Abstract
LncRNA TUG1 has not yet been reported in cerebral ischemia/reperfusion (I/R) injury. Methylcytosine dioxygenase TET2 is involved in ischemic damage. This study aimed to investigate the effects of TUG1 demethylated by TET2 on I/R-induced inflammatory response and identified its possible mechanisms.We found that TUG1 expression was significantly upregulated in oxygen-glucose deprivation and reoxygenation (OGD/R)-induced SH-SY5Y and SK-N-SH cells. Using the middle cerebral artery occlusion (MCAO) mice, we observed a similar effect. We also found that I/R injury could downregulate miR-200a-3p and upregulate NLRP3 and TET2. The knockdown of TUG1 could alleviate OGD/R-induced inflammatory response through upregulating miR-200a-3p and downregulating NLRP3 and other pro-inflammatory molecules. miR-200a-3p inhibition can partially reverse the effects of TUG1 silencing. Further experiments confirmed that TUG1 sponged miR-200a-3p to diminish miR-200a-3p and promote NLRP3 dependent inflammatory responses. Mechanically, knockdown of TET2 induced low levels of TUG1 and high levels of miR-200a-3p in both SK-N-SH and SH-SY5Y cells. IL-18, IL-1β, NLRP3, Caspase-1, and GSDMD-N were highly downregulated in OGD/R-induced SK-N-SH and SH-SY5Y cells after TET2 knockdown. TUG1 overexpression could reverse this effect. All the data indicated that TET2 could demethylate TUG1 and contribute to the inflammatory response. In additional experiments using the MCAO mice model, we confirmed knockdown of TET2 attenuated I/R-induced inflammatory response and brain injuries via decreasing TUG1 and increasing miR-200a-3p to inhibit NLRP3 expression. The demethylation of TUG1 by TET2 might aggravate I/R-induced inflammatory injury via modulating NLRP3 by miR-200a-3p. Our data confirmed that TET2 contributed to I/R-induced inflammatory response via the demethylation of TUG1 and regulated TUG1/miR-200a-3p/NLRP3 pathway.
Keywords: demethylation, ischemia/reperfusion injury, LncRNA TUG1, miR-200a-3p, OGD/R, TET2
Introduction
Stroke is the primary cause of disability and death worldwide(Campbell et al., 2019; Phipps and Cronin, 2020). Brain ischemia is characterized by the formation of vascular thrombus and blockage of blood supply to the brain (Lee et al., 2018). Cerebral ischemia/reperfusion injury (CITI) refers to the deterioration of brain tissue suffered from a period of ischemia, followed by a second injury due to the restoration of blood supply through the medical intervention (Nour et al., 2013). Although there are many signs of progress for the therapeutics on cerebral ischemic stroke, the damage caused by cerebral ischemia/reperfusion injury is still a challenging issue for a stroke patient. Therefore, it is significant to investigate the pathophysiological mechanisms of cerebral ischemia/reperfusion injury to improve stroke patients' clinical prognosis.
NLRP3 inflammasome comprises NLRP3, ASC, and pro-caspase-1. After the assembly, it can activate the release of IL-1β and IL-18 (Swanson et al., 2019). Studies showed NLRP3 had been implicated in auto-inflammatory diseases (Louvrier et al., 2020), type 2 diabetes (Lee et al., 2013), and Atherosclerosis (Wang et al., 2017). So, the NLRP3 inflammasome might be a promising target for various inflammatory diseases. We found NLRP3 was significantly upregulated during ischemia/reperfusion inflammatory response, but the definite mechanisms remain unclear.
Ten-Eleven Translocation-2 (TET2) is an evolutionarily conserved dioxygenase. Functional studies pointed out that TET2 can convert 5-methyl-cytosine to 5-hydroxymethyl-cytosine and enhance DNA demethylation (Solary et al., 2014). A previous study showed TET2 was upregulated in ischemia/reperfusion-insulted brain tissue, which led to 5-hydroxymethylcytosine (5hmC) overexpression (Miao et al., 2015). Abnormal DNA methylation had been found within some lncRNAs, such as lncRNA SNHG12 (Lu et al., 2020). Experiments showed TET2 could bind to lncRNA-ANRIL and regulated its expression, indicating lncRNAs could be regulated by methylation (Deng et al., 2016). However, whether lncRNA TUG1 is regulated by ischemia/reperfusion injury is still unclear. Using bioinformatics, we found TET2 potentially binds to TUG1. So, we proposed that the high TUG1 after ischemia/reperfusion injury was possibly due to the demethylation by TET2. Using StarBase software, we found there were potential binding sites in TUG1 and NLRP3 to miR-200a-3p. However, whether DNA demethylase TET2 regulates the TUG1 expression and participate in inflammatory response via miR-200a-3p and NLRP3 is still unclear.
In this study, we first confirmed the demethylation of the promoter region of TUG1 by the TET2 enzyme led to the high level of TUG1 during OGD/R injury. We also found the down-regulation of TUG1 attenuated the OGD/R induced inflammatory response. Mechanistically, TUG1 acts as a miRNA sponge to decrease miR-200a-3p and subsequently elevated the level of NLRP3 and promoted inflammatory response. The findings elucidated the molecular function of the TET2/TUG1/miR-200a-3p/NLRP3 signaling pathway in cerebral I/R injury and highlighted TET2 as a new therapeutic candidate to alleviate cerebral ischemia/reperfusion inflammatory injury.
Materials and Methods
Cell Cultures
SK-N-SH and its subclone SH-SY5Y cells (ATCC, Manassas, VA, USA) were routinely maintained in DMEM (HyClone, Logan, UT, USA) supplemented with 10% (v/v) FBS (Sigma– Aldrich, St. Louis, MO, USA), 100 U/ml penicillin/streptomycin (Sigma–Aldrich, St. Louis, MO, USA). Cells were kept at 37°C in 5% CO2 humidified air. The medium was changed every 3 days.
OGD/R Model
The cells were cultured in hypoxia environments, containing 1% O2, 95% N2, and 5% CO2 in serum/glucose-free DMEM medium at 37 °C for specific periods (2, 4, 8, 12 h). The medium was changed into a standard culture medium for reoxygenation, and cell plates were maintained in normoxic conditions for 24 h. In the control group, cells were kept in standard culture conditions with 5% CO2 and 95% air at 37 °C.
Measurement of Cell Viability by MTT Assay
Two thousand cells per well were seeded into 96 well plates. After 24 h, cells were subjected to OGD/R injury. The MTT assay was then performed according to the CyQUANT™ MTT Cell Viability Assay kit (V13154, Thermo Fisher Scientific, USA). The optical density was measured at 490 nm by a microplate reader (VICTOR3, Turku, Finland).
