Abstract
Background
A recent study showed that the ABO gene, chr 9q34.2, which determines blood type, may affect COVID-19 disease severity, although this result has not been reproducible. A UK study of 2200 COVID-19 patients found no relationship of ABO blood type to disease severity. A Danish study identified ABO blood group as a risk factor for SARS-CoV-2 infection but not for hospitalization or death from COVID-19.
Aim
In the current study, we wished to analyze the relationship of ABO blood group and the ABO genetic locus to COVID-19 test positivity and mortality in subjects from the UK Biobank (UKB).
Methods
ABO blood type is from UKB data field 23165. Blood type was imputed for genotyped UK Biobank participants using three SNPs (rs505922, rs8176719, and rs8176746) in the ABO gene on chromosome 9q34.2. We analyzed the chromosome 9 snp rs657152 to assess the relationship of the ABO locus to COVID-19 test positivity and mortality.
Results
COVID-19 test results (negative or positive) were not related to blood group in males (p = 0.977, two tailed Fisher exact test) or females (p = 0.548). COVID-19 outcomes (alive or died) were not related to blood group in males (p = 0.102, two tailed Fisher exact test) or females (p = 0.226). We found no significant relationship of rs657152 to COVID-19 test positivity or mortality.
Conclusion
We were not able to confirm that ABO blood group influences risk of COVID-19 infection or outcome.
Keywords: Blood groups, Covid-19, Mortality
1. Introduction
Most people infected by SARS-CoV-2 never become ill, whereas some die within days. Age and preexisting conditions, such as obesity, account for some of the disparity. But genetics plays a role.
Genome-wide association studies have found multiple genes and loci that increase risk of respiratory failure in COVID-19. Analyzing the SNP rs657152, located in the intron area of ABO, one study found that the ABO gene, chr 9q34.2, which determines blood type, may affect disease severity [1], although this result has not been reproducible. A UK study of 2200 COVID-19 patients found no relationship of ABO blood type to disease severity [2]. A Danish study identified ABO blood group as a risk factor for SARS-CoV-2 infection but not for hospitalization or death from COVID-19 [3].
In the current study, we analyzed the relationship of ABO blood types and rs657152, which has been associated with cancer and cardiocerebrovascular disease risk [4], to COVID-19 test positivity and mortality.
2. Methods
We utilized UK Biobank (UKB) data. The UKB consists of more than 500,000 community volunteers aged 40–70 years at baseline (2006–2010), living close to 22 assessment centers in England, Scotland, and Wales. Baseline assessments include demographics, lifestyle, and disease history, with linkages to electronic medical records. Our UK Biobank application was approved as UKB project 57,245 (S.L., P.H.R.). Electronic linkage between UKB records and National Health Service COVID-19 laboratory test results in England are available from March 16 to April 26, 2020, including the peak of daily COVID-19 laboratory-confirmed cases in the current outbreak. During this period, testing of older groups was largely restricted to hospital inpatients with clinical signs of infection, so test positivity is considered a good marker of severe COVID-19 [5].
ABO blood type is from UKB data field 23165. Blood type was imputed for genotyped UK Biobank participants using three SNPs (rs505922, rs8176719, and rs8176746) in the ABO gene on chromosome 9q34.2. rs8176719 deletion was taken as indicative of haplotype O; for participants with no result for rs8176719, rs505922 of T was used to indicate type O. Type B was indicated by T at rs8176746 [[6], [7], [8], [9]].
Data processing was performed on Minerva, a Linux mainframe with Centos 7.6, at the Icahn School of Medicine at Mount Sinai. We used PLINK, a whole-genome association analysis toolset, to process the UKB chromosome 9 files; and the UK Biobank Data Parser (ukbb parser), a python-based package that allows easy interfacing with the large UK Biobank dataset [10].
3. Results
We analyzed data from 12,575 subjects. The mean age was 58 ± 8 (mean ± SD). 52% were female, 48% were male. 98% were white British. 5.7% were positive for COVID-19. Overall mortality of COVID-19 test-positive subjects was 16%.
