Abstract
Protein aggregation, particularly in its prion-like form, has long been thought to be detrimental. However, recent studies have identified multiple instances where protein aggregation is important for normal physiological functions. Combining mass spectrometry and cell biological approaches, we developed a strategy for the identification of protein aggregates in cell lysates. We used this approach to characterize prion-based traits in pathogenic strains of the yeast Saccharomyces cerevisiae isolated from immunocompromised human patients. The proteins that we found, including the metabolic enzyme Cdc19, the translation elongation factor Yef3 and the fibrillarin homologue Nop1, are known to assemble under certain physiological conditions. Yet, such assemblies have not been reported to be stable or heritable. Our data suggest that some proteins which aggregate in response to stress have the capacity to acquire diverse assembled states, certain ones of which can be propagated across generations in a form of protein-based epigenetics.
This article is part of the theme issue ‘How does epigenetics influence the course of evolution?’
Keywords: prion, aggregate, chaperone, evolution, drug resistance
1. Introduction
Fungal pathogens are a leading cause of morbidity and mortality associated with hospital-acquired infections [1]. The problem is compounded by a growing immunocompromised population and a limited arsenal of antifungal drugs. Fungi are also extraordinarily adept at acquiring resistance to mainline therapies [2–4], and non-pathogenic fungi such as Saccharomyces cerevisiae can become pathogenic in immunocompromised patients. The mechanisms through which this occurs remain poorly understood.
Individual mutations that confer drug resistance have been mapped in a variety of fungal species. For example, mutations in components of the ergosterol biosynthetic pathway (e.g. ERG3 and ERG11) fuel the emergence of fluconazole resistance [2,5,6]. Other forms of genetic and epigenetic modifications, such as aneuploidy [7,8], the birth of new chromosomes [9] and epigenetic mechanisms [10], can also give rise to drug resistance. In practice, the molecular origin of drug resistance remains unknown in many clinical settings. Moreover, the frequency at which drug resistance arises is often higher than might be expected, given known mutation rates and the number of genes that, when mutated, produce resistance phenotypes [11]. These observations suggest that increases in mutation frequency, heritable epigenetic means of inducing drug resistance or both may enhance the capacity of fungal pathogens to acquire phenotypes that enable survival in the host.
Here, we examine enhanced drug resistance and other phenotypes in a recently evolved fungal pathogen: S. cerevisiae strains isolated from immunocompromised human patients. These strains, in contrast with those that are not pathogenic, very frequently displayed a high level of resistance to antifungal drugs and oxidative stress, or the capacity to rapidly evolve these and other traits. Combining genetic characterization, mass spectrometry and in vivo imaging, we establish that these properties can arise from prion-like epigenetic elements. Remarkably, these same proteins can form non-infectious aggregates in response to stress. Our data suggest that heritable remodelling of the aggregated proteome may provide a means for rapid adaptation and provide new insight into the pathogenic ‘transformation’ of this organism.
2. Material and methods
(a). Molecular biology and strain propagation
Yeast strains (electronic supplementary material, table S1) were obtained from stock centres or generously provided by the sources indicated. All strains were stored as glycerol stocks at −80°C and revived on yeast extract peptone dextrose (YPD) before testing. Yeast were grown in YPD at 30°C unless indicated otherwise. Yeast transformation was performed with a standard lithium–acetate protocol [12]. ‘Curing’ experiments were performed by transiently propagating cells on YPD medium containing 0.5 g l−1 guanidine hydrochloride as previously described [13]. Briefly, colonies were picked and re-streaked to single colonies three times (approx. 75 generations) on this medium. Then the strains were streaked to single colonies on YPG (with glycerol as a carbon source to eliminate any confounding petite colonies) before returning to standard YPD medium to restore Hsp104 function. Finally, the resulting ‘cured’ colonies were harvested and examined phenotypically.
Phenotypic profiles of each strain were performed as follows. Biological replicates of each yeast strain and its cured derivative were pre-grown in rich medium (YPD). We then plated three-fold serial dilutions of these strains onto YPD controls as well as onto plates containing the following stressors: clotrimazole (2 µg ml−1), ketoconazole (40 µg ml−1), hygromycin (20 µg ml−1) and tert-butyl hydroperoxide (2 mM). We inspected the plates daily and imaged them when the differences between the clinical isolates and their cured derivatives were largest. The number of days post-plating for each image is indicated in the figure legends. We also monitored the growth of these strains in liquid culture in 96-well microplates. Two independently cured lines of each clinical strain (two pairs of uncured and cured F1656 and F1660, respectively) and four biological replicates from each cured lineage were pre-grown in rich medium (YPD) to saturation. The saturated cultures were diluted by 500-fold and grown in 200 µl of YPD only as no stress control as well as in YPD medium containing the same stressors as in the assays on solid medium: clotrimazole (2 µg ml−1), YPD with ketoconazole (40 µg ml−1), YPD with hygromycin (20 µg ml−1) and YPD with tert-butyl hydroperoxide (2 mM). We measured optical density (OD)600 with shaking every 10 min for 48 h. We calculated the growth rate of each strain by fitting the logarithmic phase of the growth curve to an exponential growth equation (N = N0ekt).
(b). Isolation of protein aggregates
We extended a standard aggregate isolation protocol [14,15], starting from a single colony of a strain that was first inoculated in 5 ml YPD and grown overnight with aeration. The 5 ml YPD starter culture was then subcultured into 1 l of YPD. Cultures typically reached mid-exponential phase after 24 h when grown with shaking/aeration at 30°C. When the cultures reached an OD600 of 1.0, we harvested the cells at 3000g for 10 min and washed the pellet with deionized water twice before proceeding to the aggregate isolation step. One of the key difficulties in isolating protein aggregates is that membraneless organelles (e.g. stress granules) are also pelleted in simple centrifugation procedures. This differential centrifugation protocol attempts to overcome some of these drawbacks. As a point of comparison, isolations of stress granules [16,17] and ribosomes [18] typically involve the reciprocal of our centrifugation steps, taking pellets where we take the supernatant, and vice versa.
We lysed the washed yeast cells in a cryomill (Retsch) or using acid-washed glass beads (425–600 µm, Sigma-Aldrich, G8772-500G) with lysis buffer (30 mM Tris–HCl pH 7.5, 40 mM NaCl, 1 mM dithiothreitol (DTT), 3 mM CaCl2, 3 mM MgCl2, 5% glycerol, 1% triton X-100, containing EDTA-free protease inhibitor tablets (Roche complete Protease Inhibitor Cocktail, 11697498001)) at 1 ml g−1 of wet cell paste. We then spun the lysate at 800g for 10 min at 4°C to remove cell debris before transferring the supernatants to a new tube and treating with 1 µg ml−1 RNase A (Akron Biotech, 89508-840), and 20 units ml−1 DNase (TURBO Dnase, Invitrogen, AM2238) for 30 min on ice. We included RNase and DNase treatment to exclude proteins that aggregate exclusively as a result of binding nucleic acids. Following this incubation, we centrifuged the samples at 10 000g for 15 min at 4°C and kept the supernatant as the whole cell lysate (WCL) fraction. A fraction of this WCL was set aside for later analyses. The remainder was loaded on top of a 1 ml 40% sucrose cushion in an ultracentrifuge tube (Beckman Coulter Ultra-Clear Thinwall Tube, 344057). Higher molecular weight aggregates were then pelleted by ultracentrifugation at 200 000g for 1 h using Beckman SW55.1Ti or TLS55 swing-bucket rotors. We removed the top layers of supernatant carefully and then re-suspended the pellet in approximately 50 µl of the original lysis buffer. The resulting aggregate (AGG) fraction was analysed directly by immunoblot or re-solubilized in 8 M urea for further analysis by mass spectrometry.