CCK-8 Assay
We purchased a CCK-8 assay kit from Millipore Sigma (Cat# 96992) and performed the experiments according to the manufacturer's instructions. Briefly, the cells were dissociated 48h after the transfection, and 100μl of the single-cell suspension (1 × 105 cells/ml) was aliquoted into each well of the 96-well plate. After culturing for 24 h, 10 μl of CCK8 solution (0.5 mg/ml) was added into each well, and cells were incubated for a further 15 min. The absorbance was measured at 450 nm using a Synergy™ HTX Multi-Mode Microplate Reader (BioTek Instruments, CA, USA).
Cell Transfection
To investigate the functions of TUG1 and TET2, we purchased shRNA-TUG1 (shTUG1), shRNA-TET2 (shTET2) and the corresponding shRNA negative control (shNC), the miR-200a-3p mimics and inhibitor, and the corresponding negative control (mimics NC or inhibitor NC), TUG1 overexpressing plasmid (pcDNA3.1-TUG1) and the corresponding empty vector (pcDNA3.1-NC) from GenePharma (Shanghai, China). SK-N-SH or SH-SY5Y Cells were seeded into 6-well plates. At 70% confluence, shRNA-TUG1, shRNA-TET2, and the corresponding control single or together with the corresponding miRNA oligonucleotides were transfected using lipofectamine 2000 (Invitrogen). The cells were collected for subsequent experiments after 48 h.
ELISA Assay
The supernatant was collected after centrifugation to detect the levels of IL-18 and IL-1β. Then, the contents of IL-18 and IL-1β were measured using ELISA kits (BioLegend) following the instructions.
Dual-Luciferase Reporter Assay
The wild-type (WT) or mutant (MUT) binding sequences of TUG1(TUG1-WT or TUG1-Mut) or NLRP3 (NLRP3-WT or NLRP3-Mut) were introduced into the pGL3 basic vector (Promega Corporation, WI, USA) using the Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA, USA), respectively. pGL3 basic vector was a plasmid control. We co-transfected with WT TUG1, or MUT constructs to detect the luciferase activities, or together with miR-200a-3p mimics, inhibitor, or the corresponding negative control (NC). According to the manufacturer's instructions, luciferase activity was detected 48 h after transfection using a Dual-Luciferase Reporter System (Promega, Madison, WI, USA).
RNA Pull-Down and RNA Immunoprecipitation (RIP)
Briefly, cells were transfected with MS2bs-NLRP3 3′UTR WT or Mut, MS2bs-TUG1 3′UTR WT, or Mut or control vector MS2bs-Rluc. According to the instructions, cells were harvested after 48 h transfection, and RIP was performed using Protein Immunoprecipitation Kit (Millipore, USA). After RNA extraction by Trizol (Life Technologies), miR-200a-3p levels were analyzed by qRT-PCR.
Methylation-Specific Polymerase Chain Reaction (MSP)
DNA was extracted using a QIAamp DNA Mini Kit (Qiagen). Then, 1µg DNA was treated with sodium sulfite to convert unmethylated cytosine to uracil using the EpiTect Bisulfite Kit (QIAGEN, Hilden, Germany). Each bisulfite-treated sample was amplified using a specific primer for methylated versus unmethylated DNA. MSP primers for the TUG1 gene promoter were as follows: methylation, 5′-GATGGTGATAGGATTTAGTAATACG-3′ (forward), 5′- AAACTATATACACTATCGAAAACAAAACG-3′ (reverse); and unmethylation, 5′- GGTGATAGGATTTAGTAATATGAA-3′ (forward), 5′- AACTATATACACTATCAAAAACAAAACAAT′ (reverse).
Transient Focal Cerebral Ischemia/Reperfusion
Adult male C57BL/6 mice (7-8 weeks of age; body weight 23±2 g) were purchased from Guangdong Medical Laboratory Animal Center, Guangdong, China. All mice were housed in a temperature and humidity controlled environment with a 12 h light/dark cycle and ad libitum access to water and food. All experimental animal procedures were under the guideline for animal welfare and experimental conduct approved by the Institutional Animal Use and Care Committee of the Second Affiliated Hospital of Nanchang University. The transient focal cerebral ischemia/reperfusion of C57BL/6 mice at 8-9 weeks was induced by the middle cerebral artery occlusion (MCAO) surgery. C57BL/6 mice were randomly divided into four groups (n=5 per group) as follows: sham group (only filament insertion without occlusion, followed by persistent perfusion), I/R group (ischemia for 1 h and reperfusion for 24h), shNC (shRNA negative control injection before surgery + MCAO 24 h) and shTET2 (shRNA TET2 injection before surgery + MCAO 24 h). 0.5mg/kg shRNA negative control (shNC) or shRNA TET2 (shTET2) was injected into the right cerebral ventricle of mice before MCAO surgery. To generate the ischemia/reperfusion mice model, we anesthetized the mice with 5% isoflurane/100% oxygen in a chamber using a respirator (E-Z systems, Santa Ana, CA, USA). Then, the focal cerebral ischemia was induced by inserting a 6.0 intraluminal filament (Doccol Corp., Redlands, CA, USA) into the left middle cerebral artery to block the blood supply for 30 min. After the blockage, the occluding filament was removed to allow reperfusion for 24 h. The sham groups were given an insert to the left middle cerebral artery but did not block the blood supply. After the surgery, the mice were kept at 37°C until regaining full consciousness, and then their neurological deficit was evaluated using a five-point scale.
Neurological Deficit Score
After surgery, the neurological deficit score was evaluated at 24 h using a five-point scale as the previous report (Wan et al., 2019). 0, no motor deficit; 1, flexion of the forelimb contralateral to the ischemic hemisphere; 2, reduced resistance against push toward the paretic side; 3, spontaneous circling toward the paretic side; 4, unable to walk; and 5, death.
Measurement of the Infarct Volume
At 24 h after surgery, the mice were sacrificed, and the whole brains were removed. The fresh brain was sectioned at 1 mm-thickness and immersed in a 2% 2,3,5-triphenyl tetrazolium chloride (TTC)-phosphate-buffered saline (PBS) solution for 20 min at room temperature in the dark. After brief washed by 1 × PBS, the sections were fixed with 4% paraformaldehyde (Sigma, St. Louis, MO) for 30 min at room temperature. The sections were photographed using a digital camera. Each brain section and the infarct region (unstained areas) were measured using ImageJ 1.51. The infarct volume was calculated as (infarcted area/total brain area) × 100%.
Detection of Cell Apoptosis
At 24 h after reperfusion, mice were sacrificed and was perfused by precooled 4% paraformaldehyde in phosphate-buffered saline (PBS). The brains were removed and fixed in 4% paraformaldehyde-PBS solution overnight. After 3 washes by 1× PBS, the brains were immersed with 30% sucrose in 4°C PBS overnight. The brains were sectioned into 12m thickness using a Leica cryostat and stored at -20°C until use. The TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) staining was performed according to the detailed manufacture's instruction of the TUNEL detection for the detection of apoptotic cells kit (C10625, Thermo Fisher Scientific). The slides were imaged using a digital camera. The apoptotic cells were calculated as follows: the apoptotic rate (%)= the number of TUNEL positive stained cells/total cells × 100%. Three fields per slide were counted. At least five slides were measured in each treatment group.
