Summary
Genome sequencing is enabling precision medicine—tailoring treatment to the unique constellation of variants in an individual’s genome. The impact of recurrent pathogenic variants is often understood, however there is a long tail of rare genetic variants that are uncharacterized. The problem of uncharacterized rare variation is especially acute when it occurs in genes of known clinical importance with functionally consequential variants and associated mechanisms. Variants of uncertain significance (VUSs) in these genes are discovered at a rate that outpaces current ability to classify them with databases of previous cases, experimental evaluation, and computational predictors. Clinicians are thus left without guidance about the significance of variants that may have actionable consequences. Computational prediction of the impact of rare genetic variation is increasingly becoming an important capability. In this paper, we review the technical and ethical challenges of interpreting the function of rare variants in two settings: inborn errors of metabolism in newborns and pharmacogenomics. We propose a framework for a genomic learning healthcare system with an initial focus on early-onset treatable disease in newborns and actionable pharmacogenomics. We argue that (1) a genomic learning healthcare system must allow for continuous collection and assessment of rare variants, (2) emerging machine learning methods will enable algorithms to predict the clinical impact of rare variants on protein function, and (3) ethical considerations must inform the construction and deployment of all rare-variation triage strategies, particularly with respect to health disparities arising from unbalanced ancestry representation.
Genome sequencing is enabling precision medicine—tailoring treatment to the unique constellation of variants in an individual’s genome. The impact of recurrent pathogenic variants is often understood, leaving a long tail of rare genetic variants that are uncharacterized. The problem of uncharacterized rare variation is especially acute when it occurs in genes of known clinical importance with functionally consequential variants and associated mechanisms. Variants of uncertain significance (VUSs) in these genes are discovered at a rate that outpaces current ability to classify them with databases of previous cases, experimental evaluation, and computational predictors. Clinicians are thus left without guidance about the significance of variants that may have actionable consequences. Computational prediction of the impact of rare genetic variation is increasingly becoming an important capability. In this paper, we review the technical and ethical challenges of interpreting the function of rare variants in two settings: inborn errors of metabolism in newborns and pharmacogenomics. We propose a framework for a genomic learning healthcare system with an initial focus on early-onset treatable disease in newborns and actionable pharmacogenomics. We argue that (1) a genomic learning healthcare system must allow for continuous collection and assessment of rare variants, (2) emerging machine learning methods will enable algorithms to predict the clinical impact of rare variants on protein function, and (3) ethical considerations must inform the construction and deployment of all rare-variation triage strategies, particularly with respect to health disparities arising from unbalanced ancestry representation.
Introduction
We are approaching an era in which genome sequencing at birth may become a widespread practice with the potential to revolutionize healthcare. Interpretation of the genetic variants identified by sequencing, however, remains a significant challenge and limits the use of DNA sequencing as a primary diagnostic screen.1 Current algorithms used to interpret the significance of genetic mutations are not reliable enough to be used without additional clinical data.2 Yet, accumulating biomedical data enables machine learning algorithms to predict the consequence of genetic variants with increasing accuracy. The pairing of modern algorithms and widespread genome sequencing is beginning to deliver precision medicine in limited settings,3 but the broad interpretation of rare genetic variation requires both algorithmic advances and improved access to data. The identification of rare variation responsible for unusual clinical phenotypes is a particularly difficult challenge because both the responsible gene and the associated variation must be identified. A slightly more tractable problem is the identification of clinically important variants in genes that are already known to be clinically significant and have known mechanisms for influencing phenotype.
This paper focuses on two clinical domains that have known clinically important genes and in the near term should benefit greatly from improved rare variant interpretation: pharmacogenomics (PGx) and inborn errors of metabolism (IEMs). IEMs and PGx are examples of genetic practice characterized by monogenic phenotypes for which therapeutic action can be taken in response to clinically important variants in known genes. Both fields have been revolutionized by low-cost sequencing and the curation of large databases cataloging the effects of specific genetic variants. Furthermore, both fields struggle with interpretation of the phenotypic effects of rare variants that have not been clinically evaluated.
As an interdisciplinary team supported by the Chan Zuckerberg Biohub, we approach these two challenges by addressing both computational and ethical issues in order to develop a framework for genome-informed medical care that benefits all. Here, we review the current practices and limitations of variant interpretation in PGx and IEMs and highlight recent computational advances that will allow researchers to improve precision medicine. Ethical considerations include health disparities because existing genetic and genomic databases are not inclusive of individuals of diverse ancestries. As the recent strategic vision from the US National Human Genomic Research Institute (NHGRI) attests, there are significant societal implications of a genomic learning healthcare system that we cannot afford to oversimplify.4 Our focus on genes of known consequence should generalize ultimately to the more difficult cases where the gene, function, and mechanism are not well understood.
PGx and IEMs in current clinical practice
For both PGx and IEMs, our detailed understanding of the biological processes at play (the genes that are critical and how they interact) has reached a point at which routine genetic screens can inform clinical decision-making. In the United States, PGx testing is mandated by the Food and Drug Administration for a number of drugs because of safety concerns and is recommended for many others. Testing for IEMs is routine practice for nearly all newborns in the United States, but the role of genetic testing is largely limited to second-tier screens and carrier testing. These two clinical domains are linked in more ways than it may superficially appear. The clinical implications for most known PGx- and IEM-driven phenotypes are often caused by variants in a single gene. As monogenic traits, there is not only a critical importance in understanding the impact of variants in the underlying genes but also in narrowing the problem space for a tractable solution. Additionally, the mechanisms of disease and treatment response are generally understood.
PGx describes how an individual’s response to medication is influenced by genetic variation in pharmacogenes: genes encoding proteins involved in the pharmacokinetics and pharmacodynamics of a drug.5 Many pharmacogenes have common genetic variants with known clinical significance. These variants can affect the metabolism, transport, and action of drugs throughout the body and may influence efficacy or lead to adverse events. Studies have shown as many as 99.8% of individuals carry at least one genetic variant that could lead to adverse outcomes for at least one drug.6, 7, 8 In the past, clinical practice overlooked the influence of genetics on drug response and—except for several extreme case9—used a standardized dose of any particular drug for most patients, with some trial-and-adjustment to determine the ideal drug and dosage. This error-prone process can lead to decreased efficacy and increased incidence of adverse events that could be otherwise avoided.10 Clinical practice may be moving toward genetic testing prior to drug dosing, although at present, current practice is still limited to physician-guided treatment: genotyping or sequencing is ordered by a physician and carried out clinically (Figure 1A). To date, there are 60 drugs with clinical dosing guidelines published by the Clinical Pharmacogenomics Implementation Consortium (CPIC) and 94 drugs with guidelines from the Dutch Pharmacogenomics Working Group (DPWG).11 As the inexpensive interrogation of genetic information gains a foothold in clinical medicine, pharmacogenetic information will increasingly become standard care. Importantly, when genetic information is used to guide dosing, the current focus is on common polymorphisms in individuals of European ancestry. Common polymorphisms in other ancestral groups and rare variants are generally not included in current clinical dosing guidelines. This can lead to health disparities based on a patient’s ancestry and is problematic for all individuals because rare variants are estimated to contribute to as much as 50% of interindividual variation in drug response.12
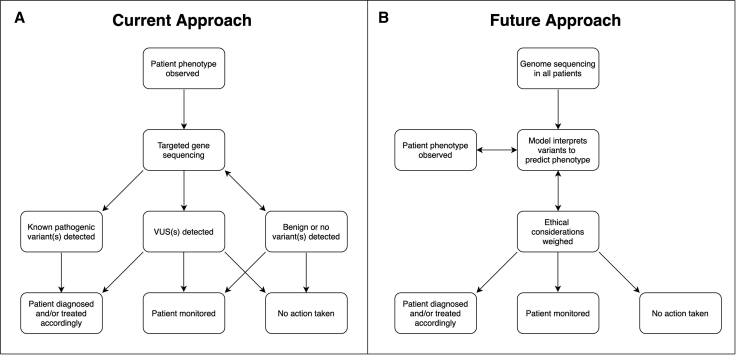
Figure 1.
Diagram of current treatment workflow and proposed workflow that integrates genomics
Simplified overview of the identification and treatment of patients with IEMs or PGx. We contrast the current approach with our proposed framework, which incorporates early sequencing and analysis of rare variants with machine learning and ethical considerations.
(A) Current practice for IEMs and PGx begins with an observable phenotype. In IEMs, this may be an altered metabolite detected by newborn screening; in PGx, perhaps an adverse event. Phenotype can also include physical examination, medical history, family history, and relevant labs or studies. Genetic sequencing is then performed, which could include targeted single-gene sequencing with copy number variant detection, gene panel, whole-exome sequencing, or in some cases, family trio sequencing to assess phasing and identify de novo variants. If annotated pathogenic variants are identified in the target gene, a patient may be diagnosed with a disease and offered preventative services (as is the case with IEMs) or given a different drug or dose adjustment (as with PGx). Identification of VUSs may result in a diagnosis, depending on the other variants identified. In this simplified figure, VUS refers to a variant in the targeted gene of interest as opposed to an incidental finding not relevant to diagnosis. Patient diagnosis can occur without DNA sequencing, as is the case with some IEMs.
