Abstract
The first animal mitochondrial genomes to be sequenced were of several vertebrates and model organisms, and the consistency of genomic features found has led to a ‘textbook description’. However, a more broad phylogenetic sampling of complete animal mitochondrial genomes has found many cases where these features do not exist, and the phylum Mollusca is especially replete with these exceptions. The characterization of full mollusc mitogenomes required considerable effort involving challenging molecular biology, but has created an enormous catalogue of surprising deviations from that textbook description, including wide variation in size, radical genome rearrangements, gene duplications and losses, the introduction of novel genes, and a complex system of inheritance dubbed ‘doubly uniparental inheritance’. Here, we review the extraordinary variation in architecture, molecular functioning and intergenerational transmission of molluscan mitochondrial genomes. Such features represent a great potential for the discovery of biological history, processes and functions that are novel for animal mitochondrial genomes. This provides a model system for studying the evolution and the manifold roles that mitochondria play in organismal physiology, and many ways that the study of mitochondrial genomes are useful for phylogeny and population biology.
This article is part of the Theo Murphy meeting issue ‘Molluscan genomics: broad insights and future directions for a neglected phylum’.
Keywords: mitochondria, mollusc, genome, evolution, doubly uniparental inheritance
1. Introduction
In the 1980s, as DNA sequencing was becoming common, the fledglings of what we now call ‘genomics’ were diminutive animal mitochondrial genomes. The first reports were of several vertebrates and model organisms, followed quickly by studies of their modes of replication, transcription, RNA processing and other aspects of molecular biology (see [1]). The consistency of genomic features found and the expectation that these studies were characteristic of all mitochondrial genomes has led to a ‘textbook description’ of mitochondrial genomes that includes a consistent size of about 16 kb, strictly maternal inheritance, a content of 37 genes (encoding 13 proteins, 2 rRNAs and 22 tRNAs) compactly organized in a nearly invariant arrangement, a single large non-coding ‘control region’ with signals for regulating replication and transcription, and transcription of a single polycistron from each strand that is processed by enzymatic removal of tRNAs into gene-specific (or, in the cases of nad4L-nad4 and/or atp8-atp6, bicistronic) mRNAs. Secondary structures were sometimes inferred for regulatory signals or to compensate for lack of tRNA genes where necessary for enzymatically separating the adjacent gene-specific transcripts.
Clearly, understanding these features is important for interpreting the patterns of evolution of these genomes, but this touches also on many other issues, including interactions with the products of nuclear genes, energy generation, wide-ranging aspects of metabolism and physiology, stress tolerance, susceptibility to oxidative stress, aspects of ecology, patterns of inheritance and population genomics. A more broad phylogenetic sampling of complete mitochondrial genomes now belies not only these general genomic features, but also makes clear that there is no potential for some of these functional molecular mechanisms.
Among bilaterian animals, the phylum Mollusca is especially replete with such examples. Due to their modest size and considerable phylogenetic information content both in gene sequences and arrangements, molluscan mitogenomes began to be studied in the early 1990s. Then, characterization of full mitogenomes required considerable effort involving challenging molecular biology including physical isolation of mitochondrial DNA (mtDNA), restriction enzyme mapping, cloning of large inserts, subcloning into a large number of separate plasmid vectors, and Sanger sequencing by directed primer walking, as evident from the first reports of molluscan mitogenomes from Mytilus edulis (Bivalvia: [2]), Katharina tunicata (Polyplacophora: [3]) and several helicid gastropods [4–6], table 1. The revolutions in genome sequencing technology since have greatly accelerated these efforts, and we now have available more than 1000 complete mitochondrial genome sequences from more than 700 species. This, plus a modest amount of work to understand the biology of these genomes, has created an enormous catalogue of surprising deviations from that textbook description, including wide variation in size, radical genome rearrangements, gene duplications and losses, the introduction of novel genes and a complex system of inheritance dubbed ‘doubly uniparental inheritance’ (DUI). This creates great potential for the discovery of biological history, processes and functions that are novel for animal mitochondrial genomes. Interestingly, expanded non-coding regions, variable repeat content, frequent gene rearrangements and large numbers of ORFans, while uncommon in other animal lineages, are frequently observed in plants [7].
Table 1.
Number of molluscan mitogenome sequences in GenBank over time. The GenBank (GB) search was structured as follows: ((‘Mollusca’[Organism]) AND (biomol_genomic[PROP] AND mitochondrion[filter] AND (‘8000’[SLEN] : ‘100000’[SLEN]) AND (‘1900/01/01’[PDAT] : ‘1999/12/31’[PDAT])). The term ‘Mollusca’ was replaced for family level searches with ‘Gastropoda; Bivalvia; Scaphopoda; Cephalopoda; Polyplacophora; Monoplacophora; Aplacophora’ and the years were adjusted for specific time intervals.
| taxon | GB/RefSeq | by 2000 | 2000–2004 | 2005–2009 | 2010–2014 | 2015–2020 |
|---|---|---|---|---|---|---|
| Gastropoda | 625/233 | 4 | 10 | 19 | 104 | 491 |
| Bivalvia | 451/186 | 0 | 4 | 45 | 130 | 272 |
| Scaphopoda | 3/2 | 1 | 1 | 0 | 0 | 1 |
| Cephalopoda | 126/50 | 0 | 1 | 7 | 53 | 65 |
| Polyplacophora | 23/13 | 1 | 0 | 0 | 2 | 20 |
| Monoplacophora | 3/2 | 0 | 0 | 0 | 0 | 3 |
| Aplacophora | 8/5 | 0 | 0 | 0 | 2 | 6 |
| Mollusca | 1239/491 | 6 | 16 | 71 | 291 | 858 |
2. Genome architecture
The first mollusc mitochondrial genome [2], sequenced nearly three decades ago with Klenow fragment of Escherichia coli DNA polymerase on polyacrylamide gels, documented unprecedented genome architectural variation compared to other metazoans and presaged the amazing variation in mollusc mtDNA genome architecture that was soon to be discovered. Several major patterns of molluscan mitochondrial genome biology were largely present, if not fully understood, in that original M. edulis mtDNA. This included, to wit, a dramatic departure in gene synteny from other invertebrate mitochondrial genomes, with all genes encoded on one strand, the presence of DUI, not recognized until 1994 [8,9], and the seemingly missing ATP synthase gene atp8 (and the subsequent question of whether bivalves actually have it [10] or not [11]).
(a). Extensive natural variation
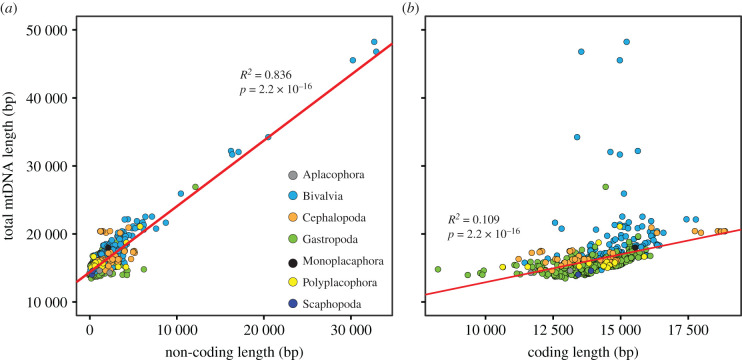
Mollusc mitochondrial genomes vary widely in size. The smallest reported so far belong to the heterobranch gastropods at approximately 13.6–14.1 kb (e.g. [4,5,12–16]) and the scaphopods [17,18]. These are only slightly larger than the smallest animal mitochondrial genomes [19], but still contain all 37 genes typical of metazoan mtDNAs, including 13 protein-coding genes, 22 tRNAs and 2 rRNAs, as well as a putative control region [12]. Not unexpectedly, these compact mitochondrial genomes feature high levels of overlapping gene boundaries. The largest mtDNAs come from the scallop Placopecten magellanicus (up to 42.0 kb, [20]) and the Arcidae clams, with Scapharca broughtonii ranging up to approximately 51.0 kb [21] and a recent report claiming that the S. kagoshimensis mitochondrial genome is approximately 56.2 kb in length [22]. The S. broughtonii mtDNA (and that of S. kagoshimensis, if verified) represents the largest animal mitochondrial genome yet recorded out of approximately 86 900 mtDNAs from more than 11 600 species present on NCBI. In both scallops and ark shells, the large genome sizes are not primarily a result of duplications or longer intergenic regions, but rather of expansion of the largest non-coding region [21,23], as is commonly the case for size variation in other mollusc mtDNAs (figure 1). These bivalves are all exceptionally long-lived, especially the Arcidae, raising the question of whether long generation times affect the pace of evolutionary change in mitochondrial genome size, although other long-lived molluscs (e.g. abalone) do not share similar expansions of their mitochondrial genomes [24].
Figure 1.
Relationship between the length of (a) non-coding and (b) coding regions on total mtDNA length in molluscan classes. Variation in non-coding length explains a greater proportion of variation in total mtDNA length compared to variation in coding length. Each circle represents a single species. When multiple mtDNAs were available for a single species, the mean across all individual records was taken as the species value. Colours represent different molluscan classes and are indicated by the key in (a).
Molluscan mitochondrial genomes have substantial variation in nucleotide composition skew asymmetry (i.e. heavy versus light strand, [25]). Strand asymmetry occurs when there are more purines (i.e. adenine and guanine) on one DNA strand than there are pyrimidines. The strand with more purines than pyrimidines is heavier and, therefore, moves farther along in caesium chloride density gradient centrifugation when separated than the complementary strand [26] and is therefore termed the heavy or ‘H’ strand, and the other the light or ‘L’ strand. This skew is thought to be caused by the bias in types of spontaneous mutations that occur in single-stranded DNA (i.e. heavy versus light strand, [25]), a condition that occurs for the displaced strand during transcription or replication (see a characterization in [27], a process known to be unusually slow for mtDNA [28]). The degree of nucleotide skew is particularly large around the control region, as this region is found in single-stranded conformation more commonly than the rest of the molecule. There have been numerous reversals of strand asymmetry in molluscs [29], likely as a result of inversions in the control region, which contains one or both origins of replication [30,31].
Molluscs have experienced many changes in the transcriptional orientations (i.e. inversions) of genes, placing them variously on strands of differing nucleotide composition skews. For example, some taxa have all genes on one strand, like all marine bivalves (e.g. scallops, oysters and clams: [32–34]) and all protein-coding genes of caenogastropods [35], while others do not, such as unionid mussels [36], heterobranchs [15], vetigastropods [37], cephalopods [38,39], scaphopods [17], aplacophorans [40] (but see [41], in which all sequenced genes of the Spathoderm clenchi mtDNA are on the same strand), monoplacophorans [42] and polyplacophorans [3]. More generally, changes in genome architectures that alter transcriptional patterns across lineages are common and appear to be largely mediated by tRNA transposition and inversion [15], as the secondary structures are hypothesized to form transcriptional barriers [43] and RNA cleavage signals [44].
Indeed, changes in the gene order are most common for tRNAs. Even families like Haliotidae that exhibit largely conserved synteny of the protein-coding genes exhibit variable tRNA locations [37]. Duplication of tRNAs appears to be a major contributor to mitochondrial genome rearrangement, as expected for the ‘duplication-random loss model’, with evidence that many molluscs contain extra tRNAs [21,32] beyond the minimal set of the 22 essential for accommodating the ‘super-wobble’ of mitochondrial translation. Interpreting this pattern of tRNA translocations is complicated by cases of remoulding of tRNA anticodons, which occurs sporadically throughout molluscs [45–47] and otherwise [48]. The cases where a single amino acid is specified by two different codon families (serine and leucine) are especially susceptible to this because a switch of anticodons alone would be sufficient since these tRNAs would each have the necessary internal signals for charging with the correct amino acid [49,50].
