Abstract
Antiviral therapeutics is one effective avenue to control and end this devastating COVID-19 pandemic. The viral RNA-dependent RNA polymerase (RdRp) of SARS-CoV-2 has been recognized as a valuable target of antivirals. However, the cell-free SARS-CoV-2 RdRp biochemical assay requires the conversion of nucleotide prodrugs into the active triphosphate forms, which regularly occurs in cells yet is a complicated multiple-step chemical process in vitro, and thus hinders the utility of this cell-free assay in the rapid discovery of RdRp inhibitors. In addition, SARS-CoV-2 exoribonuclease provides the proof-reading capacity to viral RdRp, thus creates relatively high resistance threshold of viral RdRp to nucleotide analog inhibitors, which must be examined and evaluated in the development of this class of antivirals. Here, we report a cell-based assay to evaluate the efficacy of nucleotide analog compounds against SARS-CoV-2 RdRp and assess their tolerance to viral exoribonuclease-mediated proof-reading. By testing seven commonly used nucleotide analog viral polymerase inhibitors, Remdesivir, Molnupiravir, Ribavirin, Favipiravir, Penciclovir, Entecavir and Tenofovir, we found that both Molnupiravir and Remdesivir showed the strong inhibition of SARS-CoV-2 RdRp, with EC50 value of 0.22 μM and 0.67 μM, respectively. Moreover, our results suggested that exoribonuclease nsp14 increases resistance of SARS-CoV-2 RdRp to nucleotide analog inhibitors. We also determined that Remdesivir presented the highest resistance to viral exoribonuclease activity in cells. Therefore, we have developed a cell-based SARS-CoV-2 RdRp assay which can be deployed to discover SARS-CoV-2 RdRp inhibitors that are urgently needed to treat COVID-19 patients.
Keywords: COVID-19, SARS-CoV-2, RdRp, Antiviral, High-throughput, Nucleotide analog inhibitor, Remdesivir
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes the coronavirus disease 2019 (COVID-19) pandemic, the most devastating one in the past decades with more than one million deaths and millions infected(Wu et al., 2020). The numbers of mortality and morbidity will only increase with the pandemic raging on. The mortality of SARS-CoV-2 infections is mostly due to virus-induced acute respiratory distress syndrome(Goulet et al., 2013), especially in elder patients(Chen et al., 2020; Phan et al., 2020; Remuzzi and Remuzzi, 2020). Although intensive efforts are being made to find prophylactic or therapeutic measures, currently, Remdesivir is the only drug that is approved by the FDA for the treatment of COVID-19(https://www.covid19treatmentguidelines.nih.gov/therapeutic-management/).
SARS-CoV-2 is a positive-sense single-strand RNA virus, belongs to the beta-coronavirus subgenus Sarbecovirus which also includes SARS-CoV, the causative agent of the Severe Acute Respiratory Syndrome (SARS) outbreak in 2002–2003, as well as MERS-CoV which emerged in 2012 and caused Middle East Respiratory Syndrome (MERS) (Aftab et al., 2020; Chen et al., 2020; Fehr and Perlman, 2015). SARS-CoV-2 genome seizes from 29.8 kb to 29.9 kb in length, encodes 16 non-structural proteins (nsp1 to nsp16). Some of these 16 nsps are enzymes that are essential for SARS-CoV-2 replication. These include papain-like protease (nsp3), chymotrypsin-like main protease (3CL protease, nsp5), primase complex (nsp7–nsp8), RNA-dependent RNA polymerase RdRp (nsp12), helicase (nsp13), and exoribonuclease (nsp14)(Zhu et al., 2020),(Shannon et al., 2020a), all of which are potential targets for the development of anti- SARS-CoV-2 drugs.
Among these viral enzymes, RdRp presents a valuable target due to its essential role in viral RNA synthesis, the lack of host homolog, and high sequence and structural conservation among coronaviruses (Chien et al., 2020),(Aftab et al., 2020; Hillen et al., 2020; Zhu et al., 2020). Studies have shown that the processivity of RdRp requires viral cofactors nsp7 and nsp8 (Subissi et al., 2014). This key role of nsp7 and nsp8 is highlighted in the structure of SARS-CoV-2 RdRp complex comprising the viral proteins nsp12, nsp7, nsp8, and RNA template-product duplex (Gao et al., 2020; Hillen et al., 2020). Arguably, the most promising and broad-spectrum viral RdRp inhibitors are nucleos(t)ide analogs (NAs). NAs are incorporated into the nascent viral RNA by RdRp, then either terminate RNA synthesis or slow down of RNA synthesis, thus exerting their antiviral effects (Shannon et al., 2020a; Zhu et al., 2020). Since the pandemic began, many NA inhibitors have been tested against SARS-CoV-2. Particularly, Remdesivir shows strong anti-SARS-CoV-2 activity in vitro (Hillen et al., 2020; Jorgensen et al., 2020; Norrie, 2020), and has recently been approved by the FDA in the U.S. for the treatment of COVID-19 patients (Antinori et al., 2020; Grein et al., 2020). Molnupiravir is an oral pro-drug of NHC (EIDD-2801; also known as molnupiravir or MK-4482) reduced SARS-CoV and MERS-CoV replication and pathogenesis by targeting to viral RdRp, which is well investigated in mice. Later, it was proved that molnupiravir can effeicently restrict viral replication in various animal models(Cox et al., 2021; Sheahan et al., 2020; Wahl et al., 2021). Currently, molnupiravir is under phase 2 clinical trail for prevention and treatment Of Covid-19 (https://clinicaltrials.gov/ct2/show/NCT04405739). Chien et al. recently evaluated the active nucleoside triphosphate forms of six antiviral prodrugs, Alovudine, Tenofovir alafenamide, AZT, Abacavir, Lamivudine, and Emtricitabine, by measuring their ability to terminate the polymerase reaction catalyzed by SARS-CoV-2 RdRp (Chien et al., 2020). Carolina et al. reported that both Daclatasvir and Sofosbuvir inhibit SARS-CoV-2 replication. Specifically, Daclatasvir, which targets NS5A of HCV, also inhibits SARS-CoV-2 in vitro, likely due to structural conservation between these two viral RdRps, HCV NS5B and and SARS-CoV-2 nsp12 (Sacramento et al., 2020).
It is known that all antiviral nucleoside prodrugs targeting RdRp need to be metabolized into the 5′-triphostrates after entering the host cell, then compete with endogenous nucleotide triphosphates as substrates for viral RdRp. Several studies have utilized a cell-free SARS-CoV-2 RdRp assay to assess the potency of active 5′-triphosphate NA inhibitors (Jockusch et al., 2020; Shannon et al., 2020b; Wang et al., 2020a). However, in vitro synthesis of nucleotide triphosphates is a complex and time-consuming process, which constitutes a barrier to the screening of potential NA inhibitors of RdRp using these cell-free assays. A cell-based SARS CoV-2 RdRp assay should circumvent this obstacle to directly test nucleotide prodrugs. In addition, SARS-CoV2 encodes the nsp14 exoribonuclease which is able to excise erroneous mutagenic nucleotides incorporated by nsp12 into viral RNA, thus causes resistance to nucleotide analog drugs (Smith et al., 2013). Because of this mechanism, Ribavirin is excised from growing RNA chain of CoVs, thus shows no antiviral activity(Ferron et al., 2018). Therefore, nsp14 needs to be included in the screening assay for RdRp inhibitors, which is difficult for the cell-free protocol.