(B) Hypothetical future approach to patient care in the fields of PGx and IEMs. All individuals undergo whole-genome sequencing at birth. Machine learning models use detected variants to predict phenotype (disease risk or differential drug response). Ethical considerations are addressed, and clinical action is taken accordingly.
IEMs encompass more than 1,000 genetic disorders, including organic acidemias, urea cycle defects, lysosomal storage disorders, and disorders of amino acid metabolism.13 IEMs are characterized by monogenic mutations that can affect protein function and result in altered metabolite levels. The majority are autosomal recessive disorders. Many IEMs are severe, early-onset conditions amenable to therapeutic intervention, and early treatment can lead to significantly improved clinical outcomes. Because the consequences of unrecognized IEMs in pre-symptomatic newborns can be catastrophic, detection before symptom manifestation is essential. Newborn screening (NBS), a near-universal public health practice, detects over 40 of the most common, treatable IEMs via biochemical tests performed in blood samples taken shortly after birth. IEMs occur in ∼1 in 2,000 births worldwide and are present in all ancestral groups.14 Comparing incidence across ancestry is difficult because of differences in screening between countries and the fact that ancestry is not consistently categorized within countries.15 One study of ancestrally diverse California newborns suggested that newborns with Middle Eastern ancestry had the highest incidence of IEMs (>1 in 1,000) and newborns with Japanese or Pacific Island ancestry had the lowest incidence of IEMs (<1 in 5,000).16
Presently, NBS detects IEMs by identifying elevated metabolites in blood, which is performed with tandem mass spectrometry (MS/MS), an inexpensive and rapid test. However, disorders may be missed, some analytes are non-specific, and follow-up testing may be time consuming and complex.1,17 DNA sequencing has the potential to more accurately identify disorders for which MS/MS detection is not optimal and also identify disorders for which there is no appropriate metabolite screen.
Carrier testing provides an opportunity to detect rare variants in IEMs and other disease-associated genes18 before conception. However, interpretation of genetic screening results still faces significant challenges,19 especially in cases identifying variants of uncertain significance (VUSs) where risk for inherited disease cannot be definitively assessed and actionability is questionable. The falling cost of next-generation sequencing will continue to expand the identification of genomic variants that may cause IEMs or alter drug response. Although many genetic variants have established associations with disease phenotypes or drug response, the majority are of unknown clinical consequence. Generating experimental data to validate the pathogenicity of individual variants is tedious and expensive, although recent advances have facilitated more large-scale generation of data.20 Several databases attempt to catalog variants in disease-causing genes, but there is no central catalog for associated functional data. Thus, alternative methods for determining or predicting functional effects of genetic variants are urgently needed.
At present, validation of genetic variants as causal for IEMs or important for PGx is complex, involving consideration of layers of information at the genetic, phenotypic, clinical, and familial levels.21 Variants in genes underlying IEMs frequently require functional characterization to be validated as causal. Functional validation can be carried out with a myriad of model systems, including patient-derived cells or blood, immortalized cell lines, and animal models.22 Robust functional assays suitable for the validation of variants as causal are not always available because they require a biological or biochemical measurement directly related to the function of the gene of interest. Common experimental methods to validate pathogenicity include overexpression models to assess function of the variant allele, genetic rescue whereby introduction of the wild-type allele rescues phenotype, and transgenic expression for phenotyping in model organisms such as E. coli, yeast, Drosophila, C. elegans, zebrafish, and mice.22 CRISPR-Cas9 technology allows for high-throughput functional characterization in many systems. Assays investigating mRNA and protein expression (i.e., RNA sequencing [RNA-seq] and immunoblot) can reveal variant consequences on splicing and allele expression or differential protein expression, respectively.22 The validation of clinically important variants relating to PGx is also complex. Targeted functional assays evaluating variant effects on gene function can be carried out in vitro when feasible via similar methods and models as for IEMs. Examples include enzyme activity assays23 and transporter uptake assays.24 Pharmacogenetic variation can further be validated as clinically important in pharmacokinetic/pharmacodynamic studies, whereby individuals with a particular genotype exhibit significantly different drug response compared with individuals with a different genotype for the variant in question.
Ethical considerations in rare variant interpretation
Genome-informed precision medicine must include analysis of ethical, legal, and social implications (ELSIs) in order to improve upon rather than exacerbate existing health disparities.4 We have identified six chief concerns with enhancing computational predictors for the phenotypic effects of rare variation at the scale proposed here. First, the uncertainty of results and, second, the return of clinical results can either improve or compromise clinical care. Although enhanced computational predictors for IEMs and PGx can minimize harm from the trial and error of current clinical practices, consistency in clinical education and approaches to ambiguous and incidental findings will be critical to determining societal benefit. Third, research and clinical stakeholder perspectives in approaching the classifications of VUSs can differ. Fourth, the underrepresentation of minority groups in current datasets and the underlying research that informs them needs particular attention in order to create a larger and more diverse reference genome so that biases can be reduced. Fifth, an effective genomic learning healthcare system must account for data security and privacy risks. Sixth, there needs to be transparent data sharing expectations across all levels of participation in the learning system. Building on previous ethical frameworks25,26 and the need for a nuanced approach,27 we suggest that trade-offs between ensuring individual control over data and the social obligations of individuals have yet to be resolved at the level of ethical governance provisions. Discussion of these concerns is guided by three central ethical questions, summarized in Table 1 and elaborated within the ethics spotlight sections.
Table 1.
Ethical considerations for the adoption of novel genomic technologies into learning health system practice
| Area of IEMs and PGx and ethical issues | Key question |
|---|---|
| Whole-genome sequencing for newborns: (1) uncertainty of results and (2) return of clinical results, including results from late-onset disorders | Can genome sequencing improve the uncertainty of results and return of clinical results? |
| Interpreting VUSs: (3) research and clinical divide and (4) social/racial inequity | Can we view the classification of VUSs as a social justice opportunity to close social and genetic ancestry gaps? |
| Genomic learning healthcare systems: (5) privacy risks and (6) data sharing | How can genomic learning healthcare systems ensure adequate genomic input and data governance? |
VUSs, variants of uncertain significance.
Ethics spotlight 1: Can genome sequencing improve the uncertainty of results and return of clinical results?
For the use of predictive algorithms as the primary methods of analysis for IEMs and PGx to be ethically justified, these methods must provide equal or greater certainty than current methods. Improving screening and predictive analysis for IEMs and PGx at the testing level is contingent upon the accuracy of results, the provisions around returning results, and the impact on clinical care. Even pathogenic results can have variable penetrance and/or VUSs and, given the possibility of reclassification over time, can cause significant consternation on both the part of the clinician and patient.28 Perhaps most thoroughly documented in cancer genetics,29 the clinical return of genetic results is rarely straightforward. The prohibition against the return of uncertain results, outlined by the American College of Medical Genetics and Genomics (ACMG), is such that even if there is a suspicion that an uncertain variant is pathogenic, it should conservatively be classified as a VUS because this information is used in medical decisions.2
The follow-up of uncertain results is complicated by clinician/researcher and patient expectations and understandings of actionability. Genomic literacy across different healthcare professional roles is limited.15,30 The disclosing of sequencing results should be contingent upon what has been previously explained to the patient/parent about incidental findings and potential treatments.31 As healthcare delivery is already biased with regard to decisions about referrals or withdrawals of care, including decisions made through racial discrimination, it will be challenging for algorithms to correct for existing biases in the handling of results.32 Uncertain and incidental (or secondary) results in clinical care should be considered in the context of existing slippages of fiduciary obligations—such as clinician biases and/or patient mistrust—that emerging tests may or may not be able to compensate for.33 The NHGRI has called for greater diversity among the genomic scientist workforce.4
In order to contain immediate risks around uncertainty of results and focus resources, is there a case for tiered approaches? For example, beginning with targeted sequencing and, upon accuracy improvements, expanding programs to include non-targeted sequencing, or at the individual level, only sequencing specific genes as a second-tier option if a positive test result arises in genome sequencing? Certainly, implementing genome sequencing at the routine screening level requires greater computational accuracy, accessibility, and more nuanced ethical safeguards.4,27 In the US healthcare context, it is difficult to resolve the issue of healthcare insurance coverage. Can financial disparity in the follow-up of results be partially alleviated with temporary coverage through risk-sharing agreements between payers and manufacturers of tests?34 Can ethical priorities of the clinician and patient transaction be made compatible with the needs of the genomic learning healthcare system—which must maximize scarce resources—such that genomic sequencing improves healthcare across all of society?
