Summary
The increasing scope of genetic testing allowed by next-generation sequencing (NGS) dramatically increased the number of genetic variants to be interpreted as pathogenic or benign for adequate patient management. Still, the interpretation process often fails to deliver a clear classification, resulting in either variants of unknown significance (VUSs) or variants with conflicting interpretation of pathogenicity (CIP); these represent a major clinical problem because they do not provide useful information for decision-making, causing a large fraction of genetically determined disease to remain undertreated. We developed a machine learning (random forest)-based tool, RENOVO, that classifies variants as pathogenic or benign on the basis of publicly available information and provides a pathogenicity likelihood score (PLS). Using the same feature classes recommended by guidelines, we trained RENOVO on established pathogenic/benign variants in ClinVar (training set accuracy = 99%) and tested its performance on variants whose interpretation has changed over time (test set accuracy = 95%). We further validated the algorithm on additional datasets including unreported variants validated either through expert consensus (ENIGMA) or laboratory-based functional techniques (on BRCA1/2 and SCN5A). On all datasets, RENOVO outperformed existing automated interpretation tools. On the basis of the above validation metrics, we assigned a defined PLS to all existing ClinVar VUSs, proposing a reclassification for 67% with >90% estimated precision. RENOVO provides a validated tool to reduce the fraction of uninterpreted or misinterpreted variants, tackling an area of unmet need in modern clinical genetics.
Keywords: variant interpretation, reclassification, ClinVar, VUS, machine learning
Introduction
Variant interpretation is the defining moment of genetic counseling because it provides the rationale for critical clinical decisions such as prophylactic interventions, intensive monitoring, or important suggestions regarding the patient’s life as lifestyle modifications, cascade family screening, or procreative decisions. However, despite the introduction of American College of Medical Genetics (ACMG) guidelines in 2015 and the formulation of algorithms based on these guidelines,1, 2, 3 the process of variant interpretation remains inefficient and unreliable, sometimes taking several years.4 ACMG guidelines5 provide criteria to categorize variants in five classes: pathogenic (P), likely pathogenic, variants of unknown significance (VUSs), likely benign (LB), and benign (B). The pathogenic and likely pathogenic classes are together referred to as P/LP, and the benign and likely benign classes are together referred to as B/LB. Evidence used for categorization includes features intrinsic to the variant (e.g., algorithms predicting functional impact or conservation scores); features related to the gene and its association with known diseases (e.g., presence in public databases); and clinically or genetically proven evidence on the specific phenotype-genotype association (e.g., familial segregation). Although the ACMG guidelines greatly facilitated variant interpretation providing a unified framework, clear reporting guidelines have been established only for selected genes correlated to specific diseases.6 Numerous areas of uncertainty remain, such as incidental findings (IFs) or secondary findings (SFs) and rare de novo variants in poorly characterized genes.7 In 2014, Landrum et al.8 developed ClinVar, a freely available public archive of human genomic variants and interpretation of their relationship to diseases and other conditions. The database is updated monthly and allows access to historical data. To date, more than 1,300 organizations have submitted variants to ClinVar, including clinical testing laboratories, research laboratories, single clinicians, patient registries consortia, expert panels, and other organizations.9,10 Although recognized as a key resource, the multiple stored interpretations can often be conflicting and complicate decision-making. In 2015, the NIH funded the project ClinGen11 to obtain expert-curated unified interpretation. However, manual revision is time consuming and often requires lab-based confirmation.
Existing computational tools classify known or novel variants according to the guidelines (e.g., Intervar1 or Varsome2). New variants, not yet described in ClinVar, are also characterized as VUSs because, for example, identified in laboratories that have no access to clinical evidence for phenotype-genotype correlation. Similarly problematic are “conflicting interpretation of pathogenicity” (CIP) variants, which are reported as pathogenic by some laboratories and as benign by others. Few existing tools specifically attempt the interpretation of VUSs and have relevant limitations. LEAP,12 for instance, is limited to missense and cancer-related variants and does not provide implemented code nor public interface.
Communicating uncertain genetic results is a major clinical problem13,14 and many scientific societies recommend against reporting VUSs.15 Furthermore, VUSs should be periodically reassessed, a task that necessitates automated or semi-automated procedures in order to be feasible as the number of reported variants increases.
Over time, a significant share of variants initially reported as VUSs have undergone reclassification into (likely) pathogenic or benign. These entries represent a precious source of information to define features that may help in prospective VUS reclassification. Taking advantage of the vast ClinVar database and in particular the set of VUSs reclassified over time, we trained a random forest (RF)-based algorithm, RENOVO (Reclassification Of Novel and Old Variant tOol), to solve a binary classification problem (pathogenic versus benign) and assign a continuous pathogenicity-likelihood score (PLS) to variants. This allowed us to empirically define thresholds for the identification of high-precision pathogenic (HPP) or high-precision benign (HPB) variants and thus reclassify about 67% of existing VUSs with high confidence. We externally validated our approach on datasets of variants classified through functional validation or expert consensus and developed a user-friendly public interface (see data and code availability).
Material and methods
Datasets
Training, test, and VUS/CIP validation datasets: ClinVar
The complete database of ClinVar version January 2020 was retrieved from the official ftp link. We also retrieved 24 ClinVar VCFs with reference years spanning from June 2012 to November 2019.
We first processed those datasets to standardize the nomenclature used in the classification between different years. Indeed, before 2017, all the individual classifications provided by different submitters for a single variant were reported, following the code reported in Table S1 (column “Code_OLD”). After 2017 (Table S1 [column “VCS_NEW”]), multiple classifications were collapsed into a single one, and a new class, “conflicting interpretation of pathogenicity,” was introduced to handle those variants with discordant classifications.
Starting from this classification, we developed a method to unify and simplify the assignment of a class (Table S1, column “Code_NEW”). Our method consists in reporting the most represented class whenever possible. When the same number of pathogenic (“4” or “5”) and benign (“2” or “3”) classifications are assigned to a variant, the variant is marked with “−1” to describe the ClinVar “conflicting interpretation of pathogenicity” (CIP). If the class “0” (VUS) is present but not the most represented class, we assign “−1.” In the remaining cases, even if a major class is not present, the first class found is assigned.
A total of 610,956 variants was thus retrieved and classification was collapsed in classes: pathogenic (P), likely pathogenic, likely benign (LB), benign (B), VUSs, CIP, and “other” (e.g., risk factors, protective, drug-responsive, omitted from our subsequent evaluation) according to the criteria detailed above. The pathogenic and likely pathogenic classes are together referred to as P/LP, and the benign and likely benign classes are together referred to as B/LB.
From those variants we defined:
-
•
the training set, which included all “stable” variants (n = 332,231) currently classified as P/LP or B/LB that have not changed status over time, irrespective of their time of first reporting;
-
•
the test set, which included “reclassified” variants (n = 18,312), defined as those variants that are currently classified as P/LP or B/LB but were introduced with a different status (VUSs, CIP, or “other”);
-
•
two exploratory datasets composed respectively by variants of unknown significance (VUSs, n = 216,716) and by variants with conflicting interpretation of pathogenicity (CIP variants, n = 30,440)
Validation datasets
To obtain external (i.e., non-ClinVar-based) validation, we identified datasets orthogonal to ClinVar, and variants were interpreted either through structured consensus or laboratory-based functional assays: (1) 7,460 evidence-based classified BRCA1/2 variants from the ENIGMA project16 (from this dataset we excluded 15 variants for which it was not possible to recollect their “Type” [no information about ExonicFunc.refGene or Func.refGene], obtaining a final set of 7,445 variants); (2) 3,893 BRCA1 variants17 validated in vitro through a saturation genetic assay (2,821 functional, 823 nonfunctional or “loss of function,” and 249 intermediate); (3) 893 DCM-related variants from 766 patients clinical-based classified;18 and (4) 73 variants on SCN5A,19 in vitro validated through patch clamping as benign (n = 10) or Brugada syndrome-associated (n = 63).
