Abstract
The ability to detect specific nucleic acid sequences allows for a wide range of applications such as the identification of pathogens, clinical diagnostics, and genotyping. CRISPR-Cas proteins Cas12a and Cas13a are RNA-guided endonucleases that bind and cleave specific DNA and RNA sequences, respectively. After recognition of a target sequence, both enzymes activate indiscriminate nucleic acid cleavage, which has been exploited for sequence-specific molecular diagnostics of nucleic acids. Here, we present a label-free detection approach that uses a readout based on solution turbidity caused by liquid-liquid phase separation (LLPS). Our approach relies on the fact that the LLPS of oppositely charged polymers requires polymers to be longer than a critical length. This length dependence is predicted by the Voorn-Overbeek model, which we describe in detail and validate experimentally in mixtures of polynucleotides and polycations. We show that the turbidity resulting from LLPS can be used to detect the presence of specific nucleic acid sequences by employing the programmable CRISPR-nucleases Cas12a and Cas13a. Because LLPS of polynucleotides and polycations causes solutions to become turbid, the detection of specific nucleic acid sequences can be observed with the naked eye. We furthermore demonstrate that there is an optimal polynucleotide concentration for detection. Finally, we provide a theoretical prediction that hints towards possible improvements of an LLPS-based detection assay. The deployment of LLPS complements CRISPR-based molecular diagnostic applications and facilitates easy and low-cost nucleotide sequence detection.
Significance
CRISPR-Cas proteins are highly effective for sequence-specific DNA and RNA detection. To use these proteins as molecular detectives, they need to generate a signal, typically through fluorescent molecules. Here, we have developed an alternative, label-free method for detecting specific sequences by using liquid-liquid phase separation. The detection mechanism combines CRISPR-Cas nuclease activity with the fact that certain DNA and RNA molecules phase separate in a length-dependent manner. The strategy provides a simple and cheap way to implement CRISPR-based molecular diagnostics and circumvents the chemical labeling of DNA or RNA molecules. Furthermore, the length dependence of liquid-liquid phase separation may also have implications for understanding open questions in the context of cell biology.
Introduction
CRISPR-Cas technology has revolutionized molecular biology by making genome editing (1) and nucleic acid sequence detection efficient and accessible (2, 3, 4, 5, 6). CRISPR-based detection methods rely on CRISPR RNA-guided nucleases such as Cas12a (7) (type V), Cas14a (8), and Cas13a (9) (type VI). These proteins display nonspecific cleavage activity of single-stranded DNA (ssDNA; Cas12a (5,10) and Cas14a (8)) or RNA (Cas13a (2,11,12)) upon recognition of their target sequences through diffusion and capture (13,14). The mechanisms employed by these enzymes rely on the formation of a Cas protein complex with CRISPR RNA (crRNA), which targets DNA or RNA sequences complementary to the crRNA (12,15,16). If a target is bound to the crRNA-protein complex, the enzymes display nonspecific (collateral) cleavage activity. This collateral activity is the mechanism that facilitates nucleotide detection assays in solution and on paper-based tests (2, 3, 4, 5). Previously, Cas12a and Cas13a have been employed to discriminate between two variants of the human papillomavirus (5) and between dengue and Zika viruses (3), respectively. More recently, Cas12a and Cas13a were used to detect severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in Covid-19 patients (17,18). In these studies, the detection of Cas12a and Cas13a activity relied on quenched fluorophores linked by DNA or RNA molecules (19). The proteins’ collateral endonuclease activity, which is activated after target binding, causes a fluorescent signal by cleaving the DNA or RNA linkage between a fluorophore-quencher pair (Fig. S1).
To overcome the requirement of fluorescence-based detection, we have developed a detection method based on liquid-liquid phase separation (LLPS). LLPS is a ubiquitous phenomenon in eukaryotic cells (20), in which the formation of membraneless organelles and compartments relies on the phase-separating properties of proteins and/or nucleic acids (21). Molecular and chemical properties largely determine the tendency of macromolecules to phase separate, and both in vivo and in vitro studies have contributed significantly to the current understanding of LLPS in the cellular context (22).
Building on recent advances in the fields of both LLPS and CRISPR-Cas proteins, we conceived a phase-separation-based DNA and RNA detection assay. We exploit the fact that solutions of long nucleic acid polymers and positively charged polyelectrolytes can undergo liquid-liquid phase separation into a polymer-rich (“coacervate”) phase and a polymer-depleted (“solvent”) phase (23, 24, 25). LLPS increases solution turbidity because coacervate droplets scatter incoming light, so that the onset of LLPS is visible to the naked eye (26,27). By using this effect in combination with the activity of CRISPR-Cas nucleases, it is possible to robustly couple the detection of specific DNA and RNA sequences to the turbidity of a sample solution. We first provide a mathematical model for the polymer length dependence of liquid-liquid phase separation and experimentally show that liquid-liquid phase separation critically depends on polymer length. Then, we demonstrate that LLPS can be used for nucleotide detection of specific DNA and RNA sequences with a turbidity measurement. Finally, we discuss the limitations and potential optimization steps of the assay.
Materials and methods
Length-dependent poly(dT) phase separation
Turbidity measurements to assess the length dependence of poly(dT) phase separation were performed with 0.5 mg/mL poly-L-lysine (pLL) in 1× NEB 2.1 buffer. For constant volume fraction measurements (Fig. 1 d), the concentration of poly(dT) was adjusted by keeping the overall concentration of dT monomers constant at 240 μM. The concentrations were 24, 12, 8, 6, 4.8, 4, 3.43, and 3 μM for increasing poly(dT) lengths ranging from dT10 to dT80 in steps of 10.
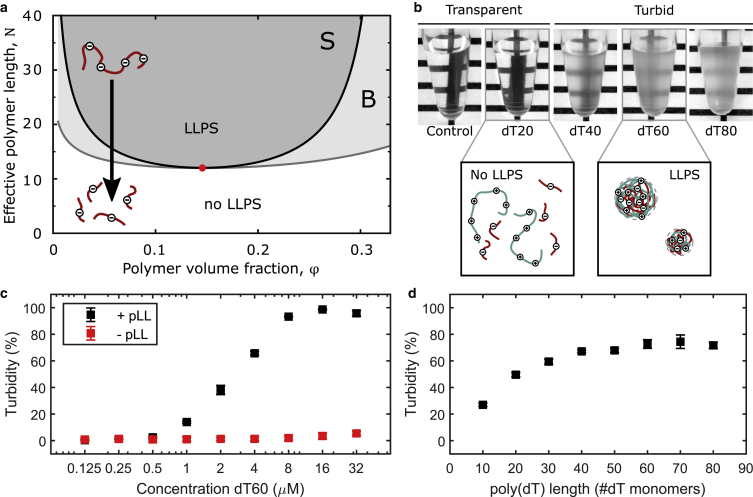
Figure 1.
