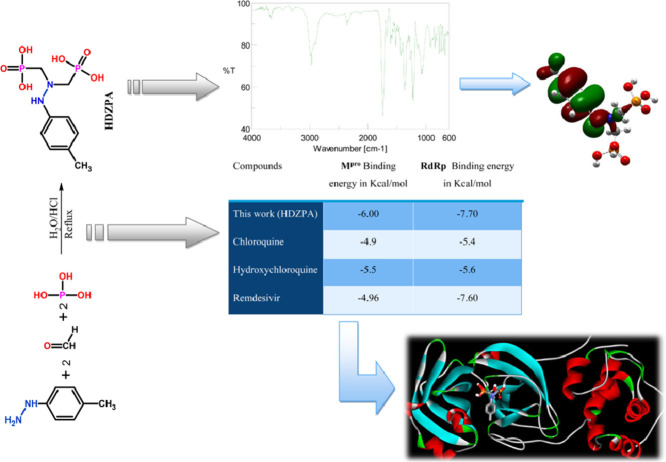
Abstract
A new α-Hydrazinophosphonic acid (HDZPA) has been synthesized and its molecular structure was determined using spectroscopic methods. The Density Functional Theory (DFT) at the B3LYP/6–31 G (d,p) level was utilized to determine the electronic properties, vibrational modes and active sites of the examined molecule. In this context, some quantum chemical parameters have been calculated in order to discuss the reactivity of the studied molecule. Also, the inhibition activity of the investigated α-Hydrazinophosphonic acid for SARS-CoV-2 main protease (Mpro) and RNA dependent RNA polymerase (RdRp) has been predicted using in silico docking.
Keywords: α-hydrazinophosphonic acid, Synthesis, Characterization, SARS-CoV-2 main protease, in silico docking, DFT
Graphical abstract
1. Introduction
The COVID-19 is an abbreviation of the Coronavirus disease 2019. At first, the COVID-19 is started as an epidemic in the Chinese city of Wuhan in December 2019, afterward it quickly propagated in Chinese territory and outside [1,2]. Two months later, the WHO declares the COVID-19 as a pandemic [3]. In general, the principal diseases caused by COVID-2019 are respiratory problems and gastrointestinal [4].
Presently, no known special, confirmed and efficient anti-coronavirus drug is discovered or developed. However, several drugs are also tested in clinical trials as effective treatments of COVID-19 patients such as the old antimalarial drug Hydroxychloroquine and the new antiviral drug developed for Ebola virus Remdesivir. In this context, a recent published paper reports that the treatment including Remdesivir and Chloroquine drugs inhibits the growth process of SARS-CoV-2 in vitro [5]. Furthermore, Remdesivir has proved an important efficacy against SARS-CoV-2 whether in vitro or in vivo and has also started its clinical experiments [6]. Also, several clinical tests realized in China show that Chloroquine presents a remarkable effect on the clinical results and viral clearance [7,8]. Another recent research suggests the use of Hydroxychloroquine as a potential treatment of COVID-19 patients [9]. In addition, the paper published by Didier Raoult et al. on March 2020 showed that the treatment containing Hydroxychloroquine and Azithromycin presents a significant reduction/disappearance of the viral load in COVID-19 patients [10].
The crystalline structure of SARS-CoV-2 main protease (Mpro) has been established at the first time by Liu et al., and the structure of Mpro is available for public access in the Protein Data Bank (PDB) [11]. Generally, this enzyme is known by its vital role in the transformation of the translated polyproteins [12]. On the other hand, RNA‐dependent RNA polymerase (RdRp) plays a pivotal role in virus replication and transcription of the viral genome [13]. So, Mpro and RdRp can be considered as targets to discover therapeutic agents to COVID-19.
In general, the physicochemical and electronic characteristics of drug molecules can affect their chemical and biological activities. Additionally, the quantum chemical calculations using Density Functional Theory (DFT) method are largely employed to determine the active sites of drugs and to correlate their activity with various quantum chemical parameters [14], [15], [16]. In this context, several biological activities of the bioactive molecules can be extensively studied using the DFT calculations [17], [18], [19].
The one-pot multicomponent reactions such as Mannich-type reaction and Kabachnik-Fields reaction are largely and efficiently used in organic synthesis to prepare the biologically active compounds [20], [21], [22], [23], [24]. Several bioactive and pharmaceutical compounds have been synthesized using one-pot multicomponent reactions including antiviral [25], anticancer [26], anti-HIV [27], antimalarial and antiinsecticidal [28], antipsychotic [29,30], antiparasitic [31], antidepressant [32], antibacterial and antimicrobial [33]. The Kabachnik-Fields reaction is a three-component reaction including a carbonyl, an amine and a dialkylephosphite or a trialkylephosphite. This reaction is very important in drug discovery studies and mainly used to synthesize α-aminophosphonates.
This paper represents the results of an experimental and theoretical study of a new α-Hydrazinophosphonic acid (HDZPA). So, the experimental part consists to synthesize the desired compound via one-pot three-component reactions and to determine its molecular structure using spectroscopic methods such as UV–Vis, FT-IR, 1H NMR, 13C NMR, and 31P NMR. On the other hand, the HDZPA has been theoretically investigated to locate their active sites, electronic and vibrational properties by means of DFT method at the B3LYP/6–31 G (d,p) level. The optimized molecular structures, the vibrational spectra, the highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO) properties, dipole moments (μ), Molecular Electrostatic Potentials maps (MEP), atomic charges, energy gap (ΔEGAP), hardness (η), local softness (σ), electronegativity (χ) and electrophilicity (ω) are calculated for of the studied molecule. Finally, the synthesized α-Hydrazinophosphonic acid was tested as a potential inhibitor for SARS-CoV-2 main protease (Mpro) and RNA dependent RNA polymerase (RdRp) using in silico docking to support drug discovery.
2. Experimental
2.1. Materials and spectroscopic details
All the chemical reagents employed for the HDZPA ligand synthesis were bought from Sigma-Aldrich and utilized without further purification. The UV–Vis spectrum of the synthesized HDZPA ligand was obtained in the range of 190–900 nm in aqueous solution by means of the Jasco V-650 UV–Vis spectrometer. In addition, the JASCO 4000 FTIR spectrometer was used to realize the FT-IR spectrum of the investigated ligand in solid state at room temperature, and the obtained vibration frequencies were listed in the range of 600–4000 cm−1. NMR spectra were recorded on a Bruker Avance 300 spectrophotometer operating at 300 MHz (1H) and 75 MHz (13C) at 298 K using tetramethylsilane (0 ppm) as the internal reference. NMR spectroscopic data were recorded in CDCl3 (δ= 7.26 ppm) using as internal standards the residual non-deuterated signal for 1H NMR and the deuterated solvent signal (δ= 77.1 ppm) for 13C NMR spectroscopy. Chemical shifts (δ) are given in ppm and coupling constants (J) are given in Hz. The following abbreviations are used for multiplicities: s = singlet, d = doublet, t = triplet, q = quartet, dd = doublet of doublets, and m = multiplet.
2.2. Synthesis of the α-Hydrazinophosphonic acid ligand
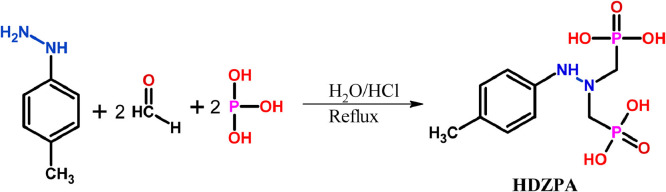
In this study, a new α-Hydrazinophosphonic acid ligand (HDZPA) was synthesized applying the Irani-Moedritzer method [34]. Accordingly, a mixture of 1.0 mmol of (4-methylphenyl)hydrazine and 2.0 mmol of H3PO3 has been dissolved in 50 ml of water and 25 ml of HCl, then the mixture was refluxed for 4 h at 100 °C (Fig. 1 ), whereas 2.0 mmol of formaldehyde 36% was added dropwise to this mixture. After the addition, the mixture was kept at the room temperature for another 3 h under reflux. Also, TLC analysis was used to check the completion of the reaction. After the reaction was completed, a rotary evaporator has been used to eliminate the solvent under reduced pressure. Finally, the obtained crude product was purified by means of chromatographic column of silica gel using ethyl acetate/MeOH (9.5:0.5, v/v). Moreover, the obtained brown solid (HDZPA) was characterized by UV–Vis, FT-IR, 1H NMR, 13C NMR, and 31P NMR.