Preprocessing and exploratory feature analysis
To analyze the data with the same pipeline, we applied preprocessing steps to different datasets. Variants were annotated with Annovar,20 and then standard names, equal across the datasets, were assigned to the features. For those features not commonly shared in all the datasets, new variables such as “Type” (i.e., combination of Func.RefGene and ExonicFunc.RefGene) and “CLNDN_dicotomize” (equal to 1 or 0 according to the presence/absence of associated diseases) were created and included in the analysis.
Variants with Type “exonic.NA” were excluded from the analysis (not counted in the dimension of training and test sets).
Both training and test sets presented a large percentage of missing values across their features, and functional scores such as MutPred, M-CAP, Mutation Assessor, SIFT, FATHMM, and PROVEAN exceeded 80% of not available values (see Figure S1). Because the presence of missing values can impact on the application of machine learning algorithms to the data, we performed a missing data imputation step: missing data for allele frequency (AF) and for functional and conservation scores were imputed with the median score for the given variant ”Type” on the training data. Scores for variant types without values on the training set were imputed with the median of the most similar classes in terms of biological function. Types were associated as displayed in Table S2.
To assess the significance of the features used to train the model, we applied the Wilcoxon rank-sum test to their distributions in B/LB and P/LP classes in the training set. Statistical significance was obtained with p value < 0.05. We also computed Spearman’s correlation among features to study feature collinearity.
RENOVO design and development
The RENOVO algorithm was developed through four main steps: (1) identification of the best machine learning algorithm for discriminating benign (B/LB) from pathogenic (P/LP) variants, with selection of random forest (RF); (2) feature selection to construct a more generalizable algorithm (RENOVO-Minimal [RENOVO-M]) compared to the initial algorithm that used the full set of features (RENOVO-Full [RENOVO-F]); (3) parameter optimization of the two models on the training set and definition of a pathogenicity likelihood score (PLS, the percentage of decision trees in the RF that classify the variant as pathogenic to rank variants); and (4) definition of PLS thresholds for confidence classes (high precision benign [HP-B], intermediate precision benign [IP-B], low precision [LP], intermediate precision pathogenic [IP-P], and high precision pathogenic [HP-P]).
We prioritized variants in the different datasets into RENOVO classes, and we used accuracy to assess goodness of the model on the training and test sets.
Preprocessing, RENOVO implementation and data analysis were performed with R (v.3.6.2.) and Python3 (packages/functions from ScikitLearn library v.0.20.3). For the implementation, we worked on a High Performance Computing (HPC) cluster (2 frontend machines with 24 cores and 128 GB ram and 12 computing nodes with 28 cores and 128 GB ram).
Additionally, to facilitate the prioritization of variants of interest with RENOVO, we developed a user-friendly web interface based on Shiny v.1.5 and on the R flexdashboard framework v.0.5.2.
Machine learning model comparison and selection of random forest
We compared the performances of different machine learning (ML) algorithms (RF, support vector machine [SVM], naive Bayes, and logistic regression) in separating benign (B/LB) versus pathogenic (P/LP) variants. We performed this analysis with Orange3; we did not separate stable and reclassified variants, and we used 66% of the total ClinVar variants as the training set and the remaining 33% as the test set. We performed a 10-fold cross validation to train the different models. We used rea under the ROC curve (AUROC), accuracy, F1 measure, precision, and recall to select the best method (see Table S3).
Feature selection
Features for RENOVO-Full (RENOVO-F) were selected manually starting from the ACMG guidelines (Table S4). It uses 25 features obtained from ClinVar and Annovar after preprocessing, listed in Table S5. Those features were selected to cover the highest number of the guidelines provided by ACMG and were shown to be able to discriminate between the two classes (significant p value ≤ 0.05 of the Wilcoxon signed-rank test, Figure S2).
On top of this model, we built a second one, RENOVO-Minimal (RENOVO-M), whose aim is to be faster and more general. Thus, it uses only a subset of the features used for RENOVO-F, chosen to reduce redundant information while covering all the guidelines. We perform this feature selection in two steps. First, we computed feature importance with SHAP (SHapley Additive exPlanations) values21 (with 10,000 samples chosen randomly from the training set, computation with SHAP Python library): the values for the 20 most important features can be seen in Figure S3. We chose to perform feature importance analysis with SHAP values because they provide a unique solution to the “impact distribution” problem among features, founded on well-defined mathematical properties (rationality, fairness, additivity22). Additionally, they represent an established method for interpretation of machine learning models.23 SHAP values compute and describe the positive or negative impact of each feature on single model predictions. Features with average SHAP values ≤ 0.01 were excluded for a total of 16 features (“Type” feature did not undergo feature selection, all its one-hot-encoded columns were retained). To cope also with feature redundancy, we computed Spearman’s correlation (Figure S4). When a correlation ≥0.85 was identified among two features of the restrained set, only the feature with highest mean SHAP value was retained. After this second step, we obtained a set of 11 features, to which we added the “Type” variable (Table S5).
Optimization of random forest parameters
We applied the same pipeline to optimize the two random forest models at the basis of RENOVO-F and RENOVO-M, RF-F and RF-M. The input dataset was divided into training and test sets (70% and 30% of the total, respectively), and optimization was performed on the parameters “n_estimators” (from 10 to 130) and “max_features” (from 3 to 8) through 5-fold cross validation and grid search. Mean computational time and accuracy were computed on the 5-folds for each optimization step (Figure S5), while total time and computational resources were computed for the complete optimization and training process (Figure S6). From results obtained in the model selection, we set min_samples_split = 5. RFs were thus trained with the optimized parameters on the complete training set: accuracy of the classification and feature importance were computed. Finally, the trained models were applied to the test set.
The parameters selected for RF-F were n_estimators = 130, max_features = 8; for RF-M, the optimized parameters were n_estimators = 70, max_features = 8 (Figure S5).
The two models were compared on the test set with ROC (receiver operating characteristics) analysis, precision-recall (PR) curves, and performance measures computed by MLeval R package. AUC of the ROC curves were compared with a chi-square test (roccomp function in STATA24).
Class definition with thresholds
RENOVO assigns a continuous pathogenicity likelihood score (PLS, the percentage of decision trees that classify the variant as pathogenic) to each variant on the basis of information features that are available in public databases (i.e., prediction scores, conservation scores, minor allele frequency) and that are collected in ClinVar. To obtain a more accurate classification in terms of pathogenicity, we drew a precision-recall (PR) curve and identified two thresholds on the PLS computed on the test set that can assure 99% and 90% of precision on recovering elements in the P/LP class. Similarly, NegativePredictiveValue-TruePositiveRate curve (NPR) was used to assess 99% and 90% of precision on B/LB class. To keep also high levels of recall, we identified the thresholds as follows: given a precision t1 (e.g., 0.90 or 0.99) and a cutoff t2 = 0.01, the threshold t is defined as t = min(x) (max(x) for NPR) where x are values on the curve PR such that precision PR(x) ≥ t1 (NPV(x) ≥ t1) and PR′(x) ≤ t2 (NPR′(x) ≤ t2). PR′(x) and NPR′(x) denote the first derivative of the curve in the x value. To avoid random fluctuation, we asked the condition on the derivative to remain valid at least for the 4 points on the curve following x. Once those four thresholds were defined, the PLS range [0,1] was separated accordingly and five new classes were associated to the new intervals: high precision benign (HP-B), intermediate precision benign (IP-B), low precision (LP), intermediate precision pathogenic (IP-P), and high precision pathogenic (HP-P) curves and related measures for this analysis were drawn with ROCR25 R package.
