Abstract
The advent of complete genomic sequencing has opened a window into genomic phenomena obscured by fragmented assemblies. A good example of these is the existence of hemizygous regions of autosomal chromosomes, which can result in marked differences in gene content between individuals within species. While these hemizygous regions, and presence/absence variation of genes that can result, are well known in plants, firm evidence has only recently emerged for their existence in metazoans. Here, we use recently published, complete genomes from wild-caught molluscs to investigate the prevalence of hemizygosity across a well-known and ecologically important clade. We show that hemizygous regions are widespread in mollusc genomes, not clustered in individual chromosomes, and often contain genes linked to transposition, DNA repair and stress response. With targeted investigations of HSP70-12 and C1qDC, we also show how individual gene families are distributed within pan-genomes. This work suggests that extensive pan-genomes are widespread across the conchiferan Mollusca, and represent useful tools for genomic evolution, allowing the maintenance of additional genetic diversity within the population. As genomic sequencing and re-sequencing becomes more routine, the prevalence of hemizygosity, and its impact on selection and adaptation, are key targets for research across the tree of life.
This article is part of the Theo Murphy meeting issue ‘Molluscan genomics: broad insights and future directions for a neglected phylum’.
Keywords: hemizygosity, mollusc, genome, presence/absence variation, structural variation, pan-genome
1. Background
The rapid development of third-generation sequencing technologies and the subsequent increase in the number of species with an assembled genome has led to extraordinary new insights into gene family evolution between related species. At the same time, the flood of new genomic information has given rise to new questions regarding the dynamics of gene family expansion and contraction, and the mechanisms that could be driving such potentially adaptive processes in disparate taxa [1,2].
With increasing depth of sequencing data, it has become apparent that not all individuals of a species possess structurally identical genomes [3,4]. While small-scale variation (single nucleotide polymorphisms, indels, duplications, inversions and translocations) between individuals was entirely expected, large-scale structural variations (SVs), incorporating gene content disparity, have come as somewhat of a surprise in metazoan lineages. Variation in DNA content between individuals of a species forms the basis of the pan-genome concept [5]. First described in bacteria, a genic pan-genome consists of a core set of genes shared by all members of a species in addition to a set of ‘dispensable’ genes which are subject to presence/absence variation (PAV) between individuals [6]. The ratio of dispensable and core genes defines whether a pan-genome is considered closed or open, with the latter requiring the sampling of a very large and undetermined number of individuals to capture the full complement of dispensable genes in a species [7]. In the context of a diploid eukaryote, dispensable genes may be present in either one, two or zero copies in any given individual, meaning that an assembled genome of a single individual may not capture the full genic complement of that species.
In plants, two recent papers on the pan-genomes of rapeseed and tomato have highlighted the extensive PAV in these species and also the functional importance of some of their dispensable genes [8,9]. In total, at least 12 plant pan-genomes have now been described (reviewed in [10]). In plants, most SVs are associated with transposons and are rich in repeat sequences.
In metazoans, information on pan-genomes is only starting to emerge with genomic variation between individuals noted in animals such as the roundworm Caenorhabditis brenneri [11], the family Caenorhabditis more generally [12] and the ascidian Ciona savignyi [13].
Analyses of humans, probably the most resequenced metazoan species, are also beginning to show evidence of a closed pan-genome (i.e. a pan-genome with a low ratio of dispensable to core genes). Based on the re-sequencing of 2504 human genomes, a total of 240 genes were found to be occasionally subject to homozygous deletions in healthy individuals (and therefore likely dispensable) [14]. Analysis of the genome from 910 individuals of African descent led to the assembly of 296 Mb of genomic sequence not included in the reference human genome [15]. In pigs, the size of the pig pan-genome, estimated based on the re-sequencing of 12 individuals, revealed the presence of 72.5 Mb of additional genomic sequence [16]. High levels of genomic heterozygosity and gene presence/absence have been recorded in the bivalve molluscs Mytilus galloprovincialis [17] and Crassostrea gigas [18]. In M. galloprovincialis, a species characterized by an open pan-genome (i.e. by a 1 : 3 dispensable to core genes ratio), this has been firmly linked to hemizygosity [17], however, it is unknown how widely this trait is shared with other animals, or with other molluscs in particular.
Hemizygosity occurs in a genome where only one of the two chromosomal pairs encodes a region or block of DNA. In mammals, the most prominent example of hemizygous DNA comes from the X chromosome when it occurs in males. Under the male condition, no homologous region for the majority of the X chromosome exists on the Y chromosome which leaves the genes encoded by these regions monoallelic. To cope with this, dosage compensation mechanisms have evolved to alleviate the issues associated with reduced transcriptional output in males [19]. Hemizygous regions can also result from insertions or deletions (indels) of blocks of DNA and can occur through potentially pathological pathways (i.e. retrovirus or transposon insertion, e.g. [20]). The detection of hemizygous DNA that encodes genes in a single individual is evidence that, at the population level, these genes are likely to be subject to PAV as individuals may possess one (hemizygous), two (homozygous) or zero copies (nullizygous) of the dispensable DNA block.
Questions arising from the increasing observation of intraspecific genomic structural variation include: how representative are the single genomes we have for most species of the population or species from which they derive? Are the gene family contractions and expansions observed in these representative genomes shared in their entirety by all other members of the species? If not, what are the implications for species-wide phenotypic variability, gametic compatibility and ecological adaptability? While some of these questions will not be resolved until data from multiple individuals per species spanning multiple phyla are available, targeted analysis of individual high-quality genomes can be used to determine how prevalent genomic structural variability and open pan-genomes may be within particular clades.
Molluscan species, which are commonly profligate broadcast spawners [21], are often found inhabiting quite variable environments [21]. Many aquatic species, particularly bivalves, are sessile filter feeders that occur in high-density beds. This places them at risk of infection and exposure to locally unfavourable conditions [22]. The maintenance of genetic variation through a pan-genome, therefore, could enable a population to adapt to changes in environment or extreme local conditions [3].
Here, we examine the hemizygous DNA complement of eight high-quality and near complete conchiferan mollusc genomes (figure 1a). We detail the impacts of widespread hemizygosity on the genomic architectures of this species-rich and ecologically important clade, and investigate the hemizygous gene complement of each species. We note that retroelement-related genes and those involved in splice repair are over-represented in our datasets. Similarly, HSP70, C1qDC, C-type lectins and immune-related GTPases are common in hemizygous regions, suggesting possible adaptive roles in stress response and immunity. Our method, using freely available public data, could be applied to any well-sampled clade. It will be broadly applicable across the tree of life as more high-quality genome sequences become available, providing a clear means of investigating this under-studied, but potentially widespread, means of genomic adaptation.
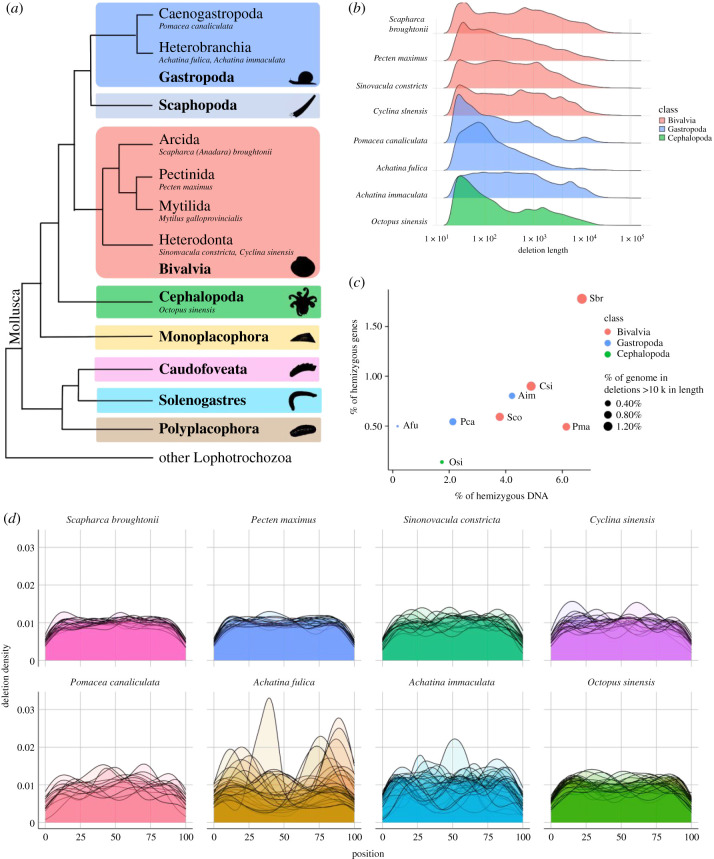
Figure 1.
Phylogeny and hemizygous loci analyses of eight molluscan species. (a) Representative cladogram of mollusc relationships after Kocot et al. [23]. Species referenced in this manuscript are shown in italics. Note, in Bivalvia and Gastropoda, numerous subclades are not shown. (b) Length distribution of hemizygous regions (deletions, log10 on both axes). (c) Percentage of each genome which is hemizygous versus the percentage of all genes which reside entirely within hemizygous DNA. The size of each point is proportional to the percentage of the genome found in large (greater than 10 kb) hemizygous regions. Species include Achatina fulica (Afu), Achatina immaculata (Aim), Cyclina sinensis (Csi), Octopus sinensis (Osi), Pecten maximus (Pma), Pomacea canaliculata (Pca), Scapharca broughtonii (Sbr) and Sinonovacula constricta (Sco). (d) Density of hemizygous loci for each species. Each chromosome is represented by an individual data series (line) which spans the beginning (0% distance) to the end (100% distance) of each chromosome. (Online version in colour.)
2. Methods
Genome assemblies and datasets used for all subsequent analyses are provided in table 1. Details on command line options for each step in the analysis pipeline can be found in electronic supplementary material, file S1. A brief description of each step involved is provided here.
Table 1.
