Abstract
Traditional molecular methods and omics-techniques across molluscan taxonomy increasingly inform biology of Mollusca. Recovery of DNA and RNA for such studies is challenged by common biological properties of the highly diverse molluscs. Molluscan biomineralization, adhesive structures and mucus involve polyphenolic proteins and mucopolysaccharides that hinder DNA extraction or copurify to inhibit enzyme-catalysed molecular procedures. DNA extraction methods that employ the detergent hexadecyltrimethylammoniumbromide (CTAB) to remove these contaminants importantly facilitate molecular-level study of molluscs. Molluscan pigments may stain DNA samples and interfere with spectrophotometry, necessitating gel electrophoresis or fluorometry for accurate quantification. RNA can reliably be extracted but the ‘hidden break’ in 28S rRNA of molluscs (like most protostomes) causes 18S and 28S rRNA fragments to co-migrate electrophoretically. This challenges the standard quality control based on the ratio of 18S and 28S rRNA, developed for deuterostome animals. High-AT content in molluscan rRNA prevents the effective purification of polyadenylated mRNA. Awareness of these matters aids the continuous expansion of molecular malacology, enabling work also with museum specimens and next-generation sequencing, with the latter imposing unprecedented demands on DNA quality. Alternative methods to extract nucleic acids from molluscs are available from literature and, importantly, from communications with others who study the molecular biology of molluscs.
This article is part of the Theo Murphy meeting issue ‘Molluscan genomics: broad insights and future directions for a neglected phylum’.
Keywords: mucopolysaccharides, CTAB, pigment, hidden break, AT rich, mollusca
1. Introduction
The public databases provide a considerable, ever expanding amount of sequence data with entries for all extant classes of the phylum Mollusca. The study of molluscan biology benefits not only from sequence data that resulted from targeted investigations aimed to resolve specific research questions, but owing to more and more common use of next-generation sequencing also from genome assemblies for molluscan species (64 listed in GenBank at the time of writing, with a bias toward bivalves and gastropods) and numerous sets of transcriptomic data (over 8000 in Genbank) that reveal gene assemblages of molluscs in response to different conditions and that aid gene discovery in general. This apparent bounty of mollusc-specific information has to be considered as modest, however, against the great biological diversity of molluscs. A greater abundance of sequence data still reflects that an initial bias of molecular studies is towards Gastropoda and Bivalvia, but more recently the other classes of the Mollusca have also been studied, including the Cephalopoda, Caudofoveata, Scaphopoda, Polyplacophora, Monoplacophora and Aplacophora [1]. With an estimated 100 000 living molluscan species that inhabit many diverse types of habits in marine, freshwater and terrestrial environments (e.g. [2]), much remains to be done to achieve an effective sampling of at least key species that begin to provide a representative sampling of the diversity of molluscan biology.
Because research publications mostly describe successful experiments, it is not directly evident from the literature that molecular studies are frequently challenged by difficulties in obtaining or working with the nucleic acids from molluscs (both DNA and RNA) that are so fundamentally needed for molecular study of molluscan biology. Informal communications among molecular malacologists, however, frequently discuss methods and suggestions for effective extraction and use of high-quality nucleic acids, needed for routine PCR as well as recent next-generation (long read) sequencing, especially for molluscan species that have not been investigated previously. This review will highlight how difficulties in obtaining genomic DNA influenced initial molecular studies of molluscs, and generally effective approaches to overcome the underlying causes that are rooted in molluscan biology. Yet additional problems may be encountered for characterization of DNA, and also of RNA. In addition to select relevant literature, informal considerations will also be presented to identify and address such challenges with a bias that reflects the experience of the author in gastropod studies.
2. Early molecular studies of molluscs
A survey of the NCBI PubMed databases shows that records of characterizations of molluscan genes were first reported in the early 1980s. At this time, a suite of routine methods had been established for research into vertebrate animals, especially focused on mammals (humans, mice). For example, in 1982, the handbook ‘Molecular cloning: A laboratory manual’ by Maniatis et al. [3] provided an extensive collection of protocols for obtaining nucleic acids and follow-up experimental analyses including restriction analyses, Southern and Northern blotting, reverse transcription, cloning, sequencing and generation of both genomic and cDNA libraries. Given the general biological features shared among animals, it stood to reason to expect that these approaches would work not only for vertebrates but could also be applied to study the molecular biology of invertebrate animals, including molluscs.
This assumption proved generally true for molluscan RNA. Biochemical approaches for isolation of ribosomes provided a means to purify ribosomal RNA components by size fractionation, allowing for experimental characterization of 5.8S rRNA using Maxam and Gilbert-like (chemical fragmentation) RNA sequencing and producing the first gene sequences reported for molluscs, including Arion rufus, Aplysia kurodai (gastropoda) and cephalopods Illex illecebrosus, and Sepia officinalis [4–7]. Similarly, because ribosomes engage with gene transcripts for protein translation, precipitation of such ribonucleoprotein complexes also provided a means to isolate mRNA that was purified by use of consecutive organic extractions with phenol and chloroform. Then, through reverse transcription to generate copy DNA (cDNA) and production of cDNA libraries, sequences of molluscan protein-encoding genes were characterized such as neuropeptides that regulate egg-laying behaviour in Aplysia [8]. Phenol extraction from cell lysates, without the need for ribosome precipitation, also yielded bulk RNA that was employed to characterize 18S gene sequences from Peltodoris nobilis (gastropod), Cryptochiton stelleri (gumboot chiton) and the Atlantic surf clam Spisula solidissima [9]. Relatively easily and reliably obtained with standard extraction protocols, more recently also with the use of guanidinium thiocyanate-based methods as first developed by Chomczynski and Sacchi in 1987 [10], RNA was used as the source nucleic acid for cDNA sequences for initial studies of molluscan genes for neuropeptides [11–13], actin [14], haemocyanin [15] and many other genes to follow.
Relative to the burgeoning insights into molluscan biology facilitated by reliable, effective extraction of RNA for molecular studies, there are remarkably few early investigations that also incorporated analyses of molluscan DNA. Evidently, the study of DNA enabled significant novel findings. A remarkable effort from 1990 described a gene encoding for cytoplasmic intermediate filament (F) proteins of the gastropod Helix aspersa as composed of 10 introns and 11 exons, spanning over 60 kb of DNA, with different gene products resulting from alternative splicing [16]. Restriction enzyme analyses demonstrated the presence of a LINE-like transposon as a frequent repeat in the genomic DNA extracted from Biomphalaria glabrata [17]. These studies, however, provide indications that obtaining DNA from these molluscs was not an easy task. Different methods were listed for extraction of genomic DNA from B. glabrata, including the use of CsCl density gradient centrifugation additional to the standard Maniatis-type methods [17]. The genomic DNA from H. aspersa was obtained (to quote Dodemont et al. [16], p.4094) ‘from several tissues using standard techniques (Maniatis et al. 1982)’ [3], but ‘despite careful preparation none of the various DNAs seemed to have a very high molecular size (greater than 100 kb) thus precluding the option of generating cosmid libraries’. Thus, while it was shown to be possible using various methods and by selective use of various tissues, molluscs did not easily yield workable DNA for molecular studies. Personal experience with initial use of ‘mammalian’-style extraction protocols, consisting of detergent-mediated cell lysis followed by organic extractions and alcohol precipitation, also amounted to (unpublished) frustrations. This approach generally leads to failure to obtain DNA from the gastropod B. glabrata, now considered a ‘model snail’ species [18], or it yielded DNA samples that were not easily amenable to simple molecular techniques such as restriction digestion.
3. Sticky proteins and slimy DNA; CTAB and alternatives
Continued research efforts to obtain quality DNA from molluscs, including the use of methods that had been developed for other difficult organisms [19,20], indicated that just like in plants, algea and fungi, problematic extraction and analyses of genomic DNA were caused by polysaccharides and polyphenolic proteins, two chemicals known to complex with DNA and to inhibit DNA-interactive enzymes, respectively [21–23]. It may seem strange that just two types of molecules hindered DNA extraction for a whole phylum with high diversity in the number of species, morphology and biological adaptations to a great variety of environments. However, recent genome-level analyses of the scaly-foot snail (Chrysomallon squamiferum), an extremophile gastropod that has adapted to life by deep-sea thermal vents, revealed genes involved in shell formation that are highly conserved among molluscs. This can be interpreted to underscore the notion that molluscs share a considerable extent of their biological features [24]. Indeed, polyphenolic proteins are thought to be involved in molluscan biomineralization [25,26]. Both polyphenolic proteins and mucopolysaccharides (long linear polysaccharides, also called glycosaminoglycans) are major components of a diversity of molluscan adhesive structures such as adhesive gels, byssal threads and mucus used both to capture food particles and as a body covering throughout the mollusca [27–33].
