Abstract
Background and Purpose:
Lactobacilli play a vital role in protecting the vagina against pathogens. Cytokines are vital components of defense against infections in women. The genital mycoplasmas, Mycoplasma genitalium and Ureaplasma urealyticum, are associated with various infectious diseases in adults and infants. The objective of our study is to identify differences in cytokine profile and Lactobacillus species dominance between a study group of non-pregnant pre-menopausal women with genital M. genitalium or U. urealyticum colonization and a control group of non-pregnant pre-menopausal women without genital M. genitalium or U. urealyticum colonization.
Methods:
A real-time polymerase chain reaction was performed to measure Lactobacillus species in vaginal swab samples. Cytokine analysis was performed using multiplex immunoassay techniques. Analysis of variance confirmed a significant difference in cytokine profiles between patient groups, with t-tests identifying the most significantly different cytokines. Categorical data analysis identified significant patterns of relative Lactobacillus species dominance in the study group.
Results:
Lactobacillus iners was the predominant Lactobacillus species in the control group (p = 0.005). There were no dominant Lactobacillus species observed in the study group. Vascular endothelial growth factor A (p = 0.002), interleukin-8 (p = 0.001), and interleukin-1β (p = 0.049) were expressed significantly higher in the study group, whereas interleukin-1 receptor antagonist (p < 0.001), interleukin-10 (p = 0.001), interleukin-12 (p = 0.002), and interferon-γ (p = 0.022) were expressed higher in the control group. Association matrices for cytokines were significantly different between two groups (p < 0.001), with mostly negative associations in the control group and mostly positive associations in the study group.
Conclusion:
Cytokine levels, their associations, and the patterns of Lactobacillus species dominance are observed to significantly diverge on the basis of M. genitalium and U. urealyticum colonization among non-pregnant pre-menopausal women.
Keywords: cytokine, genital infections, Lactobacillus, mycoplasma, vaginal microbiome
Introduction
Vulvovaginal infections have different etiologies, including fungi like Candida species; bacteria like Prevotella, Mobiluncus, Gardnerella vaginalis, and Ureaplasma, Mycoplasma; and parasites like Trichomonas vaginalis.1–3 Several factors are potentially implicated in susceptibility or resistance to vulvovaginal infection.
Lactobacilli are obligate homolactic fermenting bacteria that make up the majority of the vaginal microbiome and protect the vagina against pathogens.4 The composition of the bacteria in the vagina changes dynamically in response to several factors such as hormonal fluctuations, sexual intercourse, pregnancy, medications, and vaginal hygiene.5–8 The Lactobacillus species most commonly seen in women of childbearing age include Lactobacillus crispatus, Lactobacillus gasseri, Lactobacillus iners, and Lactobacillus jensenii.9
Together with the vaginal microbiome, careful regulation of cells of the immune system prevents infections by external pathogens.10 Depending on the vaginal environment, changes within that microbiome, that is, diminished dominance of Lactobacillus, cause a variety of immunological changes such as the production of pro-inflammatory cytokines/chemokines, greater recruitment of immune cells, and changes to the vaginal lining.10 The harmony established between the vaginal microbiota and immune system is influenced by sex hormones.10,11 Estradiol (E2) and progesterone (P) are sex hormones secreted by the ovaries which play a crucial role in the regulation of the vaginal immune system. These hormones (E2 and P) influence the epithelial cells (ECs) which stimulate immune cells to secrete cytokines.11 E2 and P function in opposition with each other. For example, E2 acts by upregulating pro-inflammatory transcription factors and alters the immune response of ECs,11 whereas P regulates transcription proteins/growth factors that decrease inflammation and promote mucosal repair.11
Mycoplasma genitalium is an emerging cause of vaginal infections and has been identified in urogenital infections of men and women.12 The genital mycoplasmas represent a complex and unique group of microorganisms associated with a wide variety of infectious diseases in adults and infants. Genital mycoplasmas refer to organisms from the genera Mycoplasma and Ureaplasma. Mycoplasmas and Ureaplasmas belong to the Mycoplasmataceae family and Mollicutes class.13 Important discoveries have revealed the role of M. genitalium in sexually transmitted diseases for men, but its role in reproductive tract diseases of women is still unclear. M. genitalium and Ureaplasma urealyticum can be detected via polymerase chain reaction (PCR).14
The pathogenicity of M. genitalium is mainly driven by virulence factors that allow the organism to bind with host epithelial cells, release specific enzymes, and avoid detection by the host’s immune system via antigenic variation.12 M. genitalium may even induce host inflammatory mediators such as tumor necrosis factor-α (TNF-α), interleukin-1α (IL-1α), IL-1β, interleukin-6 (IL-6), IL-8, and IL-10.12 Cytokines are some of the important components of defense against various infections.15,16
U. urealyticum has several studies investigating its implications in pregnancy outcomes and male infertility, but relatively limited investigations in women of childbearing age who are not pregnant.17,18 U. urealyticum has shown resistance to various antibiotics, varying by region, making closer investigation to identify potential targets for therapy crucial.18 An in-vitro investigation elucidated part of the immune defense against U. urealyticum, demonstrating adult and fetal macrophage upregulation of vascular endothelial growth factor (VEGF) and intercellular adhesion molecule-1.19
In this study, we will compare Lactobacillus spp. and cytokine profiles in non-pregnant pre-menopausal women who were and were not infected with M. genitalium or U. urealyticum. We hypothesized that differences in the centroids and/or correlation patterns of cytokine levels in the control and Mycoplasma or Ureaplasma study groups are significant and that significant patterns of Lactobacillus spp. dominance are present within and/or between the groups.
