Abstract
Formation of biomolecular condensates through liquid-liquid phase separation (LLPS) has emerged as a pervasive principle in cell biology, allowing compartmentalization and spatiotemporal regulation of dynamic cellular processes. Proteins that form condensates under physiological conditions often contain intrinsically disordered regions with low-complexity domains. Among them, the RNA-binding proteins FUS and TDP-43 have been a focus of intense investigation because aberrant condensation and aggregation of these proteins is linked to neurodegenerative diseases such as amyotrophic lateral sclerosis and frontotemporal dementia. LLPS occurs when protein-rich condensates form surrounded by a dilute aqueous solution. LLPS is per se entropically unfavorable. Energetically favorable multivalent protein-protein interactions are one important aspect to offset entropic costs. Another proposed aspect is the release of entropically unfavorable preordered hydration water into the bulk. We used attenuated total reflection spectroscopy in the terahertz frequency range to characterize the changes in the hydrogen bonding network accompanying the FUS enrichment in liquid-liquid phase-separated droplets to provide experimental evidence for the key role of the solvent as a thermodynamic driving force. The FUS concentration inside LLPS droplets was determined to be increased to 2.0 mM independent of the initial protein concentration (5 or 10 μM solutions) by fluorescence measurements. With terahertz spectroscopy, we revealed a dewetting of hydrophobic side chains in phase-separated FUS. Thus, the release of entropically unfavorable water populations into the bulk goes hand in hand with enthalpically favorable protein-protein interaction. Both changes are energetically favorable, and our study shows that both contribute to the thermodynamic driving force in phase separation.
Significance
Recently, membraneless compartmentalization via phase separation in living cells has been linked to the formation of pathological protein aggregates found in neurodegenerative diseases. Despite its significance, less is known about how liquid-liquid phase separation is triggered or prevented on a molecular scale. Two thermodynamic driving forces have been proposed: protein-protein and protein-water interactions (mostly enthalpic), as well as the release of preorganized hydration water into the bulk (mostly entropic). Whereas most studies focus on the first aspect, experimental evidence for the latter was lacking. Here, we present a first, to our knowledge, experimental study using a combination of terahertz spectroscopy and fluorescence microscopy, which reveals that both driving forces are of equal importance for biomolecule condensation.
Introduction
The spatial and temporal regulation of biochemical processes inside a cell requires compartmentalization. In addition to “classical” organelles, which are surrounded by lipid membranes, cytosolic and nuclear compartments can also form via liquid-liquid phase separation (LLPS). Multiple cellular processes, including RNA splicing, receptor-mediated signaling, mitosis, and chromatin organization, are coordinated by these membraneless compartments or biomolecular condensates (1, 2, 3, 4, 5, 6, 7, 8, 9, 10). Moreover, biomolecular condensates have been linked to the formation of pathological protein aggregates found in various neurodegenerative diseases such as Alzheimer’s disease, frontotemporal dementia, and amyotrophic lateral sclerosis (11, 12, 13, 14, 15, 16). Specifically, RNA-binding proteins such as fused in sarcoma (FUS), TAR DNA-binding protein 43 (TDP-43), and heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) are found in protein aggregates resulting from aberrant formation of stress-induced RNA granules (17). FUS is a DNA/RNA-binding protein that is involved in various cellular processes including DNA repair, RNA shearing and transport, gene transcription and regulation, translation, and micro-RNA processing (18, 19, 20, 21, 22, 23, 24). Mutations in the FUS gene have been identified in familial amyotrophic lateral sclerosis and shown to affect the subcellular localization and aggregation propensity of FUS and the reversibility of stress granules (11,25).
A key feature of proteins involved in condensate formation are multivalent interactions (26, 27, 28, 29), which are often mediated by intrinsically disordered regions or low-complexity regions (30, 31, 32, 33). For FUS, interactions are predominantly initiated by the N-terminal low-complexity region. However, pathogenic FUS mutations primarily occur in the nuclear localization signal, promoting cytosolic localization and subsequent aggregation of FUS (34,35). In addition, the methylation state of arginine residues influences LLPS of FUS, and probing with infrared spectroscopy revealed methylation-induced changes in its secondary structure (15,16,36).
So far, most studies have focused on the influence of cosolutes and physicochemical conditions on the LLPS process or protein aggregation within liquid droplets (11,15,36,37). For example, Winter and co-workers have shown pressure-dependent dissolution of LLPS droplets (36,38). Despite the recently proposed synergetic effect of water and biomolecules (39), clear experimental proof was still missing because experimental techniques to probe the solvent in LLPS formation are still rare (40, 41, 42, 43). The formation of two separate liquid-like phases intrinsically requires reorganization of the solvent, resulting in phase-separated droplets that can be considered as local solvation hotspots with properties that are distinct from those of bulk water. The open question is whether and how this promotes protein aggregation or even how the solvent can be “tuned” by addition of cosolvents to avoid protein aggregation and fibrillization (11,37).
Phase separation in biomolecular and polymer systems (aqueous two-phase systems) has been proposed to stem from enthalpically favorable water-mediated solute-solute interactions, in which structural changes in water are thought to play a key role, and some studies have found that bulk solvent properties such as dipolarity and acidity are altered in the presence of proteins and polymers (44,45). More recently, the additional need to consider the entropic contributions of solute-water interactions, i.e., hydration water, to phase separation has been recognized (39). Many theoretical and experimental studies have further revealed that water-hydrating proteins, as well as any other large solute exposing both hydrophilic and hydrophobic surface regions, show remarkably different structural, dynamical, spectroscopic, and thermodynamic properties depending on the local nanoenvironment (46, 47, 48, 49, 50, 51, 52, 53). Spatial heterogeneity in the hydration water network has been proposed to play a prominent role in protein folding and aggregation (46, 47, 48, 49). This raises the question of whether global or local hydration changes can also be relevant for LLPS.
Terahertz spectroscopy is a sensitive tool to study the changes in the water network (54) and has been shown to be especially effective in elucidating the solvation properties of water in the vicinity of complex biomolecular systems (55, 56, 57, 58). More recently, we could show that these “fingerprints” of the hydration water network can be directly linked to changes in the thermodynamics of the solvent (59,60). In particular, we unraveled the distinct thermodynamic signatures of two hydration water populations coexisting in contact with hydrophilic or hydrophobic groups of small solutes: the more tetrahedral bound water population that directly interacts with polar groups and the less tetrahedral (more disordered) wrap water population that hydrates hydrophobic groups (60). Although the bound water population shows enhanced coordination and reduced entropy with respect to bulk water, the wrap water population shows less coordination and a combination of a loss of entropy and enthalpy compared with bulk water (60). Molecular interpretation of the spectroscopic data with thermodynamic insight will help in understanding the role of the solvent in facilitating the special properties of the phase-separated liquid droplets and can help obtain a molecular picture on how this is linked to neurotoxic protein aggregation.