Real-Time Quantitative PCR (qRT-PCR)
Total RNA was isolated with TRIzol reagent (Takara Biotechnology, Dalian, China). After checking the quality, 1ug of total RNA was used to synthesize cDNA using a Reverse Transcription kit (Takara, Dalian, China). qRT-PCR was performed using SYBR Green PCR Mix kits (Applied Biosystems) on an ABI 7500 Real-Time PCR system (Applied Biosystems, Foster, CA). The relative expression levels of target genes or miRNA were calculated based on the Ct values and normalized to GAPDH or U6. Primer sequences are as follows: human TUG1, 5′- CTG AGT GTT CAC CTG GAC CT-3′ (forward), 5′- ACA GGA GTG GAG GTA AAG GC-3′ (reverse); human NLRP3, 5′- CCA CAA GAT CGT GAG AAA ACC C-3′ (forward), 5′- CGG TCC TAT GTG CTC GTC A-3′ (reverse); human TET2, 5′- GGC TAC AAA GCT CCA GAA TGG-3′ (forward), 5′- AAG AGT GCC ACT TGG TGT CTC-3′ (reverse); human Caspase-1, 5′- GAC AAG GGT GCT GAA CAA GG-3′ (forward), 5′- ATT CCC TTC TGA GCC TGA GG-3′ (reverse); human GSDMD-N, 5′-GTG TGT CAA CCT GTC TAT CAA GG-3′ (forward), 5′-CAT GGC ATC GTA GAA GTG GAA G-3′ (reverse); human IL-18, 5′- TGC ACC CCG GAC CAT ATT TA-3′ (forward), 5′- TCC TGG GAC ACT TCT CTG AA-3′ (reverse); human IL-1β, 5′- AGC TAC GAA TCT CCG ACC AC -3′ (forward), 5′- CGT TAT CCC ATG TGT CGA AGA A -3′ (reverse); human miR-200a-3p, 5′- CAG TGC AGG GTC CGA GGT-3′ (forward), 5′- AAT CGG CGT AAC ACT GTC TGG TA-3′ (reverse); Mouse TUG1, 5′- CTC TGG AGG TGG ACG TTT TGT-3′ (forward), 5′- GTG AGT CGT GTC TCT CTT TTC TC-3′ (reverse); mmu miR-200a-3p, 5′- CAG TGC AGG GTC CGA GGT-3′ (forward), 5′- AAT CGG CGT AAC ACT GTC TGG TA-3′ (reverse). Relative gene expression was calculated using the 2−ΔΔCt method.
Western Blot
The total proteins were extracted by RIPA buffer (Thermo Scientific™) supplemented with 1 mM PMSF (Thermo Scientific™). The BCA Protein Assay Kit (Pierce, Appleton, WI, USA) was applied to measure the concentrations. Then, 10 µg protein for each sample was separated on a 10% SDS-PAGE and transferred to PVDF membranes (Bio-Rad). After blocking with 5% non-fat milk (Lab Scientific), the membranes were incubated with primary antibodies overnight at 4°C. The primary antibodies were listed as follows: GSDMD-N (Abcam, ab215203, 1:1000), NLRP3 (Abcam, ab214185 1:500), Caspase-1 (Abcam, ab62698, 1:500), TET2 (Abcam, ab245287, 1:2000), IL-1β (Abcam, ab156791, 1:2000), IL-18 (Abcam, ab245697, 1:1000) and β-actin (Abcam, ab34452, 1:1000). After three washes with 1× PBST, each for 10 min, the corresponding secondary antibodies Goat Anti-Rabbit IgG H&L (HRP) (Abcam, ab205718) or Goat Anti-Mouse IgG H&L (HRP) (Abcam, ab6789) was incubated. Membranes were developed using a chemiluminescent substrate (sigma) and captured by Image Lab™ Software (Bio-Rad) 5.2.1. Image J 1.53d quantified the density of bands and the data were analyzed.
Statistical Analysis
All data are presented as Means ± SD. Data were analyzed using student's t-test or one-way analysis of variance (ANOVA) using GraphPad Prism 6 (Graph Pad Software Co., San Diego, CA, USA). p < 0.05 was considered as statistical significance.
Results
TUG1 and TET2 Were Highly Upregulated, and miR-200a-3p Was Notably Downregulated After OGD/R Injury
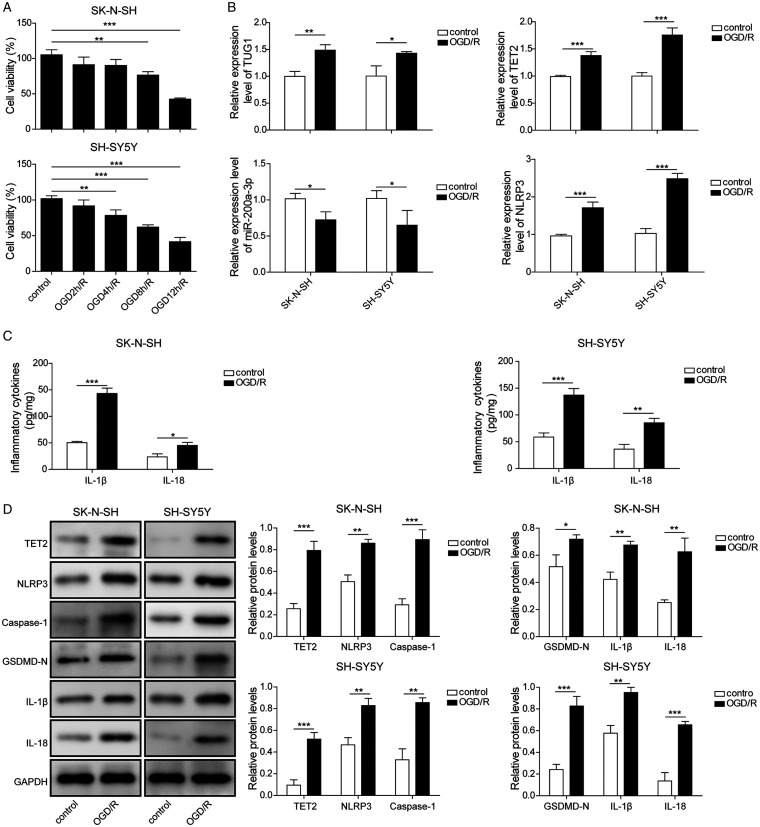
Cells were subjected to oxygen-glucose deprivation for 0, 2, 4, 8, 12 h, and recovered to 24 h under normoxic culture condition to model in vitro ischemia/reperfusion injury. Cells in normoxic conditions were considered as control. Then, the expressions of TUG1/TET2 and OGD/R injury were determined. First, the cell viabilities were detected by MTT assay. OGD12 h/R 24 h led to significant cell death (Figure 1A). We also found OGD 12 h/R 24 h induced about 50% cell death in both SK-N-SH and SH-SY5Y cells. Compared with OGD 8 h, OGD 12 h induced more significant inhibition (Figure 1A). So, OGD 12 h/R 24 h was chosen for OGD/R administration in the following experiments. Additional results showed that TUG1 was remarkably upregulated in OGD/R treated cells (Figure 1B). OGD/R exposure distinctly elevated NLRP3 and TET2 but decreased miR-200a-3p (Figure 1B). Moreover, the secretions of IL-1β and IL-18 were also remarkably enhanced (Figure 1C). Western blot assays indicated compared with control, the expressions of TET2, NLRP3, Caspase-1, GSDMD-N, and inflammation-related proteins were distinctly increased under OGD/R stimuli (Figure 1D).