The final model, RENOVO-M (RF-M + threshold definition), which uses the minimal set of features, was then applied to the VUS set, the CIP set, and the external validation sets.
To assess RENOVO improvement over existing functional and conservative scores, we compared their classification performances on the test set and on the validation sets with AUC of ROC and precision-recall curves. In addition to the scores used as RENOVO features (see Table S5), we also considered Eigen,26 CADD,27 and DeepSea.28 To not bias the results, for this analysis we used the original (not imputed) values for the existing scores. Pearson’s correlation between PLS and the other scores was also computed.
Additionally, we implemented the possibility to find PLS thresholds able to maximize specificity and sensitivity on specific genes. Once the gene of interest is identified, thresholds are selected with the R package cutpointr29 on the gene variants belonging to the test set.
Results
Feature analysis on ClinVar datasets
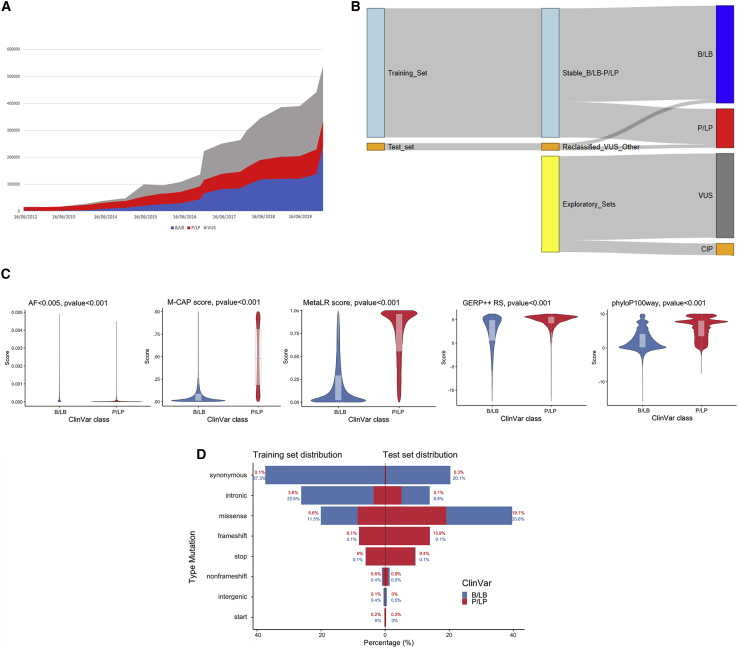
To develop our variant classification algorithm, we constructed datasets nested within the ClinVar database. We first analyzed time trends in submitted variants from 2012 to 2020 (Figure 1A). In our reference January 2020 release, containing 610,955 variants, VUS is the most represented category (35.48%); 15.56% are P/LP, 37.88% are B/LB, 5% are CIP, and 1% are classified as “other.” Total reported variants have grown following an exponential trend since 2014 (y = 3 × 10−12e(−0.0009x), with R2 = 0.94). VUS grew with the highest rate jumping from ∼50,000 in 2017 to ∼200,000 in 2020. Considering the reference release, 3% of the entire database was reclassified over years from VUS/CIP in P/LP or B/LB.
Figure 1.
ClinVar datasets overview
(A) Variants over time. Trend of the total number of variants present in ClinVar, divided by the three main categories of clinical significance: blue for benign (B/LB) variants growth, red for pathogenic (P/LP), and gray for VUSs.
(B) Sankey diagram showing the construction of the different datasets coming from ClinVar. RENOVO training and test set come both from B/LB and P/LP variants that never changed classification and that were reclassified, respectively. VUSs and conflicting interpretation of pathogenicity (CIP) variants are used as an application of RENOVO.
(C) Feature distribution: violin plots for four numerical features of the training set are displayed (AF < 0.005, M-CAP and Meta-LR functional scores, GERP++_RS and phyloP100way_vertebrate conservation scores). Blue is used for distribution in the B/LB class and red for the P/LP class. Boxplots are shown in gray. p values from Wilcoxon rank-sum test are added for each feature.
(D) Variant type distributions in training set (left) and test set (right). For each mutation type, the percentage of B/LB and P/LP variants over the total in the corresponding set is displayed. Blue is used for the B/LB class and red for the P/LP class.
On the basis of these data, we coerced P/LP and B/LB into only two categories to increase statistical power and constructed two datasets for algorithm development (Figure 1B): the training set, which includes 332,231 “stable” variants that maintained their status over time as B/LB (n = 241 416, 72.66%) or P/LP (n = 90,815, 27.33%), and the test set, which includes 18,312 variants that changed their status from VUS/CIP to B/LB (n = 9,296, 50.76%) or P/LP (n = 9,016, 49.24%).
Feature analysis on the training set revealed that distributions of allele frequency in the general population and functional and conservation scores were significantly different (Wilcoxon rank-sum test p value < 0.05) between the B/LB and P/LP classes (Figure 1C and Figure S2). Variant types were differently represented on training and test sets: in the training set, most B/LB variants were synonymous (51.31% of the B/LB) and intronic (22.1% of the B/LB), whereas P/LP variants were enriched for missense (31.22% of the P/LP) and frameshift (29.5% of P/LP), as expected; missense variants were equally distributed between B/LB (11.5%) and P/LP (8.6%) (Figure 1D).
The test set was enriched for missense variants (roughly twice as many as in the training set), as expected. Variants classified as B/LB were equally synonymous (39.61%) or missense (40.17%): within missense, B/LB and P/LP were equally distributed (20.6% and 19.1% of the total, respectively; Figure 1E).
RENOVO performance on training and test sets
We compared the performances of different ML algorithms (RF, SVM, naive Bayes, and logistic regression) in separating benign (B/LB) versus pathogenic (P/LP) variants on the training set. RF outperformed other algorithms on all metrics (Table S5) and does not require assumptions on the linearity of interactions (Table S4). Thus, we chose RF for further RENOVO development.
We first applied RENOVO with the full set of 25 features in Table S4 (called RENOVO Full or RENOVO-F) to the training and test sets. We defined the pathogenicity likelihood score (PLS) as the percentage of decision trees that classify the variant as pathogenic. The higher the PLS, the higher is the probability inferred by the RF algorithm to be in front of a pathogenic variant. The algorithm was first trained to distinguish between pathogenic (P/LP) and benign (B/LB) variants.
