Abstract
The extraordinary diversity in molluscan body plans, and the genomic mechanisms that enable it, remains one of the great questions of evolution. The eight distinct living taxonomic classes of molluscs are each unambiguously monophyletic; however, significant controversy remains about the phylogenetic relationships among those eight branches. Molluscs are the second-largest animal phylum, with over 100 000 living species with broad biological, economic and medical interest. To date, only around 53 genome assemblies have been accessioned to NCBI GenBank covering only four of the eight living molluscan classes. Furthermore, the molluscan taxa where partial or whole-genome assemblies are available are often aberrantly fast evolving or recently derived lineages. Characteristic adaptations provide interesting targets for whole-genome projects, in animals like the scaly-foot snail or octopus, but without basal-branching lineages for comparison, the context of recently derived features cannot be assessed. The currently available genomes also create a non-optimal set of taxa for resolving deeper phylogenetic branches: they are a small sample representing a large group, and those that are available come primarily from a rarefied pool. Thoughtful selection of taxa for future projects should focus on the blank areas of the molluscan tree, which are ripe with opportunities to delve into peculiarities of genome evolution, and reveal the biology and evolutionary history of molluscs.
This article is part of the Theo Murphy meeting issue ‘Molluscan genomics: broad insights and future directions for a neglected phylum’.
Keywords: Aculifera, Conchifera, Mollusca, Serialia, diversification, disparity
1. Background
Molluscs are a large and ubiquitous animal group with broad biological, economic and medical interest. Molluscs are the foundational group used to establish fundamental principles of evolutionary mechanisms [1], global biogeography [2] and past mass extinctions [3]. Bivalves are a rapidly growing aspect of global fisheries. The sea hare Aplysia and the squid Loligo have been important model organisms in neurobiological research. Molluscs also are some of the most notorious agricultural pests when they become invasive, and seashells have enthralled humans for millennia. Terrestrial and freshwater molluscs have suffered greater rates of modern extinction than any other animal group [4]. But what is most fascinating about molluscs is their extraordinary morphological diversity: the eight living taxonomic classes comprise an unparalleled diversity of surprising animal forms, from microscopic worm-molluscs to giant squid.
Given the context of a staggeringly long fossil record, and extreme morphological disparity in living species, it is perhaps unsurprising that the backbone of pan-molluscan phylogeny remains controversial despite extensive study. The eight distinct living taxonomic classes of molluscs are each unambiguously monophyletic; however, significant controversy remains about the topology of the phylogenetic relationships among those eight branches [5]. There is a high level of conflict among the results from different molecular phylogenetic approaches: ‘classical' molecular phylogenetic analyses with few gene fragments but high taxon sampling, transcriptomic approaches with intermediate data and limited taxon sampling, or true phylogenomic analyses using genome assemblies that have high data volume but very low taxon sampling. The analysis with the most comprehensive taxon sampling across all molluscs [6] recovered a topology that is largely unsupported by later transcriptomic analyses with very limited taxon sampling (e.g. [7]). In the case of molluscs, there are certain well-resolved relationships of proximity, such as a close relationship between the chitons (Polyplacophora) and aplacophoran molluscs [8], but it is unclear whether any of the conflicting phylogenies for total-group Mollusca represent a reliable reconstruction of evolutionary history [5]. More intensive work within specific clades of molluscs, and more broadly in other organismal groups, has repeatedly reaffirmed that insufficient taxon sampling can lead to inaccurate phylogenetic reconstruction, especially for complex groups [9]. The issues of molluscan phylogeny cannot therefore be solved by waiting for a new method or technology, and it is not a simple problem of conflict between traditional and revised ideas of phylogenetic relationships.
