Summary
The number and distribution of recessive alleles in the population for various diseases are not known at genome-wide-scale. Based on 6,447 exome sequences of healthy, genetically unrelated Europeans of two distinct ancestries, we estimate that every individual is a carrier of at least 2 pathogenic variants in currently known autosomal-recessive (AR) genes and that 0.8%–1% of European couples are at risk of having a child affected with a severe AR genetic disorder. This risk is 16.5-fold higher for first cousins but is significantly more increased for skeletal disorders and intellectual disabilities due to their distinct genetic architecture.
Keywords: autosomal recessive disorders, carrier frequency, pre-conception carrier screening, selection, at-risk couples
Introduction
A major public health goal is to detect at-risk couples (ARCs) for various autosomal-recessive (AR) diseases. Detecting such couples would enable them to consider reproductive choices to prevent the birth of affected children, including prenatal diagnosis (PND) and preimplantation genetic testing (PGT). Currently, the number and distribution of recessive alleles in the population is not known at genome-wide scale. Understanding the architecture of AR pathogenic variants can contribute to the knowledge base for public health policies in the preconception field and illuminate the evolution of disorders and phenotypes.
Existing estimates of the number of AR alleles carried by individuals are either derived from comparisons of the incidence of AR disorders between offspring of consanguineous and non-consanguineous couples or based on extrapolations from sequencing data of specific phenotypes and gene sets. Early calculations estimated that each individual carries at least eight heterozygous recessive pathogenic variants,1 while estimates based on consanguineous couples predicted 3–5 heterozygous recessive lethal pathogenic variants per individual.2 Later models predicted up to 100 pathogenic variants per individual.3 An analysis based on gene-dropping simulations in a well-documented isolated founder population (the Hutterites) estimated that each founder of this population carried 0.58 AR lethal pathogenic variants that lead to death between birth and reproductive age or to complete sterility.4
Sequencing-based studies of wider gene sets also yielded variable results. Screening for 437 known AR genes related to Mendelian diseases found a carrier frequency of 2.8 severe pathogenic variants per individual (range 0–7).5 In another study that used samples of various ethnicities including Caucasians, testing for a panel of 417 AR pathogenic variants found ∼0.4 AR lethal pathogenic variants per individual, leading the authors to suggest that the number for the entire genome is ∼10 times higher.6
None of the existing studies used direct gene sequencing at a genome-wide scale. Furthermore, each study used a different methodology, cohort size, and number of tested genes and variants. Therefore, current data do not allow an overall assessment of the genomic landscape of AR disease variants.
Here, we performed an exome-sequencing-based assessment of the carrier frequency of AR pathogenic or likely-pathogenic variants (PLPs), the total ARCs rate for various disorders, and the effect of different consanguinity levels on the ARCs rate for these disorders in two distinct European populations. We used direct gene sequencing and included a comprehensive set of AR genes. These analyses reveal the architecture and distribution of AR pathogenic variants throughout the genome and for different disorders. The results can inform public health policies such as design of preconception carrier screens and improve preconception counseling. Our results also provide insights into the population genetics of AR disorders, particularly regarding intellectual disabilities.
Subjects and methods
Cohorts
We analyzed two European cohorts based on Dutch and Estonian samples. For the purpose of this study, exome data were anonymized. Both populations are from Northern Europe, yet they are distant enough geographically that they can be treated as two distinct European cohorts.7,8 The Dutch cohort included 4,780 unaffected parents of children with intellectual disability (ID), tested by patient-parents trio exome sequencing. At the time of the analysis, none of the offspring had been diagnosed with an AR-ID. For 1,891 couples, the child was shown to have an autosomal-dominant (AD) de novo cause for ID. To test whether inclusion of these couples might have biased our results, we compared the average and median number of PLPs in these samples (2.37 and 2, respectively) to the average and median in all other samples (2.33 and 2, respectively). The difference between the two groups was not significantly different (t test p value = 0.34; Kruskal-Wallis test p value = 0.77). The exomes were sequenced at BGI in Copenhagen, Denmark using DNA isolated from blood. Exome capture was performed using Agilent SureSelect v4/v5 and samples were sequenced on an Illumina HiSeq instrument with 101-bp paired-end reads to a median coverage of 75×. Sequence reads were aligned to the GRCh37/hg19 reference genome using BWA v.0.5.9-r16. Variants were subsequently called by the GATK haplotyper (v.2-2) and annotated using a custom diagnostic annotation pipeline.9 The Estonian cohort included 2,356 healthy individuals, sequenced as a part of the Estonian Biobank of the Estonian Genome Center, University of Tartu (EGCUT), which is a population-based biobank, containing almost 52,000 samples of the adult population (aged ≥ 18 years), which closely reflects the age, sex, and geographical distribution of the Estonian population. WES samples DNA was enriched for target sequences (Agilent Technologies; Human All Exon V5+UTRs) according to manufacturer’s recommendations. Sequenced reads were aligned to the GRCh37/hg19 human reference genome using BWA-MEM (v.0.7.7). SAMtools (v.1.2) was applied to compress SAM to BAM (samtools view), sort (samtools sort), and index BAM (samtools index) files. PCR duplicates were then marked using Picard (v.1.136) MarkDuplicates.jar. For further BAM improvements, including realignment around known indels and base quality score recalibration, we applied GATK (v.3.4). Single sample genotypes were called by the GATK HaplotypeCaller algorithm (-ERC GVCF).
Ethical regulations
Dutch exomes were obtained as a part of an anonymized study, following guidelines for anonymized study procedures, based on approval by the institutional review board “Commissie Mensgebonden Onderzoek Regio Arnhem-Nijmegen” under number 2011/188.
Estonian exomes were obtained as a part of the Estonian Biobank project permission nr 234/T - 12 to study human exomes in the Estonian Biobank. All subjects in the Estonian Biobank have signed informed consent and the whole biobanking program is governed by special law: “Human Genes Research Act.”
Relatedness analysis
We used KING10 v.2.2 to calculate kinship coefficients and infer relationships within the cohort. For this analysis, we used 14,643 autosomal variants with a quality score of ≥1,000 located in regions covered ≥20× in all samples. KING10 inferred 30 and 52 Dutch and Estonian samples, respectively, to be related by second degree or closer. Blinded analysis in the Dutch cohort confirmed known relationships in 26 of 30 samples. There was insufficient information to determine relatedness for the other 4 samples, yet they were removed from the analysis since KING10 indicated them as having multiple second and third-degree relatives within the cohort. Overall, we excluded 17 samples from the Dutch cohort and 26 samples from the Estonian cohort by removing one individual from each pair inferred by KING10 to have a relationship of second degree or closer.
Ancestry analysis
Dutch cohort
We performed ancestry analysis for the Dutch cohort by LASER11 v.2.04, using a reference set of genotyping data from 9,608 loci on chromosome 22 of 700 samples from the Human Genome Diversity Project (HGDP).12 The reference samples are subdivided into 7 worldwide populations (Africa, America, Central/South Asia, Europe, Middle East, Oceania, East Asia) and 53 subpopulations. Based on this analysis, we excluded 643 samples of non-European descent.
Additionally, we analyzed the remaining 4,120 samples with the ADMIXTURE13 tool (v.1.3.0). We used data from the 1000 Genomes Project14 to help identify the genetic ancestry of our samples, by using samples from 5 super-populations (Africa, America, Europe, South Asia, and East Asia) and running a supervised analysis with K = 5. Next, we used the alleles frequencies as an input to a projection analysis for the samples of the cohort. ∼97% of the samples had a European component of >0.75, indicating the cohort has homogeneous European ancestry (Figure S10A) and confirming the LASER11 results.