Genomes used and basic statistics regarding assemblies.
| genome | abbreviation | bioproject | heterozygosity (%) | genome size | % hemizygosity (deletions) | % hemizygosity (deletions plus insertions) |
|---|---|---|---|---|---|---|
| Pecten maximus | Pma | PRJEB35331 | 1.69 | 918 306 378 | 6.14 | 10.68 |
| Sinonovacula constricta | Sco | PRJNA508451 | 2.42 | 1 220 848 272 | 3.78 | 7.54 |
| Cyclina sinensis | Csi | PRJNA612143 | 1.53a | 903 119 975 | 4.89 | 8.87 |
| Scapharca (Anadara) broughtonii | Sbr | PRJNA521075 | 1.79 | 884 566 040 | 6.69 | 10.81 |
| Pomacea canaliculata | Pca | PRJNA427478 | 1.92 | 440 159 624 | 2.12 | 3.91 |
| Achatina (=Lissachatina) fulica | Afu | PRJNA511624 | 0.19 | 1 855 892 613 | 0.17 | 0.37 |
| Achatina (=Lissachatina) immaculata | Aim | PRJNA561271 | 0.74 | 1 653 153 977 | 4.22 | 8.06 |
| Octopus sinensis | Osi | PRJNA541812 | 0.57 | 2 719 136 158 | 1.74 | 2.81 |
aFor C. sinensis, the heterozygosity value calculated in the original genome publication displayed as the GenomeScope model did not accurately capture the heterozygous and hemizygous peaks.
(a). Species selection
To document hemizygosity across conchiferan molluscs, we selected publicly available genomes that met three criteria: (i) the available assembly of each genome should be at or approaching chromosomal level, (ii) the sequenced individual should be wild caught and (iii) the genome assembly should be built from PacBio and Illumina datasets and these should be publicly available. These features were sought so that analyses of chromosomal distribution could be performed, artefacts from captive breeding could be avoided, so that precise boundaries of hemizygous regions could be determined and so that k-mer coverage of hemizygous regions could be performed. Hi-C Illumina short-read datasets were avoided as they do not map uniformly to their respective genomes. The Illumina datasets for Pecten maximus and Sinonovacula constricta were 10× Genomics datasets.
(b). Structural variant detection
To identify hemizygous regions, PacBio's structural variant detection pipeline pbsv v. 2.2.2 [24] was run on each of the eight genomes. The mapped long read bam file and tandem repeats annotation files required for pbsv were generated with the minimap2 [25] wrapper pbmm2 v. 1.0.0 (https://github.com/PacificBiosciences/pbmm2) and Tandem Repeats Finder v.4.09 [26] respectively. As the aim was to identify regions of the genome assemblies that were both hemizygous and contained previously annotated genes, the type of structural variants detected by pbsv that were used for subsequent analyses were limited to deletions. Detected deletions that passed pbsv's default criteria were filtered for further analysis. Insertions relative to the reference genomes were also annotated in order to determine the upper bound of hemizygosity for these individuals, however, these were not used for downstream analyses due to the absence of gene annotations within these loci. As such, unless otherwise specified, ‘hemizygous regions’ refers only to deletion-associated hemizygous regions for the remainder.
To visualize the level of hemizygosity in each species, deletions at least 10 kb in length that were not associated with tandem repeats were extracted and used as input for chromoMap v. 0.2 [27].
(c). K-mer analysis of hemizygous regions
Illumina short-read libraries were preprocessed with bbduk [28] and mapped to their respective genomes with BWA MEM v. 0.7.16 [29]. In cases where more than one dataset was used per species, mapped bam files were merged with samtools merge [30]. Mapped reads were extracted with samtools view and converted to fasta format with bbtools reformat.sh [28], and k-mer histograms (k = 21) of all mapped reads were produced with Jellyfish v. 2.3.0 [31]. This histogram was uploaded to Genomescope 2.0 [32] to obtain heterozygosity estimates for each species.
To produce histograms of k-mers from reads mapped to hemizygous regions, reads mapping to hemizygous regions were extracted with samtools view after which only those falling entirely within the defined regions were filtered with bedops bedmap v. 2.4.37 [33]. A fasta file of all filtered reads was extracted with bedtools fastaFromBed v. 2.29.0 [34] and these were used to produce k-mer histograms with Jellyfish as was performed for the whole mapped library.
(d). Nucleotide-level heterozygosity
In addition to k-mer analysis for heterozygosity estimation, we also determined nucleotide-level heterozygosity by assessing the proportion of heterozygous genotype calls for each genome assembly using bcftools [30]. Details on the command line options used for this analysis can be found in electronic supplementary material, file S1.
(e). Read coverage of hemizygous regions
The same mapped reads used for k-mer coverage were also used to determine read coverage of hemizygous regions and to compare to read coverage of the whole genomes. A sam file of all reads mapping entirely within deletions was produced and from this a bam file was produced with samtools view. The median coverage of each deletion was calculated with mosdepth v. 0.2.9 [35]. For the whole genome, bedtools genomecov v. 2.29.0 [34] was used to calculate coverage at every position in the genome and then the median coverage of every 1 000 nt (1 nt step) window was calculated.
(f). Hemizygous gene identification
Genes were extracted from hemizygous regions of the genome using bedtools intersect v. 2.29.2 [34]. Only those genes falling fully within hemizygous regions were extracted for analysis (using the -F 1 option as detailed in electronic supplementary material, file S1). Additional genes which partially overlap hemizygous regions were also identified; however, to avoid speculation as to whether these are simply disrupted or whether hemizygosity may affect gene isoforms, we decided to discard them from further analysis and to focus on just those genes that are most likely subject to PAV. From these lists, genes were identified and extracted from the full gene list for use in enrichment analyses.
(g). Protein domain and gene ontology enrichment analysis
The predicted protein translations obtained from the longest annotated isoform for each gene in the target molluscan genomes were functionally annotated with Pfam conserved domains ID and Gene Ontology terms as follows. Amino acid sequences were subject to a BLASTp analysis against UniProtKB, with an E-value threshold set to 1 × 10−5. Gene Ontology cellular component, biological process and molecular functions terms associated with the top 10 best hits were extracted and used to annotate matching query sequences. Protein sequences were also subject to conserved domain annotation. This analysis used the hmmscan module of HMMER v. 3.3.1 [36] and the search was conducted against the Pfam-A v. 33.1 database [37], annotating domains based on the default E-value threshold.
The subset of sequences associated with hemizygous regions, identified as described in the previous paragraph, were then subject to hypergeometric tests on annotations [38] using the script included in the SciPy 1.5.2 package, which were run separately for GO terms and Pfam domain IDs. We here report significantly enriched GO terms and Pfam domains, filtering out based on their over-representation in the tested subset of sequences, compared with the full genome. Namely, enriched annotations were reported for terms associated with p-values <0.05 and a difference between the number of observed and expected genes greater than or equal to 5.
(h). Phylogenetic analysis
HSP70 and C1qDC genes were identified within the genomes of the target species using genes of known homology for local BlastP searches (E-value cutoff, initially E−9) [39]. These were then reciprocally blasted against the nr database to confirm likely identity. These sequences, alongside known sequences from previous publications, were aligned using the MAFFT 7 online tool and the G-INS-i strategy [40,41]. The resulting alignments were trimmed with TrimAL v. 1.2 [42] and the ‘-gappyout’ setting.
The resulting alignments were tested for model fit using ModelFinder [43] as integrated in IQ-TREE multicore v.1.6.10 [44]. The best-fit model was used for analysis of each phylogeny as noted in the figure 4 legend. IQ-TREE multicore v.1.6.10 was used for maximum likelihood (ML) analysis with 1000 non-parametric bootstrap replicates. The resulting consensus phylogeny was then opened in FigTree v. 1.4.4 (https://github.com/rambaut/figtree/releases) for annotation and display.
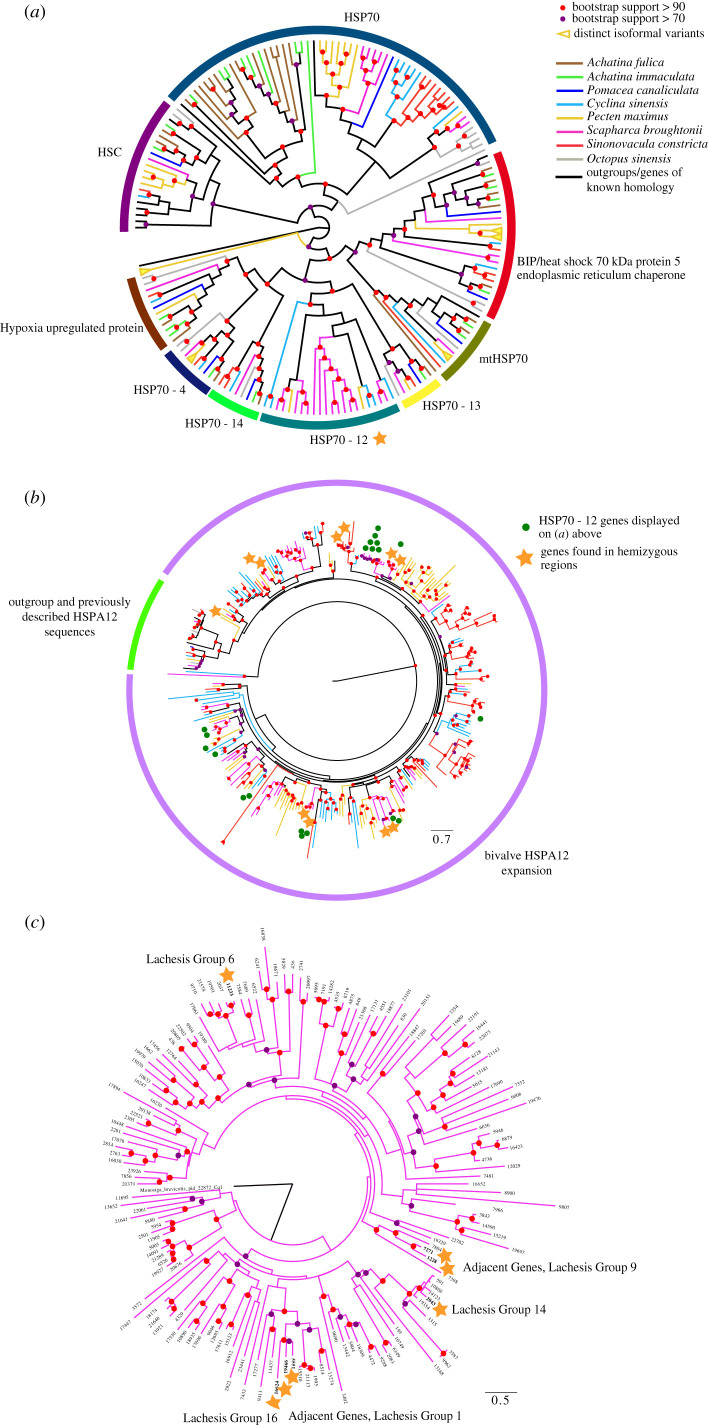
Figure 4.