Evaluating variants of uncertain significance
Variants in functionally important genes are often suspected to lead to clinical consequences. For IEMs and PGx, there are hundreds of genes in which nonsense and missense variants are associated with clinical outcomes. Although additional genetic, epigenetic, and environmental factors alter disease risk and drug response, the gene sequence is the primary determinant of phenotype for these genes. Thousands of pathogenic rare variants in these genes have been characterized with clinical consequences often well understood and cataloged. Yet exome and genome sequencing continue to identify novel variants in these genes at a rapid pace. The ACMG has developed guidelines to interpret these variants, but by design, conclusive evidence is required to assert a variant is pathogenic, even in known disease genes.2 For example, defects in PAH (MIM: 612349) cause phenylketonuria (PKU [MIM: 261600]), an IEM that can lead to severe intellectual disability and seizures when untreated. In gnomAD,35 a population database of variants seen in more than 100,000 individuals, 57% of observed protein-altering variants in PAH have unknown pathogenicity. Individuals who are homozygous for these variants at birth will have an unknown risk of developing PKU, and carriers of these variants cannot be advised of their risk of having a child with PKU. Thus, predicting the functional consequence of rare variants in IEMs and PGx is an important challenge.
To begin to address this issue, numerous publicly available databases actively catalog genetic variants and associated disease and drug response phenotypes. These databases are typically human curated and bring together information that would otherwise be dispersed across the literature, allowing researchers and clinicians to quickly access existing knowledge. Several databases focus on the pathogenicity of variants genome wide, including thousands of variants in IEM and PGx genes. These include ClinVar, ClinGen, the Human Gene Mutation Database (HGMD), and Online Mendelian Inheritance in Man (OMIM).36, 37, 38 Such platforms have a shared goal of linking genes with disease, although they take different approaches. ClinVar allows submissions from clinical laboratories, research groups, and specialized databases, presenting all submitted data through an online interface. Most submissions are not manually vetted and are presented as submitted. ClinGen and OMIM attempt to provide authoritative curation of known variants and their relationship to disease. Curators review literature and experimental data to determine pathogenicity of genetic variants. ClinVar and ClinGen share and collaboratively curate data. In addition to being used for standardizing the set of variants with known consequences, these databases are also used by researchers and clinicians to evaluate the evidence that an uncatalogued VUS causes disease based on its similarity to cataloged variants (e.g., if a VUS results in the same amino acid change as a cataloged pathogenic variant, this VUS now has strong evidence for being pathogenic).2 Similarly, efforts have been made to catalog the relationship between genetic variation and drug response, exemplified by databases including PharmVar and PharmGKB.39, 40, 41 Like ClinVar, PharmVar relies on user submissions of discovered haplotypes in genes related to pharmacogenomics.
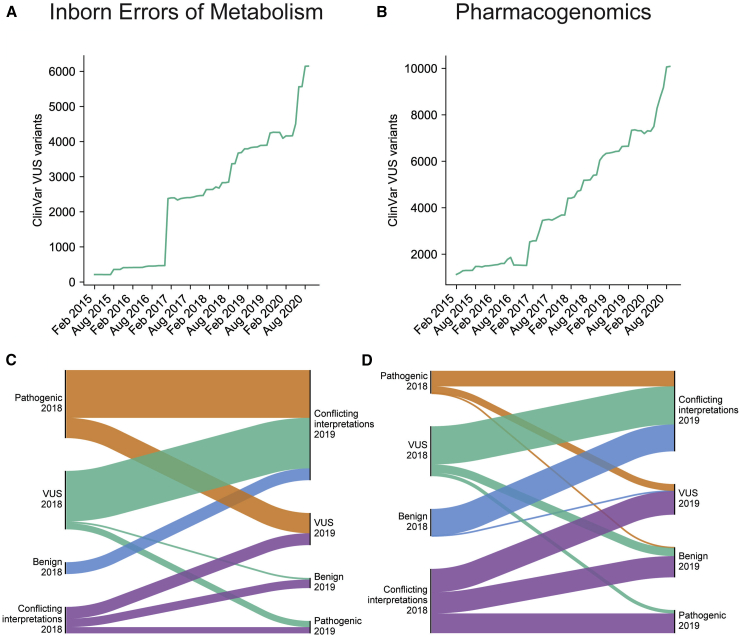
These variant databases encapsulate the combined expertise of thousands of clinical researchers across the world but also reveal a large amount of uncertainty. The majority of possible missense variants in IEM and PGx genes are classified as VUSs or are altogether missing from databases. ClinVar alone contains more than 6,000 variants classified as VUSs in IEM genes and more than 10,000 VUSs in PGx genes (Figures 2A and 2B). Variants in ClinVar change classification as researchers submit new evidence, but very few VUSs are resolved as fully pathogenic or benign (Figures 2C and 2D). Instead, many variants are subject to conflicting classifications. Indeed, 41% of IEM and PGx variants in ClinVar are of uncertain significance or have conflicting interpretations of clinical importance. For novel variants, it is often challenging to establish pathogenic certainty until they are observed by multiple clinicians who submit consistent classifications to a variant database. For VUSs without further clinical or experimental evidence, computational methods offer a possible resolution.
Figure 2.
ClinVar variants of uncertain significance in genes related to IEMs and PGx
(A–D) The number of VUSs in ClinVar between 2015 and 2020 in IEM and PGx genes, respectively (A and B). All ClinVar variants in IEM and PGx genes that were reclassified between February 2018 and February 2019 (C and D). Height of bars is proportional to number of variants reclassified. A total of 293 variant reclassifications is shown in (C), and 434 variant reclassifications are shown in (D).
Most computational approaches predict the functional impact of single-nucleotide polymorphisms (SNPs) and small insertions and deletions (INDELs) by using predictive machine learning models. The popular tool CADD uses a logistic regression model and more than 60 genomic features to learn the features that distinguish randomly generated variants from recently fixed variants in humans.42 The resulting predictor has been used to predict the pathogenicity of clinical variants and is currently used in clinical analysis pipelines.43 REVEL, a meta-predictor, uses the ensemble of scores from several prediction algorithms like CADD, each with different strengths and weaknesses, and is trained to differentiate rare unlabeled variants from HGMD pathogenic variants.44 Both CADD and REVEL are capable of predicting the effects of variants in any gene, which is typical of predictors used in clinical research. However, predictors that are gene-, gene family-, or locus-specific generally perform better for both IEMs and PGx in comparison to predictors that rely on data from the entire genome.45, 46, 47, 48, 49, 50, 51, 52 Despite their promise, such bespoke methods are constrained by the limited data available for most genes, such as the number of known pathogenic variants and associated functional data. Because these methods are designed to predict the functional impact of a variant, their predictions can be some layers removed from the clinical consequence. Additionally, pharmacogenes are not under the same evolutionary constraint as genes involved in disease, limiting the effectiveness of most predictive algorithms.47,53
To combine the best features of variant databases and computational predictors, automated systems that use both in tandem are already being tested to predict the pathogenicity of rare variants. Consider one recent study evaluating IEM detection by sequencing dried blood spots (DBSs) obtained from newborns.1 This study compared the performance of MS/MS to exome sequencing as a primary screen for IEMs on a set of 805 newborns with confirmed IEMs. Variants identified by sequencing were automatically assessed on rarity, protein consequence, and predicted pathogenicity (including CADD) and matched with cataloged pathogenic variants in ClinVar and HGMD to predict disease status. Overall, this combination was neither sufficiently sensitive nor specific compared to MS/MS, and exome sequencing notably missed a number of cases in which a pair of rare, protein-altering variants were absent from the causal gene. However, performance varied among IEMs and, in some cases, provided more specific diagnoses than conventional MS/MS analyte testing. 32% of pathogenic variants were absent from HGMD and ClinVar. Critically, sequencing led to several false positives in which an individual harbored a pair of rare, protein-altering variants in an IEM gene but did not have the associated disorder. These false positives significantly limit the ability to use DNA sequencing for screening and could be mitigated by more accurate computational methods that distinguish pathogenic from benign protein-altering variants.
Ethics spotlight 2: Can we view the classification of VUSs as a social justice opportunity?