Estonian cohort
For the Estonian samples, only VCF files were available and therefore we could not run LASER11 on this cohort. We ran ancestry analysis by ADMIXTURE,13 with the same parameters and reference samples as described for the Dutch cohort. Approximately 98% of the samples had a European component of >0.75, indicating the cohort has homogeneous European ancestry (Figure S10B). Three samples were excluded from the analysis.
After filtering the samples based on ancestry and relatedness, we had 4,120 Dutch samples and 2,327 Estonian samples of unaffected, European-descent, unrelated individuals that were used for all subsequent analyses in this study.
Assessing loss-of-function variants
Variants were annotated using the Ensembl Variant Effect Predictor (VEP)15 and LOFTEE16 tool (Loss-Of-Function Transcript Effect Estimator; Installed at 5th of January 2018, VEP v.91) with the default parameters as a part of the indel filtering process. Indels with low-confidence score (LC) by LOFTEE16 were filtered out.
Selection of genes
Genes annotated in OMIM as having only an AR phenotype(s) were automatically chosen for further analysis (1,605 genes). Genes indicated in OMIM as having both AR and AD phenotypes (AD-AR genes) were assessed by their gnomAD pLI score. This score indicates the probability that a gene is intolerant to a loss-of-function (LoF) variant. The higher the score, the more likely the gene is involved in a dominant disease, and the lower the pLI score, the more likely it is to indicate a recessive disease gene. To determine the appropriate pLI threshold for determining which AD-AR genes are more likely to be AR genes, we generated a reference list of pLI scores for manually curated 930 AR-only genes causing severe phenotypes.17,18 The 95th percentile pLI score for these AR-only genes was 0.86. Therefore, AD-AR genes with a pLI score ≤0.86 were considered as AR and added to the AR-only genes for subsequent analyses (324 genes).
The final list of AR genes included 1,929 genes (6,011 transcripts). Of these, OMIM classifies 1,605 genes as AR-only phenotypes and 324 genes as underlying both AR and AD phenotypes. To confirm that the AD-AR genes chosen by their pLI score do not bias the results, we performed the subsequent analysis both for all 1,929 genes and using only the 1,605 AR-only genes.
In addition, 1,119 out of 1,929 genes of the list were manually curated as a part of the development of an Australian PCS panel (Mackenzie’s mission project) and deemed to be associated with severe phenotypes17 (Table S1). “Severe” was defined in the Australian PCS panel design as: “The condition is one for which an “average” couple would take steps to avoid the birth of a child with that condition.”17 The Australian PCS project also includes X-linked genes that were not analyzed in this study.
Variant selection and determining variant outcome
We extracted variants located in the exons and flanking 10 bp regions of the selected genes, in regions covered ≥20× in ≥90% of the samples. Based on exon positions extracted from Ensembl15 (GRCh 37) for the selected genes, 94.3% of the coding region ±10 bp and 58.1% of the exonic regions ±10 bp (including UTRs) were covered ≥20× in ≥90% of the samples.
We used the list of transcripts from the HGMD database (v.2018.3) as a reference. If a gene had only one transcript described in the HGMD database, the outcome of the variant was defined based on this transcript. For genes with several transcripts described in the HGMD database, if >50% of the variant outcomes were LoF, missense, or other, this outcome was chosen. All other cases (1,777 and 861 variants in the Dutch and Estonian cohorts, respectively) were considered as the most severe outcome and evaluated manually if they passed the selection process.
Indel filtering
While SNV calling tools generally have good performance, the accuracy of indel calling is relatively low and prone to errors.19 In order to prevent a high incidence of false positive indels, we adjusted the selection process by using indels in autosomal-dominant genes associated with intellectual disability (AD-ID) (Table S9) as a proxy for false positives. Since our cohort includes unaffected individuals, indels found in those genes are most probably either not PLPs or false positives. In our Dutch cohort of 4,120 samples, there were 1,039 indels with ≥500 GATK quality score in 371 AD-ID genes. Raising the quality threshold to ≥1,000, removing non-LoF, common (>5% heterozygotes/>1% homozygotes in our cohort; >1% allele-frequency in gnomAD),20 longer than 10 bp, scored LC by LOFTEE,16 and adjacent indels within 10 bp range, decreased the number of AD-ID indels to 46 (4.4%). Of these remaining indels, 12 (26.1%) were in genes with non-LoF pathogenicity mechanism such as known dominant-negative or activating PLPs, 20 (43.5%) were in genes with known partial penetrance or variable expression, 9 (19.5%) were in genes lacking information about inheritance and pathogenicity mechanism, and 5 (10.9%) were in genes expected to be affected by LoF variants. Our filtering process thus significantly reduced the number of likely false positive indels and was therefore applied to indels in the analyzed gene set.
Variant selection process
For each cohort, we created a list of presumable PLPs. We included variants with ≥500 or 1,000 GATK quality score for substitutions or indels, respectively, and excluded variants with ≥5% heterozygotes or ≥1% homozygotes frequency within each cohort (Figure S1). After manual curation of the frequency drop-outs, we re-included three known pathogenic variants with >5% carrier frequency in at least one of the cohorts (Dutch or Estonian): HFE (MIM: 613609) p.Cys282Tyr (c.845G>A) (10.7% or 7.6%); BTD (MIM: 609019) p.Asp444His (c.1330G>C) (7.1% or 8.1%); and SERPINA1 (MIM: 107400) p.Glu288Val (c.863A>T) (6.4% or 3.2%). This gave rise to 91,341 (Dutch) and 45,929 (Estonian) variants in total in the cohorts.
We then selected only variants that met at least one of three criteria. (1) Classified as PLP by ClinVar21 with a review status of ≥2 stars or classified as PLP by the VKGL22 database. This curated database is publicly available and comprises DNA variant classifications established based on (former) diagnostic reports of all nine Dutch accredited laboratories. (2) Loss-of-function (LoF) variants (nonsense, frameshift, canonical splicing) with <1% or unknown frequency in gnomAD.20 Indels were filtered as described above. (3) Classified as PLP by ≥2/3 databases: InterVar23 (an automated ACMG classifier), ClinVar21 with a review status of <2 stars, and HGMD (indicated as disease causing variant by the DM flag), and does not contradict the first criterion, i.e., not classified as benign or likely-benign by Clinvar21 with a review status of ≥2 stars or by the Dutch database (Figure S1).
For the AD-AR genes, only LoF variants were included in the final PLPs list.
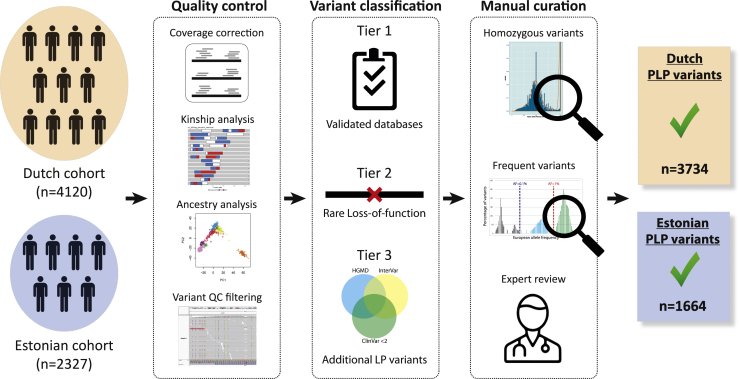
In total, the selection process filtered out >95% of the initial variants in the selected regions (Figures 1 and S1).
Figure 1.
