Abstract
Molecular biomarkers play a key role in the clinic, aiding in diagnostics and prognostics, and in the research laboratory, contributing to our basic understanding of diseases. Detecting multiple and diverse molecular biomarkers within a single accessible assay would have great utility, providing a more comprehensive picture for clinical evaluation and research, but is a challenge with standard methods. Here, we report programmable DNA nanoswitches for multiplexed detection of up to 6 biomarkers at once, with each combination of biomarkers producing a unique barcode signature among 64 possibilities. As a defining feature of our method, we show “mixed multiplexing” for simultaneous barcoded detection of different types of biomolecules – DNA, RNA, antibody and protein in a single assay. To demonstrate clinical potential, we show multiplexed detection of a prostate cancer biomarker panel in serum that includes two microRNA sequences and prostate specific antigen (PSA).
Keywords: DNA nanoswitches, DNA barcodes, biosensing, diagnostics, genotyping, multiplexed detection
Graphical Abstract
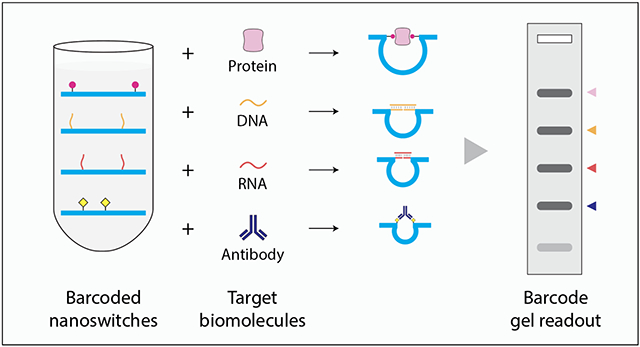
Barcodes are ubiquitous in our daily lives, as a way to reduce complex information to a simple pattern. They have also found applications in biosensing, where the study of multiple biological markers can provide useful information for understanding cellular processes as well as disease progression. Some examples of biological barcodes include hydrogel-encapsulated photonic crystals for detection of cardiovascular biomarkers,1 DNA-antibody conjugates for multiplexed protein analysis,2 duplex DNA barcodes for cell sorting,3 DNA-nanoparticle conjugates for nucleic acid4 and protein detection5 and DNA-based barcodes for nucleic acid analysis.6,7 Barcoded architectures can also enable multiplexed detection, where several biomarkers are detected in parallel in a single pot. Such strategies have used conjugated polymers,8 photonic crystals,9 carbon nanotubes,10 semiconductor quantum dots,11,12 DNA-templated silver nanoclusters,13 gold nanoparticles14 as well as DNA nanostructures.15,16 DNA nanostructures in particular are promising for molecular barcodes, as they can be designed to reconfigure in the presence of molecular biomarkers such as proteins, antibodies and nucleic acids. Here, we developed reconfigurable DNA nanoswitches that can be combined to provide barcoded detection and analysis of multiple biomarkers in a single one pot assay (Figure 1). Such a system can gather information from multiple types of biomarkers to create a single barcode that can more accurately diagnose a disease, as compared to using only a single biomarker. Studies have already shown that detecting a panel of disease biomarkers is more accurate in diagnosing specific diseases compared to individual biomarkers. For example, biomarker panels that include both the protein biomarker prostate specific antigen (PSA) and microRNAs can outperform diagnosis by PSA testing alone17,18 and profiling of both SARS-CoV-2 viral antigens and the antibody response in the blood could be useful in tracking and predicting disease progression, such as respiratory failure, in severe COVID-19 cases.19
Figure 1. DNA nanoswitch barcodes.
Schematic illustration showing construction and use of DNA nanoswitches to yield barcoded results for diagnostic assays.
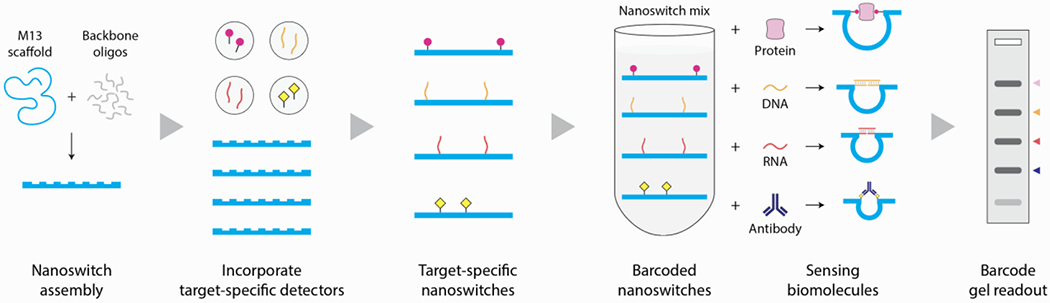
Programmable DNA nanoswitches are assembled from a long single-stranded scaffold (viral genome M13 routinely used for DNA origami) and short complementary backbone oligonucleotides.20 Pairs of backbone oligonucleotides can be modified to contain single stranded extensions (detectors) that are complementary to parts of a target nucleic acid (Figure 2a and S1). On binding the target sequence, the nanoswitch changes conformation from a linear “off” state to a looped “on” state, providing a distinct signal on an agarose gel (Figure 2a, inset). Importantly, this approach requires no complex equipment or enzymatic amplification. The signal comes from the intercalation of thousands of dye molecules from regularly used DNA gel stains (GelRed in this case). We previously used similar DNA nanoswitches for single molecule experiments,21 detection of microRNAs,22 viral RNAs,23 antigens24 and enzymes,25 as well as in molecular memory.16,26 Here, we expand the use of nanoswitches to a multiplexed DNA barcode system that can be used to detect any combination of up to 6 different biomarkers. We further show for the first time that a single barcode can be used to identify different types of biomarkers including proteins, antibodies, DNA and RNA, with clinical potential shown by detecting a prostate cancer biomarker panel in serum that includes two microRNA sequences and prostate specific antigen (PSA).