(c). Mass spectrometry and data analysis
(i). For laboratory strains
Equal amounts of aggregate (12 µg by bicinchoninic acid (BCA) assay) isolated from [RNQ+][psi−], [rnq−][psi−] and [RNQ+][PSI+] were re-suspended in sodium dodecyl sulfate (SDS)-sample buffer and run through approximately 2.5 cm of a 4–15% TGX SDS–PAGE (Bio-Rad). The top 0.5 cm gel piece near the well (containing the SDS-resistant protein species) as well as the next 2 cm gel piece (containing the SDS-soluble protein species) for each sample were excised. Each gel band was reduced with 25 mM DTT, alkylated with 10 mM iodoacetamide and then digested with trypsin (5 ng µl−1) overnight, all in the presence of 50 mM ammonium bicarbonate. The overnight trypsin digestion was quenched with 5% formic acid in 50% acetonitrile. The digested peptides were recovered from the supernatant and concentrated by speed-vac to remove the acetonitrile solvent, followed by cleaning on a C18 column. The SDS-resistant and SDS-sensitive fractions for each sample were reconstituted in 8 µl and 24 µl of 0.1% formic acid, respectively, and 2 µl of each was injected onto Orbitrap Fusion Tribrid Mass Spectrometer (Thermo Fisher) for label-free quantification. Mass spectra were analysed using Proteome Discoverer v. 2.0 (Thermo Scientific) and the Byonic v. 2.6.49 search algorithm node for peptide identification and protein inference.
Electronic supplementary material, table S2 contains a list of proteins in the AGG fractions identified in laboratory strains. Specifically, the ‘TotalProtein’ sheet contains all proteins identified in the [RNQ+][psi−], [rnq−][psi−] and [RNQ+][PSI+] strains. The protein abundance as represented by matched spectra counts (PSM) for each sample was calculated by adding the peptide spectral counts in the SDS-resistant fraction and three times the observed peptide spectral counts in the SDS-sensitive fraction (equal volumes of sample were injected for liquid chromatography with tandem mass spectrometry analysis, whereas the reconstitution volume of SDS-sensitive fraction was three times larger as discussed above). Columns with ‘PSM-Sol’ and ‘PSM-Res’ suffixes contain abundance measures for the SDS-soluble and -resistant fractions, respectively, in parts per million (ppm). The reported abundance of each protein from unification of multiple quantitative proteomic datasets is listed in [19]. The SDS-resistant enrichment score was calculated as the ratio of the relative enrichment of a protein in the SDS-resistant fraction compared to SDS-soluble fraction (SDS_resistant_ppm/SDS_soluble_ppm) between two strains. The p-value was calculated based a Fisher's exact test by constructing a contingency table on the raw spectral count data (PSM) from both the SDS-resistant and SDS-soluble fractions of the two query strains.
(ii). For clinical strains
Equal amounts of aggregate (5 µg by BCA assay) isolated from F1656, F1660 and their cured derivatives using the procedure outlined above were re-suspended in SDS-sample buffer and resolved on 4–12% TGX SDS–PAGE (Bio-Rad). The gel was then stained with imperial blue Coomassie stain (Thermo Fisher) and excised into seven equal fractions for protein digestion and peptide extraction as described in the section above (laboratory strains). The extracted peptides were cleaned with a C18 column using a stage-tip, lyophilized and reconstituted before injecting onto a VelosPro ion trap mass spectrometer. Fold changes were calculated as the ratio between spectral counts in the uncured strain relative to the cured strain normalized by the ratio of the total spectral counts of uncured strain relative to the cured strain.
(d). Microscopy and image analysis
In clinical strains, proteins of interest were expressed as C-terminal enhanced green fluorescent protein (eGFP) fusions under a constitutive glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter on a low copy number plasmid (CEN NAT-GPD-ccdB-EGFP) [20]. In the prion seeding experiment (figure 4b), proteins of interest were expressed as C-terminal eGFP fusions under an inducible galactose promoter in a low copy number vector (CEN 416GAL-ccdB-EGFP) [21] in BY4741. Strains were grown overnight in 2% raffinose to OD600 of approximately 1. Images from the overnight culture were taken as a pre-induction phase reference. The strains were then washed and diluted to OD600 = 0.1 in selective medium containing 2% galactose (inducer) as a carbon source. The strains were imaged again after 6–10 h of induction. Subsequently, the strains were washed and diluted by 200-fold in 2% raffinose and imaged after 24 h of growth (outgrowth phase). Microscopy was performed using a Leica inverted fluorescence microscope with a Hammamatsu Orca 4.0 camera. The exposure time is indicated in the figure legends. All images were adjusted to a uniform contrast and illumination corrected using standard tools in ImageJ. The background was calculated using a morphological opening with a disc of radius 75 pixels, such that the structuring element was larger than cells in the foreground. The quantification of cells with puncta was performed manually for each independent experiment. On average, 150 cells were analysed and scored for the presence of foci for each strain. For each sample, the fraction of cells with at least one ‘focus’ of fluorescence was obtained by dividing the number of cells with puncta by the number of cells with eGFP-positive signal (to ensure the EGFP-fusion protein of interest is expressed). We note that for Nop1-GFP, normal localization to the nucleolus results in a substantial number of cured cells having one focus.
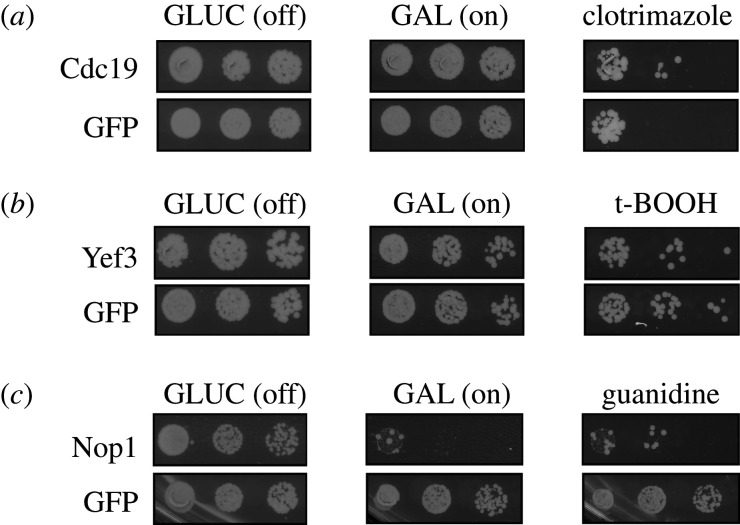
Figure 4.