Objectives
The objective of our study is to delineate the relationship between genital M. genitalium or U. urealyticum colonization in non-pregnant pre-menopausal women and variances in cytokine profiles and Lactobacillus species predominance.
Methods
Vaginal samples were collected as part of a database collection—prospective data bank creation to study vaginal conditions which was Institutional Review Board (IRB)-approved (IRB protocol# L20-225) at Texas Tech University Health Sciences Center, TX, USA. Written consent forms were obtained from all patients for this study. The samples in the database were obtained from the middle of the vagina using standardized cotton swabs.20,21 Vaginal specimens were placed into 1 mL of physiological solution (phosphate-buffered saline) and stored at −80°C.
Real-time PCR (qPCR)
The relative concentration of the vaginal flora was determined by a real-time PCR (qPCR), as described previously.22,23 The qPCR assay was performed to identify vaginal Lactobacillus spp., including L. crispatus, L. gasseri, L. iners, and L. jensenii. In addition, the presence of facultative anaerobic bacteria (G. vaginalis, Atopobium vaginae, Megasphaera spp., Eggerthella spp., Prevotella spp., U. urealyticum, Ureaplasma parvum, Mycoplasma hominis, and M. genitalium) was also determined. The qPCR analysis of gene transcripts was performed using an iCycler Real-Time PCR machine (Bio-Rad Laboratories Inc., Hercules, CA, USA) and 2× TaqMan Master Mix (Thermo Fisher Scientific Inc., Waltham, MA, USA).23 For RNA preparation, all samples were processed using TRIzol (Invitrogen, Carlsbad, CA, USA) method. The qPCR data were analyzed using the comparative ∆∆Ct method.22
Cytokine evaluation
Cytokine analysis was performed using the Bio-Plex MAGPIX Multiplex Reader instrument (Bio-Rad) and the MESO QuickPlex SQ 120 instrument (Meso Scale Discovery (MSD), Rockville, MD, USA):
The Bio-Plex Pro Human Cytokine 27-Plex Immunoassay is a 96-well kit (cat no. M500KCAF0Y, Bio-Rad) that includes magnetic beads, detection antibodies, wash buffer, sample diluent, detection antibody diluent, a 96-well flat-bottom plate, and a plastic adhesive plate seal.23 This assay detects a total of 27 cytokines in the samples. A flat-bottom 96-well plate (provided in the kit) was pre-wetted with 100 µL of Bio-Plex assay buffer. A 50 µL of working bead solution was added into each well. The plate was washed with 100 µL of Bio-Plex wash buffer (2×). Then, 50 µL of standards and 50 µL of samples were added to the appropriate wells of the plate. The plate was covered with a plastic adhesive plate sealer (to block out light) and incubated for 30 min at room temperature with shaking. The plate was then washed with 100 µL of Bio-Plex wash buffer (3×). A 25 µL of Bio-Plex detection antibody diluent was added to each well of the plate. The plate was again covered with a plastic adhesive plate seal and incubated for 30 min at room temperature with shaking. Next, the plate was washed with 100 µL of Bio-Plex wash buffer (3×). A 50 µL of Streptavidin-PE working dilution (100×) was added to each well of the plate. The plate was covered with a plastic adhesive plate seal and incubated for 10 min at room temperature with shaking. Again, the plate was washed with 100 µL of Bio-Plex wash buffer (3×). The beads were resuspended in each well with 125 µL of Bio-Plex assay buffer. The plate was covered with a plastic adhesive plate sealer and shaken at 1100 r/min. Finally, the plastic adhesive plate sealer was removed and the plate was immediately read using the Bio-Plex MAGPIX Multiplex Reader instrument.
MSD cytokine assays provide a rapid and convenient method for measuring cytokines levels of cytokines within a single, small volume sample.23 An MSD 96-well plate was pre-coated with capture antibodies on independent and well-defined spots. All vaginal swab samples were analyzed using the MSD multiplex instrument MESO QuickPlex SQ 120 (MSD).24 The MSD electrochemiluminescence (ECL) detection system has been validated for cytokine measurement in vaginal swab samples.
A total of 5 custom plates were made for the multiplex assays:
Plate 1: IFN-γ (interferon-γ), IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12p70, IL-13, TNF-α
Plate 2: GM-CSF (granulocyte-macrophage colony-stimulating factor), IL-5, IL-7, IL-15, IL-17A
Plate 3: Eotaxin, MIP-1α (macrophage inflammatory protein-1α), MIP-1β, MCP-1 (monocyte chemoattractant protein-1)
Plate 4: VEGF-A, bFGF (basic fibroblast growth factor)
Plate 5: IL-1RA, IL-9
The whole kit, including calibrators, controls, samples diluent, wash buffer as well as read buffer, detection antibody solution, and a 96-well plate (cat no. N05049A-1, MSD), was provided by MSD to perform multiplex assays and to detect a total of 23 cytokines in the samples. The plate was washed 3 times with 150 µL/well of wash buffer. Then, 50 µL of samples, calibrators, or controls was added into each well of the plate. The plate was sealed with an adhesive plate seal and incubated at room temperature for 2 h on a shaker at 700 r/min. The plate was then washed 3 times with 150 µL/well of wash buffer. A 25 µL of detection antibody solution was added to each well, sealed, and incubated at room temperature for 2 h on a shaker at 700 r/pm. The plate was then washed 3 times with 150 µL/well of wash buffer. Then, 150 µL of read buffer was added to each well. Finally, the plate was analyzed using the MSD multiplex instrument.