Liquid-liquid phase separation of oppositely charged polymers is length dependent and causes solution turbidity. (a) The phase diagram of the Flory-Huggins (28,29) model with Voorn-Overbeek (24) interaction depends on polymer length. Phase separation requires a minimal length for a given polymer volume fraction. The three regions in the diagram correspond to a homogeneous solution (white, no LLPS), a metastable regime in region B (light gray, above the binodal curve), and a regime of spontaneous phase separation in region S (dark gray, above the spinodal curve). The red dot indicates the critical point of the system. If polymers are shorter than the critical length as indicated by the critical point, phase separation does not occur, independent of polymer volume fraction. The diagram shown is for symmetric polymer solutions (see Theory). (b) Experiments confirm the predicted length-dependent phenomenon for poly(dT) of various lengths and poly-L-lysine as oppositely charged polyelectrolytes. The pictures show transparent and turbid mixtures with different poly(dT) lengths ranging from 20 to 80 nucleotides. The overall concentration of dT monomers was kept constant at 1.2 mM so that the poly(dT) volume fraction was constant in each tube. Long poly(dT) (>40 nt) leads to turbid and robustly phase-separating solutions (5 mg/mL pLL, C12RB buffer), whereas solutions with short poly(dT) (<40 nt) are transparent and do not phase separate (at equal volume fraction of nucleotides). (c) Increasing concentrations of dT60 increase the turbidity of solutions (0.5 mg/mL pLL, 1× NEB 2.1 buffer). (d) Increasing poly(dT) length increases the turbidity of solutions at constant poly(dT) volume fraction (0.5 mg/mL pLL, 1× NEB 2.1 buffer). Error bars in (c) and (d) represent standard deviation (SD) of the mean, each from three independent experiments. To see this figure in color, go online.
Turbidity assays for DNA and RNA detection
Cas12a was loaded and activated for the poly(dT) cleavage assay by mixing ∼8.6 μM Cas12a and 10 μM crRNA (Table S1) in 1× C12RB buffer and incubating for 15 min at 37°C. The Cas12a turbidity assay (Fig. 2 c) contained 187 nM of the activated Cas12a and 22 μM dT60 in 1.1× C12RB buffer. The tubes shown in the top row of Fig. 2 c furthermore contained 220 nM DNA target and were left for either 0 (left column) or 60 min (right column). For the readout, pLL was added to a final concentration of 0.5 mg/mL. The turbidity assay for Cas13a (Fig. 2 d) consisted of ∼50 nM Cas13a, ∼0.3 ng/μL Cas13a target, and 0.1 wt% poly(U) mixed in 1.1× C13RB buffer. The tubes in the upper row contained ∼0.3 ng/μL crRNA for Cas13a (Table S1), whereas the tubes in the bottom row contained no crRNA. Tubes were left for either 0 min (left column) or 60 min (right column) at 37°C, after which spermine was added to a final concentration of 1.0 wt%. Cas12a and Cas13a proteins and nucleic acids were purified and prepared as described in the Supporting materials and methods.
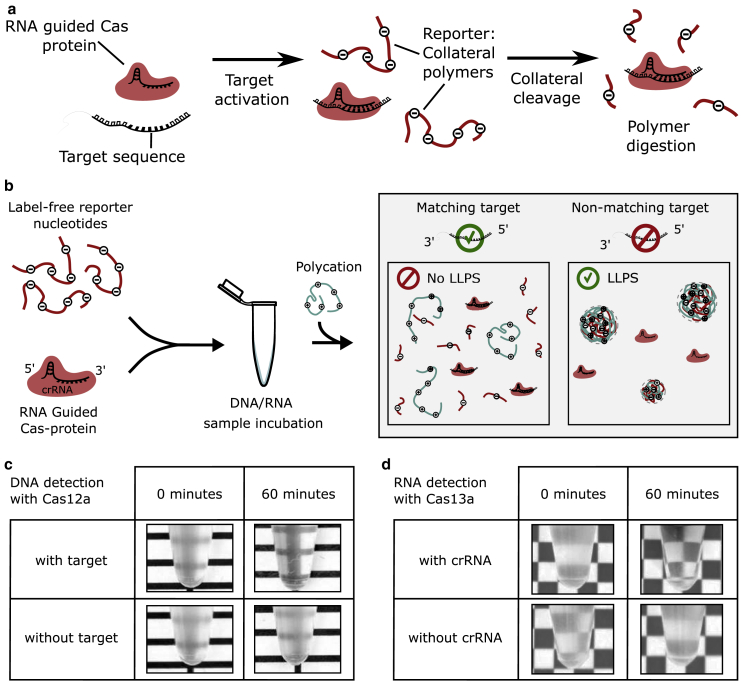
Figure 2.
Length-dependent LLPS for DNA and RNA detection. (a) Target-activated, RNA-guided Cas proteins digest reporter nucleic acid polymers such as poly(dT) and poly(U) through unspecific (collateral) cleavage activity. (b) A DNA/RNA detection assay using LLPS can be conducted as follows: RNA-guided endonuclease and collateral polymers are incubated with a sample. After incubation, the mixture is complemented with a suitable polycation to induce phase separation. In the presence of a matching target, the mixture will be transparent because collateral polymers were degraded during incubation; therefore, LLPS will not occur. In the absence of target, the mixture turns turbid due to LLPS of collateral polymer and the polycation. (c) Cas12a activity could be observed from the absence of turbidity. The enzyme was incubated with dT60, both with and without a target sequence. Phase separation was induced with poly-L-lysine. (d) Cas13a activity could be inferred by inducing LLPS of poly(U) and spermine. Cas13a was incubated with target, poly(U), and with or without crRNA (upper and lower row, respectively) for 0 or 60 min (left and right column, respectively). To see this figure in color, go online.
Poly(dT) cleavage gel
To assess the collateral activity of Cas12a on dT60 ssDNA with gel electrophoresis, we performed the following experiment in triplicate. Cas12a was loaded with crRNA by mixing ∼8.6 μM Cas12a and 20 μM crRNA in 1× NEB 2.1 buffer and incubating for 15 min at 37°C. Additionally, five tubes were prepared, each containing 40 μM dT60, of which 5 μM were labeled with Cy5 on both the 5′ and 3′ end, and 1 μM target DNA in 1× NEB 2.1. Loaded Cas12a was added to a final concentration of 1.2 μM, and the reaction was left at 37°C. After 1, 15, 30, 45, and 60 min, part of the reaction mix was extracted, loading dye was added, and the reaction was stopped by flash freezing in liquid nitrogen. The samples were stored at −80°C and quickly thawed by hand before running on a 1% agarose gel at 100 V, 90 min and 4°C (Fig. 3 a). The control sample was also left for 60 min at 37°C and lacked Cas12a. Gels were imaged using Amersham Typhoon (Cytiva, Marlborough, MA).
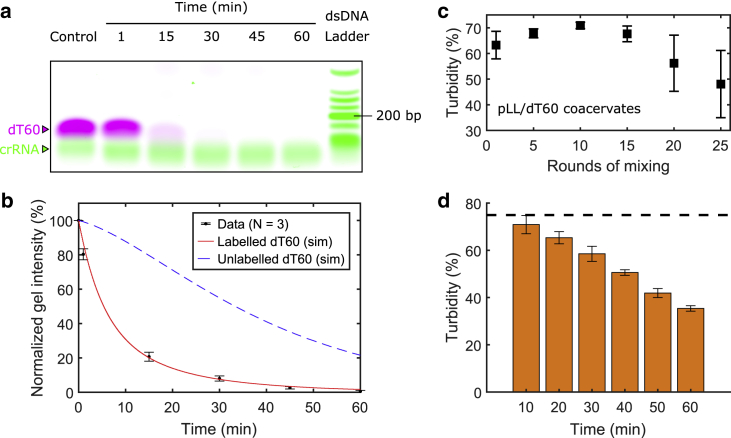
Figure 3.
