Abstract
Background
The purpose of present study was to identify the differentially expressed genes (DEGs) associated with BMP-9-induced osteogenic differentiation of mesenchymal stem cells (MSCs) by using bioinformatics methods.
Methods
Gene expression profiles of BMP-9-induced MSCs were compared between with GFP-induced MSCs and BMP-9-induced MSCs. GSE48882 containing two groups of gene expression profiles, 3 GFP-induced MSC samples and 3 from BMP-9-induced MSCs, was downloaded from the Gene Expression Omnibus (GEO) database. Then, DEGs were clustered based on functions and signaling pathways with significant enrichment analysis. Pathway enrichment analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) demonstrated that the identified DEGs were potentially involved in cytoplasm, nucleus, and extracellular exosome signaling pathway.
Results
A total of 1967 DEGs (1029 upregulated and 938 downregulated) were identified from GSE48882 datasets. R/Bioconductor package limma was used to identify the DEGs. Further analysis revealed that there were 35 common DEGs observed between the samples. GO function and KEGG pathway enrichment analysis, among which endoplasmic reticulum, protein export, RNA transport, and apoptosis was the most significant dysregulated pathway. The result of protein-protein interaction (PPI) network modules demonstrated that the Hspa5, P4hb, Sec61a1, Smarca2, Pdia3, Dnajc3, Hyou1, Smad7, Derl1, and Surf4 were the high-degree hub nodes.
Conclusion
Taken above, using integrated bioinformatical analysis, we have identified DEGs candidate genes and pathways in BMP-9 induced MSCs, which could improve our understanding of the key genes and pathways for BMP-9-induced osteogenic of MSCs.
Keywords: Differentially expressed genes, BMP-9, Mesenchymal stem cells, Enrichment analysis
Introduction
Mesenchymal stem cells (MSCs) are non-hematopoietic multipotent cells and used for bone tissue regeneration [1, 2]. MSCs can differentiate into osteoblastic, chondrogenic, and other lineages through different stimulating factors [3, 4]. Bone morphogenetic proteins (BMPs) are a group of transforming growth factor-β (TGF-β) and play an important role in embryonic development, growth, and differentiation [5]. BMP-9 (also known as growth/differentiation factor-2) has been shown to play a pivotal role in many physiological processes including neuronal and adipocyte differentiation [6]. In recent years, a series of experiments have demonstrated that BMP-9 is more osteogenic than BMP-2 and is not antagonized by noggin, which is a BMP antagonist [7, 8]. BMP-9 was reported to have a positive role in promoting the MSCs differentiation through Smad signaling pathway [9]. However, the mechanism for BMP-9-induced osteogenic differentiation was unclear. Gene expression microarray used modern research method and has been used to explore the gene network and identify the potential target gene.
The purpose of our study was to compare the gene expression of BMP-9-induced osteogenic differentiation of MSCs in the GEO database. Subsequently, Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment pathway analyses were performed to identify the potential pathway of BMP-9-induced osteogenic differentiation of MSCs. Furthermore, protein-protein interaction (PPI) was performed to identify relevant genes and pathways and molecular mechanisms.
Materials and methods
Gene expression microarray data
The microarray data GSE48882 used in our study was downloaded from Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/). GSE48882 was based on the Affymetrix GPL339 platform (Affymetrix Mouse Expression 430A Array; Affymetrix; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The GSE48882 dataset contained six samples, including three GFP1-induced MSC samples and three BMP-9-induced MSC samples.
Data processing and identification of DEGs
The original array data were performed background correction and quartile data normalization [10]. The raw data was downloaded and then put into the U-EDIT software to further analyses. Then, all of the differentially expressed genes (DEGs) between GFP-induced MSCs and BMP-9-induced MSCs were identified based on analyses GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) and bioconductor package limma [11]. The DEGs between the GFP-induced MSC samples and three BMP-9-induced MSC samples were selected (P < 0.05).
GO enrichment and KEGG pathway analysis of the DEGs
The DEGs was entered into the online software Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) [12]. The DEGs were classified into three functional groups: molecular function (MF) group, biological process (BP) group, and cellular component (CC) group.