The pioneering work of the plant and fungal research communities had found that especially a detergent called hexadecyltrimethyl-ammonium bromide (CTAB) forms complexes with polysaccharides, depending on (high) ionic strength [34]. These complexes are removed during organic extractions such that the mucopolysaccharides do not copurify upon subsequent alcohol precipitation of nucleic acids [35]. In 1990, Chapman & Brown [36] recognized that invertebrates, including molluscs, are often recalcitrant sources of DNA, but as an alternative to other (pre-existing) methods relying on SDS-mediated lysis, repeated organic extraction, and CsCl centrifugation, they suggested the use of CTAB as an alternative for DNA extraction. The broad community of molecular malacologists was more directly introduced to this approach in 1993 when Winnepenninckx and co-workers published a CTAB-based method specifically for the extraction of high molecular weight DNA from molluscs [37]. Over 850 citations to date support the notion that this method has become an important component of the molecular toolkit for the study of molluscs.
This protocol involves a modest number of steps. In short, snail tissues are mechanically disrupted in a CTAB-containing lysis buffer with proteinase K, beta-mercaptoethanol, EDTA, NaCl and TrisHCl (pH 8) for digestion at 60°C. The resulting crude lysate is extracted with chloroform followed by alcohol precipitation of DNA (and RNA) from the aqueous phase. The pellet is then rinsed and diluted in a buffer of choice. Personal experience has shown that the procedure can be scaled up or down (for extractions in 1.7 ml microcentrifuge tubes) in order to accommodate extraction of whole snails of modest size or just fractions of organs/tissues from a variety of panpulmonates and caenogastropoda. The protocol is relatively quick and robust, and is easily completed with a good outcome by novice laboratory workers including (under)graduate students in laboratory classes. Other investigators show how flexible the method is to optimize for a DNA extraction of a particular mollusc by varying incubation times, adjusting CTAB concentration, including an RNAse treatment to obtain RNA-free DNA, adding additional organic extractions/ethanol rinses, including PVP to better counter polyphenolic proteins, or using other published variations of CTAB protocols [38–42].
Whereas CTAB-based methods are generally effective, alternative methods may be considered to purify quality DNA from mucopolysaccharide-rich samples, including high-salt precipitation [43]; use of benzyl chloride with repeated extraction [44]; selective binding of DNA to chelex resin [45,46]; and differential centrifugation [47]. Noting that CTAB procedures are not always effective, Solokov developed a method that relies on careful tissue disruption in a lysis buffer with SDS, proteinase K, EDTA, NaCl and TrisHCL (pH 7.5), then adding saturated KCl to form insoluble potassium dodecyl sulfate that coprecipitates mucopolysaccharides from the tissues extract. The application of organic extraction and ethanol precipitation then yielded quality DNA from Polyplacophora, Gastropoda and Bivalves [48].
The biotechnology industry has incorporated the above insights and several commercial companies offer different kits for extraction of quality DNA from challenging biological samples that contain mucopolysaccharides (such as plant, fungi), including kits that are specific for mollusca. The literature shows that these kits are generally effective for DNA from species across molluscan taxonomy, certainly for routine processing of greater sample numbers or in combination with equipment designed for automated, standardized sample processing (for examples, see [42,49–51]). Comparisons of different methods often conclude that traditional ’home spun’ methods (like CTAB and the Sokolov method) show better yields and quality for DNA extraction relative to commercial kits [46,49]. It seems that efforts to obtain DNA from a novel mollusc could best start with a consultation with literature and other molecular malacologists to learn about likely effective methods. Recommendations may include particular methods but can extend to suggesting the use of selected tissues instead of a whole animal. In case of e.g. endangered species, destructive extraction may be avoided by non-invasive sampling of mucus secretions, as a potential source of DNA [52–56]. After initial use of more flexible laboratory-based protocols, a switch could be made to commercial kits as is convenient for yield and throughput.
4. Pigments and DNA
The extraction methods above present important developments that have enabled the study of the molecular biology of the Mollusca by helping to reduce the negative impact of polyphenolic proteins and mucopolysaccharides, yet also other sticky proteins and biochemical compounds may still co-purify with molluscan DNA. The literature does not provide clear indications that such contaminants inhibit or interfere with molecular techniques. Nevertheless, informal communications indicate that some investigators routinely dilute genomic DNA samples after extraction from molluscs 10- or 50-fold based on the practical observation that this increases the success rate of PCR reactions. Also, for some molluscan DNA templates, initially successful PCR will begin to fail with increased age of the sample, with amplification success returning only after only re-extraction of DNA sample. These observations suggest that some, as yet to be characterized, inhibitor(s) of PCR and perhaps other enzymatic reactions may co-purify when extracting DNA from molluscs.
Co-purification of some other factors along with nucleic acids is quite obvious; DNA samples extracted from molluscs often also contain pigments of various colours. In my experience, DNA pellets (obtained using a CTAB-based method) from hygrophylid and stylommatophoran gastropods are usually grey to dark in colour; the caenogastropoda Potamopygrus antipodarum (New Zealand mud snail) even yields strikingly pitch-black DNA [57]. Other communications, outside the literature, mention similar pigmentation of DNA samples from other molluscs, e.g. shades of yellow, coffee-like brown and red (ResearchGate thread started by Davide Guerra; https://www.researchgate.net/post/How-to-deal-with-pigments-that-move-with-nucleic-acids-during-DNA-extraction). Such pigments do not seem to obviously inhibit enzymatic reactions, but it does appear that these contaminants interfere with spectrophotometric quantification of molluscan DNA samples. While I am unaware of specific reports on this matter in the literature on the molecular biology of molluscs, this is suggested by the following observations. Southern blotting experiments with B. glabrata DNA (with DNA pellets, and resulting liquid-dissolved samples, routinely staining dark), using the recommended 10 mg/lane (as determined by spectrophotometry) and well-characterized gene sequences as radio-labelled probes, were prone to yield only weak banding patterns following autoradiography. Results were much improved (e.g. [58]) by effectively ignoring the spectrophotometrically indicated concentration for these DNA samples and increasing at least two-fold the amount of DNA used per lane. Analysis of DNA quantity in RNA-free samples intended for genome sequencing, by staining intensity following gel electrophoresis relative to DNA standards, indicated that spectrophotometry overestimated the concentration of (pigment-containing) B. glabrata DNA by as much as threefold. Likewise, quality checks for Illumina genome sequencing of (pigment-containing) RNA-free DNA samples from several panpulmonate gastropoda, including species of Hygrophila and Stylommatophora, provided similar discrepancies between DNA quantification by spectrophotometry versus (Qubit) fluorometry, a method that measures the fluorescent signal that results from quantitative, specific binding of marker dyes only to DNA at the exclusion of other components in the sample [59]. It may be noted that while spectrophotometry detects overall nucleic acids (failing to distinguish DNA and RNA if both are present), protein and organic content in a sample, it is best used alongside electrophoresis to inform more completely on quality, concentration, composition and degradation of any DNA sample.
Thus it is good to be aware that molluscan DNA may contain pigments and other contaminants. These may not generally prohibit the molecular study of molluscs but they can interfere with downstream analyses. In particular, spectrophotometric quantification of DNA, important for several downstream procedures including next-generation sequencing, can be challenged by molluscan pigments that copurify during the extraction of DNA from molluscs. These contaminants are not integral components of molluscan heterochromatin. Anecdotally, DNA pellets from albino B. glabrata are less intensely stained than those from wild-type pigmented snails and selective use of internal organs that are less pigmented than the ciliated epithelia that cover the snail body may also reduce the pigment content of extracted DNA samples. Extraction of DNA from whole body tissues of adult Physella acuta (Physidae, panpulmonate Gastropoda) yields dark pellets but DNA extracted from embryos (contained in egg masses, not pigmented) is clear to white (figure 1). Finally, a study involving arthropods indicates that experimentation with different extraction methods may also influence the amount of contaminants that copurify with DNA samples [60]. A general good starting point for applications that require precise amounts of input DNA, however, is to rely on fluorometry for accurate quantification of molluscan DNA.
Figure 1.
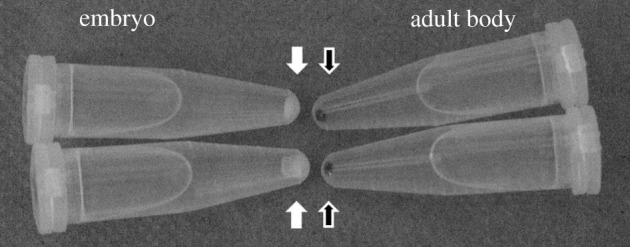
DNA from molluscs is often intensely stained. Extraction of genomic DNA from the full body soft tissues of adult (Shell length about 8 mm) Physella acuta (Gastropoda, Heterobranchia, Hygrophila and Physidae), using a CTAB-based method according to Winnepenninckx et al. [37], routinely yields darkly stained DNA pellets (two replicate DNA samples precipitated in isopropanol and pelleted by centrifugation, shown on the right). The coloration is most likely owing to a pigment of unknown nature that co-purifies with the DNA. It is not a property of the DNA itself, or of a factor that routinely associates with the DNA: the staining is not present in DNA samples extracted from embryos (egg masses) of P. acuta using the same procedure (two replicate DNA pellet samples on the left). Both types of samples can be used for restriction digestion and PCR.
