Abstract
Background
Chronic obstructive pulmonary disease (COPD) is characterized by incomplete reversible airflow limitation and chronic inflammatory response lesions. This study mainly explored whether FGFR2 and MGAT5 polymorphisms affected the risk of COPD in the Chinese people.
Methods
Five variants in FGFR2 and MGAT5 were chosen and genotyped using Agena MassARRAY platform from 315 COPD patients and 314 healthy controls. The correlation of FGFR2 and MGAT5 with COPD susceptibility was evaluated with odds ratio (OR) and 95% confidence interval (CI) via logistic regression.
Results
We found rs2420915 enhanced the risk of COPD, while rs6430491, rs2593704 reduced the susceptibility of COPD (p < 0.05). Rs2420915 could promote the incidence of COPD in the elderly and nonsmokers. Rs1907240 and rs2257129 also increased the susceptibility to COPD in nonsmokers (p < 0.05). MGAT5-rs2593704 played a protective role in COPD development in different subgroups (age ≤ 70, male, smokers, and individuals with BMI ≤ 24 kg/m2). Meanwhile, rs6430491 was linked with a lower risk of COPD in nonsmoking and BMI ≤ 24 kg/m2 subgroups.
Conclusions
We concluded that FGFR2 and MGAT5 genetic polymorphisms are correlated with the risk of COPD in the Chinese people. These data underscored the important role of FGFR2 and MGAT5 gene in the occurrence of COPD and provided new biomarkers for COPD treatment.
Trial registration: NA.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12890-021-01498-3.
Keywords: Chronic obstructive pulmonary disease, FGFR2, MGAT5, Genetic polymorphism
Background
Chronic obstructive pulmonary disease (COPD) is a common chronic disease of the respiratory system, which is mainly characterized by incomplete reversible airflow limitation and chronic inflammatory response lesions. The physiopathology of COPD were airflow limitation, gas exchange abnormalities, degeneration, necrosis and ulceration of bronchial epithelial cell, excessive expansion of lung, pale appearance and bullae of different sizes on the surface. The main clinical manifestations are cough, sputum, dyspnea, and decreased exercise endurance, which can eventually lead to pulmonary heart disease and respiratory failure. COPD has the characteristics of high morbidity, mortality and disability among the elderly. Epidemiological investigation has shown that the incidence of COPD in the Chinese population over 40 years old is approximately 13.7%, which is the third most fatal disease in the world [1]. In addition, it is reported that COPD can seriously affect the quality of life of patients [2]. According to the World Bank report, COPD is expected to account for the fifth-largest economic burden of disease worldwide by 2020. Therefore, it is important to explore the pathogenesis and etiology of COPD.
Many investigations have demonstrated that tobacco smoking is an important risk factor for COPD development [3]. Nevertheless, only 10%-20% of smokers develop COPD, and 30% of nonsmokers suffer from COPD, suggesting genetic background plays a crucial role in COPD development [4, 5]. A genome-wide study found that MAN2B, DHX15 gene were associated with COPD susceptibility in multi-ethnic populations [6]. Du et al. also showed the genetic variants of GSTP1, HO-1, and SOD-3 were correlated with COPD susceptibility [7]. Besides, other studies have been reported that PDE4D, FAM13A, CYP2B6 gene polymorphisms may exert effects on COPD susceptibility [8–10]. These findings highlighted the important role of genetic polymorphisms in the occurrence of COPD.
Fibroblast growth factor receptor 2 (FGFR2) is one of the members of the fibroblast growth factor receptor (FGFR) family, and four types, namely FGFR1, FGFR2, FGFR3, and FGFR4, have been identified. The FGFR family members are involved in a variety of physiological processes, including cell growth and migration [11]. It is reported that FGFR2 participated in lung development and it is considered as a therapeutic target for lung cancer [12, 13]. Dorry et al. [14] have shown that alveolar epithelial cell-specific FGFR2 was critical for survival in response to bleomucin-induced lung injury. Jieming et al. [15] found that FGFR2 mutants could alleviate pulmonary fibrosis of alveolar epithelial type II cells through FGF-2. These lines of evidence have led us to believe that FGFR2 may be involved in the development of lung-related disease. Nevertheless, the role of FGFR2 gene in COPD has been poorly studied.
N-acetylglucosaminyltransferaseV (MGAT5), also known as Gnt-V, catalyzes the formation of β-1,6-branched N-glycans that promote surface retention of glycoproteins [16]. MGAT5 has been reported to be involved in the proliferation, adhesion, invasion and metastasis of tumor cells [17]. Studies found that MGAT5 was highly expressed in pulmonary adenocarcinoma cells and its silence suppressed cell growth [18, 19]. In addition, Elek and colleagues illustrated that MGAT5-rs34944508 was significantly correlated with lung cancer risk [20]. These findings suggested that MGAT5 could play a key role in lung disease development. However, little is known about the role of MGAT5 in COPD development.
In this case–control study, we explored whether FGFR2 and MGAT5 genetic mutants influence the occurrence of COPD. We identified and genotyped five single nucleotide polymorphisms (SNPs) from FGFR2 and MGAT5 to evaluate the association of SNPs with COPD susceptibility. This will provide new ideas for understanding the pathological mechanism of COPD.
Methods
Study population
Our research recruited 629 subjects (315 COPD patients and 314 healthy controls) from Hainan General Hospital. Based on the Global Initiative for Chronic Obstructive Lung Disease criteria, individuals were diagnosed with COPD with the ratio of forced expiratory volume in 1 s (FEV1) /forced vital capacity (FVC) < 70% and FEV1 < 80% predicted. COPD patients with a history of bronchial asthma, tuberculosis, lung cancer, and other serious diseases were not included in this study. Controls with healthy subjects without lung dysfunction, no lung-related diseases, other chronic diseases and disorders, and severe endocrine, metabolic, and nutritional disorders from the health checkup in the same hospital during the same period. Clinical characteristics of the study subjects were collected by medical records and questionnaires, including smoking and body mass index (BMI), complications, wheeze, gasp, chest distress and respiratory infection. This study protocol received approval by the Ethics Committee of Hainan General Hospital and conformed to the declarations of Helsinki. And we also got informed consent signed by all participants.
Genotyping
Five SNPs (rs2420915, rs1907240, rs2257129, rs6430491, rs2593704) were identified and genotyped. All SNPs had a minor allele frequency in the Chinese Han Beijing population. Genomic DNA was extracted from whole blood using a DNA extraction kit (GoldMag Co. Ltd, Xi’an, China) and its concentration was detected by NanoDrop 2000 (Thermo Scientific, Waltham, USA). We applied the Agena MassARRAY platform to genotype. Data analysis and management using Agena Typer 4.0 software.
Statistical analysis
We applied for student t-test and χ2 test to assess the difference in age and gender between the cases and the control group. The Hardy–Weinberg equilibrium (HWE) of the control group was calculated by χ2 test. The relationship between genetic variants with COPD risk was evaluated with odds ratio (OR) and 95% confidence interval (CI) by logistic regression analysis. Haploview software and PLINK software were used for Haploview analysis and linkage disequilibrium [21, 22]. P value < 0.05 was considered statistically significant.