Still, there has been a large number of rearrangements of the genes encoding proteins or rRNAs, often via tandem duplication [51–53] or large-scale inversions (e.g. vetigastropods [37], versus caenogastropods [54]). In contrast with Vertebrata and Arthropoda, in which gene arrangements have remained generally very stable, extensive gene order rearrangements have been documented in every major lineage within Mollusca, including caenogastropods [55], scaphopods [18], cephalopods [38], heterobranchs [56], bivalves [57,58], aplacophorans [40,41], polyplacophorans [59] and monoplacophorans [42]. The extent of this variation has understandably added complexity to inferring ancestral gene order, as until recently many lineages were too lightly sampled to accurately infer evolutionary paths (e.g. [60], versus [61,62]).
Across animal life, in nearly all lineages, there has been strong selection to maintain the minimal set of 37 genes (but see [63]). With the possible exception of atp8 in bivalves [10,11], the genes encoding proteins or rRNAs are seldom lost and duplicates are rarely maintained for long periods in molluscs (but see [45,64,65]), and molluscan mtDNAs rarely contain fewer than the necessary minimal set of 22 tRNAs (but see [66]). There has long been speculation about the selection pressures that are responsible for this [67], including suggestions that hydrophobic proteins cannot easily move across membranes, that these proteins may be destructive in the cytoplasm, or that there is value in regulating mitochondrial function with this genome that is a remnant of its prokaryotic ancestor [67–69].
Additions to the mitochondrial genetic repertoire are uncommon but, here too, molluscs provide many of the exceptions. For example, lineage-specific open reading frames (ORFs) have been identified in bivalves that exhibit DUI [70], of which the male version in Ruditapes philippinarum was proposed to be virally derived [71]. Additionally, there is evidence of nuclear-derived genes inserting into the mitochondrial genome. For example, a novel ORF was discovered with no sequence- or domain-based homology to the rest of the mitochondrial genome of the pearl-lip oyster Pinctada maxima but has domain-based homology to the nuclear genome [72]. The mitochondrial genome of the Arcidae clam Tegillarca granosa contains 32 novel ORFs, none of which have any homology to the rest of the mitochondrial genome, and eight of which are predicted to have signal peptides, a hallmark of nuclear but not organellar genes [73].
Early studies of transcription and translation in mitochondrial systems showed cases where the adjacent gene pairs atp8-atp6 and nad4L-nad4 were not enzymatically separated as mRNAs (see more below and [74]) and, instead, were separately translated into proteins by initiation on the ribosome, sometimes at the beginning of this bicistron and other times at an internal codon [75–77]. Perhaps this is due to difficulties with translating the very small mRNAs from atp8 and nad4L. Early mitogenome sequencing revealed that these pairs were adjacent even in cases of more highly rearranged genes, suggesting this as a universal molecular process. But some molluscs do not have atp8-atp6 as adjacent [39,56,78,79] and others do not have nad4L-nad4 as adjacent (polyplacophorans [3], heterobranchs [15], scaphopods [17,18], unionid mussels [36,37], cephalopods [38,39], aplacophorans [40], monoplacophorans [42] and gastropods [56]), indicating that there must be other modes of translation and regulation.
Not only are gene rearrangements rampant in mollusc mitochondrial genomes, but even individual genes exhibit remarkable architectural variation. Perhaps most prominent among these is the splitting of the large ribosomal rRNA gene (rrnL) into two distinct genes in Crassostrea oysters [80]. The resulting transcripts do not appear to be spliced together into a single RNA, but the ribosome itself appears to be fully functional [81]. The partially duplicated rrnL and rrnS genes of the vermitid snail Thylacodes squamigerus mitochondrial genome bear a superficial resemblance to Crassostrea's split rrnL, but the fragments appear to be pseudogenes [50].
Evidence for variation in genic architecture also comes from an intriguing case of apparently convergent evolution of the male-specific version of cox2 in bivalves exhibiting DUI (see more below). In Mytilidae, cox2 is extended at the 3′ end of the transcript [82], but in some Veneridae, cox2 has a male-specific insertion in the middle of the gene [79]. It is unclear whether these cox2 modifications share similar functions, although the former was hypothesized to have a role in reproduction [83]. Finally, tRNAs are commonly found to have truncated D arms, especially in the heterobranchs [84], and there is even a case in which a tRNA has been inserted into nad5 [85]. These evident departures from the typical mode of intense purifying selection acting on mitochondrial genes likely represent lineage-specific mitochondrial adaptations and more work is required to understand their functional importance.
The largest non-coding region, inferred to perform the functions of the ‘control region’, varies widely in location also; see, for example, its varying positions in Mytilus [2] versus scallops [86], squid [87,88] and caenogastropods [89]. And the content and structure of control regions are vastly different across the major molluscan lineages, with high rates of evolutionary turnover by novel tandem duplications, often of previously duplicated regions [20,38,51,73,90,91]; transpositions, especially of tRNAs, into this region [21,32,73,92,93]; and newly evolved simple sequence repeats such as poly(AT) tracts [94,95]. Together these primary sequence features share the ability to produce secondary structures including stem-loop [34,96], cloverleaf [56,84,97,98] and cruciform [89] structures in the control region, which in other organisms appear to be related to mtDNA replication and transcription [1,56].
Some control regions provide especially valuable insight into the biology and evolution of mitochondrial genome architecture. For example, squid control regions harbour relics of tandemly triplicated whole mitochondrial genomes, followed by their subsequent loss [61,64,87,88,99]. Heterobranchs have extremely short control regions, reflecting their compact mitochondrial genomes [12], while caenogastropods have control regions of variable length with an inverted repeat interspersed by a simple sequence repeat [54,89]. Control regions of mussels exhibiting DUI have lineage-specific, tripartite control regions consisting of two variable domains interspersed by a conserved domain [93]. Recombination between the F-type and M-type control regions in which an F-type mtDNA acquires an M-type control region appears to coincide with the masculinization of F-type mtDNAs ([92,100–102]; see DUI section below for more details). Thus, although control regions are often omitted in mitochondrial genome assemblies, generally because of technical difficulties in amplifying or sequencing these regions, those that have been sequenced provide rich sources of information for understanding evolution of mitochondrial genome architecture.
(b). Moving forward to understand the processes that contribute to variation in mitochondrial genome architecture
This rich phenomenological record described above makes for an ideal system in which to investigate the underlying molecular, genetic and evolutionary mechanisms contributing to and maintaining variation in genome architecture. Based on this diversity, a few themes have emerged that warrant further investigation. First, tRNA-mediated changes in gene order have been observed across Metazoa [103]. It is hypothesized that at least part of this pattern results from accidental incorporation of tRNAs into the mtDNA when they moonlight as primers for DNA replication [48]. This hypothesis is attractive because it would also help explain why control regions often feature pseudo-tRNAs (e.g. scallops, oysters and clams [32,73,93]) and other tRNA-like secondary structures [56,84,97,98,104]. Misincorporation of tRNAs might also contribute to the high rates of evolutionary turnover in the control region, as new tRNA incorporation events push older sequences out of the control region. Complicating our understanding of this process are the evolutionary histories of tRNAs, as tRNA remoulding can obscure tRNA evolutionary history (see above). Quantifying the extent of tRNA duplication and remoulding, as well as rates and patterns of control region turnover in molluscan mitochondrial genomes, will provide valuable insight into tRNA-mediated genome architectural change.
Second, tandem duplication, which has been implicated in several molluscan genome rearrangements (e.g. [20,21], cephalopods: [38]), can happen through a variety of mechanisms [105,106] including slipped-strand mispairing [107], imprecise termination of replication [108,109], dimerization [110] and illegitimate or non-homologous recombination between repeats [111,112]. Support for the role of tandem duplication in shaping mitochondrial genomes is undermined by the scarcity of animal mitochondrial genomes that harbour duplicated copies of protein-coding genes [113]. It may be that duplicates are lost quickly, perhaps responding to selection favouring the maintenance of cytonuclear stoichiometry [114]. Evaluating these various possibilities will require better population-level sampling, especially with the help of long-read sequencing technologies like PacBio or Oxford Nanopore, which can help resolve tandem duplications [115,116].
Third, inversions are perhaps the most commonly retained form of structural rearrangements in molluscan mitochondrial genomes (see above paragraph on changes in transcriptional orientation). Inversions can arise via multiple double-stranded breaks or by inverted repeats (see [117] for the description of inverted repeat mechanisms) in which one repeat is deleted, likely via recombination [118]. However, inversions would seem to have immediately deleterious consequences for transcriptional control of mitochondrial genomes. There has been speculation of an ‘evolutionary ratchet’, whereby genes rearranging by inversions to be on a single strand would eliminate the selective pressure to maintain transcription of the other strand and, once lost, would make any further inversion of any gene immediately non-functional such that reversion to a state of genes on both strands would be highly unlikely [113]. Investigating mitochondrial transcriptional dynamics in closely related species (or M- versus F-type mtDNAs from the same species) that have inversions relative to one another might prove especially useful in understanding how inversions are able to persist longer than other types of mitochondrial genome rearrangements. How these inversions and subsequent changes in expression affect mitochondrial function and fitness will also be of broad interest to the mitochondrial community.
Fourth is the evident selective pressure for genome streamlining, both in terms of gene content and genome size. One of the more surprising observations of animal mitochondrial genomes is the degree to which genes overlap [96,113,119]. Overlapping mitochondrial ORFs often exhibit alternative reading frames [113], such that elongation of a gene via nonstop mutations may explain variation in the degree of gene overlap. Once genes do overlap, purifying selection is expected to be intense over the region, as mutations occurring in the overlap could have consequences for two separate genes. The greater degree of overlap between nad4 and nad6 in the M-type genome of Solenaia carinata compared to the F-type [96] raises the intriguing question of whether the increased intensity of selection engendered by gene overlap might compensate for the reduced efficacy of selection acting on male versus female transmitted mtDNAs [120]. Comparing whether mitochondrial genomes with high versus low Ne (e.g. F-type versus M-type mtDNAs) have lesser degrees of genic overlap and reduced rates of deleterious mutation accumulation [121] would provide a powerful test of the forces contributing to genome streamlining of animal mitochondrial genomes.
Finally, the extent to which gene order can be used as an effective phylogenetic tool for molluscs [47,61,122,123] depends upon low-level taxonomic sampling to infer rates and patterns of structural evolutionary change. The availability of more than 1000 molluscan mitochondrial genomes from over 700 different species as of September 2020 has largely solved that problem, especially for the bivalves (456 mtDNAs from 261 species), gastropods (452 mtDNAs from 358 species) and cephalopods (142 mtDNAs from 60 species). Such gene order analyses should not only take advantage of changes in major gene synteny but also of tRNA movements and inversion events. Together, these five avenues for future research represent central open questions in the evolution of mitochondrial genome architecture and should provide a framework for understanding how genome architecture contributes to mitochondrial function at molecular, cellular and organismal levels.
3. Annotation challenges
Considerable effort is required for annotation of the genes of molluscan mitogenomes. Most protein-encoding genes are easily identified with orthologues by sequencing similarity, with occasional consideration of hydrophobicity plots for atp8 and nad4L, but there are challenges with inferring the correct start codon in cases where there are multiple, closely spaced alternatives. An inference must consider the possibility of overlap with the upstream gene and the extent of evolutionary conservation of the ORF. This is confounded by the fact that molluscs employ the invertebrate mitochondrial genetic code (NCBI Genetic Code 5) that allows for alternative start codons in addition to ATG, including ATA, ATY, TTG and GTG (normally encoding for methionine, isoleucine, leucine and valine, respectively). Each of these would provide a match to at least two nucleotides of the trnM anticodon (CAU), which must do double duty in most mitochondrial systems as the tRNA for both methionine and, in the case of protein initiation, formyl-methionine.