5. RNA, the ‘not so’ hidden break
As stated above, RNA can be harvested from molluscs reliably, in a relatively straightforward manner. Beyond the use of RNA during the initial phase of molecular malacology, RNA has consistently been applied to drive broad gene discovery in molluscs. This started with expressed sequence tagging (ESTs) in the late 1990s and now includes RNA-seq data with the representation of all classes of Mollusca (e.g. [1,61–63]). Additionally, molluscan RNA continues to be used successfully for obtaining full-length cDNA sequences by rapid amplification of cDNA ends (RACE), microarray-based expression studies and annotation of exons in molluscan genomes (e.g. [64–66]). Clearly, there are ample demonstrations that molluscs do in fact yield quality RNA containing full-length mRNA transcripts that allow productive and meaningful downsteam experiments. A common standard approach, however, to test the integrity of mRNA consistently fails to confirm the quality of molluscan RNA. This can be quite vexing, in particular to novice molecular malacologists, raising doubt about methods and sample quality, especially for use in NGS RNA-seq when (commercial) facilities may be reluctant to guarantee productive sequencing for RNA samples that do not pass such quality control. As discussed below, the quality of molluscan RNA is not at issue; rather the issue arises when the method used for quality control fails to consider differences in the molecular biology of vertebrate versus (most) invertebrate animals.
First, the logical concept that underlies methods for determining mRNA quality is that transcripts must be full length to provide complete information for the transcribed gene sequence. Progressive degradation reduces particular transcripts to incomplete sequence fragments of varying length. Accordingly, electrophoretic separation of a quality RNA sample will resolve as a profile of crisp bands, each representing complete transcripts of distinct genes. By contrast, degraded RNA will form a smear owing to the many fragments of randomly overlapping sizes. Protein-coding mRNA transcripts, however, are a minority component (estimated at between 1 and 5%) among several classes that make up total cellular RNA [67]; the majority (approx. 85%) consist of ribosomal RNA (rRNA) sequences that function as structural components of ribosomes. While traditional gel electrophoresis lacks sensitivity to effectively resolve and visualize mRNA transcripts, rRNA molecules are easily detected. Thus, the presence in gel profiles of two well resolved, prominent rRNA transcripts, representing 18S (approx. 2000 nt) and 28S (approx. 4000 nt) was adopted as a proxy for overall intactness of RNA samples at a time when molecular biology was focused toward the study of vertebrate animals. This method has been refined over time by using densitometry to calculate the ratio of 28S/18S as a parameter for sample quality. The development of instruments (like the Agilent Bioanalyzer 2100) that use microcapillary electrophoresis to analyse sample composition with increased sensitivity led to the development of a calculation method that integrates the 18S and 28S signals together with additional parameters to generate an RNA integrity number (RIN) for RNA samples [68]. Ranging from 1 (degraded) to 10, RIN values of 8 or higher are considered to represent quality RNA samples.
The unequivocal presence in Mollusca of both 18S and 28S rRNA as structural components of ribosomes that are essential for protein translation evolutionarily conserved in eukaryotes is also evidenced by an abundance of sequence data (e.g. [69]). Gel analyses of molluscan RNA samples, however, consistently show only one single prominent rRNA of about 2000 nt, the size of an 18S transcript. A personal first encounter in the laboratory with this phenomenon (mid 1990s) led to discussions with other molecular malacologists and the shared conclusion that this was just how RNA samples from this gastropod, B. glabrata, looked on gel.
It is all the more noteworthy that studies by Ishikawa in the 1970s provide an explanation for the absence of the diagnostic gel band for 28S rRNA in gel profiles of molluscan RNA [70–72]. In brief, the 28S rRNA of deuterostomes (including vertebrate animals) is transcribed as one contiguous sequence whereas in most invertebrate protostomes (representing about 95% of animal diversity) including the mollusca, the 28S rDNA gene is split by a central non-transcribed region termed the ‘hidden break’. This yields two distinct, equally sized RNA subunits that combine to form a functional 28S rRNA, sticking together—not by covalent bonds—but only by weak forces. Experimental conditions (denaturation or heat) during RNA extraction commonly disrupt such weak forces (hydrogen bonds) and the functional 28S rRNA unit of 4000 nt breaks apart into the two subunits of about 2000 nt each. Owing to similar sizes, the 28S subunit fragments and the contiguous 18S rRNA transcript have the same electrophoretic mobility and resolve as a single band of approximately 2000 nt, which is not indicative of RNA degradation. Thus, the hidden break invalidates the use of the relative abundance of 18S and 28S for quality control of RNA samples.
Characterization and comparative analysis of the rDNA gene complex of B. glabrata led to the identification of a putative hidden break sequence in the 28S rDNA gene [73], predicting 28S subunits of 1791nt and 1946nt, similar in size to the 1829 nt 18S transcript. Greater than 99.76% sequence identity indicates that the hidden break impacts in the same manner the 28S genes of the sister species Biomphalaria pfeifferi, Biomphalaria sudanica and Biomphalaria choanomphala [74]. For the study of the phylogenetic distribution of the hidden break, Natsidis et al. [75] developed a computational approach to detect the hidden break from RNA-seq data, with positive prediction for 26 of 31 molluscs tested. Moreover, considering that the hidden break challenges quality control of RNA samples from protostomes (95% of animals), they argued for the development of an alternative to the standard RIN that does consider the effect of the hidden break [75]. Already, more recent equipment for evaluation of RNA quality (Agilent tapestation) employs the so-called RINe (RNA integrity number equivalent), an alternative method to assign relevant quality scores to RNA profiles based only on the 18S signal, not considering 28S [76,77].
In the realization that the presence of both 18S and 28S bands is not a valid parameter for quality control, there is no need to repeat RNA extraction efforts in hopes of avoiding the effects of the hidden break. The author does, however, still employ Bioanalyzer assays to analyse RNA samples from gastropod molluscs such as P. acuta, even though this does not provide a meaningful RIN. Samples that show relatively broad peaks (interpreted as smearing/degradation) are considered to be of lower quality. On the other hand, well-defined RNA peaks, especially for the (single) peak representing the rRNA signal and minimal signal greater than the rRNA, have been used with good results as criteria to identify quality samples (figure 2) for RT-PCR, microarray analyses and RNA-seq.
Figure 2.
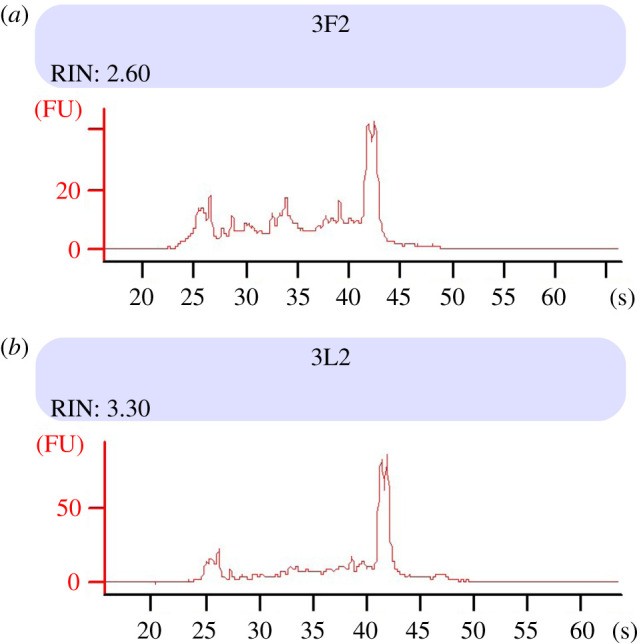
Bioanalyzer profiles of two RNA samples of individual adult Physella acuta (Gastropoda, Heterobranchia, Hygrophila, Physidae). Following Trizol extraction from whole body soft tissues, 1 µl of Turbo DNA-free treated RNA analysed using the Agilent RNA 6000 Nano kit with Agilent Bioanalyzer software v. B.02.08.SI648(S2). Note the single rRNA peak (between 40 and 45 s), representing both 18S and similarly sized subunits 28S rRNA that dissociated during extraction due to the hidden break in 28S. The automatically calculated RIN (note low value) is ignored, rather the sample is interpreted to be of good quality based on showing well-defined, narrow peaks (20–45 s), especially the rRNA peak and low signal for larger sequences (greater than 50 s). (Online version in colour.)