DEGs functional and signaling pathway enrichment were conducted using online websites of KEGG (http://www.genome.jp/) and P < 0.05 was considered to indicate a statistically significant difference [13].
Construction of the PPI Network of DEGs
We downloaded the comprehensive interaction information of BMP-9-induced osteogenic differentiation of MSCs from the Search Tool for the Retrieval of Interacting Genes (STRING) database (http://www.string-db.org/) [14]. A combined score > 0.4 was identified as experimentally validated interactions. Then, we generate network and node file and analyzed using Cytoscape software (version 3.5.1; www.cytoscape.org). P < 0.05 was considered to be a statistically significant difference [15].
Results
Identification of DEGs
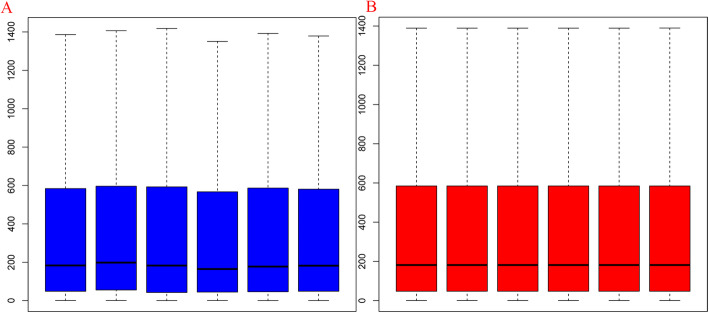
P < 0.05, logFC (fold control) > 2.0 or logFC < − 2.0 was used in current study, and we listed the first tenth DEGs in Table 1. And the samples were normalized and results were shown in Fig. 1. Among the DEGs, 1029 genes (52.31%) and 938 genes (47.69%) were upregulated and downregulated, respectively. Then, we draw a volcano plot and can be seen in Fig. 2. The top 50 upregulated and downregulated DEGs were selected to generate the heatmap and is shown in Fig. 3.
Table 1.
The differentially expressed genes (DEGs)
| Gene | logFC | AveExpr | t | P.Value | adj.P.Val | B |
|---|---|---|---|---|---|---|
| Hspa5 | 5886.413 | 8805.691 | 24.55842 | 2.89E− 06 | 0.014437 | − 4.41275 |
| Atp6v1g1 | 192.8844 | 1430.236 | 24.16697 | 3.12E− 06 | 0.014437 | − 4.41281 |
| Ntan1 | 849.4093 | 1610.547 | 22.81636 | 4.11E− 06 | 0.014437 | − 4.41304 |
| Ttc30b | − 269.874 | 376.4807 | − 22.6356 | 4.27E− 06 | 0.014437 | − 4.41307 |
| Smad7 | 1291.794 | 1054.716 | 20.67191 | 6.60E− 06 | 0.017846 | − 4.4135 |
| Wars | 382.1337 | 817.2169 | 18.04368 | 1.27E− 05 | 0.024944 | − 4.41429 |
| Hif1a | − 297.61 | 791.8699 | − 17.8299 | 1.34E− 05 | 0.024944 | − 4.41437 |
| Fbln2 | − 2928.99 | 5171.091 | − 17.4703 | 1.48E− 05 | 0.024944 | − 4.41452 |
| Klf10 | 665.8488 | 694.2806 | 16.49257 | 1.94E− 05 | 0.029027 | − 4.41495 |
| Kat2b | − 295.886 | 399.0065 | − 15.9344 | 2.29E− 05 | 0.029027 | − 4.41523 |
Fig. 1.
Box plots for expression data before normalization (a) and after normalization (b). The horizontal axis is the name of samples while the vertical axis stands for the values of expression. The black line in the cassette is the median of data in each group, which represents the degree of normalization. The black line in the right figure was almost on the same line, indicating an excellent degree of normalization
Fig. 2.
The volcano plot of differentially expressed genes. The abscissa is − log10 (P.Value) and the ordinates is logFC. The red dots stand for the up-expressed genes, green dots stand for down-expressed gene while the black dots represent genes not differentially expressed
Fig. 3.