Assay dynamics and stability. (a) Cas12a rapidly cleaves Cy5-labeled poly(dT). The control shows dT60 incubated under the same reaction conditions but lacking Cas12a (see Materials and methods). (b) Quantification of gel intensities shown in (a) is given. The normalized gel intensity is a measure for the relative amount of uncleaved poly(dT) (control 100%). The data are compared with simulation results for labeled dT60 (solid line) and are used to project the degradation of the unlabeled dT60 (dashed line). (c) Turbidity is influenced by the degree of mixing with a pipette. (d) Turbidity decreases linearly with incubation time during the first hour (1 μM target) in the Cas12a detection assay. The dashed black line indicates the turbidity of the control with no target after 1 h of incubation at 37°C. Error bars in (b)–(d) indicate SD of the mean from three independent repetitions. To see this figure in color, go online.
Stochastic simulation
We performed stochastic simulations of the poly(dT) cleavage reaction for ensembles of polymers using Gillespie’s algorithm (30). The cleavage reaction was implemented as random scission (31), which assures that every bond in the simulation has equal probability to be removed. To compare simulation data with experiments (Fig. 3 b), we monitored the polymer mass distribution over time. We mimicked the situation in the experiment with labeled dT60 by retaining only one of the two created species after each cleavage and found that this modification allows us to match the experimental data (see Document S1. Supporting materials and methods, Figs. S1–S5, and Table S1, Document S2. Article plus supporting material a). An effective cleavage rate of 8 × 10−5 bonds per second matched the data best. In the analysis of the simulation, a cut-off length was applied, below which the labeled poly(dT) polymer disappears from the gel. = 10 yielded the best agreement between the data and the simulation (see Fig. S5 b). The curves in Fig. 3 b show the polymer mass in the system with N ≥ for the situation of labeled and unlabeled poly(dT). Both simulations contained 106 dT60 oligomers as an initial condition.
Dynamics of Cas12a turbidity assay
Cas12a was loaded by mixing ∼8.6 μM Cas12a with 20 μM crRNA in 1× NEB 2.1 buffer and incubating for 15 min at 37°C. To determine how dT60 is degraded over time by activated Cas12a, we prepared a mixture containing ∼1.2 μM loaded Cas12a, 1 mM DTT, and 40 μM dT60 in 1× NEB 2.1 buffer with 1 μM single-stranded DNA target (Table S1). Every 10 min, 10 μL was taken from the incubating solution (37°C) for turbidity measurements. Subsequently, 1 μL pLL was added to a final concentration of 0.9 mg/mL. The sample was mixed by pipetting up and down eight times with a 10 μL pipette. Immediately after mixing, the absorbance was measured in a quartz cuvette (Hellma Analytics 105.211-QS with 5 μL volume and 1 mm pathlength; Müllheim, Germany) in a spectrophotometer (DeNovix DS-11+; Wilmington, DE) at 500 nm wavelength. The turbidity was calculated from the absorbance (A) using the formula (1 − 10−A) × 100%. The control was performed similarly, with a short DNA strand (1 μM) complementary to the DNA target sequence (Table S1). The absorbance was measured after 1 h incubation at 37°C.
Influence of mixing on solution turbidity
To quantify the influence of mixing on the polymer solutions, we mimicked Cas12a assay conditions by preparing a mixture containing 40 μM dT60 and 1 mM DTT in 1× NEB 2.1 buffer. From this mixture, we took samples of 10 μL, to which we added 1 μL of pLL to a final concentration of 0.9 mg/mL. The solutions were then mixed by pipetting the full volume up and down for various number of times with a 10 μL pipette tip (Fig. 3 c) before absorbance was measured as described above.
Assay optimization
Lba Cas12a (NEB) was loaded and activated by mixing ∼8.6 μM Cas12a and 10 μM Cas12a crRNALba (see Table S1) in 1× NEB 2.1 buffer and incubating for 15 min at 37°C. The Cas12a optimization assay (Fig. 5) contained 187 nM of the activated Cas12a, 220 nM ssDNA target, and a variable dT60 concentration in 1.1× NEB 2.1 buffer and was incubated for 1 h at 37°C. pLL was directly added to the tubes to a final concentration of 0.5 mg/mL after the incubation. The sample was mixed by pipetting up and down 10 times with a 10 μL pipette. Immediately after mixing, the absorbance was measured in a NanoDrop ND-1000 spectrophotometer (with 2 μL volume at a pathlength of 0.1 mm, normalized to 1 mm and blanked with 1.1× NEB 2.1 buffer; Thermo Fisher Scientific, Waltham, MA) at 500 nm wavelength by technical triplicates, for three biological replicates. The turbidity was calculated as (1 − 10−A) × 100%. The control was prepared similarly but omitted the addition of the crRNA.
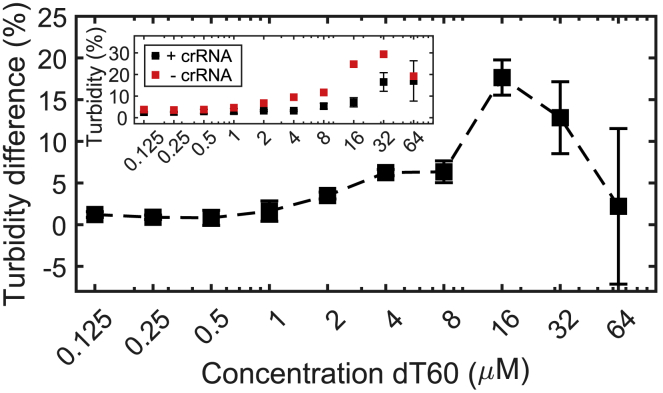
Figure 5.
Optimal dT60 concentration. To identify optimal assay conditions, we performed a dT60 titration in the Cas12a detection assay (see Materials and methods). The optimal dT60 concentration was found to be 16 μM. At that concentration, the turbidity difference between the assay and the negative control peaked at around 20%. The inset shows the raw turbidity values for the assay (+crRNA) and the negative control (−crRNA), which result in the turbidity difference of the main panel (dashed line). Error bars indicate SD of the mean from three independent repetitions of the assay. To see this figure in color, go online.
Buffer conditions and chemicals
Cas13a reactions, as well as RNA and spermine experiments, were carried out in C13RB buffer (40 mM Tris-HCl, 60 mM NaCl, 6 mM MgCl2 (pH 7.3)). Cas12a reactions and poly(dT) and pLL experiments were carried out in C12RB buffer (20 mM Tris-HCl, 66.6 mM KCl, 5 mM MgCl2, 1 mM DTT, 5% (v/v) glycerol, 50 μg/mL heparin (pH 7.5)) or 1× NEB 2.1 buffer as indicated. Poly(U) (polyuridylic acid potassium salt), spermine (spermine tetrahydrochloride), pLL (poly-L-lysine hydrobromide, 15–30 kDa), Trizma hydrochloride, sodium chloride, and magnesium dichloride were bought from Sigma-Aldrich (St. Louis, MO). Poly(dT) dT20, dT40, dT60, dT80, and Cy5-dT60-Cy5 were bought from Ella Biotech (Planegg, Germany) (Figs. 1 b and 2 c) and IDT (Coralville, IA) (dT10, dT20, dT30, dT40, dT50, dT60, dT70, and dT80) for Figs. 1, c and d, 4, and 5. RNase Alert, RNA Gel loading dye, and nuclease-free water were bought from Thermo Fisher Scientific. DNase Alert was bought from IDT. RPA kit was bought from TwistDx (Cambridge, UK).
Figure 4.