Figure 2. DNA nanoswitch overview and characterization.
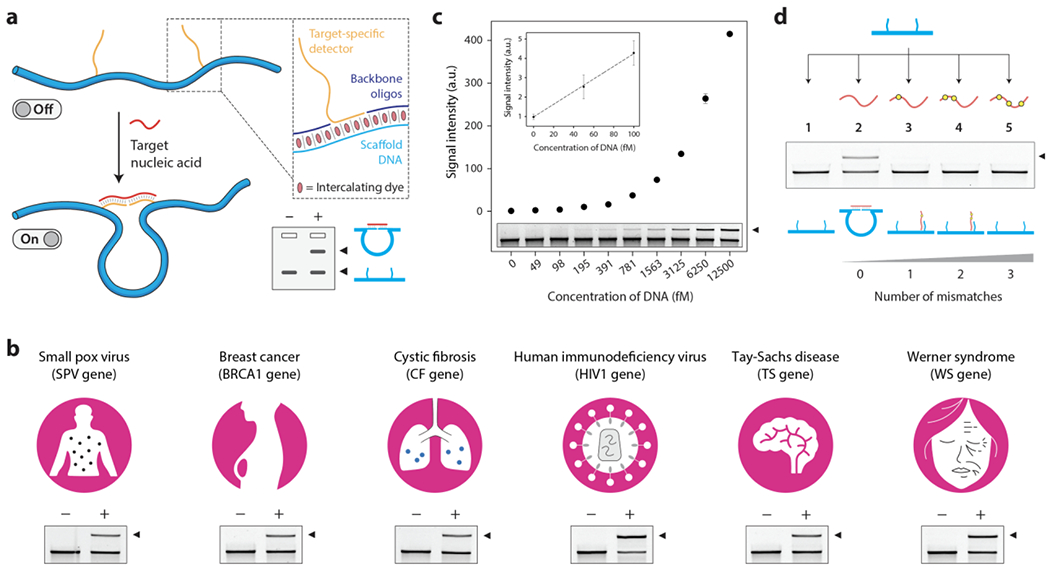
(a) Reconfiguration of the DNA nanoswitch in the presence of a nucleic acid target turning from a linear “off” state to a looped “on” state. The two states of the nanoswitch can be readout on an agarose gel (inset). (b) Demonstration of detecting different gene fragments using the DNA nanoswitch assay. (c) Sensitivity analysis of DNA nanoswitch assay for the cystic fibrosis (CF) gene fragment. (d) Specificity of DNA nanoswitches against CF gene fragments containing one to three mismatches.
To demonstrate the concept of DNA nanoswitch barcodes, we chose 6 different gene fragments corresponding to the smallpox virus gene (SP), cystic fibrosis gene (CF), Tay-Sachs disease gene (TS), breast cancer gene (BRCA1), human immunodeficiency virus gene (HIV1) and Werner syndrome gene (WS). We designed nanoswitches specific to these gene fragments and showed successful detection of a single stranded DNA oligonucleotide corresponding to the gene in each case (Figure 2b and S2).