6. AT-rich rRNA
As stated above, a total molluscan RNA sample contains multiple classes of RNA, with a great abundance (approx. 80% of the total) of rRNA sequences that originate from both nuclear and mitochondrial genomes. The use of reverse transcription with mixtures of random and poly-T primers to generate appropriate templates for sequencing will yield cDNA from mRNA and rRNA alike. The presence of many replicate copies of just these few uninformative rRNA sequences can potentially dilute the capacity of library generation to capture sequences that derive from protein-coding genes and compete with the recovery of informative data for transcriptomic studies either by EST sequencing or NGS RNA-seq. It is often suggested that the mRNA component be purified from total RNA to circumvent this problem. Unique among most (but not all) of the various RNA classes [78], mature mRNA is modified by polyadenylation for stability, i.e. a string of between 30 and 200 A residues is added at the 3′ terminus of the transcripts. Accordingly, mRNA sequence can be purified by capturing the 3′ poly(A) tail through hybridization to poly(T) oligos that are coupled to a solid carrier, at the exclusion of other classes of RNA [79].
Previous personal experience indicates that this method for enriching mRNA may be defeated by molluscan biology. A significant challenge that was not discussed in the publication presenting the gene discovery project using ORESTES for the gastropod B. glabrata [80] was that several of multiple mini cDNA libraries, generated by random primer combinations to capture open-reading frame ESTs, yielded 30–60% rRNA-derived sequences. This high percentage of non-informative sequences severely impeded progress in cataloguing the expression of protein-coding genes. Analysis of the underlying cause revealed that the contaminant rRNA sequences of both nuclear and mitochondrial origin generally had a high-AT content, with these residues frequently organized in mononucleotide sequence tracts. This sequence property caused poly(T) primers—as well as randomly designed primers that were high in T content and intended to target open-reading frames within the mRNA from protein-coding genes—to also stick (hybridize) to (A)mononucleotide tracts encompassed with rRNA sequences and initiate reverse transcription of the highly abundant rRNA sequences to cDNA at a ratio much higher than cDNA generation from mRNA transcripts. Efforts to separate polyadenylated mRNA from snail RNA by poly(T) purification did not improve the relative proportion of protein-coding gene (mRNA) sequences versus rRNA sequences. This failure to purify the mRNA was interpreted to result from binding by the poly(T) oligos of not only polyadenylated mRNA but also A-rich rRNA. To my knowledge, only few papers consider this issue as a problem that hampers effective purification of mRNA from B. glabrata, a gastropod mollusc [81,82]. Ultimately, in case of ORESTES, the rate of gene discovery was improved by excluding primer combinations that yielded high proportions of rRNA-derived sequences.
The take-home message of the previous section is that the nucleotide composition, particularly AT content, of rRNA (and potentially other non-coding genes mRNA) may render poly(T) purification ineffective for mRNA of a particular mollusc. Evaluation of available sequence data for the species of mollusc to be studied, or of close relatives, may guide the decision to employ or forgo poly(T) purification of mRNA. It is of note that mitochondrial genomes of invertebrates are generally AT-rich [83], and this may of course impact the base composition of 16S and 12S rRNA sequences. In case of the gastropod B. glabrata (family Planorbidae), both the mitochondrial and the nuclear genomes were found to be approximately 64% AT-rich [18,84]. So for instance, with the indication of similar values for P. acuta of the sister family Physidae, with AT richness of 64% for nuclear genome [85], 69% for the mitochondrial genome and 72% for the 16S rRNA gene [86], it may prove challenging to separate P. acuta mRNA from total RNA by poly(T) purification. It should be noted that this issue also impacts the study of other (invertebrate) taxa and that alternative methods are available to deplete highly abundant rRNA contaminants from mRNA samples such as subtractive hybridization and using specific guideRNAs to target rRNA sequences for CAS9-mediated degradation [87,88].
7. DNA from museum samples
Some research circumstances pose additional challenges to obtaining quality DNA from molluscan tissues. As a first example, molecular exploration of the great diversity of Mollusca may be aided by museum collections. Instead of having to undertake field collections of molluscs that are exceedingly rare or that live in isolated, inaccessible habitats (e.g. deep sea), such specimens may already have been deposited in museum collections, perhaps many years prior. Museum samples are likely to have well-documented taxonomic identifications. Additional to application in taxonomy, some museum samples may also be studied using comparative genomics [89]. Depending on the state of preservation, tissue samples that are decades old can be a source of DNA. Unfortunately, sample preservation may suffer [90]: ethanol evaporation may leave specimens in suboptimal ethanol concentrations, or even dry. A longstanding tradition of using aldehyde-containing fixatives that are good for preserving morphology is not beneficial for molecular studies: aldehyde chemically alters DNA and cross-links proteins with nucleic acids such that DNA is difficult to recover, usually yielding only small fragments. Depending on sample quality, however, commercial kits for DNA extraction have helped to characterize mitochondrial gene sequences from Deminucula atacellana, a small deep-sea bivalve [91], and from 30-year-old formaldehyde-fixed aplacophoran molluscs [92]. Recovery of nuclear genomic DNA for high-throughput sequencing remains challenging, although progress is being made with museum samples that were formalin-fixed and stored in ethanol. A test (employing reptile tissues) showed that a mild heat and alkali treatment, calibrated to minimize damage to DNA, can be applied to break protein–DNA cross-linkages caused by formalin, and DNA can then be obtained by organic extractions or commercial kits. Analysis by Illumina sequencing did provide good mitogenome sequences with high coverage but the sample was still too fragmented to provide nuclear sequences for reliable phylogenetic analysis [93]. Future improvements of such approaches may help to make molluscan samples from museum collections accessible for broader genetic analysis. DNA can be recovered from alcohol-fixed mollusc samples of up to 130 years old for PCR amplification of mitochondrial genes with several commercial extraction kits. Experimental success was generally correlated, however, with the age of the sample and the quality and consistency of fixation [90].
As an alternative to soft tissues, the shells of molluscs must also be considered as a source of DNA. By the extraction of ground up shells, Der Sarkissian and co-workers [94,95] have recovered DNA from a variety of molluscs, including museum samples, encompassing the genera Arctica, Cernuella, Crassostrea, Dreissena, Haliotis, Lymnaea, Margaritifera, Mytilus, Pecten, Ruditapes and Venerupis from marine, freshwater and terrestrial environments. Samples were suitable for high-throughput sequencing to provide sequences from the nuclear genome and complete mitogenome assemblies. Extraction is not always successful, but remarkably DNA was obtained and successfully used for genetic characterization from ancient samples of Portlandia arctica and Mytilus, bivalves recovered from permafrost and dated at ≥100 000 years old.
8. Modern DNA-omics, successes and challenges
Genome characterization is the second example of experimental work that places extra demands on obtaining high-quality DNA from molluscs. Ideally, the DNA sample represents the full organismal genome, with little fragmention and free of inhibitors. The first set of molluscan genome projects (published 2012–2017), involving Crassostrea gigas [66], Lottia gigantean [96], Octopus bimaculoides, [97], Radix auricularia [98] and B. glabrata [18], relied on Sanger sequencing and next-generation sequencing (454/Roche and Illumina), methods that produce relatively short sequence reads (≤1000 bases). As part of a strategy to aid assembly of short reads into longer genomic sequences that is still used to the present day (e.g. [99]), paired-end sequence reads were collected from the termini of size-selected genomic sequence fragments (cloned or direct), ranging up to 10kb or 40kb in length. Isolation of DNA up to this molecular weight size range was attained with either CTAB-based extraction [18] or commercial kits [66,96,97], but greater size is restricted: routine extraction generates forces that lead to mechanical shearing of DNA fragments.
High molecular weight (HMW) DNA of greater than 50 kbp in size can be obtained from molluscs by using methods that have been developed for the generation of large-insert BAC libraries. For this approach, select organs from a mollusc are disrupted into monocellular suspensions. The cells are embedded in agarose plugs that are subsequently incubated in a cell lysis buffer (containing EDTA, TrisHCl, the detergent N-Lauroyl sarcosine and proteinase K) overnight at 50°C, with gentle rotation. DNA that is released from lysed cells is protected in situ from mechanical shearing by the supporting agarose matrix. Following partial restriction digestion of the DNA held in the agarose plug, DNA fragments of up to 500 kbp can be eluted. This approach has yielded BAC libraries for B. glabrata (Gastropoda), C. gigas and Crassostrea virginica (both bivalves) containing genomic inserts with average lengths of 136 kbp, 152 kbp and 320 kbp, respectively [100,101]. DNA fragments of this size present a considerable challenge for sequencing (and assembly) with short-read approaches like first (Sanger) and second (Illumina) generation sequencing methods, but are in fact very appropriate templates for third-generation sequencing approaches like PacBio and Nanopore that routinely produce long reads from several kilobases up to a record 2.3 megabases [102]. A modified agarose plug-based approach was used effectively to prepare HMW genomic DNA for long-read sequencing toward characterization of the Pecten maximus (bivalve) genome by, in 2020 [103]. With the increasing popularity of long-read sequencing, alternative methods have been developed to minimize mechanical shearing during extraction in order to prepare HMW DNA, potentially also from molluscs [104,105], and commercial biotech companies offer an ever-growing variety of kits for extraction of HMW DNA [106,107].