Results
Characteristics of participants
The demographic and clinical features of the subjects were listed in Additional file 1: Table 1. This research included 315 COPD patients (239 males and 76 females) and 314 healthy controls (177 males and 137 females). The mean age of the case and control group was 71.23 ± 6.83 and 71.93 ± 10.11 years, respectively. Besides, there was no significant difference in age (p = 0.306) and gender (p = 0.926) distribution between the two groups.
Evaluation of COPD risk
The detailed information of SNPs in FGFR2 and MGAT5 is summarized in Table 1. HaploReg v4.1 showed that FGFR2 and MGAT5 SNPs were associated with the regulation of SiPhy cons, Enhancer histone marks, DNAse, Motifs changed, Selected eQTL hits, GRASP QTL hits, and Promoter histone marks. All SNPs conformed to HWE (p > 0.05). Our results showed that the A allele of rs2420915 near FGFR2 increased the risk of COPD (OR 1.41, 95% CI 1.12–1.77, p = 0.004). However, the A allele of rs6430491near MGAT5 (OR 0.69, 95% CI 0.55–0.87, p = 0.002) and the G allele of MGAT5-rs2593704 (OR 0.74, 95% CI 0.57–0.95, p = 0.020) were correlated to reduced risk of COPD.
Table 1.
The primary information of SNPs in FGFR2 and MGAT5
| SNP | Gene | Chr | Position | Allele A/B |
Location | MAF | HWE p |
OR (95% CI) | p | HaploReg | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Control | ||||||||||
| rs2420915 | FGFR2 | 10 | 122840277 | A/G | Near | 0.414 | 0.334 | 0.801 | 1.41 (1.12–1.77) | 0.004 | SiPhy cons, Enhancer histone marks, DNAse |
| rs1907240 | FGFR2 | 10 | 122897959 | G/A | Intron | 0.423 | 0.387 | 0.905 | 1.16 (0.93–1.46) | 0.190 | SiPhy cons, Enhancer histone marks, DNAse, Motifs changed, Selected eQTL hits |
| rs2257129 | FGFR2 | 10 | 122898697 | T/C | Intron | 0.414 | 0.387 | 0.722 | 1.12 (0.89–1.40) | 0.344 | Enhancer histone marks, Motifs changed |
| rs6430491 | MGAT5 | 2 | 134840967 | A/G | Near | 0.351 | 0.438 | 0.909 | 0.69 (0.55–0.87) | 0.002 | Enhancer histone marks, Motifs changed, GRASP QTL hits |
| rs2593704 | MGAT5 | 2 | 135005277 | G/C | Intron | 0.217 | 0.274 | 0.887 | 0.74 (0.57–0.95) | 0.020 | Promoter histone marks, Enhancer histone marks, DNAse, Proteins bound |
SNP single nucleotide polymorphism, MAF minor allele frequency, HWE Hardy–Weinberg equilibrium, OR odds ratio, 95% CI 95% confidence interval
p values were calculated from χ2 test
Bold values represent statistical significance (p < 0.05)
The relationship between SNPs and COPD risk was assessed in four genetic models. As presented in Table 2, rs2420915 was associated with a higher risk of COPD in codominant (AA: OR 1.85, 95% CI 1.12–3.07, p = 0.016; AG: OR 1.62, 95% CI 1.15–2.28, p = 0.006), dominant (OR 1.67, 95% CI 1.20–2.31, p = 0.002), and additive models (OR 1.43, 95% CI 1.13–1.81, p = 0.003). Rs6430491 decreased the susceptibility of COPD in codominant (OR 0.41, 95% CI 0.25–0.68, p = 0.0005), dominant (OR 0.70, 95% CI 0.50–0.97, p = 0.033), recessive (OR 0.47, 95% CI 0.29–0.74, p = 0.0012), and additive models (OR 0.68, 95% CI 0.54–0.86, p = 0.0012). While MGAT5-rs2593704 reduced the risk of COPD only in dominant (OR 0.70, 95% CI 0.51–0.97, p = 0.031) and additive models (OR 0.75, 95% CI 0.58–0.97, p = 0.029).
Table 2.
Associations of FGFR2 and MGAT5 genetic variants with COPD risk
| Gene | SNP | Model | Genotype | Frequency | Without adjustment | With adjustment | |||
|---|---|---|---|---|---|---|---|---|---|
| Case | Control | OR (95% CI) | pa | OR (95% CI) | pb | ||||
| FGFR2 | rs2420915 | Codominant | GG | 0.326 | 0.446 | 1.00 | 1.00 | ||
| AA | 0.153 | 0.115 | 1.83 (1.11–3.02) | 0.018 | 1.85 (1.12–3.07) | 0.016 | |||
| AG | 0.521 | 0.439 | 1.62 (1.15–2.28) | 0.006 | 1.62 (1.15–2.28) | 0.006 | |||
| Dominant | GG | 0.326 | 0.446 | 1.00 | 1.00 | ||||
| AA + AG | 0.674 | 0.554 | 1.66 (1.20–2.30) | 0.002 | 1.67 (1.20–2.31) | 0.002 | |||
| Recessive | AG + GG | 0.847 | 0.885 | 1.00 | 1.00 | ||||
| AA | 0.153 | 0.115 | 1.40 (0.88–2.22) | 0.156 | 1.42 (0.89–2.26) | 0.139 | |||
| Additive | – | – | – | 1.42 (1.12–1.80) | 0.003 | 1.43 (1.13–1.81) | 0.003 | ||
| FGFR2 | rs1907240 | Codominant | AA | 0.348 | 0.373 | 1.00 | 1.00 | ||
| GG | 0.195 | 0.146 | 1.42 (0.90–2.26) | 0.135 | 1.43 (0.90–2.27) | 0.133 | |||