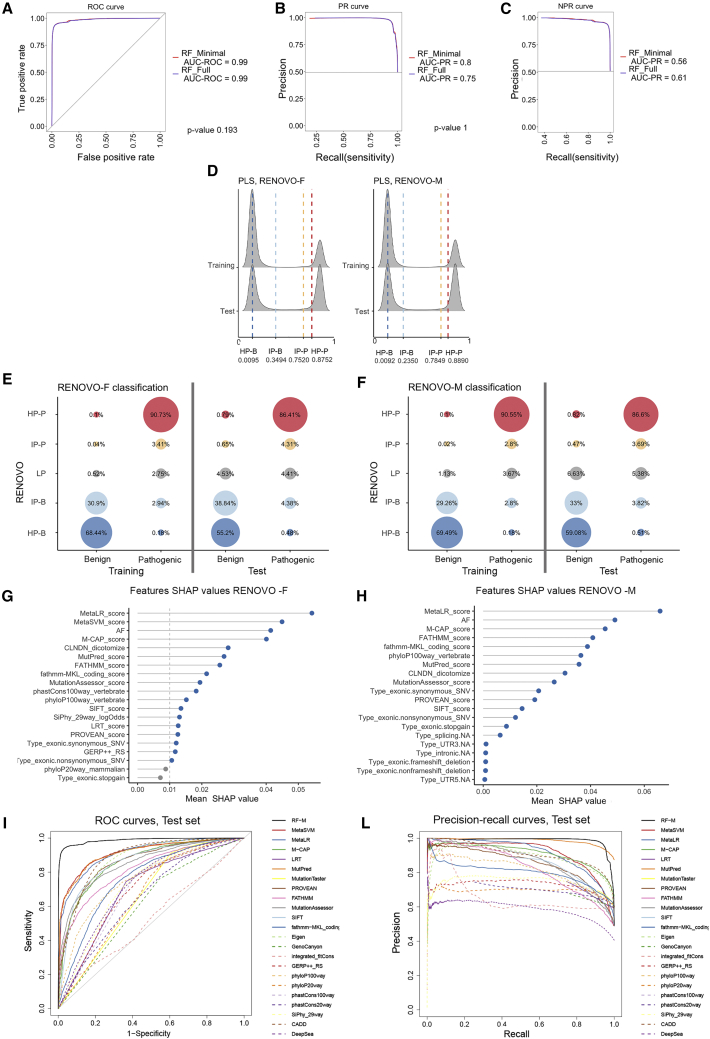
Using an arbitrary cutoff value at 0.5, such that variants with PLS ≥ 0.5 are defined as PLS-pathogenic (PLS-P) and variants with PLS < 0.5 as PLS-benign (PLS-B), RENOVO-F correctly classified 98.83% of the variants in the training set (Figure S7). On the test set, accuracy was slightly inferior at 95.18%; the relatively minor loss of accuracy despite significant differences in feature distribution between training and test sets highlights the robustness of RENOVO for heterogeneity in variant types. The AUROC measured on the test set of variants reclassified over time was extremely high at 0.99 (C.I. 0.99–0.99), indicating that RENOVO-F would have correctly classified most variants initially reported as neither P nor B (VUSs or other) (Figure 2A). Additional performance metrics highlight the good quality of RENOVO-F predictions (F1 score = 0.95, sensitivity = 0.94 [C.I. 0.94–0.95], specificity = 0.96 [C.I. 0.96–0.96], complete list in Table S6).
Figure 2.
RENOVO algorithm: Comparison between full and minimal models
(A) ROC analysis: ROC curves to evaluate performances of RENOVO-F (blue line) and RENOVO-M (red line). The curves, together with the values of the AUROCs, are showed to compare the two models. The chi-square test p value of AUROC difference is also displayed.
(B) Precision-recall curves for RENOVO-F (blue line) and RENOVO-M (red line) to evaluate the precision of the models with respect to the P/LP class. AUROCs for the two curves are reported. The chi-square test p value of AUROC difference is also displayed.
(C) Negative precision-recall curves to evaluate precision on the B/LB class: results are depicted in blue for RENOVO-F and in red for RENOVO-M. AUCs are reported for both models.
(D) Distributions of computed PLS for training and test variants for RENOVO-F (left) and RENOVO-M (right). The density plot is clearly showing a bi-modal distribution with a large separation between the two peaks, suggesting a high degree of confidence in the prediction call. Vertical lines denote the thresholds used to define RENOVO classes: blue lines define HP benign and IP benign classes and red lines HP pathogenic and IP pathogenic.
(E) RENOVO results on ClinVar datasets: prioritization results on the training (benign and pathogenic) and test (benign and pathogenic) set for RENOVO-F. Colors follow the classification provided by RENOVO: blue shades for HP and IP benign classes, red shades for HP and IP pathogenic, and gray for LP. Bubble sizes are proportional to the fractions of variants represented.
(F) RENOVO results on ClinVar datasets: prioritization results on the training (benign and pathogenic) and test (benign and pathogenic) set for RENOVO-M. Bubble colors and sizes follow the code described in (E).
(G) Feature importance with mean SHAP values retrieved for RENOVO-F. To reduce noise, only the first 20 features are shown. The vertical gray line at 0.01 represents the threshold used to keep features in the selection step: gray dots are features below this cutoff.
(H) Feature importance with mean SHAP values retrieved for RENOVO-M are displayed. To reduce noise, only the first 20 features are shown.
(I) ROC curves obtained by RENOVO-M classification (black continuous line) and by other predictive and functional scores.
(L) Precision-recall curves obtained by RENOVO-M (black continuous line) classification and by other predictive and functional scores.
We then identified PLS cutoffs, starting from the precision-recall curve for class P/LP (Figure 2B) and from the negative precision-recall curve for class B/LB (Figure 2C) as described in class definition with thresholds. The identified cutoffs (0.0095, 0.3494, 0.7520, and 0.8752) allow us to define classes with minimal precision achieved in the classification. Those precisions are equal to 99% (high precision PLS-B [HP-B] and high precision PLS-P [HP-P]), 90% (intermediate precision, IP-P and IP-B), and precision less than 90% (low precision or LP). Density plots in Figure 2D show that most of the variants in both the training and test sets are polarized in the intermediate/high precision interval. In other words, across the two datasets, 98.62% of the variants can be attributed to P or B with >90% precision. It is important to note that the RENOVO LP class is not a mere equivalent of the ClinVar VUS or CIP class: as VUSs/CIP, all variants are “lumped together” in a single class, whereas as RENOVO LP, the likelihood of pathogenicity is continuously distributed across the PLS range, providing a quantitative value that can be integrated with additional parameters (phenotype, familiarity, co-segregation, etc.) in order to refine interpretation.
Feature selection and identification of a minimal feature set
Feature SHAP weights20 in RENOVO-F are shown in Figure 2G and Figure S3. Interestingly, ensemble prediction scores (i.e., scores calculated via a combination of other scores) were identified as important features, and Meta-LR, Meta-SVM, and M-CAP appeared in the top five positions. Allele frequency (AF) in the general population was the 3rd most important feature, highlighting its key importance in discriminating variants, consistent with ACMG guidelines (Tables S4 and S5).
We identified a non-redundant feature subset that covered all the areas indicated by the guidelines (Table S5).6 We excluded scores with low or redundant information (e.g., phyloP20way_mammalian) or highly correlated with more important features (i.e., Meta-SVM score, LRT score, phastCons100way_vertebrate, GERP++, and SiPhy_29way_logOdds, Figure S4). With this restricted set of 12 features, we trained a second version of RENOVO (RENOVO Minimal, RENOVO-M). This second model achieved no significant accuracy loss on either the training set (98.79%) nor the test set (95.28%) (Figure S8). No significant difference between the ROC curves was identified (Figures 2A and 2B, chi-square test p value of AUROC difference equal to 0.193 and 1, respectively). Features most used by RENOVO-M to classify variants were Meta-LR, AF, M-CAP, and FATHMM score (see Figure 2H). Although Meta-SVM was the second feature in terms of importance for RENOVO-F, it was highly correlated (0.97) with Meta-LR,30 so it was dropped. The accuracy gained by RENOVO-M shows that the exclusion of Meta-SVM did not have a high impact on the classification. Features common in misclassified variants are detailed in the supplemental information, Figure S9, and Table S7. We recalculated thresholds for precision >99% and 90% (identified cutoffs at 0.0092, 0.235, 0.7849, and 0.8890) and re-classified variants according to RENOVO-M as above. The percentage of variants falling in each class was not significantly different than those calculated with RENOVO-F (Figures 2E and 2F).
Thus, RENOVO-M achieved a slightly better accuracy on the test set with a more parsimonious feature set, reducing the risk of model overfitting and resulting in a significant reduction of computational power: while the complete process of optimizing and training RENOVO-F required a maximum of almost 1,500 MB for a computational time of 46 min, RENOVO-M requirements were reduced to 1,300 MB and 37 min (Figure S6). For the specific optimized parameters, the average computational time to apply RENOVO dropped from 20 to 8 s when considering the minimal version (Figure S5). Furthermore, relying on fewer features has the advantage of an easier recollection and imputation of features for new variants. For these reasons, we performed all subsequent analyses with RENOVO-M, referred to in the following simply as RENOVO.