Phylogenies and cladograms of HSP70 and C1qDC superfamilies, showing the potential of hemizygous regions as a reservoir and driver of gene diversity. (a) Diagrammatic cladogram of HSP70 superfamily genes from the eight species examined here, with branches coloured according to species identity as seen in the key, and rooted with Arabidopsis thaliana HSP70 sequence. Arcs surrounding the cladogram indicate gene families. Phylogeny upon which this cladogram is based, inferred using the LG + R6 model, along with raw sequences, alignment and tree file, are available in electronic supplementary material, file S5. Note: this cladogram is not exhaustive and excludes some HSP70-related gene sequences due to alignment and trimming. (b) HSP70-12 phylogeny genes from the eight species examined here, along with outgroups and genes of known identity. Phylogeny inferred using the LG + F+R8 model. Note genes from hemizygous regions, indicated with a star. Genes also included in (a) indicated with a green dot. Phylogeny rooted with Arabidopsis thaliana HSP70 sequence. (c) C1qDC superfamily gene interrelationships in Scapharca broughtonii, displayed in a phylogeny reconstructed using the WAG + F + R6 model. Note genes from hemizygous regions, indicated with a star. The linkage groups for these genes, as assigned by Lachesis, are also noted alongside them.
3. Results
(a). Hemizygosity and heterozygosity in molluscs
Hemizygosity (flagged as deletions by pbsv) of the individual representatives of the eight molluscan species investigated here ranged from 0.17% of the total genome length in the giant African snail Achatina (=Lissachatina, [45,46]) fulica to 6.69% in the ark clam Scapharca broughtonii (table 1). If insertions relative to the reference genome are also considered, the hemizygosity of A. fulica and S. broughtonii increase to 0.37% and 10.81%, respectively (table 1). The A. fulica hemizygosity content was a clear outlier among the eight species with the next lowest belonging to the octopus Octopus sinensis at 1.74% (2.81% inclusive of insertions), while the congeneric Achatina (=Lissachatina) immaculata had 4.22% hemizygous DNA content (8.06% inclusive of insertions). The number and size (bp) of the deletion-associated hemizygous regions are shown in table 2.
Table 2.
Statistics regarding putatively hemizygous regions.
| genome | # of deletions | length of deletions | # of non-tandem deletions | length of non-tandem deletions | # of >10 k deletions | length of >10 k deletions | % hemizygous genes |
|---|---|---|---|---|---|---|---|
| Pecten maximus | 145 503 | 56 374 042 | 60 503 | 35 699 112 | 428 | 7 359 495 | 0.50 |
| Sinonovacula constricta | 108 006 | 46 151 533 | 56 831 | 34 057 642 | 410 | 12 089 568 | 0.60 |
| Cyclina sinensis | 75 880 | 44 129 845 | 46 216 | 35 875 686 | 456 | 11 645 828 | 0.91 |
| Scapharca (Anadara) broughtonii | 83 518 | 59 203 570 | 47 022 | 47 290 026 | 671 | 13 984 916 | 1.79 |
| Pomacea canaliculata | 24 569 | 9 343 212 | 15 080 | 8 489 934 | 111 | 2 830 553 | 0.55 |
| Achatina (=Lissachatina) fulica | 16 208 | 3 197 224 | 3 112 | 1 330 508 | 38 | 789 841 | 0.50 |
| Achatina (=Lissachatina) immaculata | 128 736 | 69 768 216 | 27 939 | 41 999 937 | 351 | 7 946 998 | 0.81 |
| Octopus sinensis | 153 503 | 47 178 568 | 23 187 | 24 977 873 | 131 | 2 440 767 | 0.14 |
Repetitive sequences appear to be a major component of hemizygous DNA in these species with between 39% and 85% of deletions being flagged as tandem duplication-containing by pbsv (table 2). While the length of most hemizygous regions are short (pbsv default minimum length of 20 bp), we observed between 38 and 671 hemizygous regions that exceeded 10 kb in length (figure 1b,c). The maximum length of SVs that can be annotated is directly related to the read length distribution of the mapped library, making PacBio long reads superior to Illumina short reads for SV detection [47]. Due to the limitations of pbsv, which does not annotate deletions above 100 kb in length, the maximum deletion size remains unknown for any of the eight samples. This limitation also means that the total number of deletions and total hemizygous DNA content of each sample are both likely under-estimations.
Previous work on human structural variation showed that the number of SVs was non-randomly distributed along each chromosome with the greatest density occurring within 5 Mb of the telomeric chromosomal ends [48]. The pbsv pipeline's conservative approach to annotating SVs located towards the ends of chromosomes means that putative terminal SVs are not flagged as a PASS and as such were not included for further analysis here (see Methods). This results in the appearance that hemizygosity is diminished at the terminal regions of chromosomes and highlights the fact that the numbers of hemizygous regions reported here are conservative lower estimates (figure 1d).
Focusing on deletions over 10 kb in length and which were not flagged as tandem-repeat associated by pbsv, it is evident that large hemizygous regions are not confined to particular chromosomes or chromosomal regions in any of the eight species investigated (figure 2). Although there are a large number of hemizygous regions in all species, there is a clear difference in the number of larger deletions present in the bivalves versus the gastropods and cephalopods (figures 1b and 2). This seems to also translate into the proportion of the genomes that are hemizygous in each of the three molluscan classes, however more species will need to be analysed before these trends can be confirmed (figure 1c).
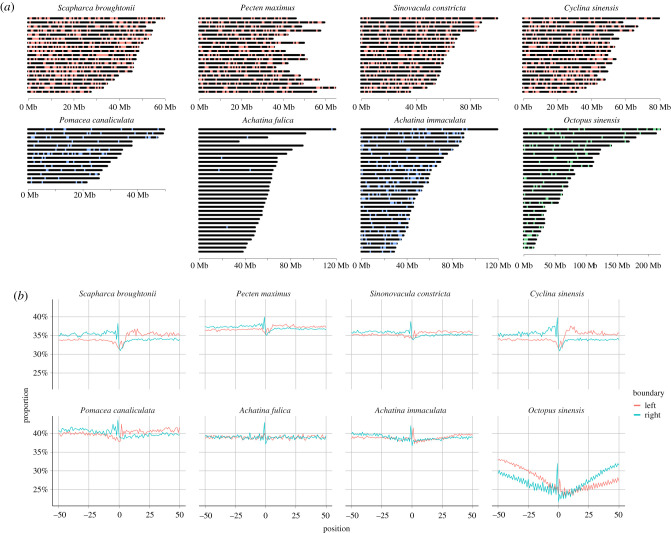
Figure 2.
Chromosomal maps of hemizygous loci and G/C content across homozygous/hemizygous boundaries. (a) Hemizygous loci greater than 10 kb in length which were not flagged by pbsv as ‘tandem repeats’. Each locus is marked as a single point which is not proportional in length to the actual size of the locus. Genomes with red loci are bivalves, those with blue loci are gastropods and the genome with green loci is a cephalopod. (b) Average G/C content spanning 50 bp downstream and 50 bp upstream of the left homozygous/hemizygous boundary or 50 bp downstream and 50 bp upstream of the right homozygous/hemizygous boundary for all annotated hemizygous loci. In each species the transition between homozygous and hemizygous DNA is marked by a G/C spike and apart from the octopus, hemizygous DNA is generally more G/C rich than the flanking homozygous regions. For octopus, hemizygous loci are more A/T rich than the flanking homozygous regions and the entire region surrounding the boundary is relatively depleted of G/C nucleotides. (Online version in colour.)
Heterozygosity of each sample as determined by Genomescope 2.0 [32] ranged from 0.19% in A. fulica to 2.41% in the razor clam S. constricta. Using a nucleotide-level heterozygosity calculation based on the proportion of heterozygous genotype calls by bcftools [30], we estimated a discrepancy when compared to the k-mer-based approach of up to 1.55% (electronic supplementary material, file S2). The difference between these two methods may result from hemizygous k-mers which fall within the ‘heterozygous’ peak. We observe limited evidence of correlation between hemizygosity and heterozygosity within molluscan classes (Bivalvia and Gastropoda), as can be seen in electronic supplementary material, figure S1. This may be due to undersampling.
(b). K-mer and read coverage of hemizygous regions
Both k-mer coverage and read coverage of the putative hemizygous regions provide strong support that these regions are in fact hemizygous. In six of the eight species, two clear peaks in both k-mer coverage (figure 3a, top rows) and read coverage (figure 3b, top rows) correspond to what are typically described as the heterozygous and homozygous peaks. The remaining two species, A. fulica and O. sinensis, have relatively low levels of both heterozygosity and hemizygosity and accordingly only have a single homozygous peak.
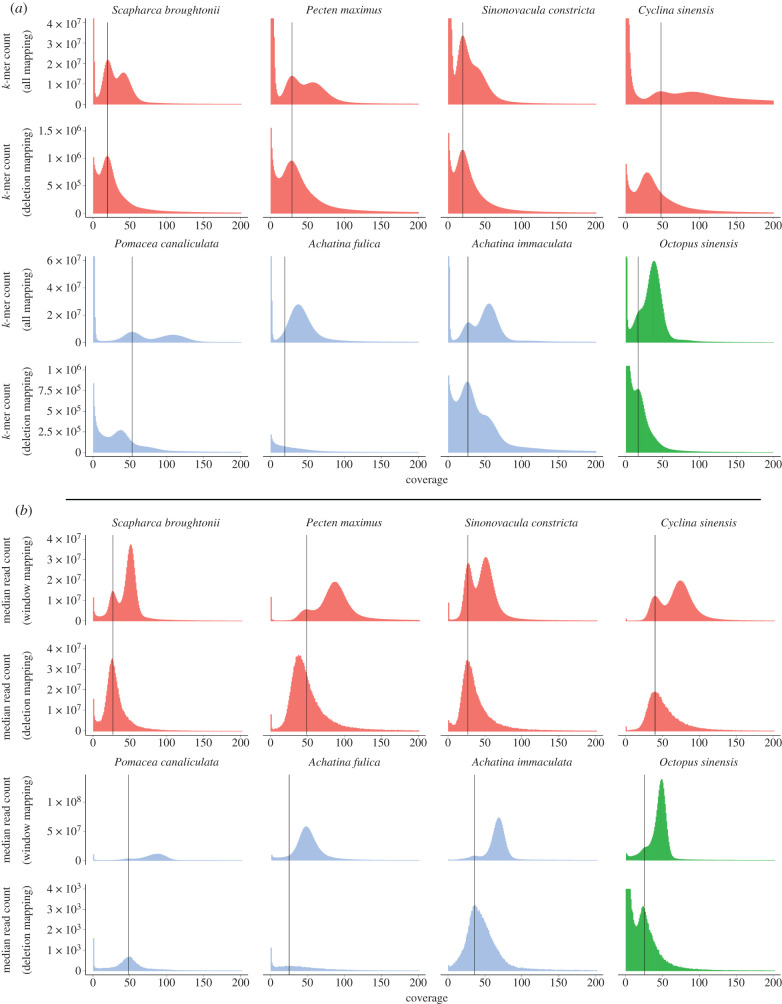
Figure 3.
k-mer and median read coverage analysis of hemizygous regions. (a) k-mer counts of all mapped reads for each genome with the corresponding k-mer counts of reads that map entirely within hemizygous regions located directly below. (b) Median read coverage of all 1000 bp sliding windows for each genome with the corresponding median read coverage of all annotated hemizygous regions located directly below. For both (a) and (b) the black vertical lines mark the ‘heterozygous’ peaks of the total mapped reads k-mer or median read coverage plots. Species are colour coded by class with red for bivalves, blue for gastropods and green for the cephalopod.