Overview of the selection of PLP variants
From left to right, variants were selected from two exome-sequencing cohorts of healthy individuals from two different European populations. Samples and variants were subjected to stringent quality control. Samples were filtered for kinship and ethnicity. Variants were filtered for quality and selected from consistently well-covered regions. For variant classification, variants were classified as PLP based on curated publicly available databases and/or on their predicted loss of function effect. For manual curation, we performed manual curation steps at both the gene and the variant level, to confirm the validity of our PLP classification selection process. Detailed information is in the subjects and methods and Figure S1.
Validation of the PLP classification process
Although we used stringent quality scores for the selection of PLPs, we performed several analyses to confirm the validity of our PLP classification process.
Manual classification
Manual classification was performed in three groups of PLP variants.
PLP variants with high-allele frequency (>1%). Within the list of 3,734 and 1,664 PLPs in the Dutch and Estonian cohorts, respectively, the majority of variants (3,686 [98.7%] and 1,613 [96.9%]) had a carrier frequency of up to 0.05%. There were 16 (0.4%) (Dutch) and 18 (1.1%) (Estonian) PLPs with more than 1% carrier frequency in the cohorts (Figure S3). Among these frequent variants, 7 variants were seen with >1% carrier frequency in both cohorts (Table S10). Manual curation of these variants showed that all common variants were previously described in European populations and/or are known to cause a mild phenotype. The observed frequency was also compared to the frequency reported in the GONL project, which is based on sequencing of a different cohort of 498 healthy Dutch individuals (Table S10). All the variants that were seen at a >1% frequency in the Dutch cohort were also reported in the GONL database, with 0.2%–5.4% allele frequency (mean 1.6%).
PLP variants in a homozygous state. For both the Dutch and Estonian cohorts, only 42 and 11 (1% and 0.5%), respectively, of the samples were homozygous for any of the PLPs. This percentage was even lower for the set of severe recessive genes (12 and 4; 0.3% and 0.2%). Overall, 19 PLPs were seen in a homozygous state in one sample or more. Manual curation of these variants showed that 11 of these have been reported to cause only a mild phenotype or even appear asymptomatic when seen in a homozygous state, and 6 have conflicting evidence about their pathogenicity.
Curation of the 214 PLPs in genes underlying deafness. All PLPs found in deafness genes were manually classified by an expert who used, among other databases, the Deafness Variation Database (DVD), a comprehensive, open-access resource that integrates all available genetic and clinical data together with expert curation.24 Of the 214 variants our selection process classified as PLP, expert manual curation classified 1 variant as likely benign, 6 variants as VUS, 174 variants as LP, and 33 variants as P. Overall, 96.7% (207 out of 214) of the variants were correctly classified as PLPs.
Assessing selection process performance for PLP variants in CFTR (MIM: 602421)
We extracted the list of variants that are classified as PLP in the CFTR2 database (v.Jan10-2020) and ran it through the selection process. The selection process classified correctly as PLP 347 out of 414 (83.8%) variants, including 113 missense variants. Most of the variants that were not classified as PLP (59 out of 67; 88%) were missense variants. Six variants were non-coding non-canonical splice site region variants, one variant was an in-frame insertion, and one variant was an intronic variant.
Analysis of selected missense variants
In order to assess the pathogenicity of missense variants classified as PLP based on tier 3 criteria of the selection process (Figure S1), we compared CADD scores of missense variants from tier 1 and tier 3 with the CADD scores of those which failed to pass the selection process (non-PLP). The CADD scores of missense variants classified as PLPs based on tier 3 criteria are similar to those of missense variants classified as PLPs based on tier 1 criteria and are significantly higher than those of missense non-PLPs (Figure S11).
Virtual matings
We simulated all possible matings within the 4,120 samples of the Dutch cohort (8,485,140 theoretical couples), and the 2,327 samples of the Estonian cohort (2,706,301 theoretical couples), irrespective of gender. Variants that were manually classified for high frequency or homozygosity and were proven to be asymptomatic or cause a very mild phenotype in the homozygous state were excluded if the virtual mating was predicted to be at risk for a homozygous offspring, yet included if the virtual mating was predicted to be at risk for a compound heterozygous offspring (Table S4).
The ARCs rates were computed by simulations rather than using allele frequencies. This method is the most accurate way to assess ARCs rates since it is based on actual genotypes from the population, whereas the calculation using allele frequencies necessarily needs to assume linkage equilibrium between all variants.
Pathogenic variants overlap simulations
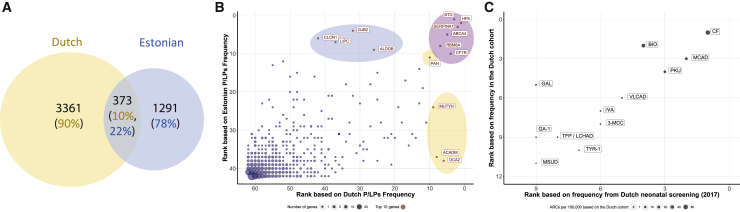
The number of shared PLPs between the Dutch and Estonian cohorts was 373 (10% of the Dutch PLPs and 22% of the Estonians PLPs) (Figure 2A). In order to determine whether this proportion of shared variants is significantly less than expected, we ran 10,000 simulations in which we randomly divided all 6,447 individuals into two groups of 4,120 and 2,327 individuals (the original sizes of the Dutch and Estonian cohorts). For each simulation we checked the proportion of shared PLPs between the two random groups. The p value was 9.99⋅10−5, and the mean number of shared variants was 950.7 (25.5% of the Dutch PLPs and 57.1% of the Estonians PLPs).
Figure 2.
PLP variants in the Dutch and Estonian cohorts: robustness of variant classification
(A) The number of PLPs in each cohort and their overlap.
(B) Genes ranked by PLP frequency: correlation between the Dutch and the Estonian cohorts. The lower the rank number, the higher the number of PLPs observed in that gene (i.e., a gene ranked 1 has the largest number of PLPs); genes with no PLPs in both cohorts were excluded (449 genes); gene names are written for genes in the top 10 ranking: blue, Estonian only top 10; orange, Dutch only top 10; purple, Dutch and Estonian top 10. The dots sizes represent the number of genes with this rankings combination. Spearman correlation coefficient: 0.69; p value < 0.00001.
(C) Comparison of disease carrier frequency estimates to published data of the Dutch neonatal screening program from 2016 and 2017. Spearman correlation coefficient 0.85; p value = 0.0005; full diseases and gene names are in Table S11.
Gene rankings
Genes in which PLPs were observed were ranked by the frequency of PLPs observed. Six genes were included in the top-10 rankings of PLPs per gene in both cohorts (Figure 2B). In order to check whether this number of genes in the top-10 rankings is statistically significant, we used Fisher exact test (p value = 1·10−5).
Consanguinity analysis
For the consanguinity calculations, we verified that variant frequencies in the two populations adhere to the Hardy-Weinberg equilibrium. We compared the observed number of wild-type (AA), heterozygotes (Aa), and homozygotes (aa) to the expected numbers (pp, 2pq, qq) using a chi-square test, showing that variants in both populations adhere to the Hardy-Weinberg equilibrium and there is no significant difference between observed and expected with a p value of 0.4 for the Dutch cohort and 0.1 for the Estonian cohort. Next, we calculated the expected risk for different degrees of consanguinity, relying on the expected proportion of shared alleles in each relationship: 1/8 for first cousins, 1/16 for first cousins once removed, 1/32 for second cousins, and 1/128 for third cousins. Therefore, the probability of a couple to be at risk in a given gene was calculated as 2pq∗<expected proportion of shared alleles>∗cohort size, where q is the sum of allele frequencies of PLP variants in a given gene, and p = 1 − q.