The great potential of the current next-generation sequencing methods is underscored by a number of studies where incorporation of positional information recorded from the organization of sequences around histones with both short-read and long-read next-generation sequence data has led to comprehensive genome assembly of Archeteuthis dux (Giant squid) [108] and yielded chromosome-level assemblies for two species of giant African land snails [109,110], P. maximus (scallop) [103] and Sinonovacula constricta (razor clam) [110], with such an assembly forthcoming for Meretrix meretrix (Venus clam; B Allam 2021, personal communication). The experimental procedures employed for these efforts (also including [41,42]) indicate that several different techniques can yield HMW DNA that is suitable for high-quality characterization of molluscan genomes, including modern extraction kits as well as traditional ‘Maniatis’-type lysis and organic extraction.
Despite the success and inherent potential of the above studies, it still seems premature to consider chromosome-level assemblies as the standard for molluscan genome characterization. Informal discussions reveal that for molluscs, HMW DNA does not necessarily equate with quality DNA that is suitable for long reads or other analysis. Using otherwise functional set-ups for long-read sequencing, analysis of genomic DNA from several gastropods and cephalopods was not productive. Sequencing failure may be owing to inhibition of PacBio chemistry by molluscan DNA. In the case of Nanopore sequencing, molluscan DNA caused extensive blockage of (sequencing) pores. Such blockage may be owing to secondary structures resulting from repetitive regions in the molluscan DNA samples, as similarly proposed for DNA from the black tiger prawn, Penaeus monodon [111]. The observation that Nanopore sequencing was able to process templates consisting of PCR-amplified sequences supports an alternative hypothesis that additional to secondary structures, inhibition could also result from unknown contaminants that associate with native DNA from (some) molluscs (E Heath-Heckman 2021, personal communication). While these obstacles to chromosome-level assemblies for some molluscs may be overcome in the future, characterization of challenging molluscan genomes, also without yielding chromosome-level assemblies, remains valid to move molecular malacology forward.
9. Concluding remarks
During the decades since the early 1980s, molecular malacology has flourished by overcoming sticky problems that impeded access and study of molluscan RNA and DNA. In hindsight, it should be recognized that these challenges represent common features of molluscan biology, often shared by other taxonomic groups including other invertebrate phyla, fungi and plants. Despite obvious common evolutionary ancestry, it may be good to realize that aspects of the biology of the vertebrate animals for which the initial molecular biology methods were developed can be quite distinct from the biology of the majority of life across phylogeny.
With a choice of CTAB-based and other DNA extraction methods to avoid sticky mucopolysccharides and inhibitory polyphenolic proteins, and recognizing the impact of sticky pigments and other factors; the complications for RNA quality control due to hidden break ‘unsticking’ of 28S rRNA subunits; and the problems for poly(T) purification of mRNA due to sticky A-rich rRNA, and even means to extract DNA from museum samples or HMW for long-read sequencing, molecular malacology can continue to explore the biology of the highly diverse Mollusca. A new challenge posed by molluscan biology, i.e. difficulties with long-read sequencing of DNA from some molluscs, does not prohibit meaningful analyses and may be resolved with future technical improvements. This mission is also supported by new analytic and bioinformatic strategies that maximize the biological information—including chromosome-level genome assemblies—that can be distilled from high-throughput (genomic) sequence data, whether long reads and short reads or only short reads if HMW DNA is unavailable. Against the background of extensive sequence data, effective isolation of nucleic acids stands at the basis of comparative genomics to reveal unique aspects of molluscan biology, such as evolutionary development, immune function, biomineralization and lineage-specific genes [1,112–115].
Of course, the experimental goals of individual investigators will legitimately range from genome assembly to PCR amplification of a single-gene sequence fragment. This defines the requirements for the quality and quantity of the nucleic acids sample to be extracted from the mollusc under investigation. The literature (in print or electronic format) gives access to a growing knowledge base that provides an extensive tool kit with alternative approaches. This benefit can be increased if future publications also include (brief) mention of extraction methods that were less productive with the mollusc of study. Importantly, however, my experience indicates that direct communication with other molecular malacologists may help the selection of the likely most successful tool to work with nucleic acids from a particular mollusc.
Acknowledgements
Many thanks to Dr Maurine Neiman and Dr Angus Davison for organizing the 2019 conference, ‘Pearls of wisdom: synergising leadership and expertise in molluscan genomics’ and editing this special issue of Philosophical Transactions B. Kevin McQuirk assisted in preparing figure 2. Dr Elisabeth Heath-Heckman and Dr Bassem Allam provided personal communications and shared experiences with long-read sequencing of molluscan DNA. Two reviewers provided comments and additional input that greatly improved the manuscript. I salute my colleagues at UNM along with the many other molecular malacologists engaged in ‘molluscanomics’ for collaborations and scientific discussions that I have enjoyed over the years.
Ethics
This article does not include research with ethical considerations.
Data accessibility
No primary data reported.
Authors' contributions
C.M.A. contributed the conception and design, drafted and revised the article and approved of the version to be published.
Competing interests
I have no competing interests.
Funding
I received no funding for this study.
References
- 1.Kocot KM, Poustka AJ, Stöger I, Halanych KM, Schrödl M. 2020. New data from Monoplacophora and a carefully-curated dataset resolve molluscan relationships. Sci. Rep. 10, 101. ( 10.1038/s41598-019-56728-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haszprunar G, Schander C, Halanych KM. 2008. Relationships of higher molluscan taxa. In Phylogeny and evolution of the mollusca (eds Ponder W, Lindberg DR), pp. 19-32. Berkeley, CA: University of California Press. [Google Scholar]
- 3.Maniatis T, Fritsch EF, Sambrook J. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- 4.Ursi D, Vandenberghe A, De Wachter R.. 1983. Nucleotide sequences of the 5.8S rRNAs of a mollusc and a porifer, and considerations regarding the secondary structure of 5.8S rRNA and its interaction with 28S rRNA. Nucleic Acids Res. 11, 8111-8120. ( 10.1093/nar/11.22.8111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walker WF, Doolittle WF. 1983. 5S rRNA sequences from four marine invertebrates and implications for base pairing models of metazoan sequences. Nucleic Acids Res. 11, 5159-5164. ( 10.1093/nar/11.15.5159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komiya H, Hasegawa M, Takemura S. 1986. Differentiation of oocyte- and somatic-type 5S rRNAs in animals. J. Biochem. 100, 369-374. ( 10.1093/oxfordjournals.jbchem.a121723) [DOI] [PubMed] [Google Scholar]
- 7.Hendriks L, De Baere R, Vandenberghe A, De Wachter R.. 1987. The nucleotide sequence of the 5S ribosomal RNA of Actinia equina and Sepia officinalis. Nucleic Acids Res. 15, 2773. ( 10.1093/nar/15.6.2773) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheller RH, Jackson JF, McAllister LB, Rothman BS, Mayeri E, Axel R. 1983. A single gene encodes multiple neuropeptides mediating a stereotyped behavior. Cell 32, 7-22. ( 10.1016/0092-8674(83)90492-0) [DOI] [PubMed] [Google Scholar]
- 9.Field KG, Olsen GJ, Lane DJ, Giovannoni SJ, Ghiselin MT, Raff EC, Pace NR, Raff RA. 1988. Molecular phylogeny of the animal kingdom. Science 239, 748-753. ( 10.1126/science.3277277) [DOI] [PubMed] [Google Scholar]
- 10.Chomczynski P, Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156-159. ( 10.1006/abio.1987.9999) [DOI] [PubMed] [Google Scholar]