Here, we addressed the role of the solvent in LLPS of FUS by using ATR (attenuated total reflection) spectroscopy in the terahertz range in combination with laser-scanning microscopy for protein aggregation. Low-frequency vibrational spectra of water in solvated FUS droplets revealed a spectroscopic signature of water within FUS droplets. We could show that the water network is distinctly different from bulk water; compared with bulk water, water molecules in phase-separated droplets showed increased tetrahedral coordination and restricted rotational (librational) motions at ambient conditions. Phase separation induced dissipation of the hydration shell, leaving only water molecules bound to the protein surface that are proposed to drive the enrichment of FUS protein inside the droplets. These findings give novel, to our knowledge, insights into the thermodynamic forces that drive LLPS.
Materials and methods
Cloning
Maltose-binding protein (MBP)-FUS-enhanced green fluorescent protein (EGFP) and MBP-FUS in a pMal vector were described previously (15). The MBP-FUS-EGFP-PLD(Y to S) construct was created by exchanging the PLD domain using MfeI and SalI cloning sites. A pUC57-Simple plasmid was obtained from GenScript Biotech (Piscataway, NJ) containing the 27 Y to S substitutions of the PLD (Y to S) sequence flanked by MfeI and SalI cloning sites. All constructs were transformed into BL21-DE3 strains, and bacterial plates were stored at 4°C until further use.
Protein expression and purification
Bacterial cultures of 1 L were grown up to an OD (600 nm) of ∼0.8 in lysogeny broth medium and protein expression was induced using 100 μM isopropyl-β-D-1-thiogalactopyranoside. After expression for 22 h at 12°C, cells were harvested by centrifugation at 8000 × g at 4°C for 20 min and washed using 1× phosphate-buffered saline buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 (pH 7.4)). The suspension was centrifuged again (3200 × g, 4°C, 10 min). The supernatant was disposed of, and the pellet was frozen at −20°C until further use.
Bacterial pellets were solubilized in resuspension (RS) buffer (50 mM Na2HPO4/NaH2PO4, 500 mM NaCl, 10 μM ZnCl2, 4 mM β-mercaptoethanol (pH 8.0)) containing 10% glycerol. During the purification process, 500 mM NaCl was present in all buffers to prevent copurification of residual RNA molecules. Lysis was performed using an SLM AMINCO French Press (Thermo Fisher Scientific, Waltham, MA), and the suspension was centrifuged for 45 min at 40,000 × g at 4°C. Protein purification was determined by tandem-affinity purification. The supernatant was filtered through a 0.2 μm sterile filter, bound at the His-Trap FF column (GE Healthcare, Chicago, IL), and washed using RS buffer containing 20 mM imidazole after being eluted by RS buffer with 200 mM imidazole. Eluted protein was applied to an MBP Trap HP column (GE Healthcare) and washed using RS buffer without imidazole. Afterwards, the protein was eluted with 20 mM maltose in RS buffer. Fractions containing protein were pooled and buffer exchanged using an Amicon column (30 kDa MWCO; Merck, Darmstadt, Germany) to RS buffer with 5% glycerol. Protein concentration was determined by using Bradford Reagent (Sigma-Aldrich, St. Louis, MO), and protein aliquots were stored at a final concentration of 2.5–3 mg/mL at −80°C until further use.
Sample preparation
Protein aliquots were thawed on ice and centrifuged at 16,000 × g at 4°C for 20 min to remove residual aggregates. The supernatant was transferred to an Amicon column (30 kDa MWCO), and the buffer was exchanged to droplet buffer (20 mM Na2HPO4/NaH2PO4 (pH 7.5), 150 mM NaCl, 2.5% glycerol, 1 mM dithiothreitol). Protein concentration was checked again using a Bradford assay with bovine serum albumin standard curves before dilution of the protein to the desired concentration and addition of tobacco etch virus (TEV) protease to a final concentration of 0.05 mg/mL. Digestion of the protein concentrations of 5 and 10 μM at room temperature was done for 20 and 40 min, respectively, before the experiments because these incubation times were shown to provide full cleavage by gel electrophoresis.
Laser-scanning microscopy
Fluorescent image data were recorded on a microscope (ELYRA PS.1; Carl-Zeiss, Oberkochen, Germany) equipped with an imaging detector (LSM 880; Carl-Zeiss). Droplet imaging was performed by using laser-scanning microscopy. For acquisition of the FUS-EGFP, a 63× NA 1.4 oil immersion objective was used to scan a stack of 67.5 × 67.5 × 5 μm and 0.330 μm for each optical section. The laser power of the argon laser source was set to 0.02% at 488 nm with a pixel dwell time of 5.71 μs. Laser power, gain, and field of view were kept constant while measuring different concentrations and kinetics of droplet formation. For the quantification of fluorescent intensities within the respective area of interest, ZEN2.1 software (Carl-Zeiss) was used to capture confocal planes with the same 16-bit settings for uncut FUS-EGFP dilution series and sample analysis. This wide dynamic range devoid of overexposure allows the calculation of concentrations in the micromolar range.
For the three-dimensional analysis of confocal z-stack images, data were imported into Imaris 9.3.1 and reconstruction of surfaces and volumes was performed using the surface module. The surface and volume data of the reconstructed droplets were transferred to GraphPad Prism (Graphpad Software, San Diego, CA) for visualization and statistical analysis.
Fluorescent recovery after photobleaching experiments were done with the ZEN2.1 bleaching and region software module and a Plan-Apochromat 100× NA 1.46 oil differential interference contrast M27 objective. Three circular areas of interest with a diameter of 12 pixels were recorded; one was bleached (red circle) with 100% laser power and a pixel dwell time of 8.71 μs, with a scan time of 111.29 ms and a pixel dwell time of 1.61 μs. For analysis and visualization, the data were transferred to GraphPad Prism.
ATR Fourier-transform infrared spectroscopy
All spectra were recorded in the range of 30–690 cm−1 with a Fourier-transform infrared (FTIR) spectrometer (Vertex 80v; Bruker, Billerica, MA) using a helium-cooled silicon bolometer (Infrared Laboratories, Tucson, AZ). In the sample compartment, a single reflection ATR unit (MVP-Pro; Harrick Scientific, Pleasantville, NY) with a diamond prism (surface area 2 × 3 mm) was placed with a custom-designed liquid sample cell on top of the crystal (see Fig. S1). The custom sample cell consists of a metal support with an o-ring and polyvinyl chloride (PVC) disk that has a 5 mm diameter hole for the liquid sample. The sample (50 μL volume) is pipetted into the PVC disk, and the PVC disk is covered and clamped to the ATR unit to create a sealed sample chamber.
To obtain the cooling spectra of water, a temperature-controlled diamond crystal (Harrick Scientific) with a diameter of 500 μm was used. The sample compartment was constantly purged with nitrogen at atmospheric pressure, and the interferometer compartment was evacuated at ∼3 mbar. Spectroscopic data were collected for a total of 60 min at 2 min intervals with an average of 64 scans and a spectral resolution of 2 cm−1. After each experiment, the diamond crystal surface was thoroughly cleaned using ultrapure water, 0.5 M NaOH, and isopropanol.