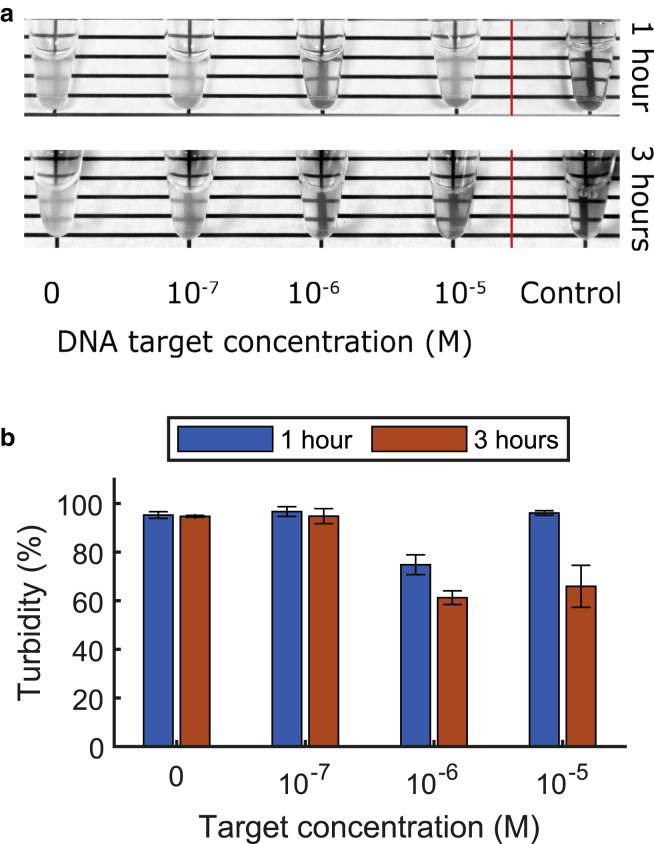
Detection limit of the DNA turbidity assay. (a) The ssDNA detection limit using Cas12a with the phase separation-based turbidity assay lies in the micromolar range. The tubes contained ∼0.86 μM activated Cas12a and 22 μM dT60 in 1× NEB 2.1 buffer and target concentrations as indicated. After 1 (top) or 3 h (bottom) of incubation at 37°C, pLL was added to a final concentration of 0.5 mg/mL. Note that these reaction conditions differ from those shown in the gel of Fig. 3a. (b) Turbidity measurements of reactions described in (a) are given. Error bars indicate SD of the mean from three independent repetitions. To see this figure in color, go online.
Theory
Model definition
Following Flory and Huggins (28,29), a polymer solution can be described using a lattice model. Each lattice site corresponds to the molecular volume of the solvent v and is occupied by one of the solution’s components (21,25). We denote the molecular volume of each component i with Vi, and the effective chain length Ni = Vi/v (for the solvent, Ns is set to 1). We further introduce the charge density σi = Zi/Ni of the polymers with valency Zi. The Flory-Huggins interaction parameter between components i and j is denoted χij. In addition, the parameter α sets the strength of electrostatic interactions (α ≈ 3.7 for aqueous solutions at room temperature (32)). Finally, ϕi denotes the volume fraction of component i. The free energy per unit volume f (in units of kBT) of a charged polymer solution with n components (including the solvent) can be written as
| (1) |
The first term of the free energy represents the entropy of mixing of the solution, which contains an explicit dependence on the effective chain length. Because of its inversely proportional relation with polymer length, this term rapidly loses significance for long polymers. The second term represents the Flory-Huggins nonionic interactions, and the third term represents the electrostatic interactions as in the Voorn-Overbeek model (24). In our system, we have three components, namely solvent molecules (index s) and two oppositely charged polymer species (indexes 1 and 2) that refer to the negatively charged polynucleotides and the positively charged poly-L-lysine.
To demonstrate how polymer length influences phase-separation behavior, we make simplifying assumptions that do not restrict the generality of our observations. First, we consider a symmetric solution, in which the oppositely charged polymers have equal length N1 = N2 = N and equal charge density σ1 = σ2 = σ and occupy equal volume fractions ϕ1 = ϕ2 = . Second, we consider the case in which the length of one polymer species, N1, is varied and the length of the second species, N2, is held constant while keeping the mixture symmetric otherwise. For both cases, we further assume that the only relevant Flory-Huggins interactions are the ones between the polymers and the solvent, so that χ1s = χ2s = χ, whereas χ12 = 0. For the first case, the free energy can be written as
| (2) |
The first two terms represent the entropy of mixing, and the last two terms represent energetic contributions due to polymer-solvent and electrostatic interactions between the two polymer species. For the second case, the length-dependent contribution to the free energy can be written as (see Supporting materials and methods for details). It should be noted here that polymer cleavage does not change the volume fraction of polymers in the system. In the case that N1 → N2, we recover the free energy of Eq. 2. For values of N2 comparable to N1, the overall qualitative behavior of the system is the same as that of the fully symmetric case, but there are quantitative differences in the binodal and spinodal curves. Below, we derive and plot the phase diagram of the symmetric case; for the case in which we vary N1, the phase diagram is plotted in Fig. S4.
Derivation of the phase diagram
The thermodynamic properties of the polymer solution are determined by the free energy in Eq. 2. The solution will exhibit a uniformly mixed phase if f(ϕ) is a convex function. For any concave regions of f(ϕ), there are two phases: a polymer-depleted phase and a polymer-enriched phase. The shape of the free energy function depends on the polymer length; f(ϕ) is convex below a critical value of Nc but becomes concave for longer polymers at a fixed value for χ. In Fig. S2, the free energy function and the chemical potential (μ = ∂f/∂ϕ) are shown for different effective chain lengths larger than, equal to, and lower than a critical length. It can be seen how the curvature of f(ϕ) changes with polymer length and thus that polymer length critically affects phase behavior.
To understand the phase behavior and stability of a polynucleotide/polycation coacervate system, it is necessary to derive the phase diagram for a specific set of parameters (see Fig. 1 a). We set χ = 0.5 and σ = 0.22 to investigate the length dependence of the model. The smallest polymer length Nc for which the solution can exhibit phase separation is called the critical point (ϕc, Nc) of the system. The critical point can be computed from the condition that both the second and the third derivative of the free energy vanish,
| (3) |
More generally, the condition that the second derivative of the free energy vanishes defines the spinodal curve (black line in Fig. 1 a),
| (4) |
Above the spinodal curve, the mixed solution is unstable and always phase separates, yielding stable coexistence of a polymer-rich liquid phase and a polymer-poor liquid phase. Whether liquid-liquid phase separation occurs in the region below the spinodal depends on the binodal curve. This curve lies below the spinodal curve and separates a region of unstable phase coexistence and a uniformly mixed solution. The binodal curve (gray line in Fig. 1 a) is given by the condition that the chemical potential and osmotic pressure of each component are equal in both phases. To find the binodal, we (numerically) solve the simultaneous equations,
| (5) |
and
| (6) |
for N ≥ Nc. In the region between the binodal and spinodal curves, two liquid phases can coexist, but the solution needs to overcome an energy barrier to demix. The theoretical model as described above, as well as the methods we used to obtain the phase diagram, are well known (24). However, only a few studies have explicitly addressed the dependence of LLPS on polymer length.