Overexpression of identified proteins is linked to clinically relevant biological traits. (a) Overexpression of Cdc19 from a galactose inducible promoter led to clotrimazole resistance relative to overexpression of a GFP control protein. (b) Overexpression of Yef3 led to sensitivity to tert-butyl hydroperoxide relative to overexpression of a GFP control. (c) Overexpression of Nop1 caused pronounced toxicity that was partially ameliorated by the addition of guanidine hydrochloride (5 g l−1) to the growth medium. Five-fold serial dilutions are shown. Drug concentrations are indicated in Material and methods.
3. Results
(a). Frequent appearance of prion-like traits in clinical isolates of Saccharomyces cerevisiae
We previously analysed hundreds of wild S. cerevisiae isolates for the presence of traits with unusual patterns of inheritance that distinguish prion-based traits from those encoded in DNA [22]. For most amyloid prions, the capacity to generate protein seeds, the ‘replicons’ of prion-based inheritance, depends upon the disaggregase known as Hsp104 [23–28]. Hsp104 is a hexameric ATPase that fragments amyloid fibres into smaller pieces. These fragments can seed new rounds of assembly in daughter cells, ensuring the faithful transmission of prion-based traits [29,30]. Consequently, transient inhibition of Hsp104 permanently eliminates prion-based phenotypes [23,26,31–33].
Transient Hsp104 inhibition impacted many traits in this collection of strains [22]. However, in most cases, these strains did not harbour an amyloid conformer of a known prion protein. That is, the molecular origin of most phenotypes with prion-like patterns of inheritance remained unknown. We and others [14,15,22,34–37] have suggested that the frequent gain and loss of prions might allow them to facilitate rapid adaptation in fluctuating environments [38,39]. Saccharomyces cerevisiae is not normally a human pathogen, but it can adapt to infect immunocompromised patients [40–42]. Because these clinical strains adopt a very different lifestyle, and often must do so on rapid timescales, we investigated whether prion biology might be associated with this process.
We tested this possibility in two clinical isolates (electronic supplementary material, table S1), transiently inhibiting Hsp104 in each strain by passaging them three times through single colony bottlenecks on a rich growth medium (YPD) containing a low concentration of the Hsp104 inhibitor guanidine hydrochloride. We then restored Hsp104 function by returning these cells first to YP-glycerol (to select against respiration-deficient colonies) and then to YPD, propagating them for an additional 50 generations in total. This regimen, which robustly eliminates known amyloid prions such as [PSI+] [26,31,32], allowed us to compare each clinical isolate with its isogenic derivative that had been ‘cured’ of amyloid prions (figure 1a). We next examined these strain pairs for phenotypes that might be relevant to survival in the human host: antifungal drug resistance and resistance to oxidative stress. These traits were common and reproducible (figure 1b; electronic supplementary material, figure S1a). For example, strain F1656, isolated from bronchoalveolar lavage, was resistant to clotrimazole and the translation inhibitor hygromycin. Strain F1660, isolated from another patient, was resistant to the oxidative stressor tert-butyl hydroperoxide (t-BOOH). Each of these traits was permanently eliminated by transient inhibition of Hsp104 (figure 1b), a behaviour that is common for prion-based traits but would never be expected had these phenotypes been generated by mutations. Several of these phenotypic transformations were also evident in liquid medium (electronic supplementary material, figure S1b). Thus, these strains possessed clinically relevant traits with prion-like features.
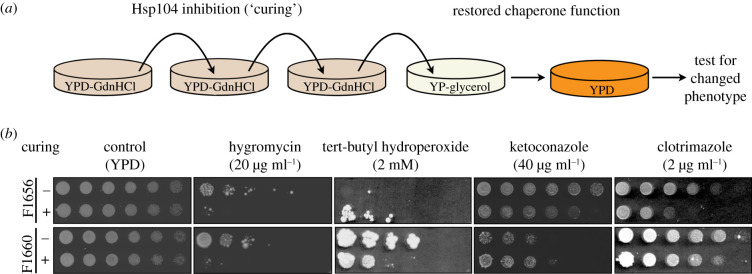
Figure 1.
Curable traits in clinical strains. (a) Schematic of the curing procedure. Strains were passaged three times on YPD medium supplemented with 0.5 g l−1 guanidine hydrochloride, followed by passage on YP-glycerol (to eliminate petites) before returning to YPD. (b) Phenotyping of the clinical strains and their cured derivatives on solid medium. All strains were inoculated in YPD and grown to OD600 approximately 1.0 before plating. Three-fold serial dilutions are shown. All drugs were supplemented in YPD plates at the concentrations indicated. The largest observed differences post-plating are shown (3 days for tert-butyl hydroperoxide, 4 days for ketoconazole and hygromycin and 9 days for clotrimazole). YPD controls confirm equivalent plating of colony-forming units. (Online version in colour.)
(b). A biochemical strategy to identify the molecular origin of the traits
We next attempted to identify proteins that were aggregated in clinical isolates, but not in their ‘cured’ derivatives. To do so we extended a differential centrifugation protocol [43,44] that physically separates amyloid protein aggregates from the soluble proteome (figure 2a). Because cells contain many large assemblies that can contaminate such purifications, we incorporated multiple centrifugation and ultracentrifugation steps (see Material and methods) that narrowed the size regime of the particles we collected to have a theoretical radius of gyration between 66 and 483 nm (particle size of approx. 160–9000 Svedberg units), which includes the most infectious prion particles [45]. We further confirmed by mass spectrometry that this fraction contains particles that are distinct from large protein complexes and membrane organelles such as ribosomes, P-bodies and stress granules. Our study had high coverage both in terms of average (32) and median (6) peptide counts per protein, resulting in identification of approximately 2600 proteins. However, markers of P-bodies (Edc3) and stress granules (Pab1, Pub1 and Ded1) were not among the most abundant proteins in the aggregate fractions (electronic supplementary material, table S2). Principle component analysis also separated the aggregated samples from total yeast proteome, establishing that this was not simply a random sampling of the proteome (electronic supplementary material, figure S2).
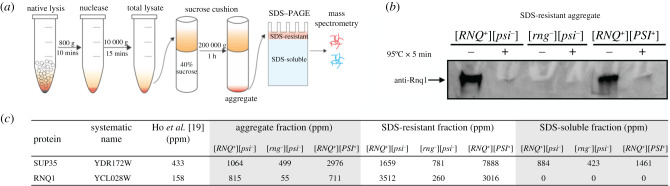
Figure 2.
Aggregate isolation protocol and validation with amyloid prion harbouring strains. (a) Aggregate isolation workflow. (b) Immunoblotting of the SDS-resistant fraction. Equal amounts of aggregates isolated from [RNQ+][psi−], [rnq−][psi−] and [RNQ+][PSI+] were re-suspended in SDS-sample buffer and split in two. One fraction was heated at 95°C for 5 min, while the other was left untreated. The samples were then resolved on SDS–PAGE and immunoblotted with an antibody against S. cerevisiae Rnq1. (c). Measured abundance of Rnq1 and Sup35 identified in the SDS-resistant and -soluble fraction of the isolated aggregates by mass spectrometry. Equal amount of aggregate isolated from [RNQ+][psi−], [rnq−][psi−] and [RNQ+][PSI+] were re-suspended in SDS-sample buffer and run through approximately 2.5 cm of a 4–15% TGX SDS–PAGE (Bio-Rad) prior to measurement by mass spectrometry. See Material and methods for further details. Expression values from total lysate across the compendium of yeast mass spectrometry experiments [19] are provided as a point of reference. (Online version in colour.)