By contrast to the whole genome datasets, k-mers and reads extracted from hemizygous regions only show a single peak of coverage that for most datasets corresponds to the ‘heterozygous’ peak of the whole genome plots (figure 3a,b, bottom rows). In the k-mer plots of Cyclina sinensis and Pomacea canaliculata, the hemizygous peaks have slightly reduced coverage relative to the whole genome heterozygous peaks, possibly due to high error rates in these datasets, and the same is true for the P. maximus read coverage datasets.
Unlike the other datasets, the A. immaculata hemizygous k-mer plot has a second small peak corresponding to the homozygous peak of the whole genome k-mer histogram. This may be explained by the fact that a member of the Achatina (actually Lissachatina, see [45]) genus underwent a whole genome duplication event prior to the divergence of A. fulica and A. immaculata [49]. Duplicated hemizygous regions would be expected to encode a proportion of identical k-mers on each of the two copies and this would result in a peak of coverage corresponding to the whole genome homozygous peak.
(c). Gene content in hemizygous regions and gene family over-representation
We have examined the gene content of hemizygous regions within our target species, directly by extracting the annotation of these genes, and more indirectly, by looking at the over-representation (enrichment) of both Pfam domains and GO terms within these gene complements. On several occasions, hemizygous genes appear to be associated with clusters of tandemly duplicated genes: for example ADAM17 in P. canaliculata, GTPase IMAP family member 9 in A. immaculata, Deoxycytidylate deaminase in O. sinensis and numerous other examples.
The number of genes associated with hemizygous regions in O. sinensis (0.14% of the total in its genome, table 2) is markedly fewer than that found in the bivalve and gastropod species. As such, the subsequent absence of GO term enrichments and the limited number of Pfam domain enrichments associated with these loci were in line with expectations. We do however see many zinc-finger domain genes in our blast results for Octopus (7/35 genes). Similarly, the gastropods A. fulica and A. immaculata display limited gene and domain enrichments which may be the result of decisions made during gene annotation in these species (see below for more details). Visual inspection of the A. immaculata hemizygous gene set revealed many repeat-containing genes with poor annotations, resulting in poor enrichment analyses, both for Pfam domains and GO terms. Interestingly, most of the annotated genes in the hemizygous regions of the Achatina species seem to be involved in disparate, unrelated processes, the functions of which can only be speculated upon.
While the genes found in hemizygous regions belonged to a variety of gene families, transposable element (TE) linked domains and GO terms were over-represented in our analyses, with a tendency for these genes to be involved in break repair/genomic stability, or immunity. We have investigated these genes in particular below, with full details of blast and enrichment analyses in the electronic supplementary material, files S3 and S4.
(i). Transposable elements
Within hemizygous regions, TE-associated genes are common in most of our target species. In P. canaliculata, for example, we see classical retrotransposon-like (gag-pol polyprotein) elements in our gene lists (electronic supplementary material, file S3). In several species, the number of hits to TEs are more striking in Pfam domain enrichment analyses, as HMM profile-based searches are much more sensitive than BLAST where clear TEs are often slightly below the E-value threshold for annotation. This is also clear in our over-representation analysis of GO terms, particularly for P. canaliculata, S. broughtonii and P. maximus. GO terms such as GO:0003964 ‘RNA-directed DNA polymerase activity’, GO:0006313 ‘transposition, DNA-mediated’ and GO:0015074 ‘DNA integration’, and associated functions, are clearly enriched in these gene sets. Pfam enrichment analyses show the same clear signal of transcriptional element over-representation. Domains such as PF13975.6 ‘gag-polyprotein putative aspartyl protease; retrotransposon-associated’, PF00078.27 ‘reverse transcriptase’ and PF03221.16 ‘Tc5 transposase DNA-binding domain’ are conspicuous in these lists of enriched domains.
Some species do not possess TE-associated genes in hemizygous regions, e.g. the two snails A. fulica and A. immaculata, and the bivalve C. sinensis. This may be a technical artefact arising from the removal of over-represented (repetitive) genes during gene annotation, or it could represent a true biological observation. Possible explanations for the association of hemizygous DNA and transposons include the use of hemizygous regions to aid transposon replication, the difficulty of hemizygous regions to purge TEs or that hemizygous regions are formed through the action of TE replication. This pattern of TE-associated gene enrichment is also seen in plant pan-genomes (e.g. [50]) suggesting a broader pattern to the link between TEs and hemizygosity.
The over-representation of Zinc-finger domains in hemizygous regions in several of our species is also interesting, given that clusters of these genes are known to be found at hotspots for copy number variation, as a means of defence against endogenous retroviruses [51]. It is possible that these genes are playing a similar role here—zinc-finger genes have a conserved role as transcriptional repressors, although their exact functionality and DNA-binding affinities have yet to be fully investigated.
(ii). DNA break repair and remodelling
We note that many of the genes found in hemizygous regions can be linked to DNA stability, repair and remodelling. GO terms such as GO:0006281 ‘DNA repair’, GO:0045739 ‘positive regulation of DNA repair’, GO:0000733 ‘DNA strand renaturation’ (P. maximus) and GO:0006310 ‘DNA recombination’ (S. broughtonii and P. canaliculata) were found to be enriched, indicating that hemizygous regions may code for their own stability. Even within the limited hemizygous-associated gene set of O. sinensis is a SETMAR orthologue which in primates is known to play a role in DNA double-strand break repair, stalled replication fork restart and DNA integration [52].
In the section above, we note the presence of Zinc-finger domain genes in hemizygous regions in several species, which are known to aid genome stability at otherwise fast-evolving sites. Several of the genes found in P. maximus also have significant homology to PIF1, RECQL and Werner syndrome ATP-dependent helicases, which are all involved in genome stability (e.g. [53]). Helicase-like domains were associated with dispensable genes in mussel M. galloprovincialis [17], although they share little to no primary sequence homology with matches in UniProt. Their possible implication in the structural aspects of hemizygous regions is an excellent target for future research.
In P. maximus, we note the presence of several G-quadruplex GO annotations (GO:0044806 ‘G-quadruplex DNA unwinding’, GO:0051880 ‘G-quadruplex DNA binding’) in addition to G/C peaks located at homozygous/hemizygous DNA transition boundaries of all species investigated here (figure 2b). G-quadruplexes are four-stranded DNA or RNA secondary structures formed from guanine tetramers. While a complete understanding of their function is still being elucidated, their presence in telomeres, promoter sequences and retroelements suggests a link to genome stability, gene regulation, transposon and retroviral biology [54–57].
(iii). Immunity
Overall, several immunity-related domains are shared in the species investigated here, but each species has its own characteristic profile. This is also the case for GO term over-representation, although we do note the importance of ontologies such as GO:0002230 ‘positive regulation of defense response to virus by host’, GO:0045087 ‘innate immune response’ and GO:0051607 ‘defense response to virus' in our enrichment analyses.
We commonly observe genes encoding immunoglobulin-domain containing proteins and C-type lectins in our hemizygous region datasets in a number of species. These have also been observed as over-represented in M. galloprovincalis [17]. The function of these is yet to be fully understood, but due to their high plasticity in protein–protein and protein–carbohydrate interactions, they have been noted elsewhere as potentially important tools for immune recognition [17].
AIG1 immunity-related GTPase genes are also observed. These are also subject to PAV in mussels. AIG1 immunity-related GTPase gene function is still obscure, but it plays an important role in host–parasite interactions in gastropods [58].
We also note the presence of defense peptides in hemizygous regions of these genomes. The presence of Stomoxyn, toxin 32 and other antimicrobial peptide (AMP) annotations might indicate components of the innate immune system are present in hemizygous regions. These are characterized by high intraspecific sequence diversity [59], and hemizygosity would result in greater variation between individual phenotypes for these genes.
(d). Individual gene families, hemizygosity and presence/absence variation
As an assay for the impact of hemizygosity on gene duplication rates and gene evolution more generally, we have studied in detail two gene families where multiple genes were found in hemizygous regions in multiple species, the HSP70 superfamily and the C1qDC containing genes. These are involved in resilience to stress and are important pattern recognition receptors in innate immunity of invertebrates, respectively [60–63], and it is possible that PAV in hemizygous regions is linked to differential adaptive capacity across the ranges of these genes.
Both HSP70 and C1qDC gene family expansions have been noted previously in bivalves [64] and we observe that their occurrences within hemizygous regions are much more prevalent in bivalves than in other molluscs, despite gastropods also possessing multiple duplicates of these genes. In figure 4a, it can be seen that multiple lineage-specific duplications have occurred in many of the genes and gene families within the HSP70 superfamily and these duplications are not limited to bivalves. A. fulica and A. immaculata in particular share many duplicate, paralogous copies of HSP70 (HSPA1), however none of these are found in hemizygous regions. By contrast, the disparate HSP70-12 (HSPA12) genes do frequently occur in hemizygous regions (figure 4b). Full sequences, alignments and alternative representations of the phylogeny for these figures (showing all bootstrap support values) can be found in the electronic supplementary material, file S5.