Results and Discussion
Nucleic acid polymer length determines the onset of LLPS
An important feature of LLPS in polymer solutions is that phase separation depends on the (effective) polymer length (33,34). Liquid-liquid phase separation occurs when the energetic penalty for mixing the two liquids is larger than the entropic gain of that mixing. Whether polymer solutions undergo liquid-liquid phase separation, i.e., spontaneously demix or not, depends both on the polymer concentration and polymer length (33, 34, 35). This means that there must be a critical polymer length above which demixing occurs at a given polymer concentration, whereas below a critical polymer length, the solution components are uniformly mixed. In the absence of an electrostatic term, the Flory-Huggins theory predicts LLPS to occur above a critical interaction strength χc = 2 at an effective chain length of N = 1 (see (21)). For polymers (i.e., N > 1), the critical interaction strength χc decreases. Therefore, phase separation can occur more readily for longer polymers. Because electrostatic interactions are expected to be significant in the case of polynucleotide LLPS, we also included electrostatic interactions into the model by following Voorn-Overbeek (24,28,29).
To illustrate the stark effect of the Voorn-Overbeek term, let us consider an example with fixed interaction strength χ = 0.5. In this case, the solution remains homogeneous up to an effective polymer length of N ≈ 104 in the absence of electrostatic interactions. However, including the electrostatic interactions decreases the critical polymer length by three orders of magnitude to Nc ≈ 12 (Fig. S2). The predicted phase diagram for charged polymer solutions is shown as a function of polymer volume fraction and polymer length in Fig. 1 a. In summary, the theory of charged polymer solutions predicts length-dependent phase behavior. The Voorn-Overbeek term in the free energy contributes significantly to the interactions necessary for phase separation to occur.
Some previous work has shown that LLPS of oppositely charged polymers depends on polymer length. For example, Spruijt et al. experimentally validated the Voorn-Overbeek model in mixtures of poly(acrylic acid) (PAA) and poly(N,N-dimethylaminoethyl methacrylate) (PDMAEMA) with different chain lengths (33). Furthermore, we recently showed that the elongation of short RNA oligomers with a template-independent RNA polymerase (polynucleotide phosphorylase) causes the onset of LLPS, and that this can be reversed by enzymatic RNA degradation (27). Finally, Saleh et al. have shown that phase-separated droplets consisting of DNA nanostars can be degraded using a DNA restriction enzyme (36). However, a direct experimental test of length-dependent LLPS in mixtures of nucleic acids and polycations is lacking.
We studied solutions of various lengths of negatively charged poly(dT) and the positively charged peptide pLL. We found that turbidity caused by phase separation of poly(dT) of 40 nucleotides (dT40) and longer was visible by the naked eye, but not for dT20 (Fig. 1 b). To quantify the extent of phase separation in poly(dT) and pLL mixtures, we performed turbidity measurements of a dT60 titration ranging from 0.125 to 32 μM. In the absence of pLL, the mixtures remain fully transparent, but in the presence of a constant amount of pLL, the mixtures turn turbid in a concentration-dependent manner (Fig. 1 c). Turbidity reached 50% at a concentration slightly above 2 μM dT60. To see how nucleotide length influence LLPS in the mixtures, it is necessary to keep the volume fraction of dT constant while changing poly(dT) length. At constant volume fraction, the solution turbidity increases with increasing poly(dT) lengths from 10 to around 40 nucleotides and then saturates at turbidity values around 70% for poly(dT) lengths up to 80 nucleotides (Fig. 1 d). These measurements are consistent with our previous observation in Fig. 1 c, in which 70% turbidity is reached at 4 μM dT60.
In summary, our experimental results for poly(dT)/pLL mixtures confirm the length dependence of LLPS of oppositely charged polymers (coacervation) in agreement with the prediction of the theoretical model and earlier work (33). We would like to point out that the agreement between the experiments and the model should be considered of a qualitative rather than quantitative nature; the assumption of symmetric solutions may not hold for mixtures of poly(dT)/pLL. Poly(dT) and pLL are different in charge density and other properties that may influence the interaction strengths between the two polymer species. Interestingly, we find that length dependence is a generic feature of the model that also holds in the case of asymmetric length dependence of solutions (see Theory and Supporting materials and methods). By establishing that LLPS of poly(dT)/pLL depends on poly(dT) length, we were able to infer that shortening of poly(dT) through Cas12a endonuclease activity would allow visual and turbidity readout for Cas12a target detection.
Inferring Cas12a and Cas13a collateral activity from solution turbidity
Next, we tested the possibility to employ LLPS as a readout mechanism for CRISPR-based nucleotide detection (schematically shown in Fig. 2, a and b). We mixed crRNA-guided Cas12a, with and without complementary single-stranded DNA target, and a 60-mer of poly(dT) (dT60). When the positively charged polyelectrolyte poly-L-lysine was added immediately after mixing, both solutions turned turbid, indicating the presence of poly(dT) through visible LLPS (Fig. 2 c, left column). However, after 60 min incubation, the solution containing the target remained transparent when pLL was added, whereas the solution containing no target turned turbid (Fig. 2 c, right column). Thus, solution transparency of the sample with target and 60 min incubation indicates the successful activation of the Cas12a and its degradation of the poly(dT) through collateral cleavage activity. This observation is quantified in the following sections using turbidity measurements.
The above approach is also feasible for specific RNA detection. We identified a system composed of poly(U)-RNA and the tetravalent polycation spermine (26) as a suitable combination for LLPS of RNA. We incubated Cas13a, the RNA target, and poly(U), with and without crRNA. When spermine was added immediately after mixing the components, both solutions turned turbid (Fig. 2 d, left column). However, when spermine was added after 60 min, the sample that was lacking crRNA underwent LLPS and turned turbid, whereas the sample that contained crRNA remained clear (Fig. 2 d, right column), indicating activation of collateral RNase activity by the Cas13a protein. This shows that LLPS-based detection is also compatible with RNA detection through Cas13a.
Our observations show that the biophysical mechanism of nucleotide-length-dependent LLPS allows for label-free DNA and RNA sequence detection. Initially, the nucleic acid polymer length exceeds the critical length for phase separation, but as degradation through Cas proteins progresses and the nucleic acid polymer lengths fall below a critical threshold, the solution loses its ability to phase separate. Thus, the absence or reduction of phase separation (as seen from solution turbidity) indicates target detection due to activated Cas12a and Cas13a proteins.
Dynamics of nucleotide degradation and solution stability
Having established that LLPS of poly(dT) and poly(U) with polycations can be exploited for CRISPR-based nucleic acid detection, we sought to quantify the dynamics of the degradation process. In diagnostic applications with CRISPR-Cas proteins and fluorescent reporter molecules, the fluorescent signal appears within minutes after the protein has been activated (Fig. S1). Quantification of the rate at which active Cas12a cleaves dT60 is more challenging, because poly(dT) cleavage cannot be observed directly. To quantify dT60 degradation we used a dT60 oligomer labeled with fluorophores (Cy5) on both the 3′ and 5′ end. Using gel electrophoresis, we observed a 20% reduction in fluorescence within 1 min after the addition of activated Cas12a and a drop to around 1% of initial fluorescence over the course of 1 h (Fig. 3, a and b). Note that because Cy5 is positively charged, the mobility of labeled dT60 is different from unlabeled dT60. We implemented a stochastic simulation of the degradation process to quantify the experiment and recovered the exponential shape of the fluorescence decay (solid line, Fig. 3 b). We found that only two parameters were necessary to accurately simulate the poly(dT) degradation, namely the molecular cleavage rate and a cut-off length below which the fluorescence of Cy5-poly(dT) oligomers disappears.