We next tested the capacity of the differential centrifugation protocol to identify known prion proteins. To do so we employed isogenic S. cerevisiae strains that differed exclusively in the presence of amyloid polymers of Sup35, which forms the [PSI+] prion, or Rnq1, which forms the [RNQ+] prion. The purification protocol and detection by mass spectrometry could easily establish that these proteins were enriched as SDS-resistant amyloids in the [PRION+] strains (figure 2b,c; electronic supplementary material, figure S3). That is, it would have been possible to identify Sup35 and Rnq1 as amyloids in these strains without prior knowledge of their prion-like properties.
(c). Aggregating proteins in Saccharomyces cerevisiae strains derived from infected human patients
We next applied this differential centrifugation protocol to the clinical strains F1656 and F1660, isolating the aggregated fraction in each of these strains and their isogenic ‘cured’ derivatives. We then identified the proteins that were uniquely aggregated in the clinical isolates, but not their cured derivatives, using shotgun mass spectrometry. This analysis identified multiple candidate proteins in each strain (electronic supplementary material, table S3). We focused on those known to form inclusions under stressful conditions. For example, the translation elongation factor Yef3 forms foci under heat stress and nutrient starvation [46]. Nutrient starvation also causes another candidate, the pyruvate kinase Cdc19, to form reversible aggregates [47]. Cdc19 was also recently reported to form amyloid-like structures in vitro, structural forms similar to those taken up by prion proteins such as Rnq1 and Sup35 [48]. Neither have been reported to aggregate in the absence of stress or in a manner that is heritable through mitotic divisions.
We next used a cell biological approach as an orthogonal test, generating carboxy-terminal eGFP fusions with each candidate protein. We expressed these fusion proteins on a low copy number plasmid under the control of a constitutive promoter in the clinical isolates and in their isogenic cured derivatives. Several of our top-ranked candidates displayed a change in their aggregation patterns after chaperone curing.
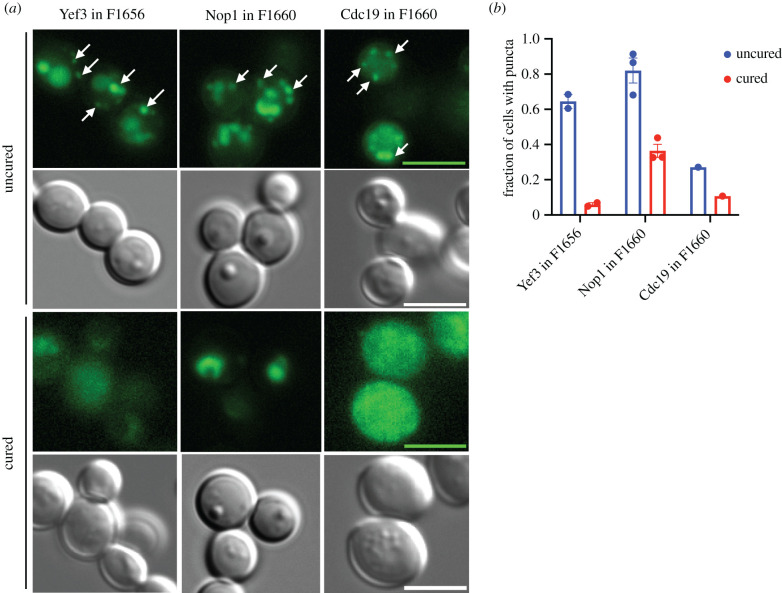
For example, Yef3-eGFP fusions formed multiple foci per cell in F1656, but in an isogenic cured strain, the localization was more diffuse. Curing reduced both Cdc19-eGFP and Nop1-eGFP foci in strain F1660 (figure 3a,b). We next tested whether these proteins influenced the phenotypes that were erased by transient guanidine hydrochloride exposure. Although the prototrophy of the clinical strains complicated direct investigation of this question, we tested how overexpression of these proteins affected the widely used laboratory strain BY4741. Overexpression of Cdc19 conferred resistance to clotrimazole and overexpression of Yef3 conferred sensitivity to t-BOOH (figure 4). The toxicity of Nop1 overexpression prevented interpretation of its phenotypic impact in the presence of stressors. However, notably, the toxicity of Nop1 overexpression was reduced in the presence of guanidine hydrochloride.
Figure 3.
Microscopy of proteins aggregated in clinical strains. (a) Proteins of interest were expressed as a C-terminal eGFP fusions under the control of a constitutive GPD promoter on a low copy number (CEN) plasmid with a nourseothricin (NAT) marker. Mid-exponential phase cells were imaged with 100× magnification. Differential interference contrast and GFP images were taken with 100 ms and 1 s exposure times, respectively. Scale bars are 5 µm. (b) Quantifications of the mean fraction of cells with puncta in each strain. Each point represents a result from an independent experiment where we counted on average approximately 150 cells per strain. Error bars represent standard error determined from three independent experiments. (Online version in colour.)
Multiple studies in the literature also support the connections between these proteins and the curable phenotypes we observed in F1656 and F1660. Classic genetic interaction studies in yeast have been limited for Cdc19 and Yef3 because of the essentiality of these proteins. However, in large-scale chemical genomics screens, heterozygous cdc19 mutants were strikingly sensitive to clotrimazole [49]. In human cancer cells, clotrimazole has been reported to reduce pyruvate kinase activity [50] to the extent that it improves the efficacy of imatinib [51] and has even been proposed as an adjuvant therapy to target the increased reliance on glycolytic metabolism that is a hallmark of cancer [52]. Yef3 and its paralogue Hef3 function as ribosomal elongation factors (eukaryotic EF3), controlling a translational response that is key to surviving oxidative stress [53,54]. Finally, Nop1 overexpression has previously been reported to slow cell cycle progression [55] and vegetative growth [56]. In sum, our combined biochemical and cell biological methodology successfully identified proteins that selectively aggregate in the clinical S. cerevisiae isolates and appear connected to at least some of the curable traits that they harbour.