In this study, we have developed a cell-based assay with Gaussia-luciferase (Gluc) as the reporter to evaluate the anti-SARS-CoV-2 RdRp activity of nucleotide prodrugs without the need of synthesizing the active nucleotide triphosphates as for the cell-free assays. In addition, using this assay, we demonstrated that the exonuclease activity of nsp14 increases the RdRp resistance to NA inhibitors.
2. Materials and methods
2.1. Cells
293T cells (ATCC CRL-3216) and A549 cells (ATCC CCL-185) were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco) with 10% (v/v) fetal bovine serum (FBS; Gibco). The cells were cultured at 37 °C in a humidified atmosphere of 5% CO2. HEK293T cells were transfected using Vigofect transfection reagents (Vigorous), according to Manufacturer's instructions.
2.2. Plasmid DNA, antibodies and reagents
The plasmids pCOVID19-nsp12, pCOVID19-nsp7, pCOVID19-nsp8, pCOVID19-nsp10, pCOVID19-nsp14 were used to express codon-optimized Flag-nsp12, Flag-nsp7, Flag-nsp8, Flag-nsp10 and Flag-nsp14, respectively, all of which contain a Flag tag at the C-terminus. For construction of nsp12 (D760A) and nsp14 (D90A & E92A) mutants, the sites-directed mutagenesis was performed using QuickMutation™ Site-Directed Mutagenesis Kit (Beyotime) following the Manufacturer's instructions. All plasmid DNA constructs were sequenced to avert unwanted mutations. In vitro CoV-Gluc mRNA was generated using the mMESSAGE mMACHINETM T7 Ultra kit under the Manufacturer's instructions.
To generate plasmid pCoV-Gluc, which produces positive-strand of vRNA encoding Gaussia luciferase (Gluc), 5′UTR-Gluc-3′UTR was first synthesized (Sangon Biotech), then inserted into the BamHl and Notl sites of pRetroX-tight-Pur vector (kindly provided by Dr.Guo Fei). The primers are Forward (5′-GGC GGA TCC ATT AAA GGT TTA TAC-3′) and Reverse (5′-TTA GCG GCC GCG TCA TTC TCC TAA GAA-3′).
Coelenterazine-h was purchased from Promega. Remdesivir (S8932), Ribavirin (S2504), Favipiravir (S7975), Entecavir (S5246), Tenofovir (S1401) and Penciclovir (S4184) were purchased from Selleck chemicals (Houston, TX, USA) and prepared in DMSO. DYKDDDDK (Flag) Tag antibody (8146) was purchased from Cell Signaling Technology, Inc (CST, USA), β-actin antibody (ab8224) from Abcam, HRP-conjugated goat anti-mouse secondary antibody (sc-2031) from Santa Cruz Co.
2.3. Western blots
Transfected HEK293T cells were collected and lysed in RIPA buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 1% NP-40, 0.1% SDS, 1 mM EDTA, 0.5% sodium deoxycholate. Cell lysates were mixed with SDS loading buffer and boiled for 20 min, followed by electrophoresis in 10% polyacrylamide-SDS gels. Proteins were transferred onto PVDF membrane. After blocking with 5% Skim milk, the membrane was blotted with anti-Flag (1:1000), or anti-β-actin (1:5000) antibodies. After incubation with HRP-conjugated secondary antibodies (1:5000), protein signals were detected with enhanced chemiluminescence.
2.4. Real-time RT-PCR
Total RNA of transfected HEK293T cells was extracted with TRIzol reagent (Life Technologies). The extracted RNA were converted to plus-strand and minus-strand cDNA with M-MLV reverse transcriptase (Promega) using primer plus Gluc-RT (5′- TGG ATC TTG CTG GCG AAT GT -3′) and minus Gluc-RT (5′- ACT GTC GTT GAC AGG ACA CG -3′) respectively for 1 h at 37 °C. The cDNAs were quantified using SsoFast EvaGreen Supermix (Bio-Rad) in applied system (Thermo Fisher Scientific). primers used for PCR are Gluc Forward (5′- CGG GTG TGA CCG AAA GGT AA -3′) and Reverse (5′- TGG ATC TTG CTG GCG AAT GT -3′). Levels of GAPDH RNA was also quantified with primers Forward (5′- GTC CAC TGG CGT CTT CAC CA -3′) and Reverse (5′- GTG GCA GTG ATG GCA TGG AC -3′), and results were used to normalize the amount of Gluc RNA.
2.5. Cell viability assay
Cell viability was examined with Cell Counting kit-8 (CCK8, Beyotime), which is a water-soluble tetrazolium salt-8 (WST-8) reagent. 1 μL of each tested compound was added to HEK293T cells and incubated for 24 h. Then 10 μL of CCk-8 reagent was added into each well and incubated for 30–90 min at 37 °C of 5% CO2. The absorbance at 450 nm was measured using the Enspire 2300 Multiable reader (PekinElmer).
2.6. Gluc activity assay
A stock of coelenterazine-h was dissolved in absolute ethyl alcohol to a concentration of 1.022 mmol/L. Right before each assay, the stock was diluted in PBS to 16.7 μM and incubated in the dark for 30 min at room temperature. For luminescence assay, 10 μL of supernatant was added to each well of a white and opaque 96-well plate, and 60 μL of 16.7 μM coelenterazine-h was injected, and luminescence was measured for 0.5 s using the Berthold Centro XS3 LB 960 microplate luminometer.
2.7. Statistical analysis
Data are presented as the means ± SD from at least three independent experiments and were analyzed with GraphPad Prism. Difference between the two indicated settings were considered statistically significant if p < 0.05(*), p < 0.01(**), p < 0.001(***). n.s indicates non-significant.