| GA | 0.457 | 0.481 | 1.02 (0.72–1.44) | 0.926 | 1.02 (0.72–1.45) | 0.901 | |||
| Dominant | AA | 0.348 | 0.373 | 1.00 | 1.00 | ||||
| GG + GA | 0.652 | 0.627 | 1.11 (0.80–1.54) | 0.525 | 1.12 (0.81–1.55) | 0.507 | |||
| Recessive | GA + AA | 0.805 | 0.854 | 1.00 | 1.00 | ||||
| GG | 0.195 | 0.146 | 1.41 (0.93–2.15) | 0.108 | 1.41 (0.93–2.15) | 0.109 | |||
| Additive | – | – | – | 1.16 (0.93–1.45) | 0.196 | 1.16 (0.93–1.45) | 0.190 | ||
| FGFR2 | rs2257129 | Codominant | CC | 0.342 | 0.370 | 1.00 | 1.00 | ||
| TT | 0.169 | 0.145 | 1.27 (0.79–2.04) | 0.332 | 1.27 (0.79–2.05) | 0.329 | |||
| TC | 0.489 | 0.486 | 1.09 (0.77–1.54) | 0.629 | 1.10 (0.77–1.55) | 0.607 | |||
| Dominant | CC | 0.342 | 0.370 | 1.00 | 1.00 | ||||
| TT + TC | 0.658 | 0.630 | 1.13 (0.81–1.57) | 0.466 | 1.14 (0.82–1.58) | 0.450 | |||
| Recessive | TC + CC | 0.831 | 0.855 | 1.00 | 1.00 | ||||
| TT | 0.169 | 0.145 | 1.21 (0.78–1.86) | 0.398 | 1.20 (0.78–1.86) | 0.402 | |||
| Additive | – | 1.12 (0.89–1.40) | 0.340 | 1.12 (0.89–1.41) | 0.333 | ||||
| MGAT5 | rs6430491 | Codominant | GG | 0.400 | 0.318 | 1.00 | 1.00 | ||
| AA | 0.102 | 0.194 | 0.42 (0.25–0.69) | 0.0006 | 0.41 (0.25–0.68) | 0.0005 | |||
| AG | 0.498 | 0.487 | 0.81 (0.58–1.15) | 0.242 | 0.81 (0.58–1.15) | 0.240 | |||
| Dominant | GG | 0.400 | 0.318 | 1.00 | 1.00 | ||||
| AA + AG | 0.600 | 0.682 | 0.70 (0.51–0.97) | 0.033 | 0.70 (0.50–0.97) | 0.033 | |||
| Recessive | AG + GG | 0.898 | 0.806 | 1.00 | 1.00 | ||||
| AA | 0.102 | 0.194 | 0.47 (0.30–0.74) | 0.0012 | 0.47 (0.29–0.74) | 0.0012 | |||
| Additive | – | – | – | 0.68 (0.54–0.86) | 0.0014 | 0.68 (0.54–0.86) | 0.0013 | ||
| MGAT5 | rs2593704 | Codominant | CC | 0.619 | 0.529 | 1.00 | 1.00 | ||
| GG | 0.054 | 0.077 | 0.60 (0.31–1.15) | 0.121 | 0.61 (0.31–1.17) | 0.136 | |||
| GC | 0.327 | 0.394 | 0.71 (0.51–0.99) | 0.045 | 0.72 (0.51–1.01) | 0.057 | |||
| Dominant | CC | 0.619 | 0.529 | 1.00 | 1.00 | ||||
| GG + GC | 0.381 | 0.471 | 0.69 (0.50–0.95) | 0.023 | 0.70 (0.51–0.97) | 0.031 | |||
| Recessive | GC + CC | 0.946 | 0.923 | 1.00 | 1.00 | ||||
| GG | 0.054 | 0.077 | 0.68 (0.36–1.29) | 0.239 | 0.69 (0.36–1.31) | 0.258 | |||
| Additive | – | – | – | 0.74 (0.57–0.96) | 0.022 | 0.75 (0.58–0.97) | 0.029 | ||
SNP single nucleotide polymorphism, OR odds ratio, 95% CI 95% confidence interval
pa values were calculated by logistic regression analysis with the comparison between COPD patients and healthy controls
pb values were calculated by logistic regression analysis with adjustment for age and gender
Bold values indicate statistical significance (p < 0.05)
Next, we evaluated the association of FGFR2 and MGAT5 variants with COPD susceptibility in different subgroups (Tables 3, 4, 5). Rs2420915 promoted the development of COPD in men, women, non-smokers, and individuals older than 70 years (p < 0.05). FGFR2-rs1907240, and -rs2257129 augmented the likelihood of COPD in non-smokers (p < 0.05). Rs6430491 in non-smokers and subjects in BMI ≤ 24 kg/m2 and rs2593704 in males, smokers, and individuals aged < 70 years and BMI ≤ 24 kg/m2 decreased the occurrence of COPD (p < 0.05).
Table 3.
The relationship of FGFR2 and MGAT5 SNPs with COPD risk stratified by age and gender
| Age | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene SNP | Model | Genotype | > 70 years | ≤ 70 years | ||||||
| Frequency in case | Frequency in control | OR (95% CI) | p | Frequency in case | Frequency in control | OR(95% CI) | p | |||
|
FGFR rs2420915 |
Allele | G | 0.581 | 0.658 | 1.00 | 0.594 | 0.675 | 1.00 | ||
| A | 0.419 | 0.342 | 1.39 (1.03–1.88) | 0.032 | 0.406 | 0.325 | 1.42 (0.99–2.02) | 0.054 | ||
| Codominant | GG | 0.301 | 0.429 | 1.00 | 0.362 | 0.467 | 1.00 | |||
| AA | 0.140 | 0.113 | 2.09 (1.03–4.25) | 0.043 | 0.173 | 0.117 | 1.79 (0.83–3.86) | 0.137 | ||
| AG | 0.559 | 0.458 | 1.87 (1.17–3.01) | 0.010 | 0.465 | 0.416 | 1.41 (0.83–2.42) | 0.207 | ||
| Dominant | GG | 0.301 | 0.429 | 1.00 | 0.362 | 0.467 | 1.00 | |||
| AA + AG | 0.699 | 0.571 | 1.91 (1.21–3.02) | 0.005 | 0.638 | 0.533 | 1.50 (0.90–2.48) | 0.119 | ||
| Recessive | AG + GG | 0.860 | 0.887 | 1.00 | 0.827 | 0.883 | 1.00 | |||
| AA | 0.140 | 0.113 | 1.43 (0.75–2.74) | 0.278 | 0.173 | 0.117 | 1.50 (0.73–3.06) | 0.269 | ||