The heat map of the differentially expressed genes (top 50 upregulated and downregulated genes). Red, upregulation; blue, downregulation
GO term enrichment analysis
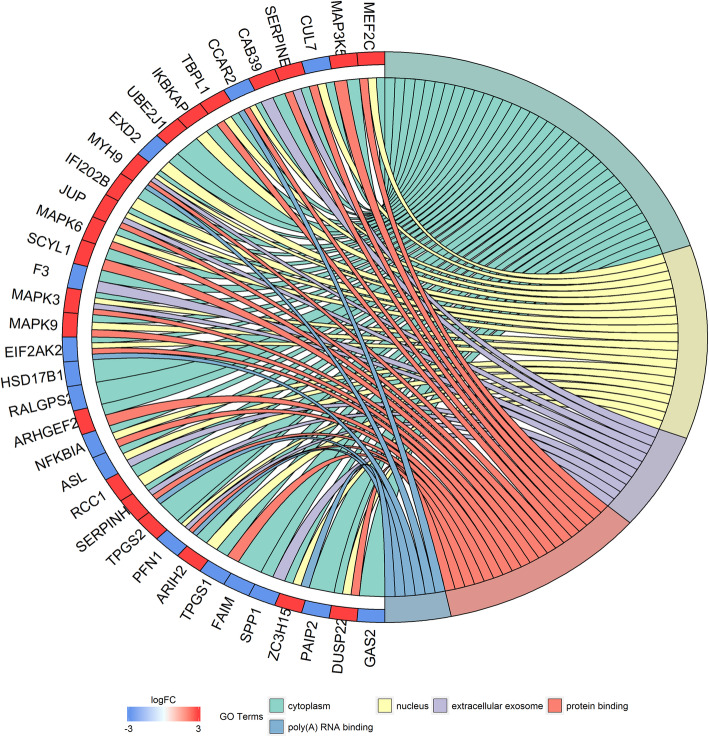
DAVID was used to carry out a gene ontology (GO) function enrichment for DEGs. Results show that the DEGs were significantly enriched in CC, including cytoplasm, nucleus, and extracellular exosome (Fig. 4). For MF, the DEGs were enriched in protein binding and poly(A) RNA binding (Fig. 4).
Fig. 4.
Go term enrichment analysis of the different genes
KEGG pathway analysis
Results show that the DEGs were enriched in protein processing in endoplasmic reticulum, protein export, RNA transport, apoptosis, Alzheimer’s disease, N-Glycan biosynthesis, Valine, leucine and isoleucine degradation, TNF signaling pathway, Toxoplasmosis, p53 signaling pathway, and AGE-RACE signaling pathway in diabetic complications (Figs. 5 and 6).
Fig. 5.
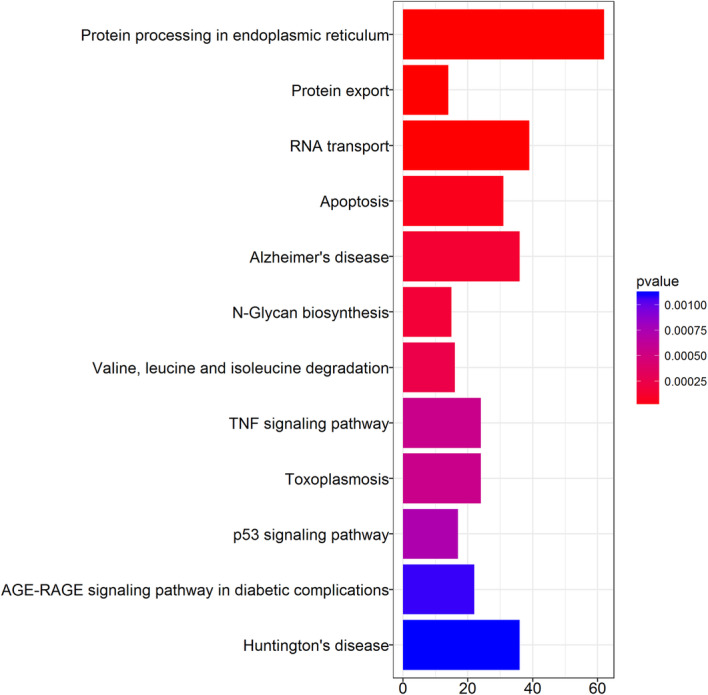
Barplot analysis of the Kegg pathway
Fig. 6.