- 11.Schaefer M, Picciotto MR, Kreiner T, Kaldany RR, Taussig R, Scheller RH. 1985. Aplysia neurons express a gene encoding multiple FMRFamide neuropeptides. Cell 41, 457-467. ( 10.1016/s0092-8674(85)80019-2) [DOI] [PubMed] [Google Scholar]
- 12.Taussig R, Scheller RH. 1986.The Aplysia FMRFamide gene encodes sequences related to mammalian brain peptides. DNA 5, 453-461. ( 10.1089/dna.1.1986.5.453) [DOI] [PubMed] [Google Scholar]
- 13.Smit AB, Vreugdenhil E, Ebberink RH, Geraerts WP, Klootwijk J, Joosse J. 1988. Growth-controlling molluscan neurons produce the precursor of an insulin-related peptide. Nature 331, 535-538. ( 10.1038/331535a0) [DOI] [PubMed] [Google Scholar]
- 14.DesGroseillers L, Auclair D, Wickham L. 1990. Nucleotide sequence of an actin cDNA gene from Aplysia californica. Nucleic Acids Res. 18, 3654. ( 10.1093/nar/18.12.3654) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang WH. 1988. cDNA cloning of the Octopus dofleini hemocyanin: sequence of the carboxyl-terminal domain. Biochemistry 27, 7276-7282. ( 10.1021/bi00419a015) [DOI] [PubMed] [Google Scholar]
- 16.Dodemont H, Riemer D, Weber K. 1990. Structure of an invertebrate gene encoding cytoplasmic intermediate filament (IF) proteins: implications for the origin and the diversification of IF proteins. EMBO J. 9, 4083-4094. ( 10.1002/j.1460-2075.1990.tb07630.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knight M, Miller A, Raghavan N, Richards C, Lewis F. 1992. Identification of a repetitive element in the snail Biomphalaria glabrata: relationship to the reverse transcriptase-encoding sequence in LINE-1 transposons. Gene 118, 181-187. ( 10.1016/0378-1119(92)90187-t) [DOI] [PubMed] [Google Scholar]
- 18.Adema CM, et al. 2017. Whole genome analysis of a schistosomiasis-transmitting freshwater snail. Nat. Commun. 8, 15451. ( 10.1038/ncomms15451). Erratum in: Nat. Commun. 8, 16153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rice EL. 1990. Nucleotide sequence of the 18S ribosomal RNA gene from the Atlantic sea scallop Placopecten magellanicus (Gmelin, 1791). Nucleic Acids Res. 18, 5551. ( 10.1093/nar/18.18.5551) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rice EL, Bird CJ. 1990. Relationships among geographically distant populations of Gracilaria verrucosa (Gracilariales, Rhodophyta) and related species. Phycologia 29, 501-510. ( 10.2216/i0031-8884-29-4-501.1) [DOI] [Google Scholar]
- 21.Aoki Y, Koshihara H. 1972. Inhibitory effects of acid polysaccharides from sea urchin embryos on RNA polymerase activity. Biochim. Biophys. Acta. 272, 33-43. ( 10.1016/0005-2787(72)90030-5) [DOI] [PubMed] [Google Scholar]
- 22.Furukawa K, Bhavanandan VP. 1983. Influences of anionic polysaccharides on DNA synthesis in isolated nuclei and by DNA polymerase α: correlation of observed effects with properties of the polysaccharides. Biochim. Biophys. Acta. 740, 466-475. ( 10.1016/0167-4781(83)90096-9) [DOI] [PubMed] [Google Scholar]
- 23.Shioda M, Murakami-Murofushi K. 1987. Selective inhibition of DNA polymerase α by a polysaccharide purified from slime of Physarum polycephalum. Biochem. Biophys. Res. Commun. 146, 61-66. ( 10.1016/0006-291x(87)90690-5) [DOI] [PubMed] [Google Scholar]
- 24.Sun J, et al. 2020. The Scaly-foot Snail genome and implications for the origins of biomineralised armour. Nat. Commun. 11, 1657. ( 10.1038/s41467-020-15522-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunt S. 1976. The gastropod operculum: a comparative study of the composition of gastropod opercular proteins. J. Molluscan Stud. 42, 251-260. ( 10.1093/oxfordjournals.mollus.a065331) [DOI] [Google Scholar]
- 26.Miserez A, Rubin D, Waite JH. 2010. Cross-linking chemistry of squid beak. J. Biol. Chem. 285, 38 115-38 124. ( 10.1074/jbc.M110.161174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grenon JF, Walker G. 1980. Biochemical and rheological properties of the pedal mucus of the limpet, Patella vulgata L. Comp. Biochem. Physiol. B 66, 451-458. [Google Scholar]
- 28.von Byern J, Klepal W.. 2006. Adhesive mechanisms in cephalopods: a review. Biofouling 22, 329-338. ( 10.1080/08927010600967840) [DOI] [PubMed] [Google Scholar]
- 29.Smith AM. 2006. The biochemistry and mechanics of gastropod adhesive gels. In Biological adhesives (eds Smith AM, Callow JA), pp. 177-192. Cham, Switzerland: Springer. ( 10.1007/978-3-319-46082-6_8) [DOI] [Google Scholar]
- 30.Silverman HG, Roberto FF. 2007. Understanding marine mussel adhesion. Mar. Biotechnol. 9, 661-681. ( 10.1007/s10126-007-9053-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao H, Waite JH. 2005. Coating proteins: structure and cross-linking in fp-1 from the green shell mussel Perna canaliculus. Biochemistry 44, 15 915-15 923. ( 10.1021/bi051530g) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Audino JA, Marian JEAR, Wanninger A, Lopes SGBC. 2015. Mantle margin morphogenesis in Nodipecten nodosus (Mollusca: Bivalvia): new insights into the development and the roles of bivalve pallial folds. BMC Dev. Biol. 15, 22. ( 10.1186/s12861-015-0074-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beninger P, St-Jean S. 1997. The role of mucus in particle processing by suspension-feeding marine bivalves: unifying principles. Mar. Biol. 129, 389-397. ( 10.1007/s002270050179) [DOI] [Google Scholar]
- 34.Murray MG, Thompson WF. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321-4325. ( 10.1093/nar/8.19.4321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heikrujam J, Kishor R, Mazumder PB. 2020. The chemistry behind plant DNA isolation protocols. In Biochemical analysis tools. methods for bio-molecules studies (eds Boldura O-M, Balta C, Awwad N), Chapter 8. IntechOpen Limited. ( 10.5772/intechopen.92206 [DOI] [Google Scholar]
- 36.Chapman RW, Brown BL. 1990. Mitochondrial DNA isolation methods. In Electrophoretic and isoelectric focusing techniques in fisheries management (ed. Whitmore DH), pp. 107-129. Boca Raton, FL: CRC Press. [Google Scholar]
- 37.Winnepenninckx B, Backeljau T, De Wachter R.. 1993. Extraction of high molecular weight DNA from molluscs. Trends Genet. 9, 407. ( 10.1016/0168-9525(93)90102-n) [DOI] [PubMed] [Google Scholar]
- 38.Mäkinen T, Panova M, André C. 2007. High levels of multiple paternity in Littorina saxatilis: hedging the bets?. J. Hered. 98, 705-711. ( 10.1093/jhered/esm097) [DOI] [PubMed] [Google Scholar]
- 39.Panova M, Johansson T, Canbäck B, Bentzer J, Rosenblad MA, Johannesson K, Tunlid A, André C. 2014. Species and gene divergence in Littorina snails detected by array comparative genomic hybridization. BMC Genomics 15, 687. ( 10.1186/1471-2164-15-687) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panova M, et al. 2016. DNA Extraction protocols for whole-genome sequencing in marine organisms. Methods Mol. Biol. 1452, 13-44. ( 10.1007/978-1-4939-3774-5_2) [DOI] [PubMed] [Google Scholar]
- 41.Arseneau JR, Steeves R, Laflamme M. 2016. Modified low-salt CTAB extraction of high-quality DNA from contaminant-rich tissues. Mol. Ecol. Resour. 17, 686-693. ( 10.1111/1755-0998.12616) [DOI] [PubMed] [Google Scholar]
- 42.Chakraborty S, Saha A, Neelavar Ananthram A. 2020. Comparison of DNA extraction methods for non-marine molluscs: is modified CTAB DNA extraction method more efficient than DNA extraction kits? 3 Biotech 10, 69. ( 10.1007/s13205-020-2051-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fang G, Hammar S, Grumet R. 1992. A quick and inexpensive method for removing polysaccharides from plant genomic DNA. Biotechniques 13, 52-56. [PubMed] [Google Scholar]
- 44.Raina K, Chandlee JM. 1996. Recovery of genomic DNA from a fungus (Sclerotinia homoeocarpa) with high polysaccharide content. Biotechniques 21, 1030-1032. ( 10.2144/96216bm14) [DOI] [PubMed] [Google Scholar]
- 45.Aranishi F, Okimoto T. 2006.A simple and reliable method for DNA extraction from bivalve mantle. J. Appl. Genet. 47, 251-254. ( 10.1007/BF03194632) [DOI] [PubMed] [Google Scholar]
- 46.Mikhailova N, Johannesson K. 1998. A comparison of different protocols for RAPD analysis of Littorina. Hydrobiologia 378, 33-42. ( 10.1023/A:1003221117784) [DOI] [Google Scholar]
- 47.Fu RZ, Wang J, Sun YR, Shaw PC. 1998. Extraction of genomic DNA suitable for PCR analysis from dried plant rhizomes/roots. Biotechniques 25, 796–798, 800–801. ( 10.2144/98255bm08) [DOI] [PubMed] [Google Scholar]
- 48.Sokolov EP. 2000. An improved method for DNA isolation from mucopolysaccharide-rich molluscan tissues. J. Molluscan Stud. 66, 573-575. ( 10.1093/mollus/66.4.573) [DOI] [Google Scholar]
- 49.Popa OP, Murariu MD, Popa LO. 2007. Comparison of four DNA extraction methods from invasive freshwater bivalve species (Mollusca: Bivalvia) in Romanian fauna. Travaux du Muséum National d'Histoire Naturelle «Grigore Antipa» Vol. L, pp. 527–536.