Fig. 1.
Synthetic rout of the studied α-Hydrazinophosphonic acid ligand (HDZPA).
Yield 81%, M.p. 79.22 °C; IR (ATR, ν(cm−1)) : 3621 (O—H), 3315 (N—H), 3250 (C—HAr), 2980 (C—HAlph), 1611 (P-OH), 1269 (N—CAr), 1202 (P = O), 1168 (N—N), 1020 (N—CAlph), 684 (C-P); 1H NMR (300 MHz, CDCl3, δ(ppm)): 2.48 (s, 3H, -CH 3), 3.12 (d, 2JH- P = 11.8 Hz, 4H, -CH 2-), 5.08 (s, 4H, -OH), 7.18 (d, J = 7.9 Hz, 2H, H-Ar), 7.71 (d, J = 7.9 Hz, 2H, H-Ar), 8.92 (s, 1H, NH); 13C{1H} NMR: (75 MHz, CDCl3, δ(ppm)): 23.9 (1C, -CH3), 67.7 (2C, d, 1JC- P = 36.8 Hz, N—CH2-), 114.1 (2C, CHAr), 129.9 (2C, CHAr), 133.3 (1C, CH3-— C Ar), 141.8 (1C, N— C Ar); 31P NMR: (121 MHz, CDCl3, δ(ppm)): 8.77 (dt, 1JP- C = 87.1 Hz, 2JP- H = 116.4 Hz); UV–Vis (H2O), λmax (nm): 244.76 and 327.25. MS (70 eV) m/z (%): 311 (M + 1, 7.51), 310 (M +·, 100), 309 (M – 1, 4.2) (see Supporting Information).
2.3. Computational details
Throughout this study, we applied the Gaussian 09 W program package to implement all quantum chemical calculations [35]. The geometry of HDZPA ligand was entirely optimized utilizing the DFT method through B3LYP hybrid functional at 6–31 G (d,p) basis set [36,37]. Also, this theory has been utilized to determine the vibrational frequencies at the obtained optimized structure of HDZPA. We chose Density Functional Theory (DFT) with B3LYP/6–31 G (d, p) level because the B3LYP functional has shown good results for organic molecules. Also, the obtained results with B3LYP/6–31 G (d, p) level are in good agreement with the experimental data, especially with ATR-FTR and UV–Vis data. The 1H, 13C and 31P NMR spectra of HDZPA in the presence of CDCl3 as solvent are predicted using the GIAO method with the hybrid B3LYP at 6–31 G (d,p) basis set, while the electronic spectrum in the water was predicted by using the Time-dependent DFT calculations (TD-DFT) with the B3LYP/6–31 G (d,p) method. The VEDA 4 program has been used to carry out various vibrational parameters and PED calculations [38]. In addition, the calculated energies of the highest occupied molecular orbital (EHOMO) and lowest unoccupied molecular orbital (ELUMO) have been utilized to determine various quantum chemical parameters such as the energy gap (ΔEGAP), dipole moments (μ), hardness (η), local softness (σ), electronegativity (χ) and electrophilicity (ω). Moreover, the following equations were applied to compute the precedent parameters [39,40]:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
2.4. Molecular docking
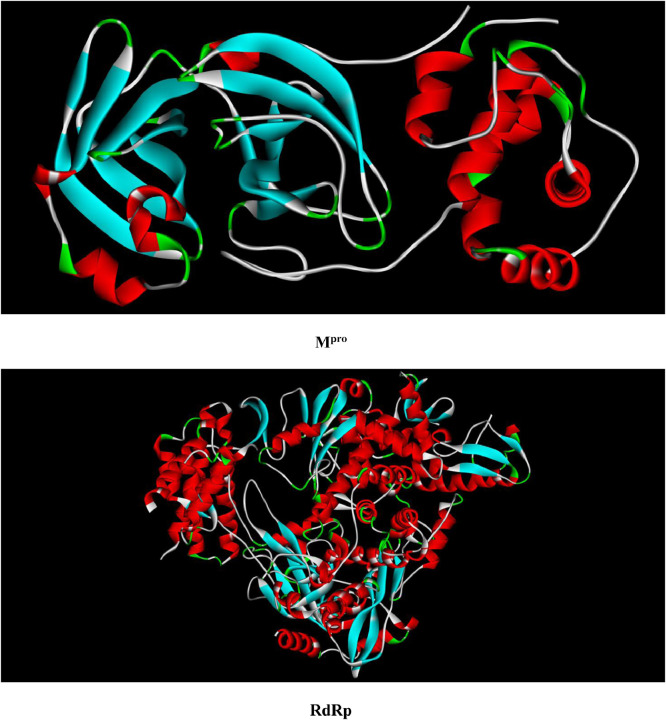
Molecular docking study was performed in order to evaluate binding affinity of the synthesized ligand against SARS-CoV-2 main protease (Mpro) and RNA dependent RNA polymerase (RdRp). The selection of Mpro as target for docking study is due to its important role in processing of translated polyproteins, while the selection of RdRp is due to its importance in replication and transcription of the viral genome. The crystal structures of Mpro and RdRp were extracted from the PDB protein database with codes of 6LU7 and 7BV2 (Fig. 2 ), respectively. For the HDZPA ligand we applied the obtained optimized geometry using DFT method at the B3LYP/6–31 G (d,p) level. So, the Autodock software version 4.2.6 has been utilized to execute the molecular docking process. The grid of 30×30×30 Ǻ3 was constructed to carry out docking simulations. The HDZPA-RdRp and HDZPA-RdRp complexes were visualized using Accelry's Discovery Studio Visualizer.
Fig. 2.
Crystal structures of Mpro and RdRp.
3. Results and discussion
3.1. Spectral study
3.1.1. UV–vis analysis
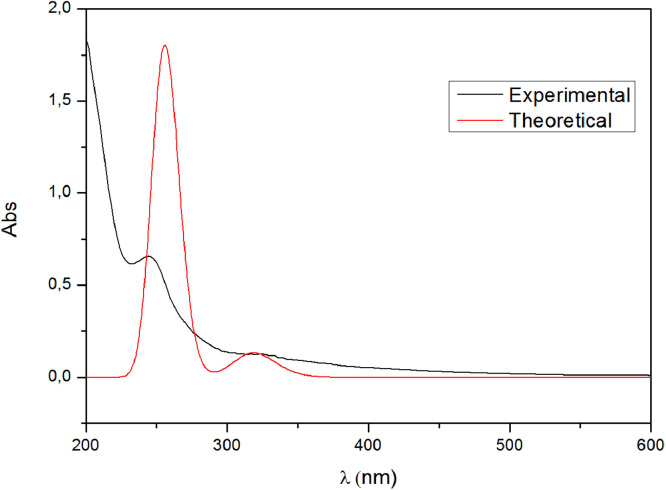
The experimental and theoretical UV–vis spectra of HDZPA (Fig. 3 ) were obtained in water at room temperature and they reveal tow absorption bands in the ultraviolet domain. The registered experimental spectrum of the investigated ligand illustrates a band at 244.76 nm related to the π→π* transitions of the C = C group of the aromatic ring, which indicates that the electron orbital jumps is from π bonding orbitals to π anti-bonding orbitals. Also, the band appeared at 327.25 nm is assigned to the n→π* transitions related to the presence of heteroatoms (O, N and P) on the molecular structure of HDZPA, which indicates that the electron orbital jumps is from non-bonding orbitals to π anti-bonding orbitals. In an atom or molecule, this type of transitions shows that electrons move from low energy levels to higher energy levels. Fig. 6 indicates that the experimental and calculated absorption spectra are in good agreement. Also, the calculated spectra shows a band at 256.07 nm associated to the π→π* transitions. In addition, the calculated band observed at 320.07 nm is related to the n→π* transitions of heteroatoms.
Fig. 3.
Experimental and calculated UV–Vis spectra of HDZPA.
Fig. 6.