Ordinarily, inferring a stop codon for any gene is straightforward but, here too, mitochondrial genomes present a challenge. In many cases, mitochondrial genomes are transcribed as a single polycistronic RNA from each strand (see [124]). The tRNA genes are then removed enzymatically, which liberates gene-specific mRNAs as proposed in the ‘punctuation model’ [44]. In the case of overlapping atp8-atp6 and of nad4L-nad4, these have been shown for yeast [74], fish [125] and mammals [126] to remain as bicistrons that are translated on mitochondrial ribosomes, sometimes from the first codon and sometimes from an internal codon that initiates the second gene. In some other cases of adjacent protein-encoding genes without an intervening tRNA, there are potential secondary structures that have been speculated to serve this function (e.g. [3]). In many other cases, it remains unknown whether these mRNAs are separated or not. The specific challenge for gene annotation from genome sequence is that, after enzymatic processing to produce gene-specific messages, some will not have a complete TAG or TAA stop codon, but may terminate on just a TA or T that is completed to a TAA stop codon by polyadenylation of the transcript [127]. Additionally, it is important to consider that some genes are known to overlap even when on the same strand, further complicating an accurate inference of the correct stop codon from genome data alone.
Of course, there are some cases where these features can be directly observed through the sequencing of expressed sequence tags (ESTs) [14], providing the sequences of the full transcripts from which the genomic boundaries can be reliably determined. This has presented some surprises. For example, ORF analysis had predicted that nad4 of the gastropod Biomphalaria glabrata mitogenome (NC_005439) was unusually long, fully overlapping with trnT, in contrast with the reported genes in the gastropods Cepaea nemoralis (NC_001816) and Albinaria caerulea (NC_001761). Independently determined EST data (AA547758) showed the cDNA for the C-terminus of nad4 to end before the downstream trnT gene, more consistent with those of the other gastropods, and to terminate on a single T nucleotide that was extended by polyadenylation to form a TAA stop codon [14].
Based on genome sequence alone, inferring the exact boundaries of rRNA genes is especially difficult. In fact, in most cases, there is simply the presumption that the rRNA gene extends to the boundary of the flanking genes, with this moderated by the extent of similarity matching to homologous genes of other organisms.
The genes for tRNAs diverge in sequence rapidly and are most commonly found by identifying potential secondary structures with a set of typical features [128–130]. Some lineages are known to have aberrant structures with some of the arms diminished or even missing, complicating this inference.
The rise of next-generation sequencing (NGS) has been a game-changer for the pace of generating complete mitogenome sequences. These methods generate an enormous number of short sequencing reads, leading to an increased reliance on computational methods for automated genome assembly. Among several alternative software packages that aim to assemble NGS data into large contigs, MITObim was specially designed for the assembly of mitogenomes [130], as well as other tools that were released more recently [131–133]. Using a provided mitochondrial genome or even a short (partial) gene sequence as an initial reference to identify sequence data of likely mitogenome origin, this program applies a strategy of BLAST and iterative mapping to select and assemble short reads from a large NGS dataset that provides adequate coverage into a linear representation of a mitogenome. Overlapping, identical sequence termini indicate that the assembly represents the full circular mitogenome. It is worth noting that reliance on computational interpretation of short sequence reads may potentially cause problems in assembling repetitive elements, such as the control region and unsuspected repetitive elements like tandem duplications or repeat regions, that may be resolved only by manual, targeted sequence characterization.
With such relative ease to derive the genome sequences, there is a greater demand for automated annotation. This need was recognized early on by the implementation of semi-automated annotation of genomes of organelles from mitochondria (and plant chloroplasts) through DOGMA (Dual Organellar GenoMe Annotator) that provided predictions of protein- and rRNA-encoding genes through BLAST similarities to previously annotated mitochondrial genomes [134] and provided tools for manually refining the beginning and end of each gene. The identification of tRNA genes employed secondary structure predictions because mitochondrial tRNA sequences share little sequence similarity among animals. Generally, computational predictions were further hindered due to the aberrant structure of several molluscan tRNAs that do not conform to the canonical cloverleaf of animal tRNAs and typically required manual validation [6]. Current utilities include AGORA (prediction of protein-coding genes in a mitogenome assembly based on BLAST similarities to a reference mitogenome; [135]), MitoZ [131–133] and MITOS [136]. The latter software performs de novo annotation of protein-encoding genes by sequence similarity and secondary structure predictions of both rRNA and tRNA. MITOS reports annotation results in the standardized format that supports the accepted, consistent nomenclature of mitochondrial genes. Updates (MITOS2 is available at http://mitos2.bioinf.uni-leipzig.de/index.py) have improved the prediction accuracy but the results still require manual curation.
Alternative start codons, the potential for incomplete stop codons and molluscan-specific tRNA structures continue to challenge automated annotation. Some possible challenges for annotation are shown in figure 2, using atp8 from gastropod mitogenomes as an example. atp8 is the shortest protein-coding gene in mitogenomes and relatively variable among gastropod species, often not detected by BLAST and thus also not recognized by MITOS. Additionally, atp8 of several gastropod species employs an alternative start codon, like ATT that normally encodes for an I (isoleucine), serving as start codon (specifying formyl-methionine) only at the initiation of protein translation. Automated gene finding and inexperienced annotators may fail to recognize ATT as a true start, choosing an upstream M-encoding nucleotide (ATG or ATA), even if part of a different gene, as an incorrect start codon. As a consequence, annotation of atp8 often requires manual inspection and comparison to atp8 from several species (figure 2).
Figure 2.
In mitogenomes of planobid gastropods, the atp8 gene is bracketed by trnN(aac) and trnL2(tta). Shaded boxes, tRNA genes; white boxes, protein-coding genes; arrowheads indicate directionality; asterisk, stop codon. ORF analyses of the mitogenome sequences that ignore the concept of tRNA gene excision from polycistronic mitogenomic transcripts frequently yield incorrect prediction of protein-coding sequence intervals. Whereas the start codon is correctly indicated, the ORF for atp8 from Biomphalaria glabrata (underlined in both nucleotide and predicted amino acid sequences, NC_005439) falls short, despite an effort to accommodate an incomplete stop codon (T--). Another issue impacts the ORF selected from the Planorbella duryi mitogenome (KY514384). It comprises a (correct) start codon and TAA stop codon but overlaps with trnL2 and yields an unusually long protein sequence. For both snail species, considering the boundaries of the (MITOS predicted) tRNA genes, the ATA is the first possible start codon downstream from trnN. At the 3′ end, a single T nucleotide remains after excision of trnL2, completed by polyadenylation to a TAA (underlined) stop codon. Such peculiarities challenge prediction of multiple genes from molluscan mitochondrial sequences, as is evidenced in several GenBank entries, despite the purported curation of submissions by this NCBI database. Re-evaluation and, if appropriate, updates by contributors of previous GenBank accessions will greatly benefit correct annotation.
A recent paper by Fourdrilis et al. [137] provides a powerful set of criteria to integrate with automated MITOS prediction for correct annotation of gastropod (molluscan) mitogenomes. These criteria include the valid insights into molluscan mitochondrial biology, including the punctuation model, as well as alternative start and stop codons. We summarize these criteria below: (i) Protein-coding genes are assumed to begin at the first eligible in-frame start codon in their 5′ end, that is, the start codon nearest to the preceding gene without overlapping with it, checking that this start codon is suitable regarding the location and gene length by aligning the derived amino acid sequence with that of closely related species. (ii) Due to transcription of mtDNA as polycistronic RNA, it is considered physically impossible to have gene overlap between two protein-coding genes encoded on the same strand and in the same open reading frame, but possible if frames are different. (iii) Protein-coding genes are assumed to end at the first in-frame full stop codon, or an abbreviated stop codon (TA- or T- in invertebrates) ending immediately before the downstream tRNA; such an abbreviated codon results from the cleavage of the transcript at the 5′ and 3′ ends of tRNAs and tRNA-like secondary structures and is subsequently completed to a TAA stop codon with A residues by polyadenylation. (iv) Putatively duplicated genes are evaluated based on quality values provided in the MITOS analysis. (v) The boundaries of tRNA genes are those predicted by MITOS. (vi) The boundaries of rRNA genes were those predicted by MITOS and not extended to flanking genes to avoid overestimating rRNA gene length.
Despite these software packages for assistance and the attention of the scientific community, the entries for mitochondrial genomes at NCBI contain a great number of easily recognized annotation errors even in the ‘Refseq’ portion. Despite having this pointed out over a decade ago with specific, simple recommendations for systematically eliminating these and conducting quality control for new entries [27], a recent study identified a great number of errors in a systematic search of complete vertebrate mitochondrial genomes at NCBI [138]. To the best of our knowledge, no such systematic study has been made of annotations for complete mollusc mitogenomes, but there is no reason to suspect that they are immune from similar errors during submission or NCBI review (e.g. [137]). Consistent, accurate, complete annotation of these genomes is critical for comparative and phylogenetic studies. We urge NCBI to implement these simple quality control measures.
4. Inheritance: doubly uniparental inheritance in bivalves
Mitochondrial genomes follow a non-Mendelian inheritance pattern of being transmitted uniparentally in most eukaryotes; in animals, mitochondrial inheritance is usually strictly maternal (from now on: strictly maternal inheritance, SMI) [139,140]. Perhaps the most striking feature of mollusc mitochondrial biology is the unique doubly-uniparental inheritance pattern so far reported in 100+ species of bivalves [141]. In species showing DUI, two sex-linked mitochondrial lineages exist: one is inherited through eggs (F-type) the other through sperm (M-type). Differently from the cases of paternal mtDNA leakage reported in several organisms [142], in DUI the sperm transmission route is stable across evolutionary time, so the F- and M-type coexist as segregated lineages for millions of years accumulating a remarkable sequence divergence. The F-M nucleotide p-distance ranges from 0.08 to 0.449, and the amino acid p-distance of mitochondrial protein-coding genes can reach 0.534 [141].
The dynamics and distribution of F- and M-type in embryos and tissues were first investigated in bivalves of the Mytilus species complex, in which DUI was observed for the first time (reviewed in [143]). Particularly interesting was the finding that in early embryos (2–8 blastomeres) sperm mitochondria stained with MitoTracker Green showed two different distribution patterns: dispersed versus aggregate. The authors were also able to show a strong link between the pattern and the sex of the progeny: females were associated with the dispersed pattern, males with the aggregated one [144,145]. These observations, together with the results of several molecular works, were used to build a first description of the mitochondrial dynamics in DUI, summarized below. Gametes are homoplasmic for the sex-specific type (F-type in eggs, M-type in spermatozoa), so upon fertilization the zygote is heteroplasmic and the fate of sperm mitochondria is tightly linked with sex. If the embryo develops into a female, the M-type mitochondria are dispersed and actively degraded as happens in some species showing SMI [146], and the animal will be homoplasmic for the F-type. Otherwise, if the embryo develops into a male, sperm mitochondria stay aggregated as they already are in the midpiece of sperm cells and are transported into the blastomere 4d, the precursor of the germline, and survive degradation; males are thus heteroplasmic, containing M-type in the germline and F-type in the somatic tissues.
The main points of this model are: (i) homoplasmy of females due to degradation of M-type; and (ii) heteroplasmy of males with retention of M-type due to the active segregation of sperm mitochondria aggregated in gonad precursors, but not in somatic tissues. A replicative advantage of M-type in males was also hypothesized, to explain its proliferation in spermatogenic tissues [145]. This is still the most commonly used description of the DUI mechanism, but some revisions have become necessary. The existence of the two patterns was confirmed in a distantly related species (divergence time 400+ Ma), the venerid clam Ruditapes philippinarum [147], but as new data were gathered and new species analysed, evidence of deviations from the mechanism as described above started emerging. The presence of M-type in male somatic tissues is now known to occur in R. philippinarum [148], Venustaconcha ellipsiformis and Utterbackia peninsularis [149] and in Mytilus galloprovincialis [150,151].
