Abstract
Covariation among traits shapes both phenotypic evolution and ecological interactions across space and time. However, rampant geographical variation in the strength and direction of such correlations can be particularly difficult to explain through generalized mechanisms. By integrating population genomics, surveys of natural history collections and spatially explicit analyses, we tested multiple drivers of trait correlations in a coral snake mimic that exhibits remarkable polymorphism in mimetic and non-mimetic colour traits. We found that although such traits co-occur extensively across space, correlations were best explained by a mixture of genetic architecture and correlational selection, rather than by any single mechanism. Our findings suggest that spatially complex trait distributions may be driven more by the simple interaction between multiple processes than by complex variation in one mechanism alone. These interactions are particularly important in mimicry systems, which frequently generate striking geographical variation and genetic correlations among colour pattern traits.
Keywords: trait correlations, colour polymorphism, mimicry, population genomics, ground snakes, Sonora
1. Background
A fundamental aim of evolutionary biology is to understand the mechanisms that generate and regulate spatial patterns of phenotypic diversity. One powerful approach to testing among mechanisms compares phenotypic covariance of two or more traits within and between populations across a wide range of taxa and traits [1,2]. Quantitative genetics has been critical for making predictions about the evolutionary dynamics of correlations between continuous traits under different generating mechanisms [3,4]. However, many trait correlations of interest to evolutionary biologists involve discrete phenotypes in natural systems not amenable to large-scale breeding or pedigree studies necessary for a quantitative genetics approach [5,6]. Spatially variable trait correlations are especially common in phenotypes with direct consequences on fitness, such as mating systems and predator defence adaptations [1,7,8], making them excellent targets for testing how traits may interact to structure phenotypic diversity across space or time.
Mimicry is an anti-predator strategy in which one organism converges on the phenotype of another to directly increase survival [9]. Trait correlations are particularly important in mimicry systems because warning signals to predators are almost always multi-trait combinations requiring the coordination of multiple genes [10,11] and/or signal modalities [12,13]. With a high selective advantage for precise mimicry, it may seem paradoxical that many mimicry systems are characterized by both colour polymorphism (in which multiple phenotypes persist in a population) and significant geographical variation in this polymorphism among populations [14–16]. However, the best-studied mimicry systems all show striking diversity in the number and spatial distribution of discrete colour traits, and many are polymorphic for colour pattern elements not involved in a warning signal (e.g. poison frogs [17], butterflies [11,18], snakes [19]). Although some studies have hypothesized why both types of traits might persist in a mimicry system [17,20,21], the relative contribution and importance of mechanisms driving correlations between high-diversity mimetic and non-mimetic traits remain unclear.
Multiple mechanisms can generate spatial correlations between mimetic and non-mimetic traits, including exogenous forces such as neutral population processes or selection and endogenous genetic mechanisms [8,22]. Trait correlations may result solely from neutral processes like genetic drift, gene flow and founder effects, although such correlations may not be stable over time or space. Alternatively, trait correlations could reflect a shared genetic mechanism, such as pleiotropy, linkage or both [8]. In the case of white-throated sparrows, for example, correlations between sexually selected colour traits and behavioural traits are underlaid by the differential expression of genes located within an inversion [23,24]. Lastly, trait correlations could be driven by correlational selection, which occurs when the co-expression of a particular combination of traits is favoured [2]. In garter snakes, for instance, predator-mediated correlational selection has led to an association between colour and behavioural traits, with different combinations having dramatic impacts on fitness [7,25]. It is important to note that these three drivers of trait correlations need not be mutually exclusive; correlational selection frequently leads traits to become genetically correlated via pleiotropy or linkage disequilibrium [26], and neutral processes are a constant presence in most natural populations.
An integrative approach comparing both phenotypic and genetic data in a system with high spatial diversity in mimetic and non-mimetic colour traits can create testable predictions about the relative impact of these mechanisms in structuring correlations across natural populations. Specifically, spatially explicit comparisons of trait frequencies, trait diversity, genetic diversity and population genetic structure should produce different combinations of observed trait distributions across space under each generating mechanism. If a correlation between mimetic and non-mimetic colour traits in a population is the product of neutral processes alone, the diversities of both trait types should also be correlated with the overall genetic diversity in that population. In addition, isolation-by-distance should shape similar patterns of phenotypic and genetic diversity across space, and phenotypic population structuring (measured as FST) should be roughly equivalent to that of neutral genetic markers. By contrast, correlational selection or shared genetic mechanisms should lead to tight spatial congruence of mimetic and non-mimetic trait diversity within and among populations, and do so independently of genetic diversity. Disentangling the relative roles of these two drivers in shaping trait correlations can be difficult using measures of population structure alone, so tests that conclusively reject one or the other are useful to identify the most likely mechanism by process of elimination.
In this study, we leveraged the geographical variation in colour polymorphism of a mimetic snake species and used population genetic analyses to test how selection and genetic architecture may shape the distribution of mimetic and non-mimetic colour traits. First, we assessed how trait frequencies vary among populations, where trait correlations are present, and how the diversity of traits is related across space. We then compared population structure and diversity of neutral genetic markers to that of colour traits to test among selective and neutral processes acting on coloration. Lastly, we modelled expected trait distributions under different genetic scenarios to infer the most likely architecture underlying the expression and distribution of mimetic and non-mimetic colour traits. By combining these approaches, we provide evidence that even though the spatial distribution of colour traits and their correlations is highly non-random with respect to genetic population structure, trait correlations in this species are better explained by a mixture of processes than by either genetic architecture or selection alone.
