involved in from Abbott, Amgen, Aralez, AstraZeneca, Bayer HealthCare Pharmaceuticals, Inc., BRAHMS, Daiichi-Sankyo, Eisai, GlaxoSmithKline, Intarcia, Janssen, MedImmune, Merck, Novartis, Pfizer, Poxel, Quark Pharmaceuticals, Roche, Takeda, The Medicines Company, and Zora Biosciences. Dr Lubitz reports grants from the US National Institutes of Health, grants from the American Heart Association, grants from Boehringer Ingelheim, grants and personal fees from Bristol-Myers Squibb/Pfizer, grants and personal fees from Bayer AG, and personal fees from Quest Diagnostics, outside the submitted work. Dr Ellinor reports grants and personal fees from Bayer AG, personal fees from Novartis, and personal fees from Quest Diagnostics, outside the submitted work. Dr Sabatine reports significant research grant support from Abbott Laboratories, Amgen, AstraZeneca, Bayer, Critical Diagnostics, Daiichi-Sankyo, Eisai, Genzyme, Gilead, GlaxoSmithKline, Intarcia, Janssen Research and Development, The Medicines Company, MedImmune, Merck, Novartis, Poxel, Pfizer, Quark Pharmaceuticals, Roche Diagnostics, and Takeda; has received modest consulting feest from Alnylam, AstraZeneca, Bristol-Myers Squibb, CVS Caremark, Dyrnamix, Esperion, IFM Pharmaceuticals, Intarcia, Ionis, Janssen Research and Development, The Medicines Company, MedImmune, Merck, MyoKardia, and Novartis; and has received significant consulting fees from Amgen. Dr Ruff reports grants from Boehringer Ingelheim, Daiichi Sankyo, MedImmune, and the National Institute of Health, as well as personal fees from Anthos, Bayer, Boehringer Ingelheim, Bristol-Myers Squibb, Daiichi Sankyo, Janssen, MedImmune, Pfizer, and Portola, outside the submitted work. The other authors report no conflicts.
Footnotes
The online-only Data Supplement is available with this article at https://www.ahajournals.org/doi/suppl/10.1161/circulationaha.119.043805.
REFERENCES
- 1.Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, de Ferranti S, Després JP, Fullerton HJ, Howard VJ, et al. ; American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Heart disease and stroke statistics–2015 update: a report from the American Heart Association. Circulation. 2015;131:e29–322. doi: 10.1161/CIR.0000000000000152 [DOI] [PubMed] [Google Scholar]
- 2.Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330:1041–1046. doi: 10.1056/NEJM199404143301503 [DOI] [PubMed] [Google Scholar]
- 3.Lloyd-Jones DM, Nam BH, D’Agostino RB Sr, Levy D, Murabito JM, Wang TJ, Wilson PW, O’Donnell CJ. Parental cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults: a prospective study of parents and offspring. JAMA. 2004;291:2204–2211. doi: 10.1001/jama.291.18.2204 [DOI] [PubMed] [Google Scholar]
- 4.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, et al. A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130. doi: 10.1038/ng.3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mega JL, Stitziel NO, Smith JG, Chasman DI, Caulfield M, Devlin JJ, Nordio F, Hyde C, Cannon CP, Sacks F, et al. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. The Lancet. 2015;385:2264–2271. doi: 10.1016/S0140-6736(14)61730-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabatine MS, Giugliano RP, Keech AC, Honarpour N, Wiviott SD, Murphy SA, Kuder JF, Wang H, Liu T, Wasserman SM, et al. ; FOURIER Steering Committee and Investigators. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med. 2017;376:1713–1722. doi: 10.1056/NEJMoa1615664 [DOI] [PubMed] [Google Scholar]
- 7.Schwartz GG, Steg PG, Szarek M, Bhatt DL, Bittner VA, Diaz R, Edelberg JM, Goodman SG, Hanotin C, Harrington RA, et al. ; ODYSSEY OUTCOMES Committees and Investigators. Alirocumab and cardiovascular outcomes after acute coronary syndrome. N Engl J Med. 2018;379:2097–2107. doi: 10.1056/NEJMoa1801174 [DOI] [PubMed] [Google Scholar]
- 8.Sabatine MS, Giugliano RP, Keech A, Honarpour N, Wang H, Liu T, Wasserman SM, Scott R, Sever PS, Pedersen TR. Rationale and design of the further cardiovascular outcomes research with PCSK9 inhibition in subjects with elevated risk trial. Am Heart J. 2016;173:94–101. doi: 10.1016/j.ahj.2015.11.015 [DOI] [PubMed] [Google Scholar]
- 9.Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, Zeng L, Ntalla I, Lai FY, Hopewell JC, et al. ; EPIC-CVD Consortium; CARDIoGRAMplusC4D; UK Biobank CardioMetabolic Consortium CHD working group. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017;49:1385–1391. doi: 10.1038/ng.3913 [DOI] [PubMed] [Google Scholar]