The ATR absorption coefficient, α(ν), was determined as shown in Eq. 1.
| (1) |
where I(ν) and I0(ν) are the intensities of the sample and reference and dp is the frequency-dependent penetration depth. Here, the bare crystal surface served as the reference. The penetration depth was determined from Eq. 2.
| (2) |
where λ is the wavelength of light; ndiamond and nsample are the refractive indices of the diamond crystal and sample, respectively; and θ is the incident angle. The incident angle was fixed at θ = 45°, ndiamond = 2.38 for the measured frequency range, and it was assumed nsample = 1.5. We note that the high protein density inside of FUS droplets could affect the refractive index of the sample relative to the dilute phase and therefore the resulting penetration depth. However, because many studies have previously shown that the condensed phase retains a large fraction of water (61,62), we have assumed here for simplicity that the refractive index of the sample is a constant value consistent with an aqueous solution.
Difference absorption spectra (Δα) were determined by subtracting the first spectrum of a 60 min series from all following spectra. 60 min was chosen as the endpoint because negligible changes in signal occurred after this point. All shown Δα spectra of the different FUS samples represent the average of at least two measurements with an average error of 5 cm−1.
Results
Water molecules in phase-separated FUS droplets show a characteristic spectral signature in the terahertz region
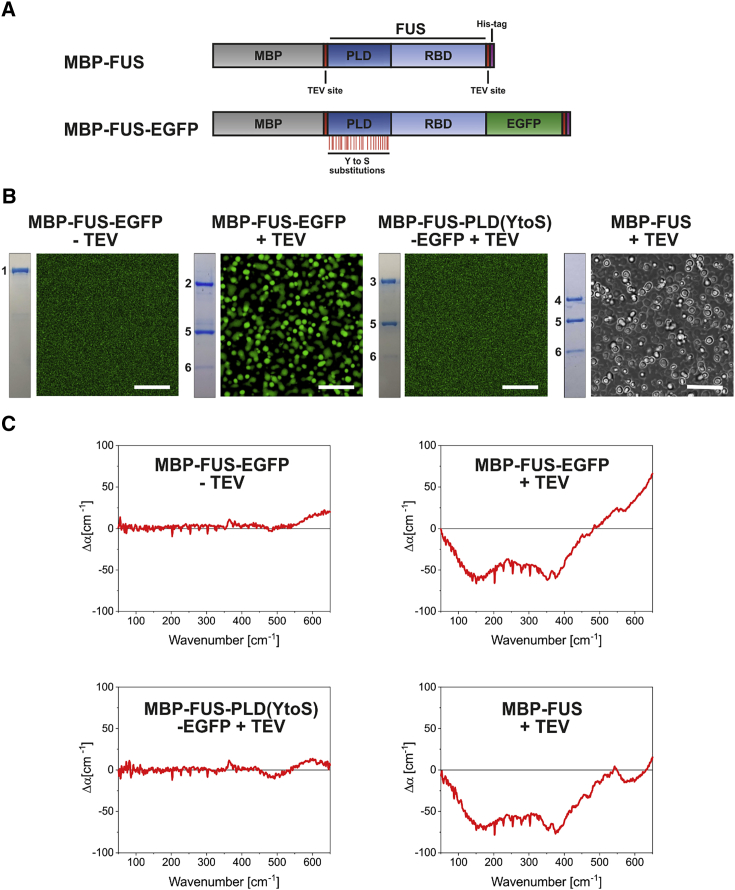
To address the role of water-water and water-protein interactions in LLPS, we employed FUS as a model protein (Fig. 1 A). A well-established approach to induce LLPS in vitro is TEV-protease-mediated cleavage of an MBP-FUS fusion protein. Formation of FUS droplets, indicative of phase separation, was only observed after TEV protease has been added to the protein sample (Fig. 1 B). Robust phase separation was observed for FUS-EGFP samples with a starting concentration of 5 μM or higher (see Fig. S2). Efficient cleavage of MBP-FUS by TEV protease was monitored by SDS-PAGE and Coomassie staining, and all measurements were carried out after full digestion was achieved. In samples treated with TEV protease, three bands were observed in the gel, corresponding to MBP (43 kDa), the TEV protease (28 kDa), and wild-type or mutant FUS with or without EGFP (82, 80, or 56 kDa). As a control for wild-type FUS undergoing LLPS, we included a previously described variant of FUS in which the tyrosine residues in the PLD were replaced by serines (Y to S). As a consequence of these mutations, multivalent interactions of the PLD with arginine residues of RNA-binding domains are abolished and phase separation is prevented (63).
Figure 1.
Terahertz fingerprint of LLPS of FUS. (A) Schematic drawing of the MBP-FUS and MBP-FUS-EGFP constructs is given. The constructs contain a His6 tag (violet) for purification and two TEV cleavage sites (red) to cleave off MBP and the His6 tag. FUS comprises the PLD (dark blue) and RBD domains (light blue). The Y to S substitutions are indicated as red lines below the PLD domain. (B) Microscopic images (50 × 50 μm; scale bar, 10 μm) of the protein samples (10 μM) are shown. The inset left of the image represents the corresponding sample analyzed by SDS-PAGE and Coomassie brilliant blue staining. Images of MBP-FUS were taken using brightfield illumination. (C) Averaged ATR-FTIR Δα spectra (50–650 cm−1) of protein samples measured 60 min after deposition on the ATR crystal surface are shown. On the left side, we show Δα spectra (50–650 cm−1) of samples that do not undergo LLPS, and on the right-hand side, spectra of samples after LLPS. The average error of all ATR-FTIR measurements is 5 cm−1. To see this figure in color, go online.
ATR-FTIR spectra in the terahertz region (50–650 cm−1) with 10 μM protein concentration were recorded. In the following, we subtracted the absorption at t = 2 min from the absorption spectra of samples at t = 60 min. The result, denoted as Δα, is plotted in Fig. 1 C. FUS-EGFP without TEV and FUS-PLD(Y to S) samples had Δα values of 0 cm−1, within experimental uncertainty (no LLPS; left side of Fig. 1 C). On the other hand, MBP-FUS-EGFP with TEV and MBP-FUS with TEV, which both underwent LLPS (right side of Fig. 1 C), showed a change in absorption, i.e., Δα is not equal to zero (see Fig. S3 and Table S1 for spectral deconvolution of observed peaks). The amplitude of Δα increased in a time-dependent manner because of the enrichment of droplets at the surface of the ATR crystal (Fig. S4). No major changes in signal were found to occur after 60 min. By comparing the spectra of FUS plus TEV protease and FUS-EGFP plus TEV protease, we found no difference in the observed bands below 500 cm−1 (right side of Fig. 1 C), implying that the ATR-FTIR spectra of phase-separated FUS in the terahertz region were not strongly influenced by the presence of EGFP. However, slight changes in the signal above 500 cm−1 in the FUS spectrum without EGFP can be seen. This observation could be due to changes in the packing density of the FUS droplets, leading to a stronger blue shift of the librational band in these samples. However, technical requirements only allowed evaluation of the spectra up to 650 cm−1. It is also important to note that the spectral contributions can be influenced by the presence of MBP, TEV protease, and His6 tag, which might be partially incorporated in the phase-separated droplets (15).
