Main text
Cells are densely packed reaction containers in which thousands of reaction processes occur specifically and simultaneously. Concepts of complex formation because of stoichiometric assembly of different building blocks as well as membrane encapsulation have long been known to scientists as means for cells to spatially regulate reactions. The last decade has moved the process of phase separation into focus as another strategy with which spatial organization can be achieved, giving rise to so-called biomolecular condensates (1). These condensates often contain multicomponent mixtures, e.g., proteins and nucleic acids, and can also provide a unique physicochemical environment. Recent evidence suggests that the composition of protein-RNA condensates plays a key role in modulating their phase behavior and in regulating biological processes. For example, in a typical transcription process, the low concentration of nascent RNAs can stimulate the initial formation of transcriptional condensates, whereas a burst of RNAs produced during elongation can dissolve the condensates, providing a feedback mechanism following the concept of reentrant phase transition (Fig. 1; (2)). Reentrant phase transition generally refers to a process in which a system transits from one state into a macroscopically similar (or identical) state via at least two-phase transitions through the variation of a single parameter, e.g., pH, temperature, ionic strength, or polymer concentration. For RNA-mediated reentrant phase transitions, we refer readers to Milin et al. for a detailed review (3). Reentrant phase transition of protein-RNA mixtures can go far beyond blob-like structures of coexisting dense and liquid phases and create layered functional topologies, which may serve as even more specialized reaction containers (4). With homotypic phase separation of e.g., an intrinsically disordered protein (IDP) being an emerging field in the life sciences, the thermodynamics behind the phase behaviors of protein-RNA mixtures remain even less well understood in the context of biomolecular mechanism and cellular function.
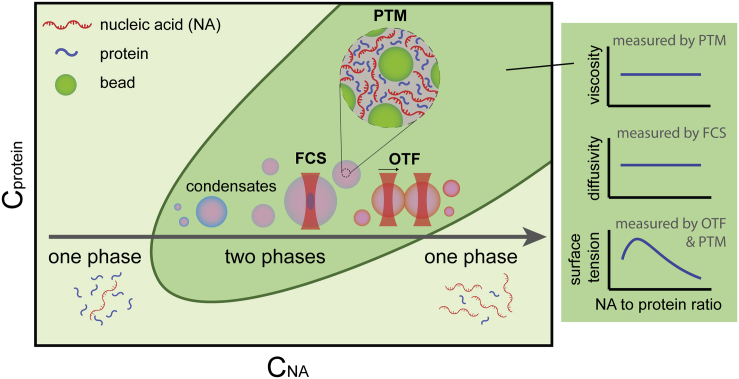
Figure 1.
Schematic phase diagram of protein-nucleic acid (NA) mixtures. Increasing the NA concentration (e.g., along the gray arrow) drives an initial phase transition from one phase to two phases and the formation of heterotypic protein-NA condensates. Further increasing the NA concentration drives the second phase transition from two phases to one phase and dissolution of the condensate. Such a process is termed as RNA-mediated reentrant phase transition, which is driven by electrostatic interactions. Right column: the interfacial tension, viscosity, and diffusivity of protein-RNA condensates across the two-phase regime can be quantified using a combined method, including PTM, FCS, and optical tweezer-induced droplet fusion (OTF). In OTF, one optically trapped condensate is forced to move toward another stationary droplet to induce droplet fusion in solution. OTF enables us to probe the ratio of viscosity over surface tension.
To quantitively understand the biophysical properties of protein-RNA condensates, in this issue of the Biophysical Journal, Alshareedah et al. make use of a clever combination of particle-tracking microrheology (PTM), optical tweezer-induced droplet fusion (OTF), and fluorescence correlation spectroscopy (FCS) to investigate the interfacial tension, viscosity, and diffusivity of protein-RNA condensates (5). Using the mixture of disordered polypeptide [RGRGG]5 and single-stranded nucleic acid (ssNA) dT40 as a simplified but powerful in vitro model system, the authors found that the interfacial tension, but not the internal viscosity or diffusivity of the multicomponent condensates, is modulated by the ssNA-to-protein stoichiometry across the entire two-phase regime. The authors suggest the existence of spatially organized mesoscale structure in protein-RNA condensates where the condensate surface, but not the condensate core, is responsive to variation in mixture composition.
The authors also noticed a twofold difference between the diffusivity and viscosity scaling probed by FCS and PTM, respectively, when tuning the salt concentration in the buffer. However, the two techniques probe different things—FCS directly quantifies the diffusion of a fluorescently labeled peptide or ssNA through a confocal volume, whereas PTM images the mobility of diffusing beads to extract the viscosity of the dense phase. Multiple effects can thus account for such a moderate discrepancy: 1) Peptide and ssNA molecules can undergo diffusion as complexes and not as free chains in the dense phase. Because of short-range cation-π interactions, RNA-protein condensates may exhibit heterogeneity in the diffusive trajectory at small length scales, which can vary with ionic strength under different experimental conditions (6). 2) The accuracy of FCS can be dependent on the ratio of the focal volume to the size of the probed object, which poses a challenge when probing diffusivity inside smaller droplets (7). 3) Because of experimental constraints on detecting rapidly moving beads, there is a size limit on how small a bead can be to give meaningful results in PTM. The authors tested two bead sizes (200 and 1000 nm) and did not find a significant difference. But to what extent the measured results can be directly extrapolated to much smaller length scales remains unknown.