These works showed also that heteroplasmy is more common than previously thought in both males and females of DUI species, and that the presence, abundance and distribution of the F- and M-types is quite variable across species, sexes and tissues. Such differences should be expected when dealing with a quantitative phenomenon like mitochondrial inheritance [139], especially across large evolutionary distances. Recently, immunohistochemistry and microscopy (both confocal and electronic) investigations on R. philippinarum showed the presence of heteroplasmy at the organelle level (both types present in the same mitochondrion) in male soma and, quite surprisingly, in undifferentiated germ cells of both sexes, while homoplasmy in both female and male gametes was confirmed [152]. According to these observations, the strict segregation of F- and M-type in gametes would be achieved during gametogenesis—thus much later in development than hypothesized before—and it was suggested that DUI is based on a mechanism of meiotic drive involving selfish genetic elements associated with mitochondria [152,153].
(a). Doubly uniparental inheritance molecular mechanism
Hybrid and triploid DUI mussels have been shown to revert to SMI [154] and the taxonomic distribution of DUI species is scattered across bivalve phylogeny, so DUI must have evolved by the modification of a mechanism of SMI, but which one? There are several different mechanisms by which SMI can be achieved [146,155], but that operating in bivalves is still unknown. Similarly to what happens in mammals, it was hypothesized that ubiquitination could be involved [156] and the results of some investigations seem to be consistent with such supposition [157–160]. A possible approach to understand which molecular mechanism is involved in DUI is to look at the differences between F- and M-type genomes, and numerous works have investigated this issue in the last 25 years. The main findings can be summarized as follows.
First, bivalve mtDNA shows an abundance of intergenic regions—or at least regions not containing known genes—and the largest are rich in genetic elements such as repeats, motifs and DNA/RNA secondary structures which differ between conspecific F and M genomes in DUI species (e.g. [93,161–164]). A strong clue supporting a role of control region elements in DUI comes from observations in the Mytilus complex. Several analyses on F- and M-type mtDNAs in Mytilus edulis, M. galloprovincialis and M. trossulus revealed the presence in male gonads of genomes having their coding sequences almost identical to those of the F genome (2–3% divergence). It was hypothesized that these genomes originated from F genomes that invaded the male germline and started to be transmitted through sperm, replacing the M-type and accumulating sequence divergence (which is initially reset to zero when the F-type replaces the M-type). This phenomenon was named ‘role-reversal’ or ‘masculinization’ (reviewed thoroughly in [143]), and the aberrant F genomes transmitted through sperm have been defined as ‘masculinized’. Following studies found that the control regions of masculinized genomes contained parts of both the typical F- and M-type mtDNAs, being actually F/M chimaeras. Role-reversal has been observed, so far, only in the Mytilus complex. These findings strongly suggest that some elements located in the control region or its proximity have a role in the inheritance mechanism. The identity and the nature of these elements are still unknown and several candidates have been proposed, including DNA and/or RNA secondary structures [162,165], specific sequences/motifs [166], or peptides encoded by ORFs located near the control region (see second point below).
The second feature that differentiates F and M genomes is the presence of lineage-specific ORFs showing no sequence similarity with known genes, and thus defined ‘ORFans’ [70,71,141,153,165,167–170]. In some cases, a protein product of these ORFans has been detected and localized [71,152,171], but their function remains unknown despite extensive in silico analyses [71,153,168–170]. Such bioinformatics work has shown that, despite high evolutionary rates and large sequence divergences, all the analysed ORFans have similar predicted structural features, supporting a similar function. The involvement of the ORFans in the DUI mechanism is still a hypothesis and their mechanism of action is an object of speculation, but it is clear that these elements are maintained in bivalve genomes and some surely produce a novel mitochondrial protein. It would be surprising if these elements turn out to be non-functional.
Third, the cytochrome c oxidase subunit 2 gene (cox2) shows curious features in bivalves, and in several DUI species, there are important differences between the F-type and M-type cox2 gene (see also §2). The cox2 gene is duplicated in the F-type of R. philippinarum [165] and the M-type of Musculista senhousia [161], with paralogous copies showing different lengths. In some other cases, cox2 has a different length in the two mtDNAs, due either to 3′ coding extensions (550 bp) or large in-frame insertions (up to 3.5 Kb) [141]. It is still not clear if such modifications of cox2 are linked to DUI for some functional reason, or are a more general feature of bivalve mtDNAs, maybe due to modifications in Complex IV of oxidative phosphorylation.
The fourth and last feature characterizing the differences between the two mitochondrial lineages concerns small non-coding RNAs (sncRNAs). Pozzi et al. [172] sequenced sncRNA libraries from gonads of R. philippinarum, and found miRNA-like sequences transcribed by intergenic regions for which a stable hairpin structure was predicted. In silico analyses showed that F and M genomes produce different mitochondrial sncRNAs with different nuclear targets. The authors hypothesized that such sncRNAs might affect nuclear gene expression through RNA interference and might influence gonad formation. More recently, Passamonti et al. [173] reported in vivo clues of the activity of two sncRNAs in R. philippinarum. Small mitochondrial RNAs have also been predicted in silico in several species of amniotes [174], and in Drosophila melanogaster, Danio rerio and Mus musculus [173].
(b). MtDNA evolutionary patterns in doubly uniparental inheritance
It is still unclear how DUI emerged and why it has been maintained for hundreds of millions of years. Traits that last so long in evolution are usually maintained by natural selection because they have a function that affects organismal fitness. For this reason, and given the tight link between mitochondrial inheritance pattern and sex in DUI species, it was hypothesized that DUI has a role in sex determination and/or gonad differentiation [143,153,157,159,171,175–178].
Studies on the patterns of molecular evolution of mitochondrial proteins in DUI bivalves clearly show that M-type evolves faster than F-type and both mtDNAs evolve faster than the mitochondrial genomes of other metazoans [143,175]. The reasons behind this pattern are the subject of debate. Relaxed selection is one possible explanation; Stewart et al. [179] suggested that F- and M-type mtDNAs evolve under different degrees of selective constraints as a consequence of different ‘selective arenas’. Supposing that F-type mtDNA is functional in all somatic tissues and the female germline, while M-type functions only in the male germline, F-type would be subject to more stringent constraints, hence the faster sequence evolution of M-type. However, the more recent findings of F- and M-type distribution across tissues (discussed above), and the findings of M-type transcriptional activity in the soma [149,180], may suggest that the above-mentioned arenas of function are not that distinct. Moreover, even if M-type mitochondria are functional only in the male germline, they have a crucial function of providing energy for sperm swimming. This is a fundamental function, especially in a broadcast spawning animal, and the relaxation of natural selection on such a trait could have long-term consequences on DUI species. Many DUI species are quite successful; for example, R. philippinarum is highly invasive, and Arctica islandica (in which DUI has been reported [181]) is the longest-living non-colonial animal known (maximum reported lifespan approximately 507 years), so it seems that DUI is not manifestly disadvantageous.
A high-throughput analysis of mtDNA single nucleotide polymorphisms (SNPs) in F- and M-type of R. philippinarum [165] revealed a similar amount of polymorphism in the two genomes, but a different distribution of allele frequencies (probably due to different bottleneck sizes), the M-type having a lower proportion of SNPs with a predicted deleterious effect. According to these data, the faster evolution of M-type is likely due to the roles of mitochondria in spermatogenesis and sperm motility, the latter being especially important in the intense sperm competition of an animal using broadcast fertilization. Indeed, one interesting feature of DUI is that mtDNA is under selection also for male functions, differently from what happens in all the SMI organisms, in which mitochondria are an evolutionary dead-end in males. This opens a series of interesting consequences and deserves thorough investigations. Recently, two comparative analyses of OXPHOS activity in gametes and somatic tissues of SMI and DUI bivalves reported a metabolic remodelling in M-type mitochondria that suggests an adaptive value of mtDNA variation, and a link between male-energetic adaptation, fertilization success and the preservation of paternally inherited mitochondria [182,183].
DUI is generally unknown or considered just a ‘freak of nature’, but it represents a unique and precious model to study mitochondrial biology and evolution. Thanks to its unusual features, it can be used as a tool to better understand mitochondrial heteroplasmy, inheritance, recombination, and the role of mitochondria in germline formation, meiosis, gametogenesis and fertilization, in some cases providing the exceptions that address general phenomena in other animal groups. Up to now, DUI has not been found outside bivalves, but, to the best of our knowledge, it has been specifically investigated in just five gastropod species [184].
5. The utility and limitations of mitochondrial genomes for phylogeny
During the last three decades, mitochondrial markers, either individually, combined or as a whole, have been commonly used for phylogenetic reconstruction within Metazoa [98,185–187]. This preference is due to several features that make mitochondrial sequences a well-suited and reliable molecular marker for phylogenetic assessment. First, all Metazoa (except some Loricifera [188]) possess a mitochondrial genome that can be obtained with relative ease compared with any particular genome region of similar size due to its high abundance and copy numbers within animal cells [98,185]. Second, gene orthology, essential for a successful phylogenetic assessment, is expected in the mitogenome, since genes from eventual duplication events shown to occur in molluscan mtDNA are rarely retained, and quickly lost or pseudogenized [98,185,187]. Furthermore, uniparental inheritance (see exception in bivalves in the DUI section above) and a general lack of recombination [189] greatly favour the reliable inference of population structure. The variable substitution rates within the different genes/regions of the mitogenome grant a range of phylogenetic signals that might potentially be useful for accessing shallow and deep relationships [98,185,187]. Mitogenomes also possess several structural features that, when thoroughly studied, can be phylogenetically informative, such as genome size, gene arrangement and content [122], as well as the presence and composition of non-coding regions and repetitive sequences, and even RNA secondary structures [185,187].
Despite the overall unarguable utility of mitogenomes for phylogenetic assessments, several limitations may affect their reliability for the same purposes. By being an ‘independent genetic unit’, that is usually uniparentally inherited with very little recombination, the mitogenome as a whole is itself a single locus that reflects the evolutionary history of the mitochondria, which for several reasons may not be the same as the species evolutionary history (e.g. due to introgression and sex-biased reproductive dispersal [187]). Furthermore, the presence of non-functional nuclear copies of mitochondrial sequences (numts) may lead to a false interpretation of phylogenetic relationships [187], particularly when single genes are amplified by PCR, and the highly variable substitution rates and base composition between taxa can make direct comparisons difficult [98,187]. Inversions can also complicate phylogenetic analysis using mtDNA gene sequences, as it is likely that genes equilibrate in nucleotide composition to their strand skew, even to the point of having convergent amino acid substitutions within physico-chemically similar groups that have arisen independently in different lineages [190].
Despite these drawbacks, overall mitogenomes represent a complete and ‘isolated’ genomic feature, easily available from a wide range of taxa, whose genetic information is comparable and compact enough to be both phylogenetically informative and investigated with low computational effort and therefore a logical choice for a comprehensive phylogenetic study. Consequently, mitochondrial DNA has been used, with a variable range of success, to assess phylogenetic relationships at several taxonomic levels ranging from shallow population-level relationships (e.g. [191]), up to phyla (Mollusca [186]; e.g. Annelida [192], Platyhelminthes [193], Rotifera [194]) and even Metazoa as a whole [98].