Comparison of RENOVO with other predictive and functional tools
We compared RENOVO classification performances on the test set with those obtained by other predictive or functional tools. We first observed that RENOVO PLS correlates strongly with scores from Meta-SVM, Meta-LR, and MutPred (Pearson’s correlation of 0.83, 0.82, and 0.75, respectively, see Figure S10). However, ROC and precision-recall analysis (Figures 2I and 2L) confirms that RENOVO outperforms the existing scores in classification tasks. Despite ensemble methods such as Meta-LR, Meta-SVM, and M-CAP’s obtaining high AUROC (∼0.9), RENOVO reached AUROC = 0.99. In precision-recall, RENOVO outperformed other tools (AUCPR = 0.99), with the exception of MutPred, which however, could be computed on only 20% of the variants in the test set (Table S8).
VUS/CIP reclassification and comparison with InterVar
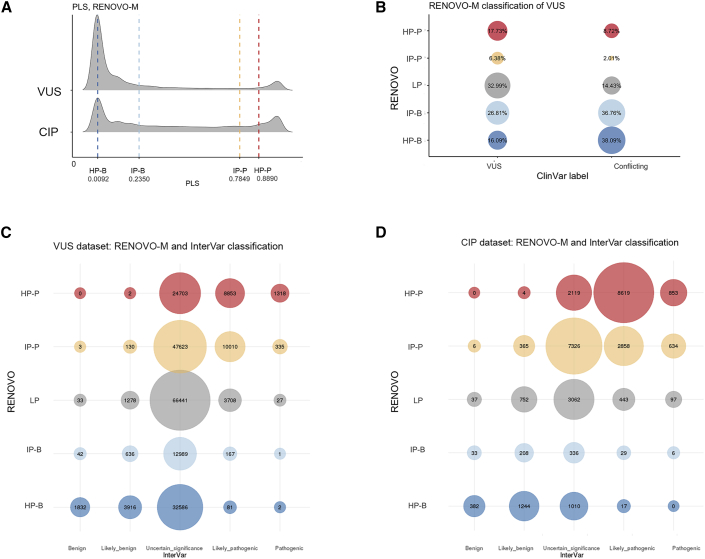
To gauge the potential of RENOVO in variant reclassification, we assessed RENOVO results on current ClinVar VUSs or CIP variants. Because VUSs and CIP variants cannot be validated by definition, as a benchmark we compared RENOVO with InterVar, which provides interpretation according to the ACMG/AMP 2015 guidelines.8 First, we verified concordance on LP/P or LB/B classes: InterVar P/LP variants were classified by RENOVO as HP/IP-P in 95.76% of the cases (93.41% HP, 2.35% IP), whereas 97.3% of the InterVar B/LB variants were classified as RENOVO HP/IP-B (73.70% HP-B, 23.65% IP-B); the high concordance rate confirms that RENOVO predictions are in agreement with ACMG/AMP guidelines. However, the two tools provided significantly different results on the 216,716 VUSs and 30,440 CIP variants, which are mostly missense (68% of the total in the VUS dataset and 48.7% in the CIP dataset, Figure S11). On VUSs, InterVar classified only 15% as either B/LB (11%) or P/LP (4%), leaving 85% of the variants as of uncertain significance (Figure S12), whereas RENOVO classified 67% of all VUSs (a total of 145,229 variants) with predicted precision >90% into either HP/IP-P (42.9%) or HP/IP-B (24.1%) classes (Figures 3A and 3B). Of the variants classified as “uncertain significance” by InterVar, RENOVO classified 39.23% as HP/IP-B and 24.72% as HP/IP-P and had an overall reclassification rate over InterVar of 63.96% (Figure 3C).
Figure 3.
RENOVO-M on VUSs and conflicting variants
(A) RENOVO-M distribution of PLS for VUSs and conflicting variants. Vertical lines represent the thresholds used to define classes: blue lines define HP benign and IP benign classes and red lines HP pathogenic and IP pathogenic.
(B) RENOVO-M classification of VUSs and conflicting variants. Bubble size is proportional to the percentage represented. Blue colors are for HP and IP benign, red for HP and IP pathogenic, and gray for the LP class.
(C) Comparison between RENOVO-M and InterVar classes on VUS ClinVar set: bubble size represents the number of common variants for each RENOVO-M class and InterVar. Colors follow the classification provided by RENOVO: blue shades for HP/IP benign classes, red shades for HP/IP pathogenic, and gray for LP.
(D) Comparison between RENOVO-M and InterVar classes on CIP ClinVar set: bubble size represents the number of common variants for each RENOVO-M class and InterVar. Colors follow the same code described in (C).
For CIP variants (Figure 3D), the results were even more clear-cut: 85.6% variants were interpreted as HP/IP-P or HP/IP-B (10.7% and 74.9%, respectively, Figure 3B), whereas InterVar classified 54.9% in P/LP or B/LB. Remaining CIP variants, classified by InterVar as VUSs (n = 13,853), were classified by RENOVO as LP only in 22.10% of the cases (Figure 3D).
RENOVO validation on external datasets
We further challenged RENOVO on external and only partially overlapping to ClinVar variant datasets in two settings of major clinical relevance: BRCA1/2 and variants associated with cardiomyopathies or channellopathies.31 In both contexts, variants were classified via either structured clinical review16,18 or functional validation through laboratory-based measurement of gene activity.17,19
In the BRCA1/2 context, we studied (1) the ENIGMA set of 38,957 BRCA1/2 variants, of which 7,460 were manually revised by the consortium, leaving only 7 VUSs and (2) a set of 3,893 BRCA1 variants functionally validated through in vitro CRISPR-Cas9 saturation screens.17
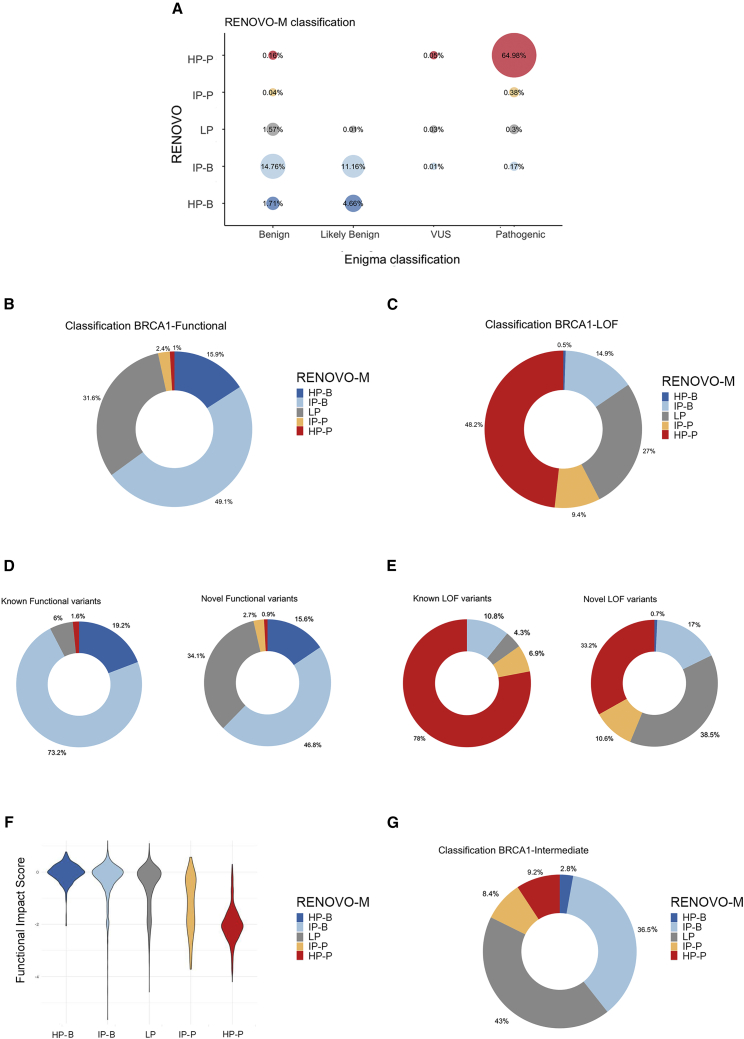
In ENIGMA,16 complete information was collected for almost all (n = 7,445) manually revised variants. RENOVO-M correctly classified 99% (4,866/4,901) of the ENIGMA pathogenic variants and 94.7% (2,404/2,537) of the benign variants (Figure 4A), similarly to InterVar (accuracy = 97.3% and 96.3% for P and B, respectively). A complete comparison of the different tools used to prioritize this set of variants (ENIGMA, RENOVO, ClinVar, and InterVar) is presented in Figure S13.