Whether the classification of VUSs and IEMs can offer a fairer distribution of the benefits of sequencing technologies across all population groups is a significant question. Most large datasets in the US contain homogeneous ancestry that is unrepresentative of the whole population.54,55 In addition to the need to improve predictive methods for IEMs, screened individuals need to be considered as part of a social group in relationship to a wider and unequal social system. The moral obligations embedded within the ethics of clinical research and practice need to be better integrated.25 For individuals seeking healthcare, polygenic risk scores are more accurate for patients of European ancestry because the data from which algorithms are trained are derived largely from individuals of European ancestry.56,57 Similarly, variant impact predictors tend to be derived from cataloged variants in databases, which are not representative of all ancestries. For example, ClinVar was recently found to be missing a large number of hearing impairment variants that primarily affect individuals of African ancestry,58 most likely indicative of a broader pattern. For variant predictors, this bias will lead to greater reliance on European ancestry variants and European genetic context, producing less accurate classification of IEM and PGx variants in other ancestral populations (e.g., African), which would only compound existing injustice in healthcare access for underrepresented populations.59,60 Disparity in ancestry representation is especially stark in data sources for genome-wide association studies, where European ancestry disproportionately represents 81% of the dataset population.54
Can we alleviate healthcare disparity by closing current ancestry gaps in genetics research? Given evidence that polygenic risk scores can be improved upon by incorporating datasets from a broader range of genetic ancestries,61 it is imperative that the genetics field strives for fairer training data. As the field matures to consider the role of genetic modifiers,62 as well as social and environmental interactions,63 genotypes of diverse individuals are needed to consider the effects of genetic modifiers and the environment on variants. Newborn screening programs, with their mandatory collection and the near universal application of testing, provide a diverse and truly representative set of individuals.16 That said, racial discrimination in healthcare and healthcare research is not simply resolvable through technical fixes. Redressing data underrepresentation and health equity in machine learning precision medicine must be viewed in the context of governance and broader social change, which we discuss in “ethics spotlight 3,” regarding questions of social obligation.
Opportunities in rare variant evaluation
In predicting the effect of a variant on gene function, we can predict its effects on the system, such as a metabolic pathway, and then on the physiology and/or pathophysiology. Cataloging observed likely clinically impactful variants in databases such as ClinVar and PharmVar38 can be effective for determining the pathogenicity of more frequent rare variants (allele frequency between 0.01% and 1%). These variants are common enough that they have been identified in multiple individuals, and therefore, the effect on phenotype can be verified. However, ultra-rare variants, defined as having an allele frequency less than 0.01%, are responsible for a large portion of rare genetic disorders. Publicly available databases of PKU patients indicate that 60% of cases involve at least one ultra-rare SNV, and in 28% of cases, the affected individual carries an ultra-rare variant on both copies of PAH. Some of these ultra-rare variants may be de novo mutations, and the individual may be the only person known to harbor that exact variant.64 The vast majority of ultra-rare variants are absent from clinical databases, indicating that the current approach of cataloguing observed genetic variants fails when allele frequencies are especially low. For PAH, which is one of the most studied metabolic genes, only 9% of possible SNVs have functional impact classified in ClinVar.
Emerging computational algorithms may serve as a means for evaluating the impact of rare variants in IEM and PGx genes. As noted above, existing algorithms have limited ability to accurately predict the impact of variants in these genes, especially among rare variants. Methods have been developed to specifically evaluate variants in pharmacogenes, but these are largely based on existing methods and may have some of the same inherent biases.47 Machine learning has revolutionized computer vision and natural language processing by effectively analyzing spatial and sequential data.65, 66, 67 Machine learning is a type of artificial intelligence in which algorithms are taught, or “trained,” to make predictions based on existing data. Machine learning forms the basis of existing variant effect prediction algorithms, where an algorithm is trained to predict whether a genetic variant will be deleterious or not on the basis of a training dataset of known deleterious and benign genetic variants. In recent years as computational power and the amount of available data has increased, a type of machine learning that uses deep neural networks, known as deep learning, has become widespread. With the rapid growth in the availability of biological data, deep learning has also been extensively used in bioinformatics,68, 69, 70, 71, 72, 73, 74, 75 including transcription factor binding site prediction,76 genome functional annotation,77 and assessment of variant function.78,79 Several methods have been developed specifically for the evaluation of alleles in pharmacogenes, namely CYP2D6 (MIM: 124030).80,81 These purpose-built models outperform existing methods and are capable of assessing the impact of any combination of variants observed in a haplotype rather than single variants. One major drawback of deep learning is that it requires an immense amount of data in order to estimate the large number of parameters required for good performance.82,83
Transfer learning offers an opportunity to leverage the power of deep learning in situations where data are limited. It is difficult to obtain sufficient data to develop phenotype prediction algorithms from genomic data via deep learning, especially when we only have tens or hundreds of individuals with both genome sequencing data and well-characterized clinical or molecular phenotypes. Transfer learning is an emerging approach for overcoming the challenge of limited data. The idea is to build models that perform a task (X) that is similar to the goal task (Y) but for which there are large amounts of relevant real or simulated data. Once the model for solving task X is performing well, it can be refined with data relevant to task Y. In the case of predicting variants, we might build a model using data from a well-studied gene (X) and then refine the model with data from a poorly studied gene (Y). The resulting model may perform very well on Y because the “lessons” learned in modeling X transfer well to Y.84, 85, 86, 87, 88, 89 There are several flavors of transfer learning that have been applied to applications in genetics and proteomics. Convolutional neural network (CNN)-based approaches pre-train weights of convolutional layers on large datasets that can be finetuned on smaller datasets.90 Transformer-based approaches, frequently used in natural language processing, have been applied to functional predictions of variants in proteins.91,92 Graph-CNNs have been used to make drug-binding predictions with protein structure data after being pre-trained with an unsupervised learning step.93 These transfer learning methods could in theory be used to create structure-based predictions of the effect of amino acid changes on drug binding. These methods combined with in silico representations of drug molecules could be used to create substrate-specific predictions of drug-protein interactions and how genetic variants may influence that behavior.
The underlying homology between gene orthologs and paralogs may allow for an increased ability to perform transfer learning. We may be able to use knowledge learned in some domains to inform others. Not surprisingly, some rare diseases have received more attention than others, often because of the incidence of the disease, serendipitous factors, and scientific opportunities. These well-studied diseases typically have significantly more variant impact data available than others. PKU has an incidence of 1 in 10,000 newborns, and there are hundreds of disease-associated cataloged variants. In comparison, tyrosine hydroxylase deficiency (THD [MIM: 605407]) affects fewer than 1 in 100,000 newborns and has been associated with fewer than 20 variants in TH (MIM: 191290). Sequencing benefits individuals with THD less simply because the disease is rarer and few known pathogenic variants exist. The chemical similarity of phenylalanine and tyrosine leads to a high degree of homology between PAH and TH, which presents an opportunity to transfer knowledge about PKU variants to better understand THD—for example, in understanding which parts of the protein may be more or less tolerant of non-synonymous mutations. However, although transfer learning may offer some advantages in the assessment of rare variation, this approach relies on the existence of genes that are similar enough to the gene of interest with sufficient data. Transfer learning may be valuable for some domains, but there is still a need to generate large amounts of high-quality data. Ideally, for knowledge to be truly transferable, data collection would be ongoing and from whole-population datasets rather than being limited to existing datasets.
Ultimately, the goal of any variant interpretation method is to improve clinical care. Integration of genetics into the clinic is already quite challenging, and integration of computational methods for predicting variant function is rife with further challenges. Learning health systems have long been proposed as models for improving healthcare,94, 95, 96 but integration of genetic data into such a system would allow for the accumulation of data to train more sophisticated predictive models as well as an opportunity to iteratively improve upon such algorithms.
A genomic learning healthcare system would allow for rapid collection and phenotyping of rare variants. Learning health systems have been proposed in healthcare since 2007, but few have fully integrated genetics to inform patient treatment.95 In existing systems, the algorithms are constantly improving on the basis of a feedback loop of data that are collected over the course of patient treatment. A genomic learning healthcare system (GLHS) would operate in much the same way, but with the addition that clinical decision support is provided on the basis of genetic data as well as clinical data.34 In this proposed system, collection, sequencing, and analysis of patient data would be required as a first step and would need to be available as part of the patient’s clinical record in the electronic health system. This would enable clinical decision support for IEM- and PGx-related conditions, providing doctors with diagnosis and treatment guidance. The algorithms underlying the clinical decision support can be evaluated regularly and updated on the basis of newly available patient data. In addition to evaluating the algorithms, sequencing and analyzing important genes for every individual treated will allow for more rapid collection and phenotyping of ultra-rare variants—if ancestrally diverse datasets are available.
The ultimate goal of a GLHS is to improve treatment for all patients by leveraging their genetic data. This includes determining the pathogenicity of rare variants that may be previously unseen in patients and potentially making clinical decisions based on their predicted impact. As a conservative first step, a genomic learning healthcare system could implement existing clinical guidance models for IEMs and PGx, such as the pharmacogenomics dosing recommendations from CPIC. Once genetic data are collected for each patient, predictive models for rare variants can be developed and implemented in clinical practice at such a time when there is sufficient confidence in the predictions of the model. Careful analysis will be needed in selecting and evaluating predictive models for both IEMs and PGx, and it is likely that gene-specific models will be needed. The specific clinical action based on a predicted phenotype will then depend on the application area and the onset and severity of the condition. Severe IEMs may require immediate intervention (such as PKU), whereas for others, a preventative approach is deployed. Some IEMs respond to pharmaceutical interventions, and genotype may predict likelihood of response to a specific medication.97,98 For late-onset IEMs, genotype may predict age of onset, which can inform appropriate patient monitoring.99 Similarly for PGx, if the potential consequences of prescribing a drug are life threatening, the clinician may select an alternate therapy. Likewise, if the consequences are mild, they may proceed with caution. We illustrate this framework in Figure 3 before turning to the ethical questions to be taken into account.