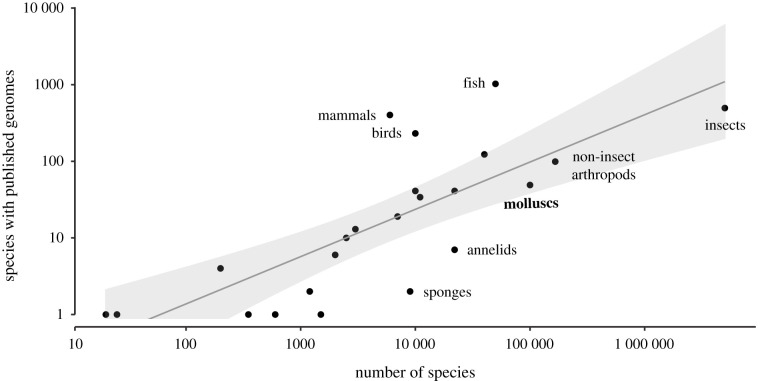
There is hope that whole-genome phylogenomics will provide the decisive tool that will produce a confident resolution of pan-molluscan branching patterns, but whole-genome assemblies are currently too sparse to provide any meaningful analysis [10]. Relatively few molluscs have been the subject of complete genomes: among around 100 000 known living species, only 53 partial or complete genomes have been made publicly available at time of writing. That sampling rate is similar to other major animal groups (figure 1), yet mollusc genomes in particular have become available only very recently: half of those (25 genomes) have been published since 2019. Our concern is that the sampling is very unevenly distributed across molluscan diversity, representing only four of the eight living classes: gastropods, bivalves, cephalopods and, most recently, the first genome of Polyplacophora [12]. More genomes are needed to progress the goal of resolving molluscan phylogeny, and those genomes must be selected strategically to represent the diverse clades within molluscs.
Figure 1.
Comparison of global animal diversity and published genome sequencing efforts. Those that fall below the regression line are comparatively under-sampled. Well-studied groups (mammals, birds) have high availability of genomic data compared to their global species richness. Updated from Sigwart et al. [11] with data from NCBI GenBank as of 1 September 2020.
Among molluscs, the reasons for the deep and unresolvable conflict between different phylogenetic hypotheses are apparently in part attributable to their extreme morphological disparity. Understanding the origins of this disparity is connected to understanding molluscan genomes; however, molluscan genomes have proven to be challenging on many different levels. There are certainly other taxa that are also complicated, in terms of morphological variation and genomics, so this is not a feature unique to this phylum. Some aspects of the complexity that confound current studies of mollusc genomes may in fact enable us to understand the very nature of molluscan diversification.
Within the eight individual classes, meanwhile, substantial progress in the last decade has established reasonably robust phylogenetic frameworks for chitons, aplacophorans, cephalopods, bivalves and many gastropod groups as well as total-group Gastropoda (e.g. [13–16]). The existing knowledge of molluscan phylogeny within each class can be used to suggest an ideal set of taxa for further genome projects (see also [5]), that would rapidly lead to a more informative phylogenomic approach for the community. As part of a strategy to eventually resolve molluscan phylogeny, we consider the potential barriers to producing new genome assemblies of diverse molluscs, and we propose a strategic set of targets that are individually interesting and would collectively advance the mission to understand animal evolution.
2. Challenges in molluscan genomes
Although not unique to molluscs, it should be noted that every step of the process in molluscan genomics is fraught with practical challenges and bottlenecks: from finding specimens and access to their habitats, to proper preservation, initial DNA extraction, sequencing, assembly and analysis, to publishing results in a landscape increasingly dominated by comparative approaches using more densely sampled data. It would be naïve to think that assembling molluscan genomes has become easy, but recent advances in methods and sequencing technology mean that it is now realistically feasible.
The rapid advancement of DNA sequencing technology has brought in third-generation sequencing approaches such as PacBio and Oxford Nanopore Technologies (ONT) that allow for long-read sequencing, but mollusc genomes are still challenging to sequence and assemble in a number of ways. To begin with, extracting high molecular weight (HMW) genomic DNA in high purity from molluscan tissue itself is still problematic, mainly due to the high mucus content and therefore polysaccharide contamination [17]. Failure to extract intact, high-quality HMW DNA inevitably results in shorter read length in the downstream sequencing. Although an additional cleaning step is recommended to remove the polysaccharides (e.g. [10]), this also causes DNA fragmentation and makes it very difficult to achieve ‘ultra-long' DNA reads as has become possible with the human genome. For instance, the longest DNA read in Mollusca so far recorded is 259.9 kb from the Mytilus coruscus (=M. unguiculatus) genome [18], an order of magnitude shorter than the human cell line world record of 2.27 Mb [19]. Optimization of HMW DNA extraction methods for molluscs is needed in order to achieve ultra-long reads.