Dotplot analysis of the Kegg pathway
PPI network of the DEGs and core genes in the PPI network
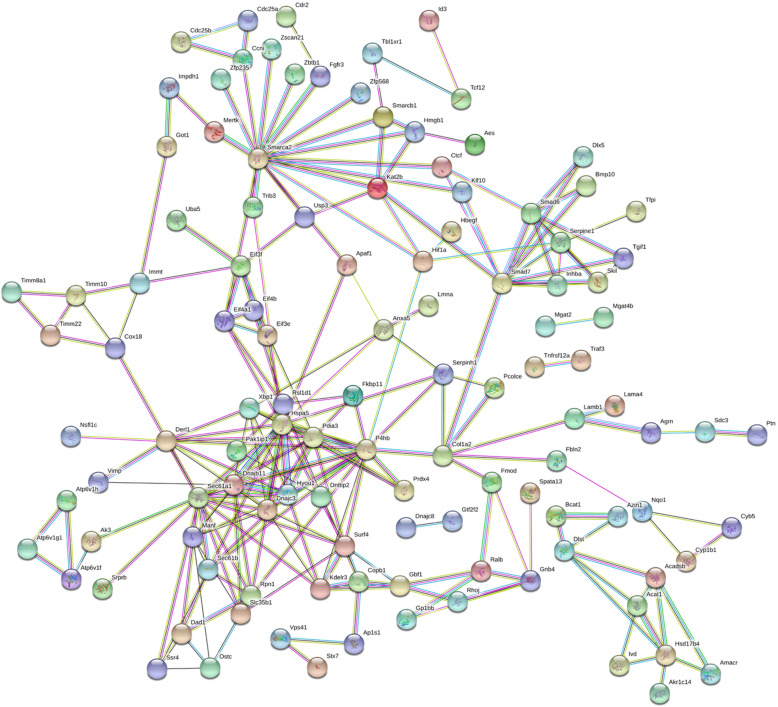
As shown in Fig. 7, the PPI network contained 197 nodes and 272 edges. The top 10 high-degree hub nodes included Hspa5, P4hb, Sec61a1, Smarca2, Pdia3, Dnajc3, Hyou1, Smad7, Derl1, and Surf4 (Fig. 8). Further analyses of the DEGs, Hspa5’s node degree score was 108 and identified as the highest node degree (Figs. 8 and 9).
Fig. 7.
PPI network of BMP-9 induced osteogenic MSCs genes. The circle stands for genes and the line indicates the interactions among genes. The interior of the circle represents the structure of proteins. The color of the line provides evidence of the different interactions among proteins. (A red line indicates the presence of fusion evidence; a green line, neighborhood evidence; a blue line, concurrence evidence; a purple line, experimental evidence; a yellow line, text mining evidence; a light blue line, database evidence; a black line, coexpression evidence)
Fig. 8.

Histogram of numbers of genes adjacent to interaction network
Fig. 9.
Interaction network of the proteins
Discussion
In the clinical application of rhBMP-2, adverse events such as tissue inflammation and bone resorption perplexed the physician [16–18]. Therefore, there is a crying need for the development of an equivalent effective and more safe bone regenerative therapies for bone regeneration. Recently, we attached importance to the rhBMP-9 since rhBMP-9 possesses different properties than rhBMP-2 [19–21]. Its precursor protein shares 50–55% amino acid sequence identity with BMP 2, 4, 5, 6, and 7. However, the present knowledge of the molecular mechanism of BMP-9-induced osteogenic differentiation of MSCs remains insufficient.