For further characterization of DNA nanoswitch detection, we chose the cystic fibrosis gene fragment. In diagnostics, sensitivity is a key parameter to detect early onset of biological or disease processes. We performed sensitivity experiments with decreasing concentrations of the DNA and found that the signal could be seen by eye at concentrations as low as 50 fM (Figure 2c and S3). Calculating the limit of detection (LOD), defined as the concentration of biomarker that yields a signal that exceeds the mean background by 3 SDs of the background, we obtained a value of ~12 fM, similar to what we previously reported for protein detection.24 We then tested specificity of the assay by challenging the CF nanoswitch with CF gene targets that contained 1-3 mutations. Results showed that for a single nucleotide mismatch, there was a 40% reduction in signal compared to the fully complementary target (Figure S4). In this assay, the nanoswitch detectors were designed to recognize the complete length of the target sequence (24-nucleotide target hybridized to two 12-nucleotide detectors). To optimize the specificity of the assay, we redesigned the nanoswitch by decreasing the length of the detector complementary to the side of the target that contains the mismatch. Using this design, we showed that the assay is highly specific, able to discriminate even a single nucleotide mismatch in the target sequence (Figure 2d and Figure S4). Next, we performed a time series experiment, showing that the assay can be performed in under an hour in most cases (Figure S5). Recognizing that gene fragments in real biological samples would be present as double stranded DNA rather than single stranded DNA, we also showed that with a heating step we could independently detect either strand in a 24 bp DNA duplex, and also detect the same CF gene target sequence in a 125 bp double stranded DNA (Figure S6).
To demonstrate barcoded detection, we used the programmability of the nanoswitch to place the detector strands along specific positions on the scaffold (Figure 3a). Nanoswitches with detectors spaced far apart will yield a longer loop on binding the target while a shorter loop will be formed when the detectors are closer together. This allows the creation of specific nanoswitches that yield different bands on the gel according to the resulting loop size (Figure 3b). For convenient construction, we designed 12 variable regions (V1-V12 in Figure 3a) for placement of detectors. We designed six nanoswitches with different separations of the two detectors along the scaffold. Each of these nanoswitches contained detectors specific to one of the six gene targets described in Figure 2b. We then prepared a mixture of these nanoswitches that can detect all six targets simultaneously: each target will trigger the specific nanoswitch and form the corresponding loop, providing a unique signature on a gel, i.e. a barcode. The separation of the detectors was chosen in a way that the six looped nanoswitches can be easily resolved on a single gel lane (Figure 