Figure 1.
Upregulation of TUG1, TET2, and Inflammation and Downregulation of miR-200a-3p Under OGD/R Conditions. A: Cell viability was determined by MTT assay after deoxygenation for 0, 2, 4, 6, 8, 12 h, then following by reoxygenation for 24 h. B: TUG1, miR-200a-3p, NLRP3, and TET2 were detected by qRT-PCR after subject to ORG 12 h/R 24 h condition. C: IL-1β and IL-18 were measured by ELISA after subject to ORG 12 h/R 24 h condition. D: TET2, NLRP3, Caspase-1, GSDMD-N, IL-18, and IL-1β were determined by western blot after subject to ORG 12 h/R 24 h condition. Four independent experiments were repeated, with three replicates each time. *p < 0.05, **p < 0.01, ***p < 0.001.
Knockdown of TUG1 Increased Expression Level of miR-200a-3p, Promoted Cell Survival, and Decreased Inflammatory Responses Under OGD/R Conditions
Given that lncRNA TUG1 was upregulated after OGD/R injury, it is crucial to investigate whether knockdown of TUG1 could influence cells' biological activities. The transfection of shTUG1 could decrease TUG1 (Figure 2A). Besides, the inactivation of TUG1 remarkably promoted the level of miR-200a-3p (Figure 2B). The CCK-8 assay showed OGD/R challenge could significantly decrease the cell viabilities in both SH-SY5Y and SK-N-SH cells. However, knockdown of TUG1 could partially attenuate the cell death induced by the OGD/R challenge (Figure 2C) and attenuated the inflammation by the decreased secretions of IL-18 and IL-1β (Figure 2D). Moreover, TUG1 silencing protected OGD/R-induced inflammation by the downregulations of NLRP3, Caspase-1, and GSDMD-N (Figure 2E).
Figure 2.
Knockdown of TUG1 Upregulated miR-200a-3p and Decreased TET2 and Inflammation. A: The expressions of TUG1 after TUG1 knockdown were measured. B: miR-200a-3p was detected by qRT-PCR under ORG 12 h/R 24 h condition. C: Cell viabilities were measured by CCK-8 assay in ORG 12 h/R 24 h -induced SH-SY5Y and SK-N-SH cells after transfection with shNC or shTUG1. D: ELISA measured IL-18 and IL-1β in ORG 12 h/R 24 h -induced cells with or without TUG1 knockdown. E: TET2 and inflammation-related proteins were detected by western blot in ORG 12 h/R 24 h -induced cells with or without TUG1 knockdown. Four independent experiments were repeated, with three replicates each time, *p < 0.05, **p < 0.01, ***p < 0.001.
NLRP3 Was a Direct Target of miR-200a-3p
The above experiment showed both miR-200a-3p and NLRP3 were remarkably changed by OGD/R injury. miR-200a-3p levels were distinctly upregulated by transfection of miR-200a-3p mimics, but significantly inhibited by miR-200a-3p inhibitor (Figure 3A). qRT-PCR analysis pointed out the expressions of NLRP3 were dramatically decreased by miR-200a-3p mimics, but its inhibitor reversed these effects (Figure 3B). To elucidate the regulatory mechanisms of miR-200a-3p in OGD/R injury, we did bioinformatics analysis and found NLRP3 3′UTR contained the possible binding sequence of miR-200a-3p (Figure 3C). By transfection of NLRP3-WT or -Mut luciferase vector together with miR-200a-3p mimics, inhibitor, or miR-NC, we found miR-200a-3p mimics inhibited the luciferase expression containing NLRP3-WT 3′UTR sequence, rather than the NLRP3-Mut 3′UTR sequence, suggesting that miR-200a-3p directly acted on NLRP3 (Figure 3D). The RIP assay further confirmed miR-200a-3p could directly interact with NLRP3. miR-200a-3p was enriched in the NLRP3 3′UTR pulled down pellet compared with that of control (Figure 3E), suggesting that miR-200a-3p directly bound to WT, not Mut NLRP3.
Figure 3.
NLRP3 Is the Direct Target of miR-200a-3p. miR-200a-3p (A) and NLRP3 (B) were detected 48hrs after transfection of miR-200a-3p mimic, inhibitor, or scramble. C: The putative complementary site in human NLRP3 3′ UTR was predicted by TargetScan software. D: The interaction between miR-200a-3p and NLRP3 3′UTR was determined by luciferase assay after transfection of NBLP3-WT or NBLP3-Mut, 24 h following cotransfection of miR-200a-3p mimic, inhibitor, or scramble. E: The physical association of NLRP3 protein and miR-200a-3p was determined by RIP assay. Four independent experiments were repeated, with three replicates each time, *p < 0.05, **p < 0.01, ***p < 0.001.
miR-200a-3p Was One of the Direct Targets of TUG1
Previous studies had confirmed lncRNAs could regulate mRNAs by sponging miRNAs. Both TUG1 and miR-200a-3p could be regulated in OGD/R injures, so we proposed that TUG1 possibly sponged miR-200a-3p. By bioinformatics analysis and further experiment, we found miR-200a-3p had a complementary sequence within TUG1 3′UTR (Figure 4A). After transfection of WT or TUG1 Mut binding sequence and miR-200a-3p inhibitor or inhibitor NC, we certified that miR-200a-3p significantly increased the luciferase activity of the TUG1 WT group, rather than the TUG1 Mut group (Figure 4B), suggesting that there was a direct interaction. Furthermore, a RIP assay further identified that the WT TUG1 sequence remarkably enriched miR-200a-3p, but not Mut TUG1(Figure 4C). By CCK-8 assay, we found that OGD/R treatment caused a significant decrease in both SH-SY5Y and SK-N-SH cell viability. However, the knockdown of TUG1 partially attenuated the OGD/R-induced decrease in cell viability. When cotransfection of miR-200a-3p inhibitor with shTUG1, miR-200a-3p inhibitor exhibited the opposite impact on the cell viability (Figure 4D). Besides, ELISA assay and western blot demonstrated that knockdown of TUG1 could attenuate inflammatory responses induced by OGD/R injury, which in turn were reversed by a miR-200a-3p inhibitor (Figure 4E and F).