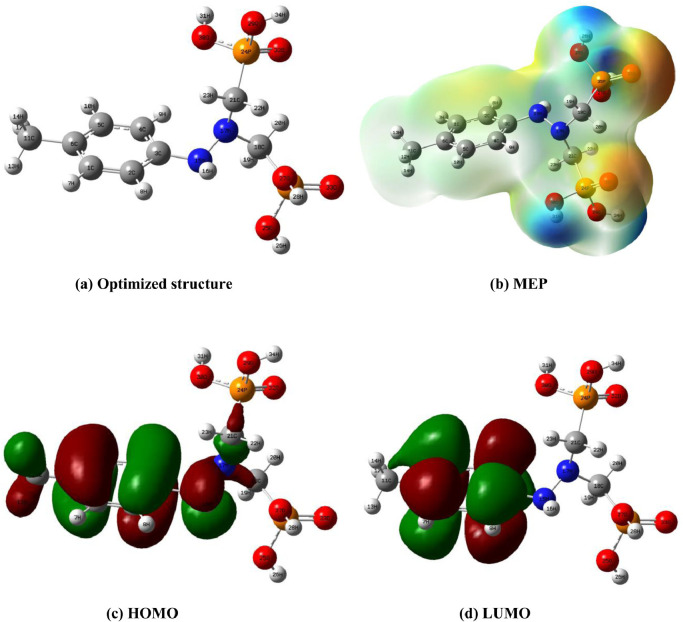
Optimized structure, MEP maps, HOMO and LUMO frontier orbitals of HDZPA.
3.1.2. Vibrational analysis
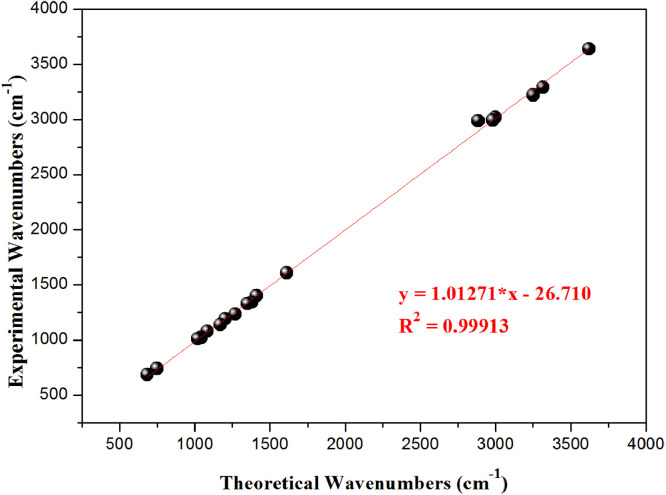
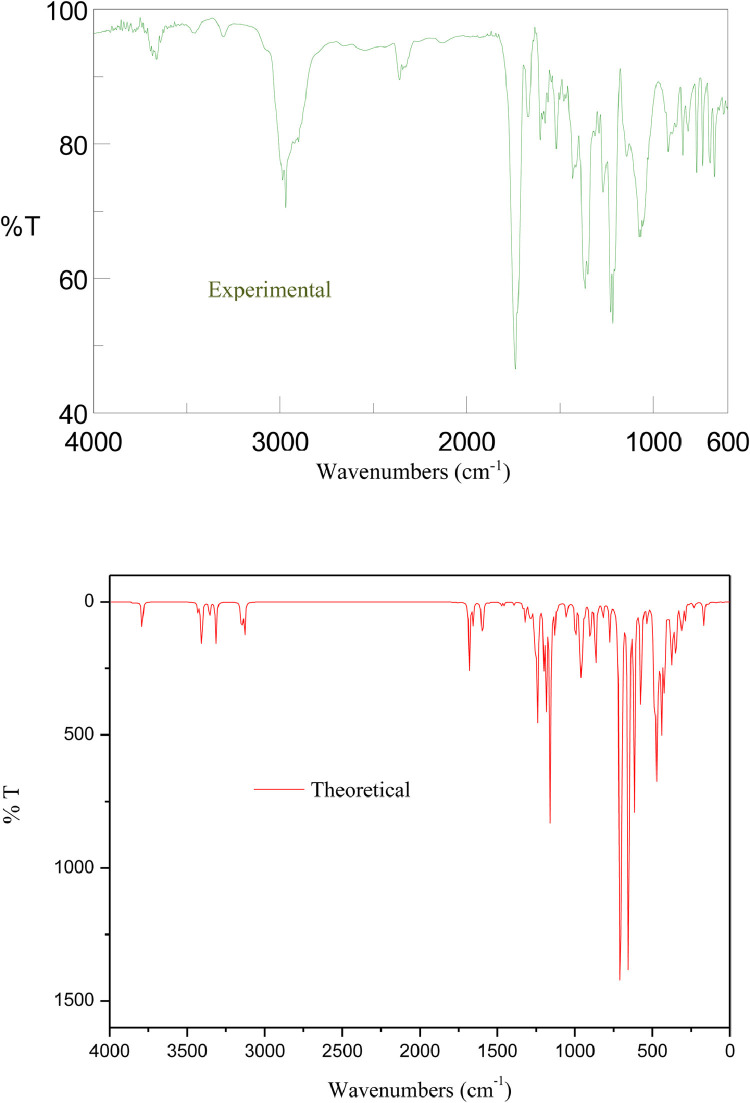
The vibrational frequencies, the vibrational mode assignments and the potential energy distributions (PED) of the characteristic groups of HDZPA are presented in Table 1 . These results were selected from the obtained experimental and calculated infrared spectra. Also, the calculated frequencies are scaled using a scaling factor of 0.9614 [41]. As a result, a good harmony has been observed between the scaling calculated frequencies and the experimental frequencies (Fig. 5). Generally, the objective of the vibrational analysis is to specify vibrational modes of a molecule. At B3LYP/6–31 G (d,p), the studied compound with 35 atoms donates (3N-6) i.e. 99 vibrational modes in the range 20.02–4126.00 cm−1. So, the attribution of these 99 vibrational modes has been performed to establish some correlation between structure and spectrum. These 99 vibrational modes distinguish 34 stretching modes, 33 bending modes and 32 torsional modes. All frequencies below 600 cm−1 are not presented in Table 1 owing to their very complex mode compositions. Therefore, the analysis of the IR spectra presented in Fig. 4 and Table 1 confirms the existence of the following vibrational modes:
Table 1.
Experimental and calculated values of wavenumber for the selected vibrations of HDZPA.
| Assignment (% PED) | Vibrational frequency (cm−1) |
||
|---|---|---|---|
| Experimental | Theoretical |
||
| Unscaled | Scaled | ||
| ν O – H (98) | 3621 (ν O – H) | 3788 | 3641 |
| ν N – H (100) | 3315 (ν N – H) | 3425 | 3293 |
| ν C – HAr (96) | 3250 (C – HAr) | 3353 | 3223 |
| νs C – HAlph (97), β H—C-H (96) | 2998 (νs C – HAlph) | 3142 | 3020 |
| νs C – HAlph (94), β HCN (24) | 2980 (νs C – HAlph) | 3117 | 2996 |
| νas C – HAlph (84), β HCC (29), ρ CCHC (45), ρout CCCH (57) | 2884 (νas C – HAlph) | 3108 | 2988 |
| ν C = C (42), ν P – OH (40), β CCH (22), ρ CCHC (14) | 1611 (ν P – OH) | 1674 | 1609 |
| γ C—H (25), β HCN (20), ρ OPHC (18) | 1411 (γ C—H) | 1459 | 1402 |
| α C—H (34), β HCN (16), β HCH (12) | 1378 (α C—H) | 1393 | 1342 |
| ω C—H (39), β HCH (10), ρ HCHP (13) | 1351 (ω C—H) | 1382 | 1328 |
| ν N – CAr (52), α C—H (31), β CCH (13), β CCN (18), β HCH (22), ρout CCCH (11) | 1269 (ν N – CAr) | 1283 | 1234 |
| ν P = O (33), γ C—H (28), γ PCH (34) | 1202 (ν P = O) | 1242 | 1194 |
| ν N – N (22), γ C—H (14), γ PCH (31), ω C—H (24) | 1168 (ν N – N) | 1186 | 1140 |
| ω C—H (24), β HCP (32), β HCH (25), ρ HCNC (16), ρ OPCN (19) | 1084 (ω C—H) | 1125 | 1081 |
| γ C—H (35), δ C—H (10), β C—N (12), ρ P-O (43) | 1041 (ρ P-O) | 1066 | 1024 |
| ν N – CAlph (34), β HCC (23), β CCC (28), ρ CCHC (10) | 1020 (ν N – CAlph) | 1052 | 1011 |
| δ C—H (41), γ C-P (27), β HCC (13), β OPC (12), β HOP (11), ρ OPCN (10) | 748 (γ C-P) | 772 | 742 |
| δ C—H (17), ν C-P (24), β HOP (26), β HCH (32), ρ OPC (14) | 684 (ν C-P) | 714 | 686 |
ν: stretching, γ: rocking, α: scissoring, δ: twisting, ω: wagging, β: in plane bending, ρ: torsion, ρout : out plane bending, s: symmetric, as: asymmetric.