3. Results
3.1. Development of the CoV-RdRp-Gluc reporter assay
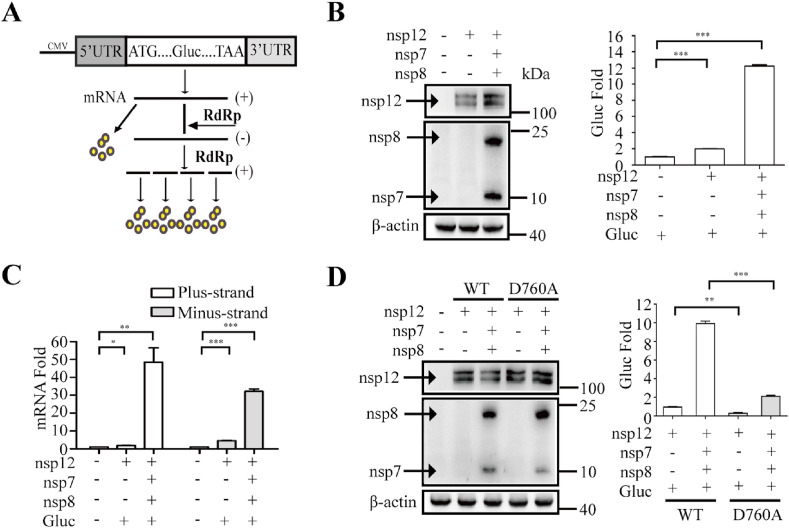
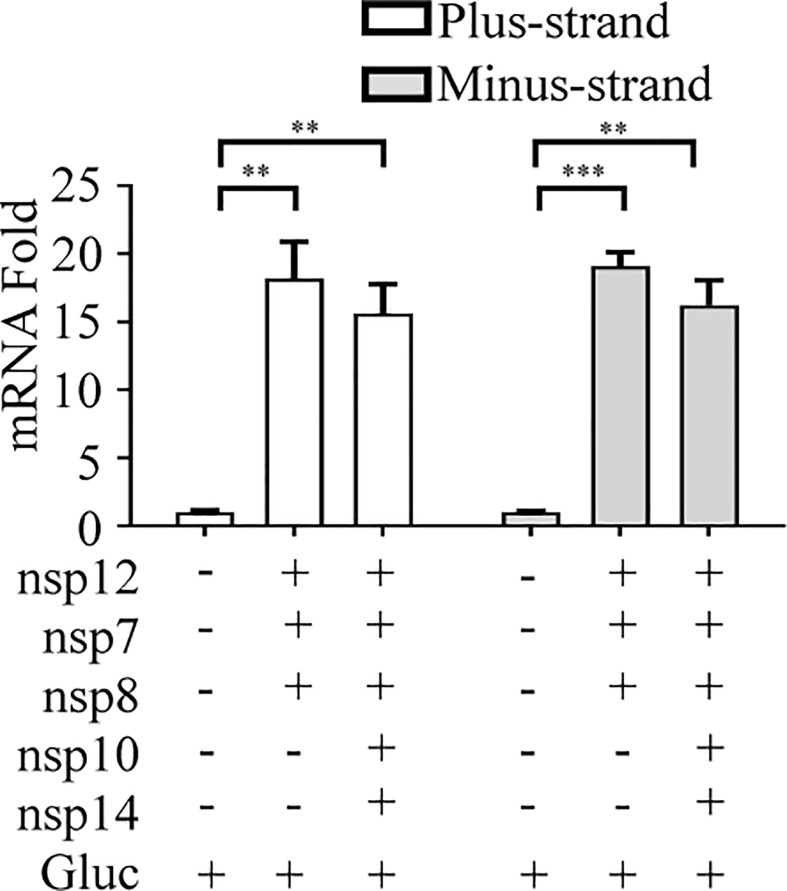
To develop the SARS-CoV-2 RdRp-dependent Gluc reporter system, named the CoV-RdRp-Gluc reporter assay for short, we first constructed the expression vector with the Gluc gene flanked by 5′ and 3’ untranslated regions (UTRs) of SARS-COV-2 (Fig. 1 A). Transcription of Gluc is initiated by the CMV (cytomegalovirus) promoter. Upon expression of SARS-CoV-2 RdRp, the Gluc RNA is further amplified by viral RdRp, resulting in the increase of Gluc mRNA and Gluc protein. Therefore, the increased Gluc activity reports the activity of SARS-CoV-2 RdRp. As expected, when the CoV-Gluc reporter-expressing cells were transfected with nsp12-expressing vector alone or together with nsp7 and nsp8, the Gluc activity increased by 2- and 12-fold, respectively, compared with that of cells expressing the reporter alone (Fig. 1B). Consistent with increased Gluc activity in the presence of RdRp, the levels of plus-strand Gluc RNA and minus-strand Gluc RNA were also significantly elevated, as determined by RT-qPCR (Fig. 1C), which verifies that RdRp increases the Gluc activity by amplifying Gluc RNA through the minus-strand stage. As a control, the RdRp mutant D760A which has one of the key catalytic residues (759-SDD-761) mutated (Subissi et al., 2014), significantly impaired the ability of stimulating Gluc expression, even together with nsp7 and nsp8 (Fig. 1D). These results demonstrate that SARS-CoV-2 RdRp is capable of stimulating the expression of the CoV-Gluc reporter in cells and that this function is markedly stimulated by viral nsp7 and nsp8, which is in agreement with early studies of coronavirus RdRp (Hillen et al., 2020; Subissi et al., 2014).
Fig. 1.
SARS-CoV-2 RdRp stimulates the expression of CoV-Gluc reporter. (A) Schematic diagram of the Gluc reporter system. The expression cassette of Gluc is in sense strand, which is flanked by the 5′ and 3′ untranslated regions (UTRs) of SARS-COV-2. The negative-sense vRNA is first synthesized by SARS-CoV-2 RdRp, followed by transcription into plus-strand RNA (mRNA) to express Gluc. (B) HEK293T cells (4 × 105) were co-transfected with the indicated plasmids (0.5 μg of each plasmid). At 48 h post-transfection, cells were collected for Western blot. Western blots were probed with anti-Flag antibody to detect nsp12 (upper panel), nsp7 and nsp8 (middle panel). β-actin was detected (lower panel) as the loading control. The supernatants of the transfected cells were subjected to Gluc activity assay. Results shown are the average of three independent experiments. (C) Levels of both plus-strand and minus-strand CoV-Gluc RNA were determined by quantitative RT-PCR from HEK293T cells that were transfected with the indicated plasmid DNA. Results shown are the average of three independent experiments. (D) The D760A RdRp mutant disables CoV-Gluc activity. HEK293T cells were transfected with the indicated plasmid DNA. Expression of RdRp, nsp7, and nsp8 was examined by Western blotting. Levels of Gluc in the culture supernatants were measured, and the level from cells expressing CoV-Gluc and wild type RdRp was arbitrarily set at 1. Results shown are the average of three independent transfection experiments.
3.2. Optimization of the CoV-RdRp-Gluc reporter assay
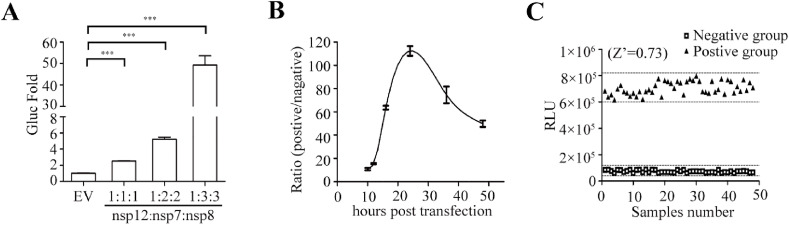
Next, we optimized this report assay to improve the sensitivity of Gluc expression to RdRp. Previous studies suggested that the active form of the SARS-CoV-2 RdRp complex consist of nsp12, nsp7 and nsp8 at a certain ratio (Gao et al., 2020; Hillen et al., 2020). This suggests that expression levels of these three viral proteins may affect the formation of active RdRp complex and subsequent viral RNA amplification. We therefore tested different ratios of the plasmid DNA expressing RdRp, nsp7 and nsp8, and observed the highest Gluc expression at the ratio of 1:3:3 (Fig. 2 A). Moreover, we measured Gluc activity in cells transfected with the three expression vectors at the above optimal ratio for different time intervals. Our data showed that Gluc activity quickly peaked at 24 h post transfection, then gradually declined. We also assessed the statistical parameters of this assay as previously described(Iversen et al., 2006), to determine its performance and robustness. The results showed that all the Z’ factor, %CV, S/B and S/N met the criteria required for High-throughput screening (HTS) assays (Iversen et al., 2006) (Fig. 2C and Table 1 ). Together, we have identified the optimal conditions for this cell-based SARS-CoV-2 RdRp assay which demonstrates its potential to be adapted for screening RdRp inhibitors.