Figure 4.
TUG1 Regulated OGD/R-Induced Inflammatory Responses by Targeting miR-200a-3p. A: The potential binding site of TUG1 and miR-200a-3p. B: Luciferase activity was analyzed 48 h after cotransfection of TUG1 WT or MUT plasmid with miR-200a-3p inhibitor or negative control. C: The RIP assay was to evaluate the enrichment of miR-200a-3p by MS2Bs-TUG1 WT, rather than MS2bs-TUG1 Mut. D: Cell viabilities were measured in ORG 12 h/R 24 h -induced SH-SY5Y and SK-N-SH cells after cotransfection with shTUG1 and miR-200a-3p inhibitor. E: IL-18 and IL-1β were measured by ELISA after TUG1 knockdown or together with a miR-200a-3p inhibition in ORG 12 h/R 24 h - injured cells. F: NLRP3 and Caspase-1 were detected by western blot after TUG1 knockdown or together with a miR-200a-3p inhibition in ORG 12 h/R 24 h -injured cells. Four independent experiments were repeated, with three replicates each time, *p < 0.05, **p < 0.01, ***p < 0.001.
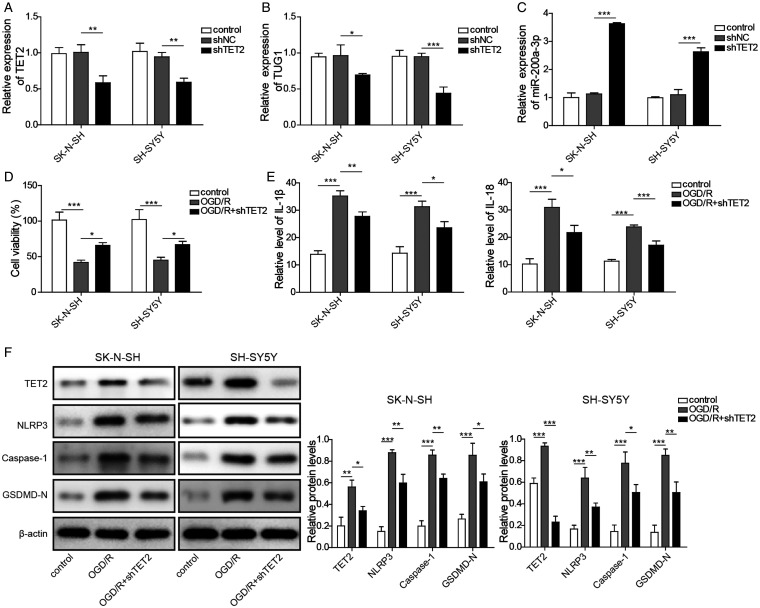
Knockdown of TET2 Decreased the TUG1 Level and Attenuated OGD/R Injury via the miR-200a-3p/NLRP3 Axis
In our experiment, we found OGD/R conditions significantly upregulated TET2. We suspected that TET2 might regulate TUG1 under OGD/R conditions. Then, we assessed the effect of TET2 knockdown on the levels of TUG1. TET2 silencing by shTET2 could efficiently decrease TET2 expression (Figure 5A). Notably, the levels of TUG1 were significantly inhibited after the knockdown of TET2 (Figure 5B). Also, we found knockdown of TET2 remarkably upregulated miR-200a-3p (Figure 5C). To investigate the role of TET2 on cell survival, we transfected the cells with shNC or shTET2 before the OGD/R challenge. Our data showed knockdown of TET2 could partially rescue OGD/R-induced significant inhibition of cell viabilities in both SH-SY5Y and SK-N-SH cells (Figure 5D). To confirm whether knockdown of TET2 could influence OGD/R induced inflammatory response, we detected inflammatory mediators and protein levels. Our data showed inhibition of TET2 significantly reduced IL-18 and IL-1β under OGD/R conditions (Figure 5E). Strikingly, it was also observed that TET2 silencing reduced the expressions of NLRP3, Caspase-1, and GSDMD-N (Figure 5F). These data suggested that knockdown of TET2 might alleviate inflammatory injury through the TUG1/miR-200a-3p/NLRP3 pathway.
Figure 5.
TET2 Silencing Decreased the TUG1 Level and Attenuated ORG 12 h/R 24 h Injury. A–C: TET2, TUG1, and miR-200a-3p expressions were detected by qRT-PCR after knockdown of TET2. D: A CCK-8 assay measured cell viabilities in ORG 12 h/R 24 h -induced SH-SY5Y and SK-N-SH cells after transfection with shNC or shTET2. E: IL-18 and IL-1β secretions were measured by ELISA in ORG 12 h/R 24 h -injured cells after knockdown of TET2. F: TET2, NLRP3, Caspase-1, and GSDMD-N expressions were determined by western blot in ORG 12 h/R 24 h -injured cells after knockdown of TET2. Four independent experiments were repeated, with three replicates each time, *p < 0.05, **p < 0.01, ***p < 0.001.
TET2 Aggravated OGD/R-Induced Inflammatory Injury Through Modulating TUG1
Regression analysis of the above results hinted that TET2 influenced the TUG1 expression. We wondered whether TET2 could directly regulate the expression of TUG1. Then, we investigated the expression of TUG1 after TET2 silencing in OGD/R conditions. Our results showed OGD/R condition significantly upregulated TUG1 levels, but knockdown of TET2 distinctly diminished OGD/R-induced TUG1 overexpression (Figure 6A). We also found OGD/R-induced high level of TUG1 was probably due to the demethylation of the promoter of TUG1, which was partially reversed by knockdown of TET2 (Figure 6B). To confirm whether TET2 regulates the cell survival in OGD/R-induced SH-SY5Y and SK-N-SH cells by TUG1, we co-transfected shTET2 single or together with pcDNA3.1-TUG1 into SH-SY5Y and SK-N-SH cells. Our results revealed that OGD/R treatment remarkably inhibited cell survival, while knockdown of TET2 significantly attenuated the OGD/R-induced cell death. However, overexpression of TUG1 by cotransfection of shTET2 and pcDNA3.1-TUG1 could alleviate the protective effects of TET2 silencing. These results indicated that knockdown of TET2 played a role in MCAO damage by regulating TUG1 expression (Figure 6C). Then, we asked whether TET2 could regulate the OGD/R-induced inflammatory response directly by regulating TUG1. We detected the releases of inflammatory factors and protein expression in OGD/R-induced cells. We found inactivation of TET2 could significantly attenuate IL-18, and IL-1β stimulated by OGF/R injury (Figure 6D) and decreased the expressions of NLRP3, GSDMD-N, and Caspase-1 (Figure 6E), but overexpression of TUG1 could antagonize the protective roles of TET2 silencing. Taken together, we confirmed TET2 upregulated TUG1 expression and contributed to inflammation through inhibition of miR-200a-3p under the OGD/R situation.
Figure 6.