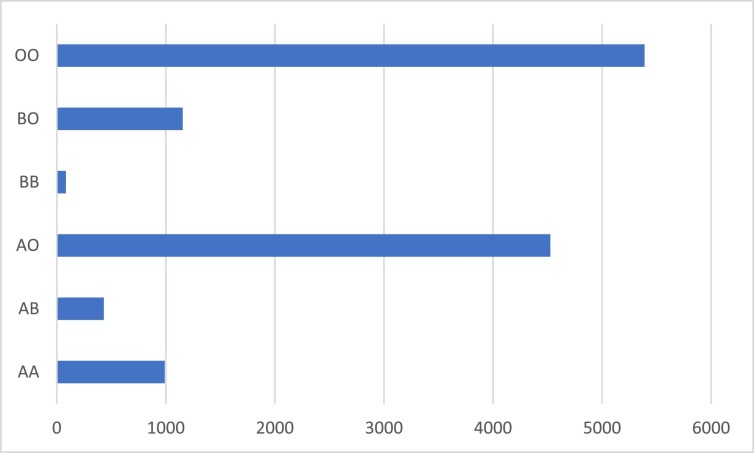
Blood group versus number of cases is shown in Fig. 1 . Type OO was most frequent.
Fig. 1.
Blood group versus number of cases, 12,575 subjects in this analysis.
COVID-19 test results (negative or positive) were not related to blood group in males (p = 0.977, two tailed Fisher exact test) or females (p = 0.548, Table 1 ). COVID-19 outcomes (alive or died) were not related to blood group in males (p = 0.102, two tailed Fisher exact test) or females (p = 0.226, Table 2 ).
Table 1.
COVID-19 test results (negative or positive) versus blood group by gender. The results for males (p = 0.977, two tailed Fisher exact test) and females (p = 0.548) were not significant.
| Gender | Blood group | Result |
Total | p value | ||
|---|---|---|---|---|---|---|
| Negative | Positive | |||||
| Female | AA | Count | 483 | 31 | 514 | 0.548 |
| % within females | 94.00% | 6.00% | 100.00% | |||
| AB | Count | 217 | 10 | 227 | ||
| % within females | 95.60% | 4.40% | 100.00% | |||
| AO | Count | 2254 | 132 | 2386 | ||
| % within females | 94.50% | 5.50% | 100.00% | |||
| BB | Count | 39 | 0 | 39 | ||
| % within females | 100.00% | 0.00% | 100.00% | |||
| BO | Count | 572 | 26 | 598 | ||
| % within females | 95.70% | 4.30% | 100.00% | |||
| OO | Count | 2646 | 152 | 2798 | ||
| % within females | 94.60% | 5.40% | 100.00% | |||
| Total | Total | Count | 6211 | 351 | 6562 | |
| % within males | 94.70% | 5.30% | 100.00% | |||
| Male | AA | Count | 449 | 27 | 476 | 0.977 |
| % within males | 94.30% | 5.70% | 100.00% | |||
| AB | Count | 191 | 13 | 204 | ||
| % within males | 93.60% | 6.40% | 100.00% | |||
| AO | Count | 2007 | 133 | 2140 | ||
| % within males | 93.80% | 6.20% | 100.00% | |||
| BB | Count | 41 | 3 | 44 | ||
| % within males | 93.20% | 6.80% | 100.00% | |||
| BO | Count | 520 | 37 | 557 | ||
| % within males | 93.40% | 6.60% | 100.00% | |||
| OO | Count | 2436 | 156 | 2592 | ||
| % within males | 94.00% | 6.00% | 100.00% | |||
| Total | Total | Count | 5644 | 369 | 6013 | |
| % within both | 93.90% | 6.10% | 100.00% | |||
Table 2.
COVID-19 outcome (alive or died) versus blood group by gender. The results for males (p = 0.102, two tailed Fisher exact test) and females (p = 0.226) were not significant.