Spectral fingerprint of LLPS scales with the total volume of droplets
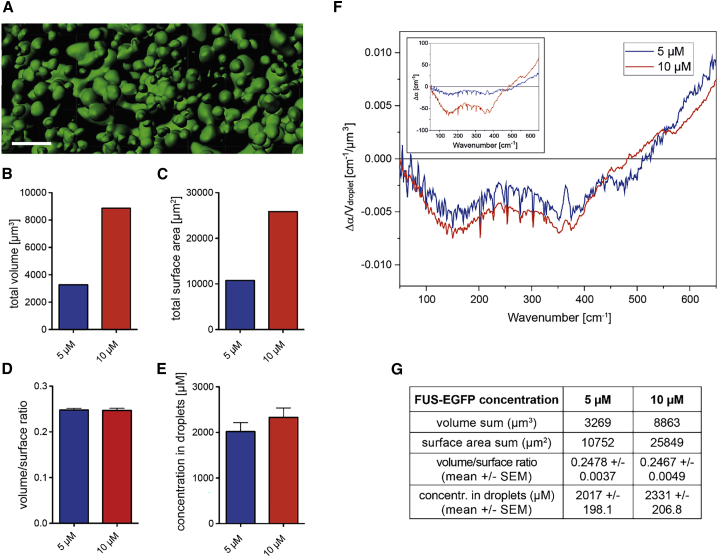
To quantify the volume and surface area upon droplet formation, we recorded images (5 and 10 μM MBP-FUS-EGFP) with confocal laser-scanning microscopy. z-stacks with a depth of 5 μm were recorded 60 min after deposition of the droplet solution on the surface (Fig. 2 A). The total volume occupied by droplets in the 10 μM solution was roughly tripled in comparison to the total droplet volume of the 5 μM solution (Fig. 2, B and G). Similarly, the total surface area occupied by droplets was determined to be 2.4 times larger in the 10 μM solution than in the 5 μM solution (Fig. 2, C and G). The average volume/surface ratio of all droplets analyzed was found to be independent of the protein concentration (Fig. 2, D and G), and mainly the number of the droplets increased with increasing protein concentration (the surface coverage roughly doubles from 5 to 10 μM). It should be noted that the confocal measurements done here are expected to represent the properties of droplets on the diamond ATR crystal because both measurements were done on hydrophobic surfaces. Although small changes in contact angle between glass and diamond surfaces could occur, it is expected that FUS droplets retain roughly the same shape between the two hydrophobic surfaces, in which a high contact angle was previously observed (65).
Figure 2.
FUS-EGFP droplet properties and influence on terahertz fingerprint. Droplet samples were analyzed 60 min after LLPS induction by TEV protease addition at concentrations of 5 (blue) and 10 μM (red). (A) Representative volumetric three-dimensional reconstruction of FUS-EGFP droplets at 10 μM is given. Reconstruction was performed using confocal laser-scanning microscopy z-stack images comprising a volume of 67.5 × 67.5 × 5.03 μm (scale bar, 5 μm). (B) Total droplet volume measured in reconstructed volumes is shown. (C) Total droplet surface area measured in reconstructed surfaces. (D) Mean volume/surface ratio of reconstructed droplets calculated from confocal z-stacks is shown. (E) Calculated protein concentration within FUS-EGFP droplets, measured according to the protocol from Zhang et al. (64), is given (see Fig. S5 for a detailed overview of the experiment). (F) Averaged 5 (blue) and 10 μM (red) FUS-EGFP Δα spectra (50–650 cm−1) after 60 min normalized to the total volume of protein droplets are shown. Inset: 5 (blue) and 10 μM (red) FUS-EGFP Δα spectra (50–650 cm−1) after 60 min and before normalization. (G) Summary of obtained values for the different biophysical properties at 5 and 10 μM is given. To see this figure in color, go online.
To determine the protein concentration inside of phase-separated droplets formed by FUS, we used confocal laser-scanning microscopy according to previously established protocols (64). Based on the fluorescence intensity calibration curve of MBP-FUS-EGFP without TEV (Fig. S5), the FUS concentration inside LLPS droplets was ∼2.0 mM (or 160 mg/mL) for both 5 and 10 μM solutions (Fig. 2, E and G), which is in line with previously reported values in literature for the LC domain of FUS (120–440 mg/mL) (33,66). It should be noted here that the derived concentration from fluorescence measurements might involve quenching of EGFP because the chemical environment, compared to a dilute solution, changes drastically. Quenching would result in an underestimation of the FUS concentration. However, a concentration in the droplets (2 mM) that significantly exceeds the concentration of the solution (10 μM) matches literature (33,61,67).
In Fig. 2 F, inset, we plot the spectral change Δα for two initial protein solutions (5 and 10 μM), for which we find an increase for the 10 μM concentration. However, when we divided Δα by the total droplet volume at a given concentration, we find that Δα/Vdroplet is the same—within our experimental uncertainty—for both solutions (Fig. 2 F). The good agreement of the spectra of 5 and 10 μM FUS normalized to total droplet volume indicates that changes in the refractive index are not responsible for the observed signal (see Fig. 2 F, inset; Fig. S6). The increase in Δα can therefore be attributed solely to an increase in the phase-separated droplet volume.
Interestingly, the spectral changes Δα are also independent of the initial protein concentration. These are attributed to changes in the hydrogen bond network upon LLPS. Because a theoretical simulation of the terahertz spectra of the water network exceeds the capability of any state-of-the-art methods, we decided to compare the experimental spectra to well-known spectra of other water phases to understand the molecular changes in the network.
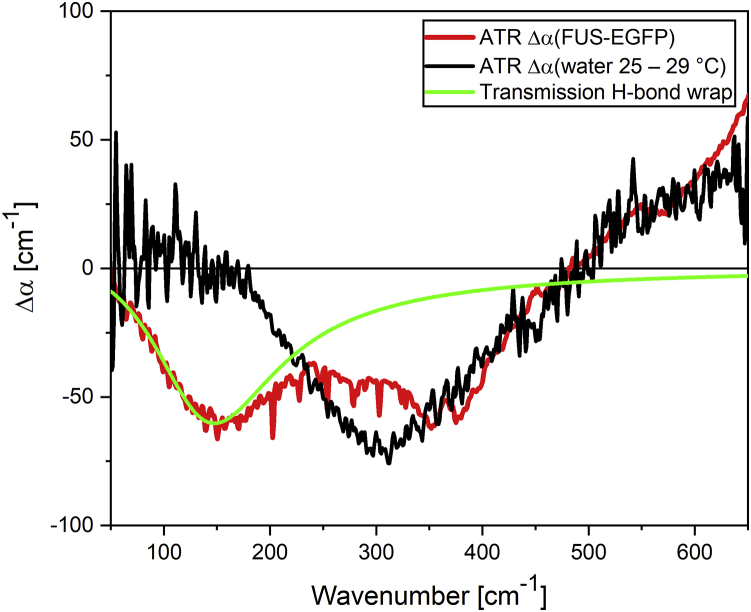
Therefore, we recorded the spectral changes of bulk water upon cooling. Any cooling of bulk water is associated with an increase in tetrahedrality of the water network (68). In Fig. 3, we plotted Δα for bulk water when decreasing the temperature from 29 to 25°C (Δα29–>25°C; see Fig. S7 and Table S2 for spectral deconvolution). In Fig. 3, we also plot Δα of the phase-separated droplets. In both cases, we observed a negative Δα in the range of 200–500 cm−1 and a positive Δα, which can both be explained by a blue shift of the librational peak centered around 600 cm−1 (69,70). Any blue shift of the librational peak, which describes the hindered rotation of a water molecule in the hydrogen bond network, indicates an even more restricted hindered rotation. Similar to the observed effect of cooling, the water inside the droplet is proposed to be more tetrahedrally coordinated than bulk water. Although this explains the spectral shape above 200 cm−1, it cannot explain the spectra below 200 cm−1.