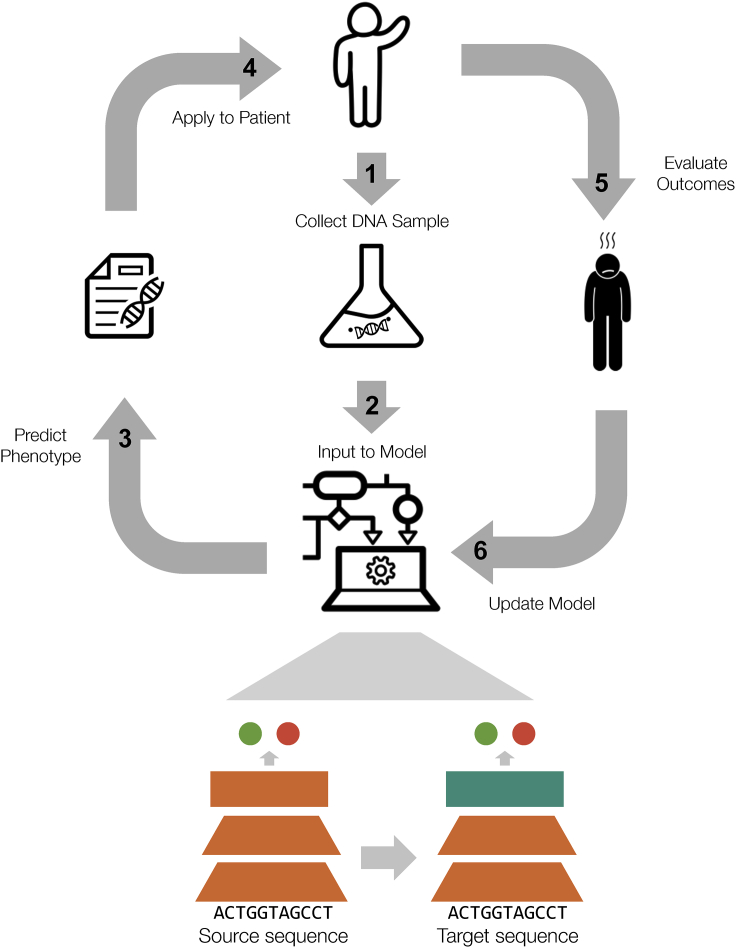
Figure 3.
Proposed workflow for a genomic learning healthcare system
Patients’ DNA samples are collected and sequenced with genomic data input to computational models. The model outputs a predicted phenotype for the patient; results are reviewed by clinicians and applied to the patient. Outcomes are evaluated and the model continues to learn from a feedback loop to improve outcomes for future patients. Icons are from The Noun Project.100, 101, 102, 103
Ethics spotlight 3: How can genomic learning healthcare systems ensure adequate genomic input and data governance?
Data governance and consent for secondary data use will significantly shape whether or not genomic learning healthcare systems can improve accuracy and reduce biases. Learning health care systems present unique ethical challenges that traditional clinical and research ethics—focusing on individual harms and a sharp research/clinical care divide—will find difficult to address.25 Data collection and input (step 1 and step 2 of Figure 3) differs between clinical and public health repositories in terms of provisions around secondary use. One option to improve data privacy and security is through the use of federated learning. This approach involves a centrally pooled dataset with non-co-located data only; data are not shared directly, and model parameters could be protected by research collaboration agreements, advances in data encryption, and a trusted third-party to oversee data access.104
The use of artificial intelligence in healthcare systems is also complicated by issues arising from the possible encoding and routinization of human bias, even with the use of seemingly neutral data sources.55 Artificial intelligence has been described as “the collective medical mind.”32 More than simply doing no harm, a GLHS should actively support greater health equity.4,105 Of central importance is whether clinical data, or model parameters if deploying a federated learning approach, could be viewed and secured as a public good insofar as all stakeholders, both healthcare and private industry, hold a moral obligation to use and share clinical data in ways that benefit society over and above individual or commercial interests.26 If viewing clinical data as a public good, determining how to deal with computational predictors and healthcare outcomes that accurately capture differences not so much resulting from human input biases but rather serving unfair social conditions would be of greatest difficulty.
For public health data use, it is important to identify and address social and political inconsistencies in the ethical oversight from institutional review boards and government bodies, particularly in regard to informed consent and anonymization of data.106 This requires careful consideration for how questions of beneficence regarding collections and distribution of quality care across populations can vary and ultimately widen health disparities.107 Taking the case of newborn DBSs, the current justification for the mandatory nature of newborn screening rests on the potential harms to the child were they not screened for these treatable conditions (see Johnston et al., 2018 for a full historical justification).27 Safeguards are needed to protect the storage and research use of genetic data, which could become more identifiable.108 With such protections, could the practice of informed consent with individuals be seen as less important than another process to ensure respect for autonomy at a group level in order to meet social obligations to contribute to both greater knowledge and efforts to reduce social inequity in health?25 Because biobanks of newborn DBSs provide a rich and unique dataset for research and improving newborn screening (and other genetic testing)—with enormous potential for contribution to a GLHS—the loss of such potential, if secondary use of newborn DBSs is only permissible on an individual consent basis, needs to be carefully weighed up against ethical concerns about respect for individual control. How do we ensure respect for individuals in a GLHS that relies on the collective contributions of entire populations in order for everyone to potentially benefit? Those implementing machine learning research in a GLHS must engage these questions directly.
Conclusion
The defining problem of the genomic age is the interpretation of human genetic variation. In reviewing computational advancements and ethical concerns, we look to develop gene-specific variant interpretation algorithms with a genomic learning healthcare system that builds from a focus on early-onset treatable disease in newborns and actionable pharmacogenomics recommendations. We seek diagnosis of IEMs and treatment for PGx that is tailored to each individual and treatment outcomes that are shared to improve treatment for future patients across all of society. The existing system is the first step toward this goal, as evidenced by confirmatory sequencing of patients and variant cataloging in databases such as ClinVar. Yet the existing system falls short because it is reactive rather than predictive and accurate treatment depends on whether the variant has been previously seen and cataloged. Importantly, it remains to be determined whether computational methods can alleviate health inequity that is reinforced by these limited variant databases. Pervasive sequencing may indeed present a social justice opportunity: to actively promote a more fair and consistent distribution of treatment across all population groups. Yet, there are many barriers blocking the way, including unrepresentative sequencing databases, secondary data use permissions, barriers to healthcare access, and existing biases at the human interface of research and caregiving.
There are technical challenges, including accurate variant classification, data limitations, and growing numbers of variants of uncertain significance. A combination of a GLHS and transfer learning can overcome existing data limitations in order to improve the computational prediction of variants. An increased understanding of each patient’s variants will enable more precise diagnosis and treatment. Most importantly, as more patients provide information into the system, lessons learned from one patient may inform the care of all patients. A dynamic and fair genomic learning healthcare system will create the greatest patient benefit from the captured genomic and phenotypic information, but this will fundamentally depend on careful consideration of societal implications.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
G.M. is supported by the Big Data to Knowledge (BD2K) from The National Institutes of Health (T32 LM012409). A.G.S. is supported by a National Science Foundation Graduate Research Fellowship under grant DGE 1752814. M.N. and B.A.K. are supported by U19HD077627. D.C.M. is supported by Stanford Clinical & Translational Science Award (NIH, NCATS: UL1TR003142). R.B.A. is supported by NIH GM102365 and HG010615. Additionally, G.M., M.L.K., J.E.H.B., M.N., S.W., K.M.G., B.A.K., R.C.G., and R.B.A. are funded by the Chan Zuckerberg Biohub.
Data and code availability
This publication did not generate datasets or code.