| Gender | Blood group | Alive | Died | p value | ||
|---|---|---|---|---|---|---|
| Females | AA | Count | 25 | 6 | 31 | |
| % within females | 80.60% | 19.40% | 100.00% | 0.226 | ||
| AB | Count | 10 | 0 | 10 | ||
| % within females | 100.00% | 0.00% | 100.00% | |||
| AO | Count | 114 | 18 | 132 | ||
| % within females | 86.40% | 13.60% | 100.00% | |||
| BO | Count | 23 | 3 | 26 | ||
| % within females | 88.50% | 11.50% | 100.00% | |||
| OO | Count | 140 | 12 | 152 | ||
| % within females | 92.10% | 7.90% | 100.00% | |||
| Total | Count | 312 | 39 | 351 | ||
| % within females | 88.90% | 11.10% | 100.00% | |||
| Males | AA | Count | 23 | 4 | 27 | 0.102 |
| % within males | 85.20% | 14.80% | 100.00% | |||
| AB | Count | 9 | 4 | 13 | ||
| % within males | 69.20% | 30.80% | 100.00% | |||
| AO | Count | 111 | 22 | 133 | ||
| % within males | 83.50% | 16.50% | 100.00% | |||
| BB | Count | 3 | 0 | 3 | ||
| % within males | 100.00% | 0.00% | 100.00% | |||
| BO | Count | 33 | 4 | 37 | ||
| % within males | 89.20% | 10.80% | 100.00% | |||
| OO | Count | 114 | 42 | 156 | ||
| % within males | 73.10% | 26.90% | 100.00% | |||
| Total | Count | 293 | 76 | 369 | ||
| % within males | 79.40% | 20.60% | 100.00% |
COVID-19 test results (negative or positive) were not related to rs657152 genotype for males (p = 0.707, two tailed Fisher exact test) or females (p = 0.875, Table 3 ). COVID-19 outcomes (alive or died) were marginally related to rs657152 genotype in males (p = 0.046, two tailed Fisher exact test); the result was insignificant when the Bonferroni correction for multiple comparisons was applied [11]. For females there was no relationship between COVID-19 outcome and rs657152 genotype (p = 0.144, Table 4 ).
Table 3.
COVID-19 test results (negative or positive) versus rs657152 genotype by gender. The results for males (p = 0.707, two tailed Fisher exact test) and females (p = 0.875) were not significant.
| Gender | rs657152 | COVID-19 |
Total | p value | ||
|---|---|---|---|---|---|---|
| Negative | Positive | |||||
| Female | AA | Count | 763 | 45 | 808 | 0.875 |
| % within females | 94.40% | 5.60% | 100.00% | |||
| AC | Count | 2833 | 155 | 2988 | ||
| % within females | 94.80% | 5.20% | 100.00% | |||
| CC | Count | 2593 | 148 | 2741 | ||
| % within females | 94.60% | 5.40% | 100.00% | |||
| Total | Count | 6189 | 348 | 6537 | ||
| % within females | 94.70% | 5.30% | 100.00% | |||
| Male | AA | Count | 690 | 47 | 737 | 0.707 |
| % within males | 93.60% | 6.40% | 100.00% | |||
| AC | Count | 2526 | 172 | 2698 | ||
| % within males | 93.60% | 6.40% | 100.00% | |||
| CC | Count | 2410 | 150 | 2560 | ||
| % within males | 94.10% | 5.90% | 100.00% | |||
| Total | Count | 5626 | 369 | 5995 | ||
| % within males | 93.80% | 6.20% | 100.00% | |||
Table 4.
COVID-19 outcome (alive or died) versus rs657152 genotype by gender. The result for males (p = 0.046, two tailed Fisher exact test) was of borderline significance, females (p = 0.144) not significant.
| Gender | rs657152 | Alive | Died | Total | p value | |
|---|---|---|---|---|---|---|
| Female | AA | Count | 39 | 6 | 45 | 0.144 |
| % within females | 86.70% | 13.30% | 100.00% | |||
| AC | Count | 133 | 22 | 155 | ||
| % within females | 85.80% | 14.20% | 100.00% | |||
| CC | Count | 137 | 11 | 148 | ||
| % within females | 92.60% | 7.40% | 100.00% | |||
| Total | Count | 309 | 39 | 348 | ||
| % within females | 88.80% | 11.20% | 100.00% | |||
| Male | AA | Count | 36 | 11 | 47 | 0.046 |
| % within males | 76.60% | 23.40% | 100.00% | |||
| AC | Count | 146 | 26 | 172 | ||
| % within males | 84.90% | 15.10% | 100.00% | |||
| CC | Count | 111 | 39 | 150 | ||
| % within males | 74.00% | 26.00% | 100.00% | |||
| Total | Count | 293 | 76 | 369 | ||
| % within males | 79.40% | 20.60% | 100.00% |
4. Discussion
Studies of blood type have reported that type O blood protects against COVID-19, whereas type A blood makes an individual more vulnerable [12]. In addition, O blood is said to provide modest protection against severe illness, whereas group A is detrimental [13,14]. Type O individuals are less prone to thrombosis and vascular dysfunction than non-O individuals and therefore could be at lesser risk in case of severe lung dysfunction [15]. We are not able to confirm these observations with UKB data. Others, as well, are skeptical of any significant role ABO blood groups might play in COVID-19 [2].