The present study performed a comprehensive analysis and built a gene interaction network based on the gene expression profiles (GSE48882) comprising three GFP-MSC samples and three BMP-9-MSC samples. The results of the analysis demonstrated that 1967 genes were identified as DEGs. Moreover, GO and KEGG pathway analyses were performed to find the interactions of DEGs. Combining with the PPI network, key potential genes and pathways may be associated with BMP-9-induced osteogenic differentiation of MSCs.
The results of GO analyses indicated cytoplasm, nucleus and extracellular exosome, protein binding and poly(A) and RNA binding. Nakamura et al. [22] revealed that BMP-9 provided comparable new bone formation and with less adipose tissues compared to BMP-2 in calvarial critical-size defects model. Recently, Sreekumar et al. [23] found that rhBMP9 induced the highest levels of osteoblast activity when compared to the clinically utilized rhBMP2 and rhBMP7. Compared with rhBMP2 and rhBMP7, rhBMP9 significantly increased osteogenic activity (ALP activity and Smad nuclear translocation).
Lin et al. [24] also found that BMP9-induced osteogenic differentiation through inhibition of the Wnt/β-catenin and P38 pathway in MSCs. And adding Wnt antagonist Dickkopf-1 (Dkk1) or P38 inhibitor SB203580 could negatively regulate BMP9-induced osteogenic differentiation. In current study, gene array indicated that BMP-9 has no effect on the Wnt/β-catenin pathway.
Chen et al. [25] found that BMP-9 induce osteogenic differentiation of MSCs through Smad pathway. When there was an overexpression of the BMP-9 in MSCs, Smad 1, Smad 7, and Smad 9 was significantly increased than controls. And insulin-like growth factor (IGF-1) could enhance BMP9-induced osteogenic differentiation in MSCs through Smad signaling pathway. The above result was similar with our results and found that BMP-9 could also increase the expression of Smad7. PPI network revealed that Smad 7 ranked the eighth of the high-degree hub nodes. Daigang et al. [26] found that BMP-9 stimulates the phosphorylation of Smad1/5/8 at a dose response manner. What is more, BMP-9 could significantly increase osteoblast differentiation genes such as Runx2, osteocalcin, and ALP.
Hspa5 was the heat shock protein and plays an important role in protein folding and transport processes. Hspa5 plays a role in facilitating the assembly of multimeric protein complexes inside the endoplasmic reticulum. And thus, BMP-9 may through protein folding and transport and promote osteogenic differentiation of MSCs. Sharff et al. [27] revealed that Hey1, through its interplay with Runx2, may play an important role in regulating BMP9-induced osteoblast lineage differentiation of MSCs. Zhou et al. [28] revealed that BMP9 reduces bone loss in ovariectomized mice by dual regulation of bone remodeling. BMP9 exerts on bone remodeling by promoting bone anabolic activity and inhibiting osteoclast differentiation in ovariectomy (OVX) mice. Hey1 and Hspa5 interact with each other and promote the expression of MSCs. And from the PPI analysis, we found that HIF-1a also plays a central role in the osteogenic differentiation of MSCs. As we know, HIF-1a can stimulate the VEGF and stimulate the angiogenesis [29–31].
Several limitations existed in our study: (1) we did not verify the key gene and pathways through PCR and Western-blot method; (2) key miRNA and lncRNA were not further analyzed and should be performed in future research; and (3) optimal dose of BMP-9 was not explored in this gene expression microarray. Future studies should be focused on the specific mechanism BMP-9-induced osteogenic differentiation of MSCs. What is more, administration with small interference RNA to further confirms our conclusion.
Conclusion
In conclusion, our study provides a comprehensive bioinformatics analysis of key genes and pathways of BMP-9 induced osteogenic differentiation of MSCs. Their related GO terms found that cytoplasm and nucleus may play important roles in BMP-9-induced osteogenic differentiation of MSCs and have the potential to be used as an potential agent for MSCs osteogenic differentiation. Further genetic and experimental studies with larger sample size are needed to confirm our results.
Acknowledgements
Not applicable.