Demethylation of TUG1 by TET2 Promoted Inflammatory Response by Regulation of NLRP3. A: TUG1 expression was measured by qRT-PCR in ORG 12 h/R 24 h -injured cells after TET2 knockdown. B: The DNA methylation and expression of TUG1 were analyzed by methylation-specific PCR in ORG 12 h/R 24 h -induced SK-N-SH cells after TET2 knockdown. C: Cell viabilities were measured in ORG 12 h/R 24 h -induced SH-SY5Y and SK-N-SH cells after cotransfection with shTET2 and pcDNA3.1-TUG1. D: IL-18 and IL-1β were assessed by ELISA in ORG 12 h/R 24 h - injured cells after TET2 knockdown single or together with TUG1 overexpression. E: NLRP3, GSDMD-N, and Caspase-1 levels were assessed by western blot in ORG 12 h/R 24 h -induced injured cells after TET2 knockdown single or together with TUG1 overexpression. Four independent experiments were repeated, with three replicates each time, *p < 0.05, **p < 0.01, ***p < 0.001.
Knockdown of TET2 Attenuated the Stroke Injury by Downregulating TUG1 and Upregulated miR-200a-3p In Vivo
To elucidate the function of TET2 in the pathological process of stroke damage, we established the MCAO mice models with knockdown of TET2 by the intra-cerebrovascular injection of TET2 lentivirus. 24 h after MCAO surgery, we measured the neurological deficit score and relative infarct volume. Our data showed that the neurological deficit score (Figure 7A) and the infarct volume (Figure 7B) in the MCAO group were significantly increased when compared with the sham group. Compared with that of MCAO + shNC group, the neurological deficit score in MCAO + shTET2 was significantly reduced. To further confirm this observation, we detected the cell apoptosis in ischemic areas by the TUNEL staining method. Our data showed that TUNEL positive cells were remarkably increased in brain infarct area sections in the MCAO group than that of the sham group (Figure 7C). However, the shTET2 group could significantly attenuate the MCAO-induced apoptotic death ratio compared with the shNC group (Figure 7C), indicating TET2 silencing had a protective role. To investigate whether TET2 knockdown could influence the expression of TUG1 and miR-200a-3p in MCAO mice models, we detected the levels of TUG1 and miR-200a-3p using the qRT-PCR method. As shown in Figure 7D, the MCAO injury could lead to a higher level of TUG1 and a lower level of miR-200a-3p than those in the sham group (Figure 7D). However, the expression of TUG1 in MCAO+shTET2 was higher than that in the MACO+shNC group. In contrast, the expression of miR-200a-3p was significantly upregulated in MCAO mice when TET2 was knockdown (Figure 7D). We also detected the protein expression of TET2, NLRP3, and other related pre-inflammation factors Caspase-1, GSDMD-N, IL-18, and IL-1β. Our data indicated that TET2, NLRP3, and other pre-inflammation factors Caspase-1, GSDMD-N, IL-18, and IL-1β were significantly upregulated in the MCAO group that in sham group. However, knockdown of TET2 exhibited a remarkably decreased level of TET2, NLRP3, and other pre-inflammation factors Caspase-1, GSDMD-N, IL-18, and IL-1β (Figure 7E). These results indicated that TET2 silencing protected MCAO injury in vivo, possibly through downregulating TUG1 and upregulating miR-200a-3p.
Figure 7.
Knockdown of TET2 Attenuated Ischemia/Reperfusion Injury by Downregulating TUG1 and Upregulating miR-200a-3p In Vivo. A: The neurologic deficit score of the MCAO mice treated with shNC or shTET2 was used to evaluate the neurologic behavior. B: The TTC staining images were used to assess the brain infarct area of the MCAO mice treated with shNC or shTET2. The relative infarct ratio was quantified to assess the effects of TET2 knockdown on brain damages induced by MCAO. C: The TUNEL staining was used to detect the levels of apoptotic cells in the MCAO mice treated with shNC or shTET2. The relative ratio of apoptotic cells was quantified in the MCAO mice treated with shNC or shTET2. D: The expression level of TUG1 and miR-200a-3p were detected using qRT-PCR in the MCAO mice treated with shNC or shTET2. E: The protein levels of TET2, NLRP3, Caspase-1, GADMDN, IL-18, and IL-1β were determined by western blot assay in the MCAO mice treated with shNC or shTET2. Three independent experiments were repeated, with five replicates each time, *p < 0.05, **p < 0.01, ***p < 0.001.
Discussion
Accumulated studies had shown epigenetic modifications played significant roles in the molecular and cellular mechanisms for brain ischemic stroke (Miao et al., 2015; Liu et al., 2020). During the past several years, the NLRP3 inflammasome is a vital regulator for cytokine secretions during ischemia/reperfusion injury. So, NLRP3 inflammasome might be a direction for the treatment for ischemic stroke. It was still unclear whether NLRP3 could be regulated by lncRNAs and be involved in the pathogenesis in ischemic stroke. Abundant studies pointed out that lncRNA TUG1 was actively involved in I/R-related inflammatory response(Chen et al., 2017; Jia et al., 2019; Su et al., 2019; Yang et al., 2019; Liang et al., 2020; Xu et al., 2020). For example, OGD/R injury could upregulate the expression of TUG1 in cultured MA-C cells and subsequently contributed to neuronal death by sponging miR-145 and directly inhibiting the expression of AQP4 (encoding aquaporin 4) (Shan et al., 2020). TUG1 could promote myocardial ischemic injury-induced cardiomyocyte cell apoptosis by directly targeting miR-9a-5p and upregulation of Krüppel-like factor 5 (KLF5) (Yang et al., 2019). The NLRP3/Caspase-1/IL-1β pathway is believed as a promising avenue against ischemic stroke (Gao et al., 2017; Zhang et al., 2017; Guo et al., 2018; Feng et al., 2020; Wang et al., 2020). Previous studies confirmed that miR-200a-3p mediated NLRP3 inflammasome activation in inflammation and brain injury (Ding et al., 2018; Yu, 2019). This study found TUG1 had a complementary binding sequence for miR-200a-3p by the TargetScan and StarBase online tools. So, we proposed that the interactions between TUG1 and miR-200a-3p participated in the OGD/R-induced inflammatory response. To illustrate this hypothesis, TUG1 and miR-200a-3p were detected after the OGD/R challenge. Our data indicated both TUG1 and miR-200a-3p had significantly been regulated by OGD/R injuries. Notably, TUG1 was upregulated, whereas miR-200a-3p was downregulated under OGD/R conditions. Meanwhile, we firstly confirmed the knockdown of TUG1 prevented OGD/R-induced injury by inhibition of inflammation-related protein expressions. miR-200a-3p was significantly increased when TUG1 was silenced. Additionally, we further demonstrated that TUG1 sponged miR-200a-3p and inhibited endogenous miR-200a-3p expression, and further increased NLRP3 expression. The overexpression of NLRP3 subsequently promoted the secretion of pro-inflammatory cytokines.