2. Methods
(a). Study system
We studied the Great Plains ground snake (Sonora episcopa), a diminutive, semi-fossorial snake found throughout the central United States and northern Mexico [27,28]. This species exhibits extraordinary colour polymorphism involving red and black pigmentation (figure 1a), such that individuals may possess a red longitudinal dorsal stripe, black crossbands, neither a red stripe nor black crossbands (resulting in a brown/grey ‘uniform’ appearance), or both a red stripe and black crossbands [29]. The latter ‘red–black banded’ phenotype is present in most Sonora species and is considered mimetic of venomous coral snakes [19,28,30]. Sonora episcopa overlaps with one species of coral snake (Micrurus tener), but the precise distribution of ground snake colour morphs is not correlated with coral snake sympatry [29]. All four colour morphs co-occur in some populations, while others exhibit one to three morphs in any combination. Previous research suggests that the evolution of these colour traits may be governed by negative frequency-dependent selection [29] and by the spatial effects of predator foraging [31]. Sexual selection is unlikely to influence colour variation in ground snakes, as there is no evidence of morph-based population structure, assortative mating or sexual dichromatism in this species [29,32], and the capacity for many snake species to even perceive colour is currently unclear [33]. While the genes that regulate colour polymorphism in ground snakes are unknown [34], analyses of morph ratios across populations demonstrate that red and black pigmentation are likely to be controlled by separate, unlinked loci [35].
Figure 1.
Proportions of polymorphic (a) mimetic and (b) non-mimetic colour phenotypes are highly variable across 49 populations of Sonora episcopa in the Great Plains of the USA and northern Mexico. Mimetic colour trait combinations (from left to right, a) include both black crossbands and a red stripe, black crossbands only, a red stripe only and neither crossbands nor red stripe (uniform). Non-mimetic colour trait combinations (from left to right, b) include both a black cap on the head and black nuchal band, a black cap only, a black nuchal band only and neither a cap nor nuchal band. The shade of grey of each population point (detection probability) corresponds to the probability of detecting a rare morph at 5% frequency given the number of individuals sampled in our dataset. See electronic supplementary material, table S1 for population details. (Online version in colour.)
In addition to being polymorphic for two mimicry-related colour traits, ground snakes are polymorphic for a black cap on the head and a black nuchal band (figure 1b). Currently, the biological function of caps and nuchal bands remains understudied, but one or both are present in numerous snake species that lack red and black banding [36]. It has been suggested that black head coloration may aid in thermoregulation [37,38], venom protection [39] or predator avoidance [40], but further study is needed. We consider the black cap and black nuchal band to be ‘non-mimetic’ colour traits in ground snakes, although we explicitly tested for a ‘melanin effect’ linking all the black traits (body crossbands, caps, nuchal bands) despite probably varying functions.
(b). Population sampling
Following Cox & Davis Rabosky [29], a ‘population’ was defined as the US county in which individuals were collected, and its ‘location’ was calculated as the centre latitude and longitude of that county (with a small number of exceptions, see electronic supplementary material, appendix S1). While we did not explicitly account for specimen collection year in our analyses, 40 of our 49 populations included at least 75% of individuals collected within a 20-year sliding time window (as in Davis Rabosky et al. [35]).
(c). Phenotypic scoring
Our phenotypic dataset consisted of 1240 individuals from 49 populations (electronic supplementary material, table S1) across the range of ground snakes and were collected by the authors or sampled from natural history collections (see Acknowledgements). All specimens were photographed and scored for the presence or absence of a red dorsal stripe, black crossbands, a black cap and a black nuchal band. These traits are generally discrete in ground snakes, making scoring relatively easy (see electronic supplementary material, appendix S2 for an alternative scoring method). However, we determined that it was impossible to truly know whether banded and mimetic individuals did or did not possess a nuchal band. In this manuscript, we present the results obtained by scoring banded and mimetic individuals as lacking the nuchal band but note that we recovered all the same significant results when scoring banded and mimetic individuals as having the nuchal band instead (electronic supplementary material, table S2). Black caps were effectively scorable as present or absent even on banded and mimetic individuals.
(d). Trait distributions and associations
To assess the spatial distribution of mimetic traits and non-mimetic traits, we calculated the relative frequency of each trait within each population and plotted all populations on a map. In order to evaluate confidence in our detection of all morphs present per population, we calculated the probability of detecting a rare morph in each population using a simple binomial probability (in which n is the sample size in each population) and assumed a frequency of 5% for rare morphs. To test for statistical associations among colour traits, we tabulated counts of each trait combination from the global dataset and created a contingency table for each pair of traits, then used a χ2-test to determine the independence of each variable.
We also used maps to visualize the spatial congruence of mimetic and non-mimetic trait diversity (Shannon's H). We tested for the relationship between mimetic H and non-mimetic H within populations using both a linear model and a nonparametric Spearman's rank correlation, as both trait types had slightly non-normal distributions. Because both non-mimetic traits involve melanin, we might predict that any spatial congruencies recovered are driven solely by crossbands, which also involve melanin. To directly test this, we removed all individuals with crossbands (banded and mimetic) from the dataset, then re-mapped and tested for the relationship between the relative frequency of the red stripe and non-mimetic H in the same manner as above. We conducted all aforementioned statistics in R v. 4.0.0 [41], and we created maps with spatially interpolated H values between point populations using the R packages ‘rangeBuilder’ [19] and ‘raster’ [42].
(e). DNA sequencing and SNP data generation
For our genetic dataset, we extracted DNA from tissue samples of 142 individuals from 32 populations across the range of ground snakes. We then conducted double digest restriction-site associated DNA sequencing (ddRADseq) following the protocol set forth in Peterson et al. [43] and using the restriction enzymes EcoR1 and MSP1. The samples were sequenced in two lanes of an Illumina HiSeq 2500 System at the University of Michigan Sequencing Core, producing 150 base pair paired-end reads.