Two main issues hinder the high-quality assembly of molluscan genomes, namely high heterozygosity and high repeat content. Researchers have often had to develop a pure line through inbreeding to reduce the heterozygosity, such as for the oyster genome project [20], which takes many years to accomplish and is impossible for species that cannot be kept and bred in captivity. The highest heterozygosity recorded to date among available molluscan genomes is 3.66% in the sea slug Elysia chlorotica, compared to human genome with usually lower than 0.1% heterozygosity [21]. The high prevalence of repeat content in molluscan genomes is also very problematic. Although long reads have been helpful in resolving repetitive elements, heterozygous regions and structural variants lead to regions that cannot be placed into an assembly. In molluscs, these regions are often longer than the DNA reads generated from molluscs, due to the complications of the high polysaccharide content mentioned above. Taking the genome sequencing of the bobtail squid Euprymna scolopes as an example, although tremendous efforts were used to combine different methods (Illumina, PacBio and Dovetail's Chicago library), one-third of the assembly was found to be ‘gaps' or unknown sequences [22].
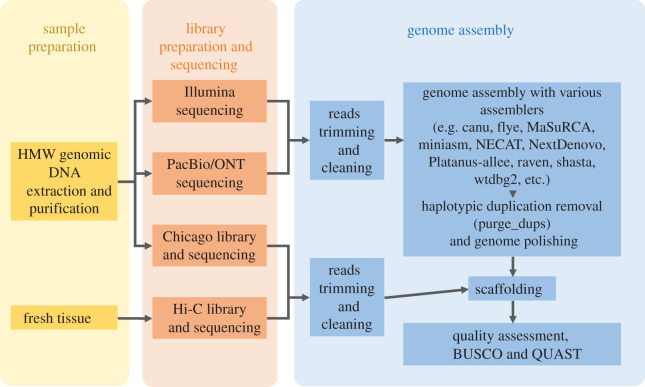
The current strategy for assembling a high-quality mollusc genome (e.g. [10]) requires the combination of multiple technologies (figure 2). Practical complications and continuously advancing technology mean that it is currently impossible to describe or propose a more straightforward analytical pipeline. The HMW DNA extracted is sequenced on the short-read Illumina platform and combined with either PacBio or ONT long-read platforms. The resultant reads are cleaned and assembled with a number of available genomic assemblers, while removing heterozygous contigs and correcting for sequencing errors. In order to achieve a chromosomal-level assembly, a Chicago library and/or Hi-C library must be prepared to scaffold the contigs. Completeness and continuity of the assembly are then assessed with QUAST and BUSCO (Benchmarking Universal Single-Copy Orthologs). The exact processes and tools to use are far from streamlined, and the pipeline used has varied from species to species in published molluscan genomes. A fully benchmarked comparison of available mollusc genomes using all available genome assemblers would be required in order to arrive at a standard process optimized for molluscs, and even then it is not clear that the currently available set of molluscan genomes would have sufficient quality or taxonomic coverage to be representative of the genomic diversity encountered more broadly across the phylum [23].
Figure 2.
A schematic showing the current practice in the assembly of a high-quality mollusc genome. (Online version in colour.)
The technical challenges and costs associated with sequencing a genome are closely linked with its size, heterozygosity and repeat contents. Estimates for these numbers are, however, sparsely available across Mollusca and appear to be greatly variable within groups [24], even for predictions of the same genus or even species. For example, the genome size of three scaphopods has been estimated to range from 2.4 to 4.5 pg [25] and that of various members within a single cyclophoroid landsnail genus Diplommatina is highly variable, between 2.73 and 7.85 pg [26,27]. The differences in techniques used, from flow cytometry to actual genome sequencing, also have differing accuracy and add to the challenge [28]. Therefore, it is often unfeasible to assess the difficulty and cost of a particular mollusc genome project before actually starting the sequencing process. Another issue is that most molluscs are small, and many are rarely collected—both these impediments are true for all living monoplacophorans, for example [29]. Currently, suitably preserved (e.g. RNAlater fixed, −80°C frozen), fresh, high-quality tissue in sufficient quantity (e.g. Hi-C empirically requires 0.1 g to over 1 g of fresh tissue) is essential to achieving a high-quality library preparation, but this is impossible for many species in Mollusca. Nevertheless, the high diversity and broad relevance of molluscs means that there are many examples of accessible, sufficiently large and individually interesting species that represent clades that have not yet been the subjects of any genome projects.