(i). HSP70-12
There is very little diversity of HSP70-12 sequence in gastropods (one copy in each of the three species examined here) or in O. sinensis (three copies). However, this family of genes has exploded in bivalves. This is especially prominent in S. broughtonii, which possesses eight copies within hemizygous regions, and 76 copies overall. S. broughtonii also possesses an HSP90 gene within a hemizygous region (EVM0009939). S. constricta (two) and P. maximus (one) also possess hemizygous copies of this gene, and more than 60 paralogues in total (figure 4b). No C. sinensis copies of this gene are within hemizygous regions, although it possesses 66 copies spread across its genome.
HSP70-12 genes have been studied in detail in scallops [63], where they are known to be protective against toxic dinoflagellates. In that study, the drastic expansion of the HSP70-12 family was observed, with a total of 47 paralogous copies of HSP70-12 noted, although the authors do not draw any link to hemizygosity. The large numbers of HSP70-12 genes observed here are, therefore, not unusual for bivalves. Given their role in protecting against specific pathogens, which may vary across the ranges of these species, PAV for HSP70-12 may provide an adaptive phenotype, although we have not formally tested this here.
In our phylogeny (figure 4b), we note that there is little or no phylogenetic signal for an evolutionarily conserved relationship between HSP70-12 genes from the hemizygous regions of different species. These are often in genes separated by a number of paralogy events, in strongly supported clades. Rather it seems that association events between HSP70-12 genes and hemizgyous regions, possibly through the action of transposons, have occurred in a lineage-specific manner as opposed to being derived from a common ancestor (electronic supplementary material, figure S2).
We observe a clear phylogenetic signal for a close relationship between pairs of hemizygous genes within species. Of the 8 copies of HSP70-12 seen in S. broughtonii, all are paired with a gene of similar sequence, indicated by a star on figure 4b. All of these paired genes, when located in the genome, were found in close proximity. These pairs are: genes 11804 and 3214, found on pseudochromosome ‘Lachesis Group 6’ 10 kb bp apart; genes 1411 and 9653, from Lachesis Group 11 (8 kb apart); 3314 and 13896, from Lachesis Group 8 (5 kb apart); and 17648 and 3510, from Contig00525, separated by only 6 kb. These paired genes are all likely tandem duplicates, potentially mediated by the action of TEs.
In the S. constricta genome, the two HSP70-12 genes seen at hemizygous loci are evm.model.Chr12.1234 and evm.model.Chr12.1237, which are nearly 50 000 base pairs apart, at sites 37 468 857–37 468 878 and 37 518 784–37 518 839 on chromosome 12, in two separate hemizygous sites.
(ii). C1qDC
We also investigated the diversity of C1qDC containing genes, as these have been noted previously as being protective against environmental and pathogenic impacts [60,61], and subject to PAV in mussels [17]. These genes are widespread in molluscs, and in bivalves in particular, with more than 100 copies commonly observed in complete genomes. We found copies of C1qDC genes in hemizygous regions (see electronic supplementary material, file S3) of C. sinensis (one copy, evm.model.Hic_asm_12.1571) and S. broughtonii (seven copies). We, therefore, chose S. broughtonii for specific investigation, as can be seen in figure 4c.
Of the seven copies of C1qDC genes found in hemizygous regions, two pairs of adjacently located genes were observed (in Lachesis Groups 1 and 9) and three single gene loci were found (in Lachesis Groups 6, 14 and 16). The pair of genes found in Lachesis Group 1, EVM0005551 and EVM0019466, are 44 kb apart from one another, at sites 21 191 896–21 194 964 and 21 238 944–21 240 116 respectively, in a single deletion (pbsv.DEL.17342). The pair in Group 9, EVM0007271 and EVM0001228, are only 24 kb apart, in a single deletion (pbsv.DEL.114852) at positions 42 253 538–42 255 235 and 42 278 930–42 291 255. Given their relatively broad spacing and the relatively long branches separating these pairs phylogenetically, these could be ancient, rather than recent, tandem duplicates, or could have come about by other processes.
The sister gene to the two genes found in Lachesis Group 1, EVM0016624, is itself found in a hemizygous region, on Lachesis Group 16. Paralogous copies of these genes are, therefore, also found in trans across the genome more broadly. It is possible their movement is mediated by the TEs enriched in hemizygous regions (see previous section), although this has not been formally tested here.
4. Discussion
(a). The prevalence and significance of hemizygosity in molluscs and across the tree of life
The detection of hemizygosity in an individual genome is indirect evidence of PAV of chromosomal regions and possibly transcriptional products within a population. This is due to the inheritance of the hemizygous region-containing chromosome pairs from the two parents which themselves may have been hemizygous, homozygous or nullizygous for the particular locus. The detection of hemizygous chromosomal regions within an individual can, therefore, provide an initial indication of the level of chromosomal and genic variability that might exist within the population or species to which it belongs.
The complete chromosomal, transcriptional and regulatory repertoire that exists within a population or species is termed its pan-genome [5]. While such a complete snapshot of a species' genetic complement can only be attained through the sequencing of multiple individuals, the detection of substantial hemizygosity within a single individual can provide strong evidence of the existence of an open pan-genome. The investigation of the prevalence and impact of hemizygosity on evolution is still in its infancy. However, given the evidence presented here, we can begin to comment on some aspects of this, and suggest fertile ground for future investigations.
Pan-genomes have been described in plants, fungi and bacteria, and it is only recently that they have been noted in metazoans [15–17,65]. To date, evidence from animals is sparse and with patchy phylogenetic distribution [10]. Here, we show that, at least among conchiferan molluscs, hemizygosity is a common phenomenon. Whether this is true of other metazoan phyla remains to be investigated. However, the read-mapping approach used here would be straightforward to apply to other taxa with little additional cost, as long as a chromosome-scale genome assembly is available, and would give an initial indication of the ubiquity of this phenomenon.
While the link between hemizygosity and pan-genomes is clear, estimating a species pan-genome size based on the hemizygosity estimates of a single individual is not possible. As such, an accurate pan-genome description can only be provided by re-sequencing of multiple individuals. In general, only traditional model organisms and humans have been the target of deep re-sequencing efforts. The most extensive re-sequencing efforts of any metazoan species have come from humans and recent results suggest that here species-wide genomic variation is associated with relatively small and rare structural variants, which account on average for 5 Mb of DNA sequence per individual, i.e. less than 0.2% of the genome size [65]. By considering population size and the level of PAV observed between the three genome assemblies in the study, it was estimated that the human pan-genome would likely include an extra 19–40 Mb of DNA relative to the reference genome, i.e. up to a 1.25% increase. However, a more recent study that resequenced 910 humans of African descent extended this to 296.5 Mb of additional DNA which equates to approximately a 10% increase on the reference human assembly [15].
By contrast with animals, in plants and fungi, hemizygosity is likely to be quite widespread across most genomes [10]. In fungi, some species have open pan-genomes, with up to 60% of coding gene sequences found to vary between individuals (e.g. Parastagonospora spp. [66]) although figures for genomic sequence variability are not yet available. In plants, figures of up to 42% of the complete genome being absent (the ‘accessory/variable/dispensable genome’) in some individuals have been reported [67]. However, these values are perhaps at the extreme end of the structural variation continuum in these clades. Core genomic retention of greater than 80% of the complete genome is more common [10].
While it is not possible to estimate pan-genome size based on hemizygosity levels within a single individual, our figures suggest that molluscs are likely to display less genomic PAV than is seen in plants but far more than has been currently observed in vertebrates, with the maximum recorded hemizygosity (inclusive of insertions) seen here 10.81%.
For each hemizygous locus observed, there are four genotypes possible for each parent and 16 possible crosses (electronic supplementary material, figure S3). Of these, 14 crosses could potentially give rise to a heterozygous (hemizygous) offspring. As both heterozygous (hemizygous) and homozygous positive individuals possess at least one copy of the locus, in the binary determination of presence or absence, both states would be counted as examples of locus presence while only those homozygous negative (nullizygous) for the locus would be counted as absence.
This means that for each hemizygous locus identified in an individual, six of the potential 14 genotype crosses involve one parent with at least one copy of the locus and the other parent with no copies of the locus. As a result, if the parents of the hemizygous individual were sampled, there would be a 3/7 chance that each locus would be subject to PAV between the two parents. If this ratio is applied to total nucleotides located within hemizygous regions for the species with the highest observed rate of hemizygosity in this study, S. broughtonii at 6.69%, the total genome percentage subject to PAV between the two parents would be 2.87%—at least an order of magnitude higher than the less than 0.2% difference between individuals observed in humans [65]. This number is also likely to be a significant underestimate as it only considers loci that are hemizygous in the offspring of the two individuals and does not take into account loci for which the sampled individual is nullizygous.
An important potential caveat of this calculation is that it relies on the assumption that the two alleles for each hemizygous locus (present and absent) exist in the population at equal frequencies. While we are unable to make such estimates for the species in question, appropriate data for genes subject to PAV in the Mediterranean mussel are available [17]. Of the 14 570 genes which were absent from at least one of 16 sampled genomes, most were found to be absent in only a small number of individuals (approx. 50% missing from between one and three of 16 sampled individuals). As the absence of a gene implies that the hemizygous locus is homozygous absent at this site (electronic supplementary material, figure S3), we calculate that for any given hemizygous locus found in the sequenced individual there is a 33.8% probability that that locus would be subject to PAV between the two parents (electronic supplementary material, file S6). Given this slightly reduced likelihood in comparison to the example in which both alleles exist at equal proportions within the population (3/7th, 42.9%), and assuming a similar situation in Scapharca, the total genome percentage subject to PAV between the two parents of the sequenced individual would be 2.25%.
While hemizygosity is indicative of PAV at the population level, this might not hold true for all loci, for example, if nullizygous individuals are inviable. However, the data collected from the Mediterranean mussel indicates that nullizygosity is broadly tolerated in this species [17]. In this case, 58% of the 12 212 genes encoded by hemizygous regions in the reference genome were subject to PAV in at least one of the 15 resequenced individuals. In mussel, PAV most often targets ‘young’ and recently expanded multigenic families, which contain a large number of dispensable genes. Nullizygosity may, therefore, be tolerated at each individual locus, even if this condition is not simultaneously tolerated at all loci, i.e. individuals are viable as long as at least one dispensable gene for each AMP family is present.