Statistical analysis
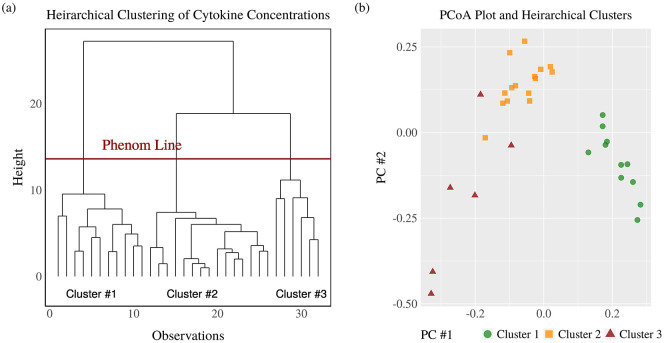
Statistical analysis was completed using R Studio with a significance level of α = 0.05 and two-sided p-values. Cytokine concentrations were standardized prior to multivariate analysis. The dendrogram and principal coordinates analysis (PCoA) plot resulting from agglomerative hierarchical clustering based on the Ward linkage and Euclidean distance shows three total clusters.
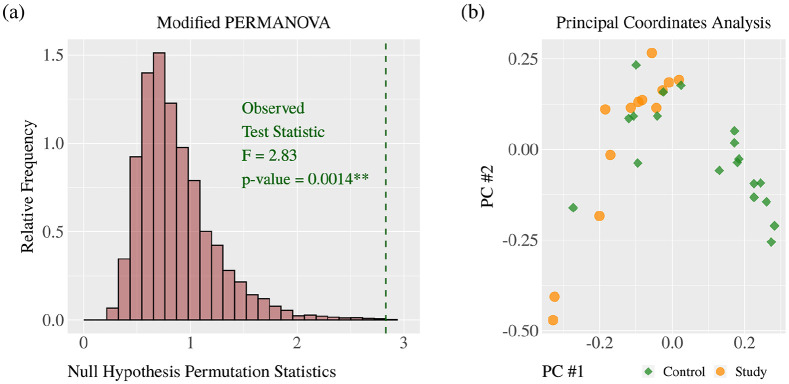
The distributions of cytokine concentrations include distributions characterized by zero-inflation, right skewness, non-normality, many outliers, unequal variances, and expression on different scales. Differences in cytokine concentrations are reported using the standardized difference of means and the permutational unequal variance t test. Acceptance of the research hypothesis was obtained using modified permutational multivariate analysis of variance (PERMANOVA) based on Euclidean distances. Modified PERMANOVA is suitable for working with zero-inflated, right-skewed, high-dimensional, and unbalanced sample data with the possibility that within-group covariance patterns are heterogeneous.25 This allows for a single multivariate p-value testing the null hypothesis of equal group centroids for cytokine concentrations. Computations were completed using the matrix operations described by Anderson et al.25 The results of PERMANOVA are visualized with a histogram of 10,000 null hypothesis permutation statistics and the observed test statistic. Differences between the groups are visualized with the PCoA plot. The multivariate pseudo-standard errors for the sample sizes were plotted to confirm the appropriateness of using PERMANOVA. The identification and measurement of individual differences in cytokine concentrations were completed using the permutational unequal variance t test and standardized difference of means (SDM).
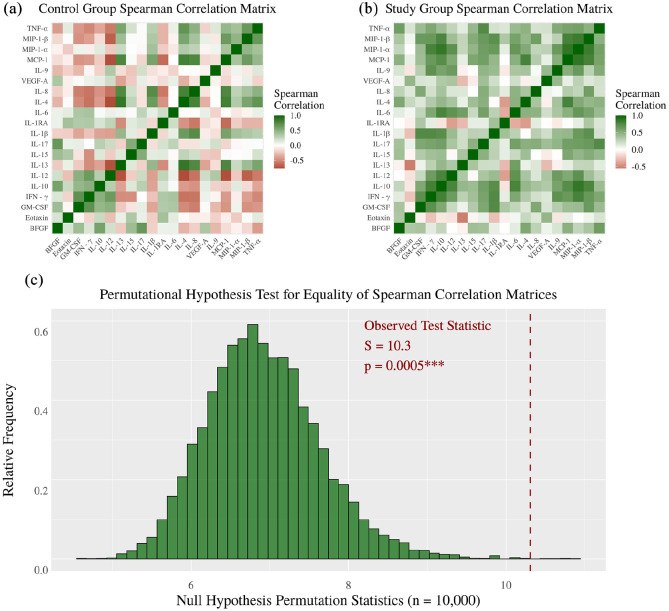
Within-group associations of cytokine concentrations are visualized with heat maps for the Spearman correlation matrix. Each heat map represents 190 different correlations. The control group association matrix is seen to have mostly negative associations while the study group association matrix is seen to have mostly positive associations. To obtain a single p-value measuring the statistical significance of these patterns, the test statistic was chosen as the Frobenius norm of the difference of sample association matrices like the S-statistic used in the study by Shipley26
This test statistic is monotone increasing in the directions toward the alternative hypothesis and has been previously used to the test hypothesis of equal correlation matrices.26,27 The p-value was computed based on 10,000 permutation distribution null statistics.
The relative dominance of different Lactobacillus spp. within each patient group are reported using percentages and statistical significance with Cochran’s Q test. Post hoc analysis is by way of the exact McNemar test and the Holm–Bonferroni adjustments. Bar plots are used to compare the frequency of relative dominance within each patient group. The power of Fisher’s exact test and the exact McNemar test for this experiment are summarized using heat maps (Supplementary Figure 1).