3b, inset). Before we tested simultaneous detection of the gene fragments, we confirmed that each individual nanoswitch does not have any cross-reactivity with non-specific targets (Figure 3c). Next, we used the nanoswitch mixture and demonstrated detection of all possible combinations of the six gene targets (a total of 64), each providing a unique barcode (Figure 3d and S7). This strategy provides a simple multiplexed assay to detect several nucleic acids in a single assay, with each recognition event translated into a unique readout. Migration of these nanoswitches in the gel is dependent primarily on the size and location of the loop, rather than on the molecular weight of the target strand (since the target strand is only 20-30 nucleotides compared to the ~7 kbp nanoswitch). Thus, the barcodes generated by the different targets is constant regardless of the target of interest, opening up the possibility of detecting different types of biomarkers simultaneously and not just nucleic acids.
Figure 3. DNA nanoswitch barcodes for multiplexed detection of nucleic acids.
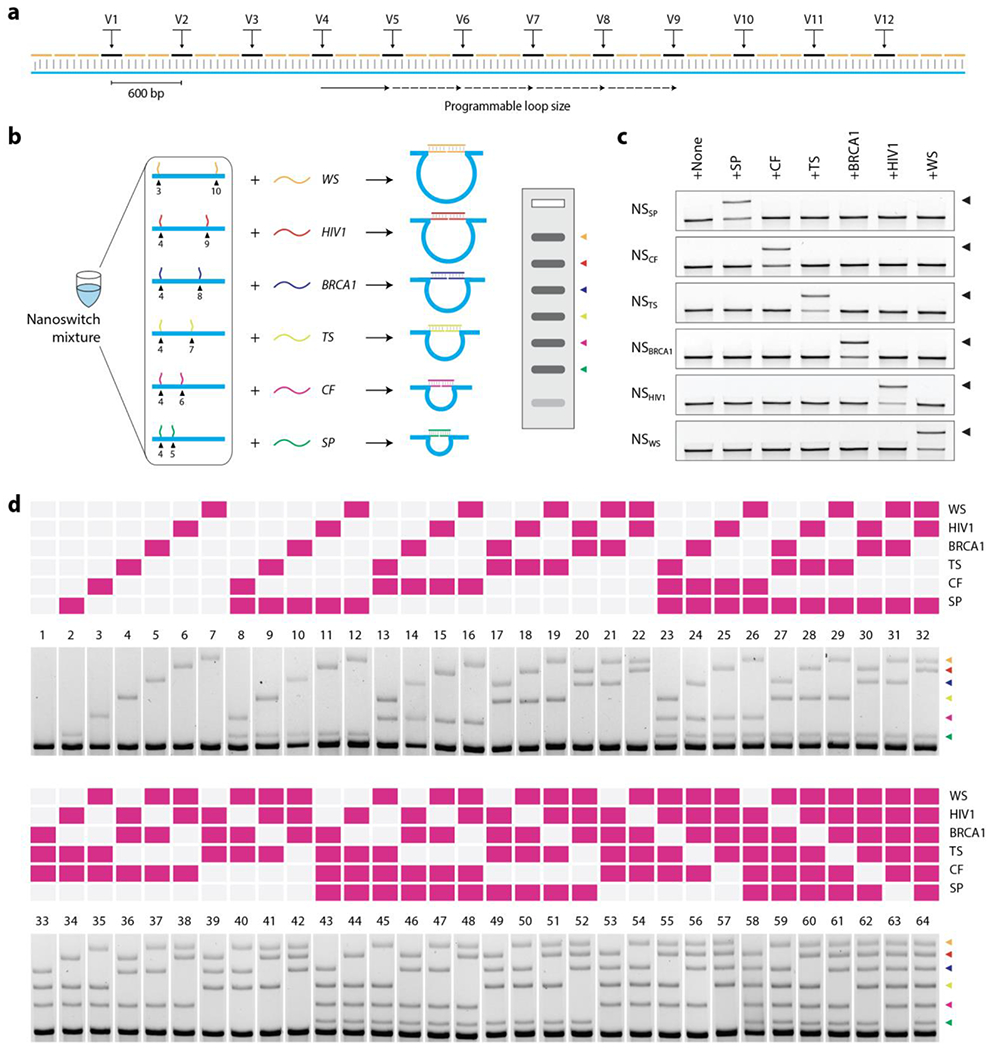
(a) Design of the DNA nanoswitch with variable regions (V1-V12) that allow programmable placement of detectors, resulting in nanoswitches with different loop sizes. (b) A mixture of nanoswitches with different loop sizes yields a multiplexed gel readout of up to 6 gene fragments (inset). Each target nucleic acid triggers the formation of a specific loop, providing a unique signal (targets are colored for clarity). (c) Demonstration of lack of cross-reactivity in nanoswitches designed for 6 different gene fragments. (d) Full set of DNA nanoswitch barcodes showing detection of all possible combinations of the 6 gene fragments.
We then extended the nanoswitch barcode design for different types of targets including proteins, antibodies, DNA and RNA. For proof-of-concept protein detection (streptavidin), we designed a nanoswitch where the detector strands were modified to contain a biotin group instead of single stranded extensions. Similarly, for detecting an antibody (anti-digoxygenin), we incorporated digoxygenin-coupled detectors in the nanoswitch (Figure 4a-b). We designed two more nanoswitches that can detect specific DNA and RNA sequences. We also designed the four nanoswitches to yield different loop sizes so that a nanoswitch mixture can provide barcoded recognition for all four targets in a single assay (Figure 4c). Again, we first confirmed that there was no cross-reactivity between these nanoswitches when tested against non-specific targets (Figure 4d). We then showed all possible detection events (16 in total for 4 targets) using this barcode (Figure 4e and S8), demonstrating that our approach can simultaneously detect nucleic acids, proteins and antibodies in a single one-pot assay. Further, to show that the individual biomarkers can be quantified, we performed a “multiplexed non-interference” analysis by changing the concentration of one biomarker while keeping the others constant. We show that the concentration series for each of the four biomarkers (anti-digoxygenin antibody, DNA, RNA and streptavidin) can be monitored in the presence of other targets (Figure 4f and S9). These results are consistent with those where the targets are present alone in a reaction when detected using the same nanoswitch mix (Figure S10).