Although mitophylogenetics have been successfully used to infer deeper evolutionary relationships within other metazoan taxa, the same success has not been achieved for the Mollusca. The reconstruction of the molluscan deep-level relationships has been extremely challenging, and consistently recovering the monophyly of the Mollusca or even of the eight molluscan classes, both presumed to be correct based on other data, has not been possible using mitochondrial markers alone [40,98,186,195,196]. Moreover, only recently and through the application of phylogenomic approaches relying on several nuclear loci, have consistent monophyletic Mollusca and monophyletic molluscan classes started to be recovered [197–200]. These studies, by contradicting the generally accepted morphocladistic Testaria hypothesis, have resulted in a fundamental reinterpretation of the phylogenetic history of Mollusca. The Testaria hypothesis placed worm-like Aplacophora (Solenogastres and Caudofoveata) as a paraphyletic basal group of the Mollusca and thus postulated a progressive evolution of body complexity, with a true shell occurring only once [200]. Conversely, all the recent phylogenomic studies unambiguously support a basal dichotomy that splits the Mollusca into two major groups, the Aculifera (including the Polyplacophora and the reciprocally monophyletic Aplacophora) and the Conchifera (including the Monoplacophora, Cephalopoda, Scaphopoda, Gastropoda and Bivalvia), thus postulating that the worm-like body plan of Aplacophora was acquired secondarily and has derived from a more complex-bodied ancestor [198,201]. However, the relationships within Conchifera are more controversial, with conflicting results regarding the positioning of Monoplacophora as either basal to all other Conchifera [201] or sister taxa to Cephalopoda [198,201], as well as the positioning of Scaphopoda as sister to Gastropoda [198,199,201] or sister to a clade composed of Gastropoda and Bivalvia [197,198,201]. Nevertheless, phylogenomic studies have been fundamental to understanding early molluscan evolution and although whole genome-scale resources are now easier to obtain, the taxon sampling is still considerably reduced when compared with the mitogenomic data already available (reviewed in [202]).
The effectiveness of mtDNA markers to infer deep Molluscan phylogeny has been a thoroughly discussed subject in recent studies [40,186,196], describing several factors that may lead to the lack of phylogenetic signal and conflicting tree topologies. Phylogenies often show long-branch attraction artefacts (LBA), with molluscan mitogenomes revealing high differentiation in nucleotide abundance and strand bias. All of these features are a probable consequence of highly frequent gene order rearrangements observed in Molluscan mitogenomes, resulting in heterogeneous substitution rates and generating systematic analytical errors (see [98,186,196,203] and references within). Furthermore, ancient (Cambrian) incomplete lineage sorting and uneven taxon sampling may also play a role in the inconsistency of the inferred phylogenetic relationships [196]. These authors also explored the phylogenetic utility of other molluscan-specific mitogenome features, such as mitogenome size variation, the highly variable (sometimes absent) protein-coding gene atp8, and even the coupling behaviour of particular genes (such as atp8-atp6 and nad4L-nad4) [196]. However, a clear phylogenetic signal is once again hindered, probably by homoplasy of these features.
Within the molluscan classes, deeper relationships based only on mitochondrial markers have also been showing a variable range of success. Recent studies on the Aculifera have expressed promising results using phylomitogenomics, supporting the usefulness of both whole mitogenome sequences and structural features [41,59,204]. For instance, new phylogenetic informative mitogenome rearrangements were detected within Polyplacophora and Caudofoveata, which along with the only Solenogastres published mitogenome, revealed a conserved protein-coding gene order likely consistent with the ancestral molluscan gene order [41,59,204]. However, mitogenome availability is still scarce for groups within the Aculifera clade. For example, mitogenome sequences for all the main lineages of the best sampled Aplacophora group, Polyplacophora (n = 18), only recently became available [204] (figure 3). Similarly, Scaphopoda, for which several phylogenetic and systematics doubts persist within its major groups, is very poorly represented regarding mitogenome availability [205]. Furthermore, although phylogenetic analysis using complete mitogenomes revealed promising results for the phylogenetic assessment within the Scaphopoda, using cox1 alone did not and, therefore, a more comprehensive and intensive whole mitogenome sequencing within the group is urgently needed [205].
Figure 3.
Top: Graphic showing the number of complete (dark colours) and partial (light colours: min. size 10 000 bp) mitogenomes available in GenBank; middle: mean, minimum and maximum size (bp) of complete mitogenomes per Mollusca class; bottom: graphic showing the percentage of total species with complete mitogenomes published in GenBank. Asterisk superscripts refer to unverified size values, due to assembly challenges; critical evaluation of these publicly available mitogenome sizes and sequence content is highly recommended.
Monoplacophoran mitogenomes have been recently sequenced to test their positioning within the Mollusca. However, consistent with the low resolution of mitochondrial markers for deep molluscan classes (see above) the results were inconclusive [42]. Nevertheless, once again unique structural features (e.g. gene arrangement and presence of large intergenic regions) that may be phylogenetically informative were detected and further sampling of the group is needed [42].
Of the three most economically important molluscan classes, Cephalopoda is the best represented in terms of mitogenome availability, which nonetheless represents only 5.5% of the total species of the group. Unlike in other molluscan classes, mitochondrial markers have shown to be informative regarding the deeper Cephalopoda phylogenetic relationships, revealing their potential to resolve long-lasting phylogenetic questions within the group [61,186].
As for the two most speciose classes of Mollusca (i.e. Bivalvia and the megadiverse Gastropoda), deep-level phylomitogenomics have been constantly inefficient. Both bivalves and gastropods have very unusual mitochondrial evolutionary patterns at both nucleotide and structural level, which render them prone to analytical inconsistencies (e.g. LBA) and hamper a consistent phylogenetic inference [186,203,206]. Inevitably, only through the application of large-scale genomic approaches are the interrelationships within both classes starting to be clarified [206–209].
Contrary to these difficulties in the resolution of deeper, older evolutionary relationships, mitochondrial genes and genomes have been much more useful in resolving more recent, intrafamilial phylogenies [210,211]. Most shallow phylogeny, phylogeographic and populations genetics studies on molluscs have relied so far on one or two mitochondrial gene fragments sometimes coupled with the same number of nuclear counterparts [212–214]. However, use of these gene fragments alone may lead to biased results and fail to reveal the mitochondrial evolutionary history of species. Furthermore, obtaining a complete mitogenome is not always a possibility, either due to the higher cost of sequencing (when compared with Sanger sequencing of a single gene) or due to logistic limitations (e.g. lack of computational resources). It is therefore important to identify the genes or regions of the mitogenome that better correspond and may be used as surrogates of the whole mitogenome evolutionary history. A study on 41 unionid bivalves statistically evaluated the coherence of the individual mitochondrial gene trees and the whole mitogenome tree, indicating that the trees using nad5 sequences were the most similar to whole mtDNA trees [215]. The results of the gene fragments more widely used in molecular studies within this bivalve taxon (i.e. cox1, rrnL and nad1) were less robust. This study also tested pairs of these widely used gene markers with much higher success, indicating that the trees constructed with the large ribosomal subunit rrnL concatenated with cox1 or nad1 are highly coherent with the whole mitogenome trees [215]. Another study within the cephalopod Octopodidae family comparing the whole mitogenome with the individual gene tree topologies also showed that the nad5 trees best represented the whole mitogenome topologies [216,217]. However, these results were obtained in specific groups of molluscs and should be tested across the Mollusca to evaluate the usefulness of individual and pairs of gene fragments in representing the whole mitochondrial genome phylogenies.
Comparisons of mitochondrial genes have great potential for revealing hidden cryptic diversity aiding in species delimitation and identification [217,218] and in understanding molluscan species phylogeographical patterns and population genetic structure, since they have already been used successfully for these purposes in other taxa [219,220]. However, to our knowledge, no comprehensive phylogeographic or population genetics study on mollusc species has used this type of marker.
In summary, studies with phylogenetic analyses of whole mitochondrial sequences and structural features of molluscs have been increasing steadily over the last decade. These studies have shown limited success in representing deeper evolutionary patterns within the Mollusca and molluscan classes. However, below the family level, robust phylogenies consistent with results of other genomic and morphological studies have been obtained. Given the high potential of whole mitogenomes for barcoding, revealing cryptic diversity and obtaining robust shallow phylogenetic relationships, it is expected that an increasing number of phylogeographic and population genetics studies using whole mitogenomes will be published shortly.
6. Summary and conclusion
Despite widespread misunderstanding based on early studies that animal mitochondrial genomes are consistent in structure, function and inheritance patterns, there is actually enormous diversity among these diminutive genomes across animal life. The phylum Mollusca, in particular, is replete with examples of extraordinary variation in genome architecture, molecular functioning and intergenerational transmission. This provides a model system for studying the evolution of these features in concert with the diverse and manifold roles of mitochondria in organismal physiology and the many ways that the study of mitochondrial genomes are useful for phylogeny and population biology.
Acknowledgements
The authors would like to thank Maurine Neiman and Angus Davison for organizing the Royal Society meeting ‘Pearls of Wisdom: Synergising Leadership and Expertise in Molluscan Genomics' and for guest-editing this issue of Philosophical Transactions B.
Data accessibility
This article has no additional data.
Authors' contributions
F.G., A.G.S., C.M.A., M.L.L., J.S. and J.B. wrote the manuscript; F.G. coordinated the work.
Competing interests
We declare we have no competing interests.
Funding
F.G. was funded by the ‘Ricerca Fondamentale Orientata’ (RFO) funding from the University of Bologna, and the Canziani bequest. A.G.S. was funded by FCT—Fundação para a Ciência e a Tecnologia—under grant SFRH/BD/137935/2018. M.L.L. was funded under project ConBiomics: the missing approach for the Conservation of Freshwater Bivalves Project no. NORTE-01-0145-FEDER-030286, co-financed by COMPETE 2020, Portugal 2020 and the European Union through the ERDF, and by FCT through national funds. J.S. was funded by the National Science Foundation (DEB – 1753695, DEB – 1753851).