Until more phyla are sampled, we cannot comment on whether the situation in molluscs or vertebrates is more representative of the norm in the Metazoa. However, it is likely that hemizygosity and the existence of open pan-genomes is much more common than we presently appreciate.
(b). Sex chromosome evolution and hemizygosity
Sex chromosomes represent a possible reservoir of hemizygosity which must be taken into consideration when presenting widespread evidence of this phenomenon across the Mollusca. In mammals, hemizygosity is largely restricted to sex chromosomes [68]. In molluscs, no sex chromosome has ever been described and consistent with their absence, hemizygous DNA appears to be fairly evenly distributed between and within all chromosomes (figures 1d and 2). Although a small amount of variability in the density of hemizygous loci can be observed across some chromosomes, we have found no evidence to suggest a link between hemizygosity and sex determination in the targeted species, which is also consistent with the observations previously collected in M. galloprovincialis [17].
In mammals, the chromosome carrying the sex-determining gene (SrY) has become progressively degenerated over time through a process of recombination suppression and genetic drift [68]. By contrast, the evidence shown here suggests mollusc autosomes have become hemizygous through transposon and/or retroviral activity.
(c). Hemizygosity and mollusc biology
Molluscs, and particularly bivalve molluscs, are commonly broadcast spawning species with high population numbers and high levels of genomic heterozygosity. This heterozygosity, as measured by k-mer distribution, can at least in part be explained by hemizygosity as k-mers from both heterozygous alleles (two distinct alleles arising from the same locus) and hemizygous alleles (present in a single copy) contribute to the putative ‘heterozygous k-mer peak' (electronic supplementary material, file S2). In the k-mer plots of total mapped reads (figure 3a, top rows), the first peak (heterozygous peak) has a coverage approximately half that of the second peak (homozygous peak); however, k-mers that come from unique (non-repetitive) hemizygous regions form coverage peaks corresponding to the whole genome heterozygous peaks and so will also contribute to this ‘heterozygous’ peak (figure 3a, bottom rows). k-mers that come from repetitive hemizygous regions (greater than two copies within hemizygous regions or one copy in a hemizygous region and at least one copy in a homozygous region) will not contribute to either the heterozygous or homozygous peaks as their coverage will be at least 1.5× that of homozygous k-mers. Finally, k-mers for which there are two copies both located within hemizygous regions would be expected to contribute to the ‘homozygous’ peak.
While the number of two copy hemizygous k-mers is expected to be low and thus comprise only a tiny portion of the homozygous peak, organisms with significant non-repetitive hemizygous DNA content should have a significant portion of their putative heterozygous peak being composed of hemizygous k-mers. Many molluscs have very high reported heterozygosity levels as determined by k-mer analysis but our findings suggest that this rate may be at least partially explained by large non-repetitive hemizygous DNA content which is impossible to discern from heterozygous DNA using a k-mer-based approach. This has also been noted elsewhere [17], but the additional evidence provided by the extra species sampled here makes this obvious.
At first sight, the M. galloprovincialis genome [17] is characterized by somewhat ‘extreme’ levels of hemizygosity and gene PAV compared to the species considered in this study. The fraction of hemizygous genomic sequence in the mussel is nearly sixfold that of S. broughtonii, the species with the highest hemizygosity among those included in this study (i.e. 36.78% versus 6.69%). Moreover, approximately 35% mussel protein-coding genes were encoded by hemizygous genomic regions, as opposed to less than 2% found in the eight molluscan species studied here (table 2). These differences may be partially attributable to the fact that the estimates provided here derive from only single individuals.
Further sampling will be needed to find the regions that are homozygous absent (nullizygous) from these specimens, but present more generally in the wider population. The size of the hemizygous regions not incorporated into these particular assemblies could be significant. Furthermore, we have not annotated ‘insertions’, regions present in these specimens but absent from the published genome assemblies. These could again represent a sizable fraction of the genome. In summary, the M. galloprovincialis pan-genome appears to have a significantly higher ‘openness’ than most other molluscs, but this could be artefactual and bears further investigation.
The very low level of heterozygosity observed in A. fulica when compared to A. immaculata and the other species examined here may be the outcome of founder effects following its unintentional introduction to China sometime prior to 1931 [69,70]. Both Achatina species are invasive in China; however, due to the sparsity of information available on the size and diversity of the introduced populations, assumptions regarding the impacts of these events on the genetic diversity of the two subsequent populations would be speculative. What is clear is that genetic diversity of the A. fulica specimen is far lower than that of the conspecific A. immaculata specimen; however, the reason for this difference is unresolved.
As noted in the results, in A. immaculata, a small peak of deletion-mapping k-mers corresponds to the homozygous peak in the whole genome k-mer histogram. If these k-mers are the result of duplicated hemizygous regions that arose and have been maintained since the whole genome duplication event that occured in an Achatina ancestor approximately 70 Ma [49], this would have significant consequences for our understanding of hemizygous DNA evolution, birth/death dynamics and long-term persistence within genomes as no reports of such long-term persistence of hemizygosity and/or PAV have yet been reported in animals.
(d). Gene content in hemizygous regions, and gene family over-representation
The idea that dispensable genes may provide improved adaptation potential, suggested ever since the first definition of the pan-genome concept in the scientific literature [71], is now broadly accepted thanks to multiple large-scale genome re-sequencing studies carried out in several prokaryotes and in a few eukaryotes. The number of pan-genomic investigations carried out in metazoans remains extremely limited, and the only molluscan species which has been so far targeted by this type of investigation is the Mediterranean mussel M. galloprovincialis. In line with the adaptive nature of prokaryotic and eukaryotic pan-genomes, mussel dispensable genes were found to be highly enriched in functions related with innate immune response and survival, which may explain the high biotic resilience and invasiveness of this species [17].
Even though the enrichment of gene families associated with adaptation were not as strong in our datasets, some notable overlaps were identified. For example, AIG1 immune-related GTPases, expanded in several stress-adapted invertebrates [62], were enriched in P. canaliculata, and recurrent annotations linked with stress-related HSP70-like proteins, as well as with C1qDC proteins and C-type lectins, which are thought to act as soluble receptors for microbe-associated molecular patterns (MAMPs) in the molluscan immune system [72] were also found.
We have investigated the contribution of HSP70-12 and C1qDC genes to hemizygous regions in particular here. In both of these families, multiple gene copies are found in hemizygous regions in bivalves, while in cephalopods and gastropods they appear to be restricted to homozygous regions. Where they are found, they commonly occur in duplicate which is likely the result of arising through TE-mediated tandem duplication.
Dispensable genes provide a significant contribution to phenotypic variability in bacteria and viruses, enabling their rapid adaptation to new ecological niches and modulating the interaction of pathogenic species with their hosts [6,73]. Similarly, the presence of open pan-genomes may explain the cosmopolitan distribution of some marine microalgae, able to thrive in largely different environmental conditions thanks to the accessory metabolic functions provided by dispensable genes [3]. Moreover, dispensable genes play a key role in the interplay between plants and associated fungi, providing improved biotic resistance to the host and enhanced pathogenicity to their parasites [74,75]. The dispensable genes in mollusc pan-genomes may likewise provide the potential for situationally, regionally and ecologically adaptive variation.
(e). How do these hemizygous structural variants come about, and why does gene presence/absence variation persist?
The most likely sources of hemizygous DNA are transposon and/or retroviral insertions. Evidence for this lies in the enrichment for retroelement-associated genes in several of the species investigated here, in addition to the conserved GC bias profile of the homozygous/hemizygous boundaries (figure 2b). Retroelements are known to encode G-rich sequences at their 5′ and/or 3′ boundaries which form four-stranded secondary structures known as G-quadruplexes [76]. G-quadruplexes have also been implicated in transposon-regulating Piwi interacting RNA (piRNA) biogenesis [77,78] and the GC enrichment observed here at the homozygous/hemizygous boundaries is consistent with transposon terminal G-quadruplexes (figure 2b). The outcome of retroelement-mediated gene duplication coupled with rapid gene turnover is likely to present as lineage-specific gene expansion as has been previously observed for both HSP70 and C1qDC (electronic supplementary material, figure S2 [64]).
Hemizygous regions likely impose a cost on the genomes that carry them. The additional load of TEs, coupled to the over-representation of DNA repair-related genes they encode, suggests that hemizygosity goes hand-in-hand with a decrease in DNA stability. Hemizygosity, if prevalent, may also interfere with homologous recombination-based DNA damage repair mechanisms through recombination suppression (electronic supplementary material, figure S4), and could potentially impact breeding between populations with high levels of haplotypic variation through post-zygotic selection, as previously suggested in M. galloprovincialis [17,79,80]. Furthermore, the insertion or deletion of large blocks of DNA, regardless of their coding capacity, is likely to impact flanking gene expression due to modification of the cis-regulatory lansdscape as was recently demonstrated in the tomato [9]. How, then, are these regions not rapidly purged from populations by natural selection?
It is possible that hemizygosity, while adding to the standing pool of genetic variation and thus adaptive at a population level, results in a larger number of errors while ‘crossing-over’ during meiosis. This could result in a need for a higher number of double-strand break repair genes (e.g. here, tankyrase, RAG51), and similar repair mechanisms. These could migrate into hemizygous regions over time (through TE/retroviral action) and be conserved by natural selection. Alternatively, new hemizygous DNA that is introduced but does not include stability genes on arrival, could be purged quickly leaving only those hemizygous regions that encode stability related genes left for us to observe. It is also possible that the large population size of many mollusc species makes these alleles (which could be rare) difficult to purge from populations as a whole. These hypotheses have not been tested rigorously here, but as additional data becomes available across the tree of life, these questions will be able to be addressed coherently.
(f). Future perspectives and open questions
There are a number of ‘known unknowns’ still to resolve regarding genes in hemizygous sites. None of the genomes investigated here have annotated long non-coding RNAs (lncRNAs) or small non-coding RNAs (snRNAs), and therefore we are unable to speculate as to whether these are also found in these regions, even though a large number of dispensable lncRNA genes have been reported in M. galloprovincialis [17].