Figure 4. Detecting multiple types of biomarkers using DNA nanoswitch barcodes.
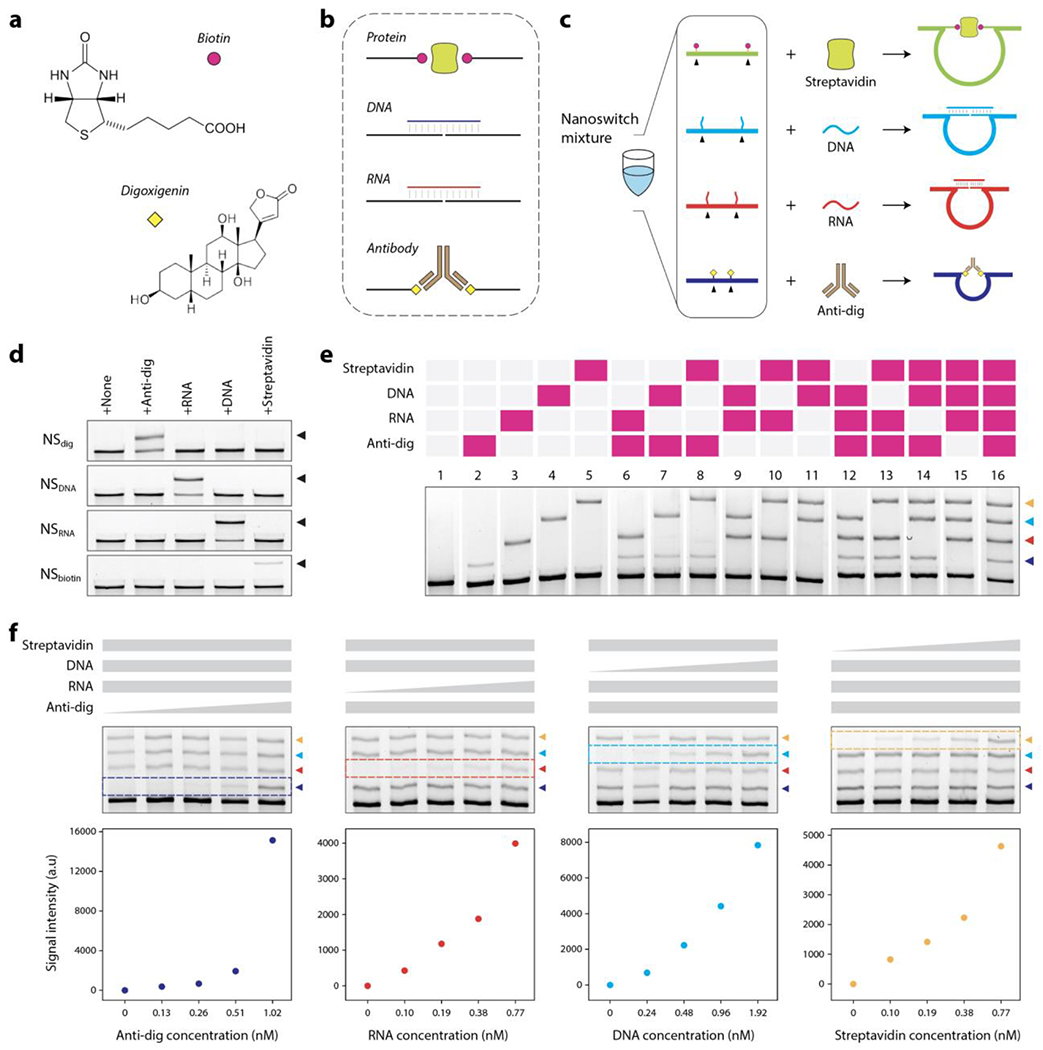
(a) Using small molecules such as biotin and digoxygenin to target macromolecules. (b) Strategy to detect nucleic acids (DNA and RNA), a protein (streptavidin) and an antibody (anti-digoxygenin). (c) A nanoswitch mixture to demonstrate simultaneous detection of four different biomarkers. (d) Demonstration of lack of crosstalk in nanoswitches designed for four different target biomolecules. (e) Full set of DNA nanoswitch barcodes showing detection of all possible combinations of anti-digoxygenin (anti-dig), an RNA sequence, a DNA sequence and streptavidin. (f) Individual targets can be analyzed even in the presence of other target biomolecules, showing a “multiplexed non-interference” in the barcoded assay.
Toward clinical relevance of the diagnostic barcodes, we then chose to detect a biomarker panel for prostate cancer (Figure 5). Detecting a panel of disease biomarkers rather than an individual biomarker can provide additional information for more accurate diagnoses.18 For example, data suggests that biomarker panels that include PSA, miR-141 and miR-30c can outperform diagnosis by PSA testing alone.17 In the multiplexed barcode demonstration above, we showed the detection of proteins and antibodies using small molecule ligands. For detecting PSA, we modified the nanoswitch detectors to contain PSA-specific antibodies. This strategy of using nanoswitches to detect antigens (which we called NLISA, nanoswitch-linked immunosorbent assay24) is similar to sandwich ELISA where a pair of antibodies are used to detect proteins of interest. In addition to PSA, we included DNA analogs of the microRNA sequences miR-30c and miR-141 based on literature demonstrating the utility of microRNA biomarkers in blood for detection of prostate cancer.17,18 To detect this panel, we designed three nanoswitches with different loop sizes and created a multiplexed nanoswitch mixture and tested detection of all biomarkers in buffer (Figure 5c). We then spiked the biomarkers in 20% serum and showed detection of individual biomarkers (Figure 5c, lanes 2-4) or simultaneous detection of all three biomarkers in a single gel lane (lane 5).
Figure 5. DNA nanoswitch barcodes for prostate cancer biomarker panel.
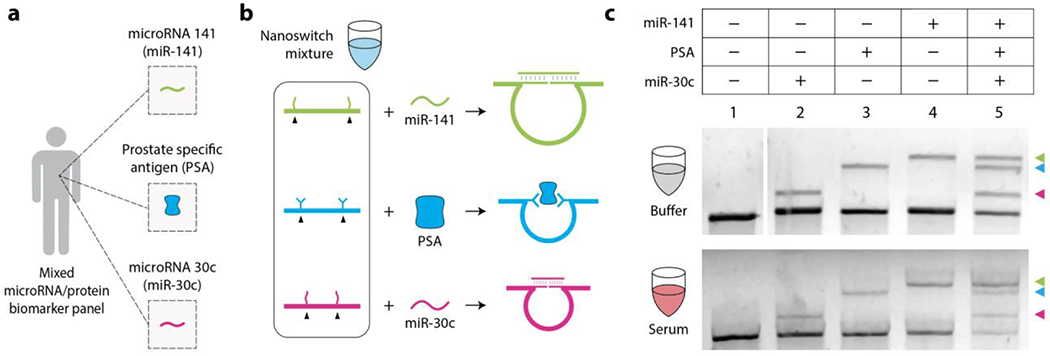
(a) An illustration of the three chosen prostate cancer biomarkers (miR-141, miR-30c and PSA). (b) DNA nanoswitch mixture for detecting prostate cancer biomarker panel. (c) Barcode detection of individual biomarkers (DNA analogs of microRNAs 141 and 30c, and PSA) and simultaneous detection of all three biomarkers in buffer and 20% fetal bovine serum (FBS).