Raw sequences were demultiplexed using the program pyRAD [44], and the resulting fast-Q files were run through the dDocent pipeline [45]. We then filtered our SNP dataset using VCFtools v. 0.1.15 [46] and vcflib (included in Freebayes [47]). Lastly, we used BayeScan v. 2.1 [48] and Fisher's exact tests in contingency analyses to identify and remove loci statistically associated with any of the four colour traits and/or likely to be under selection (electronic supplementary material, table S3). The final dataset consisted of 2125 putatively neutral SNPs from 109 individuals across 31 populations. For a more detailed description of our DNA sequencing and SNP data preparation methods, see electronic supplementary material, appendix S3 and table S4.
(f). Population clustering analyses
To assess patterns of genetic structure in ground snakes, we used STRUCTURE [49]. Our model included genetic admixture and correlated allele frequencies, and it was run for 50 000 iterations with a burn-in of 10 000 iterations. We ran this model for k-values of one through 10, with 20 independent replicates of each k. The files produced by STRUCTURE were imported to the program Structure Harvester [50], which employs the Evanno et al. [51] method to determine the k-value with the highest likelihood. We used CLUMPAK [52] and custom R scripts to graphically visualize population clustering for the most likely k-value.
(g). Inferring patterns of selection
To assess the influence of neutral processes or selection on geographical variation in colour traits, we compared population subdivision for neutral SNPs and colour traits. By treating each colour trait (crossbands, red stripe, black cap and nuchal band) as a separate dominant marker and coding the presence or absence of the trait in an individual as 1 or 0, we calculated an analogue of FST (ΦPT, hereafter referred to as FST) for each colour trait that was directly comparable to FST values of neutral genetic markers. In turn, the relationship between neutral genetic differentiation and colour trait differentiation could be used to infer the pattern of selection responsible [29,53]. If variation in colour can be explained primarily by neutral processes, FST should be roughly equivalent between neutral genetic markers and colour trait markers. By contrast, higher differentiation for neutral genetic markers than for colour trait markers would suggest balancing selection pushing populations toward similar morph compositions, while higher differentiation for colour trait markers than for neutral genetic markers would suggest diversifying selection pushing populations towards different morph compositions [29,53]. We used an analysis of molecular variance (AMOVA) in the program GenAlEx [54] to calculate FST values for all neutral SNPs (a global average), each SNP individually and each colour trait individually. We generated 95% confidence intervals by running 9999 permutations. This AMOVA also allowed us to calculate neutral genetic diversity (e.g. observed heterozygosity, unbiased expected heterozygosity, Shannon's information index) within each population and test for a positive correlation with phenotypic diversity.
(h). Modelling genetic architecture of colour traits
We followed the methodology of Davis Rabosky et al. [35] to determine the most likely genetic relationship between each pairwise combination of mimetic and non-mimetic colour traits in ground snakes. This approach was initially developed as a test to reject (or fail to reject) physical linkage between loci, but it cannot confirm the presence of linkage because simulated associations among loci could be consistent with genetic mechanisms other than linkage, such as pleiotropy. As such, we use the term ‘linkage’ here for convenience as a means to denote any sort of shared genetic mechanism. Briefly, this method involves four possible inheritance models for each trait pair (all pairwise combinations): one-locus, two-locus unlinked, two-locus with variable linkage and two-locus with strong linkage. The one-locus model assumes that the two traits are generated simply by different alleles on the same locus. The two-locus unlinked model assumes that the two traits are coded by alleles on separate loci that assort independently. For the final two models that involve linkage, we added the parameter D, a statistical measure of linkage disequilibrium. In the two-locus with variable linkage model, D is free to vary and is estimated separately for each population. In the two-locus with strong linkage model, we assume that D is close to its theoretical maximum in each population and therefore fix the relative value of D (Drel = D/Dmax) to 0.99. We applied the R scripts used in Davis Rabosky et al. [35] to fit all models to our observed trait frequencies in each population. The likelihood of models in each population was assessed by AIC scores.
3. Results
(a). Spatial variation in phenotypic frequencies
Relative frequencies of both mimetic and non-mimetic traits vary considerably across the landscape, with some populations apparently fixed for one colour trait and others exhibiting all possible combinations (figure 1). The probability of detecting a rare morph showed some geographical variation but was above 70% in most populations, suggesting that our sampling was generally sufficient. Furthermore, populations with low rare morph detection probabilities were spatially distributed such that no single region suffered from particularly low confidence. Despite the visible variation among populations in morph presence (figure 1), we found that all pairwise combinations of mimetic and non-mimetic traits co-occurred more often than expected by chance when all individuals were analysed together (all p's < 0.001; crossbands—black cap χ2 = 264.54; red stripe—black cap χ2 = 96.46; red stripe—nuchal band χ2 = 31.76; black cap—nuchal band χ2 = 50.98; crossbands—red stripe χ2 = 116.57; N = 1240). These results remain significant for red striping and non-mimetic traits even when banded and mimetic individuals are removed from analysis, suggesting that these trait co-occurrences are not driven solely by a melanin pathway (all p's < 0.001; red stripe—black cap χ2 = 17.59; red stripe—nuchal band χ2 = 73.10).
(b). Spatial patterns of neutral genetic variation
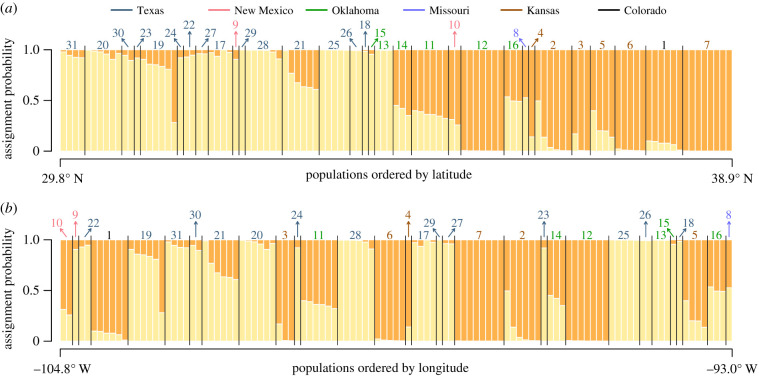
We found support for two population clusters using STRUCTURE and the Evanno et al. [51] method. These clusters correspond well with geographical regions, such that one relatively distinct cluster occurs in Kansas, Colorado and northern Oklahoma, while the other occurs in southern Oklahoma, much of Texas and southern New Mexico. Accordingly, the genetic data are much better explained by a latitudinal cline than a longitudinal one (figure 2). We also note that many populations show a significant level of admixture even when separated by several hundred kilometres, suggesting a wide area of gene flow or historical connectivity among populations. Lastly, the assignment probability of individuals to genetic clusters shows no structure by either mimetic or non-mimetic phenotype, as all morphs are represented by individuals from all genetic deme combinations (electronic supplementary material, figure S1).