Figure 4.
RENOVO-M validation in BRCA1/2-related context
(A) Comparison of RENOVO-M and ENIGMA database on the 7,445 variants reviewed by the ENIGMA Consortium. Bubble size represents the percentage of common variants for each RENOVO-M class and ENIGMA class. Colors follow the classification provided by RENOVO: blue shades for HP-B and IP-B classes, red shades for HP-P and IP-P, and gray for LP.
(B) RENOVO-M classification of in vitro functional BRCA1 variants. Dark and light blue represent variants classified as HP and IP benign, red and orange are for HP and IP pathogenic variants, and gray slices for mutations classified as LP.
(C) RENOVO-M classification of LOF in vitro BRCA1 variants; colors used as in (B).
(D) Separate view of RENOVO-M results on functional BRCA1 variants from functional assay: variants that are already present in ClinVar and on novel variants are represented on the left and the right, respectively. Color code is the same used in (B).
(E) Separate view of RENOVO-M results on LOF BRCA1 variants from functional assay: classification of variants that are already present in ClinVar is represented on the left and classification of novel variants on the right. Color code is the same used in (B).
(F) RENOVO-M pathogenicity likelihood score versus functional score defined by Findlay in the different RENOVO classes. Colors follow RENOVO-M classification.
(G) RENOVO-M classification of intermediate BRCA1 variants. Color code is described in (B).
In the Findlay dataset,16 artificially created variants were functionally scored and separated into “functional” (i.e., with no relevant loss of function, Figure 4B and Table S9), “loss-of-function” (LOF, Figure 4C), and “intermediate:” 3,347 (85%) are novel or VUS/CIP in ClinVar (Figures 4D and 4E). Findlay’s functional score was significantly higher in RENOVO HP/IP-B compared with HP/IP-P (Figure 4F). Despite the presence of ∼50% missense variants, 63.3% of the functional or LOF variants were “correctly” classified into HP/IP-B or HP/IP-P, respectively. Expectedly, performance was higher on variants already present in ClinVar (92.4% functional→HP/IP-B and 84.9% LOF→HP/IP-P) as compared to entirely novel variants (62.4% functional→HP/IP-B and 43.8% LOF→HP/IP-P) (Figures 4D and 4E). Comparatively, InterVar (Table S9) correctly classified only 35% of the functional and LOF variants (29.9% and 52.5%, respectively), leaving a high number as “uncertain significance” (60% of the functional and 45.9% of the LOF). RENOVO classified 56.9% of the intermediate variants (Figure 4G): 39.3% as HP-B/IP-B and 17.6% as HP-P/IP-P.
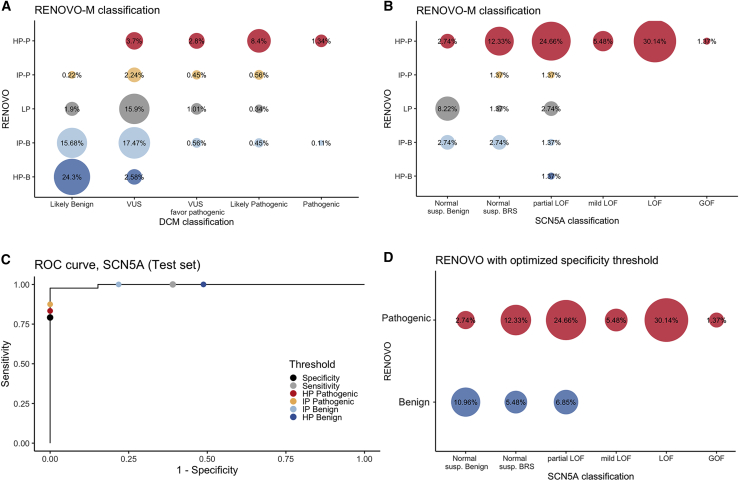
In the inherited cardiac syndrome field, we analyzed (1) the Pugh et al. dataset,18 in which 893 unique variants in 766 patients with dilated cardiomyopathy (DCM) were classified via a structured clinical-grade scoring system, and (2) the Glazer et al. dataset,19 in which high-throughput automated patch clamping was used to study the functional impact of 73 SCN5A variants candidate for Brugada syndrome. Importantly, P/B (in Pugh) or functional/LOF (in Glazer) classes were imbalanced by design in the two studies: the Pugh study aimed at increasing specificity and is enriched for B over P variants (376 versus 100); the Glazer study focused on candidate variants and is enriched for LOF (49 out of 63 total variants).
In the Pugh et al. dataset, RENOVO accuracy was high for both B/LB (357/376, 95%) and P/LP (92/100, 92%) (Figure 5A), significantly higher than InterVar for P/LP (60%) and equal for B/LB (95%). Of the 374 VUSs, RENOVO reclassified 179 as HP/IP-B and 87 as HP/-IP-P (71% in total), leaving only 38% as LP. Comparatively, InterVar reclassified only 28% of the VUSs as P/LP or B/LB, leaving 62% as “uncertain significance” (Table S10). Finally, of the 43 variants described as “VUS favor of pathogenic,” 67%29 were HP-P/IP-P (Figure 5A and Table S11).
Figure 5.
RENOVO-M validation in DCM-related context
(A) Comparison of RENOVO-M and clinical-based classification of the 893 DCM variants. Bubble size represents the percentage of common variants for each RENOVO-M class and DCM class. Colors follow the classification provided by RENOVO: blue shades for HP-B and IP-B classes, red shades for HP-P and IP-P, and gray for LP.
(B) Comparison of RENOVO-M and functional classification of 73 SCN5A variants in Glazer dataset. Bubble size represents the percentage of common variants for each RENOVO-M class and SCN5A class. Color code is defined as in (A). “Normal susp. Benign” label stays for the 10 normal suspected benign variants, while “Normal susp. BRS” for the normal suspected Brugada syndrome variants.
(C) ROC curve on the test set restricted to the SCN5A gene; effects on specificity and sensitivity of diverse PLS thresholds are represented by different colors. RENOVO-M thresholds for HP-B and IP-B are colored in dark and light blue and those for HP-P and IP-P in red and orange. PLS thresholds optimized for specificity and sensitivity are represented by black and gray dots.
(D) Comparison of RENOVO-M optimized for specificity and SCN5A database. Bubble size represents the percentage of common variants for each RENOVO-M class and DCM class. Colors follow the classification provided by RENOVO: blue for benign and red for the pathogenic class.
In the Glazer et al. dataset with variants mostly unreported or VUS/CIP in ClinVar (79%), RENOVO agreed with the functional classification in 56/63 (88.9%) variants (Figure 5B); comparatively, InterVar classified most variants (48/63, 73%) as VUSs (Tables S11 and S12).