References
- 1.Shadel GS, Clayton DA. 1997. Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 66, 409-435. ( 10.1146/annurev.biochem.66.1.409) [DOI] [PubMed] [Google Scholar]
- 2.Hoffmann RJ, Boore JL, Brown WM. 1992. A novel mitochondrial genome organization for the blue mussel, Mytilus edulis. Genetics 131, 397-412. ( 10.1093/genetics/131.2.397) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boore JL, Brown WM. 1994. Complete DNA sequence of the mitochondrial genome of the black chiton, Katharina tunicata. Genetics 138, 423-443. ( 10.1093/genetics/138.2.423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatzoglou E, Rodakis GC, Lecanidou R. 1995. Complete sequence and gene organization of the mitochondrial genome of the land snail Albinaria coerulea. Genetics 140, 1353-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Terrett JA, Miles S, Thomas RH. 1996. Complete DNA sequence of the mitochondrial genome of Cepaea nemoralis (Gastropoda: Pulmonata). J. Mol. Evol. 42, 160-168. ( 10.1007/BF02198842) [DOI] [PubMed] [Google Scholar]
- 6.Yamazaki N, et al. 1997. Evolution of pulmonate gastropod mitochondrial genomes: comparisons of gene organizations of Euhadra, Cepaea and Albinaria and implications of unusual tRNA secondary structures. Genetics 145, 749-758. ( 10.1093/genetics/145.3.749) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mower JP, Sloan DB, Alverson AJ. 2012. Plant mitochondrial genome diversity: the genomics revolution. In Plant genome diversity, vol. 1: Plant genomes, their residents, and their evolutionary dynamics (eds Wendel JF, Greilhuber J, Dolezel J, Leitch IJ), pp. 123-144. Vienna, Austria: Springer Vienna. [Google Scholar]
- 8.Skibinski DO, Gallagher C, Beynon CM. 1994. Mitochondrial DNA inheritance. Nature 368, 817-818. ( 10.1038/368817b0) [DOI] [PubMed] [Google Scholar]
- 9.Zouros E, Ball AO, Saavedra C, Freeman KR. 1994. Mitochondrial DNA inheritance. Nature 368, 818. ( 10.1038/368818a0) [DOI] [PubMed] [Google Scholar]
- 10.Breton S, Stewart DT, Hoeh WR. 2010. Characterization of a mitochondrial ORF from the gender-associated mtDNAs of Mytilus spp. (Bivalvia: Mytilidae): identification of the ‘missing’ ATPase 8 gene. Mar. Genomics 3, 11-18. ( 10.1016/j.margen.2010.01.001) [DOI] [PubMed] [Google Scholar]
- 11.Uliano-Silva M, Americo JA, Costa I, Schomaker-Bastos A, de Freitas Rebelo M, Prosdocimi F. 2016. The complete mitochondrial genome of the golden mussel Limnoperna fortunei and comparative mitogenomics of Mytilidae. Gene 577, 202-208. ( 10.1016/j.gene.2015.11.043) [DOI] [PubMed] [Google Scholar]
- 12.Kurabayashi A, Ueshima R. 2000. Complete sequence of the mitochondrial DNA of the primitive opisthobranch gastropod Pupa strigosa: systematic implication of the genome organization. Mol. Biol. Evol. 17, 266-277. ( 10.1093/oxfordjournals.molbev.a026306) [DOI] [PubMed] [Google Scholar]
- 13.Grande C, Templado J, Cervera JL, Zardoya R. 2002. The complete mitochondrial genome of the nudibranch Roboastra europaea (Mollusca: Gastropoda) supports the monophyly of opisthobranchs. Mol. Biol. Evol. 19, 1672-1685. ( 10.1093/oxfordjournals.molbev.a003990) [DOI] [PubMed] [Google Scholar]
- 14.DeJong RJ, Emery AM, Adema CM. 2004. The mitochondrial genome of Biomphalaria glabrata (Gastropoda: Basommatophora), intermediate host of Schistosoma mansoni. J. Parasitol. 90, 991-997. ( 10.1645/GE-284R) [DOI] [PubMed] [Google Scholar]
- 15.Feldmeyer B, Hoffmeier K, Pfenninger M. 2010. The complete mitochondrial genome of Radix balthica (Pulmonata, Basommatophora), obtained by low coverage shotgun next generation sequencing. Mol. Phylogenet. Evol. 57, 1329-1333. ( 10.1016/j.ympev.2010.09.012) [DOI] [PubMed] [Google Scholar]
- 16.White TR, Conrad MM, Tseng R, Balayan S, Golding R, de Frias Martins AM, Dayrat BA. 2011. Ten new complete mitochondrial genomes of pulmonates (Mollusca: Gastropoda) and their impact on phylogenetic relationships. BMC Evol. Biol. 11, 295. ( 10.1186/1471-2148-11-295) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boore JL, Medina M, Rosenberg LA. 2004. Complete sequences of the highly rearranged molluscan mitochondrial genomes of the scaphopod Graptacme eborea and the bivalve Mytilus edulis. Mol. Biol. Evol. 21, 1492-1503. ( 10.1093/molbev/msh090) [DOI] [PubMed] [Google Scholar]
- 18.Dreyer H, Steiner G. 2004. The complete sequence and gene organization of the mitochondrial genome of the gadilid scaphopod Siphonondentalium lobatum (Mollusca). Mol. Phylogenet. Evol. 31, 605-617. ( 10.1016/j.ympev.2003.08.007) [DOI] [PubMed] [Google Scholar]
- 19.Pett W, Ryan JF, Pang K, Mullikin JC, Martindale MQ, Baxevanis AD, Lavrov DV. 2011. Extreme mitochondrial evolution in the ctenophore Mnemiopsis leidyi: insight from mtDNA and the nuclear genome. Mitochondrial DNA 22, 130-142. ( 10.3109/19401736.2011.624611) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snyder M, Fraser AR, Laroche J, Gartner-Kepkay KE, Zouros E. 1987. Atypical mitochondrial DNA from the deep-sea scallop Placopecten magellanicus. Proc. Natl Acad. Sci. USA 84, 7595-7599. ( 10.1073/pnas.84.21.7595) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y-G, Kurokawa T, Sekino M, Tanabe T, Watanabe K. 2013. Complete mitochondrial DNA sequence of the ark shell Scapharca broughtonii: an ultra-large metazoan mitochondrial genome. Comp. Biochem. Physiol. D 8, 72-81. ( 10.1016/j.cbd.2012.12.003) [DOI] [PubMed] [Google Scholar]
- 22.Kong L, Li Y, Kocot KM, Yang Y, Qi L, Li Q, Halanych KM. 2020. Mitogenomics reveals phylogenetic relationships of Arcoida (Mollusca, Bivalvia) and multiple independent expansions and contractions in mitochondrial genome size. Mol. Phylogenet. Evol. 150, 106857. ( 10.1016/j.ympev.2020.106857) [DOI] [PubMed] [Google Scholar]
- 23.La Roche J, Snyder M, Cook DI, Fuller K, Zouros E. 1990. Molecular characterization of a repeat element causing large-scale size variation in the mitochondrial DNA of the sea scallop Placopecten magellanicus. Mol. Biol. Evol. 7, 45-64. [DOI] [PubMed] [Google Scholar]
- 24.Maynard BT, Kerr LJ, McKiernan JM, Jansen ES, Hanna PJ. 2005. Mitochondrial DNA sequence and gene organization in the Australian blacklip abalone Haliotis rubra (Leach). Mar. Biotechnol. 7, 645-658. ( 10.1007/s10126-005-0013-z) [DOI] [PubMed] [Google Scholar]
- 25.Francino MP, Ochman H. 1997. Strand asymmetries in DNA evolution. Trends Genet. 13, 240-245. ( 10.1016/S0168-9525(97)01118-9) [DOI] [PubMed] [Google Scholar]
- 26.Brown TA, Cecconi C, Tkachuk AN, Bustamante C, Clayton DA. 2005. Replication of mitochondrial DNA occurs by strand displacement with alternative light-strand origins, not via a strand-coupled mechanism. Genes Dev. 19, 2466-2476. ( 10.1101/gad.1352105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boore JL. 2006. Requirements and standards for organelle genome databases. OMICS 10, 119-126. ( 10.1089/omi.2006.10.119) [DOI] [PubMed] [Google Scholar]
- 28.Clayton DA. 1982. Replication of animal mitochondrial DNA. Cell 28, 693-705. ( 10.1016/0092-8674(82)90049-6) [DOI] [PubMed] [Google Scholar]
- 29.Sun S, Li Q, Kong L, Yu H. 2018. Multiple reversals of strand asymmetry in molluscs mitochondrial genomes, and consequences for phylogenetic inferences. Mol. Phylogenet. Evol. 118, 222-231. ( 10.1016/j.ympev.2017.10.009) [DOI] [PubMed] [Google Scholar]
- 30.Hassanin A, Léger N, Deutsch J. 2005. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of Metazoa, and consequences for phylogenetic inferences. Syst. Biol. 54, 277-298. ( 10.1080/10635150590947843) [DOI] [PubMed] [Google Scholar]
- 31.Fonseca MM, Harris DJ, Posada D. 2014. The inversion of the control region in three mitogenomes provides further evidence for an asymmetric model of vertebrate mtDNA replication. PLoS ONE 9, e106654. ( 10.1371/journal.pone.0106654) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith DR, Snyder M. 2007. Complete mitochondrial DNA sequence of the scallop Placopecten magellanicus: evidence of transposition leading to an uncharacteristically large mitochondrial genome. J. Mol. Evol. 65, 380-391. ( 10.1007/s00239-007-9016-x) [DOI] [PubMed] [Google Scholar]
- 33.Danic-Tchaleu G, Heurtebise S, Morga B, Lapègue S. 2011. Complete mitochondrial DNA sequence of the European flat oyster Ostrea edulis confirms Ostreidae classification. BMC Res. Notes 4, 400. ( 10.1186/1756-0500-4-400) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yuan Y, Li Q, Kong L, Yu H. 2012. The complete mitochondrial genome of the grand jackknife clam, Solen grandis (Bivalvia: Solenidae): a novel gene order and unusual non-coding region. Mol. Biol. Rep. 39, 1287-1292. ( 10.1007/s11033-011-0861-8) [DOI] [PubMed] [Google Scholar]
- 35.Márquez EJ, Castro ER, Alzate JF. 2016. Mitochondrial genome of the endangered marine gastropod Strombus gigas Linnaeus, 1758 (Mollusca: Gastropoda). Mitochondrial DNA A 27, 1516-1517. ( 10.3109/19401736.2014.953118) [DOI] [PubMed] [Google Scholar]
- 36.Doucet-Beaupré H, Breton S, Chapman EG, Blier PU, Bogan AE, Stewart DT, Hoeh WR. 2010. Mitochondrial phylogenomics of the Bivalvia (Mollusca): searching for the origin and mitogenomic correlates of doubly uniparental inheritance of mtDNA. BMC Evol. Biol. 10, 50. ( 10.1186/1471-2148-10-50) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xin Y, Ren J, Liu X. 2011. Mitogenome of the small abalone Haliotis diversicolor Reeve and phylogenetic analysis within Gastropoda. Mar. Genomics 4, 253-262. ( 10.1016/j.margen.2011.06.005) [DOI] [PubMed] [Google Scholar]
- 38.Akasaki T, Nikaido M, Tsuchiya K, Segawa S, Hasegawa M, Okada N. 2006. Extensive mitochondrial gene arrangements in coleoid Cephalopoda and their phylogenetic implications. Mol. Phylogenet. Evol. 38, 648-658. ( 10.1016/j.ympev.2005.10.018) [DOI] [PubMed] [Google Scholar]
- 39.Boore JL. 2006. The complete sequence of the mitochondrial genome of Nautilus macromphalus (Mollusca: Cephalopoda). BMC Genomics 7, 182. ( 10.1186/1471-2164-7-182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Osca D, Irisarri I, Todt C, Grande C, Zardoya R. 2014. The complete mitochondrial genome of Scutopus ventrolineatus (Mollusca: Chaetodermomorpha) supports the Aculifera hypothesis. BMC Evol. Biol. 14, 197. ( 10.1186/PREACCEPT-1371258267135966) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mikkelsen NT, Kocot KM, Halanych KM. 2018. Mitogenomics reveals phylogenetic relationships of caudofoveate aplacophoran molluscs. Mol. Phylogenet. Evol. 127, 429-436. ( 10.1016/j.ympev.2018.04.031) [DOI] [PubMed] [Google Scholar]