| Additive | – | – | – | 1.55 (1.11–2.17) | 0.010 | – | – | 1.36 (0.95–1.95) | 0.097 | |
|
MGAT5 rs2593704 |
Allele | C | 0.803 | 0.766 | 1.00 | 0.752 | 0.675 | 1.00 | ||
| G | 0.197 | 0.234 | 0.80 (0.56–1.14) | 0.223 | 0.248 | 0.325 | 0.69 (0.47–1.00) | 0.052 | ||
| Codominant | CC | 0.654 | 0.601 | 1.00 | 0.567 | 0.438 | 1.00 | |||
| GG | 0.048 | 0.069 | 0.72 (0.28–1.83) | 0.490 | 0.063 | 0.088 | 0.53 (0.20–1.43) | 0.212 | ||
| GC | 0.298 | 0.330 | 0.88 (0.55–1.41) | 0.599 | 0.370 | 0.474 | 0.60 (0.36–1.01) | 0.054 | ||
| Dominant | CC | 0.654 | 0.601 | 1.00 | 0.567 | 0.438 | 1.00 | |||
| GG + GC | 0.346 | 0.399 | 0.85 (0.55–1.33) | 0.487 | 0.433 | 0.562 | 0.59 (0.36–0.97) | 0.038 | ||
| Recessive | GC + CC | 0.952 | 0.931 | 1.00 | 0.937 | 0.912 | 1.00 | |||
| GG | 0.048 | 0.069 | 0.75 (0.30–1.89) | 0.541 | 0.063 | 0.088 | 0.68 (0.26–1.76) | 0.423 | ||
| Additive | – | – | – | 0.86 (0.60–1.24) | 0.425 | – | – | 0.67 (0.44–0.99) | 0.047 | |
| Gender | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene SNP | Model | Genotype | Male | Female | ||||||
| Frequency in case | Frequency in control | OR (95% CI) | p | Frequency in case | Frequency in control | OR (95% CI) | p | |||
| FGFR | Allele | G | 0.601 | 0.667 | 1.00 | 0.540 | 0.662 | 1.00 | ||
| rs2420915 | A | 0.399 | 0.333 | 1.33 (1.02–1.73) | 0.035 | 0.460 | 0.338 | 1.67 (1.05–2.66) | 0.029 | |
| Codominant | GG | 0.344 | 0.451 | 1.00 | 0.267 | 0.429 | 1.00 | |||
| AA | 0.143 | 0.119 | 1.63 (0.91–2.90) | 0.101 | 0.187 | 0.104 | 2.91 (1.04–8.17) | 0.043 | ||
| AG | 0.513 | 0.430 | 1.55 (1.05–2.30) | 0.027 | 0.546 | 0.467 | 1.90 (0.93–3.88) | 0.080 | ||
| Dominant | GG | 0.344 | 0.451 | 1.00 | 0.267 | 0.429 | 1.00 | |||
| AA + AG | 0.656 | 0.549 | 1.57 (1.08–2.27) | 0.017 | 0.733 | 0.571 | 2.08 (1.05–4.13) | 0.036 | ||
| Recessive | AG + GG | 0.857 | 0.881 | 1.00 | 0.813 | 0.896 | 1.00 | |||
| AA | 0.143 | 0.119 | 1.28 (0.75–2.19) | 0.370 | 0.187 | 0.104 | 1.98 (0.78–5.04) | 0.152 | ||
| Additive | – | – | – | 1.35 (1.03–1.77) | 0.030 | – | – | 1.75 (1.07–2.86) | 0.026 | |
| MGAT5 | Allele | C | 0.795 | 0.714 | 0.64 (0.48–0.87) | 0.004 | 0.743 | 0.764 | 1.11 (0.66–1.89) | 0.687 |
| rs2593704 | G | 0.205 | 0.286 | 1.00 | 0.257 | 0.236 | 1.00 | |||
| Codominant | CC | 0.645 | 0.508 | 0.54 (0.26–1.15) | 0.110 | 0.539 | 0.595 | 0.86 (0.22–3.44) | 0.832 | |
| GG | 0.054 | 0.081 | 0.59 (0.40–0.87) | 0.008 | 0.053 | 0.068 | 1.33 (0.68–2.62) | 0.407 | ||
| GC | 0.301 | 0.411 | 1.00 | 0.408 | 0.337 | 1.00 | ||||
| Dominant | CC | 0.645 | 0.508 | 0.58 (0.40–0.85) | 0.004 | 0.539 | 0.595 | 1.25 (0.66–2.40) | 0.494 | |
| GG + GC | 0.355 | 0.492 | 1.00 | 0.461 | 0.405 | 1.00 | ||||
| Recessive | GC + CC | 0.946 | 0.919 | 0.67 (0.32–1.39) | 0.279 | 0.947 | 0.932 | 0.77 (0.20–2.99) | 0.703 | |
| GG | 0.054 | 0.081 | 0.66 (0.49–0.89) | 0.007 | 0.053 | 0.068 | 1.12 (0.66–1.89) | 0.683 | ||
| Additive | – | – | – | 0.64 (0.48–0.87) | 0.004 | – | – | 1.11 (0.66–1.89) | 0.687 | |
SNP single nucleotide polymorphism, OR odds ratio, 95% CI 95% confidence interval
p values were calculated by logistic regression analysis with adjustment for age and gender
Bold values indicate statistical significance (p < 0.05)
Table 4.
The relationship of FGFR2 and MGAT5 SNPs with COPD risk stratified by smoking
| Gene SNP | Model | Genotype | Smoking | No smoking | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Frequency in case | Frequency in control | OR (95% CI) | p | Frequency in case | Frequency in control | OR(95% CI) | p | |||
|
FGFR2 rs2420915 |
Allele | G | 0.613 | 0.615 | 1.00 | 0.567 | 0.674 | 1.00 | ||
| A | 0.387 | 0.385 | 1.01 (0.64–1.60) | 0.966 | 0.433 | 0.326 | 1.58 (1.11–2.24) | 0.010 | ||
| Codominant | GG | 0.370 | 0.385 | 1.00 | 0.291 | 0.458 | 1.00 | |||
| AA | 0.144 | 0.154 | 1.07 (0.40–2.87) | 0.894 | 0.158 | 0.110 | 2.16 (0.99–4.68) | 0.052 | ||
| AG | 0.486 | 0.461 | 1.11 (0.55–2.23) | 0.781 | 0.551 | 0.432 | 1.96 (1.16–3.29) | 0.012 | ||
| Dominant | GG | 0.370 | 0.385 | 1.00 | 0.291 | 0.458 | 1.00 | |||
| AA + AG | 0.630 | 0.615 | 1.10 (0.57–2.12) | 0.785 | 0.709 | 0.542 | 2.00 (1.21–3.28) | 0.006 | ||
| Recessive | AG + GG | 0.856 | 0.846 | 1.00 | 0.842 | 0.890 | 1.00 | |||
| AA | 0.144 | 0.154 | 1.01 (0.41–2.53) | 0.977 | 0.158 | 0.110 | 1.46 (0.72–3.00) | 0.297 | ||