Because the experiment shows only the fluorescently labeled ends of dT60, we next used the simulation to project the overall poly(dT) mass (dashed line, Fig. 3 b). Notably, this quantity decreases much slower than the fluorescence signal, indicating that nucleotide detection assays using LLPS may be slow compared with experiments using fluorescent reporter molecules. The limited detection speed was indeed observed in experiments. Turbidity decreased linearly over the period of 1 h from around 70 to 30% (Fig. 3 d). The result is in stark contrast to the rapid cleavage of (Cy5-labeled) poly(dT) 60-mer substrates (Fig. 3, a and b) and fluorescence detection (Fig. S1 b). This observation can be explained by the fact that the ensemble of poly(dT) polymers needs to be degraded to resolve LLPS in the system. The change in turbidity is therefore slow compared with the degradation of individual poly(dT) polymers or fluorescence reporters. The experiments further indicate that the degree of phase separation depends on the specific composition of the ensemble of polymers in the system. Taken together, we find that the necessary number of cleavages is the limiting factor to resolve phase separation in the assay. One way to optimize the assay is therefore to minimize the number of necessary cleavages to resolve LLPS.
We also investigated the reproducibility of the mixing step, because we noted that the way the reaction mixtures and pLL are mixed influences the turbidity of solution in a more unexpected way. We find a maximal turbidity when mixing the solutions between 5 and 10 times by pipette aspiration (Fig. 3 c). While too few rounds of mixing are not sufficient to homogenize the two liquids, too many rounds of mixing lead to a significant drop in turbidity. For the nucleotide detection assay, this means that detection accuracy is influenced by how the liquids are brought together. There may be multiple reasons for this observation. First, it could be that polyelectrolytes stick to the surfaces of the pipette tip or the container, which would affect the polyelectrolyte concentration, which in turn affects LLPS. Second, it could also be that the stability of the phase-separated solution is influenced by the mechanical impact of pipetting. For the experiments in Figs. 3 and 4, solutions were consistently mixed by aspirating the liquid eight times.
A drawback of the LLPS-based nucleotide detection assay is that polycations are added after the sample has been incubated with CRISPR-Cas proteins (cf. Fig. 2 b). We explored the possibility of combining the nucleotide detection reaction and the triggering of LLPS. However, the presence of pLL as well as spermine interfered with the CRISPR-Cas proteins’ endonuclease activity (Document S1. Supporting materials and methods, Figs. S1–S5, and Table S1, Document S2. Article plus supporting material c). Although we clearly observed that spermine inhibited Cas13a, we could not find out whether this inhibition occurs at a molecular level whereby nuclease activity is molecularly blocked or whether the chemical composition of the buffer is affected in a way that reduces protein activity. One may speculate that metabolites such as spermine serve to inhibit collateral cleavage in eukaryotic cells. This view is supported by the observation that Cas13a collateral cleavage does not occur in vivo in eukaryotic cells, because activated Cas13a neither compromised cell growth, nor did it affect RNA size distributions (37).
Detection limit
To assess the detection limit of the method, we investigated the minimal amount of target that can be detected for both DNA and RNA detection. We obtained a visible readout at submicromolar concentrations for ssDNA target detection with Cas12a (Fig. 4 a), and at nanomolar concentrations for RNA target detection with Cas13a (Fig. S3). In experiments in which the incubation time was increased from 1 to 3 h, we observe a significant further reduction in turbidity (Fig. 4 b), which is in line with our conclusions from the previous sections. More cleavage steps reduce the tendency toward phase separation. In the case of the 10 μM target, turbidity barely decreased after 1 h, showing that at high target concentrations, the target also contributes to phase separation.
The detection limits we observed are similar to previous reports in the literature in the absence of amplification steps (4). To enhance the detection limit, the target sample can be amplified using standard methods such as PCR, RPA, or T7 RNA polymerase, which increases the sensitivity by orders of magnitude (3, 4, 5). By using appropriate amplification reactions, attomolar sensitivities have been shown to be possible. We expect that our method can be adapted for such high detection sensitivities when needed by using T7 or RPA amplification. However, given the complexity of these additional biochemical reactions, the experimental condition may need to be adapted to keep the phase-separating property of the solutions.
Optimal detection condition
To optimize the nucleotide detection assay, we performed additional experiments at different concentrations of dT60 and with a commercially available Lba Cas12a (NEB). At high dT60 concentrations, the Cas12a cleavage activity is not sufficient to cleave the necessary amounts of dT60 to inhibit phase separation. At low concentrations of dT60, phase separation does not occur because of insufficient amounts of dT60. Therefore, there is an optimal concentration of dT60 at which the difference in turbidity between the assay and control is maximized. To find the optimal dT60 concentration, we measured the turbidity difference between a negative control (−crRNA) and the assay (+crRNA). As shown in Fig. 5, the maximal turbidity difference between assay and negative control occurs at a concentration of 16 μM dT60. We note, however, that this optimum depends on buffer conditions as well as on polycation concentration and should be assessed before the assay.
Theoretical predictions for future developments
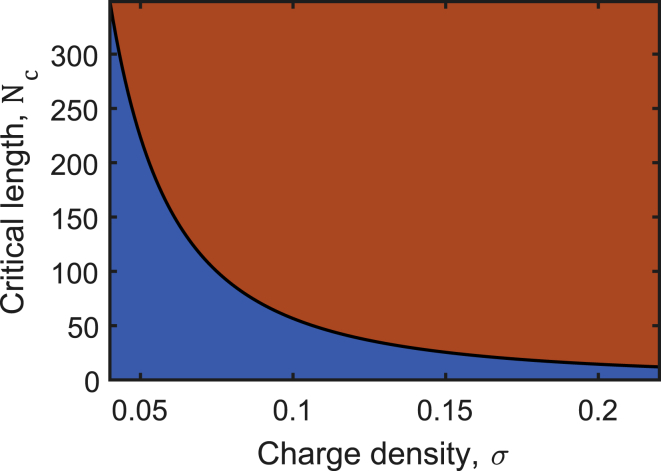
Finally, we investigated how the assay could be improved from a chemical point of view, based on the understanding we gained so far. The performed experiments have shown that one of the limitations of the assay in its current form is that a large number of cleavage reactions is necessary to inhibit phase separation. A solution to this problem would be to find conditions in which the critical polymer size Nc is larger than in the poly(dT)/pLL mixtures we studied. In such a system, fewer cleavage reactions could be sufficient to reduce the average polymer size below the threshold of phase separation more quickly and maybe more pronouncedly. To this end, we investigated the properties of the critical point as derived in the Theory section above. The charge density σ is the chemical property of the polymers that mainly determines their electrostatic interactions. Therefore, we chose to parametrically evaluate the critical point as a function of the charge density, Nc = Nc(σ). Fig. 6 shows that at lower charge densities, the critical polymer length Nc increases. For a phase-separating system of polyelectrolytes, this observation means that reducing the polymer charge density at constant polymer length leads to the loss of the phase-separating property. Similarly, a non-phase-separating solution of polyelectrolytes at a given charge density can phase-separate by simply elongating the polymers beyond the critical length Nc.
Figure 6.
Polymer length and charge density delimit the regime of phase separation. The solid line shows the critical polymer length Nc at the critical volume fraction ϕc as a function of the polymer charge density σ. The lighter gray region (orange online) indicates the regime in which the system can undergo phase separation. For a given charge density, the system phase separates for polymers of length N > Nc. For a given polymer length N, the system phase separates if the charge density lies above a critical value σ > σc, i.e., to the right of the solid line. LLPS-based detection assays can be optimized by utilizing the shown relationship. Specifically, longer polymers with lower charge density may perform better in the assay. To see this figure in color, go online.