A 3p21.31 6 gene cluster is a genetic susceptibility locus in patients with Covid-19 and respiratory failure [1]. This locus came from Neanderthals through interbreeding with Homo sapiens tens of thousands of years ago [16]. It appears in 16% of Europeans and 50% of south Asians but is absent from most people of African descent. UKB did not have the SNPS that were used to associate this locus with COVID-19.
Other Covid-19 susceptibility genes have been identified, for example, IFNAR2, which codes for a cell receptor for interferon. A variant of IFNAR2 found in one in four Europeans raises the risk of severe COVID-19 by 30%. The gene DPP9 codes for an enzyme active in lung disease. Another, gene, TYK2, encodes a signaling protein involved in inflammation. Drugs that target DPP9 and TYK2 proteins are already in use: inhibitors of DPP9 for diabetes, and baricitinib, which blocks TYK2, for arthritis. Baricitinib is in early clinical testing for COVID-19 [2].
One weakness in our study is immortal time bias [17]. During the period of observation, an interval exists during which the outcome event cannot occur. The research participants are immortal because they must survive long enough for the outcome event being evaluated. In the current study the outcome events would be COVID-19 test positivity and death. The possibility exists that we have not been able to substantiate the role of the ABO locus 9q34.2 and ABO blood groups in COVID-19 due to immortal time bias.
The impact of genetic risk factors on COVID-19 seems modest at best. The massive vaccination campaign now underway, combined with increasing population immunity, will no doubt end the current epidemic.
Funding sources
None.
CRediT authorship contribution statement
Dr. Lehrer and Dr. Rheinstein contributed equally to the conception, writing, and data analysis of this study.
Declaration of competing interest
None.
Footnotes
This work was supported in part through the computational resources and staff expertise provided by Scientific Computing at the Icahn School of Medicine at Mount Sinai.
Research reported in this paper was also supported by the Office of Research Infrastructure of the National Institutes of Health under award numbers S10OD018522 and S10OD026880. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
References
- 1.Group SCG, Ellinghaus D., Degenhardt F., Bujanda L., Buti M., Albillos A., Invernizzi P., Fernandez J., Prati D., Baselli G., Asselta R., Grimsrud M.M., Milani C., Aziz F., Kassens J., May S., Wendorff M., Wienbrandt L., Uellendahl-Werth F., Zheng T., Yi X., de Pablo R., Chercoles A.G., Palom A., Garcia-Fernandez A.E., Rodriguez-Frias F., Zanella A., Bandera A., Protti A., Aghemo A., Lleo A., Biondi A., Caballero-Garralda A., Gori A., Tanck A., Carreras Nolla A., Latiano A., Fracanzani A.L., Peschuck A., Julia A., Pesenti A., Voza A., Jimenez D., Mateos B., Nafria Jimenez B., Quereda C., Paccapelo C., Gassner C., Angelini C., Cea C., Solier A., Pestana D., Muniz-Diaz E., Sandoval E., Paraboschi E.M., Navas E., Garcia Sanchez F., Ceriotti F., Martinelli-Boneschi F., Peyvandi F., Blasi F., Tellez L., Blanco-Grau A., Hemmrich-Stanisak G., Grasselli G., Costantino G., Cardamone G., Foti G., Aneli S., Kurihara H., ElAbd H., My I., Galvan-Femenia I., Martin J., Erdmann J., Ferrusquia-Acosta J., Garcia-Etxebarria K., Izquierdo-Sanchez L., Bettini L.R., Sumoy L., Terranova L., Moreira L., Santoro L., Scudeller L., Mesonero F., Roade L., Ruhlemann M.C., Schaefer M., Carrabba M., Riveiro-Barciela M., Figuera Basso M.E., Valsecchi M.G., Hernandez-Tejero M., Acosta-Herrera M., D’Angio M., Baldini M., Cazzaniga M., Schulzky M., Cecconi M., Wittig M., Ciccarelli M., Rodriguez-Gandia M., Bocciolone M., Miozzo M., Montano N., Braun N., Sacchi N., Martinez N., Ozer O., Palmieri O., Faverio P., Preatoni P., Bonfanti P., Omodei P., Tentorio P., Castro P., Rodrigues P.M., Blandino Ortiz A., de Cid R., Ferrer R., Gualtierotti R., Nieto R., Goerg S., Badalamenti S., Marsal S., Matullo G., Pelusi S., Juzenas S., Aliberti S., Monzani V., Moreno V., Wesse T., Lenz T.L., Pumarola T., Rimoldi V., Bosari S., Albrecht W., Peter W., Romero-Gomez M., D’Amato M., Duga S., Banales J.M., Hov J.R., Folseraas T., Valenti L., Franke A., Karlsen T.H. Genomewide association study of severe covid-19 with respiratory failure. N. Engl. J. Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaiser J. Found: genes that sway the course of the coronavirus. Science. 2020;370:275–276. doi: 10.1126/science.370.6514.275. [DOI] [PubMed] [Google Scholar]