- 50.Gebhardt K, Knebelsberger T. 2015. Identification of cephalopod species from the North and Baltic Seas using morphology, COI and 18S rDNA sequences. Helgol. Mar. Res. 69, 259-271. ( 10.1007/s10152-015-0434-7) [DOI] [Google Scholar]
- 51.Pereira JC, Chaves R, Bastos E, Leitão A, Guedes-Pinto H. 2011. An efficient method for genomic DNA extraction from different molluscs species. Int. J. Mol. Sci. 12, 8086-8095. ( 10.3390/ijms12118086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawai K, Shimizu M, Hughes R, Takenaka O. 2004. A non-invasive technique for obtaining DNA from marine intertidal snails. J. Mar. Biol. Assn. UK 84, 773-774. ( 10.1017/S0025315404009907h) [DOI] [Google Scholar]
- 53.Armbruster GFJ, Koller B, Baur B. 2005. Foot mucus and periostracum fraction as nondestructive source of DNA in the land snail Arianta arbustorum, and the development of new microsatellite loci. Conserv. Genet. 6, 313-316. ( 10.1007/s10592-004-7823-9) [DOI] [Google Scholar]
- 54.Palmer ANS, Styan CA, Shearman DCA. 2008. Foot mucus is a good source for nondestructive genetic sampling in Polyplacophora. Conserv. Genet. 9, 229-231. ( 10.1007/s10592-007-9320-4) [DOI] [Google Scholar]
- 55.Régnier C, Gargominy O, Falkner G, Puillandre N. 2011. Foot mucus stored on FTA® cards is a reliable and non-invasive source of DNA for genetics studies in molluscs. Conserv. Genet. Resour. 3, 377-382. ( 10.1007/s12686-010-9345-8) [DOI] [Google Scholar]
- 56.Morinha F, Travassos P, Carvalho D, Magalhães P, Cabral JA, Bastos E. 2014. DNA sampling from body swabs of terrestrial slugs (Gastropoda: Pulmonata): a simple and noninvasive method for molecular genetics approaches. J. Molluscan Stud. 80, 99-101. ( 10.1093/mollus/eyt045) [DOI] [Google Scholar]
- 57.Adema CM, Lun CM, Hanelt B, Seville RS. 2009. Digenean trematode infections of native freshwater snails and invasive Potamopyrgus antipodarum in the Grand Teton National Park/John D. Rockefeller Memorial Parkway Area. J. Parasitol. 95, 224-227. ( 10.1645/GE-1614.1) [DOI] [PubMed] [Google Scholar]
- 58.Zhang SM, Loker ES. 2004. Representation of an immune responsive gene family encoding fibrinogen-related proteins in the freshwater mollusc Biomphalaria glabrata, an intermediate host for Schistosoma mansoni. Gene 341, 255-266. ( 10.1016/j.gene.2004.07.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singer VL, Jones LJ, Yue ST, Haugland RP. 1997. Characterization of PicoGreen reagent and development of a fluorescence-based solution assay for double-stranded DNA quantitation. Anal. Biochem. 249, 228-238. ( 10.1006/abio.1997.2177) [DOI] [PubMed] [Google Scholar]
- 60.Chen H, Rangasamy M, Tan SY, Wang H, Siegfried BD. 2010. Evaluation of five methods for total DNA extraction from western corn rootworm beetles. PLoS ONE 5, e11963. ( 10.1371/journal.pone.0011963) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Knight M, Miller AN, Geoghagen NSM, Lewis FA, Kerlavage AR. 1998. Expressed sequence tags (ESTs) of Biomphalaria glabrata, an intermediate snail host of Schistosoma mansoni: use in the identification of RFLP markers. Malacologia 39, 175-182. [Google Scholar]
- 62.Rafferty GP, Powell R. 2002. Identification of genes expressed in the gill tissue of the Pacific oyster (Crassostrea gigas) using expressed-sequence tags. J. Molluscan Stud. 68, 397-399. ( 10.1093/mollus/68.4.397) [DOI] [Google Scholar]
- 63.Kocot KM, et al. 2011. Phylogenomics reveals deep molluscan relationships. Nature 477, 452-456. ( 10.1038/nature10382) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu D, Hu B, Wen C, Lin G, Tao Z, Hu X, Xie Y. 2013. Gene identification and recombinant protein of a lysozyme from freshwater mussel Cristaria plicata. Fish Shellfish Immunol. 34, 1033-1041. ( 10.1016/j.fsi.2012.12.009) [DOI] [PubMed] [Google Scholar]
- 65.Saavedra C, Milan M, Leite RB, Cordero D, Patarnello T, Cancela ML, Bargelloni LA. 2017. Microarray study of carpet-shell clam (Ruditapes decussatus) shows common and organ-specific growth-related gene expression differences in gills and digestive gland. Front. Physiol. 8, 943. ( 10.3389/fphys.2017.00943) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang G, et al. 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49-54. ( 10.1038/nature11413) [DOI] [PubMed] [Google Scholar]
- 67.Eddy SR. 2001. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2, 919-929. ( 10.1038/35103511) [DOI] [PubMed] [Google Scholar]
- 68.Schroeder A, et al. 2006. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7, 3. ( 10.1186/1471-2199-7-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Petrov NB, Vladychenskaia NS. 2005. [Phylogeny of protostome moulting animals (Ecdysozoa) inferred from 18 and 28S rRNA gene sequences]. Mol. Biol. (Mosk). 39, 590-601. In Russian. [PubMed] [Google Scholar]
- 70.Ishikawa H, Newburgh RW.1972. Studies of the thermal conversion of 28 s RNA of Galleria mellonella (L.) to an 18 s product. J. Mol. Biol. 64, 135-144. ( 10.1016/0022-2836(72)90325-7) [DOI] [PubMed] [Google Scholar]
- 71.Ishikawa H. 1973. Comparative studies on the thermal stability of animal ribosomal RNA's. Comp. Biochem. Physiol. B 46, 217-227. ( 10.1016/0305-0491(73)90312-x) [DOI] [PubMed] [Google Scholar]
- 72.Ishikawa H. 1977. Evolution of ribosomal RNA. Comp. Biochem. Physiol. B 58, 1-7. ( 10.1016/0305-0491(77)90116-x) [DOI] [PubMed] [Google Scholar]
- 73.Adema C. 2006. rDNA gene region (18S-ITS1–5.8S-ITS2–28S) of Biomphalaria glabrata. Albuquerque, NM: University of New Mexico. ( 10.25827/45GK-6J98) [DOI] [Google Scholar]
- 74.Zhang SM, Bu L, Laidemitt MR, Lu L, Mutuku MW, Mkoji GM, Loker ES. 2018. Complete mitochondrial and rDNA complex sequences of important vector species of Biomphalaria, obligatory hosts of the human-infecting blood fluke, Schistosoma mansoni. Sci. Rep. 8, 7341. ( 10.1038/s41598-018-25463-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Natsidis P, Schiffer PH, Salvador-Martínez I, Telford MJ. 2019. Computational discovery of hidden breaks in 28S ribosomal RNAs across eukaryotes and consequences for RNA Integrity Numbers. Sci. Rep. 9, 19477. ( 10.1038/s41598-019-55573-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Padmanaban A. 2012. RNA quality control using the Agilent 2200 TapeStation system- Assessment of the RINe quality metric. See http://hpst.cz/sites/default/files/oldfiles/rna-quality-control-using-agilent-2200-tapestation-systemassessment-rine-quality-metric.pdf (accessed 24 Feb 2021).