Figure 2.
Genetic population structure inferred from 2125 neutral SNPs shows an isolation-by-distance cline when ordered by (a) latitude but not by (b) longitude. The most likely number of population clusters (k) was found to be two, and most populations have assignment probability mixtures of these two demes. Black lines separate populations along each x-axis, and the number above each population can be matched to corresponding population names in electronic supplementary material, table S1. (Online version in colour.)
(c). Spatial correlations in trait diversity
Neutral genetic diversity within populations did not predict phenotypic diversity, as observed heterozygosity was not correlated with either mimetic (F1,24 = 3.113, p = 0.090; Spearman's ρ = −0.197, p = 0.336) or non-mimetic trait diversity (F1,24 = 0.404, p = 0.531; Spearman's ρ = −0.209, p = 0.305; electronic supplementary material, figure S2). However, we also found that mimetic and non-mimetic trait diversity (H) were variable across the landscape, with areas of both positive and negative correlations (figure 3a,b). While mimetic H was positively correlated with non-mimetic H overall (F1,47 = 8.218, p = 0.006; figure 3c), the areas of matching and mismatching had some spatial structure. Mapping trait diversity revealed areas of high mimetic H and high non-mimetic H (e.g. Kansas, central-western Oklahoma), areas of low mimetic H and low non-mimetic H (e.g. Colorado, New Mexico, western Texas), and areas of low mimetic H and high non-mimetic H (e.g. central Texas, southeastern Oklahoma), but very little evidence of areas with high mimetic H and low non-mimetic H. When we removed individuals with bands from analysis, we recovered similar spatial patterns of non-random associations (figure 3d,e), and the relative frequency of red stripe remained significantly correlated with non-mimetic H (F1,47 = 8.054, p = 0.007; figure 3f). Similar to the test of global trait correlations above, significant correlations in the absence of banded individuals suggest that these trait associations are not driven solely by a melanin pathway.
Figure 3.
Mimetic and non-mimetic trait diversity (H) vary non-randomly across space (a,b) and are positively correlated overall (c). Using all individuals in the dataset, spatial concordance of similar diversity indices (high–high, low–low) occurs everywhere except the Edwards Plateau region of central Texas and northward into southern Oklahoma. After removing all banded and mimetic individuals from analysis (d–f), we continue to show a positive correlation and non-random association across space with the same general area of mismatch, suggesting that the patterns observed in (a–c) are not driven by a shared melanin mechanism uniting crossbands and non-mimetic traits. To enhance visualization of correlations across maps, populations with higher H values (or relative frequencies) are represented by darker points and lower values by lighter points. Between populations, higher interpolated values are shown in orange and lower values in blue. Note that in (d–f), the Otero County, Colorado population was removed because all sampled individuals possessed crossbands.
(d). Strength of selection
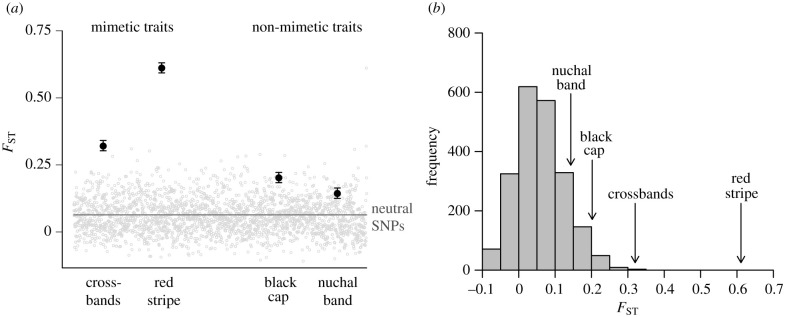
We found strong evidence of selection on mimetic traits but weaker evidence of selection on non-mimetic traits. Global FST was relatively low for neutral genetic markers (FST = 0.064), while FST for mimetic traits and non-mimetic traits were all two to ten times higher (figure 4a; crossbands FST = 0.320; red stripe FST = 0.611; black cap FST = 0.202; nuchal band FST = 0.143; all p-values < 0.05). When FST was analysed on a per-locus basis, both mimetic and non-mimetic colour traits were located in the extreme end of the FST frequency distribution (figure 4b). However, both mimetic colour traits had significantly higher FST values than either of the non-mimetic colour traits (p < 0.05), suggesting that mimetic traits may be under stronger selection.
Figure 4.
All four colour traits have higher FST than the mean FST of neutral SNPs, but the two mimetic colour traits (crossbands, red stripe) have much higher FST than either of the two non-mimetic traits (black cap, nuchal band). Note that we use FST for ease of comparisons among colour traits and neutral genetic markers, but this value is actually ΦPT (an analogue of FST) for colour traits. (a) Mean FST values for all four colour traits (black points) plotted over the distribution of 2125 neutral ddRAD markers (grey points). The grey line represents the mean FST value for all neutral markers. Error bars represent 95% confidence intervals after 9999 permutations, giving an assessment of the repeatability of our calculated means. (b) Mimetic colour traits had FST values in the trailing edge of the frequency distribution of all observed FST values, while non-mimetic traits had values that were closer to the mode of the distribution.