Web resources
Chan Zuckerberg Biohub, https://www.czbiohub.org/
ClinGen, https://clinicalgenome.org/
ClinVar, https://www.ncbi.nlm.nih.gov/clinvar/
OMIM, https://www.omim.org/
PharmGKB, https://www.pharmgkb.org/
PharmVar, https://www.pharmvar.org/
References
- 1.Adhikari A.N., Gallagher R.C., Wang Y., Currier R.J., Amatuni G., Bassaganyas L., Chen F., Kundu K., Kvale M., Mooney S.D. The role of exome sequencing in newborn screening for inborn errors of metabolism. Nat. Med. 2020;26:1392–1397. doi: 10.1038/s41591-020-0966-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kather J.N., Pearson A.T., Halama N., Jäger D., Krause J., Loosen S.H., Marx A., Boor P., Tacke F., Neumann U.P. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat. Med. 2019;25:1054–1056. doi: 10.1038/s41591-019-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green E.D., Gunter C., Biesecker L.G., Di Francesco V., Easter C.L., Feingold E.A., Felsenfeld A.L., Kaufman D.J., Ostrander E.A., Pavan W.J. Strategic vision for improving human health at The Forefront of Genomics. Nature. 2020;586:683–692. doi: 10.1038/s41586-020-2817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lavertu A., McInnes G., Daneshjou R., Whirl-Carrillo M., Klein T.E., Altman R.B. Pharmacogenomics and big genomic data: from lab to clinic and back again. Hum. Mol. Genet. 2018;27(R1):R72–R78. doi: 10.1093/hmg/ddy116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Driest S.L., Shi Y., Bowton E.A., Schildcrout J.S., Peterson J.F., Pulley J., Denny J.C., Roden D.M. Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clin. Pharmacol. Ther. 2014;95:423–431. doi: 10.1038/clpt.2013.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reisberg S., Krebs K., Lepamets M., Kals M., Mägi R., Metsalu K., Lauschke V.M., Vilo J., Milani L. Translating genotype data of 44,000 biobank participants into clinical pharmacogenetic recommendations: challenges and solutions. Genet. Med. 2019;21:1345–1354. doi: 10.1038/s41436-018-0337-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McInnes G., Lavertu A., Sangkuhl K., Klein T.E., Whirl-Carrillo M., Altman R.B. Pharmacogenetics at scale: An analysis of the UK Biobank. Clin. Pharmacol. Ther. 2020 doi: 10.1002/cpt.2122. Published online November 25, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin M.A., Klein T.E., Dong B.J., Pirmohamed M., Haas D.W., Kroetz D.L., Clinical Pharmacogenetics Implementation Consortium Clinical pharmacogenetics implementation consortium guidelines for HLA-B genotype and abacavir dosing. Clin. Pharmacol. Ther. 2012;91:734–738. doi: 10.1038/clpt.2011.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Institute of Medicine . The National Academies Press; Washington, DC: 2000. To Err Is Human: Building a Safer Health System. [PubMed] [Google Scholar]
- 11.Bank P.C.D., Caudle K.E., Swen J.J., Gammal R.S., Whirl-Carrillo M., Klein T.E., Relling M.V., Guchelaar H.-J. Comparison of the Guidelines of the Clinical Pharmacogenetics Implementation Consortium and the Dutch Pharmacogenetics Working Group. Clin. Pharmacol. Ther. 2018;103:599–618. doi: 10.1002/cpt.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingelman-Sundberg M., Mkrtchian S., Zhou Y., Lauschke V.M. Integrating rare genetic variants into pharmacogenetic drug response predictions. Hum. Genomics. 2018;12:26. doi: 10.1186/s40246-018-0157-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreira C.R., van Karnebeek C.D.M., Vockley J., Blau N. A proposed nosology of inborn errors of metabolism. Genet. Med. 2019;21:102–106. doi: 10.1038/s41436-018-0022-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Waters D., Adeloye D., Woolham D., Wastnedge E., Patel S., Rudan I. Global birth prevalence and mortality from inborn errors of metabolism: a systematic analysis of the evidence. J. Glob. Health. 2018;8:021102. doi: 10.7189/jogh.08.021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Popejoy A.B., Crooks K.R., Fullerton S.M., Hindorff L.A., Hooker G.W., Koenig B.A., Pino N., Ramos E.M., Ritter D.I., Wand H. Clinical Genetics Lacks Standard Definitions and Protocols for the Collection and Use of Diversity Measures. Am. J. Hum. Genet. 2020;107:72–82. doi: 10.1016/j.ajhg.2020.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feuchtbaum L., Carter J., Dowray S., Currier R.J., Lorey F. Birth prevalence of disorders detectable through newborn screening by race/ethnicity. Genet. Med. 2012;14:937–945. doi: 10.1038/gim.2012.76. [DOI] [PubMed] [Google Scholar]
- 17.Azzopardi P.J., Upshur R.E.G., Luca S., Venkataramanan V., Potter B.K., Chakraborty P.K., Hayeems R.Z. Health-care providers’ perspectives on uncertainty generated by variant forms of newborn screening targets. Genet. Med. 2020;22:566–573. doi: 10.1038/s41436-019-0670-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Azimi M., Schmaus K., Greger V., Neitzel D., Rochelle R., Dinh T. Carrier screening by next-generation sequencing: health benefits and cost effectiveness. Mol. Genet. Genomic Med. 2016;4:292–302. doi: 10.1002/mgg3.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kraft S.A., Duenas D., Wilfond B.S., Goddard K.A.B. The evolving landscape of expanded carrier screening: challenges and opportunities. Genet. Med. 2019;21:790–797. doi: 10.1038/s41436-018-0273-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fowler D.M., Fields S. Deep mutational scanning: a new style of protein science. Nat. Methods. 2014;11:801–807. doi: 10.1038/nmeth.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duzkale H., Shen J., McLaughlin H., Alfares A., Kelly M.A., Pugh T.J., Funke B.H., Rehm H.L., Lebo M.S. A systematic approach to assessing the clinical significance of genetic variants. Clin. Genet. 2013;84:453–463. doi: 10.1111/cge.12257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodenburg R.J. The functional genomics laboratory: functional validation of genetic variants. J. Inherit. Metab. Dis. 2018;41:297–307. doi: 10.1007/s10545-018-0146-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suiter C.C., Moriyama T., Matreyek K.A., Yang W., Scaletti E.R., Nishii R., Yang W., Hoshitsuki K., Singh M., Trehan A. Massively parallel variant characterization identifies NUDT15 alleles associated with thiopurine toxicity. Proc. Natl. Acad. Sci. USA. 2020;117:5394–5401. doi: 10.1073/pnas.1915680117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oshiro C., Mangravite L., Klein T., Altman R. PharmGKB very important pharmacogene: SLCO1B1. Pharmacogenet. Genomics. 2010;20:211–216. doi: 10.1097/FPC.0b013e328333b99c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Faden R.R., Kass N.E., Goodman S.N., Pronovost P., Tunis S., Beauchamp T.L. An ethics framework for a learning health care system: a departure from traditional research ethics and clinical ethics. Hastings Cent. Rep. 2013;(Spec No):S16–S27. doi: 10.1002/hast.134. [DOI] [PubMed] [Google Scholar]
- 26.Larson D.B., Magnus D.C., Lungren M.P., Shah N.H., Langlotz C.P. Ethics of Using and Sharing Clinical Imaging Data for Artificial Intelligence: A Proposed Framework. Radiology. 2020;295:675–682. doi: 10.1148/radiol.2020192536. [DOI] [PubMed] [Google Scholar]
- 27.Johnston J., Lantos J.D., Goldenberg A., Chen F., Parens E., Koenig B.A., members of the NSIGHT Ethics and Policy Advisory Board Sequencing Newborns: A Call for Nuanced Use of Genomic Technologies. Hastings Cent. Rep. 2018;48(Suppl 2):S2–S6. doi: 10.1002/hast.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Couzin-Frankel J. Unknown significance. Science. 2014;346:1167–1170. doi: 10.1126/science.346.6214.1167. [DOI] [PubMed] [Google Scholar]
- 29.Vineis P. Ethical issues in genetic screening for cancer. Ann. Oncol. 1997;8:945–949. doi: 10.1023/a:1008296719733. [DOI] [PubMed] [Google Scholar]
- 30.Hippman C., Nislow C. Pharmacogenomic Testing: Clinical Evidence and Implementation Challenges. J. Pers. Med. 2019;9:40. doi: 10.3390/jpm9030040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCullough L.B., Brothers K.B., Chung W.K., Joffe S., Koenig B.A., Wilfond B., Yu J.-H., Clinical Sequencing Exploratory Research (CSER) Consortium Pediatrics Working Group Professionally Responsible Disclosure of Genomic Sequencing Results in Pediatric Practice. Pediatrics. 2015;136:e974–e982. doi: 10.1542/peds.2015-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Char D.S., Shah N.H., Magnus D. Implementing Machine Learning in Health Care - Addressing Ethical Challenges. N. Engl. J. Med. 2018;378:981–983. doi: 10.1056/NEJMp1714229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez-Martin N., Magnus D. Privacy and ethical challenges in next-generation sequencing. Expert Rev. Precis. Med. Drug Dev. 2019;4:95–104. doi: 10.1080/23808993.2019.1599685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu C.Y., Williams M.S., Ginsburg G.S., Toh S., Brown J.S., Khoury M.J. A proposed approach to accelerate evidence generation for genomic-based technologies in the context of a learning health system. Genet. Med. 2018;20:390–396. doi: 10.1038/gim.2017.122. [DOI] [PubMed] [Google Scholar]