To extend the assay further toward clinical utility, we demonstrated multiplexed detection of PSA and miR-141 at 100 pM and 300 fM, respectively, more closely mimicking clinical levels (Figure S11). While the clinical threshold for PSA is well-established at ~200 pM,27 absolute quantification of microRNA levels is less established. Representative moderate- to low-abundance microRNAs exist in human plasma with copy numbers corresponding to tens to hundreds of fM, while highly expressed microRNAs are in the 1-10 pM range.28
In previous works we have made detailed comparisons of nanoswitch-based detection of microRNAs22 and proteins24 with more established techniques like qPCR and ELISA, respectively. For microRNA, our analytical sensitivity outperforms Northern blotting and microarray, but not qPCR which can in principle detect single copies. Our analytical specificity is as good or better than other techniques, reaching 1 nucleotide, which in this context could be useful in detecting single nucleotide polymorphisms (SNPs).29 For proteins, the nanoswitch assay can outperform commercially available rapid sandwich ELISA assays in both analytical sensitivity and specificity, each by about an order of magnitude.24 Outside of performance metrics, the nanoswitch assay really shines in its simplicity and minimalism. Once the nanoswitches are made, the reaction is a simple mix step followed by a gel readout. For both proteins and microRNAs the whole end-to-end process has been demonstrated in less than 1 hour, but is more typically performed in a few hours. Unlike many other methods, the nanoswitch assay has no wash steps, no enzymes, and no expensive equipment requirements. The nanoswitches can be dried and stored for later use, and are stable after drying and retain their functionality to provide detection barcodes (Figure S12). Conveniently, our strategy uses gel electrophoresis for readout, which can be conducted outside of a laboratory setting, using, for example, an electronic buffer-less gel system (ThermoFisher).
Here we have shown that DNA nanoswitch barcodes can be used to detect up to six biomarkers in a single assay, and even a mixture of protein, antibody and nucleic acids in a single pot with a common consolidated workflow. By optimizing the placement of the detector strands or affinity reagents, multiplexing of the assay could be expanded to more biomarkers, largely limited by the resolution of the gel. These multiplexed assays retain both the features and performance of the traditional nanoswitch assays. As with the singleplex assays, they can detect protein and nucleic acid biomarkers at biologically relevant concentrations, and the assay is compatible with biological fluids such as serum. The barcoded assay enables all-at-once detection of biomarker panels, potentially reducing the number of steps and consequentially the cost, time, effort, and opportunity for error. Furthermore, our method provides direct detection without amplification, which makes absolute quantification more straightforward. The multiplexing also allows flexibility to include built in controls or references. Further development could also allow for more diverse types of readouts for nanoswitch barcodes, including nanopore30 or microfluidic chip31 based readouts or integration into sensor arrays to enable macroscopic graphical readouts.32
Supplementary Material
ACKNOWLEDGEMENT
Research reported in this publication was supported by the NIH through NIGMS under award R35GM124720 to K.H. and NCI under award R21 CA212827 to W.P.W. and K.H. Additional funding was provided by the Boston Children’s Hospital Technology Development Fund (W.P.W.)
Footnotes
SUPPORTING INFORMATION
Supporting information can be found online. Detailed experimental methods, additional results, full sequences used in the study.
CONFLICT OF INTEREST
The authors declare the following competing financial interest(s): A.R.C., C.H.H., D.Y., W.P.W., and K.H. are inventors on patent applications covering aspects of this work.