Ischemic stroke is one of the most common reasons for the global mortality burden (Bejot et al., 2016). Several studies had proven that stroke is a comprehensive and multifactorial disease with abundant genetic modifications (Miao et al., 2015; Tang and Zhuang, 2019; Sharma et al., 2020). DNA methylation contributed directly or indirectly to the development of ischemic brain damage. Recent studies showed the TET2 enzyme could regulate lncRNAs via the binding to the promoter region of lncRNAs. For example, TET2 is bound to the promoter region of lncRNA-ANRIL and regulates its transcription (Deng et al., 2016). Hence, we hoped to clarify whether TET2 could regulate TUG1 and participate in I/R injury. In our study, we found TUG1 was significantly decreased when TET2 was silenced. However, miR-200a-3p expression was remarkably upregulated after TET2 silencing. Additionally, knockdown of TET2 decreased OGD/R-induced inflammation. These results hinted that knockdown of TET2 downregulated TUG1, upregulated miR-200a-3p, and suppressed inflammatory response by inactivating the NLRP3/Caspase-1/IL-1β signaling pathway. However, we were still unclear about whether TET2 directly regulated TUG1 in OGD/R-induced injury. So, the expression and methylation level of TUG1 were detected after the knockdown of TET2 under OGD/R condition. We observed that knockdown of TET2 decreased TUG1 expression and methylation of the TUG1 gene in OGD/R condition, which meant demethylation of TUG1 by TET2 was responsible for the elevated level of TUG1 in OGD/R condition. Then we hope to elucidate whether demethylation of TUG1 by TET2 was involved in the inflammatory injury. By cotransfection of shTET2 with TUG1 overexpression or control vector, we found TET2 knockdown decreased inflammatory response, but overexpression of TUG1 could partially antagonize the effects of TET2 knockdown. Taken together, we firstly confirmed TET2 acted as a transcription regulator that promoted OGD/R-induced inflammation through the demethylation of the TUG1 gene and influenced its downstream molecules. We demonstrated that the interactions between TET2 and TUG1 might be involved in cerebral ischemia/reperfusion inflammatory response. To confirm whether TET2 contributed to the inflammatory response via regulating TUG1 and its downstream, we established the MCAO mice model and investigated the role of TET2 in brain inflammation. We found MCAO could significantly induce neuronal death and cause brain infarct. MCAO also remarkably increased TUG1 expression and decreased miR-200a-3p. However, knockdown of TET2 could partially attenuate MCAO-induced brain injuries, possibly through downregulating TUG1 and upregulating miR-200-3p. This article elaborated on the pathophysiological role of the TET2/TUG1/miR-200a-3p axis in inflammatory injury and delineated the possible cellular and molecular mechanisms.
In conclusion, we first demonstrated that TUG1 was directly regulated by demethylase TET2 and confirmed a functional interaction between TUG1 and TET2. TUG1 as a central regulator promoted OGD/R-induced inflammatory injury directly via regulating the miR-200a-3p/NLRP3 signaling pathway. Epigenetic modification of lncRNAs through demethylation by TET2 might be a new target to treat cerebral ischemia/reperfusion injury. In the future, additional in vivo experiments and clinical trials should be applied to explore the cellular mechanisms of the TET2/TUG1 axis in I/R injury.
Summary Statement
We first demonstrated that TUG1 was directly regulated by demethylase TET2 and confirmed a functional interaction between TUG1 and TET2. Significantly, TUG1 as a central regulator promoted OGD/R-induced inflammatory injury directly via regulating the miR-200a-3p/NLRP3 signaling pathway.
Supplemental Material
Supplemental material, sj-pdf-1-asn-10.1177_17590914211003247 for LncRNA TUG1 Demethylated by TET2 Promotes NLRP3 Expression, Contributes to Cerebral Ischemia/Reperfusion Inflammatory Injury by Min Yin, Wei-Ping Chen, Xiao-Ping Yin, Jiang-Long Tu, Na Hu and Zheng-Yu Li in ASN Neuro
Footnotes
Author's Contributions: Conception and study design: MY; Data acquisition: WPC; Data analysis: XPY, JLT; Manuscript drafting: NH; Manuscript revising: ZYL. All authors have read and approved the final version of this manuscript to be published.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the foundation project of the Natural Science Foundation of Jiangxi Province (grant no. 20171BAB205026 and 20181BAB205029), hospital project of the Second Affiliated Hospital of Nanchang University (grant no. 2019YNQN12012), and The National Natural Science Foundation of China (grant no. 8176050182).
ORCID iD: Zheng-Yu Li https://orcid.org/0000-0001-8892-3851
Supplemental material: Supplementary material for this article is available online.
References
- Bejot Y., Daubail B., Giroud M. (2016). Epidemiology of stroke and transient ischemic attacks: Current knowledge and perspectives. Rev Neurol (Paris), 172, 59–68. [DOI] [PubMed] [Google Scholar]
- Campbell B. C. V., De Silva D. A., Macleod M. R., Coutts S. B., Schwamm L. H., Davis S. M., Donnan G. A. (2019). Ischaemic stroke. Nat Rev Dis Primers, 5, 70. [DOI] [PubMed] [Google Scholar]
- Chen S., Wang M., Yang H., Mao L., He Q., Jin H., Ye Z. M., Luo X. Y., Xia Y. P., Hu B. (2017). LncRNA TUG1 sponges microRNA-9 to promote neurons apoptosis by up-regulated Bcl2l11 under ischemia. Biochem Biophys Res Commun, 485, 167–173. [DOI] [PubMed] [Google Scholar]
- Deng W., Wang J., Zhang J., Cai J., Bai Z., Zhang Z. (2016). TET2 regulates LncRNA-ANRIL expression and inhibits the growth of human gastric cancer cells. IUBMB Life, 68, 355–364. [DOI] [PubMed] [Google Scholar]
- Ding X. Q., Wu W. Y., Jiao R. Q., Gu T. T., Xu Q., Pan Y., Kong L. D. (2018). Curcumin and allopurinol ameliorate fructose-induced hepatic inflammation in rats via miR-200a-mediated TXNIP/NLRP3 inflammasome inhibition. Pharmacol Res, 137, 64–75. [DOI] [PubMed] [Google Scholar]
- Feng Y. S., Tan Z. X., Wang M. M., Xing Y., Dong F., Zhang F. (2020). Inhibition of NLRP3 inflammasome: A prospective target for the treatment of ischemic stroke. Front Cell Neurosci, 14, 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L., Dong Q., Song Z., Shen F., Shi J., Li Y. (2017). NLRP3 inflammasome: A promising target in ischemic stroke. Inflamm Res, 66, 17–24. [DOI] [PubMed] [Google Scholar]