3. A framework for sampling molluscan genomes
The number of available molluscan genomes is now increasing so rapidly that any account of current coverage is likely to be obsolete within a few months. However, there are major gaps that may not be filled for a significant span of time, and as we approach a major transition in the technological availability of molluscan genomes, it is useful to reflect on what has been done and where research activities should be directed in coming years. The current coverage (table 1) includes up to four of eight classes at the time of writing. The three classes that are covered by multiple genomes—bivalves, gastropods and cephalopods—are the most familiar molluscs and the most economically important. However, these are not the largest classes: Polyplacophora in fact is more species rich than Cephalopoda in terms of living species.
Table 1.
Summary data on the taxonomic distribution of genome resources across the phylum Mollusca. Each class is divided into orders and major clades, indicating the approximate number of described species, the average C-value for members of the clade where any data have been published, and number of species with published genomes as of 1 September 2020. Clades noted in italic text are those without any published genomes that are of critical relevance to molluscan phylogeny and/or clades with more than 500 species and no published genomes.
| class | order (or clade) | known species | C-value average (from n spp.) | species with published genomes | |
|---|---|---|---|---|---|
| Bivalvia | Adapedonta | 120 | 2 | ||
| Anomalodesmata | 550 | ||||
| Arcida | 350 | 1.67 (3) | 1 | ||
| Cardiida | 1000 | ||||
| Carditida | 150 | ||||
| Galeommatida | 250 | ||||
| Gastrochaenida | 15 | ||||
| Limida | 300 | 1.40 (2) | |||
| Lucinida | 250 | ||||
| Myida | 250 | 1.38 (5) | 3 | ||
| Mytilida | 300 | 1.76 (31) | 5 | ||
| Nuculanida | 400 | ||||
| Nuculida | 200 | 4.25 (2) | |||
| Ostreida | 200 | 1.16 (19) | 4 | ||
| Palaeoheterodonta | 900 | 3.10 (2) | 1 | ||
| Pectinida | 500 | 1.69 (1) | 4 | ||
| Solemyida | 50 | 2.10 (1) | |||
| Venerida | 1000 | 1.70 (42) | 4 | ||
| Caudofoveata | Chaetodermatida | 150 | |||
| Cephalopoda | Nautilida | 5 | |||
| Myopsida | 50 | 3.65 (2) | 1 | ||
| Oegopsida | 250 | ||||
| Sepiida | 200 | 3.75 (1) | 1 | ||
| Spirulida | 1 | ||||
| Octopoda | 250 | 4.13 (3) | 3 | ||
| Vampyromorpha | 1 | ||||
| Monoplacophora | Tryblidiida | 40 | |||
| Polyplacophora | Chitonida | 700 | 1.57 (9) | 1 | |
| Lepidopleurida | 200 | ||||
| Scaphopoda | Dentaliida | 300 | 4.01 (2) | ||
| Gadilida | 300 | 2.41 (1) | |||
| Solenogastres | Cavibelonia | 200 | 0.64 (5) | ||
| Neomeniamorpha | 50 | 0.30 (1) | |||
| Pholidoskepia | 60 | ||||
| Sterrofustia | 20 | ||||
| Gastropoda | Patellida | 400 | 1 | ||
| Neomphalida | 50 | 0.44 (1) | 1 | ||
| Cocculinida | 60 | ||||
| Pleurotomariida | 70 | ||||
| Trochida | 2400 | 1.56 (5) | |||
| Lepetellida | 900 | 1.53 (15) | 3 | ||
| Seguenziida | 700 | ||||
| Cycloneritida | 1000 | ||||
| Hypsogastropoda | 13 000 | 1.19 (19) | |||
| Cerithiimorpha | 1500 | 2.10 (15) | |||
| Campanilimorpha | 20 | ||||
| Neogastropoda | 17 000 | 3.10 (26) | 5 | ||
| Architaenioglossa | 4000 | 4.05 (29) | 4 | ||
| Acteonimorpha | 250 | ||||
| Ringiculida | 80 | ||||
| Nudibranchia | 2500 | 1.16 (5) | |||
| Pleurobranchida | 90 | ||||
| Runcinida | 60 | ||||
| Cephalaspidea | 800 | 1.60 (1) | |||
| Umbraculida | 10 | ||||
| Aplysiida | 90 | 1.90 (3) | 1 | ||
| Pteropoda | 150 | 1 | |||
| Sacoglossa | 350 | 2.83 (1) | 2 | ||
| Acochlidia | 50 | ||||
| Siphonarimorpha | 100 | ||||
| Hygrophila | 900 | 1.34 (12) | 4 | ||
| Systellommatophora | 200 | ||||
| Stylommatophora | 20 000 | 2.86 (8) | 1 | ||
| Ellobiida | 300 | ||||
| ‘Lower Heterobranchia’ | 500 | ||||