(d). Cdc19 aggregates are heritable across cell divisions
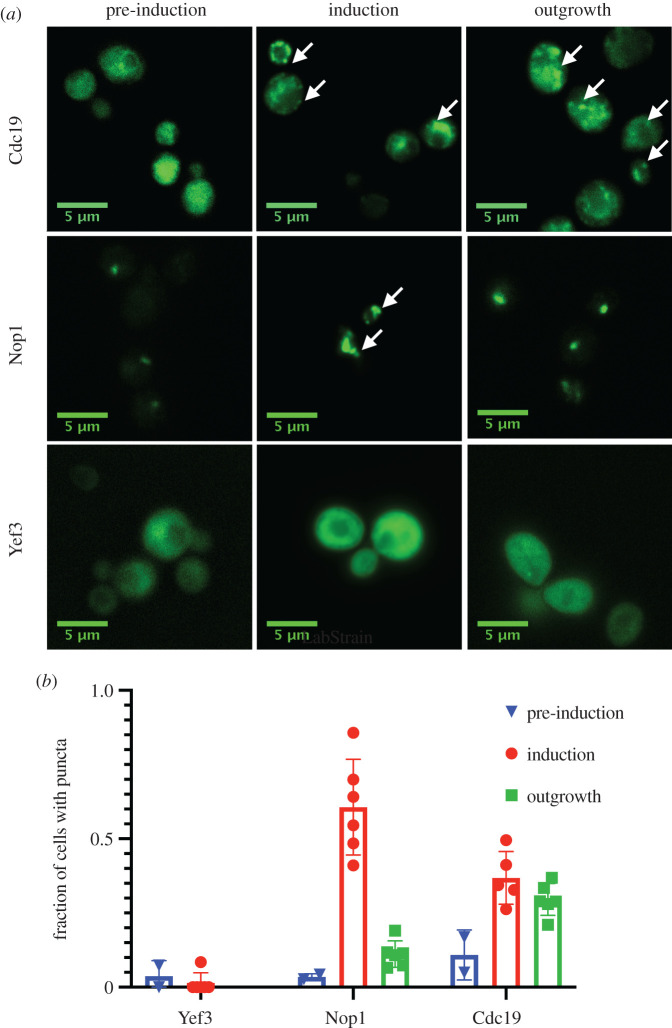
Our observation that assemblies formed by Yef3, Nop1 and Cdc19 in clinical strains were permanently eliminated by transient exposure to guanidine hydrochloride suggested that they might have prion-like properties. However, these proteins do not have any discernable N/Q-rich prion-like domains, and only Nop1 had a large disordered region [57] (electronic supplementary material, figure S4). We, therefore, examined their capacity to self-template experimentally. To do so, we used a seeding assay in which transient overexpression of the protein-eGFP fusion is used to spark a change in the distribution of GFP fluorescence [40,58]. We then examined whether this altered pattern of GFP fluorescence could be maintained over many generations, even after reduction in the levels of protein expression to an amount that previously produced diffuse patterns of fluorescence. This assay allowed us to test whether the proteins could propagate as aggregates over long biological timescales, or would instead return to a diffuse state, as would be expected if there was no prion-like hysteresis or ‘memory’ within the system (figure 5a).
Figure 5.
Heritable prion-like aggregation of candidate proteins. (a) Proteins of interest were expressed as C-terminal eGFP fusions under an inducible galactose promoter in a low copy number (CEN) plasmid in the laboratory strain BY4741. Strains were grown overnight to stationary phase in SRaffinose-URA. Images from the overnight culture were taken as a ‘pre-induction’ view. The strains were then washed and diluted to OD600 = 0.1 in 2% galactose (inducer). After 10 h, ‘induced’ images were taken. The strains were then washed and diluted by 200-fold in 2% raffinose and imaged after 24 h of ‘outgrowth’, by which time they had achieved an OD600 of approximately 1.0. (b) Quantifications of the mean fraction of cells with puncta in each strain. Each point represents a result from an independent experiment where we counted on average approximately 150 cells per strain. Error bars represent standard error determined from three independent experiments. (Online version in colour.)
In the laboratory strain in which we carried out these experiments (BY4741), Yef3 did not form visible aggregates (figure 5b,c). It is thus likely that strain F1656 harbours additional factors, perhaps conceptually akin to [PIN+] (for PSI-inducible, such as [RNQ+]) [59–62], that enable the aggregation of this protein. Multiple blots for amyloid conformers of Rnq1 in both F1656 and F1660 were negative [22]. By contrast, Nop1 and Cdc19 were diffuse at low levels of expression but formed aggregates when induced. Remarkably, Cdc19 continued to propagate as foci after withdrawal of the inducer. This was true even after substantial numbers of cell divisions over which, even assuming an infinite half-life of the Cdc19-GFP fusion, the originally overproduced protein would have been diluted substantially. These observations, combined with the chaperone dependence of the aggregates, suggest that Cdc19 can acquire a heritable, prion-like conformation in strains that have evolved to infect human hosts.
4. Discussion
Prions were originally conceived of as an explanation for a devastating neurodegenerative disease with baffling patterns of inheritance [63–68]. Although this form of transgenerational information transfer is paradigm shifting, it was also considered to be rare [69]. We previously discovered that prions imparting strong biological phenotypes, such as [PSI+] and [MOT3+], are present in wild strains of S. cerevisiae, including in clinical isolates from immunocompromised patients [22]. However, these experiments also revealed something even more surprising: abundant biological phenotypes with prion-like patterns of transmission that did not arise from any known prion protein. Here, we investigated the molecular origin of such phenotypes. Our findings suggest that S. cerevisiae may commonly employ protein aggregation, some of it prion-like, as it adapts to new environments [70–72], including human hosts.
Harbouring one prion can increase the likelihood of acquiring another [60–62] and several, such as [PSI+] and [RNQ+], commonly co-occur [22,73]. This combinatorial interdependence, along with the fact that ‘curing’ eliminates multiple prions, can complicate efforts to induce such aggregates in laboratory strains. Although we could easily observe amyloid conformers of relatively rare proteins such as Rnq1 (approx. 1000 molecules per cell [74]), like all proteomic approaches, our method is fundamentally limited by a protein's natural abundance. Obtaining high coverage will thus be required to identify prion proteins that are expressed at low levels.
Cdc19 has previously been shown to form reversible aggregates in vivo, regulated in part by phosphorylation that, in turn, exerts cell cycle control [47]. In vitro, it forms amyloid-like fibrillar aggregates [48]. Our data here demonstrated that in clinical fungal isolates, Cdc19 can form stable aggregates that self-template. We do not yet know whether or how these heritable aggregates in clinical strains might differ from physiological aggregation of the protein. But Cdc19 appears to commonly aggregate in clinical isolates, emerging in both that we studied here and two other clinical isolates YJM428 and YJM653 previously examined [43]. We also note that Pma1 (found in both F1656 and F1660 in this study) appeared in the aggregate fraction of other different strains that have previously been studied (DK365, I14, F1535 and YJM653) Pma1 aggregation is associated with the [GAR+] prion [75] and we have previously observed that some natural soil isolates of S. Cerevisiae naturally harbour this element [76]. In future work it will be interesting whether [GAR+] also plays an important role in clinical strains.
Transient inhibition of Hsp104 cures most amyloid prions [29,30], but we and others have recently found other types of prion-like aggregation that do not involve the formation of large amyloid aggregates [77–79]. Further refinements will be necessary to develop proteomic strategies that identify such assemblies and ideally also characterize their conformational diversity. In this regard, integration with structural proteomic strategies such as limited proteolysis coupled mass spectrometry [80] may provide a particularly promising route forward. Because prions' adaptive benefit is derived from their heritability and reversibility, it will also be important to choose a diverse array of strains so that it is possible to ‘catch’ these protein-based heritable elements.