Gene panels
All 1,929 genes were divided into panels based on their related disorders (Table S1). The ID-related genes were divided into two panels: ID and metabolic-ID. The ID panel includes genes that are related to syndromic and non-syndromic ID but does not include metabolic genes. All genes that are related to metabolic disorders with or without phenotypes other than ID were included in the metabolic panel. An additional gene group, multisystem disorders, comprised all genes underlying more than one phenotype, excluding intellectual disability (ID) and metabolic disorders (Table S1).
Consanguinity ratio
In order to examine whether the differences between the consanguinity ratio (CR) scores are statistically significant, we ran 5,000 simulations per panel for each cohort. In each simulation, we randomly assigned genes into panels of same total bp length as the original panels (Figure S6). We computed the p values based on how many times the simulated CR was different from the actual CR for each panel using a two-tailed test with a Bonferroni correction (Figure S6; Table S12).
Analysis for genetic selection
To determine whether groups of genes show different patterns of selection, we used three different scores that serve as a proxy of purifying selection. We calculated the normalized gene singleton density, the coding regions singleton density (coding singleton density),25 and the residual variation intolerance score (RVIS)26 for Europeans samples on the 1000 Genomes data14 (Table S14).
We compared the scores of gene sets described in this study to reference lists of genes that show different signatures of purifying selection: “essential” genes27 and “haploinsufficiency severe” genes28 as target of strong purifying selection, and genes labeled as “non essential”27 and “olfactory” receptors29 as controls (no strong selection signatures) (Table S14). Statistical significance of the differences between the medians was evaluated using the Wilcoxon-rank sum test for the gene panel ID/skeletal compared to all other disorders (Figures 5 and S8).
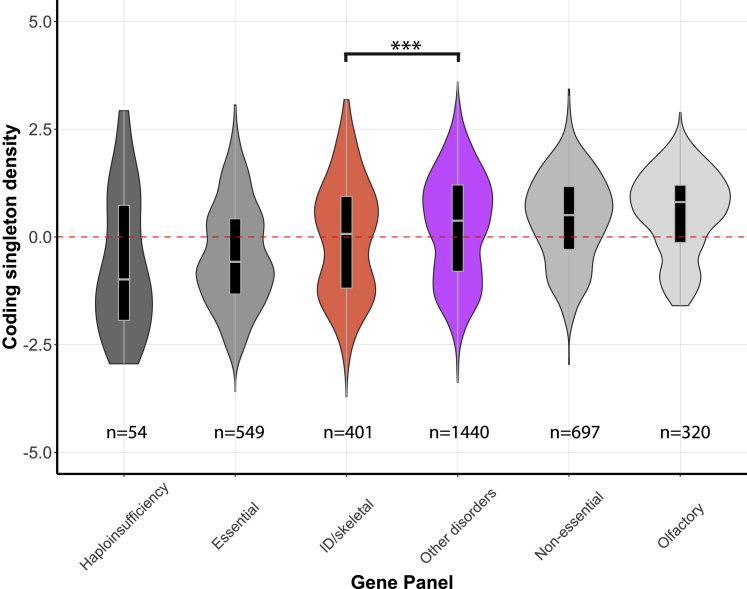
Figure 5.
Coding singleton density in coding regions based on 1000 Genomes data (European populations) for the different gene panels
Violin plots of the distribution of coding singleton density scores for different gene panels. Gray colors indicate external reference gene sets, whereas colored violin plots are gene sets as defined in this manuscript. Tested differences of the median are indicated by braces, giving rise to a p value of 1.8·10−4 for ID/skeletal gene set compared to “other disorders” gene set, using a Wilcoxon rank sum test with Bonferroni correction (Table S14).
Next, we simulated a large meta-population with ten subpopulations with an effective population size (Ne) of 10,000 each. Each population could exchange migrants with a rate of 1% in each generation. We simulated the possibility of a locus to mutate and its increase in frequency in different scenarios: no selection (s = 0) and increased purifying selection against the heterozygotes (s > 0), with a total negative selection against homozygotes (s = 1). The simulated selection coefficients are linked to the percentage of reduction of an individual to have offspring (for s = 0.05 heterozygous carriers have 5% less probability to mate and have offspring). The simulations were done using simuPOP30 v.1.1.7 (Figure S7).
Results
We gathered exome samples from two cohorts of healthy individuals of Dutch (n = 4,120) and Estonian (n = 2,327) populations and filtered these for quality, kinship, and ethnicity (Figure 1). For these cohorts we analyzed a set of 1,929 AR disease genes (subjects and methods; Table S1), including a subset of 1,119 genes that were previously categorized as associated with severe phenotypes by manual expert curation as part of the development of an Australian pre-conception screening (PCS) panel17 (subjects and methods). In these genes, we selected all pathogenic and likely pathogenic variants (PLPs) based on existing classifications from databases and ACMG guidelines31 (subjects and methods). The filtering process was applied to a total of 91,341 and 45,929 variants for the Dutch and Estonian cohorts, respectively, and excluded >95% of the variants as either benign or likely benign or variants of unknown significance (VUS), resulting in 3,734 and 1,664 PLPs for the Dutch and Estonian cohorts, respectively (Figures 1 and S1, subjects and methods).
Pathogenic variants
More than half of the PLPs (55.2% and 59.1% in the Dutch and Estonian cohorts) are rare loss-of-function (LoF) variants not previously described in Clinvar21 or the Dutch society of laboratory specialists initiative for data sharing of variant classifications (VKGL database22) (classified as PLP by tier 2 criteria) (Figure S1). About a third are known PLPs in the VKGL22 database and/or classified as PLP by ClinVar21 with a status review of 2 or more stars (34.6% and 27.6% in the Dutch and Estonian cohorts) (tier 1 criteria). The remaining 10.1% and 13.2% are variants classified as PLP by ≥2/3 databases (tier 3 criteria) (subjects and methods, Figure S1).
Since the classification of variants as PLPs forms the basis of this study, we performed several analyses that confirmed the validity of our classification methodology (see subjects and methods, Figure S1).
When comparing the two cohorts, we found that only 373 PLPs (10% of the Dutch PLPs and 22% of the Estonian PLPs) were shared between both cohorts (Figure 2A). The 90% (n = 3,361) of unique Dutch PLPs had an average allele frequency of 3·10−6, and the 78% (n = 1,291) of unique Estonian PLPs had an average allele frequency of 2·10−6 (Figure 2A). This highlights the difference in the underlying genetic architecture between the two populations and suggests that results obtained for each of these cohorts are largely independent, in that they are not based on the same genetic variation. This was to be expected since the Dutch and Estonian populations are separated geographically with limited interaction over recent history.7
Carrier frequency in the European population
On average, each individual carries 2.3 (range 0–11) (Dutch) or 2.0 (range 0–9) (Estonian) PLPs for the set of 1,929 AR genes (median 2/2; Table 1). For the subset of 1,119 recessive genes that are associated with severe phenotypes, the mean number of PLPs per individual is 1.5 (range 0–8) and 1.1 (range 0–6) in the Dutch and Estonian cohorts, respectively (median 1/1; Table 1). In the cohorts, there were 397 (9.6%) Dutch and 315 (13.5%) Estonian individuals with no PLPs and 144 (3.5%) Dutch and 29 (1.2%) Estonian individuals with more than 5 PLPs (Figure S2). Overall, our results establish that on average, Europeans carry at least 1 PLP variant for a severe AR disorder and ∼2 PLPs for any AR disorder.
Table 1.