REFERENCES
- (1).Ji J; Lu W; Zhu Y; Jin H; Yao Y; Zhang H; Zhao Y Porous Hydrogel-Encapsulated Photonic Barcodes for Multiplex Detection of Cardiovascular Biomarkers. ACS Sens . 2019, 4 (5), 1384–1390. 10.1021/acssensors.9b00352. [DOI] [PubMed] [Google Scholar]
- (2).Agasti SS; Liong M; Peterson VM; Lee H; Weissleder R Photocleavable DNA Barcode–Antibody Conjugates Allow Sensitive and Multiplexed Protein Analysis in Single Cells. J. Am. Chem. Soc . 2012, 134 (45), 18499–18502. 10.1021/ja307689w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Gentile SD; Griebel ME; Anderson EW; Underhill GH Click Chemistry-Based DNA Labeling of Cells for Barcoding Applications. Bioconjugate Chem . 2018, 29 (8), 2846–2854. 10.1021/acs.bioconjchem.8b00435. [DOI] [PubMed] [Google Scholar]
- (4).Cui H-F; Xu T-B; Sun Y-L; Zhou A-W; Cui Y-H; Liu W; Luong JHT Hairpin DNA as a Biobarcode Modified on Gold Nanoparticles for Electrochemical DNA Detection. Anal. Chem . 2015, 87 (2), 1358–1365. 10.1021/ac504206n. [DOI] [PubMed] [Google Scholar]
- (5).Stoeva SI; Lee J-S; Smith JE; Rosen ST; Mirkin CA Multiplexed Detection of Protein Cancer Markers with Biobarcoded Nanoparticle Probes. J. Am. Chem. Soc . 2006, 128 (26), 8378–8379. 10.1021/ja0613106. [DOI] [PubMed] [Google Scholar]
- (6).Li Y; Cu YTH; Luo D Multiplexed Detection of Pathogen DNA with DNA-Based Fluorescence Nanobarcodes. Nature Biotechnology 2005, 23 (7), 885–889. 10.1038/nbt1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Zhou Z; Luo G; Wulf V; Willner I Application of DNA Machineries for the Barcode Patterned Detection of Genes or Proteins. Anal. Chem . 2018, 90 (11), 6468–6476. 10.1021/acs.analchem.7b04916. [DOI] [PubMed] [Google Scholar]
- (8).Zheng W; He L Label-Free, Real-Time Multiplexed DNA Detection Using Fluorescent Conjugated Polymers. J. Am. Chem. Soc . 2009, 131 (10), 3432–3433. 10.1021/ja809175q. [DOI] [PubMed] [Google Scholar]
- (9).Meade SO; Chen MY; Sailor MJ; Miskelly GM Multiplexed DNA Detection Using Spectrally Encoded Porous SiO2 Photonic Crystal Particles. Anal. Chem . 2009, 81 (7), 2618–2625. 10.1021/ac802538x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Chen J; Huang Y; Shi M; Zhao S; Zhao Y Highly Sensitive Multiplexed DNA Detection Using Multi-Walled Carbon Nanotube-Based Multicolor Nanobeacon. Talanta 2013, 109, 160–166. 10.1016/j.talanta.2013.02.003. [DOI] [PubMed] [Google Scholar]
- (11).Liu X; Freeman R; Golub E; Willner I Chemiluminescence and Chemiluminescence Resonance Energy Transfer (CRET) Aptamer Sensors Using Catalytic Hemin/G-Quadruplexes. ACS Nano 2011, 5 (9), 7648–7655. 10.1021/nn202799d. [DOI] [PubMed] [Google Scholar]
- (12).Zhang C; Hu J Single Quantum Dot-Based Nanosensor for Multiple DNA Detection. Anal. Chem . 2010, 82 (5), 1921–1927. 10.1021/ac9026675. [DOI] [PubMed] [Google Scholar]
- (13).Zhang Y; Zhu C; Zhang L; Tan C; Yang J; Chen B; Wang L; Zhang H DNA-Templated Silver Nanoclusters for Multiplexed Fluorescent DNA Detection. Small 2015, 11 (12), 1385–1389. 10.1002/smll.201402044. [DOI] [PubMed] [Google Scholar]
- (14).Jang K-J; Lee H; Jin H-L; Park Y; Nam J-M Restriction-Enzyme-Coded Gold-Nanoparticle Probes for Multiplexed DNA Detection. Small 2009, 5 (23), 2665–2668. 10.1002/smll.200901105. [DOI] [PubMed] [Google Scholar]
- (15).Lertanantawong B; Krissanaprasit A; Chaibun T; Gothelf KV; Surareungchai W Multiplexed DNA Detection with DNA Tweezers in a One-Pot Reaction. Materials Science for Energy Technologies 2019, 2 (3), 503–508. 10.1016/j.mset.2019.05.001. [DOI] [Google Scholar]