Results
The characteristics of the patient groups are summarized in Supplementary Table 1. Age and parity were not significant between the groups. All patients were seen in Odessa, TX, comprised of a primarily Hispanic population in West Texas.
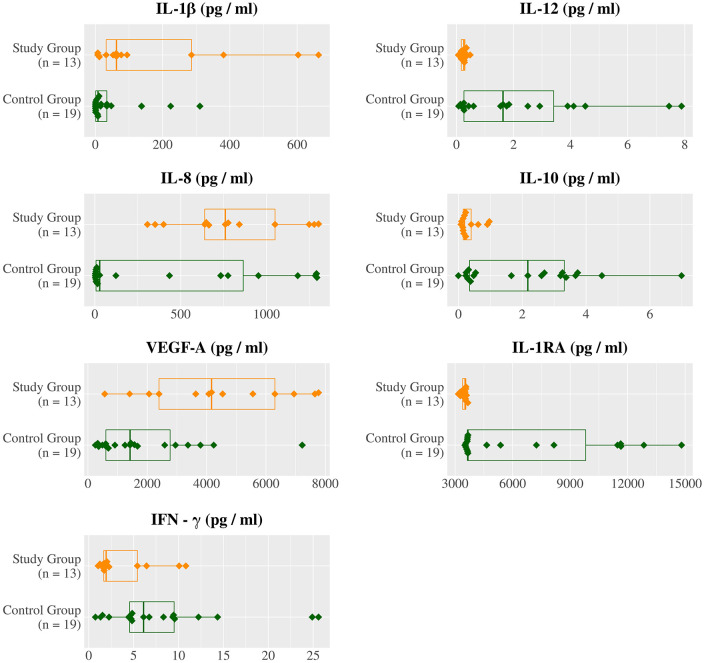
Agglomerative hierarchical clustering supports the research hypothesis that cytokine levels diverge on the basis M. genitalium or U. urealyticum colonization with the dendrogram (Figure 1(a)) and PCoA plot (Figure 1(b)), resulting in three clusters with one of the clusters only present in the control group (Table 1). Acceptance of the research hypothesis was attained through PERMANOVA (p = 0.0014*) (Figure 4(a)) with a PCoA plot used for visualization (Figure 4(b)). The largest and statistically most significant differences in individual cytokine concentrations were observed for VEGF-A (p = 0.002, SDM = +1.06), IL-8 (p = 0.020, SDM = +0.73), and IL-1β (p = 0.049, SDM = +0.79) which were higher in the study group, and IL-1RA (p < 0.001, SDM = −0.91), IL-10 (p < 0.001, SDM = −1.04), IL-12 (p = 0.002, SDM = −0.95), and IFN-γ (p = 0.022, SDM = −0.73) which were lower in the study group (Figure 2, Figure 3, Table 2).
Figure 1.
Dendrogram and principal coordinate analysis plot for clustering by cytokine concentration.
Dendrogram and principal coordinate analysis plot showing three well-defined clusters of patients based on cytokine concentrations.
Table 1.
Contingency table for agglomerative hierarchical clustering.
| Cluster 1 | Cluster 2 | Cluster 3 | |
|---|---|---|---|
| Control group | 11 | 6 | 2 |
| Study group | 0 | 9 | 4 |
A total of three clusters were measured in the control and Mycoplasma or Ureaplasma study group. Cluster 1 was only found in control group where other two clusters were found in both groups.
Figure 4.
Cytokine levels analyzed using modified PERMANOVA and principal coordinate analysis.
There is a statistically significant difference of cytokine levels between the control and Mycoplasma or Ureaplasma study group (**p = 0.0013). Principal coordinate analysis was performed to visually confirm shifts in cytokine levels between two groups.
Figure 2.
Distributions of cytokine concentrations in control group and study group.
VEGF-A, IL-8, and IL-1β are expressed significantly higher in the Mycoplasma or Ureaplasma study group, whereas IL-1RA, IL-10, IL-12, and IFN-γ expressed significantly higher in the control group.
Figure 3.
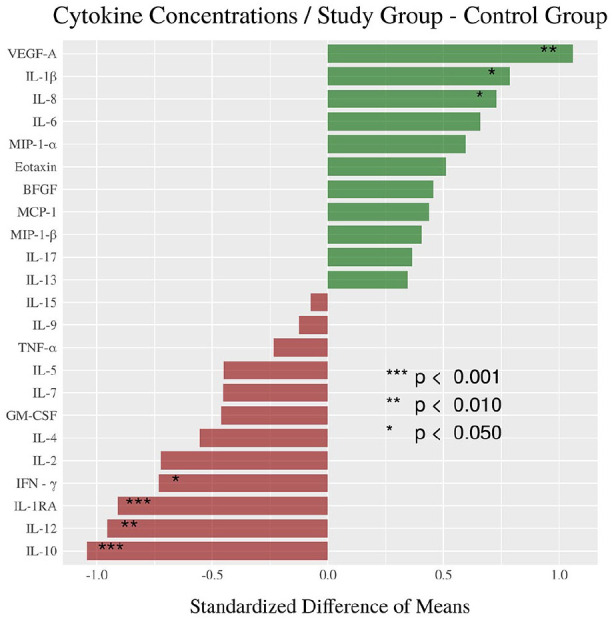
Standardized difference of means for cytokine concentrations.
The standardized difference of means of cytokine concentrations describe and measure the differences between the control and Mycoplasma or Ureaplasma study group (*p < 0.05; **p < 0.01; ***p < 0.001).
Table 2.