Figure 3.
Δα spectrum of 10 μM FUS-EGFP shows two distinct features corresponding to hydration water (low-frequency part) and increased tetrahedral coordination (high-frequency part). In red, we plotted the ATR Δα spectrum of FUS-EGFP. In black, we show the difference of the spectrum of bulk water at 25°C minus bulk water at 29°C, and in green, the difference of the simulated spectrum of bulk water minus the water network spectrum of the water population around hydrophobes, called H-bond wrap. For details, see Conti Nibali et al. (60). To see this figure in color, go online.
To explain the observed spectra in the range below 200 cm−1, we compare the observed Δα to the spectrum of a two-dimensional water network at a hydrophobic surface, the so-called H-bond wrap population, as reported by Conti Nibali and co-workers (Fig. 3; (60)). Based on the similarity in spectral line shape, we attribute the negative Δα observed at 155 cm−1 to a change in the innermost hydration water layer upon formation of phase-separated droplets. However, we should note that although the H-bond wrap absorption peak was recorded in transmission, these data are recorded with ATR, which results in a red shift of spectral features compared to transmission spectra (Fig. S8). It is worth mentioning that copartitioning of MBP, TEV protease, and His6 tag to FUS droplets cannot be fully excluded, but based on the observed strong changes in the water network, we still expect them to be mainly due to formation of LLPS by FUS.
Discussion
We estimate an increase in the concentration of FUS in the droplets to ∼2 mM (160 mg/mL), which is consistent with the range reported in literature for the LC domain of FUS in droplets (120–440 mg/mL) (Fig. 2 E; (33,66)). This indicates that phase-separated proteins are in a highly condensed state.
After 60 min, up to 27% of the probed volume is occupied by droplets. At the same time, we find an overall maximal signal loss of the H-bond stretching feature at 155 cm−1 of 1.5–2% (Fig. S9). With a 2 mM protein concentration (or 160 mg/mL, protein volume fraction 12%) inside the droplets, a loss of ∼3% water signal is expected, taking into account only pure volume exclusion of water by proteins. The fact that a smaller percentage of water was lost than expected from pure volume exclusion suggests that the droplets retain a large fraction of water and is consistent with previous studies of Ddx4, which found that the condensed phase has a high water content (∼73%) (66).
A major driving force of LLPS of FUS seems to be based on the intermolecular intertwining of PLD and RBD domains (63). According to this proposed mechanism, there will be variances in the local protein and water densities, leading to pockets of confined water molecules trapped in between protein molecules.
With a very rough approximation assuming a hard sphere with a radius of gyration of 40.6 Å in a uniformly spaced cubic lattice, FUS molecules are 93 Å apart for a 2 mM solution, which corresponds to ∼3 layers of water molecules between each FUS protein (71,72). These crowding conditions can result in a loss of degrees of freedom in the hydration water, i.e., a loss of entropy.
Water near FUS molecules will be strongly bound to polar amino acids and experience a loss of configurational degrees of freedom because of this coordination and by confinement effects. Moreover, nonpolar amino acid side chains are expected to form direct interactions because of their high local concentrations. This mechanism is energetically favorable because hydrating hydrophobic residues is entropically unfavorable, whereby desolvation of nonpolar regions would lead to a minimization of solvent-exposed hydrophobic surfaces (46). Recently, a study by the Han group similarly found that dehydration plays an essential role in LLPS, in which addition of polyethylene glycol was found to draw water to its hydration shell and subsequently decrease hydration water content in polymer coacervates by ∼10% (62).
Fluorescent recovery after photobleaching experiments after 1 h revealed no loss of dynamics inside FUS droplets (Fig. S10). This suggests that FUS molecules were in a highly dynamic state and maturation of the droplets into a gel-like or aggregated state did not occur during the timescale of the ATR-FTIR measurements (≤1 h). The volume/surface ratio was similar for FUS droplets formed in a 5 or 10 μM FUS solution (Fig. 2 D), which is consistent with a similar protein concentration inside the droplets (Fig. 2, E and G).
Though the performed ATR-FTIR measurements can only reveal information on the averaged ensemble of all water molecules, they still show changes in two parts of the spectrum, i.e., centered at 155 cm−1 and around 300 cm−1. Whereas the proposal of distinct water populations, i.e., the more ordered hydration water and bulk water, to be key to phase separation is not new (39), unanswered questions concerning the structure of these water populations and how these are affected by other compounds remain. Our THz fingerprints (negative 155 cm−1 band) of the water network inside the FUS droplets have now revealed two changes: a reduced wrap water population, previously shown to be hydrating hydrophobic patches (60), and more water molecules with constrained librational motions are simultaneously observed, as expected for more tetrahedral water, inside the highly concentrated droplets. More hindered librations are expected for bound as well as for confined water. Although our technique is not able to distinguish between contributions from water interacting with the surface of droplets and water inside of the droplets, the latter likely dominates the observed spectral changes because of the overall high water content inside of FUS droplets. Upon overlap of hydration shells of FUS inside the droplets, we expect the less-bound wrap waters to be released and the more strongly bound water molecules to remain.
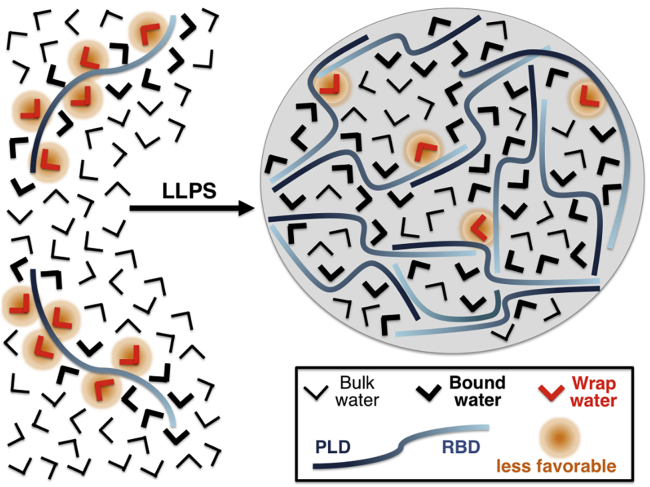
Based on the microscopic and spectroscopic evidence presented in this work, we propose a molecular mechanism involved in formation of LLPS droplets (Fig. 4). The key element of this concept is the existence of distinct hydration water populations around proteins having distinct thermodynamic properties. We propose that the thermodynamic driving force for LLPS is the minimization of the solvation or desolvation of hydrophobic patches, similar to the case of protein folding (46). This theory is in line with previous studies on elastin-like polypeptides and polymers, in which entropically favorable exclusion of water from the hydration shell was thought to play a central role in LLPS (62,73).