5. Conclusion
Two proteins that we identified, the pyruvate kinase Cdc19 and the translation elongation factor Yef3, have previously been found to aggregate in response to stress. Cdc19 forms large assemblies in response to nutrient deprivation [47]; Yef3 does so in response to heat stress [46]. Also, by homology to fibrillarin [81], Nop1 is likely to form a phase-separated liquid droplet. None of these assemblies have been reported to be heritable. Even the archetypical yeast prion protein, the translation termination factor Sup35, can form stress-induced aggregates that do not appear to be infectious [82].
Our observations suggest that the capacity to form multiple types of aggregated species, some of which are heritable, and some of which are not, may be a common property of such proteins (figure 6). Understanding the interconversion among different aggregated forms, and its potential regulation, stands as a goalpost for future studies. Regardless, our data suggest that S. cerevisiae can harness this diversity of conformations—and the phenotypes they encode—as an epigenetic strategy to adapt to new niches.
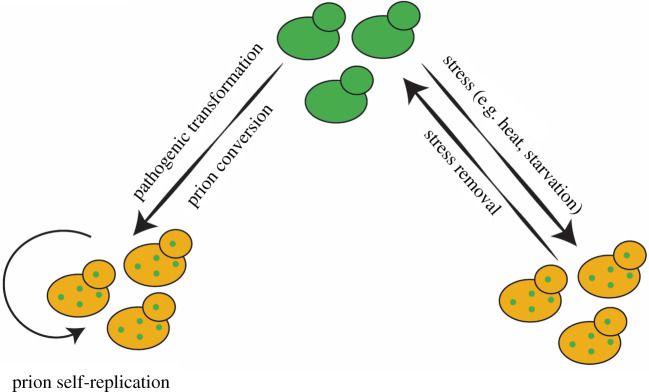
Figure 6.
Model for reversible versus heritable protein aggregation in the context of evolutionary adaptation. When stresses are transient, reversible adaptation via protein aggregation is favoured. However, when stresses become more constant, such as during pathogenic transformation, proteins that formerly aggregated reversibly may acquire the capacity to do so heritably. (Online version in colour.)
Acknowledgements
We thank Lucy Xie, Shady Saad, Anne Brunet, Itamar Harel and Gene Yeo for helpful discussions and critical review of the manuscript. We also thank Soledad Larios for providing a continuous supply of media to support the work.
Data accessibility
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary materials. Any additional supporting materials or information are available from the corresponding author, D.F.J., upon reasonable request.
Authors' contributions
Y.R.C., I.Z. and D.F.J. conceived of the study. Y.R.C., I.Z., K.S. and J.E.E. performed the mass spectrometry experiments. Y.R.C. carried out all additional experiments and data analysis. Y.R.C. and D.F.J. wrote the manuscript.
Competing interests
We declare we have no competing interests.
Funding
Y.R.C. was supported by a Stanford Graduate Fellowship. This work was also supported by National Institutes of Health (DP2-GM119140 and RF1-AG057334) and National Science Foundation (NSF-MCB116762) grants to D.F.J. as well as by seed grants from the Stanford Alzheimer's Disease Research Center (funded by 5P50AG047366-02 and by generous support from the Zaffaroni Family). D.F.J. also received support from a Science and Engineering Fellowship of the David and Lucile Packard Foundation, the Glenn Foundation for Medical Research and the Bert and Kuggie Vallee Foundation.
References
- 1.Pfaller MA, Diekema DJ. 2010. Epidemiology of invasive mycoses in North America. Crit. Rev. Microbiol. 36, 1-53. ( 10.3109/10408410903241444) [DOI] [PubMed] [Google Scholar]
- 2.Cowen LE, Lindquist S. 2005. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science 309, 2185-2189. ( 10.1126/science.1118370) [DOI] [PubMed] [Google Scholar]
- 3.Cowen LE. 2008. The evolution of fungal drug resistance: modulating the trajectory from genotype to phenotype. Nat. Rev. Microbiol. 6, 187-198. ( 10.1038/nrmicro1835) [DOI] [PubMed] [Google Scholar]
- 4.Shapiro RS, Robbins N, Cowen LE. 2011. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol. Mol. Biol. Rev. 75, 213-267. ( 10.1128/MMBR.00045-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson JB, Sirjusingh C, Parsons AB, Boone C, Wickens C, Cowen LE, Kohn LM. 2003. Mode of selection and experimental evolution of antifungal drug resistance in Saccharomyces cerevisiae. Genetics 163, 1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cowen LE, Carpenter AE, Matangkasombut O, Fink GR, Lindquist S. 2006. Genetic architecture of Hsp90-dependent drug resistance. Eukaryot. Cell 5, 2184-2188. ( 10.1128/EC.00274-06). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selmecki AM, Dulmage K, Cowen LE, Anderson JB, Berman J. 2009. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet. 5, e1000705. ( 10.1371/journal.pgen.1000705). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ford CB, et al. 2015. The evolution of drug resistance in clinical isolates of Candida albicans. eLife 4, e00662. ( 10.7554/eLife.00662). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmad KM, Ishchuk OP, Hellborg L, Jorgensen G, Skvarc M, Stenderup J, Jorck-Ramberg D, Polakova S, Piskur J. 2013. Small chromosomes among Danish Candida glabrata isolates originated through different mechanisms. Antonie Van Leeuwenhoek 104, 111-122. ( 10.1007/s10482-013-9931-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben-Ami R, Zimmerman O, Finn T, Amit S, Novikov A, Wertheimer N, Lurie-Weinberger M, Berman J. 2016. Heteroresistance to fluconazole is a continuously distributed phenotype among Candida glabrata clinical strains associated with in vivo persistence. mBio 7, e00655-16. ( 10.1128/mBio.00655-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hillenmeyer ME, et al. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320, 362-365. ( 10.1126/science.1150021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gietz RD, Schiestl RH, Willems AR, Woods RA. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355-360. ( 10.1002/yea.320110408) [DOI] [PubMed] [Google Scholar]
- 13.Alberti S, Halfmann R, Lindquist S. 2010. Biochemical, cell biological, and genetic assays to analyze amyloid and prion aggregation in yeast. Methods Enzymol. 470, 709-734. ( 10.1016/S0076-6879(10)70030-6) [DOI] [PubMed] [Google Scholar]
- 14.Kryndushkin D, Pripuzova N, Burnett BG, Shewmaker F. 2013. Non-targeted identification of prions and amyloid-forming proteins from yeast and mammalian cells. J. Biol. Chem. 288, 27 100-27 111. ( 10.1074/jbc.M113.485359). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kryndushkin D, Pripuzova N, Shewmaker FP. 2017. Isolation and analysis of prion and amyloid aggregates from yeast cells. Cold Spring Harb Protoc 2017, pdb prot089045. ( 10.1101/pdb.prot089045) [DOI] [PubMed] [Google Scholar]
- 16.Wheeler JR, Jain S, Khong A, Parker R. 2017. Isolation of yeast and mammalian stress granule cores. Methods 126, 12-17. ( 10.1016/j.ymeth.2017.04.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, Parker R. 2017. The stress granule transcriptome reveals principles of mRNA accumulation in stress granules. Mol. Cell 68, 808-820. ( 10.1016/j.molcel.2017.10.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. 2012. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. Protoc. 7, 1534-1550. ( 10.1038/nprot.2012.086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho B, Baryshnikova A, Brown GW. 2018. Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Syst. 6, 192-205. ( 10.1016/j.cels.2017.12.004) [DOI] [PubMed] [Google Scholar]