(e). Potential scenarios of genetic architecture
Our modelling of different genetic relationships between pairs of traits suggests that the co-occurrence of at least some mimetic and non-mimetic traits may result from genetic mechanisms (electronic supplementary material, table S5). For all pairwise trait combinations, the two-locus model was strongly favoured over the one-locus model (all ΔAIC > 40), suggesting that these traits are more likely to be controlled by separate loci than a single locus. However, we also found support for differing levels of non-random associations among traits, including some trait combinations that strongly rejected the hypothesis of genetic linkage among loci. In particular, the associations of (i) the red stripe and nuchal band and (ii) crossbands and the red stripe both showed no evidence of linkage disequilibrium (consistent with Davis Rabosky et al. [35]). By contrast, complete independent assortment was not supported for crossbands and black caps, with all models that incorporate a linkage parameter being strongly favoured over an unlinked model for this trait combination (all ΔAIC > 70). The final two trait combinations (red stripe—black cap and black cap—nuchal band) also favoured models with a linkage parameter, but more conclusively (i) rejected models of strong linkage than (ii) supported linkage over independent assortment. Although these models are better designed to reject a hypothesis of genetic linkage among loci rather than confirm it, our results overall are consistent with the potential for linkage disequilibrium among some loci, especially for the traits we measured involving melanism (crossbands, caps and nuchal bands). Nevertheless, our results also strongly suggest that neither linkage, nor being controlled by a single locus, can fully explain trait associations in this system.
4. Discussion
We found evidence that mimetic and non-mimetic trait correlations are best explained by a mixture of endogenous genetic mechanisms and exogenous selective forces in S. episcopa rather than by either process alone. Across the landscape, mimetic and non-mimetic trait frequencies are highly variable, but their diversities are correlated and distributed in a non-random fashion. Neither mimetic nor non-mimetic trait diversity is predicted by genetic diversity, ruling out neutral processes as the sole drivers of these spatial patterns. Measures of population structuring (FST) are higher for both types of phenotypic traits than for neutral genetic markers, implying that these colour traits are indeed under selection. While these phenotypic FST values do not allow us to fully separate the relative roles of shared genetic mechanisms and correlational selection in shaping trait correlations, our modelling scenarios indicate that some correlations, especially those among melanistic traits, may be underlaid by shared genetic architecture. By contrast, other trait correlations, namely those that involve the red stripe, do not appear to share a genetic mechanism, suggesting that these traits may instead be associated as a result of correlational selection.
Our support for shared genetic mechanisms underlying melanistic trait correlations (those involving the black cap, crossbands and nuchal band) is consistent with previous research showing that in many taxa, colour polymorphism is maintained by a single gene or set of linked genes that produces multiple phenotypes through differential expression or modulation [11,18,55,56]. In fact, we may have an a priori expectation for melanistic traits to show evidence of pleiotropy or linkage disequilibrium in systems where colour morphs exist as reduced subsets of one another, such as in ground snakes, where the three melanistic traits could be considered subsets of a head-to-tail banded phenotype. In such ‘reduced subset’ polymorphic systems, a single melanin synthesis gene or network of linked genes may determine whether melanin is produced, while cis- and/or trans-regulatory elements determine where on the body and to what extent the melanistic pattern is present [56–58]. Reduced subset colour polymorphism is common in many taxa (e.g. snails [59], fruit flies [56], lizards [60], frogs [17]), but the role of shared genetic mechanisms in driving the spatial patterns of colour morphs is not often explicitly tested. In particular, polymorphic systems in which colour trait distributions do not follow a predicted environmental cline may be prime candidates for tests of genetic linkage among traits.
Our observed patterns of correlations between the red stripe and melanistic traits are better explained by correlational selection than by shared genetic mechanisms, as evidenced by our modelling scenarios and consistent with previous research in ground snakes suggesting that at least some red and black traits are controlled by separate, unlinked loci [35]. Furthermore, if all mimetic and non-mimetic traits shared a common genetic mechanism, we would expect tight positive correlations among those traits in all populations, yet we uncovered a non-uniform landscape of trait correlations with several areas of mismatch. This mosaic pattern of trait correlations could have been generated by geographical variation in selection and localized adaptations/stochastic processes [61,62], a phenomenon that is particularly common in systems where colour polymorphism is driven by interspecific interactions [63,64]. In the case of ground snakes, the combination of red and black mimics the colour pattern of venomous coral snakes, deceitfully signalling to potential predators that the snakes should be avoided [30]. Because both red and black are necessary to convey such a signal, coral snake mimicry is probably driven by correlational selection. As the predator communities vary across space, so too might the correlational selective regimes, thus generating a complex geographical mosaic of trait distributions and correlations like the one recovered here.
Lastly, our findings have important implications for species that were associated with mimicry at some point in their evolutionary history but currently do not exhibit mimetic traits. While the ground snake that we studied is highly polymorphic in both mimetic and non-mimetic traits, the congeneric Taylor's ground snake (Sonora taylori) is fixed for the uniform phenotype but polymorphic for the black cap [65]. As many of the extant species and recent ancestors of Sonora are thought to be coral snake mimics or at least contain mimetic morphs [19], it is possible that non-mimetic polymorphic traits were initially maintained by correlations with mimetic polymorphic traits, but the two types of traits have since become decoupled in Taylor's ground snakes. In essence, after losing mimetic phenotypes, the entire lineage is now left only with polymorphic black caps. This outcome would suggest that correlations between mimetic and non-mimetic colour traits could serve as a potential explanation for the persistence of non-mimetic colour polymorphism in species that have evolutionarily lost their association with mimicry. Insights such as these can only be gleaned from spatially explicit tests of the drivers of colour trait distributions and correlations.