Figure 4.
Scheme showing the proposed water-mediated contribution of FUS to LLPS. Formation of phase-separated droplets is supported by an increase in tetrahedral coordination of water molecules (bound water; thick black) and minimization of less favorable water interactions (wrap water; red). To see this figure in color, go online.
Here, we are able to build on these previous studies by revealing that bound waters inside the droplet, stabilized by protein-water interactions, are the most abundant hydration water population, coexisting with water molecules in a bulk-like state. We speculate that from the point of view of the solvent, LLPS is thermodynamically promoted by the release of “unhappy” wrap water molecules from the protein hydration layer to the bulk.
The existence of distinct water populations hydrating polar and hydrophobic patches in the proteins hydration layer is not a unique feature of the studied system; it is, rather, a general characteristic of biomolecules (46,48,50). Therefore, the proposed microscopic mechanism for the water contribution to LLPS based on distinct water populations is expected to be generalizable. However, the final balance between the water populations will depend on specific properties of the system, e.g., relative abundance of hydrophilic and hydrophobic residues on the surface, their spatial distribution, surface morphology, surface charge, and presence of ions or cosolutes. An interesting perspective would therefore be to investigate up to what extent the balance between bound and wrap populations, and consequently the role of water in LLPS, can be manipulated by tuning these specific properties.
In general, the following thermodynamic driving forces have to be considered: protein-protein and protein-water interactions, as well as the changes in the water-water interactions. Our study focuses on the latter, previously less accessible part. Our results now provide an experimental handle to monitor how both forces act in a concerted manner (39). Previous work regarding LLPS of proteins (63,74,75) used a model of stickers and spacers to describe the main structural features of phase-separating protein systems, extending the classical Flory-Stockmayer theory based on polymers. Within the scope of the studied FUS protein system and based on the proposed molecular picture for the water contribution, we expect that bound waters will be found at locations in the droplet where interactions between arginine and tyrosine residues are formed. At the same time, wrap water molecules will be removed from hydrophobic patches of the protein. Combining our study and the sticker-spacer approach could allow us in the near future to link contributions from protein-protein, protein-water, and water-water interactions. Additionally, the techniques introduced here will allow future examination of the link between cosolutes and the solvent in influencing LLPS. Cosolutes affect the ability of proteins to undergo LLPS, and hence must participate in driving the relative balance of bound, wrap, and bulk water.
Conclusion
In conclusion, our spectroscopic measurements provide novel, to our knowledge, insight into the influence of LLPS on the hydrogen bond network. We propose a model for condensation that is based on a minimization of “wrap water molecules” and an increase in constrained water molecules compared to the bulk. This goes along with an increased protein concentration, favoring protein-protein interactions. Whereas the first process is favorable in terms of entropy, the latter process is favorable in terms of enthalpy. Thus, both processes contribute to a decrease in free energy promoting the formation of the droplets. Our results provide experimental evidence for a synergic effect of the solvent and FUS proteins in phase separation. This is anticipated to be a more general principle implicated in biomolecular condensate formation, which is of general biological relevance because of the relevance of assembling of membraneless compartments.
Author contributions
M.H. and J.T. designed research. J.A., E.M.A., and V.B. performed research and contributed analytic tools. E.M.A., J.A., and V.B. analyzed data. S.P. contributed to the interpretation. All authors discussed the results and wrote the manuscript.
Acknowledgments
We thank Dr. Dorothee Dormann for providing the FUS plasmids. We also thank Barbara Kachholz for helping with recombinant protein purification and Alejandro Colchero for designing the ATR liquid sample cell.
This work was funded by the Deutsche Forschungsgemeinschaft (German Research Foundation) under Germany’s Excellence Strategy EXC 2033–390677874–RESOLV and TA 167/6-3 (to J.T.) and DFG WI/2111-6 (to K.F.W.). M.H. acknowledges the financial support by the ERC Advanced grant “THz calorimetry.” Confocal laser-scanning and SR-SIM microscopy were funded by the German Research Foundation and the State Government of North Rhine-Westphalia (INST 213/840-1 FUGG).
Footnotes
J. Ahlers, E. M. Adams, and V. Bader contributed equally to this work.
Editor: Rohit Pappu.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.01.019.
Supporting citations
References (76, 77, 78, 79) appear in the Supporting material.
Supporting material
References
- 1.Riback J.A., Katanski C.D., Drummond D.A. Stress-triggered phase separation is an adaptive, evolutionarily tuned response. Cell. 2017;168:1028–1040.e19. doi: 10.1016/j.cell.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shin Y., Brangwynne C.P. Liquid phase condensation in cell physiology and disease. Science. 2017;357:eaaf4382. doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 3.Strom A.R., Emelyanov A.V., Karpen G.H. Phase separation drives heterochromatin domain formation. Nature. 2017;547:241–245. doi: 10.1038/nature22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang A., Conicella A.E., Fawzi N.L. A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBO J. 2018;37:e97452. doi: 10.15252/embj.201797452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woodruff J.B. Assembly of mitotic structures through phase separation. J. Mol. Biol. 2018;430:4762–4772. doi: 10.1016/j.jmb.2018.04.041. [DOI] [PubMed] [Google Scholar]
- 6.Molliex A., Temirov J., Taylor J.P. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–133. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larson A.G., Elnatan D., Narlikar G.J. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature. 2017;547:236–240. doi: 10.1038/nature22822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franzmann T.M., Jahnel M., Alberti S. Phase separation of a yeast prion protein promotes cellular fitness. Science. 2018;359:eaao5654. doi: 10.1126/science.aao5654. [DOI] [PubMed] [Google Scholar]