Previous work by Zhang et al. also employed PTM to measure the viscosity shift of protein-RNA condensates when tuning the concentration of CLN3 RNA in its mixture with the polyQ-expansion protein Whi3 and yet found that the viscosity of the protein-RNA condensates can be significantly increased by adding mRNA (8). Whi3 has disordered motifs as well as RNA-binding motifs, which may feature homotypic (protein-protein or RNA-RNA) and heterotypic interactions (protein-RNA) and thus deviate from obligate heterotypic interactions. Besides, Alshareedah et al. (5) used shorter ssNA as a well-defined model, whereas Zhang et al. used a thousand bp-long structured mRNA CLN3, which may also complicate the simple picture. We simply do not yet know how such different length scales and biological systems affect the physicochemical behavior of reentrant phase separation. In the future, the use of e.g., active microrheology (9) could be crucial to better study and understand the viscosity behavior of the protein-RNA condensates, especially in living cells. In contrast to passive microrheology, in active microrheology, the system is driven out of nonequilibrium, which enables us to probe dynamics.
Another exciting suggestion by the authors is that the internal viscosity of the heterotypic condensates might remain insensitive to stoichiometric alterations. Considering that biochemical reaction rates can be limited by diffusive encounters of biomolecular reactants, reentrant phase separation of protein-RNA mixtures offers a potential mechanism to spatially regulate diffusion-controlled reactions in the condensates. Reinforcing the importance of viscosity regulation in cells, Persson et al. recently discovered a form of cellular adaption that enables budding yeast to maintain a near-constant intracellular viscosity despite changes in temperature and energy status of the cell (10).
Alshareedah et al. also report that condensates with excess ssNAs show a significant repulsive force preceding their coalescence (5). We wonder if the changes in condensate surface composition would provide a mechanism for spatially segregating the reaction environments or if the surface would serve as a barrier for transport across the interface. So far, a direct quantitative link between the biological functions and biophysical properties of protein-RNA condensates has not been fully established, but the systematic approach described by Alshareedah et al. (5) provides now a quantitative path to study and potentially reveal biological mechanisms to create spatially segregated environments for better control of biomolecular reaction conditions.
Editor: Jason Kahn.
References
- 1.Banani S.F., Lee H.O., Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henninger J.E., Oksuz O., Young R.A. RNA-mediated feedback control of transcriptional condensates. Cell. 2021;184:207–225.e24. doi: 10.1016/j.cell.2020.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Milin A.N., Deniz A.A. Reentrant phase transitions and non-equilibrium dynamics in membraneless organelles. Biochemistry. 2018;57:2470–2477. doi: 10.1021/acs.biochem.8b00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alshareedah I., Moosa M.M., Banerjee P.R. Phase transition of RNA-protein complexes into ordered hollow condensates. Proc. Natl. Acad. Sci. USA. 2020;117:15650–15658. doi: 10.1073/pnas.1922365117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alshareedah I., Thurston G.M., Banerjee P.R. Quantifying viscosity and surface tension of multicomponent protein-nucleic acid condensates. Biophys. J. 2021;120:1161–1169. doi: 10.1016/j.bpj.2021.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shakya A., King J.T. Non-Fickian molecular transport in protein–DNA droplets. ACS Macro Lett. 2018;7:1220–1225. doi: 10.1021/acsmacrolett.8b00565. [DOI] [PubMed] [Google Scholar]
- 7.Gennerich A., Schild D. Anisotropic diffusion in mitral cell dendrites revealed by fluorescence correlation spectroscopy. Biophys. J. 2002;83:510–522. doi: 10.1016/S0006-3495(02)75187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H., Elbaum-Garfinkle S., Gladfelter A.S. RNA controls PolyQ protein phase transitions. Mol. Cell. 2015;60:220–230. doi: 10.1016/j.molcel.2015.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jawerth L.M., Ijavi M., Fischer-Friedrich E. Salt-dependent rheology and surface tension of protein condensates using optical traps. Phys. Rev. Lett. 2018;121:258101. doi: 10.1103/PhysRevLett.121.258101. [DOI] [PubMed] [Google Scholar]
- 10.Persson L.B., Ambati V.S., Brandman O. Cellular control of viscosity counters changes in temperature and energy availability. Cell. 2020;183:1572–1585.e16. doi: 10.1016/j.cell.2020.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]