| Additive | – | – | – | 1.05 (0.66–1.68) | 0.836 | – | – | 1.60 (1.10–2.31) | 0.013 | |
|
FGFR2 rs1907240 |
Allele | A | 0.617 | 0.577 | 1.00 | 0.542 | 0.640 | 1.00 | ||
| G | 0.383 | 0.423 | 0.85 (0.54–1.33) | 0.470 | 0.458 | 0.360 | 1.50 (1.07–2.11) | 0.020 | ||
| Codominant | AA | 0.407 | 0.327 | 1.00 | 0.301 | 0.381 | 1.00 | |||
| GG | 0.172 | 0.173 | 0.92 (0.35–2.40) | 0.859 | 0.217 | 0.102 | 2.63 (1.22–5.68) | 0.014 | ||
| GA | 0.421 | 0.500 | 0.68 (0.33–1.38) | 0.282 | 0.482 | 0.517 | 1.19 (0.70–2.01) | 0.525 | ||
| Dominant | AA | 0.407 | 0.327 | 1.00 | 0.301 | 0.381 | 1.00 | |||
| GG + GA | 0.593 | 0.673 | 0.73 (0.37–1.44) | 0.366 | 0.699 | 0.619 | 1.43 (0.86–2.35) | 0.165 | ||
| Recessive | GA + AA | 0.828 | 0.827 | 1.00 | 0.783 | 0.898 | 1.00 | |||
| GG | 0.172 | 0.173 | 1.14 (0.48–2.72) | 0.769 | 0.217 | 0.102 | 2.38 (1.17–4.81) | 0.016 | ||
| Additive | – | – | – | 0.90 (0.58–1.41) | 0.645 | – | – | 1.51 (1.06–2.14) | 0.023 | |
|
FGFR2 rs2257129 |
Allele | C | 0.627 | 0.577 | 1.00 | 0.552 | 0.640 | 1.00 | ||
| T | 0.373 | 0.423 | 0.81 (0.52–1.28) | 0.371 | 0.448 | 0.360 | 1.45 (1.03–2.04) | 0.035 | ||
| Codominant | CC | 0.390 | 0.327 | 1.00 | 0.303 | 0.381 | 1.00 | |||
| TT | 0.137 | 0.173 | 0.75 (0.28–2.02) | 0.572 | 0.200 | 0.102 | 2.40 (1.11–5.23) | 0.027 | ||
| TC | 0.473 | 0.500 | 0.79 (0.39–1.61) | 0.517 | 0.497 | 0.517 | 1.21 (0.72–2.05) | 0.468 | ||
| Dominant | CC | 0.390 | 0.327 | 1.00 | 0.303 | 0.381 | 1.00 | |||
| TT + TC | 0.610 | 0.673 | 0.78 (0.40–1.54) | 0.474 | 0.697 | 0.619 | 1.41 (0.86–2.33) | 0.177 | ||
| Recessive | TC + CC | 0.863 | 0.827 | 1.00 | 0.800 | 0.898 | 1.00 | |||
| TT | 0.137 | 0.173 | 0.86 (0.35–2.11) | 0.742 | 0.200 | 0.102 | 2.14 (1.05–4.36) | 0.036 | ||
| Additive | – | – | – | 0.85 (0.53–1.36) | 0.499 | – | – | 1.46 (1.02–2.09) | 0.039 | |
|
MGAT5 rs6430491 |
Allele | G | 0.588 | 0.606 | 1.00 | 0.699 | 0.576 | 1.00 | ||
| A | 0.412 | 0.394 | 1.08 (0.68–1.70) | 0.757 | 0.301 | 0.424 | 0.59 (0.41–0.83) | 0.003 | ||
| Codominant | GG | 0.333 | 0.365 | 1.00 | 0.452 | 0.364 | 1.00 | |||
| AA | 0.156 | 0.154 | 1.11 (0.42–2.91) | 0.839 | 0.054 | 0.212 | 0.21 (0.09–0.49) | 0.0003 | ||
| AG | 0.511 | 0.481 | 1.28 (0.63–2.62) | 0.495 | 0.494 | 0.424 | 0.92 (0.55–1.55) | 0.758 | ||
| Dominant | GG | 0.333 | 0.365 | 1.00 | 0.452 | 0.364 | 1.00 | |||
| AA + AG | 0.667 | 0.635 | 1.24 (0.63–2.42) | 0.537 | 0.548 | 0.636 | 0.69 (0.42–1.12) | 0.136 | ||
| Recessive | AG + GG | 0.844 | 0.846 | 1.00 | 0.946 | 0.788 | 1.00 | |||
| AA | 0.156 | 0.154 | 0.96 (0.40–2.32) | 0.927 | 0.054 | 0.212 | 0.22 (0.10–0.49) | 0.0002 | ||
| Additive | – | – | – | 1.10 (0.68–1.76) | 0.704 | – | – | 0.58 (0.40–0.83) | 0.003 | |
|
MGAT5 rs2593704 |
Allele | C | 0.789 | 0.635 | 1.00 | 0.777 | 0.770 | 1.00 | ||
| G | 0.211 | 0.365 | 0.46 (0.29–0.76) | 0.002 | 0.223 | 0.230 | 0.96 (0.64–1.43) | 0.834 | ||
| Codominant | CC | 0.639 | 0.404 | 1.00 | 0.602 | 0.609 | 1.00 | |||
| GG | 0.061 | 0.135 | 0.37 (0.12–1.16) | 0.088 | 0.048 | 0.070 | 0.73 (0.26–2.04) | 0.544 | ||
| GC | 0.300 | 0.461 | 0.42 (0.21–0.84) | 0.014 | 0.350 | 0.321 | 1.09 (0.65–1.82) | 0.756 | ||
| Dominant | CC | 0.639 | 0.404 | 1.00 | 0.602 | 0.609 | 1.00 | |||
| GG + GC | 0.361 | 0.596 | 0.41 (0.21–0.79) | 0.008 | 0.398 | 0.391 | 1.02 (0.63–1.67) | 0.930 | ||
| Recessive | GC + CC | 0.939 | 0.865 | 1.00 | 0.952 | 0.930 | 1.00 | |||
| GG | 0.061 | 0.135 | 0.54 (0.18–1.62) | 0.268 | 0.048 | 0.070 | 0.71 (0.26–1.95) | 0.501 | ||
| Additive | – | – | – | 0.53 (0.32–0.87) | 0.012 | – | – | 0.96 (0.65–1.43) | 0.850 | |
SNP single nucleotide polymorphism, OR odds ratio, 95% CI 95% confidence interval
p values were calculated by logistic regression analysis with adjustment for age and gender
Bold values indicate statistical significance (p < 0.05)
Table 5.
The relationship of FGFR2 and MGAT5 SNPs with COPD risk stratified by BMI
| Gene | SNP | Model | Genotype | BMI ≤ 24 | |||
|---|---|---|---|---|---|---|---|
| Frequency in case | Frequency in control | OR (95% CI) | p | ||||
| FGFR2 | rs2420915 | Allele | G | 0.586 | 0.627 | 1.00 | |
| A | 0.414 | 0.373 | 1.19 (0.80–1.76) | 0.392 | |||
| Codominant | GG | 0.320 | 0.373 | 1.00 | |||
| AA | 0.148 | 0.120 | 1.58 (0.64–3.92) | 0.324 | |||