- 10.Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–1224. doi: 10.1038/s41588-018-0183-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fulcher J, O’Connell R, Voysey M, Emberson J, Blackwell L, Mihaylova B, Simes J, Collins R, Kirby A, Colhoun H, et al. ; Cholesterol Treatment Trialists’ (CTT) Collaboration. Efficacy and safety of LDL-lowering therapy among men and women: meta-analysis of individual data from 174,000 participants in 27 randomised trials. Lancet. 2015;385:1397–1405. doi: 10.1016/S0140-6736(14)61368-4 [DOI] [PubMed] [Google Scholar]
- 13.Fuchsberger C, Abecasis GR, Hinds DA. minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–784. doi: 10.1093/bioinformatics/btu704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das S, Forer L, Schönherr S, Sidore C, Locke AE, Kwong A, Vrieze SI, Chew EY, Levy S, McGue M, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48:1284–1287. doi: 10.1038/ng.3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.NHLBI Trans-Omics for Precision Medicine. University of Washington, Seattle WA. 2019. https://www.nhlbiwgs.org. Accessed August 20th, 2019. [Google Scholar]
- 16.Loh PR, Danecek P, Palamara PF, Fuchsberger C, A Reshef Y, K Finucane H, Schoenherr S, Forer L, McCarthy S, Abecasis GR, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loh PR, Palamara PF, Price AL. Fast and accurate long-range phasing in a UK Biobank cohort. Nat Genet. 2016;48:811–816. doi: 10.1038/ng.3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR; 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arnett DK, Blumenthal RS, Albert MA, Buroker AB, Goldberger ZD, Hahn EJ, Himmelfarb CD, Khera A, Lloyd-Jones D, McEvoy JW, et al. 2019 ACC/AHA Guideline on the Primary Prevention of Cardiovascular Disease: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. 2019;140:e596–e646. doi: 10.1161/CIR.0000000000000678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith SC Jr, Benjamin EJ, Bonow RO, Braun LT, Creager MA, Franklin BA, Gibbons RJ, Grundy SM, Hiratzka LF, Jones DW, et al. ; World Heart Federation and the Preventive Cardiovascular Nurses Association. AHA/ACCF Secondary Prevention and Risk Reduction Therapy for Patients with Coronary and other Atherosclerotic Vascular Disease: 2011 update: a guideline from the American Heart Association and American College of Cardiology Foundation. Circulation. 2011;124:2458–2473. doi: 10.1161/CIR.0b013e318235eb4d [DOI] [PubMed] [Google Scholar]
- 22.D’Agostino RB Sr, Vasan RS, Pencina MJ, Wolf PA, Cobain M, Massaro JM, Kannel WB. General cardiovascular risk profile for use in primary care: the Framingham Heart Study. Circulation. 2008;117:743–753. doi: 10.1161/CIRCULATIONAHA.107.699579 [DOI] [PubMed] [Google Scholar]
- 23.Goff DC Jr, Lloyd-Jones DM, Bennett G, Coady S, D’Agostino RB, Gibbons R, Greenland P, Lackland DT, Levy D, O’Donnell CJ, et al. ; American College of Cardiology/American Heart Association Task Force on Practice Guidelines. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation. 2014;129(25 Suppl 2):S49–S73. doi: 10.1161/01.cir.0000437741.48606.98 [DOI] [PubMed] [Google Scholar]
- 24.Pereira A, Mendonca MI, Borges S, Sousa AC, Freitas S, Henriques E, Rodrigues M, Freitas AI, Guerra G, Freitas C, et al. Additional value of a combined genetic risk score to standard cardiovascular stratification. Genet Mol Biol. 2018;41:766–774. doi: 10.1590/1678-4685-GMB-2017-0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khera AV, Kathiresan S. Genetics of coronary artery disease: discovery, biology and clinical translation. Nat Rev Genet. 2017;18:331–344. doi: 10.1038/nrg.2016.160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Natarajan P, Young R, Stitziel NO, Padmanabhan S, Baber U, Mehran R, Sartori S, Fuster V, Reilly DF, Butterworth A, et al. Polygenic risk score identifies subgroup with higher burden of atherosclerosis and greater relative benefit from statin therapy in the primary prevention setting. Circulation. 2017;135:2091–2101. doi: 10.1161/CIRCULATIONAHA.116.024436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabatine MS, De Ferrari GM, Giugliano RP, Huber K, Lewis BS, Ferreira J, Kuder JF, Murphy SA, Wiviott SD, Kurtz CE, et al. Clinical benefit of evolocumab by severity and extent of coronary artery disease. Circulation. 2018;138:756–766. doi: 10.1161/CIRCULATIONAHA.118.034309 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.