Mean cytokine concentrations (pg/mL) and statistical significance.
| Cytokine | Control group | Study group | SDM | p-value |
|---|---|---|---|---|
| IL-1RA | 6526.02 | 3462.78 | −0.91 | <0.001 *** |
| IL-10 | 2.12 | 0.33 | −1.04 | 0.001 *** |
| IL-12 | 2.23 | 0.25 | −0.95 | 0.002** |
| VEGF-A | 1881.88 | 4389.49 | 1.06 | 0.002 *** |
| IL-8 | 429.68 | 789.82 | 0.73 | 0.020* |
| IFN-γ | 8.21 | 3.68 | −0.73 | 0.022* |
| IL-1β | 45.27 | 180.15 | 0.79 | 0.049* |
| IL-4 | 0.23 | 0.11 | −0.55 | 0.087 |
| IL-6 | 1.56 | 6.38 | 0.66 | 0.112 |
| IL-2 | 6.30 | 1.27 | −0.72 | 0.123 |
| MIP-1α | 8.09 | 19.46 | 0.59 | 0.130 |
| IL-7 | 3.37 | 1.10 | −0.45 | 0.159 |
| EOTAXIN | 4.51 | 6.78 | 0.511 | 0.189 |
| GM-CSF | 2.92 | 1.22 | −0.46 | 0.199 |
| IL-5 | 0.01 | 0.00 | −0.45 | 0.255 |
| MCP-1 | 14.59 | 28.67 | 0.44 | 0.302 |
| MIP-1β | 20.95 | 36.61 | 0.41 | 0.354 |
| BFGF | 2.84 | 8.90 | 0.46 | 0.439 |
| IL-13 | 4.00 | 6.18 | 0.35 | 0.446 |
| TNF-α | 2.58 | 1.62 | −0.24 | 0.497 |
| IL-17 | 4.97 | 16.08 | 0.36 | 0.664 |
| IL-9 | 1.92 | 1.73 | −0.13 | 0.734 |
| IL-15 | 0.26 | 0.20 | −0.08 | 0.941 |
SDM: standardized difference of means; IL: interleukin; VEGF-A: vascular endothelial growth factor A; MIP: macrophage inflammatory protein; GM-CSF: granulocyte-macrophage colony-stimulating factor; MCP: monocyte chemoattractant protein; BFGF: basic fibroblast growth factor; IFN: interferon.
Identification and measurement of differences in each cytokine concentration were determined using SDM. IL-10, IL-12, and IL-1RA were significantly higher in control group, whereas VEGF-A, IFN-γ, and IL-8 were significantly elevated in Mycoplasma or Ureaplasma study group (*p < 0.05; **p < 0.01; ***p < 0.001).
The control group cytokine associations are mostly negative (Figure 5(a)) while study group cytokine associations are mostly positive (Table 3, Figure 5(b)). The significance of this pattern was confirmed with a permutation test (p < 0.0005) (Figure 5(c)) indicating a multivariate cytokine response to M. genitalium or U. urealyticum colonization as opposed to only separate independent univariate cytokine responses.
Figure 5.
Association of cytokine levels using Spearman correlation.
There is a statistically significant difference of within-group cytokine-level associations. The control group associations are mostly negative, and Mycoplasma or Ureaplasma study group associations are mostly positive (***p < 0.0001).
Table 3.
Comparison of Spearman correlation for cytokine concentrations.
| Patient group | Cytokine | Cytokine | Spearman ρ | p-value |
|---|---|---|---|---|
| Control | MIP-1α | IL-12 | −0.7531 | 0.0002*** |
| Study | MIP-1α | IL-12 | +0.5462 | 0.0535 |
| Control | VEGF-A | IFN-γ | −0.5088 | 0.0278* |
| Study | VEGF-A | IFN-γ | +0.5612 | 0.0460* |
| Control | TNF-α | GM-CSF | −0.4642 | 0.0463* |
| Study | TNF-α | GM-CSF | +0.6446 | 0.0174* |
| Control | IL-1β | GM-CSF | −0.3647 | 0.1140 |
| Study | IL-1β | GM-CSF | +0.7455 | 0.0003*** |
MIP: macrophage inflammatory protein; IL: interleukin; VEGF-A: vascular endothelial growth factor A; IFN : interferon; TNF-α: tumor necrosis factor-α; GM-CSF: granulocyte-macrophage colony-stimulating factor.
Spearman correlation was calculated to determine the association of cytokine in the control and Mycoplasma or Ureaplasma study group (*p < 0.05; ***p < 0.001).
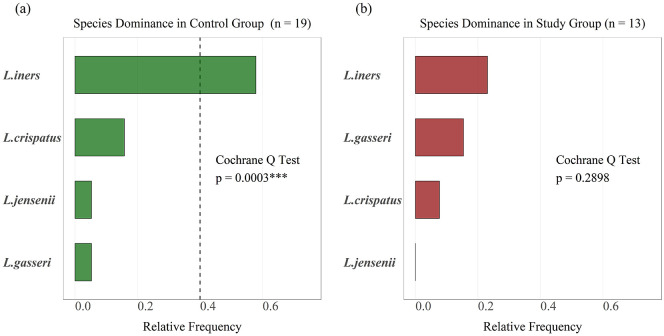
Using Cochran’s Q test (p < 0.0003, Table 5 and Table 6), L. iners was identified to be the predominant species in the control group (Figure 6(a)) while no species predominance was confirmed in study group’s vaginal flora (p = 0.2898) (Figure 6(b), Table 4).
Table 5.