- 42.Stöger I, Kocot KM, Poustka AJ, Wilson NG, Ivanov D, Halanych KM, Schrödl M. 2016. Monoplacophoran mitochondrial genomes: convergent gene arrangements and little phylogenetic signal. BMC Evol. Biol. 16, 274. ( 10.1186/s12862-016-0829-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fernández-Silva P, Enriquez JA, Montoya J. 2003. Replication and transcription of mammalian mitochondrial DNA. Exp. Physiol. 88, 41-56. ( 10.1113/eph8802514) [DOI] [PubMed] [Google Scholar]
- 44.Ojala D, Montoya J, Attardi G. 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature 290, 470-474. ( 10.1038/290470a0) [DOI] [PubMed] [Google Scholar]
- 45.Wu X, Li X, Li L, Xu X, Xia J, Yu Z. 2012. New features of Asian Crassostrea oyster mitochondrial genomes: a novel alloacceptor tRNA gene recruitment and two novel ORFs. Gene 507, 112-118. ( 10.1016/j.gene.2012.07.032) [DOI] [PubMed] [Google Scholar]
- 46.Wu X, Li X, Yu Z. 2015. The mitochondrial genome of the scallop Mimachlamys senatoria (Bivalvia, Pectinidae). Mitochondrial DNA 26, 242-244. ( 10.3109/19401736.2013.823181) [DOI] [PubMed] [Google Scholar]
- 47.Guerra D, Bouvet K, Breton S. 2018. Mitochondrial gene order evolution in Mollusca: inference of the ancestral state from the mtDNA of Chaetopleura apiculata (Polyplacophora, Chaetopleuridae). Mol. Phylogenet. Evol. 120, 233-239. ( 10.1016/j.ympev.2017.12.013) [DOI] [PubMed] [Google Scholar]
- 48.Cantatore P, Gadaleta MN, Roberti M, Saccone C, Wilson AC. 1987. Duplication and remoulding of tRNA genes during the evolutionary rearrangement of mitochondrial genomes. Nature 329, 853-855. ( 10.1038/329853a0) [DOI] [PubMed] [Google Scholar]
- 49.Higgs PG, Jameson D, Jow H, Rattray M. 2003. The evolution of tRNA-Leu genes in animal mitochondrial genomes. J. Mol. Evol. 57, 435-445. ( 10.1007/s00239-003-2494-6) [DOI] [PubMed] [Google Scholar]
- 50.Rawlings TA, MacInnis MJ, Bieler R, Boore JL, Collins TM. 2010. Sessile snails, dynamic genomes: gene rearrangements within the mitochondrial genome of a family of caenogastropod molluscs. BMC Genomics 11, 440. ( 10.1186/1471-2164-11-440) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu X, Wu X, Yu Z. 2010. The mitogenome of Paphia euglypta (Bivalvia: Veneridae) and comparative mitogenomic analyses of three venerids. Genome 53, 1041-1052. ( 10.1139/G10-096) [DOI] [PubMed] [Google Scholar]
- 52.Nolan JR, Bergthorsson U, Adema CM. 2014. Physella acuta: atypical mitochondrial gene order among panpulmonates (Gastropoda). J. Molluscan Stud. 80, 388-399. ( 10.1093/mollus/eyu025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xie G-L, Köhler F, Huang X-C, Wu R-W, Zhou C-H, Ouyang S, Wu X-P. 2019. A novel gene arrangement among the Stylommatophora by the complete mitochondrial genome of the terrestrial slug Meghimatium bilineatum (Gastropoda, Arionoidea). Mol. Phylogenet. Evol. 135, 177-184. ( 10.1016/j.ympev.2019.03.002) [DOI] [PubMed] [Google Scholar]
- 54.Bandyopadhyay PK, Stevenson BJ, Cady MT, Olivera BM, Wolstenholme DR. 2006. Complete mitochondrial DNA sequence of a Conoidean gastropod, Lophiotoma (Xenuroturris) cerithiformis: gene order and gastropod phylogeny. Toxicon 48, 29-43. ( 10.1016/j.toxicon.2006.04.013) [DOI] [PubMed] [Google Scholar]
- 55.Rawlings TA, Collins TM, Bieler R. 2001. A major mitochondrial gene rearrangement among closely related species. Mol. Biol. Evol. 18, 1604-1609. ( 10.1093/oxfordjournals.molbev.a003949) [DOI] [PubMed] [Google Scholar]
- 56.Grande C, Templado J, Zardoya R. 2008. Evolution of gastropod mitochondrial genome arrangements. BMC Evol. Biol. 8, 61. ( 10.1186/1471-2148-8-61) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zheng R, Li J, Niu D. 2010. The complete DNA sequence of the mitochondrial genome of Sinonovacula constricta (Bivalvia: Solecurtidae). Acta Oceanol. Sin. 29, 88-92. ( 10.1007/s13131-010-0026-y) [DOI] [Google Scholar]
- 58.Lopes-Lima M, et al. 2017. The first Margaritiferidae male (M-type) mitogenome: mitochondrial gene order as a potential character for determining higher-order phylogeny within Unionida (Bivalvia). J. Molluscan Stud. 83, 249-252. ( 10.1093/mollus/eyx009) [DOI] [Google Scholar]
- 59.Irisarri I, Eernisse DJ, Zardoya R. 2014. Molecular phylogeny of Acanthochitonina (Mollusca: Polyplacophora: Chitonida): three new mitochondrial genomes, rearranged gene orders and systematics. J. Nat. Hist. 48, 2825-2853. ( 10.1080/00222933.2014.963721) [DOI] [Google Scholar]
- 60.Yokobori S-I, Fukuda N, Nakamura M, Aoyama T, Oshima T. 2004. Long-term conservation of six duplicated structural genes in cephalopod mitochondrial genomes. Mol. Biol. Evol. 21, 2034-2046. ( 10.1093/molbev/msh227) [DOI] [PubMed] [Google Scholar]
- 61.Uribe JE, Zardoya R. 2017. Revisiting the phylogeny of Cephalopoda using complete mitochondrial genomes. J. Molluscan Stud. 83, 133-144. ( 10.1093/mollus/eyw052) [DOI] [Google Scholar]
- 62.Luo Y-J, Satoh N, Endo K. 2015. Mitochondrial gene order variation in the brachiopod Lingula anatina and its implications for mitochondrial evolution in lophotrochozoans. Mar. Genomics 24, 31-40. ( 10.1016/j.margen.2015.08.005) [DOI] [PubMed] [Google Scholar]
- 63.Lavrov DV, Pett W. 2016. Animal mitochondrial DNA as we do not know it: mt-genome organization and evolution in nonbilaterian lineages. Genome Biol. Evol. 8, 2896-2913. ( 10.1093/gbe/evw195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kawashima Y, Nishihara H, Akasaki T, Nikaido M, Tsuchiya K, Segawa S, Okada N. 2013. The complete mitochondrial genomes of deep-sea squid (Bathyteuthis abyssicola), bob-tail squid (Semirossia patagonica) and four giant cuttlefish (Sepia apama, S. latimanus, S. lycidas and S. pharaonis), and their application to the phylogenetic analysis of Decapodiformes. Mol. Phylogenet. Evol. 69, 980-993. ( 10.1016/j.ympev.2013.06.007) [DOI] [PubMed] [Google Scholar]
- 65.Williams ST, Foster PG, Hughes C, Harper EM, Taylor JD, Littlewood DTJ, Dyal P, Hopkins KP, Briscoe AG. 2017. Curious bivalves: systematic utility and unusual properties of anomalodesmatan mitochondrial genomes. Mol. Phylogenet. Evol. 110, 60-72. ( 10.1016/j.ympev.2017.03.004) [DOI] [PubMed] [Google Scholar]
- 66.Wu X, Xu X, Yu Z, Kong X. 2009. Comparative mitogenomic analyses of three scallops (Bivalvia: Pectinidae) reveal high level variation of genomic organization and a diversity of transfer RNA gene sets. BMC Res. Notes 2, 69. ( 10.1186/1756-0500-2-69) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adams KL, Palmer JD. 2003. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol. Phylogenet. Evol. 29, 380-395. ( 10.1016/S1055-7903(03)00194-5) [DOI] [PubMed] [Google Scholar]
- 68.Allen JF. 2003. The function of genomes in bioenergetic organelle. Phil. Trans. R. Soc. Lond. B Biol. Sci. 358, 19-38. ( 10.1098/rstb.2002.1191) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Timmis JN, Ayliffe MA, Huang CY, Martin W. 2004. Endosymbiotic gene transfer: organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123-135. ( 10.1038/nrg1271) [DOI] [PubMed] [Google Scholar]
- 70.Breton S, Ghiselli F, Passamonti M, Milani L, Stewart DT, Hoeh WR. 2011. Evidence for a fourteenth mtDNA-encoded protein in the female-transmitted mtDNA of marine mussels (Bivalvia: Mytilidae). PLoS ONE 6, e19365. ( 10.1371/journal.pone.0019365) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Milani L, Ghiselli F, Maurizii MG, Nuzhdin SV, Passamonti M. 2014. Paternally transmitted mitochondria express a new gene of potential viral origin. Genome Biol. Evol. 6, 391-405. ( 10.1093/gbe/evu021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wu X, Li X, Li L, Yu Z. 2012. A unique tRNA gene family and a novel, highly expressed ORF in the mitochondrial genome of the silver-lip pearl oyster, Pinctada maxima (Bivalvia: Pteriidae). Gene 510, 22-31. ( 10.1016/j.gene.2012.08.037) [DOI] [PubMed] [Google Scholar]
- 73.Sun S, Kong L, Yu H, Li Q. 2015. The complete mitochondrial DNA of Tegillarca granosa and comparative mitogenomic analyses of three Arcidae species. Gene 557, 61-70. ( 10.1016/j.gene.2014.12.011) [DOI] [PubMed] [Google Scholar]
- 74.Simon M, Faye G. 1984. Organization and processing of the mitochondrial oxi3/oli2 multigenic transcript in yeast. Mol. Gen. Genet. 196, 266-274. ( 10.1007/BF00328059) [DOI] [PubMed] [Google Scholar]
- 75.Zeng X, Hourset A, Tzagoloff A. 2007. The Saccharomyces cerevisiae ATP22 gene codes for the mitochondrial ATPase subunit 6-specific translation factor. Genetics 175, 55-63. ( 10.1534/genetics.106.065821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rak M, Tzagoloff A. 2009. F1-dependent translation of mitochondrially encoded Atp6p and Atp8p subunits of yeast ATP synthase. Proc. Natl Acad. Sci. USA 106, 18 509-18 514. ( 10.1073/pnas.0910351106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barros MH, Tzagoloff A. 2017. Aep3p-dependent translation of yeast mitochondrial ATP8. Mol. Biol. Cell 28, 1426-1434. ( 10.1091/mbc.e16-11-0775) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.He C-B, Wang J, Gao X-G, Song W-T, Li H-J, Li Y-F, Liu W-D, Su H. 2011. The complete mitochondrial genome of the hard clam Meretrix meretrix. Mol. Biol. Rep. 38, 3401-3409. ( 10.1007/s11033-010-0449-8) [DOI] [PubMed] [Google Scholar]
- 79.Bettinazzi S, Plazzi F, Passamonti M. 2016. The complete female- and male-transmitted mitochondrial genome of Meretrix lamarckii. PLoS ONE 11, e0153631. ( 10.1371/journal.pone.0153631) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Milbury CA, Gaffney PM. 2005. Complete mitochondrial DNA sequence of the eastern oyster Crassostrea virginica. Mar. Biotechnol. 7, 697-712. ( 10.1007/s10126-005-0004-0) [DOI] [PubMed] [Google Scholar]
- 81.Milbury CA, Lee JC, Cannone JJ, Gaffney PM, Gutell RR. 2010. Fragmentation of the large subunit ribosomal RNA gene in oyster mitochondrial genomes. BMC Genomics 11, 485. ( 10.1186/1471-2164-11-485) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Curole JP, Kocher TD. 2002. Ancient sex-specific extension of the cytochrome c oxidase II gene in bivalves and the fidelity of doubly-uniparental inheritance. Mol. Biol. Evol. 19, 1323-1328. ( 10.1093/oxfordjournals.molbev.a004193) [DOI] [PubMed] [Google Scholar]
- 83.Chakrabarti R, et al. 2007. Reproductive function for a C-terminus extended, male-transmitted cytochrome c oxidase subunit II protein expressed in both spermatozoa and eggs. FEBS Lett. 581, 5213-5219. ( 10.1016/j.febslet.2007.10.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sevigny JL, et al. 2015. The mitochondrial genomes of the nudibranch mollusks, Melibe leonina and Tritonia diomedea, and their impact on gastropod phylogeny. PLoS ONE 10, e0127519. ( 10.1371/journal.pone.0127519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun S, Kong L, Yu H, Li Q. 2015. The complete mitochondrial genome of Scapharca kagoshimensis (Bivalvia: Arcidae). Mitochondrial DNA 26, 957-958. ( 10.3109/19401736.2013.865174) [DOI] [PubMed] [Google Scholar]