PLP variants and ARCs for AR genes in the study cohorts
| Mean/median PLPs per sample | ARCs %(N) (Dutch total: 8,485,140; Estonian total: 2,706,301) | |
|---|---|---|
| 1,929 genes | ||
| Dutch cohort (N = 4,120) | 2.3/2 | 1.5% (124,722) |
| Estonian cohort (N = 2,327) | 2/2 | 1.3% (34,570) |
| 1,119 severe genes | ||
| Dutch cohort (N = 4,120) | 1.5/1 | 1% (83,878) |
| Estonian cohort (N = 2,327) | 1.1/1 | 0.8% (20,710) |
Frequency of PLPs per gene
Having established the number of PLPs that an individual carries for recessive disorders, we wanted to investigate which genes have the highest carrier frequencies and have the largest effect on ARCs rates. Most variants in both cohorts are rare, with a carrier frequency of up to 0.05% (Figure S3). As a result, at the gene level, 96.6% of 1,929 tested genes had a total PLP carrier frequency of no more than 0.5% in both cohorts (Figure S4). Of these, 589 (30.5%) and 1,012 (52.5%) genes in the Dutch and Estonian cohorts, respectively, did not have any PLP carriers. At the other end of the distribution, there were 30 (1.6%) and 24 (1.3%) genes with more than 1% PLP carrier frequency in the Dutch and Estonian cohorts, respectively (Figure S4). There is an overlap between common genes in both populations, as 23 genes have more than 0.5% carrier frequency in both populations (Table S2).
Ranking genes by the frequency of PLP carriers demonstrated good correlation between the two populations (Spearman’s correlation coefficient Rho = 0.69, p value = 5.05·10−276). The exceptions were 8 genes in which recurrent PLPs are very common in one population and not in the other (Figure 2B). Six genes were shared in the top-10 rankings of both cohorts (p value = 0.0001, permutation test; subjects and methods).
In conclusion, although the cohorts are independent and each cohort has its own unique variants, the patterns of gene carrier frequencies are similar for both cohorts (Figure 2B).
To validate our per-gene carrier frequency estimates, we compared our results to the published 2016–2017 data of the Dutch neonatal screening program. We compared the rankings based on the observed frequency of the tested disorders to our data-based estimates and found an essentially complete correlation (Figure 2C, Spearman’s correlation coefficient Rho = 0.99, Table S3).
ARCs in the European population
To determine the rate of ARCs, we simulated all possible virtual matings among the Dutch cohort (n = 8,485,140 matings) and among the Estonian cohort (n = 2,706,301 matings; subjects and methods). Simulations for all 1,929 AR genes resulted in 124,722 (1.5%) and 34,570 (1.3%) ARCs in the Dutch and Estonian cohorts, respectively (Table 1), representing virtual matings in which both partners carried a PLP variant in the same gene. Simulations that excluded 324 genes associated with both AD and AR phenotypes, leaving the subset of 1,605 AR-only genes (Table S1), yielded very similar results with 121,607 (1.4%) ARCs in the Dutch cohort. Couples in which both partners carried PLPs that are known to cause mild or asymptomatic phenotype in homozygotes were excluded from this analysis (Table S4). Simulations of the subset of 1,119 severe genes yielded 83,878 (1%) (Dutch) and 20,710 (0.8%) (Estonian) ARCs (Table 1). Therefore, we estimate that 0.8%–1% of European couples are at risk for a child with a severe AR condition, and at least 1.3%–1.5% of couples are at risk for any AR condition.
Considering all 1,929 genes, 90% of the ARCs are explained by the 115 and 84 most frequent genes in the Dutch and Estonian cohorts, respectively. For the 1,119 genes that are associated with severe disease, 90% of the ARCs are explained by the 70 (Dutch) and 57 (Estonian) most frequent genes in the cohorts (Figure S9; Table S5). Since most ARCs are explained by a limited number of genes, adding more genes to existing PCS panels for non-consanguineous couples is not expected to substantially increase the PCS yield, due to diminishing marginal returns.
Effect of consanguinity on ARCs in the European population
Consanguineous unions are not common in European populations but are common worldwide and are increasing in Europe due to immigration. Previously, consanguinity has been estimated to occur in ∼0.06% of couples of Dutch descent,32 similar to other European countries.33,34 Consanguineous couples are typically the first to be referred for preconception screening, because of their increased risk for recessive disease. However, the precise magnitude of this increased risk is unclear, and it is unknown whether it is the same for different disorders.
We simulated consanguineous matings based on the Hardy-Weinberg principles and calculated the expected risks for different degrees of consanguinity, relying on the count of shared alleles that is expected by the relationship (subjects and methods).
We estimate that for any AR disorder, the rate of ARCs is 20.9%–24.9% for first cousins, 10.4%–12.4% for first cousins once removed, and 5.2%–6.2% for second cousins. The ARCs rate for third cousins is 1.3%–1.6%, i.e., not different from that of non-consanguineous unions (Table S6).
For first-cousin unions, considering all 1,929 genes, 90% of the ARCs are explained by the 749 and 540 most frequent genes in the Dutch and Estonian cohorts, respectively (Table S5). This shows that diminishing marginal returns effect is not seen in consanguineous couples, and therefore these couples are expected to derive a greater yield from an exome-based PCS, in comparison to non-consanguineous couples.
ARCs per phenotype in consanguineous and non-consanguineous matings
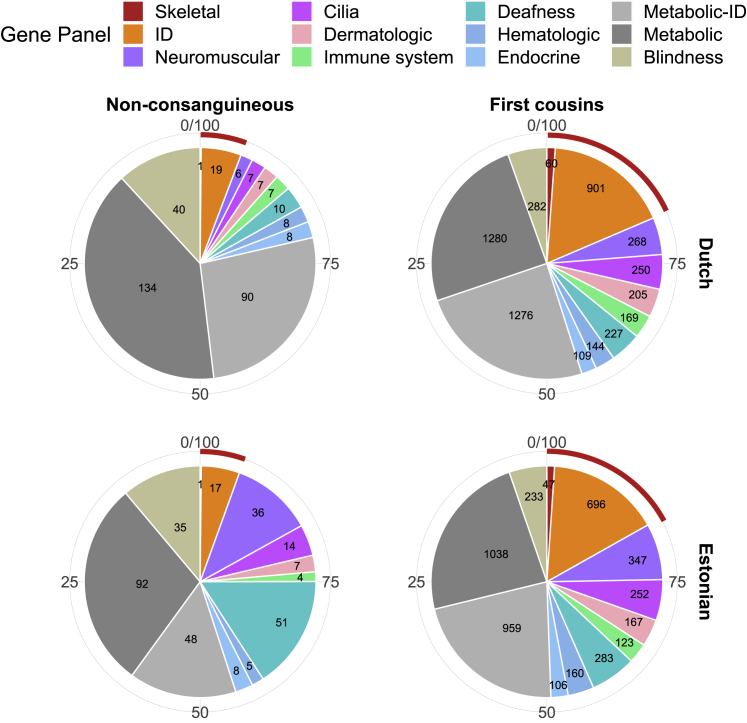
We compared ARCs rates for consanguineous versus non-consanguineous matings for all genes (1,929 genes) and for the sub-group of severe genes (1,119 genes). We also performed this comparison for gene-groups based on diagnostic gene panels corresponding to 12 different types of disorders (Table S1).