Supplementary Material
Acknowledgements
The authors would like to thank the following institutions for providing specimens, tissues and/or photographs for use in this study: Arizona State University, University of Arizona, California Academy of Sciences, University of California, Berkeley, San Diego Natural History Museum, Fort Hayes State University, University of Kansas, New Mexico State University, University of New Mexico, University of Oklahoma, University of Texas at Arlington, University of Texas at Austin and University of Texas at El Paso. We would also like to thank John Schenk and Lance McBrayer for thoughtful comments during the development of this project, as well as Brianna Mims, Peter Cerda, Taylor West, Hayley Crowell, Jenna Crowe-Riddell, Molly A. Hirst, Eric Gulson-Castillo, Teresa Pegan, Dan Rabosky, Pascal Title and anonymous reviewers for their feedback on the manuscript.
Ethics
Specimens added to the dataset of Cox & Davis Rabosky [29] were collected under permit no. 16834 from the Missouri Department of Conservation and permit no. 3630 from the New Mexico Department of Game and Fish.
Data accessibility
Data are accessible from the Dryad Digital Repository: https://dx.doi.org/10.5061/dryad.7h44j0zt7 [66].
Authors' contributions
J.D.C., A.R.D.R. and C.L.C. designed the study, collected/procured samples and scored and photographed specimens. J.D.C. and T.J.R. analysed specimen photographs. J.D.C. and I.A.H. prepared samples for sequencing and assembled the results. J.D.C., A.R.D.R. and C.L.C. analysed and interpreted the results. J.D.C., I.A.H. and A.R.D.R. constructed the figures. J.D.C. wrote the first draft of the manuscript, and all authors contributed substantially to revisions.
Competing interests
We declare we have no competing interests.
Funding
Funding for this project was provided by grants from Georgia Southern University's Graduate Student Organization to J.D.C., a National Science Foundation Postdoctoral Fellowship in Biology (DBI-0906046) to A.R.D.R. and funds from the University of Michigan to J.D.C. and A.R.D.R.
References
- 1.Roff DA. 1996. The evolution of genetic correlations: an analysis of patterns. Evolution 50, 1392-1403. ( 10.1111/j.1558-5646.1996.tb03913.x) [DOI] [PubMed] [Google Scholar]
- 2.Lande R, Arnold SJ. 1983. The measurement of selection on correlated characters. Evolution 37, 1210-1226. ( 10.1111/j.1558-5646.1983.tb00236.x) [DOI] [PubMed] [Google Scholar]
- 3.Mackay TFC. 2001. The genetic architecture of quantitative traits. Annu. Rev. Genet. 35, 303-339. ( 10.1146/annurev.genet.35.102401.090633) [DOI] [PubMed] [Google Scholar]
- 4.Hill WG. 2010. Understanding and using quantitative genetic variation. Phil. Trans. R. Soc. B 365, 73-85. ( 10.1098/rstb.2009.0203) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kruuk LE. 2004. Estimating genetic parameters in natural populations using the ‘animal model’. Phil. Trans. R. Soc. B 359, 873-890. ( 10.1098/rstb.2003.1437) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gienapp P, Fior S, Guillaume F, Lasky JR, Sork VL, Csilléry K. 2017. Genomic quantitative genetics to study evolution in the wild. Trends Ecol. Evol. 32, 897-908. ( 10.1016/j.tree.2017.09.004). [DOI] [PubMed] [Google Scholar]
- 7.Brodie ED III. 1989. Genetic correlations between morphology and antipredator behaviour in natural populations of the garter snake Thamnophis ordinoides. Nature 342, 542-543. ( 10.1038/342542a0) [DOI] [PubMed] [Google Scholar]
- 8.Saltz JB, Hessel FC, Kelly MW. 2017. Trait correlations in the genomics era. Trends Ecol. Evol. 32, 279-290. ( 10.1016/j.tree.2016.12.008) [DOI] [PubMed] [Google Scholar]
- 9.Endler JA. 1981. An overview of the relationships between mimicry and crypsis. Biol. J. Linn. Soc. 16, 25-31. ( 10.1111/j.1095-8312.1981.tb01840.x) [DOI] [Google Scholar]
- 10.Charlesworth D. 2016. The status of supergenes in the 21st century: recombination suppression in Batesian mimicry and sex chromosomes and other complex adaptations. Evol. Appl. 9, 74-90. ( 10.1111/eva.12291) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joron M, et al. 2011. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature 477, 203-206. ( 10.1038/nature10341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rowe C, Halpin C. 2013. Why are warning displays multimodal? Behav. Ecol. Sociobiol. 67, 1425-1439. ( 10.1007/s00265-013-1515-8) [DOI] [Google Scholar]
- 13.Dalziell AH, Welbergen JA. 2016. Mimicry for all modalities. Ecol. Lett. 19, 609-619. ( 10.1111/ele.12602) [DOI] [PubMed] [Google Scholar]
- 14.Gray SM, McKinnon JS. 2006. Linking color polymorphism maintenance and speciation. Trends Ecol. Evol. 22, 71-79. ( 10.1016/j.tree.2006.10.005) [DOI] [PubMed] [Google Scholar]
- 15.Joron M, Mallet J. 1998. Diversity in mimicry: paradox or paradigm? Trends Ecol. Evol. 13, 461-466. ( 10.1016/S0169-5347(98)01483-9) [DOI] [PubMed] [Google Scholar]
- 16.Farallo VR, Forstner MR. 2012. Predation and the maintenance of color polymorphism in a habitat specialist squamate. PLoS ONE 7, e30316. ( 10.1371/journal.pone.0030316) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang IJ, Shaffer HB. 2008. Rapid color evolution in an aposematic species: a phylogenetic analysis of color variation in the strikingly polymorphic strawberry poison-dart frog. Evolution 62, 2742-2759. ( 10.1111/j.1558-5646.2008.00507.x) [DOI] [PubMed] [Google Scholar]