Fig. 2.
Improvement of the CoV-RdRp-Gluc reporter assay. (A) Effect of different ratios of nsp12, nsp7 and nsp8 on CoV-Gluc expression.HEK 293T cells were transfected with nsp12, nsp7 and nsp8 plasmid DNA. Levels of Gluc in the culture supernatant were measured, and the level from transfection of CoV-Gluc alone was arbitrarily set at 1. Data shown are the average of three independent experiments. (B) Expression of CoV-Gluc at different time intervals after transfection of HEK293T cells with nsp12, nsp7 and nsp8 at the ratio of 1:3:3. (C) Determination of the Z′ factor of the CoV-RdRp-Gluc reporter assay.
Table 1.
Statistical parameters to determine the HTS property.
| Statistical parameter | Value | Criteria |
|---|---|---|
| Z′ | 0.73 | >0.5 |
| %CV | 7 | <20 |
| S/B | 9.6 | >3 |
| S/N | 12.86 | >10 |
a: Z’ = 1–3(STD positive groups +STD negative groups)/(MEAN positive groups + MEAN negative groups).
b: %CV (coefficient of variation) = STD positive groups/MEAN positive groups × 100%.
c: S/B (signal-to-background ratio) = MEAN positive groups/MEAN negative groups.
d:S/N(signal-to-noise ratio)=(MEAN positive groups-MEAN negative groups)/((STD positive groups)2+(STD negative groups)2)1/2.
3.3. Determine the anti-RdRp activity of nucleotide analog prodrugs using the CoV-RdRp-Gluc reporter assay
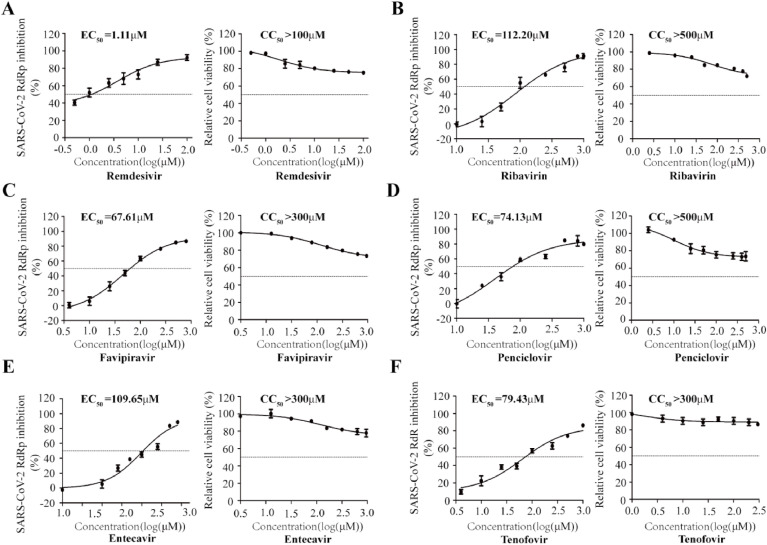
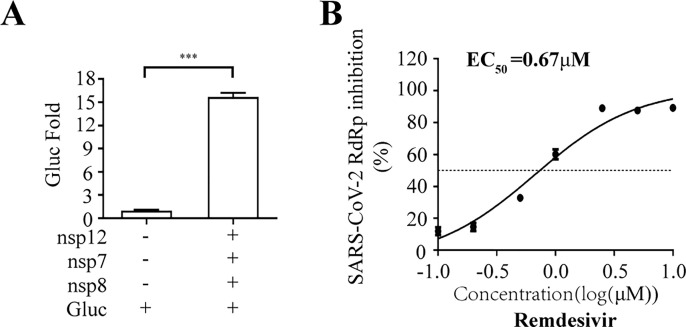
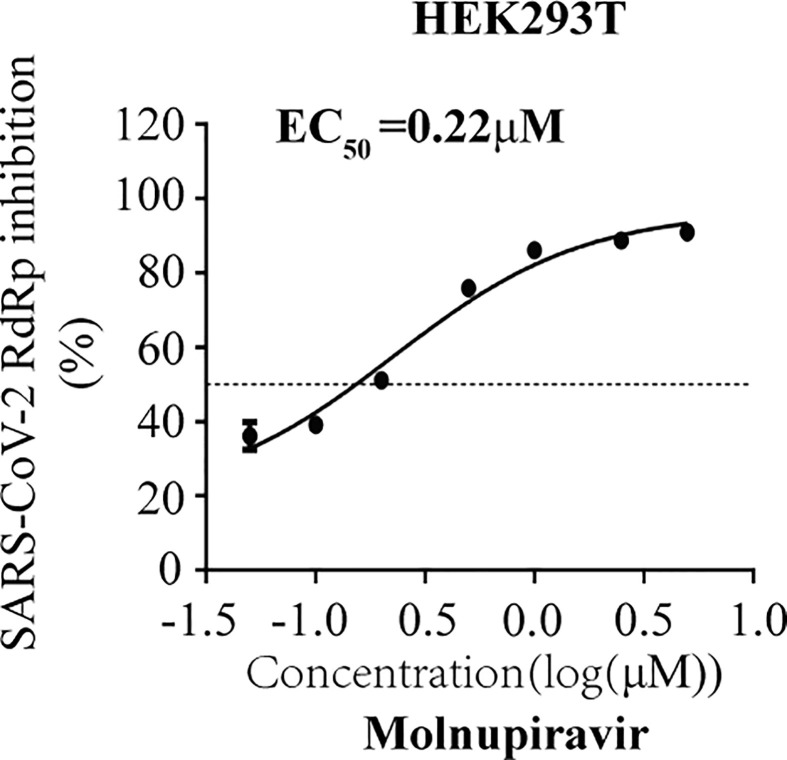
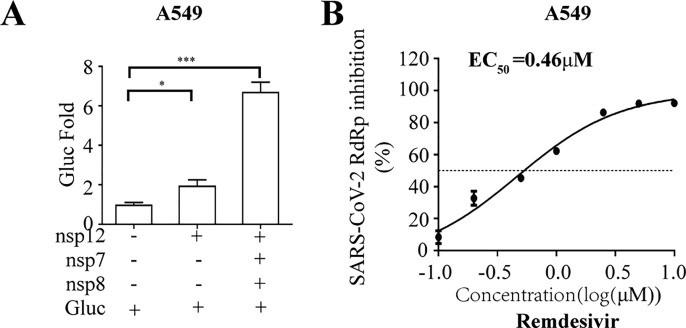
To demonstrate the utility of this assay in studying RdRp inhibitors, we tested seven well-known NAs targeting viral DNA or RNA polymerase, including Remdesivir, Ribavirin, Favipiravir, Penciclovir, Entecavir and Tenofovir (Jockusch et al., 2020), in their non-triphosphorylation forms. The data showed that Remdesivir efficiently inhibited SARS-CoV-2 RdRp activity with an EC50 value of 1.11 μM, and its CC50 value is above 100 μM (Fig. 3 A). More interestingly, Molnupiravir presented strong inhibition of SARS-CoV-2 RdRp activity with an EC50 value of 0.22 μM (Fig. S1). The broad spectrum antiviral agent Ribavirin suppresses HCV RdRp, and recently was suggested for emergency use to treat COVID-19 patients(Te et al., 2007),(Khalili et al., 2020). In our cell-based assay, Ribavirin and other NA inhibitors exhibited limited inhibition SARS-CoV-2 RdRp, with high EC50 values ranging from 70 to 110 μM (Fig. 3B to F). Furthermore, to exclude any influence on the CMV promoter-driven RNA expression step by NA candidates, we re-transfected with CoV-Gluc mRNA into cells co-transfected with the plasmids (nsp12, nsp7 and nsp8) at 12h post transfection. When co-transfected with nsp12, nsp7 and nsp8, the Gluc activity increased by 15- folds, compared with that of cells expressing the reporter alone (Fig. S2A). And Remdesivir efficiently inhibited SARS-CoV-2 RdRp activity with an EC50 value of 0.67 μM (Fig. S2B). Next, to validate this cell-based assay with a disease-relevant human lung cell line (A549), the Gluc activity in A549 increased by 2- and 7- fold, respectively, compared with that of cells expressing the reporter alone (Fig. S3A). Surprisingly, Remdesivir efficiently inhibited SARS-CoV-2 RdRp activity with an EC50 value of 0.46 μM (Fig. S3B). It is worth to note that their EC50 values against RdRp activity herein are very close to their EC50 values against SARS-CoV-2 infection in cells reported previously(Wang et al., 2020c). Together, these data support the use of this cell-based assay in screening for NA inhibitors of SARS-CoV-2 RdRp.