Abbreviations
- DEGs
Differentially expressed genes
- MSCs
Mesenchymal stem cells
- GEO
Gene Expression Omnibus
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PPI
Protein-protein interaction
- BMPs
Bone morphogenetic proteins
- TGF-β
Transforming growth factor-β
- GO
Gene Ontology
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- MF
Molecular function
- BP
Biological process
- CC
Cellular component
- STRING
Search Tool for the Retrieval of Interacting Genes
- Dkk1
Dickkopf-1
- IGF-1
Insulin-like growth factor
Authors’ contributions
JQW, LBM, and LFL design the study and conducted the bioinformatic analysis. YML, JW, and JY perform the statistically analyses. FHZ and TYL write the draft. LY and JQW edited and confirmed the final manuscript. All authors read and approved the final manuscript.
Funding
There is no funding for this article.
Availability of data and materials
We state that the data will not be shared since all the raw data are present in the figures included in the article.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Bhuiyan DB, Middleton JC, Tannenbaum R, Wick TM. Bone regeneration from human mesenchymal stem cells on porous hydroxyapatite-PLGA-collagen bioactive polymer scaffolds. Bio-Med Materials Eng. 2017;28(6):671–685. doi: 10.3233/BME-171703. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Wang Y, Gou W, Lu Q, Peng J, Lu S. Role of mesenchymal stem cells in bone regeneration and fracture repair: a review. Int Orthop. 2013;37(12):2491–2498. doi: 10.1007/s00264-013-2059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito H. Chemokines in mesenchymal stem cell therapy for bone repair: a novel concept of recruiting mesenchymal stem cells and the possible cell sources. Modern Rheumatol. 2011;21(2):113–121. doi: 10.3109/s10165-010-0357-8. [DOI] [PubMed] [Google Scholar]
- 4.Rapp AE, Bindl R, Heilmann A, et al. Systemic mesenchymal stem cell administration enhances bone formation in fracture repair but not load-induced bone formation. Eur Cells Materials. 2015;29:22–34. doi: 10.22203/eCM.v029a02. [DOI] [PubMed] [Google Scholar]
- 5.Moon SH, Kim I, Kim SH. Mollugin enhances the osteogenic action of BMP-2 via the p38-Smad signaling pathway. Archives of pharmacal research. 2017;40(11):1328–1335. doi: 10.1007/s12272-017-0964-4. [DOI] [PubMed] [Google Scholar]
- 6.Lopez-Coviella I, Berse B, Krauss R, Thies RS, Blusztajn JK. Induction and maintenance of the neuronal cholinergic phenotype in the central nervous system by BMP-9. Science (New York, N.Y.) 2000;289(5477):313–316. doi: 10.1126/science.289.5477.313. [DOI] [PubMed] [Google Scholar]
- 7.Kang Q, Sun MH, Cheng H, Peng Y, Montag AG, Deyrup AT, Jiang W, Luu HH, Luo J, Szatkowski JP, Vanichakarn P, Park JY, Li Y, Haydon RC, He TC. Characterization of the distinct orthotopic bone-forming activity of 14 BMPs using recombinant adenovirus-mediated gene delivery. Gene therapy. 2004;11(17):1312–1320. doi: 10.1038/sj.gt.3302298. [DOI] [PubMed] [Google Scholar]
- 8.Cheng H, Jiang W, Phillips FM, et al. Osteogenic activity of the fourteen types of human bone morphogenetic proteins (BMPs). J Bone Joint Surg. 2003;(85-a, 8):1544–52. [DOI] [PubMed]