Fig. 5.
Correlation diagram between the calculated and experimental wavenumbers of HDZPA.
Fig. 4.
Experimental and calculated IR spectra of HDZPA.
3.2.1. Phosphonic acid group (-PO(OH)2) vibrations
The phosphonic acid group (-PO(OH)2) presents several vibrational modes such as ν(P = O), ν(–OH), ν(P–O), and ν(C–P). Generally, the vibrational modes related with this group are observed as medium to strong absorption peaks in IR spectra. The peak which is highest in wavenumber appeared at 3620 cm−1 is attributed to the O—H stretching vibrations. Also, the P = O stretching vibrations is observed at 1202 cm−1, while the intense peak appeared at 1611 cm−1 is related to the P–O stretching vibrations. The stretching vibrations of C–P are located at 684 cm−1. Also, the presence of hydrogen bonds in the molecular structure of HDZPA can expanded the characteristic peaks of ν(P = O) and ν(P–O) and there corresponding frequencies can be down-shifted below expected regions. The symmetric and asymmetric stretch modes of the (P-O) are appeared at 1020 and 1102 cm−1, respectively. In addition, the deformation mode of the (P-O-H) appears clearly at 983 cm−1.
3.2.2. N—H stretching vibrations
In general, the N—H stretching vibrations are facilely determined by the apparition of a sharp peak at highest wavenumbers. For the investigated molecule, the asymmetric and symmetric stretching vibrations of N—H are located at 3315 cm−1. Also, the N—H out of plane bending vibrations are situated at 621 cm−1. The N—H bending vibrations of the secondary amine (N—H wagging) are observed between 652 and 910 cm−1.
3.2.3. C—H vibrations
The molecular structure of HDZPA indicates the presence of the aromatic and aliphatic C—H stretching vibrations. So, the obtained peak between 3005 and 3250 cm−1 can be attributed to the aromatic C—H asymmetric and symmetric stretching vibrations. Also, the asymmetric and symmetric stretching vibrations of the aliphatic C—H (CH2 and CH3 groups) are obtained at 2980 and 2848 cm−1, respectively. Also, the out of plane and in plane bending vibrations of the aromatic C—H have been specified and presented in Table 1. In addition, all peaks appeared between 1400 and1250 cm−1 are assigned to the deformation modes of C—H. In experimental spectra, the CH2 scissoring is attributed to 1495–1445 cm−1, while the CH2 wagging is assigned to 1358–1322 cm−1.
3.2.5. C—N vibrations
The attributed peaks of the aromatic and aliphatic C—N stretching vibrations of the studied ligand are situated at 1269 cm−1 and 1020 cm−1, respectively. The thin peak located at 822 cm−1 may be due to C—N band vibration. Also, the calculated in plane bending vibrations of C—N are observed at 468 and 497 cm−1.
3.2.7. N—N vibrations
The characteristic peak of the stretching vibrations of N—N group is appeared with a medium intensity at 1168 cm−1. Also, the appeared peak at 791 cm−1 is associated to the symmetric wag. Moreover, the peak situated at 885 cm−1 may be assigned to the in-plane bending vibrations of C—N-N.
3.2.8. C—C and c = c vibrations
Generally, the C—C stretching modes of aromatic rings are appeared between 1400 and 1650 cm−1. Consequently, for the studied compound, the C—C stretching vibrations are observed at 1520 and 1742 cm−1 in the experimental IR spectrum. Also, the aromatic C = C semicircular stretching vibrations are appeared between 1625 and 1430 cm−1.
3.1.3. NMR analysis
The analysis of the experimental 1H NMR spectrum of HDZPA permits to determine the following characteristic signals: a singular signal located at 2.48 ppm is related to the three protons of – CH 3 group. The 4H of the two aliphatic – CH 2 – groups are appeared as a doublets signal at 3.12 ppm and this can be proved by the obtained values of integration (3.9 ≈ 4H) and coupling constant (J = 11.8 Hz) of the corresponded signal. Also, the 4H of the phosphonic acid groups ( – OH) are observed as a singular signal at 5.08 ppm. Moreover, the observed signals in the region of 7.05–7.15 ppm are attributed to the aromatic protons ( – CH Ar –, 4H). The proton of the – NH group is appeared as a singular signal at 8.92 ppm.
Furthermore, the elucidation of the obtained experimental 13C NMR spectrum of HDZPA shows the presence of the following characteristic signals: the carbon of the – CH3 group was observed as a singular signal at 23.89 ppm. The tow signals situated at 67.70 ppm and 68.19 ppm can be associated to the carbons of the two N – CH2 – groups. The carbons of the aromatic ring are observed between 114.14 ppm and 133.37 ppm.
The experimental 31P NMR spectrum of HDZPA shows two signals at 8.42 ppm and 9.13 ppm, which confirms the presence of the phosphorus atoms in the molecular structure of ligand. Also, the obtained signals of the phosphorus atoms are revealed as a triplet due to their coupling with the two protons of the methylene group (N – CH 2 – P). Indeed, in the case where the molecule contains a phosphorus atom, then this one couples with the nuclei of spin ½. The NMR signals are therefore duplicated. Thus we will observe coupling constants of the 3 J and 4 J type.
Thus, for HDZPA all the carbons and protons shown will couple with phosphorus. The most striking example is that of carbons of methylene groups (N – CH2 – P) for which there will be a coupling constant J = 13.9 Hz.
On the other hand, the results presented in Table 2 show that the calculated NMR chemical shifts for HDZPA are in agreement with the experimental chemical shifts. On the other hand, we observe better correlations are obtained for the H and P atoms in solvent than the C atoms. Also, we observe in the 1H NMR results that the theoretical values were larger than those obtained in the experimental measurements.
Table 2.
Experimental and calculated NMR chemical shifts (δ in ppm) for HDZPA.
|
1H NMR |
13C NMR |
31P NMR |
|||
|---|---|---|---|---|---|
| Experimental | Calculated | Experimental | Calculated | Experimental | Calculated |
| 2.48 | 2.10 | 23.84 | 12.55 | 8.42 | 8.94 |
| 3.12 | 3.90 | 67.95 | 52.03 | 9.13 | 8.95 |
| 5.08 | 5.10 | 68.2 | 57.34 | ||
| 7.05 | 8.50 | 114.14 | 113.76 | ||
| 7.08 | 8.58 | 120.36 | 115.22 | ||
| 7.12 | 8.58 | 122.15 | 115.31 | ||
| 7.15 | 8.61 | 129.35 | 115.79 | ||
| 8.92 | 8.80 | 129.67 | 125.11 | ||
| 137.37 | 130.98 | ||||
3.2. DFT study
3.2.1. Optimized molecular structures
In Fig. 2(a) we present the obtained optimized molecular geometry of HDZPA ligand. Additionally, the calculated value of total energy for this molecule at the optimal structure was −43,437.641 eV, which signifies the energy of the more stable conformation of the studied ligand.
3.2.2. Frontier molecular orbital analysis
Generally, the reactivity and the stability of drug molecules can be described significantly using the HOMO and LUMO orbitals and their energies. Also, the aptitude of a molecule to contribute electrons to an electrophilic species can be examined by HOMO, while the ability of a molecule to accept electrons from nucleophilic species may be determined using LUMO [42]. So, the capacity of a molecule to give electrons to an acceptor species is favored by the high values of EHOMO [43], whereas the ability of a molecule to take electrons is preferred by the low values of ELUMO [44]. On the other hand, the energy gap (ΔEGAP) of drug molecules is an important parameter to determine their reactivity and stability [45]. Additionally, the calculated value of ΔEGAP can explain the charge transport interactions in the molecule. In general, ΔEGAP represents the necessary energy to excite the electrons of a molecule. Also, molecules can be highly chemically reactive, unstable and excited easily when ΔEGAP is smaller, while it can be very stable and less chemically reactive if ΔEGAP is very large [46].