Cochran’s Q test for frequency of dominance of Lactobacillus species.
| DF | Q-statistic | p-value | Post hoc analysis (exact McNemar test) | |
|---|---|---|---|---|
| Control | 3 | 18.55 | 0.0003*** | (Lactobacillus jensenii, Lactobacillus gasseri, Lactobacillus crispatus) < (Lactobacillus iners) |
| Study | 3 | 3.75 | 0.2898 | − |
Using Cochran’s Q test, L. iners was identified as a predominant species in the control group (***p < 0.001), whereas none of Lactobacillus species was found predominant in Mycoplasma or Ureaplasma study group.
Table 6.
Post hoc analysis for Cochran’s Q test in control group.
| Lactobacillus spp. | Lactobacillus spp. | Exact McNemar test (p) | Holm–Bonferroni adjustment |
|---|---|---|---|
| Lactobacillus iners | Lactobacillus jensenii | 0.0010*** | α / 3 = 0.0167 |
| L. iners | Lactobacillus gasseri | 0.0127* | α / 2 = 0.0250 |
| L. iners | Lactobacillus crispatus | 0.0127* | α / 1 = 0.0500 |
The proportions of Lactobacillus species dominance are compared in control group using post hoc analysis for Cochran’s Q test. L. iners was the most frequently predominant species in control group (*p < 0.05; ***p < 0.001).
Figure 6.
Within-group Lactobacillus spp. dominance measured by Cochran’s Q test.
Lactobacillus iners was confirmed as a predominant Lactobacillus spp. in control group (***p < 0.0003) and no Lactobacillus spp. predominance was confirmed in Mycoplasma or Ureaplasma study group (p = 0.2898).
Table 4.
Comparison of each Lactobacillus species in control and Mycoplasma or Ureaplasma study group.
| Lactobacillus spp. | Control group (n = 19) | Study group (n = 13) | Fisher’s test (p) |
|---|---|---|---|
| Lactobacillus iners | 57.9% | 23.1% | 0.0751 |
| Lactobacillus gasseri | 5.3% | 15.4% | 0.5518 |
| Lactobacillus crispatus | 15.8% | 7.7% | 0.6291 |
| Lactobacillus jensenii | 5.3% | 0.0% | 1.0000 |
Each Lactobacillus species was compared between control and Mycoplasma or Ureaplasma study group using Fisher’s test. L. iners found higher in control group (p < 0.09).
Discussion
Concentrations of cytokines of IL-1RA, IL-10, IL-12, VEGF-A, IL-8, IFN-γ, and IL-1β were significantly different between the study and control groups. Some of these results were consistent with other studies examining defense mechanisms and responses against Mycoplasma and Ureaplasma infections. Cytokines have pro- and anti-inflammatory functions and are involved in mediating cytotoxic, humoral, and cellular immunity.28 We saw significantly elevated level of VEGF-A in our study group compared with control. Other studies elucidated the immunology of Ureaplasma serovars in amniotic epithelial cells and fluid by comparing cytokine profiles in Ureaplasma-infected individuals versus other bacteria.29–31 Those studies found that Ureaplasma serovars initiate an innate immune response that led to elevated concentrations of IL-1β, IL-8, TNF-α, and VEGF-A.29–31 A study in 2020 observed elevated IL-8 levels in in-vitro cervico-epithelial cells when cultured with U. urealyticum compared to when cultured with commensal L. crispatus.32 In in-vitro studies of M. genitalium, there were significantly elevated levels of secreted IL-6, IL-7, IL-8, MCP-1, and GM-CSF by endocervical epithelial cells when inoculated with M. genitalium.33 There were also significantly lower levels of IL-10, IL-12, and IL-1RA in our study group compared with the control group. IL-10 and IL-1RA are anti-inflammatory cytokines, and IL-12 signals for lymphocyte differentiation.28,34 In an in-vitro study, there were statistically significant increases in IL-12p40, IL-10, and IL-1RA cytokine mRNA but no significant concentrations of IL-12p40, IL-10, and IL-1RA in response to U. urealyticum.35 The in-vitro study did not show a statistically significant decrease in IL-10 or IL-1RA or significant changes in those cytokine concentrations compared with control where the adult monocyte was exposed to broth.35 The cytokine elevation and depressions seen in prior studies compared with ours suggest further investigation is needed to verify the cytokines involved in U. urealyticum and M. genitalium infections. A limitation to our study includes the inconsistencies observed in elevations or depressions in specific cytokines compared with other studies, which is potentially due to differences in sample preparation.
The microbiome plays a vital role in vaginal health. The metabolic activity of vaginal microbes maintains healthy ecosystems in anatomic niches throughout the vagina, and changes in microbial composition may result in a diseased state.36 Previous studies have shown that vaginal communities divide into five different types, in which the four dominant species are L. iners, L. crispatus, L. gasseri, or L. jensenii.9 Multiple species cannot continuously inhabit the same niche forever in the vagina, as one will compete with others.37 In our study, L. iners was the predominant species in the control group while no Lactobacillus spp. was dominant in the Mycoplasma or Ureaplasma study group. The observed dominance of L. iners in the control group might depend on factors such as age, Caucasian population, and West Texas location (Supplementary Table 1). Our findings for the control group are consistent with other studies finding healthy women of childbearing age having L. iners as a predominant species.38 Our study is also in agreement with the previous studies that L. iners and L. crispatus are Lactobacillus spp. native to the healthy human vagina.37
While there was no significant species dominance in our Mycoplasma or Ureaplasma study group, some studies suggest that L. iners is capable of surviving unfavorable conditions such as during infection or when given treatment for infections compared with L. crispatus.36,38 L. iners is able to do that because it has more complicated nutritional requirements and a smaller genome indicative of a symbiotic lifestyle compared with L. crispatus, L. jensenii, and L. gasseri.39
L. iners may have clonal variants capable of withstanding normal and dysbiotic conditions.36,38 Further studies should be done to explore whether there are potential protective effects that all Lactobacillus spp. in the vaginal microbiome can offer by maintaining dominance during infection and affecting cytokine expression.