- 35.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alföldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., Genome Aggregation Database Consortium The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rehm H.L., Berg J.S., Brooks L.D., Bustamante C.D., Evans J.P., Landrum M.J., Ledbetter D.H., Maglott D.R., Martin C.L., Nussbaum R.L., ClinGen ClinGen—the clinical genome resource. N. Engl. J. Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaedigk A., Ingelman-Sundberg M., Miller N.A., Leeder J.S., Whirl-Carrillo M., Klein T.E., PharmVar Steering Committee The Pharmacogene Variation (PharmVar) Consortium: Incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database. Clin. Pharmacol. Ther. 2018;103:399–401. doi: 10.1002/cpt.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaedigk A., Sangkuhl K., Whirl-Carrillo M., Twist G.P., Klein T.E., Miller N.A., PharmVar Steering Committee The Evolution of PharmVar. Clin. Pharmacol. Ther. 2019;105:29–32. doi: 10.1002/cpt.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whirl-Carrillo M., McDonagh E.M., Hebert J.M., Gong L., Sangkuhl K., Thorn C.F., Altman R.B., Klein T.E. Pharmacogenomics knowledge for personalized medicine. Clin. Pharmacol. Ther. 2012;92:414–417. doi: 10.1038/clpt.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47(D1):D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Velde K.J., Kuiper J., Thompson B.A., Plazzer J.-P., van Valkenhoef G., de Haan M., Jongbloed J.D.H., Wijmenga C., de Koning T.J., Abbott K.M., InSiGHT Group Evaluation of CADD Scores in Curated Mismatch Repair Gene Variants Yields a Model for Clinical Validation and Prioritization. Hum. Mutat. 2015;36:712–719. doi: 10.1002/humu.22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Q., Liu X., Gibbs R.A., Boerwinkle E., Polychronakos C., Qu H.-Q. Gene-specific function prediction for non-synonymous mutations in monogenic diabetes genes. PLoS ONE. 2014;9:e104452. doi: 10.1371/journal.pone.0104452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hamasaki-Katagiri N., Salari R., Wu A., Qi Y., Schiller T., Filiberto A.C., Schisterman E.F., Komar A.A., Przytycka T.M., Kimchi-Sarfaty C. A gene-specific method for predicting hemophilia-causing point mutations. J. Mol. Biol. 2013;425:4023–4033. doi: 10.1016/j.jmb.2013.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Y., Mkrtchian S., Kumondai M., Hiratsuka M., Lauschke V.M. An optimized prediction framework to assess the functional impact of pharmacogenetic variants. Pharmacogenomics J. 2019;19:115–126. doi: 10.1038/s41397-018-0044-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adhikari A.N. Gene-specific features enhance interpretation of mutational impact on acid α-glucosidase enzyme activity. Hum. Mutat. 2019;40:1507–1518. doi: 10.1002/humu.23846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lal D., May P., Perez-Palma E., Samocha K.E., Kosmicki J.A., Robinson E.B., Møller R.S., Krause R., Nürnberg P., Weckhuysen S. Gene family information facilitates variant interpretation and identification of disease-associated genes in neurodevelopmental disorders. Genome Med. 2020;12:28. doi: 10.1186/s13073-020-00725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heyne H.O., Baez-Nieto D., Iqbal S., Palmer D.S., Brunklaus A., May P., Johannesen K.M., Lauxmann S., Lemke J.R., Møller R.S. Predicting functional effects of missense variants in voltage-gated sodium and calcium channels. Sci. Transl. Med. 2020;12:eaay6848. doi: 10.1126/scitranslmed.aay6848. [DOI] [PubMed] [Google Scholar]
- 51.Clerx M., Heijman J., Collins P., Volders P.G.A. Predicting changes to INa from missense mutations in human SCN5A. Sci. Rep. 2018;8:12797. doi: 10.1038/s41598-018-30577-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li B., Mendenhall J.L., Kroncke B.M., Taylor K.C., Huang H., Smith D.K., Vanoye C.G., Blume J.D., George A.L., Jr., Sanders C.R., Meiler J. Predicting the Functional Impact of KCNQ1 Variants of Unknown Significance. Circ Cardiovasc Genet. 2017;10:e001754. doi: 10.1161/CIRCGENETICS.117.001754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jorge L.F., Eichelbaum M., Griese E.U., Inaba T., Arias T.D. Comparative evolutionary pharmacogenetics of CYP2D6 in Ngawbe and Embera Amerindians of Panama and Colombia: role of selection versus drift in world populations. Pharmacogenetics. 1999;9:217–228. [PubMed] [Google Scholar]
- 54.Popejoy A.B., Fullerton S.M. Genomics is failing on diversity. Nature. 2016;538:161–164. doi: 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Obermeyer Z., Powers B., Vogeli C., Mullainathan S. Dissecting racial bias in an algorithm used to manage the health of populations. Science. 2019;366:447–453. doi: 10.1126/science.aax2342. [DOI] [PubMed] [Google Scholar]
- 56.Duncan L., Shen H., Gelaye B., Meijsen J., Ressler K., Feldman M., Peterson R., Domingue B. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun. 2019;10:3328. doi: 10.1038/s41467-019-11112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin A.R., Gignoux C.R., Walters R.K., Wojcik G.L., Neale B.M., Gravel S., Daly M.J., Bustamante C.D., Kenny E.E. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am. J. Hum. Genet. 2017;100:635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chakchouk I., Zhang D., Zhang Z., Francioli L.C., Santos-Cortez R.L.P., Schrauwen I., Leal S.M. Disparities in discovery of pathogenic variants for autosomal recessive non-syndromic hearing impairment by ancestry. Eur. J. Hum. Genet. 2019;27:1456–1465. doi: 10.1038/s41431-019-0417-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perera M.A., Cavallari L.H., Johnson J.A. Warfarin pharmacogenetics: an illustration of the importance of studies in minority populations. Clin. Pharmacol. Ther. 2014;95:242–244. doi: 10.1038/clpt.2013.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amendola C.P., Silva-Jr J.M., Carvalho T., Sanches L.C., Silva U.V.A.E., Almeida R., Burdmann E., Lima E., Barbosa F.F., Ferreira R.S. Goal-directed therapy in patients with early acute kidney injury: a multicenter randomized controlled trial. Clinics (São Paulo) 2018;73:e327. doi: 10.6061/clinics/2018/e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rahit K.M.T.H., Tarailo-Graovac M. Genetic Modifiers and Rare Mendelian Disease. Genes (Basel) 2020;11:239. doi: 10.3390/genes11030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hunter D.J. Gene-environment interactions in human diseases. Nat. Rev. Genet. 2005;6:287–298. doi: 10.1038/nrg1578. [DOI] [PubMed] [Google Scholar]
- 64.Blau N., Shen N., Carducci C. Molecular genetics and diagnosis of phenylketonuria: state of the art. Expert Rev. Mol. Diagn. 2014;14:655–671. doi: 10.1586/14737159.2014.923760. [DOI] [PubMed] [Google Scholar]
- 65.Hinton G., Deng L., Yu D., Dahl G., Mohamed A.-R., Jaitly N., Senior A., Vanhoucke V., Nguyen P., Kingsbury B. Deep neural networks for acoustic modeling in speech recognition. IEEE Signal Process. Mag. 2012;29:82–97. [Google Scholar]
- 66.Kiros R., Zhu Y., Salakhutdinov R.R., Zemel R., Urtasun R., Torralba A., Fidler S. Skip-Thought Vectors. In: Cortes C., Lawrence N.D., Lee D.D., Sugiyama M., Garnett R., editors. Advances in Neural Information Processing Systems 28. Curran Associates, Inc.; 2015. pp. 3294–3302. [Google Scholar]
- 67.Collobert R., Weston J. Proceedings of the 25th International Conference on Machine Learning. New York, NY, USA: ACM; 2008. A Unified Architecture for Natural Language Processing: Deep Neural Networks with Multitask Learning; pp. 160–167. [Google Scholar]
- 68.Ching T., Himmelstein D.S., Beaulieu-Jones B.K., Kalinin A.A., Do B.T., Way G.P., Ferrero E., Agapow P.-M., Zietz M., Hoffman M.M. Opportunities and obstacles for deep learning in biology and medicine. J. R. Soc. Interface. 2018;15:20170387. doi: 10.1098/rsif.2017.0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Min S., Lee B., Yoon S. Deep learning in bioinformatics. Brief. Bioinform. 2017;18:851–869. doi: 10.1093/bib/bbw068. [DOI] [PubMed] [Google Scholar]