- 9.Banjade S., Rosen M.K. Phase transitions of multivalent proteins can promote clustering of membrane receptors. eLife. 2014;3:e04123. doi: 10.7554/eLife.04123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banani S.F., Lee H.O., Rosen M.K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017;18:285–298. doi: 10.1038/nrm.2017.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patel A., Lee H.O., Alberti S. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 12.Conicella A.E., Zerze G.H., Fawzi N.L. ALS mutations disrupt phase separation mediated by α-helical structure in the TDP-43 low-complexity C-terminal domain. Structure. 2016;24:1537–1549. doi: 10.1016/j.str.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo L., Kim H.J., Shorter J. Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell. 2018;173:677–692.e20. doi: 10.1016/j.cell.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wegmann S., Eftekharzadeh B., Hyman B.T. Tau protein liquid-liquid phase separation can initiate tau aggregation. EMBO J. 2018;37:e98049. doi: 10.15252/embj.201798049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofweber M., Hutten S., Dormann D. Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell. 2018;173:706–719.e13. doi: 10.1016/j.cell.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 16.Qamar S., Wang G., St George-Hyslop P. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell. 2018;173:720–734.e15. doi: 10.1016/j.cell.2018.03.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alberti S., Dormann D. Liquid-liquid phase separation in disease. Annu. Rev. Genet. 2019;53:171–194. doi: 10.1146/annurev-genet-112618-043527. [DOI] [PubMed] [Google Scholar]
- 18.Guo W., Naujock M., Van Den Bosch L. HDAC6 inhibition reverses axonal transport defects in motor neurons derived from FUS-ALS patients. Nat. Commun. 2017;8:861. doi: 10.1038/s41467-017-00911-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamelgarn M., Chen J., Zhu H. ALS mutations of FUS suppress protein translation and disrupt the regulation of nonsense-mediated decay. Proc. Natl. Acad. Sci. USA. 2018;115:E11904–E11913. doi: 10.1073/pnas.1810413115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morlando M., Dini Modigliani S., Bozzoni I. FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J. 2012;31:4502–4510. doi: 10.1038/emboj.2012.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rulten S.L., Rotheray A., Caldecott K.W. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res. 2014;42:307–314. doi: 10.1093/nar/gkt835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang W.-Y., Pan L., Tsai L.-H. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat. Neurosci. 2013;16:1383–1391. doi: 10.1038/nn.3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yasuda K., Zhang H., Mili S. The RNA-binding protein Fus directs translation of localized mRNAs in APC-RNP granules. J. Cell Biol. 2013;203:737–746. doi: 10.1083/jcb.201306058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang T., Wu Y.-C., Wang J. FUS regulates activity of MicroRNA-mediated gene silencing. Mol. Cell. 2018;69:787–801.e8. doi: 10.1016/j.molcel.2018.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mateju D., Franzmann T.M., Alberti S. An aberrant phase transition of stress granules triggered by misfolded protein and prevented by chaperone function. EMBO J. 2017;36:1669–1687. doi: 10.15252/embj.201695957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang J., Lim L., Song J. A unified mechanism for LLPS of ALS/FTLD-causing FUS as well as its modulation by ATP and oligonucleic acids. PLoS Biol. 2019;17:e3000327. doi: 10.1371/journal.pbio.3000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murthy A.C., Fawzi N.L. The (un)structural biology of biomolecular liquid-liquid phase separation using NMR spectroscopy. J. Biol. Chem. 2020;295:2375–2384. doi: 10.1074/jbc.REV119.009847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dao T.P., Kolaitis R.-M., Castañeda C.A. Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol. Cell. 2018;69:965–978.e6. doi: 10.1016/j.molcel.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conicella A.E., Dignon G.L., Fawzi N.L. TDP-43 α-helical structure tunes liquid-liquid phase separation and function. Proc. Natl. Acad. Sci. USA. 2020;117:5883–5894. doi: 10.1073/pnas.1912055117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato M., Han T.W., McKnight S.L. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nott T.J., Petsalaki E., Baldwin A.J. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 2015;57:936–947. doi: 10.1016/j.molcel.2015.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murray D.T., Kato M., Tycko R. Structure of FUS protein fibrils and its relevance to self-assembly and phase separation of low-complexity domains. Cell. 2017;171:615–627.e16. doi: 10.1016/j.cell.2017.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murthy A.C., Dignon G.L., Fawzi N.L. Molecular interactions underlying liquid-liquid phase separation of the FUS low-complexity domain. Nat. Struct. Mol. Biol. 2019;26:637–648. doi: 10.1038/s41594-019-0250-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dormann D., Rodde R., Haass C. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010;29:2841–2857. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marrone L., Drexler H.C.A., Sterneckert J. FUS pathology in ALS is linked to alterations in multiple ALS-associated proteins and rescued by drugs stimulating autophagy. Acta Neuropathol. 2019;138:67–84. doi: 10.1007/s00401-019-01998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cinar S., Cinar H., Winter R. Pressure-sensitive and osmolyte-modulated liquid-liquid phase separation of eye-lens γ-crystallins. J. Am. Chem. Soc. 2019;141:7347–7354. doi: 10.1021/jacs.8b13636. [DOI] [PubMed] [Google Scholar]
- 37.Maharana S., Wang J., Alberti S. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science. 2018;360:918–921. doi: 10.1126/science.aar7366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cinar H., Cinar S., Winter R. Pressure-induced dissolution and reentrant formation of condensed, liquid-liquid phase-separated elastomeric α-elastin. Chemistry. 2018;24:8286–8291. doi: 10.1002/chem.201801643. [DOI] [PubMed] [Google Scholar]
- 39.Ribeiro S.S., Samanta N., Marcos J.C. The synergic effect of water and biomolecules in intracellular phase separation. Nat. Rev. Chem. 2019;3:552–561. [Google Scholar]
- 40.Reddy G., Straub J.E., Thirumalai D. Dry amyloid fibril assembly in a yeast prion peptide is mediated by long-lived structures containing water wires. Proc. Natl. Acad. Sci. USA. 2010;107:21459–21464. doi: 10.1073/pnas.1008616107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tros M., Zheng L., Woutersen S. Picosecond orientational dynamics of water in living cells. Nat. Commun. 2017;8:904. doi: 10.1038/s41467-017-00858-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arya S., Singh A.K., Mukhopadhyay S. Femtosecond hydration map of intrinsically disordered α-synuclein. Biophys. J. 2018;114:2540–2551. doi: 10.1016/j.bpj.2018.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fisette O., Päslack C., Schäfer L.V. Hydration dynamics of a peripheral membrane protein. J. Am. Chem. Soc. 2016;138:11526–11535. doi: 10.1021/jacs.6b07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaslavsky B.Y., Ferreira L.A., Uversky V.N. The solvent side of proteinaceous membrane-less organelles in light of aqueous two-phase systems. Int. J. Biol. Macromol. 2018;117:1224–1251. doi: 10.1016/j.ijbiomac.2018.06.030. [DOI] [PubMed] [Google Scholar]
- 45.Zaslavsky B.Y., Uversky V.N. In aqua veritas: the indispensable yet mostly ignored role of water in phase separation and membrane-less organelles. Biochemistry. 2018;57:2437–2451. doi: 10.1021/acs.biochem.7b01215. [DOI] [PubMed] [Google Scholar]
- 46.Chandler D. Interfaces and the driving force of hydrophobic assembly. Nature. 2005;437:640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- 47.Sarupria S., Garde S. Quantifying water density fluctuations and compressibility of hydration shells of hydrophobic solutes and proteins. Phys. Rev. Lett. 2009;103:037803. doi: 10.1103/PhysRevLett.103.037803. [DOI] [PubMed] [Google Scholar]