In order to make reliable quantitative comparative assessments of hemizygosity between genomes, future comparative studies should use genomes built with consistent assembly and annotation pipelines. Independently built genomes like those assessed here, which used a range of source data and assembly methods (see electronic supplementary material, file S7), likely suffer from assumptions made by the underlying software regarding how to treat hemizygous regions. Under some scenarios, longer (but lower coverage) alleles might be preferred while other pipelines may prioritize more consistent higher coverage, with low coverage alleles excluded from the final assembly. Crucially for gene enrichment analyses, custom repeat libraries might flag repetitive transposon-associated genes, marking them for exclusion from final gene sets. This would result in their exclusion from subsequent enrichment analyses and may explain why enrichment for TE-associated genes was not universally detected here. The utilization of previously assembled genomes in this study may also mask errors associated with false duplications of large highly heterozygous genomic regions. As recently discussed in a preprint by the Vertebrate Genome Project consortium, highly heterozygous genomes are subject to false duplications and, in the present study, these would manifest as false-positive hemizygous calls [81]. Standardized assembly and annotation pipelines would aid in dealing with these issues.
The widespread presence of hemizygosity in mollusc genomes also suggests that some modern assembly algorithms may need adjustment to take into account the prevalence of hemizygous regions. Upon encountering hemizygous regions with coverage significantly below that of the remainder of the assembly, it is plausible that some algorithms may break contigs at the point of low coverage in the assumption that the low coverage region corresponds to contamination or other artefact. Haplotype-aware assembly algorithms will likely cope with this in many cases. However, along with haplotype-blind assembly methods, even haplotype-aware assemblers may ignore the ‘deletion’ genotype, particularly when generating a haploid approximation of the full diploid genome sequence.
Re-sequencing of multiple individuals will be important to obtain a truer picture of the complete pan-genomic complement, and to determine how common particular dispensable genes and genomic regions are within the species. Genotyping tools such as Paragraph [82] or vg [83] could also be used to note the presence or absence of predefined hemizygous regions. When performing re-sequencing experiments, it would be interesting to contrast wild-caught individuals and those that have gone through bottlenecks (domesticated/island effect) to test the impact of these on genomic evolution.
5. Conclusion
In this work, we have put forward the first systematic investigation of the prevalence of hemizygosity across a metazoan clade. We have found that a number of recently sequenced conchiferan molluscs show widespread hemizygosity at multiple loci across their genomes. Bivalves, in particular, have a striking pattern of hemizygosity, which may reflect the broadcast spawning life cycle of the species sequenced. Genes found in these regions in the mollusc species examined are enriched for functions related to transposition, DNA repair and stress response, suggesting that these loci could be both linked to repetitive elements and could provide adaptive potential under specific environmental circumstances.
This approach, which is both cost-effective and broadly applicable, will be useful for assaying for the presence and utility of hemizygosity more generally across the tree of life. This phenomenon remains under-investigated, but may have profound implications for our understanding of genomic evolution at both the population and species level.
Acknowledgements
Our thanks to Dr Angus Davison and Dr Maurine Neiman for organizing the Pearls of Wisdom symposium and curating this Theo Murphy issue. We thank the members of our laboratories for their support in our work. In particular, we thank Samuele Greco of the Gerdol lab for his assistance. We acknowledge and thank Noah Schlottman, Casey Dunn, Nobu Tamura, T. Michael Keesey, Scott Hartmann, Katie S. Collins, and Brockhaus and Efron for their Phylopic images (http://creativecommons.org/licenses/by-sa/3.0/).
Ethics
No ethical permissions are required for the work carried out in this manuscript. This work does not include human tissue, vertebrate animals, fieldwork or museum specimens in its analyses.
Data accessibility
The datasets and code supporting this article have been uploaded as part of the electronic supplementary material. All genomic sequences are available from the original sources with accession numbers as cited in the manuscript.
Authors' contributions
A.D.C. conceived of the study and designed experiments. A.D.C., N.J.K. and M.G. undertook the data analysis and drafted the manuscript. All authors gave final approval for publication and agree to be held accountable for the work performed therein.
Competing interests
The authors declare they have no competing interests.
Funding
N.J.K.'s attendance of this symposium was supported by the US National Science Foundation Theo Murphy Meeting Participation Award.
References
- 1.Han MV, Thomas GWC, Lugo-Martinez J, Hahn MW. 2013. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol. Biol. Evol. 30, 1987-1997. ( 10.1093/molbev/mst100) [DOI] [PubMed] [Google Scholar]
- 2.Moyers BA, Zhang J. 2016. Evaluating phylostratigraphic evidence for widespread de novo gene birth in genome evolution. Mol. Biol. Evol. 33, 1245-1256. ( 10.1093/molbev/msw008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Read BA, et al. 2013. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 499, 209-213. ( 10.1038/nature12221) [DOI] [PubMed] [Google Scholar]
- 4.McCarthy CGP, Fitzpatrick DA. 2019. Pan-genome analyses of model fungal species. Microb. Genom. 5, e000243. ( 10.1099/mgen.0.000243) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tettelin H, et al. 2005. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial ‘pan-genome’. Proc. Natl Acad. Sci. USA 102, 13 950-13 955. ( 10.1073/pnas.0506758102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McInerney JO, McNally A, O'Connell MJ. 2017. Why prokaryotes have pangenomes. Nat. Microbiol. 2, 17040. ( 10.1038/nmicrobiol.2017.40) [DOI] [PubMed] [Google Scholar]
- 7.Vernikos G, Medini D, Riley DR, Tettelin H. 2015. Ten years of pan-genome analyses. Curr. Opin. Microbiol. 23, 148-154. ( 10.1016/j.mib.2014.11.016) [DOI] [PubMed] [Google Scholar]
- 8.Song J-M, et al. 2020. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants 6, 34-45. ( 10.1038/s41477-019-0577-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alonge M, et al. 2020. Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell 182, 145-161.e23. ( 10.1016/j.cell.2020.05.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golicz AA, Bayer PE, Bhalla PL, Batley J, Edwards D. 2020. Pangenomics comes of age: from bacteria to plant and animal applications. Trends Genet. 36, 132-145. ( 10.1016/j.tig.2019.11.006) [DOI] [PubMed] [Google Scholar]
- 11.Dey A, Chan CKW, Thomas CG, Cutter AD. 2013. Molecular hyperdiversity defines populations of the nematode Caenorhabditis brenneri. Proc. Natl Acad. Sci. USA 110, 11 056-11 060. ( 10.1073/pnas.1303057110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crombie TA, et al. 2019. Deep sampling of Hawaiian Caenorhabditis elegans reveals high genetic diversity and admixture with global populations. eLife 8, e50465. ( 10.7554/elife.50465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Small KS, Brudno M, Hill MM, Sidow A. 2007. Extreme genomic variation in a natural population. Proc. Natl Acad. Sci. USA 104, 5698-5703. ( 10.1073/pnas.0700890104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sudmant PH, et al. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526, 75-81. ( 10.1038/nature15394) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherman RM, et al. 2019. Assembly of a pan-genome from deep sequencing of 910 humans of African descent. Nat. Genet. 51, 30-35. ( 10.1038/s41588-018-0273-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tian X, et al. 2020. Building a sequence map of the pig pan-genome from multiple de novo assemblies and Hi-C data. Sci. China Life Sci. 63, 750-763. ( 10.1007/s11427-019-9551-7) [DOI] [PubMed] [Google Scholar]
- 17.Gerdol M, et al. 2020. Massive gene presence-absence variation shapes an open pan-genome in the Mediterranean mussel. Gen. Biol. 21, 275. ( 10.1186/s13059-020-02180-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rosa RD, Alonso P, Santini A, Vergnes A, Bachère E. 2015. High polymorphism in big defensin gene expression reveals presence–absence gene variability (PAV) in the oyster Crassostrea gigas. Dev. Comp. Immunol. 49, 231-238. ( 10.1016/j.dci.2014.12.002) [DOI] [PubMed] [Google Scholar]
- 19.Shevchenko AI, Dementyeva EV, Zakharova IS, Zakian SM. 2019. Diverse developmental strategies of X chromosome dosage compensation in eutherian mammals. Int. J. Dev. Biol. 63, 223-233. ( 10.1387/ijdb.180376as) [DOI] [PubMed] [Google Scholar]
- 20.Quadrana L, et al. 2019. Transposition favors the generation of large effect mutations that may facilitate rapid adaption. Nat. Commun. 10, 3421. ( 10.1038/s41467-019-11385-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gosling E. 2015. Marine bivalve molluscs, 2nd edn. Hoboken, NJ: John Wiley & Sons. [Google Scholar]
- 22.Zannella C, Mosca F, Mariani F, Franci G, Folliero V, Galdiero M, Tiscar PG, Galdiero M. 2017. Microbial diseases of bivalve mollusks: infections, immunology and antimicrobial defense. Mar. Drugs 15, 182. ( 10.3390/md15060182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kocot KM, Poustka AJ, Stöger I, Halanych KM, Schrödl M. 2020. New data from Monoplacophora and a carefully-curated dataset resolve molluscan relationships. Sci. Rep. 10, 101. ( 10.1038/s41598-019-56728-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biosciences P. 2017. pbsv. Menlo Park, CA: Github. See https://github.com/PacificBiosciences/pbsv.
- 25.Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094-3100. ( 10.1093/bioinformatics/bty191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573-580. ( 10.1093/nar/27.2.573) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anand L, Rodriguez Lopez CM. 2020. chromoMap: an R package for interactive visualization and annotation of chromosomes. bioRxiv 605600. ( 10.1101/605600) [DOI] [PMC free article] [PubMed]
- 28.Bushnell B. 2014. BBMap. See https://sourceforge.net/projects/bbmap/.
- 29.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 1303.3997.