Fig. 3.
Dose-dependent inhibition CoV-Gluc. HEK293T cells were transfected with CoV-Gluc, nsp12, nsp7, and nsp8 plasmid DNA at the ratio of 1:10:30:30. 12 h post transfection, cells were re-seeded in 96-well plates (104/well), and treated with serially diluted Remdesivir (A), Ribavirin (B), Favipiravir (C), Penciclovir (D), Entecavir (E), Tenofovir (F). After 24 h incubation, Gluc activity in supernatants was determined. To assess cell viability, HEK293T cells (104/wells) were seeded in 96-well plates, and treated with these inhibitors as indicated above. The CC50 values were measured with the cell counting kit-8. Results shown are the average of three independent experiments.
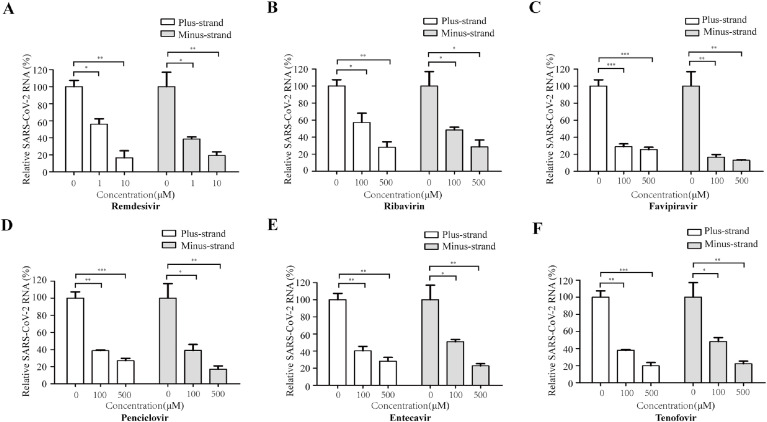
We further examined the RNA synthesis efficiency of SARS-CoV-2 RdRp by qualifying the levels of plus-strand RNA and minus-strand RNA of Gluc after treatment with each of the six NA inhibitors. Remdesivir diminished the levels of both plus-strand RNA and minus-strand Gluc RNA in a dose-dependent manner, with a reduction of 60% of minus-strand RNA at 1 μM concentration (Fig. 4 A). Other NA inhibitors significantly reduced levels of Gluc RNA at concentrations above 100 μM (Fig. 4B–F). It is noted that for each NA tested, similar folds of inhibition on Gluc activity (Fig. 3) and RNA synthesis (Fig. 4) were observed at the same concentration of the drug. These data further support the feasibility of this assay in discovering SARS-CoV-2 RdRp inhibitors without need of preparing the triphosphate forms of NAs.
Fig. 4.
Inhibition of CoV-Gluc RNA expression by NAs. HEK293T cells were transfected with CoV-Gluc, nsp12, nsp7, nsp8 plasmid DNA at the ration of 1:10:30:30. 6 h post transfection, supernatants were replaced with fresh medium containing Remdesivir (A), Ribavirin(B), Favipiravir(C), Penciclovir(D), Entecavir(E), Tenofovir(F), respectively. Cells were cultured for 24 more hours, total cellular RNA was extracted, levels of CoV-Gluc were determined by real-time RT-PCR.
3.4. Exoribonuclease nsp14 increases resistance of SARS-CoV-2 RdRp to nucleotide analog inhibitors
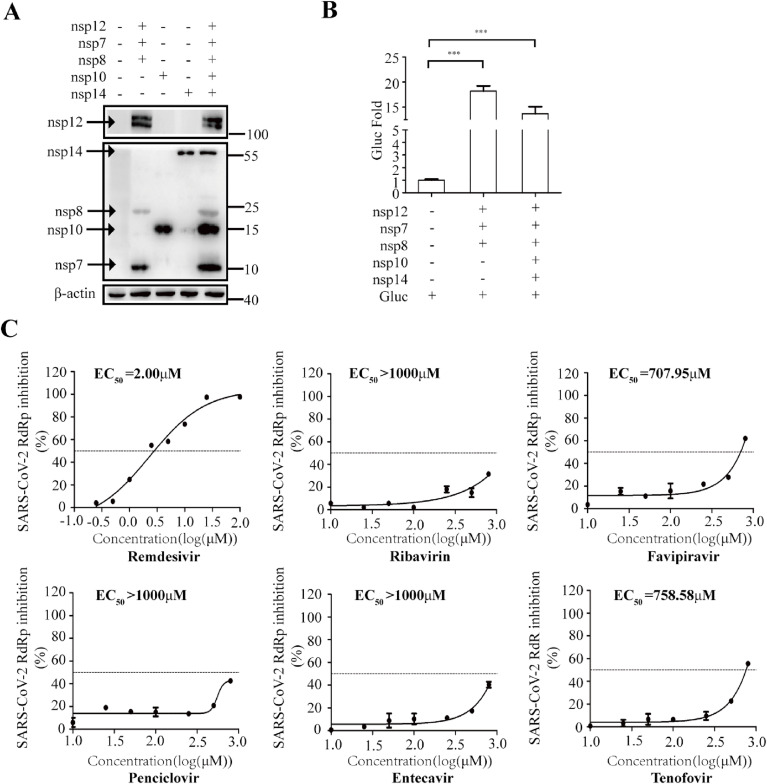
Exoribonuclease nsp14 in complex with its activator nsp10 provides the proofreading function during coronavirus replication (Ma et al., 2015). To explore whether the inhibition activity of NA inhibitors is affected by this proofreading function, we introduced nsp14 and nsp10 into the cell-based CoV-RdRp-Gluc assay. With expression of nsp14 and nsp10, Gluc activity decreased by 30% (Fig. 5 A and B). Besides, presence of nsp14 and nsp10 also slightly decrease efficiency of both plus-strand and minus-strand CoV-Gluc mRNA synthesis (Fig. S4). Then, we tested the sensitivity of SARS-CoV-2 RdRp to the NA inhibitors with expression of nsp14 and nsp10. Compared with the results without nsp14 and nsp10 (Fig. 3), expression of nsp14 and nsp10 increased the EC50 value of all NA inhibitors, ranging from 1.8 to 15 folds (Fig. 5C and Table 2 ). Among these NA inhibitors, Remdesivir was the least sensitive to the proofreading activity by the nsp14/nsp10 exoribonuclease complex, since the EC50 value increased by 1.8 folds, less than that of all other inhibitors. Therefore, the exoribonuclease complex csp14/nsp10 augments the resistance property of SARS-CoV-2 RdRp to NA inhibitors.