For this specific dataset, we tested the effect of moving the PLS cutoff value to favor specificity over sensitivity (i.e., identifying true benign variants accepting a loss of true pathogenic). A PLS threshold value of 0.9068 achieved 80% specificity and 85.7% sensitivity (Figure 5C), allowing the correct identification of 8/10 benign variants while misclassifying 9/63 pathogenic variants (Figure 5D and Table S13). This shows that RENOVO can also provide acceptable estimates on rare variant datasets where achieving large sample size is impossible.
Comparison of RENOVO with functional and predictive scores on the validation sets (Table S14) confirms that RENOVO overall outperforms other tools: AUROC and AUC-PR from RENOVO obtained high values and are classified as the best ones in three out of four cases (Enigma, DCM, and SCN5A) and the second best, after Eigen, for the BRCA dataset.
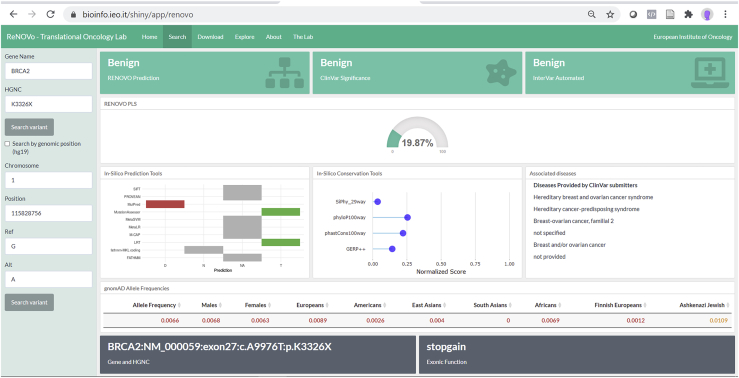
RENOVO user interface
RENOVO graphical interface (Figure 6) allows one to (1) search for variants of interest through genomics or cDNA or protein nomenclature, (2) display the classification of ClinVar, InterVar, and RENOVO (Figure 6), (3) rank the features by weight in the interpretation, and (4) explore the entire gene and reported variants with RENOVO classification.
Figure 6.
Dashboard
RENOVO web interface: example of results provided by our RENOVO web app when a variant is searched. Variants can be searched with HGVSc; HGVSp entire or partial nomenclatures (e.g., c.A9976T or p.Lys3326∗); or chromosome, position, reference, and alternative (e.g., 13-32972626-A-T). Interpretations taken from ClinVar and Intervar for the same variants are also displayed, as well as the values of the features used by RENOVO to classify the variant. In this figure, the BRCA2 variant p.Lys3326∗, which was initially associated with risk of breast and ovarian cancer, then considered as a VUS in agreement with ACMG criteria, and finally reclassified as benign in ClinVar, is reported.
For each variant, it is also possible to choose, identify, and download a much larger number of features from different databases, all aimed at supporting the classification of the variant. Finally, from this interface users can search for the variant or the associated disease on PubMed, LitVar, ClinVar, and OMIM.
Discussion
In this work, we provide a computational tool to improve genetic variant interpretation and minimize the fraction of variants with uncertain significance or conflicting interpretation. As our historical analysis shows, the VUS class represents the largest and fastest growing variant class in ClinVar, and there is no trend for decrease. This has been facilitated by the rapid surge in sequencing volume by large molecular genetics laboratories with no direct contact with the patient and therefore insufficient information to appropriately assess co-segregation or phenotype-genotype correlation. Interpretation according to ACMG guidelines remains highly operator dependent, as demonstrated by a survey of nine large genetics laboratories that evaluated 99 variants with a mere 34% agreement rate.15 Our RENOVO algorithm avoids altogether the possibility of VUSs, constraining a decision between pathogenic or not and assigning a likelihood score (the PLS) to this binary classification. The PLS can be treated as a quantitative measure of uncertainty and as a diagnostic test whose cutoff can be calibrated differently according to the specific clinical need. For instance, in complex multifactorial contexts such as genetic cardiomyopathies and channelopathies,31 it may be preferable to favor specificity (i.e., the accuracy of calling benign variants) at the expenses of sensitivity (i.e., the accuracy of calling pathogenic variants) to identify the genetic marker to test in asymptomatic family members. Often in such contexts, the development of a dedicated pathogenicity prediction tool is rendered impossible by the limited sample size allowed by the rarity or small number of candidate variants. It becomes particularly useful when a specific score recalibration is allowed by available modest-sized datasets, as in the SCN5A case study provided here. For low-precision variants, integration with the clinical picture, including co-segregation considerations,13 is essential, and the availability of a continuous score may facilitate interpretation. Also, despite the increasing use of direct functional assessment of variants through laboratory assays, efficient techniques, such as genetic manipulation with CRISPR-Cas9 or disease modeling with human induced pluripotent stem cells,32,33 remain difficult to apply to the routine clinical context, in which quick answers are needed. The high concordance of RENOVO with two different types of functional assays suggests that the PLS can be used as a short-term surrogate while proper evaluation makes its course.
Limitations of RENOVO include the lack of gene- and disease-specific optimization and uneven performance across variant classes, especially for missense variants. These performance losses are expected because RENOVO attempts to provide a generalized framework for variant interpretation; focusing on specific contexts would have significantly reduced overall statistical power and the possibility to generalize its application. Moreover, variant types that are not represented in ClinVar, such as those situated on gene enhancers or promoters, are not likely to be adequately classified with RENOVO because they are not included in the training set and are likely to be associated with significantly different features. Furthermore, RENOVO clearly suffers from the same ethnicity biases associated with ClinVar and as such is likely to underperform in non-Caucasian populations.34 These considerations will constitute the basis for future improvement.
SHAP analysis demonstrated the higher impact of “engineered features” (e.g., Meta-LR) after removing collinearity in the variant prioritization, with respect to raw features (e.g., mutation type). In this respect, RENOVO can be considered a meta-learner that combines the predictive ability of different learners30 and takes advantage of the work done in the past to create functional and predictive scores, which are combined with qualitative/quantitative variables proper of the variants, such as AF or type. This training strategy, together with the implementation of the missing values imputation step, enhances RENOVO performances and makes its application more generalizable than other tools: indeed, several scores could not be calculated for a large set of variants, while RENOVO is able to infer missing values on the basis of similar mutations and perform classification.
The online RENOVO interface allows us to rapidly interpret variants already reported at least in dbNSFP35 and compare each variant with ClinVar and InterVar classification. In addition, with the online RENOVO interface, all the features drawn from different public databases can be visualized in a single display, greatly facilitating clinicians in their genetics consultations.
Declaration of interests
The authors declare no competing interests.
Acknowledgments
V.F. and L.M. are supported by grant GR-2016-02361272 from the Italian Ministry of Health. G.V. and E.B. are PhD students at the European School of Molecular Medicine (SEMM), Milan, Italy. G.V.’s PhD fellowship is supported by Alliance Against Cancer funds. This work was partially supported by the Italian Ministry of Health with Ricerca Corrente and 5x1000 funds.
Published: March 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.03.010.
Data and code availability
RENOVO web application is available at https://bioserver.ieo.it/shiny/app/renovo. RENOVO code with pre-computed model weights are available at https://github.com/mazzalab-ieo/renovo.