Available genomes either represent interesting adaptive features that capture the public imagination (for example the scaly-foot snail and octopus, [10,30]), or species with importance for fisheries (for example oysters and scallops, [20,31]), and potentially other economic impacts such as biomineralization and bio-materials. There is a comparatively high number of genomes in Pectinoidea (Pectinida + Ostreida), due to the economic importance of these bivalves as well as the independent origin of a dispersed visual system [32]. These selections reflect a research landscape where sequencing and assembling one molluscan genome is challenging and resource intensive, and one strategically selected high-quality genome can produce a high impact publication. The increasing number of molluscan genomes already available now opens the possibility of important comparative genomics, using tools that have been developed to study the ecology and evolution in other animal groups.
The most obvious and significant gaps are the four missing classes: the two aplacophoran classes, scaphopods and monoplacophorans, with chitons represented by a single species [12]. All of these groups suffer from inaccessibility of the animals and a dearth of taxonomic experts. Equally important but less often noted is that a broad lack of familiarity with the animals among potential reviewers may be a significant barrier to funding and publication, so these projects are stuck in a near ‘Catch-22' paradox that stymies new research.
There are also important accidental gaps created by the tendency to select individually interesting species, and this will collectively undermine efforts at whole-genome phylogenomics unless it is confronted now. To obtain taxon sampling that is relevant for large-scale phylogenetic reconstruction, it is critically important to supply genomes that represent the early diverging branches within major clades. For example, although there are now more than five cephalopod genomes, pan-molluscan phylogenomics would benefit most from a nautilid genome, to anchor the symplesiomorphic characters of cephalopods within the molluscan tree. Likewise, it is extremely important to have any chiton genome, but pan-molluscan phylogeny will eventually require the representation of the order Lepidopleurida which retains more plesiomorphic chiton characters. Among gastropods, a number of phylogenetically crucial clades such as Cycloneritida and Cocculinida are still lacking genomic data. Similarly, in bivalves, some major clades such as Anomalodesmata and the protobranchs (Nuculanida, Nuculida and Solemyida) are so far completely absent. Furthermore, some groups are currently only represented by highly fragmented, relatively incomplete genomes (e.g. pteropod gastropods represented by Limacina bulimoides with 60.2% BUSCO score and palaeoheterodont bivalves represented by Venustaconcha ellipsiformis with 68% BUSCO score, [33,34]) and would benefit from high-quality genome assemblies. We suggest that the main priority in selecting new genome sequencing projects should be to fill these gaps across Mollusca as a whole and to increase the taxon sampling group by group.
In selecting focal species, we urge researchers to consider the phylogenetic context of each species, instead of only the individual features within a species lineage. If the goal is to fill in the blanks by selecting a species that can represent one of many clades that are still missing, we note that all lineages are equally representative and that all living species have been evolving for the same length of time. If additional information were available in advance, one might consider the range of genome sizes or GC content in many species across a clade and then select a representative example in the middle of the range of values. Heterozygosity is also not apparently predictable a priori, since high heterozygosity would be expected in broadcast spawners [35] but the highest heterozygosity so far encountered is in a sacoglossan, Elysia chloritica, which has internal fertilization. In reality, taxon selection has to be guided by practical issues, including access to appropriate material.