- 9.Cao J, Wei Y, Lian J, Yang L, Zhang X, Xie J, Liu Q, Luo J, He B, Tang M. Notch signaling pathway promotes osteogenic differentiation of mesenchymal stem cells by enhancing BMP9/Smad signaling. Int J Mol Med. 2017;40(2):378–388. doi: 10.3892/ijmm.2017.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiang W, Zhang B, Lv F, Feng G, Chen L, Yang F, Zhang K, Cao C, Wang P, Chu M. The potential regulatory mechanisms of the gonadotropin-releasing hormone in gonadotropin transcriptions identified with bioinformatics analyses. Reproductive biology and endocrinology : RB&E. 2017;15(1):46. doi: 10.1186/s12958-017-0264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei Z, Qi X, Chen Y, et al. Bioinformatics method combined with logistic regression analysis reveal potentially important miRNAs in ischemic stroke. Bioscience Reports. 2020;40(8):BSR20201154. [DOI] [PMC free article] [PubMed]
- 12.Li F, Jin Y, Pei X, Guo P, Dong K, Wang H, Chen Y, Guo P, Meng LB, Wang Z. Bioinformatics analysis and verification of gene targets for renal clear cell carcinoma. Comput Biol Chem. 2021;92:107453. doi: 10.1016/j.compbiolchem.2021.107453. [DOI] [PubMed] [Google Scholar]
- 13.Hermawan A, Ikawati M, Jenie RI, Khumaira A, Putri H, Nurhayati IP, Angraini SM, Muflikhasari HA. Identification of potential therapeutic target of naringenin in breast cancer stem cells inhibition by bioinformatics and in vitro studies. Saudi Pharm J. 2021;29(1):12–26. doi: 10.1016/j.jsps.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Wang L. Screening and identification of potential peripheral blood biomarkers for Alzheimer's disease based on bioinformatics analysis. Med Sci Monit. 2020;26:e924263. doi: 10.12659/MSM.924263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ning YL, Yang ZQ, Xian SX, Lin JZ, Lin XF, Chen WT. Bioinformatics analysis identifies hub genes and molecular pathways involved in sepsis-induced myopathy. Med Sci Monit. 2020;26:e919665. doi: 10.12659/MSM.919665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perri B, Cooper M, Lauryssen C, Anand N. Adverse swelling associated with use of rh-BMP-2 in anterior cervical discectomy and fusion: a case study. Spine J. 2007;7(2):235–239. doi: 10.1016/j.spinee.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 17.Smucker JD, Rhee JM, Singh K, Yoon ST, Heller JG. Increased swelling complications associated with off-label usage of rhBMP-2 in the anterior cervical spine. Spine. 2006;31(24):2813–2819. doi: 10.1097/01.brs.0000245863.52371.c2. [DOI] [PubMed] [Google Scholar]
- 18.McClellan JW, Mulconrey DS, Forbes RJ, Fullmer N. Vertebral bone resorption after transforaminal lumbar interbody fusion with bone morphogenetic protein (rhBMP-2) J Spinal Disord Tech. 2006;19(7):483–486. doi: 10.1097/01.bsd.0000211231.83716.4b. [DOI] [PubMed] [Google Scholar]
- 19.Rosen V. BMP and BMP inhibitors in bone. Ann New York Acad Sci. 2006;1068(1):19–25. doi: 10.1196/annals.1346.005. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura T, Shinohara Y, Momozaki S, Yoshimoto T, Noguchi K. Co-stimulation with bone morphogenetic protein-9 and FK506 induces remarkable osteoblastic differentiation in rat dedifferentiated fat cells. Biochem Biophys Res Commun. 2013;440(2):289–294. doi: 10.1016/j.bbrc.2013.09.073. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Hong S, Li M, Zhang J, Bi Y, He Y, Liu X, Nan G, Su Y, Zhu G, Li R, Zhang W, Wang J, Zhang H, Kong Y, Shui W, Wu N, He Y, Chen X, Luu HH, Haydon RC, Shi LL, He TC, Qin J. Noggin resistance contributes to the potent osteogenic capability of BMP9 in mesenchymal stem cells. J Orthop Res. 2013;31(11):1796–1803. doi: 10.1002/jor.22427. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura T, Shirakata Y, Shinohara Y, Miron RJ, Hasegawa-Nakamura K, Fujioka-Kobayashi M, Noguchi K. Comparison of the effects of recombinant human bone morphogenetic protein-2 and -9 on bone formation in rat calvarial critical-size defects. Clin Oral Investig. 2017;21(9):2671–2679. doi: 10.1007/s00784-017-2069-3. [DOI] [PubMed] [Google Scholar]