Figs. 6(c) and 6(d) illustrate the calculated HOMO and LUMO orbitals for the HDZPA ligand. Generally, the green color characterizes the negative phases, whereas the red color indicates the positive phases [39]. So, we observe from Figs. 6(c) and 6(d) that the HOMO and LUMO are frequently located on the aromatic ring, the amino and methylene groups. On the outer hand, we observe from Table 3 that the HDZPA molecule has an elevated value of EHOMO and ELUMO, which signifies that the HDZPA can liberate electrons to an acceptor molecule. Moreover, the calculated value of ΔEGAP indicates the stability and the reactive of HDZPA.
Table 3.
Calculated quantum chemical parameters of HDZPA using DFT/B3LYP 6–31 G (d,p) method.
| Quantum chemical parameters | HDZPA |
|---|---|
| ETot (eV) | – 43437.641 |
| EHOMO (eV) | – 7.9536200 |
| ELUMO (eV) | – 0.1333359 |
| ΔEGAP (eV) | 7.8202841 |
| μ (Debye) | 4.87960000 |
| η (eV) | 3.91014205 |
| Σ | 0.25574518 |
| χ (eV) | 4.04347795 |
| ω | 2.09068030 |
3.2.3. Molecular surface electrostatic potential (MEP)
One of the helpful ways of quantum chemical calculations used to determine the active sites of drug molecules is the measure of their molecular electrostatic potential (MEP) maps. In general, the MEP of molecules is associated to their electronic densities and can elucidate their chemical activities, electrostatic effects and partial charges. Also, MEP map is a visual method which we can used largely to identify the relative polarity of molecules and to establish their negative and positive electrostatic potentials [47]. Additionally, the total electrons density plotted with electrostatic potential surface can be used to characterize the charge density, the dimension and the form of the active sites and to locate the place of the chemical reactivity of molecules. So, the 3D maps presentation of the electrostatic potential variation is represented by a gradient of colors. In principle, the negative electrostatic potentials regions related to the electrophilic reactions are graphically presented in yellow and red colors, while the positive electrostatic potentials corresponding to the nucleophilic reactions sites are illustrated in blue color. Moreover, the zones of nil potential are presented in green color and the evolution of the potential obeys the sequence red<orange<yellow<green<blue [48].
The calculated 3D MEP maps of HDZPA by means of DFT method are displayed in Fig. 6(b). The examination of the achieved MEP map elucidates that the yellow and red zones are situated on the O33, O32, O27, O29, O30, and O25, which proves that these atoms are the probable sites of the electrophilic reactions. Also, the aromatic ring is accounted as negative regions. Conversely, the green and blue colors are observed in the region of the carbon and hydrogen atoms, which represent the positive sites designated for the nucleophilic reactions.
3.2.4. Mulliken atomic charges
The estimation of the partial atomic charges of molecules can be provided by Mulliken charges analysis. Furthermore, the adsorptive sites of drug molecules can be proved by determination of their atomic Mulliken charges. In this context, the obtained values of the atomic Mulliken charges of HDZPA are regrouped in Table 4 . The examination of the obtained results indicates that the oxygen and nitrogen atoms have the most negative charges, which is due to the molecular relaxation. As well, the hydrogen atoms cover the positive charges. In particular, the largest part of negative charges are localized on the O27, O25, O29, O30, O32, O33, N15 and N17 atoms of HDZPA, which are possibly the active centers of adsorption [49]. Generally, the heteroatoms (N and O) can share their pairs of electrons with acceptor molecules. In addition, the P24 and P35 atoms of HDZPA have the majority positive charges.
Table 4.
Calculated Mullikan atomic charges of HDZPA.
| Atom | Mulliken Charge |
|---|---|
| C1 | - 0.1602500 |
| C2 | - 0.2060730 |
| C3 | 0.2726000 |
| C4 | - 0.1716900 |
| C5 | - 0.1688970 |
| C6 | - 0.0401750 |
| H7 | 0.1619580 |
| H8 | 0.1675950 |
| H9 | 0.1784940 |
| H10 | 0.1600150 |
| C11 | - 0.3415360 |
| H12 | 0.1276730 |
| H13 | 0.1287390 |
| H14 | 0.1330520 |
| N15 | - 0.5272320 |
| H16 | 0.3072080 |
| N17 | - 0.4714520 |
| C18 | - 0.2857870 |
| H19 | 0.1834720 |
| H20 | 0.1942020 |
| C21 | - 0.2843220 |
| H22 | 0.1780410 |
| H23 | 0.1889790 |
| P24 | 1.5113980 |
| O25 | - 0.7163540 |
| H26 | 0.4000000 |
| O27 | - 0.7174640 |
| H28 | 0.4056340 |
| O29 | - 0.7042910 |
| O30 | - 0.6919460 |
| H31 | 0.4038440 |
| O32 | - 0.7710550 |
| O33 | - 0.7617180 |
| H34 | 0.4031740 |
| P35 | 1.5141630 |
3.2.5. Dipole moment
The dipole moment (µ) can be used to evaluate the chemical reactivity of molecules. In general, the value µ designates the polarity of drug molecules, which is correlated to the fractional electric charge distribution in these molecules [50]. As well, the importance of µ illustrates in the mechanism of reactions and designates the ability of molecules to interact with other molecular species. In Table 3 we present the calculated value of µ for the examined ligand. The examination of this value indicates that the HDZPA ligand offer high abilities to interact with surrounding medium.
3.2.6. Global reactivity descriptors
The calculated values of EHOMO and ELUMO have been used to calculate the global reactivity parameters such as local softness (σ), hardness (η), electrophilicity (ω) and electronegativity (χ). Also, the relation between the stability of molecules and their global chemical reactivity can be determined using these parameters [51]. Generally, the chemical reactivity and stability of molecules can be evaluated measuring their hardness and local softness. In addition, the resistance of drug molecules against the deformation of their electron clouds or polarization can be measured by the hardness. Moreover, hard molecules are described by the elevated values of ΔEGAP, whereas the soft molecules are expressed by the low values of ΔEGAP. On the outer hand, the high gap energy value indicates that the molecule is more stable and low reactive, while the small gap energy value is related to high reactivity and low stability of the molecule. So, the ΔEGAP is an important parameter to determine the stability and the reactivity of molecules. From the presented values of η and σ in Table 3, we observe that HDZPA has an elevated value of σ and least value of η, which indicates its high reactivity. Furthermore, the calculated value of χ indicates that HDZPA is an electronegative species.
The electrophilicity of molecules estimate their abilities to take electrons. As well, the elevated values of ω prove the better electrophility, whereas the low values of ω signify a poor electrophile [52]. According to the electrophilicity value, we can classify the organic molecules into three categories: marginal electrophiles with ω< 0.8 eV, moderate electrophiles with 0.8 <ω< 1.5 eV and strong electrophiles with ω> 1.5 eV [53]. From Table 3 we note that the examined ligand is a marginal electrophile with ω <0.8 eV.
We can observe clearly a good correlation between MEP analysis results and the calculated values of global reactivity descriptors.
3.3. Molecular docking analysis
The molecular docking between HDZPA ligand and the Mpro and RdRp receptors was executed to define the appropriate conformation of the HDZPA in the receptor and the secondary forces resulting between HDZPA and the active amino acids of the receptor. This leads to the development of new drug designs. Based on the minimum binding energy, the non-covalent bonds, π-π* and π-σ interactions between the active amino acids of the Mpro and RdRp target receptors and the HDZPA ligand were tested.