| AG | 0.532 | 0.507 | 1.19 (0.65–2.17) | 0.574 | |||
| Dominant | GG | 0.320 | 0.373 | 1.00 | |||
| AA + AG | 0.680 | 0.627 | 1.26 (0.71–2.24) | 0.429 | |||
| Recessive | AG + GG | 0.852 | 0.880 | 1.00 | |||
| AA | 0.148 | 0.120 | 1.43 (0.62–3.31) | 0.406 | |||
| Additive | – | – | – | 1.24 (0.81–1.88) | 0.318 | ||
| FGFR2 | rs1907240 | Allele | A | 0.574 | 0.597 | 1.00 | |
| G | 0.426 | 0.403 | 1.10 (0.75–1.62) | 0.632 | |||
| Codominant | AA | 0.348 | 0.328 | 1.00 | |||
| GG | 0.200 | 0.135 | 1.43 (0.60–3.41) | 0.419 | |||
| GA | 0.452 | 0.537 | 0.79 (0.43–1.46) | 0.452 | |||
| Dominant | AA | 0.348 | 0.328 | 1.00 | |||
| GG + GA | 0.652 | 0.672 | 0.92 (0.51–1.64) | 0.771 | |||
| Recessive | GA + AA | 0.800 | 0.865 | 1.00 | |||
| GG | 0.200 | 0.135 | 1.64 (0.75–3.60) | 0.215 | |||
| Additive | – | – | – | 1.10 (0.75–1.63) | 0.626 | ||
| FGFR2 | rs2257129 | Allele | C | 0.586 | 0.597 | 1.00 | |
| T | 0.414 | 0.403 | 1.05 (0.71–1.54) | 0.824 | |||
| Codominant | CC | 0.341 | 0.328 | 1.00 | |||
| TT | 0.169 | 0.135 | 1.23 (0.51–2.97) | 0.641 | |||
| TC | 0.490 | 0.537 | 0.86 (0.47–1.59) | 0.641 | |||
| Dominant | CC | 0.341 | 0.328 | 1.00 | |||
| TT + TC | 0.659 | 0.672 | 0.94 (0.52–1.68) | 0.827 | |||
| Recessive | TC + CC | 0.831 | 0.865 | 1.00 | |||
| TT | 0.169 | 0.135 | 1.35 (0.61–2.99) | 0.466 | |||
| Additive | – | – | – | 1.05 (0.70–1.57) | 0.811 | ||
| MGAT5 | rs6430491 | Allele | G | 0.639 | 0.604 | 1.00 | |
| A | 0.361 | 0.396 | 0.86 (0.58–1.27) | 0.456 | |||
| Codominant | GG | 0.386 | 0.403 | 1.00 | |||
| AA | 0.108 | 0.194 | 0.48 (0.21–1.08) | 0.075 | |||
| AG | 0.506 | 0.403 | 1.19 (0.65–2.20) | 0.572 | |||
| Dominant | GG | 0.386 | 0.403 | 1.00 | |||
| AA + AG | 0.614 | 0.597 | 0.96 (0.54–1.69) | 0.888 | |||
| Recessive | AG + GG | 0.892 | 0.806 | 1.00 | |||
| AA | 0.108 | 0.194 | 0.43 (0.20–0.920 | 0.029 | |||
| Additive | – | – | – | 0.78 (0.51–1.18) | 0.231 | ||
| MGAT5 | rs2593704 | Allele | C | 0.791 | 0.657 | 1.00 | |
| G | 0.209 | 0.343 | 0.51 (0.33–0.77) | 0.001 | |||
| Codominant | CC | 0.633 | 0.433 | 1.00 | |||
| GG | 0.052 | 0.119 | 0.31 (0.12–0.83) | 0.020 | |||
| GC | 0.315 | 0.448 | 0.52 (0.29–0.94) | 0.031 | |||
| Dominant | CC | 0.633 | 0.433 | 1.00 | |||
| GG + GC | 0.367 | 0.567 | 0.48 (0.27–0.83) | 0.009 | |||
| Recessive | GC + CC | 0.948 | 0.881 | 1.00 | |||
| GG | 0.052 | 0.119 | 0.41 (0.16–1.05) | 0.064 | |||
| Additive | – | – | – | 0.54 (0.36–0.83) | 0.005 | ||
SNP single nucleotide polymorphism, OR odds ratio, 95% CI 95% confidence interval
p values were calculated by logistic regression analysis with adjustment for age and gender
Bold values indicate statistical significance (p < 0.05)
Haplotype analysis
We further analyzed the haplotype and linkage disequilibrium of FGFR2 and MGAT5 variants in cases and control group. The results in Fig. 1 showed that an LD plot consisted of two SNPs (rs1907240 and rs2257129). And there was no correlation of haplotypes with COPD risk (p > 0.05, Table 6).
Fig. 1.
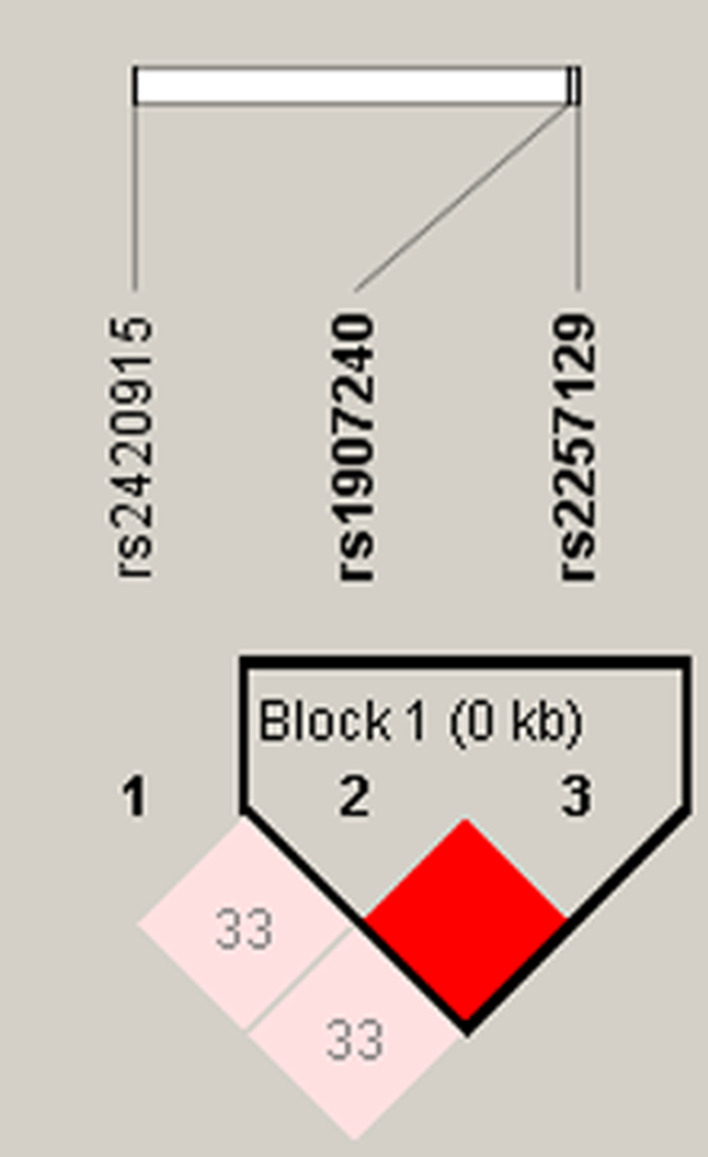
Haplotype block map for SNPs of FGFR2. Block 1 includes rs1907240, and rs2257129. The numbers inside the diamonds indicate the D’ for pairwise analyses
Table 6.
Haplotype association of FGFR2 polymorphisms with COPD risk
| SNP | Haplotype | Frequency in cases | Frequency in controls | Without adjustment | With adjustment | ||
|---|---|---|---|---|---|---|---|
| OR (95% CI) | p | OR (95% CI) | p | ||||
| rs1907240|rs2257129 | GT | 0.410 | 0.388 | 1.10 (0.88–1.38) | 0.416 | 1.10 (0.88–1.38) | 0.409 |
| rs1907240|rs2257129 | AC | 0.423 | 0.388 | 1.16 (0.92–1.45) | 0.207 | 1.16 (0.92–1.45) | 0.203 |
SNP single nucleotide polymorphism, OR odds ratio, CI confidence interval
Discussion
We assessed the correlation between FGFR2 and MGAT5 mutants and COPD susceptibility in the Chinese population. The results revealed that rs2420915 increased the incidence of COPD, while rs6430491, rs2593704 reduced the risk of COPD. In addition, rs2420915, rs1907240, rs2257129, rs6430491, and rs2593704 were associated with COPD susceptibility in different subgroups. These data emphasized the crucial role of FGFR2 and MGAT5 in the pathogenesis of COPD, and provide new biomarkers for the treatment and diagnosis of COPD.