Fig. 5.
Effect of nsp14 and nsp10 on the responses of CoV-RdRp-Gluc to NA inhibitors. (A, B) HEK293T cells were transfected with CoV-Gluc, nsp12, nsp7, nsp8, nsp10, and nsp14 plasmid DNA at the ratio of 1:10:30:30:10:90. At 48 h post transfection, cell lysates were analyzed by Western blots using anti-Flag or anti-β-actin antibodies (A). Gluc activity in supernatants was determined and the results are shown in (B). (C) HEK293T cells were transfected with plasmids as described above. At 12 h post transfection, cells were re-seeded in 96-well plates (104/well), and treated with serially diluted NA inhibitors including Remdesivir, Ribavirin, Favipiravir, Penciclovir, Entecavir, and Tenofovir. After 24 h, Gluc activity in supernatants were quantified. Results shown are the average of three independent experiments.
Table 2.
EC50 values of NA inhibitors against SARS-CoV-2 RdRp.
| Compounds | nsp12 (EC50/μM) | nsp12 + 14 (EC50/μM) | nsp12 + 14(mutant)(EC50/μM) | Ratio of EC50 (12 + 14/12) | Ratio of EC50 (12 + 14(mutant)/12) |
|---|---|---|---|---|---|
| Remdesivir | 1.11 | 2.00 | 1.52 | 1.80 | 1.37 |
| Ribavirin | 112.20 | >1000 | 120.05 | >8.91 | 1.07 |
| Penciclovir | 74.13 | >1000 | 87.55 | >13.49 | 1.18 |
| Favipiravir | 67.61 | 707.95 | 88.30 | 10.47 | 1.31 |
| Entecavir | 109.65 | >1000 | 116.50 | 9.12 | 1.06 |
| Tenofovir | 79.43 | 758.58 | 107.50 | 9.55 | 1.35 |
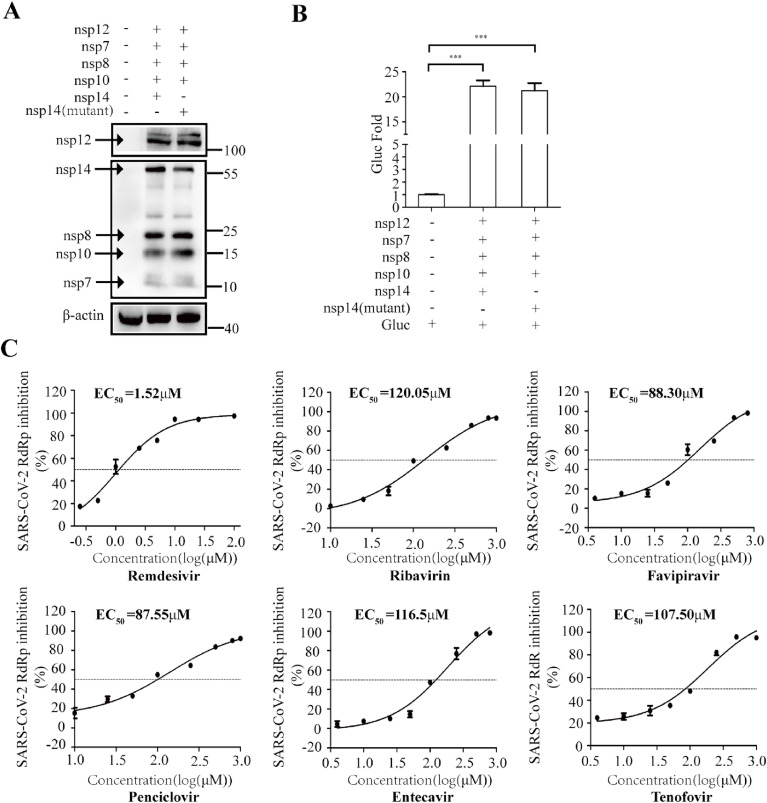
To demonstrate that the increased EC50 values of the NA inhibitors are due to the proofreading function of the exoribonuclease activity (Fig. 5), we examined the effect of the exoribonuclease-deficient mutant of nps14 on the response of viral RdRp to NA inhibitors. To this end, we generated the D90A and E92A mutations of the catalytic residues in nsp14, which have been shown to abrogate the exoribonuclease activity(Romano et al., 2020). Similar levels of Gluc activity were detected with the expression of either wild-type or mutated nsp14 (Fig. 6 A and B). However, as opposed to the wild type nps14 that markedly increased the EC50 values of NA inhibitors, the mutated nsp14 only marginally affected the EC50 values, from 1.07 to 1.37 folds (Fig. 6C and Table 2). Together, our data demonstrate that the exoribonuclease complex nsp14/nap10 renders SARS-CoV-2 RdRp resistant to NA inhibitors, which highlights the necessity of including nsp14 and nsp10 in the cell-based CoV-RdRp-Gluc assay to identify effective RdRp inhibitors.
Fig. 6.
The nsp14 mutant does not affect NA inhibition of CoV-RdRp-Gluc reporter. (A, B) HEK 293T cells were transfected with CoV-Gluc, nsp12, nsp7, nsp8, nsp10, nsp14 or nps14 (D90A&E92A) at the ratio of 1:10:30:30:10:90. Western blotting was performed with anti-Flag or anti-β-actin antibodies, results are shown in (A). Gluc activity in supernatants was measured, results are shown in (B). (C) At 12 h post transfection, cells were re-seeded in 96-well plates (104/wells), and treated with NA inhibitors Remdesivir, Ribavirin, Favipiravir, Penciclovir, Entecavir, or Tenofovir. After 24 h, supernatants were analyzed for Gluc activity. Results shown are the average of three independent experiments.