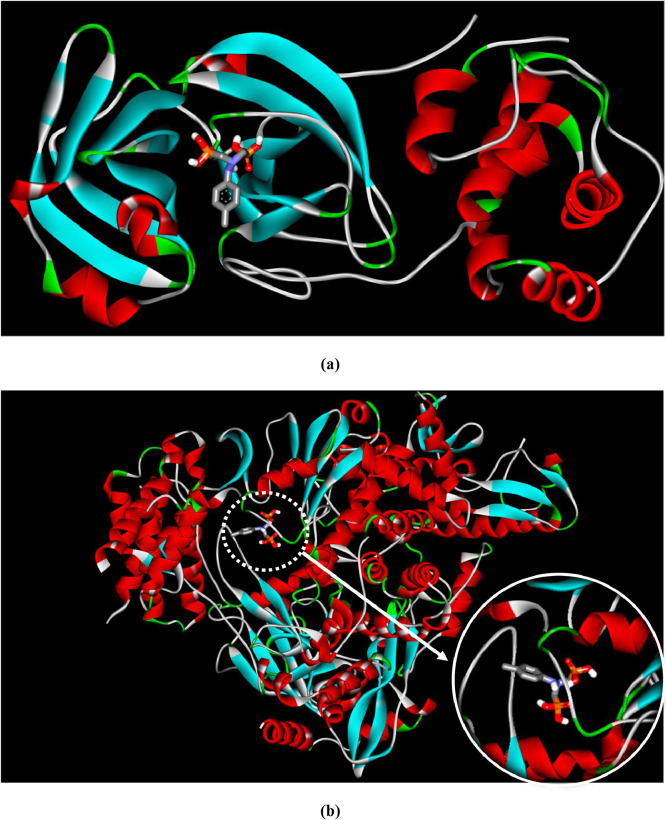
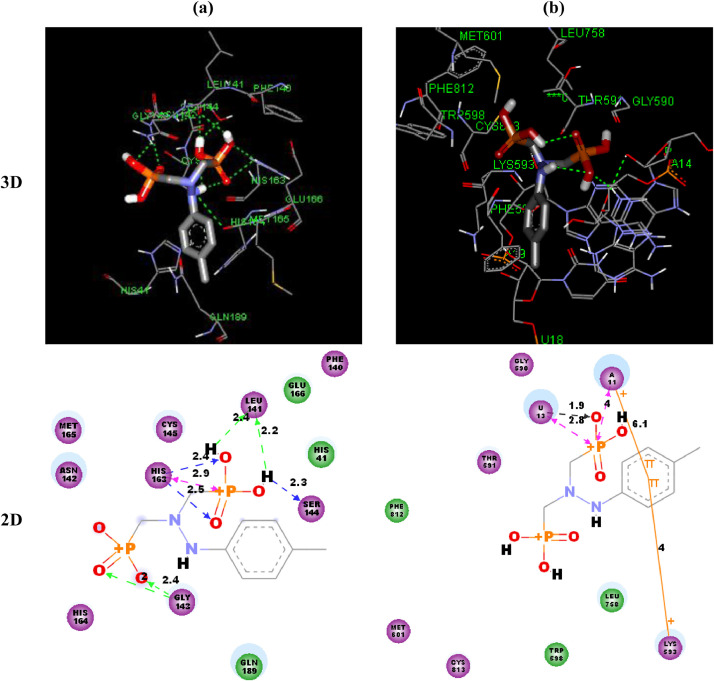
Fig. 7 represents crystal structures of the best docked modes of Mpro-HDZPA and RdRp-HDZPA complexes. We can observe from Figs. 7(a) and 7(b) that the HDZPA ligand prefers to bind in the outer structure of Mpro, while it favors to bind in the inner pocket of RdRp.
Fig. 7.
Best docked model visualization of HDZPA ligand with SARS-CoV-2 main protease (a) and RNA dependent RNA polymerase (b).
Fig. 8 illustrates the detailed presentation of interactions of the investigated ligand with Mpro and RdRp. Generally, the HDZPA binds with Mpro and RdRp by means of various hydrogen and Van der Waals bonding interactions. For Mpro, we can observe from Fig. 8(a) that the HDZPA interacts through H-bonding with HIS163, GLY143, SER144 and LEU141 amino acids whereas bends with van der Waals forces with some amino acids such as GLN189, HIS41, HIS164, MET165, GLU166 and PHE140. Generally, the hydrophilic interactions included H-bonding whereas hydrophobic interactions implicated van der Waals forces. In case of RdRp, we can see from Fig. 8(b) that the HDZPA interacts with just one hydrogen bond with the A13 Nucleotide. Concerning van der Waals forces, HDZPA connects with GLY590, THR591, LEU758, TRP598, PHE812, MET601, CYS813, PHE594 and LYS593. The calculated values of the binding energy of HDZPA and some drugs have been presented in Table 5 . Comparing the obtained values of binding energy of HDZPA with those of some drugs reported in the literature, we observe that our molecule presents the lowest values of bending energy in molecular interactions with Mpro and RdRp, which indicates that HDZPA has the better binding affinity and the Mpro-HDZPA and RdRp-HDZPA complexes are more stable than these formed for Chloroquine, Hydroxychloroquine and Remdesivir.
Fig. 8.
3D and 2D Binding-interaction diagrams of HDZPA ligand with SARS-CoV-2 main protease (a) and RNA dependent RNA polymerase (b).
Table 5.
Molecular docking results of HDZPA and some drugs with Mpro and RdRp.
In conclusion, the in silico docking results revealed that HDZPA is expected as a potential compound to treat COVID-19. On the other hand, we recommend researchers to complete the in vitro and in vivo studies of HDZPA with novel corona virus in order to confirm the obtained docking results and to know the exact impact of the studied compound.
4. Conclusion
In this research, the α-Hydrazinophosphonic acid has been synthesized in good yield using one-pot three-component reactions and characterized using spectroscopic methods. Also, the quantum chemical study of HDZPA ligand has been performed applying DFT method at B3LYP/6–31 G (d,p) basis set. In this context, the optimal structures, the vibrational spectra, the 3D MEP map, HOMO and LUMO orbitals, ΔEGAP, dipole moments, Milliken atomic charges, hardness, local softness, electrophilicity and electronegativity have been calculated for the examined molecule. So, the essential findings are described below. Firstly, the experimental and theoretical IR spectra of the investigated molecule were completely analyzed and the vibrational modes have been attributed. The calculated values of EHOMO and ELUMO show that the HDZPA share their electrons to an acceptor species. Also, the obtained values of ΔEGAP indicate that the HDZPA is a reactive and instable species. The 3D MEP maps illustrate that the possibly sites of the electrophilic reactions are situated on the O33, O32, O27, O29, O30, and O25 atoms. According the Mulliken charges results, the O27, O25, O29, O30, O32, O33, N15 and N17 atoms of HDZPA are possibly the active sites of interaction. The obtained values of the global reactivity descriptors demonstrate that the HDZPA ligand is a powerful electrophile and electrons donor. The interaction of HDZPA with Mpro and RdRp revealed that the investigated ligand can be binds to Mpro and RdRp by means of various bonding contacts. The calculated binding energies of Mpro-HDZPA and RdRp-HDZPA complexes indicated the ability of HDZPA to inhibit SARS-CoV-2. Finally, the in silico docking results suggest that the studied α-Hydrazinophosphonic acid has potential to be developed as a therapeutic agent against SARS-CoV-2.
Author Statement
Khalissa Benbouguerra: Writing, Synthesis and characterization, Data curation, Formal analysis, Methodology and Resources, Theoretical studies and Review, Resources and Investigation.
Nadjib Chafai: Conceptualization, Methodology, Validation, Supervision, Project administration, Resources and Investigation, Theoretical studies and Review.
Salah Chafaa: Project administration, review & editing, Supervision, Investigation, Funding acquisition and Validation.
Youcef Islam Touahria: Data curation.
Hamida Tlidjane: Data curation.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported by the General Directorate for Scientific Research and Technological Development (DGRSDT), Algerian Ministry of Scientific Research, Laboratory of Electrochemistry of Molecular Materials and Complex (LEMMC), Ferhat ABBAS University of Sétif.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.molstruc.2021.130480.
Appendix. Supplementary materials
References
- 1.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang L., Wang Y., Ye D., Liu Q. Review of the 2019 Novel Coronavirus (SARS-CoV-2) based on current evidence. Int J Antimicrob Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO Director-General's opening remarks at the media briefing on COVID-19–1 1 March 2020. [https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-I-media-briefing-on-covid-19—11-march-2020].