- 20.Newby GA, et al. 2017. A genetic tool to track protein aggregates and control prion inheritance. Cell 171, 966-979. ( 10.1016/j.cell.2017.09.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alberti S, Gitler AD, Lindquist S. 2007. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 24, 913-919. ( 10.1002/yea.1502) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. 2012. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature 482, 363-368. ( 10.1038/nature10875) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. 1995. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 268, 880-884. ( 10.1126/science.7754373) [DOI] [PubMed] [Google Scholar]
- 24.Patino MM, Liu JJ, Glover JR, Lindquist S. 1996. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 273, 622-626. ( 10.1126/science.273.5275.622) [DOI] [PubMed] [Google Scholar]
- 25.Glover JR, Lindquist SL. 1998. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell 94, 73-82. ( 10.1016/S0092-8674(00)81223-4) [DOI] [PubMed] [Google Scholar]
- 26.Ferreira PC, Ness F, Edwards SR, Cox BS, Tuite MF. 2001. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol. Microbiol. 40, 1357-1369. ( 10.1046/j.1365-2958.2001.02478.x) [DOI] [PubMed] [Google Scholar]
- 27.Kryndushkin DS, Alexandrov IM, Ter-Avanesyan MD, Kushnirov VV. 2003. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 278, 49 636-49 643. ( 10.1074/jbc.M307996200) [DOI] [PubMed] [Google Scholar]
- 28.Shorter J, Lindquist SL. 2004. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science 304, 1793-1797. ( 10.1126/science.1098007) [DOI] [PubMed] [Google Scholar]
- 29.Chernoff YO. 2007. Stress and prions: lessons from the yeast model. FEBS Lett. 581, 3695-3701. ( 10.1016/j.febslet.2007.04.075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sweeny EA, Shorter J. 2008. Prion proteostasis: Hsp104 meets its supporting cast. Prion 2, 135-140. ( 10.4161/pri.2.4.7952) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cox BS, Tuite MF, Mundy CJ. 1980. Reversion from suppression to nonsuppression in SUQ5 [psi+] strains of yeast: the classification of mutations. Genetics 95, 589-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eaglestone SS, Ruddock LW, Cox BS, Tuite MF. 2000. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI(+)] of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 97, 240-244. ( 10.1073/pnas.97.1.240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jung G, Masison DC. 2001. Guanidine hydrochloride inhibits Hsp104 activity in vivo: a possible explanation for its effect in curing yeast prions. Curr. Microbiol. 43, 7-10. ( 10.1007/s002840010251) [DOI] [PubMed] [Google Scholar]
- 34.Byers JS, Jarosz DF. 2014. Pernicious pathogens or expedient elements of inheritance: the significance of yeast prions. PLoS Pathog. 10, e1003992. ( 10.1371/journal.ppat.1003992) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.True HL, Lindquist SL. 2000. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407, 477-483. ( 10.1038/35035005) [DOI] [PubMed] [Google Scholar]
- 36.Lancaster AK, Bardill JP, True HL, Masel J. 2010. The spontaneous appearance rate of the yeast prion [PSI+] and its implications for the evolution of the evolvability properties of the [PSI+] system. Genetics 184, 393-400. ( 10.1534/genetics.109.110213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia DM, Jarosz DF. 2014. Rebels with a cause: molecular features and physiological consequences of yeast prions. FEMS Yeast Res. 14, 136-147. ( 10.1111/1567-1364.12116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Itakura AK, Chakravarty AK, Jakobson CM, Jarosz DF. 2020. Widespread prion-based control of growth and differentiation strategies in Saccharomyces cerevisiae. Mol. Cell 77, 266-278. ( 10.1016/j.molcel.2019.10.027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chakravarty AK, Smejkal T, Itakura AK, Garcia DM, Jarosz DF. 2020. A non-amyloid prion particle that activates a heritable gene expression program. Mol. Cell 77, 251-265. ( 10.1016/j.molcel.2019.10.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Byers JS, Jarosz DF. 2017. High-throughput screening for protein-based inheritance in S. cerevisiae. J. Vis. Exp. 126, 56069. ( 10.3791/56069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jain S, Wheeler JR, Walters RW, Agrawal A, Barsic A, Parker R. 2016. ATPase-modulated stress granules contain a diverse proteome and substructure. Cell 164, 487-498. ( 10.1016/j.cell.2015.12.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Llanos R, Llopis S, Molero G, Querol A, Gil C, Fernandez-Espinar MT.. 2011. In vivo virulence of commercial Saccharomyces cerevisiae strains with pathogenicity-associated phenotypical traits. Int. J. Food Microbiol. 144, 393-399. ( 10.1016/j.ijfoodmicro.2010.10.025) [DOI] [PubMed] [Google Scholar]
- 43.King OD, Masel J. 2007. The evolution of bet-hedging adaptations to rare scenarios. Theor. Popul. Biol. 72, 560-575. ( 10.1016/j.tpb.2007.08.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lancaster AK, Masel J. 2009. The evolution of reversible switches in the presence of irreversible mimics. Evolution 63, 2350-2362. ( 10.1111/j.1558-5646.2009.00729.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marchante R, Beal DM, Koloteva-Levine N, Purton TJ, Tuite MF, Xue WF. 2017. The physical dimensions of amyloid aggregates control their infective potential as prion particles. eLife 6, e27109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wallace EW, et al. 2015. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162, 1286-1298. ( 10.1016/j.cell.2015.08.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saad S, Cereghetti G, Feng Y, Picotti P, Peter M, Dechant R. 2017. Reversible protein aggregation is a protective mechanism to ensure cell cycle restart after stress. Nat. Cell Biol. 19, 1202-1213. ( 10.1038/ncb3600) [DOI] [PubMed] [Google Scholar]
- 48.Grignaschi E, Cereghetti G, Grigolato F, Kopp MR. G., Caimi S, Faltova L, Saad S, Peter M, Arosio P. 2018. A hydrophobic low-complexity region regulates aggregation of the yeast pyruvate kinase Cdc19 into amyloid-like aggregates in vitro. J. Biol. Chem. 293, 11 424-11 432. ( 10.1074/jbc.RA117.001628) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee AY, et al. 2014. Mapping the cellular response to small molecules using chemogenomic fitness signatures. Science 344, 208-211. ( 10.1126/science.1250217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Furtado CM, Marcondes MC, Sola-Penna M, de Souza ML, Zancan P.. 2012. Clotrimazole preferentially inhibits human breast cancer cell proliferation, viability and glycolysis. PLoS ONE 7, e30462. ( 10.1371/journal.pone.0030462) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Motawi TM, Sadik NA, Fahim SA, Shouman SA. 2015. Combination of imatinib and clotrimazole enhances cell growth inhibition in T47D breast cancer cells. Chem. Biol. Interact. 233, 147-156. ( 10.1016/j.cbi.2015.03.028) [DOI] [PubMed] [Google Scholar]