We first investigated the allele counts for PLPs per gene per panel. We found striking differences in the distribution of allele counts between the different disorders (Figure S5). For example, only a small fraction of the genes for ID has high (>10) allele counts (8% in the Dutch and 1% in the Estonian cohorts), compared to other panels. In comparison, many more deafness genes have high allele counts (24% and 11% in the Dutch and Estonian cohorts). Next, we calculated the expected number of ARCs per panel for first cousins versus non-consanguineous couples. For each disorder, the fold-increase in ARCs due to consanguinity (first cousins) is indicated as the consanguinity ratio (CR). The CR was 16 for all genes combined, indicating a 16-fold higher risk for first cousins than for unrelated couples across the entire dataset (Figure 3C). The CR for different phenotypic groups of disorders is consistent between the two populations (Spearman correlation 0.5; rising to 0.75 when excluding two common Estonian variants in CLCN1 [MIM: 118425] and GJB2 [MIM: 121011]) (Figure 3D). Notably, we find that the CR is significantly higher for ID and skeletal disorders compared to the average of all genes, in both cohorts (Figures 3 and S6; Table S12). Thus, while consanguinity generally elevates the risk for an affected child with all AR conditions, this elevated risk is not the same for different disorders (Figure 3C). To test whether inclusion of genes with both AD and AR phenotypes biases this analysis, we calculated the CRs separately for the 1,605 AR-only genes and obtained similar results (Table S13).
Figure 3.
Effect of consanguinity on ARCs rates for different disease categories
(A) Rates of ARCs per 100,000 couples (on the y axis) for different disorders (on the x axis) in the Dutch cohort (orange) and Estonian cohort (blue) for non-consanguineous couples.
(B) Same as in (A) but for first-cousin couples.
(C) Consanguinity-ratio (CR) scores. Scores in red are significantly higher compared to a random set of AR genes with the same coding bp length.
(D) Correlation of first-cousin CRs between the Dutch and Estonian cohorts for different disorders. x axis shows the rank in the Dutch cohort; y axis shows the rank in the Estonian cohort. The lower the rank number, the higher the CR in that gene panel (i.e., a gene-panel ranked 1 has the highest CR). The size of the dots indicates the number of genes in the gene panel. The red color indicates gene panels with statistically significant higher CRs compared to a random set of AR genes with the same coding bp length. Rankings excluding common variants in GJB2 (deafness) and CLCN1 (neuromuscular) are marked with an asterisk.
Based on the allele counts and CR scores, we calculated the expected distribution of disorders among affected children (Figure 4). In the Dutch cohort, metabolic disorders and blindness constitute 79% of expected disorders for affected children to non-consanguineous parents, while they constitute only 55% for affected children to parents who are first cousins. Other phenotypes like ID and skeletal disorders are expected to be very rare in affected children to non-consanguineous parents, but much more common in children to parents who are first cousins (Figure 4).
Figure 4.
Effect of consanguinity on affected offspring rates for different disorders
The expected number of affected children for different disorders per 100,000 births, in non-consanguineous and first-cousin couples. The red outer arch represents disorders with significantly higher CR scores.
The difference between the cohorts for the metabolic gene panel is mostly attributed to a high carrier frequency in two genes (CFTR and SERPINA1) in the Dutch cohort.
Heterozygote selection as a possible cause for the differences of PLPs patterns among disorders
A possible reason for the difference in the genetic architecture of ID and skeletal disorders compared to other disorders might be a fitness effect for heterozygous carriers of pathogenic variants in ID/skeletal genes. It is well known that in some AR diseases there is indeed a phenotypic manifestation in heterozygotes.35,36 Simulations show that even if heterozygosity for deleterious AR alleles reduces fitness only mildly, this would greatly reduce the frequency of variants in recessive genes for ID and skeletal disorders. In particular, if heterozygotes for a PLP variant have 0.5% less offspring (reduced fitness), in a large population this PLP variant will not have a frequency higher than 0.09% (Figure S7).37 Based on this hypothesis, we investigated the density of coding singleton variants (i.e., coding variants reported in only one individual) for the different gene panels in the 1000 Genomes dataset14 (Figure 5). This dataset contains genome-sequences samples from five different European populations (GBR, TSI, FIN, IBS, and CEU). We found that among these various European populations, the ID and skeletal disorders genes show a decreased number of coding singletons compared to the other gene sets and are more similar to a set of essential genes that includes genes that are more likely to be under selection (Figure 5). Similar patterns were observed for the singleton density across the entire gene and for the RVIS (residual variation intolerance score) that is based on the number of functional variants in a gene (Figure S8). In conclusion, these results suggest that genes in the ID and skeletal panels are subject to increased selection pressures in the European population.
Discussion
We found that almost all individuals (>85%) carry at least one PLP variant, with an average of at least 1.3 PLPs for a severe AR disorder and 2.2 PLPs for any AR disorder. We believe that this represents a lower bound-estimate because of our stringent selection criteria. Exact estimation of the upper bound would require various assumptions on the number of missed PLPs in our gene panels, the number of undiscovered AR genes, and the frequency distribution of PLPs in those undiscovered genes. However, we find that under realistic conditions, it is unlikely that the number of PLPs will exceed 8 per individual. If novel AR genes that have not yet been discovered contribute less PLPs than those that have been discovered recently, then the upper bound for the estimate is more in the range of 4–5 PLPs per individual (Figure S12; supplemental subjects and methods).
Analysis of virtual matings for each population shows that in the absence of consanguinity, the rate of ARCs is 0.8%–1% for a severe AR disorder. This translates to ∼225 newborns with a severe AR disorder per 100,000 births. We believe these should be considered as minimal estimates. First, we analyzed only the genes that are currently known as AR disease genes, while many new AR genes are still being discovered. Second, we took great care to avoid (likely) benign variants and VUS in our analysis, with the likely result of having excluded some variants that are actually pathogenic. This applies mainly to missense variants for which it is most challenging to predict their phenotypic effect and to hypomorphic variants. Lastly, in our analysis we did not consider variants in regions that are poorly covered by exome sequencing, and other types of variation that are difficult to identify using exome sequencing, such as intronic variants and copy number variation. Notably, the common SMN1 (MIM: 600354) exon 7 and 8 deletion variant is not present in our data. Future analyses of whole-genome sequencing data may give us the opportunity to obtain even more comprehensive estimates, although the systematic interpretation for these other types of variation will pose a significant challenge.
Crucial to our approach is the fact that we employed expert manual revision of classified variants. The current ACMG-based variant classification scheme is focused on pathogenicity but does not consider the degree of pathogenic effect. Thus, two variants classified as “pathogenic” in the same gene may have very different phenotypic effects. For example, in CFTR, both deltaF508 and R117H are classified as pathogenic, but whereas deltaF508 will result in classic cystic fibrosis, R117H may remain undetected or lead to mild disease. To avoid such problems, expert manual revision of classified variants cannot be spared from the classification process in individual cases.
Our results also underline the importance of population-specific databases. As seen in Table S7, up to 30% of PLPs in four major gene panels (deafness, blindness, ID, and metabolic disease), largely rare missense variants, were recognized only when we included information from the Dutch VKGL22 database. As expected, the number of PLPs added based on this part of the selection process was higher for the Dutch population than for the Estonian population (Table S8). General worldwide databases currently do not include population-specific, unique, rare missense variants and thus local databases are required for accurate classification of a significant proportion of PLPs.
Based on our results, we expect first-cousin consanguineous couples to be at 16 times higher risk for a child with an AR disorder compared to non-consanguineous couples. This translates to ∼3,400 newborns with a severe AR disorder per 100,000 births for first cousins. As expected, the risks gradually decreased for more distant relationships and the risk for third cousins was similar to that for non-consanguineous couples at ∼0.9% for a severe AR disorder and 1.4% for any AR disorder (Table S6). These results provide empirical evidence for the common assumption that a third-degree cousin relationship is similar in risk of AR diseases to random mating within an outbred population.