- 30.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078-2079. ( 10.1093/bioinformatics/btp352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marçais G, Kingsford C. 2011. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764-770. ( 10.1093/bioinformatics/btr011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ranallo-Benavidez TR, Jaron KS, Schatz MC. 2020. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 11, 1432. ( 10.1038/s41467-020-14998-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neph S, et al. 2012. BEDOPS: high-performance genomic feature operations. Bioinformatics 28, 1919-1920. ( 10.1093/bioinformatics/bts277) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841-842. ( 10.1093/bioinformatics/btq033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pedersen BS, Quinlan AR. 2018. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics 34, 867-868. ( 10.1093/bioinformatics/btx699) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29-W37. ( 10.1093/nar/gkr367) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finn RD, et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42, D222-D230. ( 10.1093/nar/gkt1223) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falcon S, Gentleman R. 2008. Hypergeometric testing used for gene set enrichment analysis. In Bioconductor case studies (eds Hahne F, Huber W, Gentleman R, Falcon S), pp. 207-220. Berlin, Germany: Springer. [Google Scholar]
- 39.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215, 403-410. ( 10.1016/S0022-2836(05)80360-2) [DOI] [PubMed] [Google Scholar]
- 40.Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160-1166. ( 10.1093/bib/bbx108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772-780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972-1973. ( 10.1093/bioinformatics/btp348) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587-589. ( 10.1038/nmeth.4285) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268-274. ( 10.1093/molbev/msu300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fontanilla IKC. 2010. Achatina (Lissachatina) fulica Bowdich: its molecular phylogeny, genetic variation in global populations, and its possible role in the spread of the rat lungworm Angiostrongylus cantonensis (Chen). PhD thesis, University of Nottingham, UK. [Google Scholar]
- 46.Fontanilla IKC, Sta Maria IMP, Garcia JRM, Ghate H, Naggs F, Wade CM. 2014. Restricted genetic variation in populations of Achatina (Lissachatina) fulica outside of East Africa and the Indian Ocean Islands points to the Indian Ocean Islands as the earliest known common source. PLoS ONE 9, e105151. ( 10.1371/journal.pone.0105151) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahmoud M, Gobet N, Cruz-Dávalos DI, Mounier N, Dessimoz C, Sedlazeck FJ. 2019. Structural variant calling: the long and the short of it. Genome Biol. 20, 246. ( 10.1186/s13059-019-1828-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Audano PA, et al. 2019. Characterizing the major structural variant alleles of the human genome. Cell 176, 663-675.e19. ( 10.1016/j.cell.2018.12.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu C, et al. 2020. Giant African snail genomes provide insights into molluscan whole-genome duplication and aquatic-terrestrial transition. bioRxiv 2020.02.02.930693. ( 10.1101/2020.02.02.930693) [DOI] [PubMed]
- 50.Morgante M, De Paoli E, Radovic S. 2007. Transposable elements and the plant pan-genomes. Curr. Opin. Plant Biol. 10, 149-155. ( 10.1016/j.pbi.2007.02.001) [DOI] [PubMed] [Google Scholar]
- 51.Lukic S, Nicolas J-C, Levine AJ. 2014. The diversity of zinc-finger genes on human chromosome 19 provides an evolutionary mechanism for defense against inherited endogenous retroviruses. Cell Death Differ. 21, 381. ( 10.1038/cdd.2013.150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaheen M, Williamson E, Nickoloff J, Lee S-H, Hromas R. 2010. Metnase/SETMAR: a domesticated primate transposase that enhances DNA repair, replication, and decatenation. Genetica 138, 559-566. ( 10.1007/s10709-010-9452-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alicia K, Byrd KDR. 2017. Structure and function of Pif1 helicase. Biochem. Soc. Trans. 45, 1159. ( 10.1042/BST20170096) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bochman ML, Paeschke K, Zakian VA. 2012. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 13, 770-780. ( 10.1038/nrg3296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boán F, Gómez-Márquez J. 2010. In vitro recombination mediated by G-quadruplexes. Chembiochem 11, 331-334. ( 10.1002/cbic.200900612) [DOI] [PubMed] [Google Scholar]
- 56.Puterova J, Lexa M, Kejnovsky E. 2018. Quadruplex DNA in long terminal repeats in maize LTR retrotransposons inhibits the expression of a reporter gene in yeast. BMC Genom. 19, 184. ( 10.1186/s12864-018-4547-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tassinari M, Perrone R, Nadai M, Richter SN. 2019. Stable and conserved G-quadruplexes in the long terminal repeat promoter of retroviruses. ACS Infect. Dis. 5, 1150-1159. ( 10.1021/acsinfecdis.9b00011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu L, Loker ES, Zhang S-M, Buddenborg SK, Bu L. 2020. Genome-wide discovery, and computational and transcriptional characterization of an AIG gene family in the freshwater snail Biomphalaria glabrata, a vector for Schistosoma mansoni. BMC Genom. 21, 1-20. ( 10.1186/s12864-019-6419-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tennessen JA. 2005. Molecular evolution of animal antimicrobial peptides: widespread moderate positive selection. J. Evol. Biol. 18, 1387-1394. ( 10.1111/j.1420-9101.2005.00925.x) [DOI] [PubMed] [Google Scholar]
- 60.Liu H-H, Xiang L-X, Shao J-Z. 2014. A novel C1q-domain-containing (C1qDC) protein from Mytilus coruscus with the transcriptional analysis against marine pathogens and heavy metals. Dev. Comp. Immunol. 44, 70-75. ( 10.1016/j.dci.2013.11.009) [DOI] [PubMed] [Google Scholar]
- 61.Gerdol M, Manfrin C, De Moro G, Figueras A, Novoa B, Venier P, Pallavicini A. 2011. The C1q domain containing proteins of the Mediterranean mussel Mytilus galloprovincialis: a widespread and diverse family of immune-related molecules. Dev. Comp. Immunol. 35, 635-643. ( 10.1016/j.dci.2011.01.018) [DOI] [PubMed] [Google Scholar]
- 62.Guerin MN, Weinstein DJ, Bracht JR. 2019. Stress adapted Mollusca and Nematoda exhibit convergently expanded Hsp70 and AIG1 gene families. J. Mol. Evol. 87, 289-297. ( 10.1007/s00239-019-09900-9) [DOI] [PubMed] [Google Scholar]
- 63.Hu B, et al. 2019. Diverse expression regulation of Hsp70 genes in scallops after exposure to toxic Alexandrium dinoflagellates. Chemosphere 234, 62-69. ( 10.1016/j.chemosphere.2019.06.034) [DOI] [PubMed] [Google Scholar]
- 64.Takeuchi T, et al. 2016. Bivalve-specific gene expansion in the pearl oyster genome: implications of adaptation to a sessile lifestyle. Zool. Lett. 2, 3. ( 10.1186/s40851-016-0039-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li R, et al. 2010. Building the sequence map of the human pan-genome. Nat. Biotechnol. 28, 57-63. ( 10.1038/nbt.1596) [DOI] [PubMed] [Google Scholar]
- 66.Syme RA, Tan K-C, Rybak K, Friesen TL, McDonald BA, Oliver RP, Hane JK. 2018. Pan-Parastagonospora comparative genome analysis—effector prediction and genome evolution. Genome Biol. Evol. 10, 2443-2457. ( 10.1093/gbe/evy192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou P, et al. 2017. Exploring structural variation and gene family architecture with de novo assemblies of 15 Medicago genomes. BMC Genom. 18, 261. ( 10.1186/s12864-017-3654-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wright AE, Dean R, Zimmer F, Mank JE. 2016. How to make a sex chromosome. Nat. Commun. 7, 12087. ( 10.1038/ncomms12087) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jarrett VHC. 1931. The spread of the snail Achatina fulica to South China. Hong Kong Naturalist. 2, 262-264. [Google Scholar]
- 70.Mead AR. 1961. The giant African snail: a problem in economic malacology. Chicago, Ill: University of Chicago Press. [Google Scholar]
- 71.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. 2005. The microbial pan-genome. Curr. Opin. Genet. Dev. 15, 589-594. ( 10.1016/j.gde.2005.09.006) [DOI] [PubMed] [Google Scholar]
- 72.Gerdol M et al. 2018. Immunity in molluscs: recognition and effector mechanisms, with a focus on Bivalvia. In Advances in comparative immunology, pp. 225-341. Cham, Switzerland: Springer. [Google Scholar]
- 73.Wang L, Luo Y, Zhao Y, Gao GF, Bi Y, Qiu H-J. 2020. Comparative genomic analysis reveals an ‘open’ pan-genome of African swine fever virus. Transbound. Emerg. Dis. 67, 1553-1562. ( 10.1111/tbed.13489) [DOI] [PubMed] [Google Scholar]
- 74.Hübner S, et al. 2019. Sunflower pan-genome analysis shows that hybridization altered gene content and disease resistance. Nat. Plants 5, 54-62. ( 10.1038/s41477-018-0329-0) [DOI] [PubMed] [Google Scholar]
- 75.Plissonneau C, Hartmann FE, Croll D. 2018. Pangenome analyses of the wheat pathogen Zymoseptoria tritici reveal the structural basis of a highly plastic eukaryotic genome. BMC Biol. 16, 5. ( 10.1186/s12915-017-0457-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kejnovsky E, Tokan V, Lexa M. 2015. Transposable elements and G-quadruplexes. Chromosome Res. 23, 615-623. ( 10.1007/s10577-015-9491-7) [DOI] [PubMed] [Google Scholar]
- 77.Vourekas A, Zheng K, Fu Q, Maragkakis M, Alexiou P, Ma J, Pillai RS, Mourelatos Z, Wang PJ. 2015. The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev. 29, 617-629. ( 10.1101/gad.254631.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Calcino AD, Fernandez-Valverde SL, Taft RJ, Degnan BM. 2018. Diverse RNA interference strategies in early-branching metazoans. BMC Evol. Biol. 18, 160. ( 10.1186/s12862-018-1274-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bierne N, Bonhomme F, Boudry P, Szulkin M, David P. 2006. Fitness landscapes support the dominance theory of post-zygotic isolation in the mussels Mytilus edulis and M. galloprovincialis. Proc. Biol. Sci. 273, 1253-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.El Ayari T, Trigui El Menif N, Hamer B, Cahill AE, Bierne N. 2019. The hidden side of a major marine biogeographic boundary: a wide mosaic hybrid zone at the Atlantic-Mediterranean divide reveals the complex interaction between natural and genetic barriers in mussels. Heredity 122, 770-784. ( 10.1038/s41437-018-0174-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rhie A, et al. 2020 Towards complete and error-free genome assemblies of all vertebrate species. bioRxiv 110833. ( 10.1101/2020.05.22.110833) [DOI] [PMC free article] [PubMed]
- 82.Chen S, et al. 2019. Paragraph: a graph-based structural variant genotyper for short-read sequence data. Genome Biol. 20, 291. ( 10.1186/s13059-019-1909-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hickey G, et al. 2020. Genotyping structural variants in pangenome graphs using the vg toolkit. Genome Biol. 21, 35. ( 10.1186/s13059-020-1941-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets and code supporting this article have been uploaded as part of the electronic supplementary material. All genomic sequences are available from the original sources with accession numbers as cited in the manuscript.