4. Discussion
To accelerate drug discovery for the fight against the COVID-19 pandemic, efficient high-throughput screening systems are urgently needed to rapidly test a large number of prodrugs targeting SARS-CoV-2 RdRp. In this study, we have established a cell-based assay for this purpose (Fig. 1, Fig. 2). Specifically, in contrast to the cell-free biochemical RdRp assays that can only test NA triphosphates, this cell-based assay is capable of detecting the inhibition activity of NA prodrug forms against SARS-CoV-2 RdRp by simply measuring Gluc activity in culture supernatants. We have tested the non-triphosphate forms of six commonly used NA inhibitors, including Remdesivir, Ribavirin, Favipiravir, Penciclovir, Entecavir, Tenofovir (Fig. 3, Fig. 4) and Molnupiravir (Fig. S1), and found that their EC50 values against RdRp activity herein are very close to their EC50 values against SARS-CoV-2 infection in cells reported previously (Wang et al., 2020b, 2020c). These data provide further evidence supporting the feasibility of using the assay in screen of SARS-CoV-2 RdRp inhibitors.
In this assay, the activity of SARS-CoV-2 RdRp is markedly stimulated by viral nsp7 and nsp8 up to 6-fold, suggesting that nsp8 and nsp7 acts as the co-factors of SARS-CoV-2 RdRp to promote gene synthesis. This is not only in agreement with early studies of coronavirus RdRp (Hillen et al., 2020; Subissi et al., 2014), also supports that the assay functionally mimics the RNA synthesis process driven by SARS-CoV-2 RdRp. During the preparation of this manuscript, a cell-based reporter assay for MERS-CoV RdRp activity has been recently reported to discover viral polymerase inhibitors(Min et al., 2020). Surprisingly, the nsp8 and nsp7 were shown no effect on the activity of MERS-CoV RdRp in the cell-based system(Min et al., 2020). The discrepancy may reflect either the different mechanism underlying coronavirus RdRp, or different experimental approach to establish the reporter system.
In addition, we also demonstrated that viral exoribonuclease nsp14 enables SARS-CoV-2 RdRp resistant to NA inhibitors. Co-expression of nsp14 and nsp10 increased the EC50 values of all the NA inhibitors tested in this study (Fig. 5, Fig. 6). These data partially explain the poor inhibition activity of NA inhibitors against SARS-CoV-2 that have been reported both in vitro and in vivo, which may present a challenge for the development of strong SARS-CoV-2 RdRp inhibitors. Besides, as Agostini et al. reported that murine hepatitis virus (MHV) mutant lacking ExoN proofreading was more sensitive to Remdesivir, which has similar EC50 against MHV as SARS-CoV and MERS-CoV (Agostini et al., 2018). This study and our data together raise the necessity of including nsp14 and nsp10 in the CoV-RdRp-Gluc assay to ensure the discovery of NA inhibitors that potently inhibit SARS-CoV-2 RdRp even in the presence of the nsp14 and nsp10 exoribonuclease complex, thus can inhibit SARS-CoV-2 infection. Moreover, among these NA drugs, to a certain degree, Penciclovir,Entecavir and Tenofovir also have the anti-SARS-CoV-2 RdRp activity in our assay. However, it is still subject to controversy that the anti-SARS-CoV-2 activity links to inhibition viral RdRp, due to the limited animal or clinical data accordingly.
Remdesivir is an adenosine analog for treating Ebola virus infection as a chain terminator of viral RdRp. Remdesivir is shown to inhibit potently SARS-CoV-2 activity in vitro (Hillen et al., 2020; Jorgensen et al., 2020; Norrie, 2020), and has recently been approved by the FDA for the treatment of COVID-19 patients (Antinori et al., 2020; Grein et al., 2020; https://www.covid19treatmentguidelines.nih.gov/therapeutic-management/). Among all NA drugs tested herein, except Molnupiravir, Remdesivir was the most potent compound against SARS-CoV-2 RdRp in our assay, providing the first evidence supporting that Remdesivir acts as a direct inhibitor of SARS-CoV-2 RdRp activity expressed in cells. Molnupiravir is an oral pro-drug of NHC (EIDD-2801; also known as molnupiravir or MK-4482) inhibit SARS-CoV and MERS-CoV replication and pathogenesis by targeting to viral RdRp, which is well investigated, with the strongest inhibition of SARS-CoV-2 RdRp activity in our cell-based assay, is currently under phase 2 clinical trail(https://clinicaltrials.gov/ct2/show/NCT04405739; Sheahan et al., 2020; Wahl et al., 2021).
In summary, we have developed the cell-based CoV-RdRp-Gluc assay for the discovery of SARS-CoV-2 RdRp inhibitors. Our results showed that all the Z’ factor and other parameters of this reporter system met the criteria required for High-throughput screening (HTS) assays (Fig. 2C and Table 1), supporting its use in screening SARS-CoV-2 RdRp RdRp inhibitors. To reduce the variation and improve convenience of the current format based on transient transfection, we are in the process to establish a stable cell lines co-expressing all the components, which empower this assay to be easily formatted into HTS, thus is expected to expedite the finding of effective COVID-19 treatments.