Another practical concern is the return on investment for working on a challenging species, and the potential for results to become obsolete because of advances in technology or duplication by another parallel effort. We offer two observations of the lived experience working on mollusc genomes. First, mollusc species dramatically outnumber the people working on them, and so increasing the taxonomic spread of genome sequencing efforts is very unlikely to produce major duplication of efforts even if there is no communication or coordination among research groups. Second, while work on molluscs is primarily limited to one genome per species, in other organismal groups, genome science has progressed to issues of comparative genomics and population genomics, and this demonstrates that having multiple genomes per species is an aspiration.
Several other organismal groups have established genome consortium projects. In molluscs, there are currently two dedicated public databases that present comparative analyses of a subset of mollusc genomes [36,37]. These projects, like NCBI, currently only report successful completed projects, whereas the community would arguably benefit more from open sharing of projects that have already been attempted and failed. In 2018, an earlier version of the figure presented here (figure 1) showed that mollusc genomes were dramatically under-sampled compared to other major clades with research consortia ([11], fig. 1). The rapid increase in new mollusc genome publications is clearly attributable to methodological and technological advances. A consortium would be welcome, but a lack of such organization does not appear to have hindered progress in molluscan genomics.
4. Conclusion
New technology has very recently enabled the assembly of high-quality genomes for many mollusc species, yet most of molluscan diversity remains unsampled. The goal of assembling data for molluscan genomics is far too ambitious for one research group, and, as in other animal groups with more accessible resources for comparative genomics, redundancy can be an advantage. Although we identify a number of critical gaps in the taxon sampling within molluscs, our view is that there is no conflict if as many research groups as possible target these gaps. More anomalodesmatan genomes are better than zero.
We need broad and dense sampling to resolve molluscan phylogeny. The extreme disparity and diversity in molluscs have shown us repeatedly that it is not sufficient to sample what is easy and dismiss what does not fit. Molluscan oddities are essential to the identity of Mollusca. Complex questions cannot be answered by sampling the same groups repeatedly with tokenistic input from some additional diversity. To understand the evolution of molluscan body forms—indeed of all animal body forms—we must embrace the full pantheon of challenging molluscan diversity.
Acknowledgements
We are grateful to Angus Davison and Maurine Neiman for the invitation to participate in the meeting ‘Pearls of wisdom: synergising leadership and expertise in molluscan genomics'. Comments from Tilman Schell, Chris Laumer and an anonymous referee all improved an earlier version of this paper.
Data accessibility
The data used in this paper are accessible through publicly available databases cited herein.
Authors' contributions
J.D.S. and D.R.L. conceived this study; all authors contributed to writing the manuscript.
Competing interests
We have no competing interests.
Funding
This work was supported by the Hong Kong Branch of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou).