- 3.Barnkob M.B., Pottegård A., Støvring H., Haunstrup T.M., Homburg K., Larsen R., Hansen M.B., Titlestad K., Aagaard B., Møller B.K., Barington T. Reduced prevalence of SARS-CoV-2 infection in ABO blood group O. Blood Adv. 2020;4:4990–4993. doi: 10.1182/bloodadvances.2020002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y., Liu L., Huang Y., Zheng H., Li L. Association of ABO polymorphisms and pancreatic cancer/cardiocerebrovascular disease: a meta-analysis. BMC Med. Genet. 2020;21:1–10. doi: 10.1186/s12881-020-0975-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atkins J.L., Masoli J.A., Delgado J., Pilling L.C., Kuo C.-L., Kuchel G.A., Melzer D. Preexisting comorbidities predicting COVID-19 and mortality in the UK Biobank community cohort. J. Gerontol. A Biol. Sci. Med. Sci. 2020 doi: 10.1093/gerona/glaa183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melzer D., Perry J.R., Hernandez D., Corsi A.-M., Stevens K., Rafferty I., Lauretani F., Murray A., Gibbs J.R., Paolisso G. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paré G., Chasman D.I., Kellogg M., Zee R.Y., Rifai N., Badola S., Miletich J.P., Ridker P.M. Novel association of ABO histo-blood group antigen with soluble ICAM-1: results of a genome-wide association study of 6,578 women. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolpin B.M., Kraft P., Gross M., Helzlsouer K., Bueno-de-Mesquita H.B., Steplowski E., Stolzenberg-Solomon R.Z., Arslan A.A., Jacobs E.J., LaCroix A. Pancreatic cancer risk and ABO blood group alleles: results from the pancreatic cancer cohort consortium. Cancer Res. 2010;70:1015–1023. doi: 10.1158/0008-5472.CAN-09-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groot H.E., Villegas Sierra L.E., Said M.A., Lipsic E., Karper J.C., van der Harst P. Genetically determined ABO blood group and its associations with health and disease. Arterioscler. Thromb. Vasc. Biol. 2020;40:830–838. doi: 10.1161/ATVBAHA.119.313658. [DOI] [PubMed] [Google Scholar]
- 10.Zhu A., Salminen L.E., Thompson P.M., Jahanshad N. Organization for Human Brain Mapping; Rome, Italy: 2019. The UK Biobank Data Parser: A Tool With Built in and Customizable Filters for Brain Studies. (June 9–13, 2019) [Google Scholar]
- 11.Weisstein E.W. Bonferroni Correction. 2004. https://mathworld.wolfram.com/
- 12.Zhao J., Yang Y., Huang H., Li D., Gu D., Lu X., Zhang Z., Liu L., Liu T., Liu Y. Relationship between the ABO blood group and the COVID-19 susceptibility. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dai X. ABO blood group predisposes to COVID-19 severity and cardiovascular diseases. Eur. J. Prev. Cardiol. 2020;27:1436–1437. doi: 10.1177/2047487320922370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muñiz-Diaz E., Llopis J., Parra R., Roig I., Ferrer G., Grifols J., Millán A., Ene G., Ramiro L., Maglio L. Relationship between the ABO blood group and COVID-19 susceptibility, severity and mortality in two cohorts of patients. Blood Transfus. 2021;19:54. doi: 10.2450/2020.0256-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pendu J.L., Breiman A., Rocher J., Dion M., Ruvoen-Clouet N. ABO blood types and COVID-19: spurious, anecdotal, or truly important relationships? A reasoned review of available data. Viruses. 2021;13 doi: 10.3390/v13020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeberg H., Pääbo S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature. 2020;587:610–612. doi: 10.1038/s41586-020-2818-3. [DOI] [PubMed] [Google Scholar]
- 17.Yadav K., Lewis R.J. Immortal time bias in observational studies. JAMA. 2021;325:686–687. doi: 10.1001/jama.2020.9151. [DOI] [PubMed] [Google Scholar]