- 77. Comparison of RIN and RINeAlgorithms for the Agilent 2100 Bioanalyzer and the Agilent 2200 TapeStation systems. https://www.agilent.com/cs/library/technicaloverviews/Public/5991–3426EN.pdf . (accessed 30 Jan 2021)
- 78.Amaral PP, Mattick JS. 2008. Noncoding RNA in development. Mamm. Genome 19, 454-492. ( 10.1007/s00335-008-9136-7) [DOI] [PubMed] [Google Scholar]
- 79.Aviv H, Leder P. 1972. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc. Natl Acad. Sci. USA 69, 1408-1412. ( 10.1073/pnas.69.6.1408) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hanelt B, Lun CM, Adema CM. 2008. Comparative ORESTES-sampling of transcriptomes of immune-challenged Biomphalaria glabrata snails. J. Invertebr. Pathol. 99, 192-203. ( 10.1016/j.jip.2008.06.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nowak TS, Woodards AC, Jung Y, Adema CM, Loker ES. 2004. Identification of transcripts generated during the response of resistant Biomphalaria glabrata to Schistosoma mansoni infection using suppression subtractive hybridization. J. Parasitol. 90, 1034-1040. ( 10.1645/GE-193R1) [DOI] [PubMed] [Google Scholar]
- 82.Lockyer AE, Spinks JN, Walker AJ, Kane RA, Noble LR, Rollinson D, Dias-Neto E, Jones CS. 2007. Biomphalaria glabrata transcriptome: identification of cell-signalling, transcriptional control and immune-related genes from open reading frame expressed sequence tags (ORESTES). Dev. Comp. Immunol. 31, 763-782. ( 10.1016/j.dci.2006.11.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Albu M, Min XJ, Hickey D, Golding B. 2008. Uncorrected nucleotide bias in mtDNA can mimic the effects of positive Darwinian selection. Mol. Biol. Evol. 25, 2521-2524. ( 10.1093/molbev/msn224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.DeJong RJ, Emery AM, Adema CM. 2004. The mitochondrial genome of Biomphalaria glabrata (Gastropoda: Basommatophora), intermediate host of Schistosoma mansoni. J. Parasitol. 90, 991-997. ( 10.1645/GE-284R) [DOI] [PubMed] [Google Scholar]
- 85.Schultz JH, Bu L, Kamel B, Adema CM. 2020. RNA-seq: The early response of the snail Physella acuta to the digenetic trematode Echinostoma paraensei. J. Parasitol. 106, 490-505. ( 10.1645/19-36) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nolan JR, Bergthorsson U, Adema CM. 2014. Physella acuta: atypical mitochondrial gene order among panpulmonates (Gastropoda). J. Molluscan Stud. 80, 388-399. ( 10.1093/mollus/eyu025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gu W, Crawford ED, O'Donovan BD, Wilson MR, Chow ED, Retallack H, DeRisi JL. 2016. Depletion of Abundant Sequences by Hybridization (DASH): using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. 17, 41. ( 10.1186/s13059-016-0904-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim IV, Ross EJ, Dietrich S, Döring K, Sánchez Alvarado A, Kuhn CD. 2019. Efficient depletion of ribosomal RNA for RNA sequencing in planarians. BMC Genomics 20, 909. ( 10.1186/s12864-019-6292-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rowe KC, Singhal S, Macmanes MD, Ayroles JF, Morelli TL, Rubidge EM, Bi K, Moritz CC. 2011. Museum genomics: low-cost and high-accuracy genetic data from historical specimens. Mol. Ecol. Resour. 11, 1082-1092. ( 10.1111/j.1755-0998.2011.03052.x) [DOI] [PubMed] [Google Scholar]
- 90.Jaksch K, Eschner A, Rintelen TV, Haring E.2016. DNA analysis of molluscs from a museum wet collection: a comparison of different extraction methods. BMC Res. Notes 9, 348. ( 10.1186/s13104-016-2147-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chase MR, Etter RJ, Rex MA, Quattro JM. 1998. Extraction and amplification of mitochondrial DNA from formalin-fixed deep-sea mollusks. Biotechniques 24, 243-247. ( 10.2144/98242bm16) [DOI] [PubMed] [Google Scholar]
- 92.Schander C, Halanych KM. 2003. DNA, PCR and formalinized animal tissue – a short review and protocols. Org. Divers. Evol. 3, 195-205, ( 10.1078/1439-6092-00071) [DOI] [Google Scholar]
- 93.Hykin SM, Bi K, McGuire JA. 2015. Fixing formalin: a method to recover genomic-scale DNA sequence data from formalin-fixed museum specimens using high-throughput sequencing. PLoS ONE 10, e0141579. ( 10.1371/journal.pone.0141579) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Der Sarkissian C, et al. 2017. Ancient DNA analysis identifies marine mollusc shells as new metagenomic archives of the past. Mol. Ecol. Resour. 17, 835-853. ( 10.1111/1755-0998.12679) [DOI] [PubMed] [Google Scholar]
- 95.Der Sarkissian C, Möller P, Hofman CA, Ilsøe P, Rick TC, Schiøtte T, Sørensen MV, Dalén L, Orlando L.. 2020. Unveiling the ecological applications of ancient DNA from mollusk shells. Front. Ecol. Evol. 8, 37. ( 10.3389/fevo.2020.00037) [DOI] [Google Scholar]
- 96.Simakov O, et al. 2013. Insights into bilaterian evolution from three spiralian genomes. Nature 493, 526-531. ( 10.1038/nature11696) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Albertin CB, Simakov O, Mitros T, Wang ZY, Pungor JR, Edsinger-Gonzales E, Brenner S, Ragsdale CW, Rokhsar DS. 2015. The octopus genome and the evolution of cephalopod neural and morphological novelties. Nature 524, 220-224. ( 10.1038/nature14668) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schell T, et al. 2017. An annotated draft genome for Radix auricularia (Gastropoda, Mollusca). Genome Biol. Evol. 9, 32. ( 10.1093/gbe/evx032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Choo LQ, Bal TMP, Choquet M, Smolina I, Ramos-Silva P, Marlétaz F, Kopp M, Hoara G, Peijnenburg KTCA. 2020. Novel genomic resources for shelled pteropods: a draft genome and target capture probes for Limacina bulimoides, tested for cross-species relevance. BMC Genomics 21, 11. ( 10.1186/s12864-019-6372-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Adema CM, et al. 2006. A bacterial artificial chromosome library for Biomphalaria glabrata, intermediate snail host of Schistosoma mansoni. Mem. Inst. Oswaldo Cruz. 101(Suppl. 1), 167-177. ( 10.1590/s0074-02762006000900027) [DOI] [PubMed] [Google Scholar]
- 101.Cunningham C, et al. 2006. New resources for marine genomics: bacterial artificial chromosome libraries for the Eastern and Pacific oysters (Crassostrea virginica and C. gigas). Mar. Biotechnol. (NY) 8, 521-533. ( 10.1007/s10126-006-6013-9) [DOI] [PubMed] [Google Scholar]
- 102.Amarasinghe SL, Su S, Dong X, Zappia L, Ritchie ME, Gouil Q. 2020. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 21, 30. ( 10.1186/s13059-020-1935-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kenny NJ, et al. 2020. The gene-rich genome of the scallop Pecten maximus. Gigascience 9, giaa037. ( 10.1093/gigascience/giaa037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang M, Zhang Y, Scheuring CF, Wu CC, Dong JJ, Zhang HB. 2012. Preparation of megabase-sized DNA from a variety of organisms using the nuclei method for advanced genomics research. Nat. Protoc. 7, 467-478. ( 10.1038/nprot.2011.455) [DOI] [PubMed] [Google Scholar]
- 105.Vilanova S, et al. 2020. SILEX: a fast and inexpensive high-quality DNA extraction method suitable for multiple sequencing platforms and recalcitrant plant species. Plant Methods 16, 110. ( 10.1186/s13007-020-00652-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Abdel-Latif A, Osman G. 2017. Comparison of three genomic DNA extraction methods to obtain high DNA quality from maize. Plant Methods 13, 1. ( 10.1186/s13007-016-0152-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mayjonade B, Gouzy J, Donnadieu C, Pouilly N, Marande W, Callot C, Langlade N, Muños S. 2016. Extraction of high-molecular-weight genomic DNA for long-read sequencing of single molecules. Biotechniques 61, 203-205. ( 10.2144/000114460) [DOI] [PubMed] [Google Scholar]
- 108.Da Fonseca RR, et al. 2020. A draft genome sequence of the elusive giant squid, Architeuthis dux. Gigascience 1, giz152. ( 10.1093/gigascience/giz152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Guo Y, et al. 2019. A chromosomal-level genome assembly for the giant African snail Achatina fulica. Gigascience 8, giz124. ( 10.1093/gigascience/giz124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ran Z, Li Z, Yan X, Liao K, Kong F, Zhang L, Cao J, Zhou C, Zhu P, He S, Huang W, Xu J. 2019. Chromosome-level genome assembly of the razor clam Sinonovacula constricta (Lamarck, 1818). Mol. Ecol. Resour. 19, 1647-1658. ( 10.1111/1755-0998.13086) [DOI] [PubMed] [Google Scholar]
- 111.Van Quyen D, Gan HM, Lee YP, Nguyen DD, Nguyen TH, Tran XT, Nguyen VS, Khang DD, Austin CM.. 2020. Improved genomic resources for the black tiger prawn (Penaeus monodon). Mar. Genomics 52, 100751. ( 10.1016/j.margen.2020.100751) [DOI] [PubMed] [Google Scholar]
- 112.Baron OL, Deleury E, Reichhart JM, Coustau C. 2016. The LBP/BPI multigenic family in invertebrates: Evolutionary history and evidences of specialization in mollusks. Dev. Comp. Immunol. 57, 20-30. ( 10.1016/j.dci.2015.11.006) [DOI] [PubMed] [Google Scholar]
- 113.Gorbushin AM. 2018. Immune repertoire in the transcriptome of Littorina littorea reveals new trends in lophotrochozoan proto-complement evolution. Dev. Comp. Immunol. 84, 250-263. ( 10.1016/j.dci.2018.02.018) [DOI] [PubMed] [Google Scholar]
- 114.Aguilera F, McDougall C, Degnan BM. 2017. Co-option and de novo gene evolution underlie molluscan shell diversity. Mol. Biol. Evol. 34, 779-792. ( 10.1093/molbev/msw294) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Clark MS, et al. 2020. Deciphering mollusc shell production: the roles of genetic mechanisms through to ecology, aquaculture and biomimetics. Biol. Rev. Camb. Phil. Soc. 95, 1812-1837. ( 10.1111/brv.12640) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No primary data reported.