Conclusions
There are a variety of (bio)chemical mechanisms that influence the phase separation of nucleotides under specific conditions (21). Nucleotide length has been recognized as one of the factors driving phase separation in polymer solutions (33), as well as in the cellular context (20). For example, length-dependent LLPS has been described for the cyclic GMP-AMP synthase, which forms liquid condensates with double-stranded DNA depending on DNA length (38).
Here, we have exploited the length dependence of LLPS for designing a visual readout for in vitro detection of specific oligonucleotides. The mechanism of the method is supported by a mathematical model that demonstrates theoretically that LLPS of interacting polymers depends on polymer length. The model also allowed us to predict that a reduced polymer charge density will improve the assay performance. Given that the presented theoretical model is a simplification of the experimental situation, it will be interesting to study more quantitative versions of the model in the future. Consider, for example, two different polymer species or the effect of salts. Experiments with (single-stranded) poly(dT) of various length and poly-L-lysine confirmed the length-dependent transition between phase-separated and homogeneously mixed polymer solutions. We have used the length dependence of LLPS to describe a method for the label-free detection of short nucleotide sequences through transparency/turbidity of sample solutions by using CRISPR-Cas nucleases Cas12a and Cas13a. To detect specific DNA/RNA sequences, samples were incubated with Cas12a/Cas13a and collateral polymer (poly(dT)/poly(U)) and then complemented with polycations to either trigger LLPS or not. If a preprogrammed target is detected, the collateral polymer is digested, LLPS does not occur, and the solution remains transparent. If no target is recognized, the solution turns turbid through interactions between nucleic acid polymers and the added polycations, leading to LLPS and increased solution turbidity. The assay can be run within 1 h with a detection limit for single-stranded DNA and RNA targets in the micromolar and nanomolar range, respectively. Lastly, we found that the poly(dT) concentration is a critical factor. At a dT60 concentration of 16 μM, the strongest signal could be recorded relative to the negative control.
Existing nucleotide detection applications with Cas12a and Cas13a use chemically labeled nucleotides and fluorescence detection equipment or implementation on a paper strip. There are also a growing number of applications in which nanoparticles are used in conjunction with Cas12a or Cas13a detection. For example, Cas12a cleavage can be utilized to trigger a single-stranded DNA polymerization reaction, which subsequently allows the formation of bright copper nanoparticles (39). Furthermore, DNA-functionalized gold nanoparticles can be used in various ways to generate bright signals after the target activation of Cas12a and its indiscriminate cleavage activity. This can be achieved through metal-enhanced fluorescence (40) or by inhibiting the assembly of DNA-functionalized plasmonic nanoparticles (41). The abovementioned assays outperform our method in terms of lower detection limits, which are 10 pM (−PCR), femtomolar (−PCR), and 10 aM (+PCR), respectively. It is possible to improve the detection limit of our assay by including amplification steps. The readouts of nanoparticle-based detection methods are as simple as turbidity. They require either a ultraviolet light source (39) or the measurement of color changes (40,41). The production of nanoparticles, however, is arguably more complicated.
Length-dependent LLPS allows for a nucleotide detection assay without chemically labeled nucleotides and with limited equipment requirements. Because of the lack of chemical labels or metals, the assay is more environmentally friendly and cheaper than other methods that use CRISPR-Cas proteins. Key ingredients of the assay are polyelectrolytes, which are either abundant in nature or can be synthesized enzymatically. Polycations such as the protein protamine or metabolites like spermine are ubiquitous in certain cell types, from which they can be recovered sustainably. Furthermore, poly(dT) and poly(U) can be synthesized with the enzymes TdT (42) and PNPase (43), respectively, thereby avoiding the environmental impact of traditional chemical DNA and RNA synthesis.
Because transparency and turbidity of solution are the readout of our assay, it is possible to assess the test outcome with the unaided eye or with a simple device that measures optical solution properties. One limitation of the presented technique is that phase separation is highly dependent on solution composition. Therefore, a readout will only be conclusive if there is a clearly visible difference between the sample and a negative control. Nevertheless, we anticipate that LLPS-based nucleotide detection assays will be useful in various high-throughput or low-cost point-of-care diagnostic applications (44), especially when the availability and operability of high-end laboratory equipment is limited.
More generally, we speculate that a better understanding of length-dependent LLPS may provide insights into specific cellular mechanisms of LLPS (20,38). One example is RNA repeat disorders, in which a critical number of RNA repeats determines the onset of phase separation through multivalent interactions (34). Similarly, multivalent protein-protein interactions (45), as well as amino-acid sequences (46,47), are determinants of LLPS in vivo and in vitro. It appears that the mechanisms of phase separation based on polymer length and multivalent interactions have in common that longer polymer size and additional multivalent interactions both lead to an increased tendency for phase separation. Establishing a unified understanding of these observations remains a task for future research (48).
Author contributions
L.R., W.K.S., and S.J.J.B. conceived the study. W.K.S. and J.M.J. performed experiments. R.G.-L. performed the optimization experiment. L.R., F.L.N., and A.C.H. contributed to initial experiments. A.C.H. purified Cas12a. W.K.S., L.R., and T.I., developed the theoretical model. L.R. performed stochastic simulations. W.K.S., J. M. J., R.G.-L., F.L.N., M.D., D.H.M., T.I., S.J.J.B., and L.R. analyzed and discussed the data. L.R., W.K.S., and J.M.J. wrote the manuscript with input from all authors.
Acknowledgments
We thank members of the iGEM team 2017 from Delft University of Technology for their contributions: Aafke van Aalst, Kimberley Barentsen, Kelly Hamers, Floor de Jong, Fiona Murphy (also for carefully checking the manuscript), Guillermo Serena Ruiz, Gabriella Tany, Isabell Trinh, Jasper Veerman, Amária Vledder, and Hielke Walinga and team supervisors Aljoscha Wahl, Cristóbal Almendros Romero, Anthony Birnie, Hirad Daneshpour, David Foschepoth, Patrick de Jonge, Sebastian Kieper, Benjamin Lehner, Rebecca McKenzie, Jochem Vink, and Esengül Yildirim.
The plasmids encoding Cas12a and Cas13a were a kind gift from the Feng Zhang lab (Broad Institute). This work was supported by The Netherlands Organization for Scientific Research (NWO/OCW) as part of the Frontiers of Nanoscience program and the Fonds Ernest Solvay managed by the Koning Boudewijnstichting. F.L.N. was supported by an NWO Veni Grant 016.Veni.181.092 and by the Academic Health Sciences Centre with the AHSCC-19Engage prize, S.J.J.B. was supported by the NWO VICI (VI.C.182.027), and L.R. was supported by FOM programme nr. 110 (NWO). The Delft University of Technology has filed a patent application (no. WO2019078722A1) pertaining to this manuscript, on which L.R., W.K.S., and S.J.J.B. are co-inventors.
Editor: Jason Kahn.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.02.013.