For couples in an outbred population, expanding the scope of PCS to wider panels/exome sequencing is not expected to raise the number of ARCs significantly, due to diminishing marginal returns. A modest number of genes accounts for the majority of ARCs (Figure S9, Table S5), while genes with rare PLPs hardly impact the ARC rate. This assessment includes ARCs for variable severity phenotypes. In contrast, consanguineous couples will benefit from a wider scope of PCS due to the significant influence of genes with rare PLPs on the ARC rate for these couples (Figure S9, Table S5). Therefore, PCS by extensive gene panel or exome sequencing is especially relevant to consanguineous couples.
To assess the effects of consanguinity, we devised the CR score, which indicates the increased risk for an AR disorder due to consanguinity. We found that while consanguinity generally increases the risk for an affected child with an AR condition by about 16-fold, this additional risk is not the same for different disorders. Our data show that for consanguineous couples, the relative risk for AR-ID and AR skeletal disorders is significantly higher than for other disorders. Whereas about 1 in 4 individuals carries a PLP variant in a gene for ID, we calculate an expected incidence in the Dutch cohort of only 19 per 100,000 (0.02%) AR ID in offspring of unrelated parents because couples who are both carriers for PLPs in the same gene are rare. For consanguineous couples, this rises more than 45-fold to 901 per 100,000 (0.9%). In contrast, for other disorders such as metabolic disorders, the expected incidence is 134 per 100,000 (0.13%) in offspring of unrelated parents and 1,280 per 100,000 (1.3%) in offspring of consanguineous couples. We find that these striking differences are due to differences in the distribution and frequencies of PLPs among different disorders, i.e., differences in their genetic architecture. While in non-consanguineous couples only frequent variants have a strong impact on the ARCs rate, in consanguineous couples even rare variants can have a strong impact on the ARCs rate.
The results in both the Dutch and Estonian cohorts show that ∼25% carries a PLP variant in an ID gene, almost all of these variants being rare. There is a single common allele (0.5%) in the Estonian cohort in CRADD (MIM: 603454) which is a well-known cause of AR syndromic ID and likely represents a Northern Scandinavian (Finnish) founder mutation.38 These observations are in line with previous studies on individuals with ID and other neurodevelopmental disorders (NDDs) from outbred populations, which showed a very small (2%–3%) contribution of AR variants to ID, with de novo pathogenic variants explaining the majority of affected individuals.39, 40, 41 In consanguineous couples, a much higher proportion of NDD-affected individuals is explained by AR inheritance.39
The unique genetic architecture observed for the ID and skeletal disorders compared to other disorders could be explained by a small negative effect on fitness for heterozygous carriers of PLPs in these genes. Our results suggest that there is indeed stronger purifying selection on the ID and skeletal disorders genes with respect to other groups of genes, with selection patterns that are more similar to those of essential genes (Figures 5, S8). Elucidation of the magnitude and mechanisms of such negative fitness effects will require analysis of large population samples with relevant phenotypic readouts.
We analyzed two distinct Northern European populations, with no geographical relation between them, and found remarkably consistent results. Although these populations have distinct PLPs and common alleles, our estimates of the overall carrier frequency per sample, the most frequently mutated genes, ARC rates, and CRs are very similar. This resemblance may in part be due to shared or similar selection pressures.
This study provides an estimate for the overall burden of AR PLP variants in two European populations. Our approaches can be applied to other populations, in order to establish their specific AR architecture. Such results can be used by clinicians for baseline risk calculations and be incorporated in PCS guidelines. Given that the majority (>85%) of the population carries at least 1 disease allele for any AR disorder, and that 1 in ∼4 carries an allele for AR ID, it should now be feasible to study the aggregate effects of these PLPs in terms of development, health, and disease at the population level.
Declaration of interests
S.C. is a paid consultant to MyHeritage. All other authors declare no competing interests.
Acknowledgments
C.T.-S. and Y.X. were supported by Wellcome grant number 098051. A.M. was partially supported by the EU through the ERD Fund, Project No. 2014-2020.4.01.15-0012 “Gentransmed.” S.C. thanks the Israel Science Foundation grant 407/17 and the Abisch-Frenkel Foundation. Inclusion of RadboudUMC data was in part supported by the Solve-RD project that has received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 779257. This work was in part financially supported by grants from the Netherlands Organization for Scientific Research (917-17-353 to C.G.). We thank Maartje van de Vorst and Karolis Sablauskas for helping with data analysis.
Published: March 18, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.03.004.
Contributor Information
Christian Gilissen, Email: christian.gilissen@radboudumc.nl.
Han G. Brunner, Email: han.brunner@radboudumc.nl.
Data and code availability
The published article includes all datasets generated or analyzed during this study. All variants that were classified as PLP in this study are listed in Table S15. The code generated during this study are available at https://github.com/hilafrid/AR_PLPs.
Web resources
CFTR2 database, https://cftr2.org/
Dutch neonatal screening results 2016, https://www.rivm.nl/documenten/monitor-van-neonatale-hielprikscreening-2016
Dutch neonatal screening results 2017, https://www.rivm.nl/documenten/monitor-van-neonatale-hielprikscreening-2018
gnomAD Browser, https://gnomad.broadinstitute.org/
GoNL (Genomes of the Netherlands), http://www.nlgenome.nl/search/
Human Gene Mutation Database, http://www.hgmd.cf.ac.uk/ac/index.php
OMIM, https://www.omim.org/
VKGL Dutch database, https://vkgl.molgeniscloud.org/
Supplemental information
References
- 1.Muller H.J. Our load of mutations. Am. J. Hum. Genet. 1950;2:111–176. [PMC free article] [PubMed] [Google Scholar]
- 2.Morton N.E., Crow J.F., Muller H.J. AN ESTIMATE OF THE MUTATIONAL DAMAGE IN MAN FROM DATA ON CONSANGUINEOUS MARRIAGES. Proc. Natl. Acad. Sci. USA. 1956;42:855–863. doi: 10.1073/pnas.42.11.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kondrashov A.S. Contamination of the genome by very slightly deleterious mutations: why have we not died 100 times over? J. Theor. Biol. 1995;175:583–594. doi: 10.1006/jtbi.1995.0167. [DOI] [PubMed] [Google Scholar]