In conclusion, our data suggest that Lactobacillus spp. profile was significantly different in both groups. L. iners was the predominant Lactobacillus species in the control group. There was no significantly dominant Lactobacillus species in the M. genitalium or U. urealyticum colonization group. There was a statistically significant difference regarding cytokines level between the control and Mycoplasma or Ureaplasma study group. IL-1β, IL-8, and VEGF-A were expressed significantly higher in the Mycoplasma or Ureaplasma study group while IL-1RA, IL-10, IL-12, and IFN-γ were expressed higher in the control group.
Supplemental Material
Supplemental material, sj-jpg-1-whe-10.1177_17455065211009181 for Cytokine profiles and Lactobacillus species presence in pre-menopausal subjects with genital Mycoplasma genitalium or Ureaplasma urealyticum colonization by John Garza, Kushal Gandhi, Sarah Choi, Asley Sanchez and Gary Ventolini in Women’s Health
Acknowledgments
We are grateful to the Texas Tech University Health Sciences Center, Permian Basin, TX, and Ailena Mulkey, Evangelina Santiago, and Jammie Holland from Clinical Research Institute, TX, for their excellent support on this study. We also appreciate the kind efforts of Mr. Erik Wilkinson (Regional Director of the TTUHSC – Library of the Health Sciences at the Permian Basin, Odessa, TX) in the literature search and review. In addition, we would like to acknowledge Dr. Scott Gygax and his team for their assistance in the quantification of the vaginal flora by qPCR analysis.
Footnotes
Contributorship: Clinical Research Institute nurses consulted the patients and G.V. collected the vaginal swab samples from the patients. K.G. and A.S. carried out the analyses. J.G., K.G., A.S., S.C., and G.V. analyzed the data. All the authors contributed in manuscript writing and editing. All the authors approved the final version of the manuscript.
Declaration of conflicting interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Guarantor: G.V. is the guarantor of this article.
ORCID iD: Gary Ventolini  https://orcid.org/0000-0001-8067-7342
https://orcid.org/0000-0001-8067-7342
Patients’ consent: Consent forms were obtained from all patients.
Supplemental material: Supplemental material for this article is available online.
References
- 1. Matthews N, Wong V, Brooks J, et al. Genital diseases in the mature woman. Clin Dermatol 2018; 36(2): 208–221. [DOI] [PubMed] [Google Scholar]
- 2. Sobel JD. Recurrent vulvovaginal candidiasis. Am J Obstet Gynecol 2016; 214(1): 15–21. [DOI] [PubMed] [Google Scholar]
- 3. Paladine HL, Desai UA. Vaginitis: diagnosis and treatment. Am Fam Physician 2018; 97(5): 321–329. [PubMed] [Google Scholar]
- 4. Tachedjian G, Aldunate M, Bradshaw CS, et al. The role of lactic acid production by probiotic Lactobacillus species in vaginal health. Res Microbiol 2017; 168(9-10): 782–792. [DOI] [PubMed] [Google Scholar]
- 5. Plummer EL, Vodstrcil LA, Fairley CK, et al. Sexual practices have a significant impact on the vaginal microbiota of women who have sex with women. Sci Rep 2019; 9(1): 19749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. van der Veer C, Bruisten SM, van Houdt R, et al. Effects of an over-the-counter lactic-acid containing intra-vaginal douching product on the vaginal microbiota. BMC Microbiol 2019; 19(1): 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. MacIntyre DA, Chandiramani M, Lee YS, et al. The vaginal microbiome during pregnancy and the postpartum period in a European population. Sci Rep 2015; 5: 8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Song SD, Acharya KD, Zhu JE, et al. Daily vaginal microbiota fluctuations associated with natural hormonal cycle, contraceptives, diet, and exercise. Am Soc Microbiol 2020; 5(4): e00593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ravel J, Gajer P, Abdo Z, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci U S A 2011; 108(Suppl. 1): 4680–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Torcia MG. Interplay among vaginal microbiome, immune response and sexually transmitted viral infections. Int J Mol Sci 2019; 20(2): 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barrios De Tomasi J, Opata MM, Mowa CN. Immunity in the cervix: interphase between immune and cervical epithelial cells. J Immunol Res 2019; 2019: 7693183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sethi S, Singh G, Samanta P, et al. Mycoplasma genitalium: an emerging sexually transmitted pathogen. Indian J Med Res 2012; 136(6): 942–955. [PMC free article] [PubMed] [Google Scholar]
- 13. Gdoura R, Kchaou W, Chaari C, et al. Ureaplasma urealyticum, Ureaplasma parvum, Mycoplasma hominis and Mycoplasma genitalium infections and semen quality of infertile men. BMC Infect Dis 2007; 7: 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Patel MA, Nyirjesy P. Role of Mycoplasma and Ureaplasma species in female lower genital tract infections. Curr Infect Dis Rep 2010; 12(6): 417–422. [DOI] [PubMed] [Google Scholar]
- 15. Espinosa V, Rivera A. Cytokines and the regulation of fungus-specific CD4 T cell differentiation. Cytokine 2012; 58(1): 100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Netea MG, Joosten LA, van der Meer JW, et al. Immune defence against Candida fungal infections. Nat Rev Immunol 2015; 15(10): 630–642. [DOI] [PubMed] [Google Scholar]