- 4.Gobato R., Mitra A. The Inside Story of Coronavirus Pandemic. Parana Journal of Science and Education (PJSE) 2020;6:93–100. doi: 10.13140/RG.2.2.24852.04489. [DOI] [Google Scholar]
- 5.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and Chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Wit E., Feldmann F., Cronin J., Jordan R., Okumura A., Thomas T., Scott D., Cihlar T., Feldmann H. Prophy-lactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection. Proc Natl Acad Sci. 2020;117:6771–6776. doi: 10.1073/pnas.1922083117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao J., Tian Z., Yang X. Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 8.Chinese Clinical Trial Registry. http://www.chictr.org.cn/searchproj.aspx?title=%E6%B0%AF%E5%96%B9&officialname=ðylam=&secondaryid=&applier=&studyleader=ðicalcommitteesanction=&sponsor=&studyailment=&studyailmentcode=&studytype=0&studystage=0&studydesign=0&minstudyexecutetime=&maxstudyexecutetime=&recruitmentstatus=0&gender=0&agreetosign=&secsponsor=®no=®status=0&country=&province=&city=&institution=&institutionlevel=&measure=ðylam=&sourceofspends=&createyear=0&isuploadrf=&whetherpublic=&btngo=btn&verifycode=&page=1.
- 9.Colson P., Rolain J.-.M., Raoult D. Chloroquine for the 2019 novel coronavirus SARS-CoV-2. Int J Antimicrob Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gautret P., Lagier Jean-Christophe, Parola P., Van Thuan Hoang L.Meddeb, Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T., Honoré S., Colson P., Chabrière E., La Scola B., Rolain Jean-Marc, Brouqui P., Raoult D. Hydroxychloroquine and ethyl amino as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int J Antimicrob Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.X. Liu, B. Zhang, Z. Jin, H. Yang, Z. Rao, The Crystal Structure of 2019-nCoV Main Protease in Complex with an Inhibitor N3. Deposited: 2020-01-26 Released: 2020-02-05. doi: 10.2210/pdb6LU7/pdb Available from: https://www.rcsb.org/structure/6LU7.
- 12.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S., Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent Sci. 2020;6:315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oftadeh M., Madadi Mahani N., Hamadanian M. Density functional theory study of the local molecular properties of acetamide derivatives as anti-HIV drugs. Res Pharm Sci. 2013;8:285–297. PMID: 24082898 PMCID: PMC3757594. [PMC free article] [PubMed] [Google Scholar]
- 15.Behzadi H., Roonasi P., Taghipour Khatoon Assle, Van der Spoel D., Manzetti S. Relationship between electronic properties and drug activity of seven quinoxaline compounds: a DFT study. J. Mol. Struct. 2015;1091:196–202. doi: 10.1016/j.molstruc.2015.03.001. [DOI] [Google Scholar]
- 16.Tariq A., Nazir S., Arshad A.W., Nawaz F., Ayub K., Iqbal J. DFT study of the therapeutic potential of phosphorene as a new drug-delivery system to treat cancer. RSC Adv. 2019;9:24325–24332. doi: 10.1039/C9RA02778E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hellal A., Chafaa S., Chafai N., Touafri L. Synthesis, antibacterial screening and DFT studies of series of α-amino-phosphonates derivatives from aminophénols. J. Mol. Struct. 2017;1134:217–225. doi: 10.1016/j.molstruc.2016.12.079. [DOI] [Google Scholar]
- 18.Mehri M., Chafai N., Ouksel L., Benbouguerra K., Hellal A., Chafaa S. Synthesis, electrochemical and classical evaluation of the antioxidant activity of three α-aminophosphonic acids: experimental and theoretical investigation. Journal J. Mol. Struct. 2018;1171:179–189. doi: 10.1016/j.molstruc.2018.05.074. [DOI] [Google Scholar]
- 19.Hellal A., Chafaa S., Chafai N. Synthesis, Antibacterial and Antifungal Screening of Three new of Alpha-aminophosphonic acids. International Journal of Scientific & Engineering Research. 2015;6:1622–1627. [Google Scholar]
- 20.Shahrisa A., Teimuri-Mofrad R., Gholamhosseini-Nazari M. Synthesis of a new class of Betti bases by the Mannich-type reaction: efficient, facile, solvent-free and one-pot protocol. Mol Divers. 2015;19:87–101. doi: 10.1007/s11030-014-9559-x. [DOI] [PubMed] [Google Scholar]
- 21.Salter M.M., Kobayashi J., Shimizu Y., Kobayashi S. Direct-Type Catalytic Three-Component Mannich Reactions Leading to an Efficient Synthesis of α,β-Diamino Acid Derivatives. Org. Lett. 2006;8:3533–3536. doi: 10.1021/ol0613012. [DOI] [PubMed] [Google Scholar]
- 22.Bhagat S., Chakraborti A.K. An Extremely Efficient Three-Component Reaction of Aldehydes/Ketones, Amines, and Phosphites (Kabachnik-Fields Reaction) for the Synthesis of α-Aminophosphonates Catalyzed by Magnesium Perchlorate. J. Org. Chem. 2007;72:1263–1270. doi: 10.1021/jo062140i. [DOI] [PubMed] [Google Scholar]
- 23.Wu J., Sun W., Xia H.G., Sun X. A facile and highly efficient route to α-amino phosphonates via three-component reactions catalyzed by Mg(ClO4)2 or molecular iodine. Org. Biomol. Chem. 2006;4:1663–1666. doi: 10.1039/B602536F. [DOI] [PubMed] [Google Scholar]
- 24.Ranu B.C., Hajra A., Jana U. A simple, efficient, and general one-pot reaction of aldehydes and ketones with amines in the presence of indium(III) chloride as a catalyst provides α-amino phosphonates. Sonication accelerates the reaction. Org. Lett. 1999;1:1141–1143. doi: 10.1021/ol990079g. [DOI] [Google Scholar]
- 25.Weng J., Li Y.B., Wang R.B., Li F.Q., Liu C., Chan A.S., Lu G. A practical and azide-free synthetic approach to oseltamivir from diethyl d-tartrate. J Org Chem. 2010;75:3125–3128. doi: 10.1021/jo100187m. [DOI] [PubMed] [Google Scholar]
- 26.Davis T.A., Johnston J.N. Catalytic, Enantioselective Synthesis of Stilbene cis-Diamines: a Concise Preparation of (-)-Nutlin-3, a Potent p53/MDM2 Inhibitor. Chem Sci. 2011;2:1076–1079. doi: 10.1039/C1SC00061F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie H., Zhang Y., Zhang S., Chen X., Wang W. Bifunctional Cinchona alkaloid thiourea catalyzed highly efficient, enantioselective aza-Henry reaction of cyclic trifluoromethyl ketimines: synthesis of anti-HIV drug DPC 083. Angew Chem Int Ed Engl. 2011;50:11773–11776. doi: 10.1002/anie.201105970. [DOI] [PubMed] [Google Scholar]
- 28.Jakubec P., Hawkins A., Felzmann W., Dixon D.J. Total synthesis of manzamine A and related alkaloids. J Am Chem Soc. 2012;134:17482–17485. doi: 10.1021/ja308826x. [DOI] [PubMed] [Google Scholar]
- 29.Handa S., Gnanadesikan V., Matsunaga S., Shibasaki M. Heterobimetallic transition metal/rare earth metal bifunctional catalysis: a Cu/Sm/Schiff base complex for syn-selective catalytic asymmetric nitro-Mannich reaction. J Am Chem Soc. 2010;132:4925–4934. doi: 10.1021/ja100514y. [DOI] [PubMed] [Google Scholar]
- 30.Davis T.A., Danneman M.W., Johnston J.N. Chiral proton catalysis of secondary nitroalkane additions to azomethine: synthesis of a potent GlyT1 inhibitor. Chem Commun (Camb) 2012;48:5578–5580. doi: 10.1039/C2CC32225K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choudhary M.K., Das A., Kureshy R.I., Kumar M., Noorul H.K., Abdi S.H., Bajaj H.C. Chiral Cu (I,I)-amino alcohol based complexes for asymmetric aza-henry reaction of N-Ts imines. Catal. Sci. Technol. 2014;4:548–555. doi: 10.1039/C3CY00774J. [DOI] [Google Scholar]
- 32.Hynes P.S., Stupple P.A., Dixon D.J. Organocatalytic asymmetric total synthesis of (R)-rolipram and formal synthesis of (3S,4R)-paroxetine. Org Lett. Apr. 2008;10:1389–1391. doi: 10.1021/ol800108u. [DOI] [PubMed] [Google Scholar]
- 33.Jakubec P., Cockfield D.M., Dixon D.J. Total synthesis of (-)-nakadomarin A. J Am Chem Soc. 2009;131:16632–16633. doi: 10.1021/ja908399s. [DOI] [PubMed] [Google Scholar]
- 34.Moedritzer K., Irani R.R. The direct synthesis of α-aminomethylphosphonic acids. Mannich-type reactions with orthophosphorous acid. J. Org. Chem. 1966;31:1603–1607. doi: 10.1021/jo01343a067. [DOI] [Google Scholar]
- 35.M.J. Frisch, G.W. Trucks, H.B. Schlegel, G.E. Scuseria, M.A. Robb, J.R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G.A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H.P. Hratchian, A.F. Izmaylov, J. Bloino, G. Zheng, J.L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J.A. Montgomery Jr., J.E. Peralta, F. Ogliaro, M. Bearpark, J.J. Heyd, E. Brothers, K.N. Kudin, V.N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J.C. Burant, S.S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J.M. Millam, M. Klene, J.E. Knox, J.B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R.E. Stratmann, O. Yazyev, A.J. Austin, R. Cammi, C. Pomelli, J.W. Ochterski, R.L. Martin, K. Morokuma, V.G. Zakrzewski, G.A. Voth, P. Salvador, J.J. Dannenberg, S. Dapprich, A.D. Daniels, O. Farkas, J.B. Foresman, J.V. Ortiz, J. Cioslowski, D.J. Fox, Gaussian 09, Revision A.02, Gaussian, Inc., Wallingford, CT, 2009.