- 4.Gao Z., Waggoner D., Stephens M., Ober C., Przeworski M. An estimate of the average number of recessive lethal mutations carried by humans. Genetics. 2015;199:1243–1254. doi: 10.1534/genetics.114.173351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell C.J., Dinwiddie D.L., Miller N.A., Hateley S.L., Ganusova E.E., Mudge J., Langley R.J., Zhang L., Lee C.C., Schilkey F.D. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci. Transl. Med. 2011;3:65ra4. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lazarin G.A., Haque I.S., Nazareth S., Iori K., Patterson A.S., Jacobson J.L., Marshall J.R., Seltzer W.K., Patrizio P., Evans E.A., Srinivasan B.S. An empirical estimate of carrier frequencies for 400+ causal Mendelian variants: results from an ethnically diverse clinical sample of 23,453 individuals. Genet. Med. 2013;15:178–186. doi: 10.1038/gim.2012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nelis M., Esko T., Mägi R., Zimprich F., Zimprich A., Toncheva D., Karachanak S., Piskácková T., Balascák I., Peltonen L. Genetic structure of Europeans: a view from the North-East. PLoS ONE. 2009;4:e5472. doi: 10.1371/journal.pone.0005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Novembre J., Johnson T., Bryc K., Kutalik Z., Boyko A.R., Auton A., Indap A., King K.S., Bergmann S., Nelson M.R. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lelieveld S.H., Reijnders M.R.F., Pfundt R., Yntema H.G., Kamsteeg E.J., de Vries P., de Vries B.B.A., Willemsen M.H., Kleefstra T., Löhner K. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci. 2016;19:1194–1196. doi: 10.1038/nn.4352. [DOI] [PubMed] [Google Scholar]
- 10.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C., Zhan X., Liang L., Abecasis G.R., Lin X. Improved ancestry estimation for both genotyping and sequencing data using projection procrustes analysis and genotype imputation. Am. J. Hum. Genet. 2015;96:926–937. doi: 10.1016/j.ajhg.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg N.A., Pritchard J.K., Weber J.L., Cann H.M., Kidd K.K., Zhivotovsky L.A., Feldman M.W. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 13.Alexander D.H., Lange K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics. 2011;12:246. doi: 10.1186/1471-2105-12-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zerbino D.R., Achuthan P., Akanni W., Amode M.R., Barrell D., Bhai J., Billis K., Cummins C., Gall A., Girón C.G. Ensembl 2018. Nucleic Acids Res. 2018;46(D1):D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirk E.P., Ong R., Boggs K., Hardy T., Righetti S., Kamien B., Roscioli T., Amor D.J., Bakshi M., Chung C.W.T. Gene selection for the Australian Reproductive Genetic Carrier Screening Project (“Mackenzie’s Mission”) Eur. J. Hum. Genet. 2021;29:79–87. doi: 10.1038/s41431-020-0685-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuller Z.L., Berg J.J., Mostafavi H., Sella G., Przeworski M. Measuring intolerance to mutation in human genetics. Nat. Genet. 2019;51:772–776. doi: 10.1038/s41588-019-0383-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang H., Wu Y., Narzisi G., O’Rawe J.A., Barrón L.T.J., Rosenbaum J., Ronemus M., Iossifov I., Schatz M.C., Lyon G.J. Reducing INDEL calling errors in whole genome and exome sequencing data. Genome Med. 2014;6:89. doi: 10.1186/s13073-014-0089-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Velde K.J., Imhann F., Charbon B., Pang C., van Enckevort D., Slofstra M., Barbieri R., Alberts R., Hendriksen D., Kelpin F. MOLGENIS research: advanced bioinformatics data software for non-bioinformaticians. Bioinformatics. 2019;35:1076–1078. doi: 10.1093/bioinformatics/bty742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q., Wang K. InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines. Am. J. Hum. Genet. 2017;100:267–280. doi: 10.1016/j.ajhg.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zazo Seco C., Wesdorp M., Feenstra I., Pfundt R., Hehir-Kwa J.Y., Lelieveld S.H., Castelein S., Gilissen C., de Wijs I.J., Admiraal R.J.C. The diagnostic yield of whole-exome sequencing targeting a gene panel for hearing impairment in The Netherlands. Eur. J. Hum. Genet. 2017;25:308–314. doi: 10.1038/ejhg.2016.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mezzavilla M., Cocca M., Guidolin F., Gasparini P. A population-based approach for gene prioritization in understanding complex traits. Hum. Genet. 2020;139:647–655. doi: 10.1007/s00439-020-02152-4. [DOI] [PubMed] [Google Scholar]
- 26.Petrovski S., Wang Q., Heinzen E.L., Allen A.S., Goldstein D.B. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hart T., Tong A.H.Y., Chan K., Van Leeuwen J., Seetharaman A., Aregger M., Chandrashekhar M., Hustedt N., Seth S., Noonan A. Evaluation and Design of Genome-Wide CRISPR/SpCas9 Knockout Screens. G3 (Bethesda) 2017;7:2719–2727. doi: 10.1534/g3.117.041277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rehm H.L., Berg J.S., Brooks L.D., Bustamante C.D., Evans J.P., Landrum M.J., Ledbetter D.H., Maglott D.R., Martin C.L., Nussbaum R.L., ClinGen ClinGen--the Clinical Genome Resource. N. Engl. J. Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mainland J.D., Li Y.R., Zhou T., Liu W.L.L., Matsunami H. Human olfactory receptor responses to odorants. Sci. Data. 2015;2:150002. doi: 10.1038/sdata.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng B., Kimmel M. simuPOP: a forward-time population genetics simulation environment. Bioinformatics. 2005;21:3686–3687. doi: 10.1093/bioinformatics/bti584. [DOI] [PubMed] [Google Scholar]
- 31.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ten Kate L.P., Teeuw M.E., Henneman L., Cornel M.C. Consanguinity and endogamy in the Netherlands: demographic and medical genetic aspects. Hum. Hered. 2014;77:161–166. doi: 10.1159/000360761. [DOI] [PubMed] [Google Scholar]
- 33.Fuster V., Colantonio S.E. Inbreeding coefficients and degree of consanguineous marriages in Spain: a review. Am. J. Hum. Biol. 2003;15:709–716. doi: 10.1002/ajhb.10198. [DOI] [PubMed] [Google Scholar]
- 34.Jorde L.B., Pitkänen K.J. Inbreeding in Finland. Am. J. Phys. Anthropol. 1991;84:127–139. doi: 10.1002/ajpa.1330840203. [DOI] [PubMed] [Google Scholar]
- 35.Miller A.C., Comellas A.P., Hornick D.B., Stoltz D.A., Cavanaugh J.E., Gerke A.K., Welsh M.J., Zabner J., Polgreen P.M. Cystic fibrosis carriers are at increased risk for a wide range of cystic fibrosis-related conditions. Proc. Natl. Acad. Sci. USA. 2020;117:1621–1627. doi: 10.1073/pnas.1914912117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mullin S., Hughes D., Mehta A., Schapira A.H.V. Neurological effects of glucocerebrosidase gene mutations. Eur. J. Neurol. 2019;26:388–e29. doi: 10.1111/ene.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amorim C.E.G., Gao Z., Baker Z., Diesel J.F., Simons Y.B., Haque I.S., Pickrell J., Przeworski M. The population genetics of human disease: The case of recessive, lethal mutations. PLoS Genet. 2017;13:e1006915. doi: 10.1371/journal.pgen.1006915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Polla D.L., Rahikkala E., Bode M.K., Määttä T., Varilo T., Loman T., Philips A.K., Kurki M., Palotie A., Körkkö J. Phenotypic spectrum associated with a CRADD founder variant underlying frontotemporal predominant pachygyria in the Finnish population. Eur. J. Hum. Genet. 2019;27:1235–1243. doi: 10.1038/s41431-019-0383-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin H.C., Jones W.D., McIntyre R., Sanchez-Andrade G., Sanderson M., Stephenson J.D., Jones C.P., Handsaker J., Gallone G., Bruntraeger M., Deciphering Developmental Disorders Study Quantifying the contribution of recessive coding variation to developmental disorders. Science. 2018;362:1161–1164. doi: 10.1126/science.aar6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Ligt J., Willemsen M.H., van Bon B.W., Kleefstra T., Yntema H.G., Kroes T., Vulto-van Silfhout A.T., Koolen D.A., de Vries P., Gilissen C. Diagnostic exome sequencing in persons with severe intellectual disability. N. Engl. J. Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 41.Gilissen C., Hehir-Kwa J.Y., Thung D.T., van de Vorst M., van Bon B.W., Willemsen M.H., Kwint M., Janssen I.M., Hoischen A., Schenck A. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated or analyzed during this study. All variants that were classified as PLP in this study are listed in Table S15. The code generated during this study are available at https://github.com/hilafrid/AR_PLPs.