- 17. Capoccia R, Greub G, Baud D. Ureaplasma urealyticum, Mycoplasma hominis and adverse pregnancy outcomes. Curr Opin Infect Dis 2013; 26(3): 231–240. [DOI] [PubMed] [Google Scholar]
- 18. Huang C, Zhu HL, Xu KR, et al. Mycoplasma and Ureaplasma infection and male infertility: a systematic review and meta-analysis. Andrology 2015; 3(5): 809–816. [DOI] [PubMed] [Google Scholar]
- 19. Glaser K, Silwedel C, Waaga-Gasser AM, et al. Ureaplasma isolates differentially modulate growth factors and cell adhesion molecules in human neonatal and adult monocytes. Cytokine 2018; 105: 45–48. [DOI] [PubMed] [Google Scholar]
- 20. Ventolini G. Measuring treatment outcomes in women with vulvodynia. J Clin Med Res 2011; 3(2): 59–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ventolini G, Barhan S, Duke J. Vulvodynia, a step-wise therapeutic prospective cohort study. J Obstet Gynaecol 2009; 29(7): 648–650. [DOI] [PubMed] [Google Scholar]
- 22. Ventolini G, Gygax SE, Adelson ME, et al. Vulvodynia and fungal association: a preliminary report. Med Hypotheses 2013; 81(2): 228–230. [DOI] [PubMed] [Google Scholar]
- 23. Gandhi K, Gutierrez P, Garza J, et al. Vaginal Lactobacillus species and inflammatory biomarkers in pregnancy. Minerva Ginecol 2020; 72: 299–309. [DOI] [PubMed] [Google Scholar]
- 24. Taylor BD, Holzman CB, Fichorova RN, et al. Inflammation biomarkers in vaginal fluid and preterm delivery. Hum Reprod 2013; 28(4): 942–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Anderson MJ, Walsh DC, Robert Clarke K, et al. Some solutions to the multivariate Behrens–Fisher problem for dissimilarity-based analyses. Austral New Zealand J Stat 2017; 59(1): 57–79. [Google Scholar]
- 26. Shipley B. A permutation procedure for testing the equality of pattern hypotheses across groups involving correlation or covariance matrices. Stat Comput 2000; 10(3): 253–257. [Google Scholar]
- 27. Cui X, Li C, Zhao J, et al. Covariance structure regularization via Frobenius-norm discrepancy. Linear Algebra Appl 2016; 510: 124–145. [Google Scholar]
- 28. Borish LC, Steinke JW. 2. Cytokines and chemokines. J Allergy Clin Immunol 2003; 111(2 Suppl): S460–S475. [DOI] [PubMed] [Google Scholar]
- 29. Triantafilou M, De Glanville B, Aboklaish AF, et al. Synergic activation of toll-like receptor (TLR) 2/6 and 9 in response to Ureaplasma parvum & urealyticum in human amniotic epithelial cells. PLoS ONE 2013; 8(4): e61199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li YH, Brauner A, Jensen JS, et al. Induction of human macrophage vascular endothelial growth factor and intercellular adhesion molecule-1 by Ureaplasma urealyticum and downregulation by steroids. Biol Neonate 2002; 82(1): 22–28. [DOI] [PubMed] [Google Scholar]
- 31. Kacerovsky M, Celec P, Vlkova B, et al. Amniotic fluid protein profiles of intraamniotic inflammatory response to Ureaplasma spp. and other bacteria. PLoS ONE 2013; 8(3): e60399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cavanagh M, Amabebe E, Anumba DOC. Differential cytokine and metabolite production by cervicovaginal epithelial cells infected with Lactobacillus crispatus and Ureaplasma urealyticum. Anaerobe 2020; 62: 102101. [DOI] [PubMed] [Google Scholar]
- 33. McGowin C, Radtke AL, Abraham K, et al. Mycoplasma genitalium infection activates cellular host defense and inflammation pathways in a 3-dimensional human endocervical epithelial cell model. J Infect Dis 2013: 1857–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Opal SM, DePalo VA. Anti-inflammatory cytokines. Chest 2000; 117(4): 1162–1172. [DOI] [PubMed] [Google Scholar]
- 35. Glaser K, Silwedel C, Fehrholz M, et al. Ureaplasma species differentially modulate pro- and anti-inflammatory cytokine responses in newborn and adult human monocytes pushing the state toward pro-inflammation. Front Cell Infect Microbiol 2017; 7: 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mayer BT, Srinivasan S, Fiedler TL, et al. Rapid and profound shifts in the vaginal microbiota following antibiotic treatment for bacterial vaginosis. J Infect Dis 2015; 212(5): 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. France MT, Mendes-Soares H, Forney LJ. Genomic comparisons of Lactobacillus crispatus and Lactobacillus iners reveal potential ecological drivers of community composition in the vagina. Appl Environ Microbiol 2016; 82(24): 7063–7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Spear GT, Gilbert D, Landay AL, et al. Pyrosequencing of the genital microbiotas of HIV-seropositive and -seronegative women reveals Lactobacillus iners as the predominant Lactobacillus Species. Appl Environ Microbiol 2011; 77(1): 378–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Petrova MI, Reid G, Vaneechoutte M, et al. Lactobacillus iners: friend or foe? Trends Microbiol 2017; 25(3): 182–191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-jpg-1-whe-10.1177_17455065211009181 for Cytokine profiles and Lactobacillus species presence in pre-menopausal subjects with genital Mycoplasma genitalium or Ureaplasma urealyticum colonization by John Garza, Kushal Gandhi, Sarah Choi, Asley Sanchez and Gary Ventolini in Women’s Health