The FGFR2 gene belongs to the fibroblast growth factor receptor family and is located on chromosome 10q26.13 in humans. FGFR2 gene has been reported to encode FGFR2b in epithelial cells and FGFR2c in mesenchymal cells [23]. Yu et al. [24] have found that FGFR2 mutant attenuated lung fibrosis by inhibiting α-smooth muscle actin and collagen deposit. Furthermore, Masunaga et al. [25] indicated that the expression of FGFR2 was highly expressed in pulmonary papillary adenoma cells compared with nontumorous lung. Besides, FGFR2b signaling facilitated alveolar epithelial regeneration through bronchial epithelial stem cells after lung injury [26]. These findings demonstrated that FGFR2 gene played a crucial role in lung disease. However, there are no reports on rs2420915, rs1907240, and rs2257129 in lung disease and COPD. In our study, we first investigated the impact of rs2420915, rs1907240, rs2257129 on the occurrence of COPD. The results indicated that rs2420915, FGFR2-rs1907240, and -rs2257129 were risk factors for COPD development. These data suggested that FGFR2 variants may be involved in COPD development, and it provided new clues for individualized treatment of COPD patients.
MGAT5, a typical cancer-associated glycosyltransferase, is located in 2q21.2-q21.3. It is closely associated with the growth, migration, and invasion of tumor cells [27, 28]. Dosaka-Akita et al. [29] found that MGAT5 is associated with histology and prognosis in non-small cell lung cancers. Similarly, Zhou et al. [18] reported that MGAT5 was overexpressed in pulmonary adenocarcinoma cells, and knockdown of MGAT5 could suppress cell growth both in vitro and in vivo. Moreover, Elek et al. [20] demonstrated that the allele frequencies of rs34944508 in the 3′-UTR of MGAT5 gene were significantly different among control, COPD, lung cancer, and comorbid COPD and lung cancer, and indicated that rs34944508 might influence lung cancer risk in Caucasian. Nevertheless, no studies focused on the role of rs6430491, and rs2593704 in COPD development. We, for the first time, found rs6430491, and MGAT5-rs2593704 are correlated with a decreased risk of COPD, and illustrated that MGAT5 gene has a potential role in the pathogenesis of COPD.
Some research has shown that the intronic SNPs can modify gene function by altering the expression of gene [30, 31]. In our research, rs1907240, rs2257129, rs2593704 are located in the intron region of FGFR2 and MGAT5 gene. Combining previous studies and database predictions, we hypothesize that FGFR2 and MGAT5 intron SNPs cause changes in FGFR2 and MGAT5 expression and activity via influencing mRNA splicing and ultimately affect disease susceptibility. In subsequent experiments, we will examine the functional consequences of the intronic polymorphisms to support our hypothesis in vitro and ex vivo, focusing on the regulation of gene expression and splicing. In addition, rs2420915 and rs6430491 were associated with the regulation of SiPhy cons, enhancer histone marks, DNAse, motifs changed, GRASP QTL hits. These functions could affect the expression of gene, and ultimately alter the susceptibility of COPD.
Although the interesting results on the relationship of FGFR2 and MGAT5 polymorphisms with COPD susceptibility, several limitations of this study need to be stated. Firstly, we only genotyped three SNPs in FGFR2 and two SNPs in MGAT5, more SNPs of these two genes are needed to investigate. Secondly, the selection bias is inevitable when all the study individuals are enrolled from the same hospital. Thirdly, the molecular mechanism of FGFR2 and MGAT5 to COPD susceptibility remains unknown and should be studied in further study.
Conclusions
Our results suggested that FGFR2 and MGAT5 genetic polymorphisms are correlated with the risk of COPD in the Chinese Han people. These data underscored the important role of FGFR2 and MGAT5 gene in the occurrence of COPD and provided new biomarkers for COPD treatment.
Supplementary Information
Additional file 1: Supplemental table 1. Demographic and clinical characteristics of study populations.
Acknowledgements
We sincerely thank all participators in this study.
Abbreviations
- COPD
Chronic obstructive pulmonary disease
- FEV1
Forced expiratory volume in one second
- FVC
Forced vital capacity
- BMI
Body mass index
- SNP
Single nucleotide polymorphism
- HWE
Hardy–Weinberg equilibrium
- OR
Odd ratio
- CI
95% Confidence intervals
- FGFR2
Fibroblast growth factor receptor 2
- MGAT5
N-acetylglucosaminyltransferaseV
Authors' contributions
XL drafted the manuscript. GZ performed the experiments. XT analyzed the data. GL edited manuscript. YD designed and supervised the study. All authors read and approved the final manuscript.
Funding
This study supported by the National Natural Science Foundation of China (No. 81660013 and No. 81860015) and Hainan Provincial Social Development Project (SF201402).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Declarations
Ethics approval and consent to participate
Written informed consents were required from study populations before they got involved in the study. All procedures were in accordance with the Declaration of Helsinki, and this study was approved by the ethics committee of Hainan General Hospital.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaobo Li and Guangyu Zhou have contributed equally to this work
References
- 1.Flattet Y, Garin N, Serratrice J, Perrier A, Stirnemann J, Carballo S. Determining prognosis in acute exacerbation of COPD. Int J Chron Obstruct Pulmon Dis. 2017;12:467–475. doi: 10.2147/COPD.S122382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donaldson GC, Wilkinson TM, Hurst JR, Perera WR, Wedzicha JA. Exacerbations and time spent outdoors in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2005;171(5):446–452. doi: 10.1164/rccm.200408-1054OC. [DOI] [PubMed] [Google Scholar]
- 3.Salvi S. Tobacco smoking and environmental risk factors for chronic obstructive pulmonary disease. Clin Chest Med. 2014;35(1):17–27. doi: 10.1016/j.ccm.2013.09.011. [DOI] [PubMed] [Google Scholar]
- 4.Eisner MD, Anthonisen N, Coultas D, Kuenzli N, Perez-Padilla R, Postma D, et al. An official American Thoracic Society public policy statement: novel risk factors and the global burden of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2010;182(5):693–718. doi: 10.1164/rccm.200811-1757ST. [DOI] [PubMed] [Google Scholar]
- 5.Shukla RK, Kant S, Bhattacharya S, Mittal B. Association of genetic polymorphism of GSTT1, GSTM1 and GSTM3 in COPD patients in a north Indian population. COPD. 2011;8(3):167–172. doi: 10.3109/15412555.2011.560128. [DOI] [PubMed] [Google Scholar]