- Guo M., Wang X., Zhao Y., Yang Q., Ding H., Dong Q., Chen X., Cui M. (2018). Ketogenic diet improves brain ischemic tolerance and inhibits NLRP3 inflammasome activation by preventing Drp1-mediated mitochondrial fission and endoplasmic reticulum stress. Front Mol Neurosci, 11, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia H., Ma H., Li Z., Chen F., Fang B., Cao X., Chang Y., Qiang Z. (2019). Downregulation of LncRNA TUG1 inhibited TLR4 signaling pathway-mediated inflammatory damage after spinal cord ischemia reperfusion in rats via suppressing TRIL expression. J Neuropathol Exp Neurol, 78, 268–282. [DOI] [PubMed] [Google Scholar]
- Lee H. M., Kim J. J., Kim H. J., Shong M., Ku B. J., Jo E. K. (2013). Upregulated NLRP3 inflammasome activation in patients with type 2 diabetes. Diabetes, 62, 194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. H. C., Lee M. H. H., Wu C. Y. C., Couto E. S. A., Possoit H. E., Hsieh T. H., Minagar A., Lin H. W. (2018). Cerebral ischemia and neuroregeneration. Neural Regen Res, 13, 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Li F., Li H., Wang R., Du M. (2020). Overexpression of lncRNA HULC attenuates myocardial ischemia/reperfusion injury in rat models and apoptosis of hypoxia/reoxygenation cardiomyocytes via targeting miR-377-5p through NLRP3/caspase1/IL1beta signaling pathway inhibition. Immunol Invest, 1–14. [DOI] [PubMed] [Google Scholar]
- Liu X., Fan B., Chopp M., Zhang Z. (2020). Epigenetic mechanisms underlying adult post stroke neurogenesis. Int J Mol Sci, 21, 6197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvrier C., Assrawi E., El Khouri E., Melki I., Copin B., Bourrat E., Lachaume N., Cador-Rousseau B., Duquesnoy P., Piterboth W., Awad F., Jumeau C., Legendre M., Grateau G., Georgin-Lavialle S., Karabina S. A., Amselem S., Giurgea I. (2020). NLRP3-associated autoinflammatory diseases: Phenotypic and molecular characteristics of germline versus somatic mutations. J Allergy Clin Immunol, 145, 1254–1261. [DOI] [PubMed] [Google Scholar]
- Lu C., Wei Y., Wang X., Zhang Z., Yin J., Li W., Chen L., Lyu X., Shi Z., Yan W., You Y. (2020). DNA-methylation-mediated activating of lncRNA SNHG12 promotes temozolomide resistance in glioblastoma. Mol Cancer, 19, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z., He Y., Xin N., Sun M., Chen L., Lin L., Li J., Kong J., Jin P., Xu X. (2015). Altering 5-hydroxymethylcytosine modification impacts ischemic brain injury. Hum Mol Genet, 24, 5855–5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nour M., Scalzo F., Liebeskind D. S. (2013). Ischemia-reperfusion injury in stroke. Interv Neurol, 1, 185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipps M. S., Cronin C. A. (2020). Management of acute ischemic stroke. Br Med J, 368, l6983. [DOI] [PubMed] [Google Scholar]
- Shan W., Chen W., Zhao X., Pei A., Chen M., Yu Y., Zheng Y., Zhu S. (2020). Long noncoding RNA TUG1 contributes to cerebral ischaemia/reperfusion injury by sponging mir-145 to up-regulate AQP4 expression. J Cell Mol Med, 24, 250–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A. R., Shashikiran U., Uk A. R., Shetty R., Satyamoorthy K., Rai P. S. (2020). Aberrant DNA methylation and miRNAs in coronary artery diseases and stroke: A systematic review. Brief Funct Genomics, 19, 259–285. [DOI] [PubMed] [Google Scholar]
- Solary E., Bernard O. A., Tefferi A., Fuks F., Vainchenker W. (2014). The Ten-Eleven Translocation-2 (TET2) gene in hematopoiesis and hematopoietic diseases. Leukemia, 28, 485–496. [DOI] [PubMed] [Google Scholar]
- Su Q., Liu Y., Lv X. W., Ye Z. L., Sun Y. H., Kong B. H., Qin Z. B. (2019). Inhibition of lncRNA TUG1 upregulates miR-142-3p to ameliorate myocardial injury during ischemia and reperfusion via targeting HMGB1- and Rac1-induced autophagy. J Mol Cell Cardiol, 133, 12–25. [DOI] [PubMed] [Google Scholar]
- Swanson K. V., Deng M., Ting J. P. (2019). The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat Rev Immunol, 19, 477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Zhuang S. (2019). Histone acetylation and DNA methylation in ischemia/reperfusion injury. Clin Sci (Lond), 133, 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J. J., Wang P. Y., Zhang Y., Qin Z., Sun Y., Hu B. H., Su D. F., Xu D. P., Liu X. (2019). Role of acute-phase protein ORM in a mice model of ischemic stroke. J Cell Physiol, 234, 20533–20545. [DOI] [PubMed] [Google Scholar]
- Wang H., Chen H., Jin J., Liu Q., Zhong D., Li G. (2020). Inhibition of the NLRP3 inflammasome reduces brain edema and regulates the distribution of aquaporin-4 after cerebral ischaemia-reperfusion. Life Sci , 251, 117638. [DOI] [PubMed] [Google Scholar]
- Wang R., Wang Y., Mu N., Lou X., Li W., Chen Y., Fan D., Tan H. (2017). Activation of NLRP3 inflammasomes contributes to hyperhomocysteinemia-aggravated inflammation and atherosclerosis in apoE-deficient mice. Lab Invest, 97, 922–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Niu Y., Li H., Pan G. (2020). Downregulation of lncRNA TUG1 attenuates inflammation and apoptosis of renal tubular epithelial cell induced by ischemia-reperfusion by sponging miR-449b-5p via targeting HMGB1 and MMP2. Inflammation, 43, 1362–1374. [DOI] [PubMed] [Google Scholar]
- Yang D., Yu J., Liu H. B., Yan X. Q., Hu J., Yu Y., Guo J., Yuan Y., Du Z. M. (2019). The long non-coding RNA TUG1-miR-9a-5p axis contributes to ischemic injuries by promoting cardiomyocyte apoptosis via targeting KLF5. Cell Death Dis, 10, 908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, J. C. J., Yang H., Chen S., Wang Z. (2019). Overexpression of miR‑200a‑3p promoted inflammation in sepsis‑induced brain injury through ROS‑induced NLRP3. Int J Mol Med, 45, 1811–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Jiang L., Che F., Lu Y., Xie Z., Wang H. (2017). Arctigenin attenuates ischemic stroke via SIRT1-dependent inhibition of NLRP3 inflammasome. Biochem Biophys Res Commun, 493, 821–826. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-asn-10.1177_17590914211003247 for LncRNA TUG1 Demethylated by TET2 Promotes NLRP3 Expression, Contributes to Cerebral Ischemia/Reperfusion Inflammatory Injury by Min Yin, Wei-Ping Chen, Xiao-Ping Yin, Jiang-Long Tu, Na Hu and Zheng-Yu Li in ASN Neuro