References
- 1.Vermeij GJ. 1987. Escalation and evolution. Cambridge, MA: Harvard University Press. [Google Scholar]
- 2.Roy K, Jablonski D, Valentine JW, Rosenberg G. 1998. Marine latitudinal diversity gradients: tests of causal hypotheses. Proc. Natl Acad. Sci. USA 95, 3699-3702. ( 10.1073/pnas.95.7.3699) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sepkoski JJ. 1986. Phanerozoic overview of mass extinction. In Patterns and processes in the history of life (eds DM Raup, D Jablonski), pp. 277-295. Berlin, Germany: Springer. [Google Scholar]
- 4.Lydeard C, et al. 2004. The global decline of nonmarine mollusks. BioScience 54, 321-330. ( 10.1641/0006-3568(2004)054[0321:TGDONM]2.0.CO;2) [DOI] [Google Scholar]
- 5.Sigwart JD, Lindberg DR. 2015. Consensus and confusion in molluscan trees: evaluating morphological and molecular phylogenies. Syst. Biol. 64, 384-395. ( 10.1093/sysbio/syu105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stöger I, Sigwart JD, Kano Y, Knebelsberger T, Marshall BA, Schwabe E, Schrödl M. 2013. The continuing debate on deep molluscan phylogeny: evidence for Serialia (Mollusca, Monoplacophora + Polyplacophora). BioMed Res. Int. 2013, 407072. ( 10.1155/2013/407072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith SA, Wilson NG, Goetz FE, Feehery C, Andrade SC, Rouse GW, Giribet G, Dunn CW. 2011. Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature 480, 364-367. ( 10.1038/nature10526) [DOI] [PubMed] [Google Scholar]
- 8.Sigwart JD, Sutton MD. 2007. Deep molluscan phylogeny: synthesis of palaeontological and neontological data. Proc. R. Soc. B 274, 2413-2419. ( 10.1098/rspb.2007.0701) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pollock DD, Zwickl DJ, McGuire JA, Hillis DM. 2002. Increased taxon sampling is advantageous for phylogenetic inference. Syst. Biol. 51, 664. ( 10.1080/10635150290102357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun J, et al. 2020. The Scaly-foot Snail genome and implications for the origins of biomineralised armour. Nat. Commun. 11, 1657. ( 10.1038/s41467-020-15522-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sigwart JD, Bennett KD, Edie SM, Mander L, Okamura B, Padian K, Wheeler Q, Winston JE, Yeung NW.. 2018. Measuring biodiversity and extinction–present and past. Integr. Comp. Biol. 58, 1111-1117. ( 10.1093/icb/icy113). [DOI] [PubMed] [Google Scholar]
- 12.Varney RM, Speiser DI, McDougall C, Degnan BM, Kocot KM. 2021. The iron-responsive genome of the chiton Acanthopleura granulata . Genome Biol. Evol . 13 , evaa263. ( 10.1093/gbe/evaa263) [DOI] [PMC free article] [PubMed]
- 13.Sigwart JD, Stoeger I, Knebelsberger T, Schwabe E. 2013. Chiton phylogeny (Mollusca: Polyplacophora) and the placement of the enigmatic species Choriplax grayi (H. Adams & Angas). Invertebr. Syst. 27, 603-621. ( 10.1071/IS13013) [DOI] [Google Scholar]
- 14.Mikkelsen NT, Kocot KM, Halanych KM. 2018. Mitogenomics reveals phylogenetic relationships of caudofoveate aplacophoran molluscs. Mol. Phylogenet. Evol. 127, 429-436. ( 10.1016/j.ympev.2018.04.031) [DOI] [PubMed] [Google Scholar]
- 15.Lemer S, Bieler R, Giribet G. 2019. Resolving the relationships of clams and cockles: dense transcriptome sampling drastically improves the bivalve tree of life. Proc. R. Soc. B 286, 20182684. ( 10.1098/rspb.2018.2684) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uribe JE, Irisarri I, Templado J, Zardoya R. 2019. New patellogastropod mitogenomes help counteracting long-branch attraction in the deep phylogeny of gastropod mollusks. Mol. Phylogenet. Evol. 133, 12-23. ( 10.1016/j.ympev.2018.12.019) [DOI] [PubMed] [Google Scholar]
- 17.Jaksch K, Eschner A, Rintelen TV, Haring E. 2016. DNA analysis of molluscs from a museum wet collection: a comparison of different extraction methods. BMC Res. Notes 9, 348. ( 10.1186/s13104-016-2147-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li R, Zhang W, Lu J, Zhang Z, Mu C, Song W, Migaud H, Wang C, Bekaert M. 2020. The whole-genome sequencing and hybrid assembly of Mytilus coruscus. Front. Genetics 11, 440. ( 10.3389/fgene.2020.00440) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Payne A, Holmes N, Rakyan V, Loose M. 2018. BulkVis: a graphical viewer for Oxford nanopore bulk FAST5 files. Bioinformatics 35, 2193-2198. ( 10.1093/bioinformatics/bty841) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang G, et al. 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49-54. ( 10.1038/nature11413) [DOI] [PubMed] [Google Scholar]