- 36.Chafai N., Chafaa S., Benbouguerra K., Hellal A., Mehri M. Synthesis, spectral analysis, anti-corrosive activity and theoretical study of an aromatic hydrazone derivative. J. Mol. Struct. 2019;1181:83–92. doi: 10.1016/j.molstruc.2018.12.073. [DOI] [Google Scholar]
- 37.Benbouguerra K., Chafaa S., Chafai N., Mehri M., Moumeni O., Hellal A. Synthesis, spectroscopic characterization and a comparative study of the corrosion inhibitive efficiency of an α-aminophosphonate and Schiff base derivatives: experimental and theoretical investigations. J. Mol. Struct. 2018;1157:165–176. doi: 10.1016/j.molstruc.2017.12.049. [DOI] [Google Scholar]
- 38.Jamroz M.H. Vibrational energy distribution analysis (VEDA): scopes and limitations. Spectrochim. Acta - Part A Mol. Biomol. Spectrosc. 2013;114:220–230. doi: 10.1016/j.saa.2013.05.096. [DOI] [PubMed] [Google Scholar]
- 39.Chafai N., Chafaa S., Benbouguerra K., Daoud D., Hellal A., Mehri M. Synthesis, characterization and the inhibition activity of a new α-aminophosphonic derivative on the corrosion of XC48 carbon steel in 0.5 M H2SO4: experimental and theoretical studies. J. Taiwan Inst. Chem. Eng. 2017;70:331–344. doi: 10.1016/j.jtice.2016.10.026. [DOI] [Google Scholar]
- 40.Djenane M., Chafaa S., Chafai N., Kerkour R., Hellal A. Synthesis, spectral properties and corrosion inhibition efficiency of new ethylhydrogen[(methoxyphenyl)(ethyl amino)methyl]phosphonate derivatives: experimental and theoretical investigation. J. Mol. Struct. 2018;1175:398–413. doi: 10.1016/j.molstruc.2018.07.087. [DOI] [Google Scholar]
- 41.Jeffrey P.M., Damian M., Leo R. An evaluation of harmonic vibrational frequency scale factors. J. Phys. Chem. 2007;111(45):11683–11700. doi: 10.1021/jp073974n. [DOI] [PubMed] [Google Scholar]
- 42.Hong L.X., Ru L.X., Zhou Z.X. Calculation of vibrational spectroscopic and NMR parameters of 2-Dicyanovinyl-5-(4-N,N-dimethylaminophenyl) thiophene by ab initio HF and density functional methods. Comput. Theor.Chem. 2011;969:27–34. doi: 10.1016/j.comptc.2011.05.010. [DOI] [Google Scholar]
- 43.Herrag L., Hammouti B., Elkadiri S., Aouniti A., Jama C., Vezin H., Bentiss F. Adsorption properties and inhibition of mild steel corrosion in hydrochloric solution by some newly synthesized diamine derivatives: experimental and theoretical investigations. Corros. Sci. 2010;52:3042–3051. doi: 10.1016/j.corsci.2010.05.024. [DOI] [Google Scholar]
- 44.Khaled K.F. Studies of iron corrosion inhibition using chemical, electrochemical and computer simulation techniques. Electrochim Acta. 2010;55:6523–6532. doi: 10.1016/j.electacta.2010.06.027. [DOI] [Google Scholar]
- 45.Lgaz H., Bhat K.S., Salghi R., Shubhalaxmi S.Jodeh, Algarra M., Hammouti B., Ali I.H., Essamri A. Insights into corrosion inhibition behavior of three chalcone derivatives for mild steel in hydrochloric acid solution. J. Mol. Liq. 2017;238:71–83. doi: 10.1016/j.molliq.2017.04.124. [DOI] [Google Scholar]
- 46.Fliszar S. Springer; New York: 1983. Charge Distributions and Chemical Effects. [Google Scholar]
- 47.Silverstein R.M., Webster F.X. Wiley; New York: 1998. Spectroscopic Identification of Organic Compounds. [Google Scholar]
- 48.Yesilkaynak T., Binzet G., Emen F.M., Florke U., Kulcu N., Arslan H. Theoretical and experimental studies on N-(6-methylpyridin-2-yl-carbamothioyl)biphenyl-4-carboxamide. Eur. J. Chem. 2010;1:1–5. doi: 10.5155/eurjchem.1.1.1-5.3. [DOI] [Google Scholar]
- 49.Moumeni O., Chafaa S., Kerkour R., Benbouguerra K., Chafai N. Synthesis, structural and anticorrosion properties of diethyl(phenylamino)methyl) phosphonate derivatives: experimental and theoretical study. J. Mol. Struct. 2020;1206 doi: 10.1016/j.molstruc.2020.127693. [DOI] [Google Scholar]
- 50.Issa R.M., Awad M.K., Atlam F.M. Quantum chemical studies on the inhibition of corrosion of copper surface by substituted uracils. Appl. Surf. Sci. 2008;255:2433–2441. doi: 10.1016/j.apsusc.2008.07.155. [DOI] [Google Scholar]
- 51.Vijayaraj R., Subramanian V., Chattaraj P.K. Comparison of Global Reactivity Descriptors Calculated Using Various Density Functionals: a QSAR Perspective. J. Chem. Theory Comput. 2009;5:2744–2753. doi: 10.1021/ct900347f. [DOI] [PubMed] [Google Scholar]
- 52.Hellal A., Chafaa S., Chafai N. Synthesis, characterization and computational studies of three α-amino-phosphonic acids derivatives from Meta, Ortho and Para aminophenol. J. Mol. Struct. 2016;1103:110–124. doi: 10.1016/j.molstruc.2015.08.070. [DOI] [Google Scholar]
- 53.Domingo L.R., Aurell M.J., Pérez P., Contreras R. Quantitative characterization of the global electrophilicity power of common diene/dienophile pairs in Diels-Alder reactions. Tetrahedron. 2002;58:4417–4423. doi: 10.1016/S0040-4020(02)00410-6. [DOI] [Google Scholar]
- 54.Nallusamy S., Mannu J., Ravikumar C., Angamuthu K., Nathan B., Nachimuthu K., Ramasamy G., Muthurajan R., Subbarayalu M., Neelakandan K. Shortlisting Phytochemicals Exhibiting Inhibitory Activity against Major Proteins of SARS-CoV-2 through Virtual Screening. Research Square. 2020:1–26. doi: 10.21203/rs.3.rs-31834/v1. [DOI] [Google Scholar]
- 55.Hagar M., Ahmed H.A., Aljohani G., Alhaddad O.A. Investigation of Some Antiviral N-Heterocycles as COVID 19 Drug: molecular Docking and DFT Calculations. Int. J. Mol. Sci. 2020;21:3922. doi: 10.3390/ijms21113922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elfiky A.A., Ribavirin Remdesivir, Sofosbuvir Galidesivir. Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): a molecular docking study. Life Sci. 2020;253 doi: 10.1016/j.lfs.2020.117592. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.