- 18.Kunte K, Zhang W, Tenger-Trolander A, Palmer DH, Martin A, Reed RD, Mullen SP, Kronforst MR. 2014. doublesex is a mimicry supergene. Nature 507, 229-232. ( 10.1038/nature13112) [DOI] [PubMed] [Google Scholar]
- 19.Davis RAR, Cox CL, Rabosky DL, Title PO, Holmes IA, Feldman A, McGuire JA. 2016. Coral snakes predict the evolution of mimicry across New World snakes. Nat. Commun. 7, 1-9. ( 10.1038/ncomms11484) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohsaki N. 1995. Preferential predation of female butterflies and the evolution of Batesian mimicry. Nature 378, 173-175. ( 10.1038/378173a0) [DOI] [Google Scholar]
- 21.Wang IJ. 2011. Inversely related aposematic traits: reduced conspicuousness evolves with increased toxicity in a polymorphic poison-dart frog. Evolution 65, 1637-1649. ( 10.1111/j.1558-5646.2011.01257.x) [DOI] [PubMed] [Google Scholar]
- 22.Nei M. 1967. Modification of linkage intensity by natural selection. Genetics 57, 625-641. ( 10.1093/genetics/57.3.625) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas JW, Cáceres M, Lowman JJ, Morehouse CB, Short ME, Baldwin EL, Maney DL, Martin CL. 2008. The chromosomal polymorphism linked to variation in social behavior in the white-throated sparrow (Zonotrichia albicollis) is a complex rearrangement and suppressor of recombination. Genetics 179, 1455-1468. ( 10.1534/genetics.108.088229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zinzow-Kramer WM, Horton BM, McKee CD, Michaud JM, Tharp GK, Thomas JW, Tuttle EM, Yi S, Maney DL. 2015. Genes located in a chromosomal inversion are correlated with territorial song in white-throated sparrows. Genes Brain Behav. 14, 641-654. ( 10.1111/gbb.12252) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brodie ED III. 1992. Correlational selection for color pattern and antipredator behavior in the garter snake Thamnophis ordinoides. Evolution 46, 1284-1298. ( 10.1111/j.1558-5646.1992.tb01124.x) [DOI] [PubMed] [Google Scholar]
- 26.Sinervo B, Svensson E. 2002. Correlational selection and the evolution of genomic architecture. Heredity 89, 329-338. ( 10.1038/sj.hdy.6800148) [DOI] [PubMed] [Google Scholar]
- 27.Frost DR. 1983. Sonora semiannulata. Cat. Am. Amphib. Reptiles 333, 1-4. [Google Scholar]
- 28.Cox CL, Davis Rabosky AR, Holmes IA, Reyes-Velasco J, Roelke CE, Smith EN, Flores-Villela O, McGuire JA, Campbell JA. 2018. Synopsis and taxonomic revision of three genera in the snake tribe Sonorini. J. Nat. Hist. 52, 945-988. ( 10.1080/00222933.2018.1449912) [DOI] [Google Scholar]
- 29.Cox CL, Davis Rabosky AR. 2013. Spatial and temporal drivers of phenotypic diversity in polymorphic snakes. Am. Nat. 182, E40-E57. ( 10.1086/670988) [DOI] [PubMed] [Google Scholar]
- 30.Savage JM, Slowinski JB. 1992. The colouration of the venomous coral snakes (family Elapidae) and their mimics (families Aniliidae and Colubridae). Biol. J. Linn. Soc. 45, 235-254. ( 10.1111/j.1095-8312.1992.tb00642.x) [DOI] [Google Scholar]
- 31.Holmes IA, Grundler MR, Davis Rabosky AR. 2017. Predator perspective drives geographic variation in frequency-dependent polymorphism. Am. Nat. 190, E78-E93. ( 10.1086/693159) [DOI] [PubMed] [Google Scholar]
- 32.Cox CL, Chippindale PT. 2014. Patterns of genetic diversity in the polymorphic ground snake (Sonora semiannulata). Genetica 142, 361-370. ( 10.1007/s10709-014-9780-7) [DOI] [PubMed] [Google Scholar]
- 33.Simões BF, et al. 2016. Visual pigments, ocular filters and the evolution of snake vision. Mol. Biol. Evol. 33, 2483-2495. ( 10.1093/molbev/msw148) [DOI] [PubMed] [Google Scholar]
- 34.Cox CL, Davis Rabosky AR, Chippindale PT. 2013. Sequence variation in the Mc1r gene for a group of polymorphic snakes. Gene 513, 282-286. ( 10.1016/j.gene.2012.10.065) [DOI] [PubMed] [Google Scholar]
- 35.Davis Rabosky AR, Cox CL, Rabosky DL. 2016. Unlinked Mendelian inheritance of red and black pigmentation in snakes: implications for Batesian mimicry. Evolution 70, 944-953. ( 10.1111/evo.12902) [DOI] [PubMed] [Google Scholar]
- 36.Uetz P, Freed P, Hošek J. 2020. The reptile database. See http://www.reptile-database.org/.
- 37.Luiselli L. 1992. Reproductive success in melanistic adders: a new hypothesis and some considerations on Andrén and Nilson's (1981) suggestions. Oikos 64, 601-604. ( 10.2307/3545182) [DOI] [Google Scholar]
- 38.Bittner TD, King RB, Kerfin JM. 2002. Effects of body size and melanism on the thermal biology of garter snakes (Thamnophis sirtalis). Copeia 2002, 477-482. ( 10.1643/0045-8511(2002)002[0477:Eobsam]2.0.Co;2) [DOI] [Google Scholar]
- 39.Pough FH, Kwiecinski G, Bemis W. 1978. Melanin deposits associated with the venom glands of snakes. J. Morphol. 155, 63-71. ( 10.1002/jmor.1051550105) [DOI] [PubMed] [Google Scholar]
- 40.Stevens M. 2007. Predator perception and the interrelation between different forms of protective coloration. Proc. R. Soc. B 274, 1457-1464. ( 10.1098/rspb.2007.0220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.R Core Team. 2020. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 42.Hijmans RJ. 2020. raster: geographic data analysis and modeling. R package version 3.1-5 ed.