Supporting material
References
- 1.Wright A.V., Nuñez J.K., Doudna J.A. Biology and applications of CRISPR systems: harnessing nature’s toolbox for genome engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 2.East-Seletsky A., O’Connell M.R., Doudna J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538:270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gootenberg J.S., Abudayyeh O.O., Zhang F. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356:438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gootenberg J.S., Abudayyeh O.O., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360:439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J.S., Ma E., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360:436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li L., Li S., Wang J. HOLMESv2: a CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth. Biol. 2019;8:2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- 7.Zetsche B., Gootenberg J.S., Zhang F. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrington L.B., Burstein D., Doudna J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362:839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shmakov S., Abudayyeh O.O., Koonin E.V. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li S.-Y., Cheng Q.-X., Wang J. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 2018;28:491–493. doi: 10.1038/s41422-018-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abudayyeh O.O., Gootenberg J.S., Zhang F. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu L., Li X., Wang Y. The molecular architecture for RNA-guided RNA cleavage by Cas13a. Cell. 2017;170:714–726.e10. doi: 10.1016/j.cell.2017.06.050. [DOI] [PubMed] [Google Scholar]
- 13.Jeon Y., Choi Y.H., Bae S. Direct observation of DNA target searching and cleavage by CRISPR-Cas12a. Nat. Commun. 2018;9:2777. doi: 10.1038/s41467-018-05245-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reithmann E., Reese L., Frey E. Quantifying protein diffusion and capture on filaments. Biophys. J. 2015;108:787–790. doi: 10.1016/j.bpj.2014.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swarts D.C., van der Oost J., Jinek M. Structural basis for guide RNA processing and seed-dependent DNA targeting by CRISPR-Cas12a. Mol. Cell. 2017;66:221–233.e4. doi: 10.1016/j.molcel.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swarts D.C., Jinek M. Mechanistic insights into the cis- and trans-acting DNase activities of Cas12a. Mol. Cell. 2019;73:589–600.e4. doi: 10.1016/j.molcel.2018.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broughton J.P., Deng X., Chiu C.Y. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020;38:870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joung J., Ladha A., Zhang F. Point-of-care testing for COVID-19 using SHERLOCK diagnostics. medRxiv. 2020 doi: 10.1101/2020.05.04.20091231. [DOI] [Google Scholar]
- 19.Kellner M.J., Koob J.G., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14:2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banani S.F., Lee H.O., Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brangwynne C.P., Tompa P., Pappu R.V. Polymer physics of intracellular phase transitions. Nat. Phys. 2015;11:899–904. [Google Scholar]
- 22.Dignon G.L., Best R.B., Mittal J. Biomolecular phase separation: from molecular driving forces to macroscopic properties. Annu. Rev. Phys. Chem. 2020;71:53–75. doi: 10.1146/annurev-physchem-071819-113553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bungenberg de Jong H.G., Kruyt H.R. Coacervation (partial miscibility in colloid systems) Proc. R. Acad. Amsterdam. 1929;32:849–856. [Google Scholar]
- 24.Overbeek J.T.G., Voorn M.J. Phase separation in polyelectrolyte solutions; theory of complex coacervation. J. Cell. Physiol. Suppl. 1957;49(Suppl 1):7–22. discussion, 22–26. [PubMed] [Google Scholar]
- 25.Hyman A.A., Weber C.A., Jülicher F. Liquid-liquid phase separation in biology. Annu. Rev. Cell Dev. Biol. 2014;30:39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 26.Aumiller W.M., Jr., Pir Cakmak F., Keating C.D. RNA-based coacervates as a model for membraneless organelles: formation, properties, and interfacial liposome assembly. Langmuir. 2016;32:10042–10053. doi: 10.1021/acs.langmuir.6b02499. [DOI] [PubMed] [Google Scholar]
- 27.Spoelstra W.K., van der Sluis E.O., Reese L. Nonspherical coacervate shapes in an enzyme-driven active system. Langmuir. 2020;36:1956–1964. doi: 10.1021/acs.langmuir.9b02719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flory P.J. Thermodynamics of high polymer solutions. J. Chem. Phys. 1942;10:51–61. [Google Scholar]
- 29.Huggins M.L. Some properties of solutions of long-chain compounds. J. Phys. Chem. 1942;46:151–158. [Google Scholar]
- 30.Gillespie D.T. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 1977;81:2340–2361. [Google Scholar]
- 31.Krapivsky P.L., Redner S., Ben-Naim E. Cambridge University Press; Cambridge, UK: 2010. A Kinetic View of Statistical Physics. [Google Scholar]
- 32.Qin J., Priftis D., de Pablo J.J. Interfacial tension of polyelectrolyte complex coacervate phases. ACS Macro Lett. 2014;3:565–568. doi: 10.1021/mz500190w. [DOI] [PubMed] [Google Scholar]
- 33.Spruijt E., Westphal A.H., van der Gucht J. Binodal compositions of polyelectrolyte complexes. Macromolecules. 2010;43:6476–6484. [Google Scholar]
- 34.Jain A., Vale R.D. RNA phase transitions in repeat expansion disorders. Nature. 2017;546:243–247. doi: 10.1038/nature22386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li L., Srivastava S., Tirrell M.V. Phase behavior and salt partitioning in polyelectrolyte complex coacervates. Macromolecules. 2018;51:2988–2995. [Google Scholar]
- 36.Saleh O.A., Jeon B.J., Liedl T. Enzymatic degradation of liquid droplets of DNA is modulated near the phase boundary. Proc. Natl. Acad. Sci. USA. 2020;117:16160–16166. doi: 10.1073/pnas.2001654117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abudayyeh O.O., Gootenberg J.S., Zhang F. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du M., Chen Z.J. DNA-induced liquid phase condensation of cGAS activates innate immune signaling. Science. 2018;361:704–709. doi: 10.1126/science.aat1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bogers J.F.M., Berghuis N.F., van Leeuwen H.C. Bright fluorescent nucleic acid detection with CRISPR-Cas12a and poly(thymine) templated copper nanoparticles. Biol. Methods Protoc. 2021;6:bpaa020. doi: 10.1093/biomethods/bpaa020. Published online October 8, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi J.H., Lim J., Choi J.W. CRISPR-Cas12a-based nucleic acid amplification-free DNA biosensor via Au nanoparticle-assisted metal-enhanced fluorescence and colorimetric analysis. Nano Lett. 2021;21:693–699. doi: 10.1021/acs.nanolett.0c04303. [DOI] [PubMed] [Google Scholar]
- 41.Li Y., Mansour H., Li F. Naked-eye detection of grapevine red-blotch viral infection using a plasmonic CRISPR Cas12a assay. Anal. Chem. 2019;91:11510–11513. doi: 10.1021/acs.analchem.9b03545. [DOI] [PubMed] [Google Scholar]
- 42.Bollum F.J. Thermal conversion of nonpriming deoxyribonucleic acid to primer. J. Biol. Chem. 1959;234:2733–2734. [PubMed] [Google Scholar]
- 43.Yehudai-Resheff S., Hirsh M., Schuster G. Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol. Cell. Biol. 2001;21:5408–5416. doi: 10.1128/MCB.21.16.5408-5416.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Dongen J.E., Berendsen J.T.W., Segerink L.I. Point-of-care CRISPR/Cas nucleic acid detection: recent advances, challenges and opportunities. Biosens. Bioelectron. 2020;166:112445. doi: 10.1016/j.bios.2020.112445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li P., Banjade S., Rosen M.K. Phase transitions in the assembly of multivalent signalling proteins. Nature. 2012;483:336–340. doi: 10.1038/nature10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pak C.W., Kosno M., Rosen M.K. Sequence determinants of intracellular phase separation by complex coacervation of a disordered protein. Mol. Cell. 2016;63:72–85. doi: 10.1016/j.molcel.2016.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang J., Choi J.M., Hyman A.A. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 2018;174:688–699.e16. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berry J., Brangwynne C.P., Haataja M. Physical principles of intracellular organization via active and passive phase transitions. Rep. Prog. Phys. 2018;81:046601. doi: 10.1088/1361-6633/aaa61e. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.