Web resources
ClinVar ftp site, ftp://ftp.ncbi.nlm.nih.gov/pub/clinvar/vcf_GRCh37
Orange3, https://orangedatamining.com/
MLeval R package, https://github.com/crj32/MLeval
Supplemental information
References
- 1.Li Q., Wang K. InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines. Am. J. Hum. Genet. 2017;100:267–280. doi: 10.1016/j.ajhg.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kopanos C., Tsiolkas V., Kouris A., Chapple C.E., Albarca Aguilera M., Meyer R., Massouras A. VarSome: the human genomic variant search engine. Bioinforma Oxf Engl. 2019;35:1978–1980. doi: 10.1093/bioinformatics/bty897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tavtigian S.V., Greenblatt M.S., Harrison S.M., Nussbaum R.L., Prabhu S.A., Boucher K.M., Biesecker L.G., ClinGen Sequence Variant Interpretation Working Group (ClinGen SVI) Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genet. Med. 2018;20:1054–1060. doi: 10.1038/gim.2017.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Esterling L., Wijayatunge R., Brown K., Morris B., Hughes E., Pruss D., Manley S., Bowles K.R., Ross T.S. Impact of a Cancer Gene Variant Reclassification Program Over a 20-Year Period. JCO Precis. Oncol. 2020;4:944–954. doi: 10.1200/PO.20.00020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalia S.S., Adelman K., Bale S.J., Chung W.K., Eng C., Evans J.P., Herman G.E., Hufnagel S.B., Klein T.E., Korf B.R. Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): a policy statement of the American College of Medical Genetics and Genomics. Genet. Med. 2017;19:249–255. doi: 10.1038/gim.2016.190. [DOI] [PubMed] [Google Scholar]
- 7.ACMG Board of Directors The use of ACMG secondary findings recommendations for general population screening: a policy statement of the American College of Medical Genetics and Genomics (ACMG) Genet. Med. 2019;21:1467–1468. doi: 10.1038/s41436-019-0502-5. [DOI] [PubMed] [Google Scholar]
- 8.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Landrum M.J., Kattman B.L. ClinVar at five years: Delivering on the promise. Hum. Mutat. 2018;39:1623–1630. doi: 10.1002/humu.23641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rehm H.L., Berg J.S., Brooks L.D., Bustamante C.D., Evans J.P., Landrum M.J., Ledbetter D.H., Maglott D.R., Martin C.L., Nussbaum R.L. ClinGen–the Clinical Genome Resource. N. Engl. J. Med. 2015;372:2235–2242. doi: 10.1056/NEJMsr1406261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai C., Zimmer A.D., O’Connor R., Kim S., Chan R., van den Akker J., Zhou A.Y., Topper S., Mishne G. LEAP: Using machine learning to support variant classification in a clinical setting. Hum. Mutat. 2020;41:1079–1090. doi: 10.1002/humu.24011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarvik G.P., Browning B.L. Consideration of Cosegregation in the Pathogenicity Classification of Genomic Variants. Am. J. Hum. Genet. 2016;98:1077–1081. doi: 10.1016/j.ajhg.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Federici G., Soddu S. Variants of uncertain significance in the era of high-throughput genome sequencing: a lesson from breast and ovary cancers. J. Exp. Clin. Cancer Res. 2020;39:46. doi: 10.1186/s13046-020-01554-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amendola L.M., Jarvik G.P., Leo M.C., McLaughlin H.M., Akkari Y., Amaral M.D., Berg J.S., Biswas S., Bowling K.M., Conlin L.K. Performance of ACMG-AMP Variant-Interpretation Guidelines among Nine Laboratories in the Clinical Sequencing Exploratory Research Consortium. Am. J. Hum. Genet. 2016;98:1067–1076. doi: 10.1016/j.ajhg.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsons M.T., Tudini E., Li H., Hahnen E., Wappenschmidt B., Feliubadaló L., Aalfs C.M., Agata S., Aittomäki K., Alducci E., KConFab Investigators Large scale multifactorial likelihood quantitative analysis of BRCA1 and BRCA2 variants: An ENIGMA resource to support clinical variant classification. Hum. Mutat. 2019;40:1557–1578. doi: 10.1002/humu.23818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Findlay G.M., Daza R.M., Martin B., Zhang M.D., Leith A.P., Gasperini M., Janizek J.D., Huang X., Starita L.M., Shendure J. Accurate classification of BRCA1 variants with saturation genome editing. Nature. 2018;562:217–222. doi: 10.1038/s41586-018-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pugh T.J., Kelly M.A., Gowrisankar S., Hynes E., Seidman M.A., Baxter S.M., Bowser M., Harrison B., Aaron D., Mahanta L.M. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet. Med. 2014;16:601–608. doi: 10.1038/gim.2013.204. [DOI] [PubMed] [Google Scholar]
- 19.Glazer A.M., Wada Y., Li B., Muhammad A., Kalash O.R., O’Neill M.J., Shields T., Hall L., Short L., Blair M.A. High-Throughput Reclassification of SCN5A Variants. Am. J. Hum. Genet. 2020;107:111–123. doi: 10.1016/j.ajhg.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundberg S., Lee S.-I. A Unified Approach to Interpreting Model Predictions. arXiv. 2017:170507874. https://arxiv.org/abs/1705.07874 [Google Scholar]
- 22.Kuhn H.W., Tucker A.W. Princeton University Press; 2016. Contributions to the Theory of Games (AM-28), Volume II.https://books.google.it/books?id=Pd3TCwAAQBAJ [Google Scholar]
- 23.Thorsen-Meyer H.-C., Nielsen A.B., Nielsen A.P., Kaas-Hansen B.S., Toft P., Schierbeck J., Strøm T., Chmura P.J., Heimann M., Dybdahl L. Dynamic and explainable machine learning prediction of mortality in patients in the intensive care unit: a retrospective study of high-frequency data in electronic patient records. Lancet Digit Health. 2020;2:e179–e191. doi: 10.1016/S2589-7500(20)30018-2. [DOI] [PubMed] [Google Scholar]
- 24.DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 25.Sing T., Sander O., Beerenwinkel N., Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 26.Ionita-Laza I., McCallum K., Xu B., Buxbaum J.D. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Genet. 2016;48:214–220. doi: 10.1038/ng.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou J., Troyanskaya O.G. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods. 2015;12:931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thiele C., Hirschfeld G. cutpointr: Improved Estimation and Validation of Optimal Cutpoints in R. arXiv. 2020 https://arxiv.org/abs/2002.09209 200209209. [Google Scholar]
- 30.Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K., Liu X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ackerman M.J., Priori S.G., Willems S., Berul C., Brugada R., Calkins H., Camm A.J., Ellinor P.T., Gollob M., Hamilton R., Heart Rhythm Society (HRS) European Heart Rhythm Association (EHRA) HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Europace. 2011;13:1077–1109. doi: 10.1093/europace/eur245. [DOI] [PubMed] [Google Scholar]
- 32.Findlay G.M., Boyle E.A., Hause R.J., Klein J.C., Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513:120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma N., Zhang J.Z., Itzhaki I., Zhang S.L., Chen H., Haddad F., Kitani T., Wilson K.D., Tian L., Shrestha R. Determining the Pathogenicity of a Genomic Variant of Uncertain Significance Using CRISPR/Cas9 and Human-Induced Pluripotent Stem Cells. Circulation. 2018;138:2666–2681. doi: 10.1161/CIRCULATIONAHA.117.032273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Popejoy A.B., Ritter D.I., Crooks K., Currey E., Fullerton S.M., Hindorff L.A., Koenig B., Ramos E.M., Sorokin E.P., Wand H., Clinical Genome Resource (ClinGen) Ancestry and Diversity Working Group (ADWG) The clinical imperative for inclusivity: Race, ethnicity, and ancestry (REA) in genomics. Hum. Mutat. 2018;39:1713–1720. doi: 10.1002/humu.23644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X., Wu C., Li C., Boerwinkle E. dbNSFP v3.0: A One-Stop Database of Functional Predictions and Annotations for Human Nonsynonymous and Splice-Site SNVs. Hum. Mutat. 2016;37:235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RENOVO web application is available at https://bioserver.ieo.it/shiny/app/renovo. RENOVO code with pre-computed model weights are available at https://github.com/mazzalab-ieo/renovo.