- 6.Manichaikul A, Hoffman EA, Smolonska J, Gao W, Cho MH, Baumhauer H, et al. Genome-wide study of percent emphysema on computed tomography in the general population. The Multi-Ethnic Study of Atherosclerosis Lung/SNP Health Association Resource Study. Am J Respir Crit Care Med. 2014;189(4):408–418. doi: 10.1164/rccm.201306-1061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du Y, Zhang H, Xu Y, Ding Y, Chen X, Mei Z, et al. Association among genetic polymorphisms of GSTP1, HO-1, and SOD-3 and chronic obstructive pulmonary disease susceptibility. Int J Chron Obstruct Pulmon Dis. 2019;14:2081–2088. doi: 10.2147/COPD.S213364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang B, Liang B, Yang J, Xiao J, Ma C, Xu S, et al. Association of FAM13A polymorphisms with COPD and COPD-related phenotypes in Han Chinese. Clin Biochem. 2013;46(16–17):1683–1688. doi: 10.1016/j.clinbiochem.2013.07.013. [DOI] [PubMed] [Google Scholar]
- 9.Yoon HK, Hu HJ, Rhee CK, Shin SH, Oh YM, Lee SD, et al. Polymorphisms in PDE4D are associated with a risk of COPD in non-emphysematous Koreans. COPD. 2014;11(6):652–658. doi: 10.3109/15412555.2014.898045. [DOI] [PubMed] [Google Scholar]
- 10.Ding Y, Li Q, Feng Q, Xu D, Wu C, Zhao J, et al. CYP2B6 genetic polymorphisms influence chronic obstructive pulmonary disease susceptibility in the Hainan population. Int J Chron Obstruct Pulmon Dis. 2019;14:2103–2115. doi: 10.2147/COPD.S214961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beenken A, Mohammadi M. The FGF family: biology, pathophysiology and therapy. Nat Rev Drug Discov. 2009;8(3):235–253. doi: 10.1038/nrd2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Theelen WS, Mittempergher L, Willems SM, Bosma AJ, Peters DD, van der Noort V, et al. FGFR1, 2 and 3 protein overexpression and molecular aberrations of FGFR3 in early stage non-small cell lung cancer. J Pathol Clin Res. 2016;2(4):223–233. doi: 10.1002/cjp2.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan T, Klinkhammer K, Lyu H, Gao S, Yuan J, Hopkins S, et al. Temporospatial expression of Fgfr1 and 2 during lung development, homeostasis, and regeneration. Front Pharmacol. 2020;11:120. doi: 10.3389/fphar.2020.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorry SJ, Ansbro BO, Ornitz DM, Mutlu GM, Guzy RD. FGFR2 is required for AEC2 homeostasis and survival after bleomycin-induced lung injury. Am J Respir Cell Mol Biol. 2020;62(5):608–621. doi: 10.1165/rcmb.2019-0079OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jieming G, Liu C, Yang Y, Mo S, Yang X, Wang J. Inhibitory effects of msFGFR2c on the epithelial-to-mesenchymal transition of AE2 cells in pulmonary fibrosis. Biotech Lett. 2020;42(6):1061–1070. doi: 10.1007/s10529-020-02852-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nabi IR, Shankar J, Dennis JW. The galectin lattice at a glance. J Cell Sci. 2015;128(13):2213–2219. doi: 10.1242/jcs.151159. [DOI] [PubMed] [Google Scholar]
- 17.Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, et al. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129(1):123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 18.Zhou X, Chen H, Wang Q, Zhang L, Zhao J. Knockdown of Mgat5 inhibits CD133+ human pulmonary adenocarcinoma cell growth in vitro and in vivo. Clin Invest Med. 2011;34(3):E155–E162. doi: 10.25011/cim.v34i3.15188. [DOI] [PubMed] [Google Scholar]
- 19.Cai J, Huang J, Wang W, Zeng J, Wang P. miR-124-3p regulates FGF2-EGFR pathway to overcome pemetrexed resistance in lung adenocarcinoma cells by targeting MGAT5. Cancer Manag Res. 2020;12:11597–11609. doi: 10.2147/CMAR.S274192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elek Z, Kovács Z, Keszler G, Szabó M, Csanky E, Luo J, et al. High throughput multiplex SNP-analysis in chronic obstructive pulmonary disease and lung cancer. Curr Mol Med. 2020;20(3):185–193. doi: 10.2174/1566524019666191017123446. [DOI] [PubMed] [Google Scholar]
- 21.Du J, Jin T, Cao Y, Chen J, Guo Y, Sun M, et al. Association between genetic polymorphisms of MMP8 and the risk of steroid-induced osteonecrosis of the femoral head in the population of northern China. Medicine. 2016;95(37):e4794. doi: 10.1097/MD.0000000000004794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Q, Liao S, Wei H, Liu D, Li J, Zhang X, et al. CDKN2BAS polymorphisms are associated with coronary heart disease risk a Han Chinese population. Oncotarget. 2016;7(50):82046–82054. doi: 10.18632/oncotarget.12575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katoh M. Cancer genomics and genetics of FGFR2 (Review) Int J Oncol. 2008;33(2):233–237. [PubMed] [Google Scholar]
- 24.Yu ZH, Wang DD, Zhou ZY, He SL, Chen AA, Wang J. Mutant soluble ectodomain of fibroblast growth factor receptor-2 IIIc attenuates bleomycin-induced pulmonary fibrosis in mice. Biol Pharm Bull. 2012;35(5):731–736. doi: 10.1248/bpb.35.731. [DOI] [PubMed] [Google Scholar]
- 25.Masunaga A, Nagashio R, Iwamoto S, Takeyama N, Sato Y, Miyazaki A, et al. A case of pulmonary papillary adenoma: possible relationship between tumor histogenesis/tumorigenesis and fibroblast growth factor receptor 2 IIIb. Pathol Int. 2012;62(9):640–645. doi: 10.1111/j.1440-1827.2012.02841.x. [DOI] [PubMed] [Google Scholar]
- 26.Yuan T, Volckaert T, Redente EF, Hopkins S, Klinkhammer K, Wasnick R, et al. FGF10-FGFR2B signaling generates basal cells and drives alveolar epithelial regeneration by bronchial epithelial stem cells after lung injury. Stem Cell Rep. 2019;12(5):1041–1055. doi: 10.1016/j.stemcr.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li B, Su S, Zhang MY, He L, Wang QD, He K. Effect of GnT-V knockdown on the proliferation, migration and invasion of the SMMC7721/R human hepatocellular carcinoma drug-resistant cell line. Mol Med Rep. 2016;13(1):469–476. doi: 10.3892/mmr.2015.4492. [DOI] [PubMed] [Google Scholar]
- 28.Hassani Z, Saleh A, Turpault S, Khiati S, Morelle W, Vignon J, et al. Phostine PST3.1a targets MGAT5 and inhibits glioblastoma-initiating cell invasiveness and proliferation. Mol Cancer Res MCR. 2017;15(10):1376–1387. doi: 10.1158/1541-7786.MCR-17-0120. [DOI] [PubMed] [Google Scholar]
- 29.Dosaka-Akita H, Miyoshi E, Suzuki O, Itoh T, Katoh H, Taniguchi N. Expression of N-acetylglucosaminyltransferase v is associated with prognosis and histology in non-small cell lung cancers. Clin Cancer Res Off J Am Assoc Cancer Res. 2004;10(5):1773–1779. doi: 10.1158/1078-0432.CCR-1047-3. [DOI] [PubMed] [Google Scholar]
- 30.Zhao H, Yang W, Qiu R, Li J, Xin Q, Wang X, et al. An intronic variant associated with systemic lupus erythematosus changes the binding affinity of Yinyang1 to downregulate WDFY4. Genes Immun. 2012;13(7):536–542. doi: 10.1038/gene.2012.33. [DOI] [PubMed] [Google Scholar]
- 31.Seo S, Takayama K, Uno K, Ohi K, Hashimoto R, Nishizawa D, et al. Functional analysis of deep intronic SNP rs13438494 in intron 24 of PCLO gene. PLoS ONE. 2013;8(10):e76960. doi: 10.1371/journal.pone.0076960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Supplemental table 1. Demographic and clinical characteristics of study populations.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