- 48.Jamadagni S.N., Godawat R., Garde S. Hydrophobicity of proteins and interfaces: insights from density fluctuations. Annu. Rev. Chem. Biomol. Eng. 2011;2:147–171. doi: 10.1146/annurev-chembioeng-061010-114156. [DOI] [PubMed] [Google Scholar]
- 49.Monroe J.I., Shell M.S. Computational discovery of chemically patterned surfaces that effect unique hydration water dynamics. Proc. Natl. Acad. Sci. USA. 2018;115:8093–8098. doi: 10.1073/pnas.1807208115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Monroe J., Barry M., Shell M.S. Water structure and properties at hydrophilic and hydrophobic surfaces. Annu. Rev. Chem. Biomol. Eng. 2020;11:523–557. doi: 10.1146/annurev-chembioeng-120919-114657. [DOI] [PubMed] [Google Scholar]
- 51.Xi E., Venkateshwaran V., Garde S. Hydrophobicity of proteins and nanostructured solutes is governed by topographical and chemical context. Proc. Natl. Acad. Sci. USA. 2017;114:13345–13350. doi: 10.1073/pnas.1700092114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cyran J.D., Donovan M.A., Backus E.H.G. Molecular hydrophobicity at a macroscopically hydrophilic surface. Proc. Natl. Acad. Sci. USA. 2019;116:1520–1525. doi: 10.1073/pnas.1819000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shin S., Willard A.P. Characterizing hydration properties based on the orientational structure of interfacial water molecules. J. Chem. Theory Comput. 2018;14:461–465. doi: 10.1021/acs.jctc.7b00898. [DOI] [PubMed] [Google Scholar]
- 54.Conti Nibali V., Havenith M. New insights into the role of water in biological function: studying solvated biomolecules using terahertz absorption spectroscopy in conjunction with molecular dynamics simulations. J. Am. Chem. Soc. 2014;136:12800–12807. doi: 10.1021/ja504441h. [DOI] [PubMed] [Google Scholar]
- 55.Ebbinghaus S., Meister K., Havenith M. Antifreeze glycoprotein activity correlates with long-range protein-water dynamics. J. Am. Chem. Soc. 2010;132:12210–12211. doi: 10.1021/ja1051632. [DOI] [PubMed] [Google Scholar]
- 56.Png G.M., Falconer R.J., Abbott D. Tracking aggregation and fibrillation of globular proteins using terahertz and far-infrared spectroscopies. IEEE Trans. Terahertz Sci. Technol. 2016;6:45–53. [Google Scholar]
- 57.Novelli F., Ostovar Pour S., Davis J.A. Time-domain THz spectroscopy reveals coupled protein-hydration dielectric response in solutions of native and fibrils of human lysozyme. J. Phys. Chem. B. 2017;121:4810–4816. doi: 10.1021/acs.jpcb.7b02724. [DOI] [PubMed] [Google Scholar]
- 58.Adams E.M., Lampret O., Havenith M. Solvent dynamics play a decisive role in the complex formation of biologically relevant redox proteins. Phys. Chem. Chem. Phys. 2020;22:7451–7459. doi: 10.1039/d0cp00267d. [DOI] [PubMed] [Google Scholar]
- 59.Böhm F., Schwaab G., Havenith M. Mapping hydration water around alcohol chains by THz calorimetry. Angew. Chem. Int. Ed. Engl. 2017;56:9981–9985. doi: 10.1002/anie.201612162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Conti Nibali V., Pezzotti S., Havenith M. Wrapping up hydrophobic hydration: locality matters. J. Phys. Chem. Lett. 2020;11:4809–4816. doi: 10.1021/acs.jpclett.0c00846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brady J.P., Farber P.J., Kay L.E. Structural and hydrodynamic properties of an intrinsically disordered region of a germ cell-specific protein on phase separation. Proc. Natl. Acad. Sci. USA. 2017;114:E8194–E8203. doi: 10.1073/pnas.1706197114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park S., Barnes R., Han S. Dehydration entropy drives liquid-liquid phase separation by molecular crowding. Commun. Chem. 2020;3:83. doi: 10.1038/s42004-020-0328-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang J., Choi J.-M., Hyman A.A. A molecular grammar governing the driving forces for phase separation of prion-like RNA binding proteins. Cell. 2018;174:688–699.e16. doi: 10.1016/j.cell.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zeng M., Chen X., Zhang M. Reconstituted postsynaptic density as a molecular platform for understanding synapse formation and plasticity. Cell. 2018;174:1172–1187.e16. doi: 10.1016/j.cell.2018.06.047. [DOI] [PubMed] [Google Scholar]
- 65.Jo Y., Jung Y. Interplay between intrinsically disordered proteins inside membraneless protein liquid droplets. Chem. Sci. (Camb.) 2020;11:1269–1275. doi: 10.1039/c9sc03191j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burke K.A., Janke A.M., Fawzi N.L. Residue-by-residue view of in vitro FUS granules that bind the C-terminal domain of RNA polymerase II. Mol. Cell. 2015;60:231–241. doi: 10.1016/j.molcel.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wei M.-T., Elbaum-Garfinkle S., Brangwynne C.P. Phase behaviour of disordered proteins underlying low density and high permeability of liquid organelles. Nat. Chem. 2017;9:1118–1125. doi: 10.1038/nchem.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Morawietz T., Marsalek O., Markland T.E. The interplay of structure and dynamics in the Raman spectrum of liquid water over the full frequency and temperature range. J. Phys. Chem. Lett. 2018;9:851–857. doi: 10.1021/acs.jpclett.8b00133. [DOI] [PubMed] [Google Scholar]
- 69.Maréchal Y. Infrared spectra of water. I. Effect of temperature and of H/D isotopic dilution. J. Chem. Phys. 1991;95:5565–5573. [Google Scholar]
- 70.Maréchal Y. The molecular structure of liquid water delivered by absorption spectroscopy in the whole IR region completed with thermodynamics data. J. Mol. Struct. 2011;1004:146–155. [Google Scholar]
- 71.Myatt D.P., Hatter L., Clifton L.A. Monomeric green fluorescent protein as a protein standard for small angle scattering. Biomed. Spectrosc. Imaging. 2017;6:123–134. [Google Scholar]
- 72.Bernadó P., Blackledge M. A self-consistent description of the conformational behavior of chemically denatured proteins from NMR and small angle scattering. Biophys. J. 2009;97:2839–2845. doi: 10.1016/j.bpj.2009.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reichheld S.E., Muiznieks L.D., Sharpe S. Direct observation of structure and dynamics during phase separation of an elastomeric protein. Proc. Natl. Acad. Sci. USA. 2017;114:E4408–E4415. doi: 10.1073/pnas.1701877114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choi J.-M., Holehouse A.S., Pappu R.V. Physical principles underlying the complex biology of intracellular phase transitions. Annu. Rev. Biophys. 2020;49:107–133. doi: 10.1146/annurev-biophys-121219-081629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Choi J.-M., Dar F., Pappu R.V. LASSI: a lattice model for simulating phase transitions of multivalent proteins. PLoS Comput. Biol. 2019;15:e1007028. doi: 10.1371/journal.pcbi.1007028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schindelin J., Arganda-Carreras I., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schwaab G., Sebastiani F., Havenith M. Ion hydration and ion pairing as probed by THz spectroscopy. Angew. Chem. Int. Ed. Engl. 2019;58:3000–3013. doi: 10.1002/anie.201805261. [DOI] [PubMed] [Google Scholar]
- 78.Bertie J.E., Lan Z. Infrared intensities of liquids XX: the intensity of the OH stretching band of liquid water revisited, and the best current values of the optical constants of H2O(l) at 25°C between 15,000 and 1 cm−1. Appl. Spectrosc. 1996;50:1047–1057. [Google Scholar]
- 79.Ebbinghaus S., Kim S.J., Havenith M. An extended dynamical hydration shell around proteins. Proc. Natl. Acad. Sci. USA. 2007;104:20749–20752. doi: 10.1073/pnas.0709207104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.