- 43.Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. 2012. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS ONE 7, e37135. ( 10.1371/journal.pone.0037135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eaton DAR. 2014. PyRAD: assembly of de novo RADseq loci for phylogenetic analyses. Bioinformatics 30, 1844-1849. ( 10.1093/bioinformatics/btu121) [DOI] [PubMed] [Google Scholar]
- 45.Puritz JB, Hollenbeck CM, Gold JR. 2014. dDocent: a RADseq, variant-calling pipeline designed for population genomics of non-model organisms. PeerJ 2, e431. ( 10.7717/peerj.431) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Danecek P, et al. 2011. The variant call format and VCFtools. Bioinformatics 27, 2156-2158. ( 10.1093/bioinformatics/btr330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garrison E, Marth G. 2012. Haplotype-based variant detection from short-read sequencing. arXiv: 12073907.
- 48.Foll M, Gaggiotti O. 2008. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180, 977-993. ( 10.1534/genetics.108.092221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Earl DA, VonHoldt BM. 2011. Structure harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv. Genet. Resour. 4, 359-361. ( 10.1007/s12686-011-9548-7) [DOI] [Google Scholar]
- 51.Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611-2620. ( 10.1111/j.1365-294X.2005.02553.x) [DOI] [PubMed] [Google Scholar]
- 52.Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. 2015. CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 15, 1179-1191. ( 10.1111/1755-0998.12387) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gillespie RG, Oxford GS. 1998. Selection on the color polymorphism in Hawaiian happy-face spiders: evidence from genetic structure and temporal fluctuations. Evolution 52, 775-783. ( 10.1111/j.1558-5646.1998.tb03701.x) [DOI] [PubMed] [Google Scholar]
- 54.Peakall R, Smouse PE. 2006. GenAlEx 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 6, 288-295. ( 10.1111/j.1471-8286.2005.01155.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vestergaard JS, Twomey E, Larsen R, Summers K, Nielsen R. 2015. Number of genes controlling a quantitative trait in a hybrid zone of the aposematic frog Ranitomeya imitator. Proc. R. Soc. B 282, 20141950. ( 10.1098/rspb.2014.1950) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wittkopp PJ, Carroll SB, Kopp A. 2003. Evolution in black and white: genetic control of pigment patterns in Drosophila. Trends Genet. 19, 495-504. ( 10.1016/s0168-9525(03)00194-x) [DOI] [PubMed] [Google Scholar]
- 57.Rogers WA, Grover S, Stringer SJ, Parks J, Rebeiz M, Williams TM. 2014. A survey of the trans-regulatory landscape for Drosophila melanogaster abdominal pigmentation. Dev. Biol. 385, 417-432. ( 10.1016/j.ydbio.2013.11.013) [DOI] [PubMed] [Google Scholar]
- 58.Gompel N, Prud'homme B, Wittkopp PJ, Kassner VA, Carroll SB. 2005. Chance caught on the wing: cis-regulatory evolution and the origin of pigment patterns in Drosophila. Nature 433, 481-487. ( 10.1038/nature03235) [DOI] [PubMed] [Google Scholar]
- 59.Ożgo M, Komorowska A. 2009. Shell banding polymorphism in Cepaea vindobonensis in relation to habitat in southeastern Poland. Malacologia 51, 81-88. ( 10.4002/040.051.0105) [DOI] [Google Scholar]
- 60.Chapple DG, Hutchinson MN, Maryan B, Plivelich M, Moore JA, Keogh JS. 2008. Evolution and maintenance of colour pattern polymorphism in Liopholis (Squamata: Scincidae). Aust. J. Zool. 56, 103-115. ( 10.1071/ZO08040) [DOI] [Google Scholar]
- 61.McLean CA, Stuart-Fox D. 2014. Geographic variation in animal colour polymorphisms and its role in speciation. Biol. Rev. 89, 860-873. ( 10.1111/brv.12083) [DOI] [PubMed] [Google Scholar]
- 62.Svensson E, Sinervo B. 2004. Spatial scale and temporal component of selection in side-blotched lizards. Am. Nat. 163, 726-734. ( 10.1086/383592) [DOI] [PubMed] [Google Scholar]
- 63.Forsman A, Ahnesjö J, Caesar S, Karlsson M. 2008. A model of ecological and evolutionary consequences of color polymorphism. Ecology 89, 34-40. ( 10.1890/07-0572.1) [DOI] [PubMed] [Google Scholar]
- 64.Brodie ED Jr, Ridenhour BJ, Brodie ED III. 2002. The evolutionary response of predators to dangerous prey: Hotspots and coldspots in the geographic mosaic of coevolution between garter snakes and newts. Evolution 56, 2067-2082. ( 10.1111/j.0014-3820.2002.tb00132.x) [DOI] [PubMed] [Google Scholar]
- 65.Mulaik S, Mulaik D. 1941. Variation in Sonora taylori. Copeia 1941, 263. ( 10.2307/1437478) [DOI] [Google Scholar]
- 66.Curlis JD, Davis Rabosky AR, Holmes IA, Renney TJ, Cox CL. 2021. Data from: Genetic mechanisms and correlational selection structure trait variation in a coral snake mimic. Dryad Digital Repository. ( 10.5061/dryad.7h44j0zt7) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Curlis JD, Davis Rabosky AR, Holmes IA, Renney TJ, Cox CL. 2021. Data from: Genetic mechanisms and correlational selection structure trait variation in a coral snake mimic. Dryad Digital Repository. ( 10.5061/dryad.7h44j0zt7) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Data are accessible from the Dryad Digital Repository: https://dx.doi.org/10.5061/dryad.7h44j0zt7 [66].