- (16).Halvorsen K; Wong WP Binary DNA Nanostructures for Data Encryption. PLOS ONE 2012, 7 (9), e44212. 10.1371/journal.pone.0044212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Kachakova D; Mitkova A; Popov E; Popov I; Vlahova A; Dikov T; Christova S; Mitev V; Slavov C; Kaneva R Combinations of Serum Prostate-Specific Antigen and Plasma Expression Levels of Let-7c, MiR-30c, MiR-141, and MiR-375 as Potential Better Diagnostic Biomarkers for Prostate Cancer. DNA and Cell Biology 2014, 34 (3), 189–200. 10.1089/dna.2014.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Hao Y; Zhao Y; Zhao X; He C; Pang X; Wu T-C; Califano JA; Gu X Improvement of Prostate Cancer Detection by Integrating the PSA Test With MiRNA Expression Profiling. Cancer Investigation 2011, 29 (4), 318–324. 10.3109/07357907.2011.554477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Ogata AF; Maley AM; Wu C; Gilboa T; Norman M; Lazarovits R; Mao C-P; Newton G; Chang M; Nguyen K; Kamkaew M; Zhu Q; Gibson TE; Ryan ET; Charles RC; Marasco WA; Walt DR Ultra-Sensitive Serial Profiling of SARS-CoV-2 Antigens and Antibodies in Plasma to Understand Disease Progression in COVID-19 Patients with Severe Disease. Clin Chem. 10.1093/clinchem/hvaa213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Koussa MA; Halvorsen K; Ward A; Wong WP DNA Nanoswitches: A Quantitative Platform for Gel-Based Biomolecular Interaction Analysis. Nature Methods 2015, 12 (2), 123–126. 10.1038/nmeth.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Halvorsen K; Schaak D; Wong WP Nanoengineering a Single-Molecule Mechanical Switch Using DNA Self-Assembly. Nanotechnology 2011, 22 (49), 494005. 10.1088/0957-4484/22/49/494005. [DOI] [PubMed] [Google Scholar]
- (22).Chandrasekaran AR; MacIsaac M; Dey P; Levchenko O; Zhou L; Andres M; Dey BK; Halvorsen K Cellular MicroRNA Detection with MiRacles: MicroRNA- Activated Conditional Looping of Engineered Switches. Science Advances 2019, 5 (3), eaau9443. 10.1126/sciadv.aau9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zhou L; Chandrasekaran AR; Punnoose JA; Bonenfant G; Charles S; Levchenko O; Badu P; Cavaliere C; Pager CT; Halvorsen K Programmable Low-Cost DNA-Based Platform for Viral RNA Detection. Science Advances 2020, eabc6246. 10.1126/sciadv.abc6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Hansen CH; Yang D; Koussa MA; Wong WP Nanoswitch-Linked Immunosorbent Assay (NLISA) for Fast, Sensitive, and Specific Protein Detection. PNAS 2017, 114 (39), 10367–10372. 10.1073/pnas.1708148114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Chandrasekaran AR; Trivedi R; Halvorsen K Ribonuclease-Responsive DNA Nanoswitches. Cell Reports Physical Science 2020, 1 (7), 100117. 10.1016/j.xcrp.2020.100117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Chandrasekaran AR; Levchenko O; Patel DS; MacIsaac M; Halvorsen K Addressable Configurations of DNA Nanostructures for Rewritable Memory. Nucleic Acids Res 2017, 45 (19), 11459–11465. 10.1093/nar/gkx777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Thompson IM; Pauler DK; Goodman PJ; Tangen CM; Lucia MS; Parnes HL; Minasian LM; Ford LG; Lippman SM; Crawford ED; Crowley JJ; Coltman CA Prevalence of Prostate Cancer among Men with a Prostate-Specific Antigen Level ≤4.0 Ng per Milliliter. New England Journal of Medicine 2004, 350 (22), 2239–2246. 10.1056/NEJMoa031918. [DOI] [PubMed] [Google Scholar]
- (28).Mitchell PS; Parkin RK; Kroh EM; Fritz BR; Wyman SK; Pogosova-Agadjanyan EL; Peterson A; Noteboom J; O’Briant KC; Allen A; Lin DW; Urban N; Drescher CW; Knudsen BS; Stirewalt DL; Gentleman R; Vessella RL; Nelson PS; Martin DB; Tewari M Circulating MicroRNAs as Stable Blood-Based Markers for Cancer Detection. PNAS 2008, 105 (30), 10513–10518. 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Chen X; Sullivan PF Single Nucleotide Polymorphism Genotyping: Biochemistry, Protocol, Cost and Throughput. The Pharmacogenomics Journal 2003, 3 (2), 77–96. 10.1038/sj.tpj.6500167. [DOI] [PubMed] [Google Scholar]
- (30).Beamish E; Tabard-Cossa V; Godin M Programmable DNA Nanoswitch Sensing with Solid-State Nanopores. ACS Sens . 2019, 4 (9), 2458–2464. 10.1021/acssensors.9b01053. [DOI] [PubMed] [Google Scholar]
- (31).Zhang Y; Sun J; Zou Y; Chen W; Zhang W; Xi JJ; Jiang X Barcoded Microchips for Biomolecular Assays. Anal. Chem . 2015, 87 (2), 900–906. 10.1021/ac5032379. [DOI] [PubMed] [Google Scholar]
- (32).Chandrasekaran AR Processing DNA-Based Molecular Signals into Graphical Displays. ACS Synth. Biol . 2020, 9 (7), 1490–1498. 10.1021/acssynbio.0c00246. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