- 52.Kadavakollu S, Stailey C, Kunapareddy CS, White S. 2014. Clotrimazole as a cancer drug: a short review. Med. Chem. (Los Angeles) 4, 722-724. ( 10.4172/2161-0444.1000219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goscinska K, Shahmoradi Ghahe S, Domogala S, Topf U. 2020. Eukaryotic elongation factor 3 protects Saccharomyces cerevisiae yeast from oxidative stress. Genes (Basel) 11, 1432-1446. ( 10.3390/genes11121432) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shenton D, Smirnova JB, Selley JN, Carroll K, Hubbard SJ, Pavitt GD, Ashe MP, Grant CM. 2006. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J. Biol. Chem. 281, 29 011-29 021. ( 10.1074/jbc.M601545200) [DOI] [PubMed] [Google Scholar]
- 55.Stevenson LF, Kennedy BK, Harlow E. 2001. A large-scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes. Proc. Natl Acad. Sci. USA 98, 3946-3951. ( 10.1073/pnas.051013498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yoshikawa K, Tanaka T, Ida Y, Furusawa C, Hirasawa T, Shimizu H. 2011. Comprehensive phenotypic analysis of single-gene deletion and overexpression strains of Saccharomyces cerevisiae. Yeast 28, 349-361. ( 10.1002/yea.1843) [DOI] [PubMed] [Google Scholar]
- 57.Jones DT, Cozzetto D. 2015. DISOPRED3: precise disordered region predictions with annotated protein-binding activity. Bioinformatics 31, 857-863. ( 10.1093/bioinformatics/btu744) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chakrabortee S, et al. 2016. Intrinsically disordered proteins drive heritable transformations of biological traits. Cell 167, 369-381. ( 10.1016/j.cell.2016.09.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Derkatch IL, Bradley ME, Masse SV, Zadorsky SP, Polozkov GV, Inge-Vechtomov SG, Liebman SW. 2000. Dependence and independence of [PSI(+)] and [PIN(+)]: a two-prion system in yeast? EMBO J. 19, 1942-1952. ( 10.1093/emboj/19.9.1942) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Derkatch IL, Bradley ME, Hong JY, Liebman SW. 2001. Prions affect the appearance of other prions: the story of [PIN(+)]. Cell 106, 171-182. ( 10.1016/S0092-8674(01)00427-5) [DOI] [PubMed] [Google Scholar]
- 61.Patel BK, Liebman SW. 2007. ‘Prion-proof’ for [PIN+]: infection with in vitro-made amyloid aggregates of Rnq1p-(132–405) induces [PIN+]. J. Mol. Biol. 365, 773-782. ( 10.1016/j.jmb.2006.10.069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vitrenko YA, Gracheva EO, Richmond JE, Liebman SW. 2007. Visualization of aggregation of the Rnq1 prion domain and cross-seeding interactions with Sup35NM. J. Biol. Chem. 282, 1779-1787. ( 10.1074/jbc.M609269200) [DOI] [PubMed] [Google Scholar]
- 63.Prusiner SB. 1982. Novel proteinaceous infectious particles cause scrapie. Science 216, 136-144. ( 10.1126/science.6801762) [DOI] [PubMed] [Google Scholar]
- 64.Prusiner SB, Gajdusek C, Alpers MP. 1982. Kuru with incubation periods exceeding two decades. Ann. Neurol. 12, 1-9. ( 10.1002/ana.410120102) [DOI] [PubMed] [Google Scholar]
- 65.Gajdusek DC, Zigas V. 1957. Degenerative disease of the central nervous system in New Guinea; the endemic occurrence of kuru in the native population. N. Engl. J. Med. 257, 974-978. ( 10.1056/NEJM195711142572005) [DOI] [PubMed] [Google Scholar]
- 66.Hulland TJ. 1960. Scrapie and kuru. Can. Vet. J. 1, 496. [PMC free article] [PubMed] [Google Scholar]
- 67.Griffith JS. 1967. Self-replication and scrapie. Nature 215, 1043-1044. ( 10.1038/2151043a0) [DOI] [PubMed] [Google Scholar]
- 68.Alper T, Cramp WA, Haig DA, Clarke MC. 1967. Does the agent of scrapie replicate without nucleic acid? Nature 214, 764-766. ( 10.1038/214764a0) [DOI] [PubMed] [Google Scholar]
- 69.Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. 2005. Yeast prions URE3] and [PSI+] are diseases. Proc. Natl Acad. Sci. USA 102, 10 575-10 580. ( 10.1073/pnas.0504882102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Du Z, Park KW, Yu H, Fan Q, Li L.. 2008. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat. Genet. 40, 460-465. ( 10.1038/ng.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Du Z, Crow ET, Kang HS, Li L.. 2010. Distinct subregions of Swi1 manifest striking differences in prion transmission and SWI/SNF function. Mol. Cell. Biol. 30, 4644-4655. ( 10.1128/MCB.00225-10) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Harvey ZH, Chen Y, Jarosz DF. 2018. Protein-based inheritance: epigenetics beyond the chromosome. Mol. Cell 69, 195-202. ( 10.1016/j.molcel.2017.10.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Saupe SJ, Jarosz DF, True HL. 2016. Amyloid prions in fungi. Microbiol. Spectr. 4, 29-42. ( 10.1128/Microbiolspec.Funk-0029-2016) [DOI] [PubMed] [Google Scholar]
- 74.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. 2003. Global analysis of protein expression in yeast. Nature 425, 737-741. ( 10.1038/nature02046) [DOI] [PubMed] [Google Scholar]
- 75.Brown JC, Lindquist S. 2009. A heritable switch in carbon source utilization driven by an unusual yeast prion. Genes Dev. 23, 2320-2332. ( 10.1101/gad.1839109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jarosz DF, Lancaster AK, Brown JC, Lindquist S. 2014. An evolutionarily conserved prion-like element converts wild fungi from metabolic specialists to generalists. Cell 158, 1072-1082. ( 10.1016/j.cell.2014.07.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jarosz DF, et al. 2014. Cross-kingdom chemical communication drives a heritable, mutually beneficial prion-based transformation of metabolism. Cell 158, 1083-1093. ( 10.1016/j.cell.2014.07.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garcia DM, Dietrich D, Clardy J, Jarosz DF. 2016. A common bacterial metabolite elicits prion-based bypass of glucose repression. eLife 5, e17978. ( 10.7554/eLife.17978) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Harvey ZH, Chakravarty AK, Futia RA, Jarosz DF. 2020. A prion epigenetic switch establishes an active chromatin state. Cell 180, 928-940. ( 10.1016/j.cell.2020.02.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Feng Y, et al. 2014. Global analysis of protein structural changes in complex proteomes. Nat. Biotechnol. 32, 1036-1044. ( 10.1038/nbt.2999) [DOI] [PubMed] [Google Scholar]
- 81.Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP. 2016. Coexisting liquid phases underlie nucleolar subcompartments. Cell 165, 1686-1697. ( 10.1016/j.cell.2016.04.047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Franzmann TM, et al. 2018. Phase separation of a yeast prion protein promotes cellular fitness. Science 359, aao5654. ( 10.1126/science.aao5654) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article and/or its supplementary materials. Any additional supporting materials or information are available from the corresponding author, D.F.J., upon reasonable request.