- 21.Cai H, et al. 2019. A draft genome assembly of the solar-powered sea slug Elysia chlorotica. Sci. Data 6, 190022. ( 10.1038/sdata.2019.22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Belcaid M, et al. 2019. Symbiotic organs shaped by distinct modes of genome evolution in cephalopods. Proc. Natl Acad. Sci. 116, 3030-3035. ( 10.1073/pnas.1817322116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun J, Li R, Chen C, Sigwart JD, Kocot KM. 2021. Benchmarking Oxford nanopore read assemblers for high-quality molluscan genomes. Phil. Trans. R. Soc. B 376, 20200160. ( 10.1098/rstb.2020.0160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murgarella M, Puiu D, Novoa B, Figueras A, Posada D, Canchaya C. 2016. A first insight into the genome of the filter-feeder mussel Mytilus galloprovincialis. PLoS ONE 11, e0151561. ( 10.1371/journal.pone.0151561) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kocot KM, Jeffery NW, Mulligan K, Halanych KM, Gregory TR. 2016. Genome size estimates for Aplacophora, Polyplacophora and Scaphopoda: small solenogasters and sizeable scaphopods. J. Molluscan Stud. 82, 216-219. ( 10.1093/mollus/eyv054) [DOI] [Google Scholar]
- 26.Ieyama H, Tada A. 1991. Chromosomal studies and the quantitative evaluation of nuclear images stained with Feulgen dye in the Diplommatidinidae. Venus 50, 68-78. [Google Scholar]
- 27.Ieyama H, Ogaito H. 1998. Chromosomes and nuclear DNA contents of two subspecies in the Diplommatinidae. Venus 57, 133-136. [Google Scholar]
- 28.Gregory TR. 2020. Animal genome size database. See http://www.genomesize.com.
- 29.Sigwart JD, Wicksten MK, Jackson MG, Herrera S. 2019. Deep-sea video technology tracks a monoplacophoran to the end of its trail (Mollusca, Tryblidia). Mar. Biodivers. 49, 825-832. ( 10.1007/s12526-018-0860-2) [DOI] [Google Scholar]
- 30.Albertin CB, Simakov O, Mitros T, Wang ZY, Pungor JR, Edsinger-Gonzales E, Brenner S, Ragsdale CW, Rokhsar DS. 2015. The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 524, 220-224. ( 10.1038/nature14668) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kenny NJ, et al. 2020. The gene-rich genome of the scallop Pecten maximus. GigaScience 9, giaa037. ( 10.1093/gigascience/giaa037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kingston AC, Chappell DR, Miller HV, Lee SJ, Speiser DI. 2017. Expression of G proteins in the eyes and parietovisceral ganglion of the bay scallop Argopecten irradians. Biol. Bull. 233, 83-95. ( 10.1086/694448) [DOI] [PubMed] [Google Scholar]
- 33.Renaut S, Guerra D, Hoeh WR, Stewart DT, Bogan AE, Ghiselli F, Milani L, Passamonti M, Breton S. 2018. Genome survey of the freshwater mussel Venustaconcha ellipsiformis (Bivalvia: Unionida) using a hybrid de novo assembly approach. Genome Biol. Evol. 10, 1637-1646. ( 10.1093/gbe/evy117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choo LQ, Bal TM, Choquet M, Smolina I, Ramos-Silva P, Marlétaz F, Kopp M, Hoarau G, Peijnenburg KT. 2020. Novel genomic resources for shelled pteropods: a draft genome and target capture probes for Limacina bulimoides, tested for cross-species relevance. BMC Genomics 21, 1-14. ( 10.1186/s12864-019-6419-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ellegren H, Galtier N. 2016. Determinants of genetic diversity. Nat. Rev. Genet. 17, 422-433. ( 10.1038/nrg.2016.58) [DOI] [PubMed] [Google Scholar]
- 36.Edinburgh University. 2020. MolluscDB ENSEMBL. See https://ensembl.molluscdb.org/index.html (accessed 14 Nov 2020).
- 37.Liu F, Li Y, Yu H, Zhang L, Hu J, Bao Z, Wang S. 2020. MolluscDB: an integrated functional and evolutionary genomics database for the hyper-diverse animal phylum Mollusca. Nucleic Acids Res. 49, D988-D997. ( 10.1093/nar/gkaa918) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used in this paper are accessible through publicly available databases cited herein.