- 23.Sreekumar V, Aspera-Werz RH, Tendulkar G, Reumann MK, Freude T, Breitkopf-Heinlein K, Dooley S, Pscherer S, Ochs BG, Flesch I, Hofmann V, Nussler AK, Ehnert S. BMP9 a possible alternative drug for the recently withdrawn BMP7? New perspectives for (re-)implementation by personalized medicine. Arch Toxicol. 2017;91(3):1353–1366. doi: 10.1007/s00204-016-1796-6. [DOI] [PubMed] [Google Scholar]
- 24.Lin L, Qiu Q, Zhou N, Dong W, Shen J, Jiang W, Fang J, Hao J, Hu Z. Dickkopf-1 is involved in BMP9-induced osteoblast differentiation of C3H10T1/2 mesenchymal stem cells. BMB reports. 2016;49(3):179–184. doi: 10.5483/BMBRep.2016.49.3.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen L, Zou X, Zhang RX, Pi CJ, Wu N, Yin LJ, Deng ZL. IGF1 potentiates BMP9-induced osteogenic differentiation in mesenchymal stem cells through the enhancement of BMP/Smad signaling. BMB Reports. 2016;49(2):122–127. doi: 10.5483/BMBRep.2016.49.2.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daigang L, Jining Q, Jinlai L, Pengfei W, Chuan S, Liangku H, Ding T, Zhe S, Wei W, Zhong L, Kun Z. LPS-stimulated inflammation inhibits BMP-9-induced osteoblastic differentiation through crosstalk between BMP/MAPK and Smad signaling. Exp Cell Res. 2016;341(1):54–60. doi: 10.1016/j.yexcr.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Sharff KA, Song WX, Luo X, Tang N, Luo J, Chen J, Bi Y, He BC, Huang J, Li X, Jiang W, Zhu GH, Su Y, He Y, Shen J, Wang Y, Chen L, Zuo GW, Liu B, Pan X, Reid RR, Luu HH, Haydon RC, He TC. Hey1 basic helix-loop-helix protein plays an important role in mediating BMP9-induced osteogenic differentiation of mesenchymal progenitor cells. J Biol Chem. 2009;284(1):649–659. doi: 10.1074/jbc.M806389200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou YM, Yang YY, Jing YX, Yuan TJ, Sun LH, Tao B, Liu JM, Zhao HY. BMP9 reduces bone loss in ovariectomized mice by dual regulation of bone remodeling. J Bone Miner Res. 2020;35(5):978–993. doi: 10.1002/jbmr.3957. [DOI] [PubMed] [Google Scholar]
- 29.Yoon DK, Park JS, Rho GJ, Lee HJ, Sung IY, Son JH, Park BW, Kang YH, Byun SH, Hwang SC, Woo DK, Cho YC, Byun JH. The involvement of histone methylation in osteoblastic differentiation of human periosteum-derived cells cultured in vitro under hypoxic conditions. Cell Biochem Function. 2017;35(7):441–452. doi: 10.1002/cbf.3302. [DOI] [PubMed] [Google Scholar]
- 30.Hulley PA, Bishop T, Vernet A, Schneider JE, Edwards JR, Athanasou NA, Knowles HJ. Hypoxia-inducible factor 1-alpha does not regulate osteoclastogenesis but enhances bone resorption activity via prolyl-4-hydroxylase 2. J Pathol. 2017;242(3):322–333. doi: 10.1002/path.4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou N, Hu N, Liao JY, Lin LB, Zhao C, Si WK, Yang Z, Yi SX, Fan TX, Bao W, Liang X, Wei X, Chen H, Chen C, Chen Q, Lin X, Huang W. HIF-1alpha as a regulator of BMP2-induced chondrogenic differentiation, osteogenic differentiation, and endochondral ossification in stem cells. Cell Physiol Biochem. 2015;36(1):44–60. doi: 10.1159/000374052. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We state that the data will not be shared since all the raw data are present in the figures included in the article.