Author contributions
S.C.and X.L. designed the study; J.Z., S.G and. D.Y.performed the main part of the experimental study; Q.L. and L.M. conducted the bioinformatics analysis; D.Y., J.W., Y.Z, S.G. and F.G. contributed to data analysis; F.G. contributed Key reagents; Z.L. drafted the paper; S.C., R.L. and C.L edited the manuscript. All authors analyzed the results and approved the final version of the manuscript.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgments
This work was supported by The National Mega-Project for Infectious Disease [2018ZX10301408], The National Mega-Project for Significant new drug discovery [2018ZX09711003-002-002],The National Key Research and Development program of China [2018YFE0107600 and 2016YFD0500307], CAMS Innovation Fund for Medical Sciences [2018-I2M-3–004 and 2020–12M-2–010], the Fundamental Research Funds for the Central Universities [33320200046] and the National Science and Technology Infrastructure of China [NPRC-32]. We thank National Infrastructure of Microbial Resources [NIMR-2014-3]and CAMS Collection Center of Pathogenic Micro-organisms [CAMS-CCPM-A] for providing valuable reagents.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.antiviral.2021.105078.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
References
- Aftab S.O., Ghouri M.Z., Masood M.U., Haider Z., Khan Z., Ahmad A., Munawar N. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J. Transl. Med. 2020;18:275. doi: 10.1186/s12967-020-02439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., et al. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9 doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antinori S., Cossu M.V., Ridolfo A.L., Rech R., Bonazzetti C., Pagani G., Gubertini G., Coen M., Magni C., Castelli A., et al. Compassionate remdesivir treatment of severe Covid-19 pneumonia in intensive care unit (ICU) and Non-ICU patients: clinical outcome and differences in post-treatment hospitalisation status. Pharmacol. Res. 2020;158 doi: 10.1016/j.phrs.2020.104899. 104899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., Qiu Y., Wang J., Liu Y., Wei Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. 10223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien M., Anderson T.K., Jockusch S., Tao C., Li X., Kumar S., Russo J.J., Kirchdoerfer R.N., Ju J. Nucleotide analogues as inhibitors of SARS-CoV-2 polymerase, a key drug target for COVID-19. J. Proteome Res. 2020;19(11):4690–4697. doi: 10.1021/acs.jproteome.0c00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- clinicaltrials https://clinicaltrials.gov/ct2/show/NCT04405739
- covid19treatmentguidelines https://www.covid19treatmentguidelines.nih.gov/therapeutic-management/
- Cox R.M., Wolf J.D., Plemper R.K. Therapeutically administered ribonucleoside analogue MK-4482/EIDD-2801 blocks SARS-CoV-2 transmission in ferrets. Nat. Microbiol. 2021;6:11–18. doi: 10.1038/s41564-020-00835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol. Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron F., Subissi L., Silveira De Morais A.T., Le N.T.T., Sevajol M., Gluais L., Decroly E., Vonrhein C., Bricogne G., Canard B., et al. Structural and molecular basis of mismatch correction and ribavirin excision from coronavirus RNA. Proc. Natl. Acad. Sci. U.S.A. 2018;115:E162–E171. doi: 10.1073/pnas.1718806115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Yan L., Huang Y., Liu F., Zhao Y., Cao L., Wang T., Sun Q., Ming Z., Zhang L., et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science. 2020;368:779–782. doi: 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet M.L., Olagnier D., Xu Z., Paz S., Belgnaoui S.M., Lafferty E.I., Janelle V., Arguello M., Paquet M., Ghneim K., et al. Systems analysis of a RIG-I agonist inducing broad spectrum inhibition of virus infectivity. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003298. e1003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.X., et al. Compassionate use of remdesivir for patients with severe covid-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillen H.S., Kokic G., Farnung L., Dienemann C., Tegunov D., Cramer P. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- Iversen P.W., Eastwood B.J., Sittampalam G.S., Cox K.L. A comparison of assay performance measures in screening assays: signal window, Z' factor, and assay variability ratio. J. Biomol. Screen. 2006;11:247–252. doi: 10.1177/1087057105285610. [DOI] [PubMed] [Google Scholar]
- Jockusch S., Tao C., Li X., Anderson T.K., Chien M., Kumar S., Russo J.J., Kirchdoerfer R.N., Ju J. A library of nucleotide analogues terminate RNA synthesis catalyzed by polymerases of coronaviruses that cause SARS and COVID-19. Antivir. Res. 2020;180 doi: 10.1016/j.antiviral.2020.104857. 104857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen S.C.J., Kebriaei R., Dresser L.D. Remdesivir: review of pharmacology, pre-clinical data, and emerging clinical experience for COVID-19. Pharmacotherapy. 2020;40:659–671. doi: 10.1002/phar.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalili J.S., Zhu H., Mak N.S.A., Yan Y., Zhu Y. Novel coronavirus treatment with ribavirin: groundwork for an evaluation concerning COVID-19. J. Med. Virol. 2020;92:740–746. doi: 10.1002/jmv.25798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Wu L., Shaw N., Gao Y., Wang J., Sun Y., Lou Z., Yan L., Zhang R., Rao Z. Structural basis and functional analysis of the SARS coronavirus nsp14-nsp10 complex. Proc. Natl. Acad. Sci. U.S.A. 2015;112:9436–9441. doi: 10.1073/pnas.1508686112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J.S., Kim G.W., Kwon S., Jin Y.H. A cell-based reporter assay for screening inhibitors of MERS coronavirus RNA-dependent RNA polymerase activity. J. Clin. Med. 2020;9 doi: 10.3390/jcm9082399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrie J.D. Remdesivir for COVID-19: challenges of underpowered studies. Lancet. 2020;395:1525–1527. doi: 10.1016/S0140-6736(20)31023-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L.T., Nguyen T.V., Luong Q.C., Nguyen T.V., Nguyen H.T., Le H.Q., Nguyen T.T., Cao T.M., Pham Q.D. Importation and human-to-human transmission of a novel coronavirus in vietnam. N. Engl. J. Med. 2020;382(9):872–874. doi: 10.1056/NEJMc2001272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remuzzi A., Remuzzi G. COVID-19 and Italy: what next? Lancet. 2020;395:1225–1228. doi: 10.1016/S0140-6736(20)30627-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. 2020. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacramento C.Q., Fintelman-Rodrigues N., Temerozo J.R., da Silva Gomes Dias S., Ferreira A.C., Mattos M., Pão C.R.R., de Freitas C.S., Soares V.C., Bozza F.A., et al. bioRxiv; 2020. The <em>in Vitro</em> Antiviral Activity of the Anti-hepatitis C Virus (HCV) Drugs Daclatasvir and Sofosbuvir against SARS-CoV-2. 2020.2006.2015.153411. [Google Scholar]
- Shannon A., Le N.T., Selisko B., Eydoux C., Alvarez K., Guillemot J.C., Decroly E., Peersen O., Ferron F., Canard B. Remdesivir and SARS-CoV-2: structural requirements at both nsp12 RdRp and nsp14 Exonuclease active-sites. Antivir. Res. 2020;178 doi: 10.1016/j.antiviral.2020.104793. 104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon A., Selisko B., Le N.T., Huchting J., Touret F., Piorkowski G., Fattorini V., Ferron F., Decroly E., Meier C., et al. Rapid incorporation of Favipiravir by the fast and permissive viral RNA polymerase complex results in SARS-CoV-2 lethal mutagenesis. Nat. Commun. 2020;11:4682. doi: 10.1038/s41467-020-18463-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Zhou S., Graham R.L., Pruijssers A.J., Agostini M.L., Leist S.R., Schafer A., Dinnon K.H., 3rd, Stevens L.J., et al. An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abb5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.C., Blanc H., Surdel M.C., Vignuzzi M., Denison M.R. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003565. e1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subissi L., Posthuma C.C., Collet A., Zevenhoven-Dobbe J.C., Gorbalenya A.E., Decroly E., Snijder E.J., Canard B., Imbert I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E3900–E3909. doi: 10.1073/pnas.1323705111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Te H.S., Randall G., Jensen D.M. Mechanism of action of ribavirin in the treatment of chronic hepatitis C. Gastroenterol. Hepatol. 2007;3:218–225. [PMC free article] [PubMed] [Google Scholar]
- Wahl A., Gralinski L.E., Johnson C.E., Yao W., Kovarova M., Dinnon K.H., 3rd, Liu H., Madden V.J., Krzystek H.M., De C., et al. SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature. 2021;591:451–457. doi: 10.1038/s41586-021-03312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C., et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428 e413. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Wu J., Wang H., Gao Y., Liu Q., Mu A., Ji W., Yan L., Zhu Y., Zhu C., et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell. 2020;182:417–428 e413. doi: 10.1016/j.cell.2020.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Anirudhan V., Du R., Cui Q., Rong L. RNA-dependent RNA polymerase of SARS-CoV-2 as a therapeutic target. J. Med. Virol. 2020;93(1):300–310. doi: 10.1002/jmv.26264. [DOI] [PubMed] [Google Scholar]
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Chen C.Z., Gorshkov K., Xu M., Lo D.C., Zheng W. SLAS Discov; 2020. RNA-dependent RNA Polymerase as a Target for COVID-19 Drug Discovery. 2472555220942123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.