- 70.Zou J., Huss M., Abid A., Mohammadi P., Torkamani A., Telenti A. A primer on deep learning in genomics. Nat. Genet. 2019;51:12–18. doi: 10.1038/s41588-018-0295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Libbrecht M.W., Noble W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015;16:321–332. doi: 10.1038/nrg3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yue T., Wang H. Deep Learning for Genomics: A Concise Overview. arXiv. 2018 https://arxiv.org/abs/1802.00810 1802.00810. [Google Scholar]
- 73.Angermueller C., Pärnamaa T., Parts L., Stegle O. Deep learning for computational biology. Mol. Syst. Biol. 2016;12:878. doi: 10.15252/msb.20156651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ma J., Yu M.K., Fong S., Ono K., Sage E., Demchak B., Sharan R., Ideker T. Using deep learning to model the hierarchical structure and function of a cell. Nat. Methods. 2018;15:290–298. doi: 10.1038/nmeth.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kelley D.R., Snoek J., Rinn J.L. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 2016;26:990–999. doi: 10.1101/gr.200535.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shen Z., Bao W., Huang D.-S. Recurrent Neural Network for Predicting Transcription Factor Binding Sites. Sci. Rep. 2018;8:15270. doi: 10.1038/s41598-018-33321-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Khodabandelou G., Mozziconacci J., Routhier E. Genome functional annotation using deep convolutional neural network. bioRxiv. 2018 doi: 10.1101/330308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Quang D., Chen Y., Xie X. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. 2015;31:761–763. doi: 10.1093/bioinformatics/btu703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou J., Troyanskaya O.G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods. 2015;12:931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.McInnes G., Dalton R., Sangkuhl K., Whirl-Carrillo M., Lee S.-B., Tsao P.S., Gaedigk A., Altman R.B., Woodahl E.L. Transfer learning enables prediction of CYP2D6 haplotype function. PLoS Comput. Biol. 2020;16:e1008399. doi: 10.1371/journal.pcbi.1008399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van der Lee M., Allard W.G., Rolf H.A., Baak-Pablo R.F., Menafra R., Birgit A.L., Deenen M.J., Neven P., Johansson I., Gastaldello S. A unifying model to predict variable drug response for personalised medicine. bioRxiv. 2020 doi: 10.1101/2020.03.02.967554. [DOI] [Google Scholar]
- 82.Erhan D., Bengio Y., Courville A., Manzagol P.-A., Vincent P., Bengio S. Why Does Unsupervised Pre-training Help Deep Learning? J. Mach. Learn. Res. 2010;11:625–660. [Google Scholar]
- 83.Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014;15:1929–1958. [Google Scholar]
- 84.Pan S.J., Yang Q. A Survey on Transfer Learning. IEEE Trans. Knowl. Data Eng. 2010;22:1345–1359. [Google Scholar]
- 85.Shao L., Zhu F., Li X. Transfer learning for visual categorization: a survey. IEEE Trans. Neural Netw. Learn. Syst. 2015;26:1019–1034. doi: 10.1109/TNNLS.2014.2330900. [DOI] [PubMed] [Google Scholar]
- 86.Weiss K., Khoshgoftaar T.M., Wang D. A survey of transfer learning. J. Big Data. 2016;3:9. [Google Scholar]
- 87.Zamir A.R., Sax A., Shen W., Guibas L.J., Malik J., Savarese S. Taskonomy: Disentangling task transfer learning. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition. 2018:3712–3722. [Google Scholar]
- 88.Yosinski J., Clune J., Bengio Y., Lipson H. How transferable are features in deep neural networks? In: Ghahramani Z., Welling M., Cortes C., Lawrence N.D., Weinberger K.Q., editors. Advances in Neural Information Processing Systems 27. Curran Associates, Inc.; 2014. pp. 3320–3328. [Google Scholar]
- 89.Taroni J.N., Grayson P.C., Hu Q., Eddy S., Kretzler M., Merkel P.A., Greene C.S. MultiPLIER: A Transfer Learning Framework for Transcriptomics Reveals Systemic Features of Rare Disease. Cell Syst. 2019;8:380–394.e4. doi: 10.1016/j.cels.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McInnes G., Dalton R., Sangkuhl K., Whirl-Carrillo M., Lee S.-B., Tsao P.S., Gaedigk A., Altman R.B., Woodahl E.L. Transfer learning enables prediction of CYP2D6 haplotype function. bioRxiv. 2020 doi: 10.1101/684357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rao R., Bhattacharya N., Thomas N., Duan Y., Chen P., Canny J., Abbeel P., Song Y. Evaluating Protein Transfer Learning with TAPE. In: Wallach H., Larochelle H., Beygelzimer A., d’Alché-Buc F., Fox E., Garnett R., editors. Advances in Neural Information Processing Systems 32. Curran Associates, Inc.; 2019. pp. 9689–9701. [PMC free article] [PubMed] [Google Scholar]
- 92.Elnaggar A., Heinzinger M., Dallago C., Rehawi G., Wang Y., Jones L., Gibbs T., Feher T., Angerer C., Steinegger M. ProtTrans: Towards Cracking the Language of Life’s Code Through Self-Supervised Deep Learning and High Performance Computing. arXiv. 2020 doi: 10.1109/TPAMI.2021.3095381. https://arxiv.org/abs/2007.06225 2007.06225. [DOI] [PubMed] [Google Scholar]
- 93.Torng W., Altman R.B. Graph Convolutional Neural Networks for Predicting Drug-Target Interactions. J. Chem. Inf. Model. 2019;59:4131–4149. doi: 10.1021/acs.jcim.9b00628. [DOI] [PubMed] [Google Scholar]
- 94.Guise J.-M., Savitz L.A., Friedman C.P. Mind the Gap: Putting Evidence into Practice in the Era of Learning Health Systems. J. Gen. Intern. Med. 2018;33:2237–2239. doi: 10.1007/s11606-018-4633-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Etheredge L.M. A rapid-learning health system: what would a rapid-learning health system look like, and how might we get there? Health Aff. (Millwood) 2007;26:w107–w118. doi: 10.1377/hlthaff.26.2.w107. [DOI] [PubMed] [Google Scholar]
- 96.Greene S.M., Reid R.J., Larson E.B. Implementing the learning health system: from concept to action. Ann. Intern. Med. 2012;157:207–210. doi: 10.7326/0003-4819-157-3-201208070-00012. [DOI] [PubMed] [Google Scholar]
- 97.Leuders S., Wolfgart E., Ott T., du Moulin M., van Teeffelen-Heithoff A., Vogelpohl L., Och U., Marquardt T., Weglage J., Feldmann R., Rutsch F. Influence of PAH Genotype on Sapropterin Response in PKU: Results of a Single-Center Cohort Study. JIMD Rep. 2014;13:101–109. doi: 10.1007/8904_2013_263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yıldız Y., Talim B., Haliloglu G., Topaloglu H., Akçören Z., Dursun A., Sivri H.S., Coşkun T., Tokatlı A. Determinants of Riboflavin Responsiveness in Multiple Acyl-CoA Dehydrogenase Deficiency. Pediatr. Neurol. 2019;99:69–75. doi: 10.1016/j.pediatrneurol.2019.06.015. [DOI] [PubMed] [Google Scholar]
- 99.Grünert S.C. Clinical and genetical heterogeneity of late-onset multiple acyl-coenzyme A dehydrogenase deficiency. Orphanet J. Rare Dis. 2014;9:117. doi: 10.1186/s13023-014-0117-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lamm V. People Drawn Thin Collection, Person. From thenounproject.com.
- 101.Alberto Gongora, H. Big Data Collection, Machine Learning. From thenounproject.com.
- 102.Wray A. Gene Testing. From thenounproject.com.
- 103.ProSymbols, U.S. STEM Elements Line Icons Collections, DNA.
- 104.Rieke N., Hancox J., Li W., Milletarì F., Roth H.R., Albarqouni S., Bakas S., Galtier M.N., Landman B.A., Maier-Hein K. The future of digital health with federated learning. NPJ Digit Med. 2020;3:119. doi: 10.1038/s41746-020-00323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rajkomar A., Hardt M., Howell M.D., Corrado G., Chin M.H. Ensuring Fairness in Machine Learning to Advance Health Equity. Ann. Intern. Med. 2018;169:866–872. doi: 10.7326/M18-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.O’Doherty K.C., Christofides E., Yen J., Bentzen H.B., Burke W., Hallowell N., Koenig B.A., Willison D.J. If you build it, they will come: unintended future uses of organised health data collections. BMC Med. Ethics. 2016;17:54. doi: 10.1186/s12910-016-0137-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Green A.R., Tan-McGrory A., Cervantes M.C., Betancourt J.R. Leveraging quality improvement to achieve equity in health care. Jt. Comm. J. Qual. Patient Saf. 2010;36:435–442. doi: 10.1016/s1553-7250(10)36065-x. [DOI] [PubMed] [Google Scholar]
- 108.Shringarpure S.S., Bustamante C.D. Privacy Risks from Genomic Data-Sharing Beacons. Am. J. Hum. Genet. 2015;97:631–646. doi: 10.1016/j.ajhg.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This publication did not generate datasets or code.