- 86.Rigaa A, Monnerot M, Sellos D. 1995. Molecular cloning and complete nucleotide sequence of the repeated unit and flanking gene of the scallop Pecten maximus mitochondrial DNA: putative replication origin features. J. Mol. Evol. 41, 189-195. ( 10.1007/BF00170672) [DOI] [PubMed] [Google Scholar]
- 87.Sasuga J, Yokobori S, Kaifu M, Ueda T, Nishikawa K, Watanabe K. 1999. Gene contents and organization of a mitochondrial DNA segment of the squid Loligo bleekeri. J. Mol. Evol. 48, 692-702. ( 10.1007/PL00006513) [DOI] [PubMed] [Google Scholar]
- 88.Tomita K, Yokobori S-I, Oshima T, Ueda T, Watanabe K. 2002. The cephalopod Loligo bleekeri mitochondrial genome: multiplied noncoding regions and transposition of tRNA genes. J. Mol. Evol. 54, 486-500. ( 10.1007/s00239-001-0039-4) [DOI] [PubMed] [Google Scholar]
- 89.McComish BJ, Hills SFK, Biggs PJ, Penny D. 2010. Index-free de novo assembly and deconvolution of mixed mitochondrial genomes. Genome Biol. Evol. 2, 410-424. ( 10.1093/gbe/evq029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Simison WB, Lindberg DR, Boore JL. 2006. Rolling circle amplification of metazoan mitochondrial genomes. Mol. Phylogenet. Evol. 39, 562-567. ( 10.1016/j.ympev.2005.11.006) [DOI] [PubMed] [Google Scholar]
- 91.Zbawicka M, Wenne R, Burzyński A. 2014. Mitogenomics of recombinant mitochondrial genomes of Baltic Sea Mytilus mussels. Mol. Genet. Genomics 289, 1275-1287. ( 10.1007/s00438-014-0888-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Breton S, Burger G, Stewart DT, Blier PU. 2006. Comparative analysis of gender-associated complete mitochondrial genomes in marine mussels (Mytilus spp.). Genetics 172, 1107-1119. ( 10.1534/genetics.105.047159) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cao L, Ort BS, Mizi A, Pogson G, Kenchington E, Zouros E, Rodakis GC. 2009. The control region of maternally and paternally inherited mitochondrial genomes of three species of the sea mussel genus Mytilus. Genetics 181, 1045-1056. ( 10.1534/genetics.108.093229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brauer A, et al. 2012. The mitochondrial genome of the venomous cone snail Conus consors. PLoS ONE 7, e51528. ( 10.1371/journal.pone.0051528) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gao B, Peng C, Chen Q, Zhang J, Shi Q. 2018. Mitochondrial genome sequencing of a vermivorous cone snail Conus quercinus supports the correlative analysis between phylogenetic relationships and dietary types of Conus species. PLoS ONE 13, e0193053. ( 10.1371/journal.pone.0193053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang X-CC, Rong J, Liu Y, Zhang M-HH, Wan Y, Ouyang S, Zhou C-HH, Wu X-PP. 2013. The complete maternally and paternally inherited mitochondrial genomes of the endangered freshwater mussel Solenaia carinatus (Bivalvia: Unionidae) and implications for Unionidae taxonomy. PLoS ONE 8, e84352. ( 10.1371/journal.pone.0084352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhu HC, Shen HD, Zheng P, Zhang Y. 2012. Complete mitochondrial genome of the jackknife clam Solen grandis (Veneroida, Solenidae). Mitochondrial DNA 23, 115-117. ( 10.3109/19401736.2011.653803) [DOI] [PubMed] [Google Scholar]
- 98.Bernt M, Braband A, Schierwater B, Stadler PF. 2013. Genetic aspects of mitochondrial genome evolution. Mol. Phylogenet. Evol. 69, 328-338. ( 10.1016/j.ympev.2012.10.020) [DOI] [PubMed] [Google Scholar]
- 99.Jiang L, Ge C, Liu W, Wu C, Zhu A. 2016. Complete mitochondrial genome of the Loligo duvaucelii. Mitochondrial DNA A 27, 2723-2724. ( 10.3109/19401736.2015.1046164) [DOI] [PubMed] [Google Scholar]
- 100.Zouros E. 2000. The exceptional mitochondrial DNA system of the mussel family Mytilidae. Genes Genet. Syst. 75, 313-318. ( 10.1266/ggs.75.313) [DOI] [PubMed] [Google Scholar]
- 101.Burzynski A, Zbawicka M, Skibinski DO, Wenne R. 2003. Evidence for recombination of mtDNA in the marine mussel Mytilus trossulus from the Baltic. Mol. Biol. Evol. 20, 388-392. ( 10.1093/molbev/msg058) [DOI] [PubMed] [Google Scholar]
- 102.Stewart DT, Breton S, Blier PU, Hoeh WR. 2009. Masculinization events and doubly uniparental inheritance of mitochondrial DNA: a model for understanding the evolutionary dynamics of gender-associated mtDNA in mussels. In Evolutionary biology: concept, modeling, and application (ed. Pontarotti P), pp. 163-173. Berlin, Germany: Springer. [Google Scholar]
- 103.Rawlings TA, Collins TM, Bieler R. 2003. Changing identities: tRNA duplication and remolding within animal mitochondrial genomes. Proc. Natl Acad. Sci. USA 100, 15 700-15 705. ( 10.1073/pnas.2535036100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Brown GG, Gadaleta G, Pepe G, Saccone C, Sbisà E. 1986. Structural conservation and variation in the D-loop-containing region of vertebrate mitochondrial DNA. J. Mol. Biol. 192, 503-511. ( 10.1016/0022-2836(86)90272-X) [DOI] [PubMed] [Google Scholar]
- 105.Boore JL. 2000. The duplication/random loss model for gene rearrangement exemplified by mitochondrial genomes of deuterostome animals. In Comparative genomics: empirical and analytical approaches to gene order dynamics, map alignment and the evolution of gene families (eds Sankoff D, Nadeau JH), pp. 133-147. Dordrecht, The Netherlands: Springer Netherlands. [Google Scholar]
- 106.Ludwig A, May B, Debus L, Jenneckens I. 2000. Heteroplasmy in the mtDNA control region of sturgeon (Acipenser, Huso and Scaphirhynchus). Genetics 156, 1933-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Levinson G, Gutman GA. 1987. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol. Biol. Evol. 4, 203-221. [DOI] [PubMed] [Google Scholar]
- 108.Stanton DJ, Daehler LL, Moritz CC, Brown WM. 1994. Sequences with the potential to form stem-and-loop structures are associated with coding-region duplications in animal mitochondrial DNA. Genetics 137, 233-241. ( 10.1093/genetics/137.1.233) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Macey JR, Larson A, Ananjeva NB, Fang Z, Papenfuss TJ. 1997. Two novel gene orders and the role of light-strand replication in rearrangement of the vertebrate mitochondrial genome. Mol. Biol. Evol. 14, 91-104. ( 10.1093/oxfordjournals.molbev.a025706) [DOI] [PubMed] [Google Scholar]
- 110.Raimond R, Marcadé I, Bouchon D, Rigaud T, Bossy JP, Souty-Grosset C. 1999. Organization of the large mitochondrial genome in the isopod Armadillidium vulgare. Genetics 151, 203-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mita S, Rizzuto R, Moraes CT, Shanske S, Arnaudo E, Fabrizi GM, Koga Y, DiMauro S, Schon EA. 1990. Recombination via flanking direct repeats is a major cause of large-scale deletions of human mitochondrial DNA. Nucleic Acids Res. 18, 561-567. ( 10.1093/nar/18.3.561) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kajander OA, Rovio AT, Majamaa K, Poulton J, Spelbrink JN, Holt IJ, Karhunen PJ, Jacobs HT. 2000. Human mtDNA sublimons resemble rearranged mitochondrial genomes found in pathological states. Hum. Mol. Genet. 9, 2821-2835. ( 10.1093/hmg/9.19.2821) [DOI] [PubMed] [Google Scholar]
- 113.Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27, 1767-1780. ( 10.1093/nar/27.8.1767) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sharbrough J, Cruise JL, Beetch M, Enright NM, Neiman M. 2017. Genetic variation for mitochondrial function in the New Zealand freshwater snail Potamopyrgus antipodarum. J. Hered. 108, 759-768. ( 10.1093/jhered/esx041) [DOI] [PubMed] [Google Scholar]
- 115.Ji H, et al. 2019. Using high-resolution annotation of insect mitochondrial DNA to decipher tandem repeats in the control region. RNA Biol. 16, 830-837. ( 10.1080/15476286.2019.1591035) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Calcino A, Baranyi C, Wanninger A. 2020. The expanded plant-like mitogenome of the invasive quagga mussel, Dreissena rostriformis. bioRxiv, 310516. ( 10.1101/2020.09.23.310516) [DOI]
- 117.Zampini É, Lepage É, Tremblay-Belzile S, Truche S, Brisson N. 2015. Organelle DNA rearrangement mapping reveals U-turn-like inversions as a major source of genomic instability in Arabidopsis and humans. Genome Res. 25, 645-654. ( 10.1101/gr.188573.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lobachev KS, Shor BM, Tran HT, Taylor W, Keen JD, Resnick MA, Gordenin DA. 1998. Factors affecting inverted repeat stimulation of recombination and deletion in Saccharomyces cerevisiae. Genetics 148, 1507-1524. ( 10.1093/genetics/148.4.1507) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cheng R, Zheng X, Ma Y, Li Q. 2013. The complete mitochondrial genomes of two octopods Cistopus chinensis and Cistopus taiwanicus: revealing the phylogenetic position of the genus Cistopus within the order Octopoda. PLoS ONE 8, e84216. ( 10.1371/journal.pone.0084216) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Camus MF, Clancy DJ, Dowling DK. 2012. Mitochondria, maternal inheritance, and male aging. Curr. Biol. 22, 1717-1721. ( 10.1016/j.cub.2012.07.018) [DOI] [PubMed] [Google Scholar]
- 121.Neiman M, Taylor DR. 2009. The causes of mutation accumulation in mitochondrial genomes. Proc. R. Soc. B 276, 1201-1209. ( 10.1098/rspb.2008.1758) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Boore JL, Brown WM. 1998. Big trees from little genomes: mitochondrial gene order as a phylogenetic tool. Curr. Opin. Genet. Dev. 8, 668-674. ( 10.1016/S0959-437X(98)80035-X) [DOI] [PubMed] [Google Scholar]
- 123.Allcock AL, Cooke IR, Strugnell JM. 2011. What can the mitochondrial genome reveal about higher-level phylogeny of the molluscan class Cephalopoda? Zool. J. Linn. Soc. 161, 573-586. ( 10.1111/j.1096-3642.2010.00656.x) [DOI] [Google Scholar]
- 124.D'Souza AR, Minczuk M. 2018. Mitochondrial transcription and translation: overview. Essays Biochem. 62, 309-320. ( 10.1042/EBC20170102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zardoya R, Pérez-Martos A, Bautista JM, Montoya J. 1995. Analysis of the transcription products of the rainbow trout (Oncorynchus mykiss) liver mitochondrial genome: detection of novel mitochondrial transcripts. Curr. Genet. 28, 67-70. ( 10.1007/BF00311883) [DOI] [PubMed] [Google Scholar]
- 126.Fearnley IM, Walker JE. 1986. Two overlapping genes in bovine mitochondrial DNA encode membrane components of ATP synthase. EMBO J. 5, 2003-2008. ( 10.1002/j.1460-2075.1986.tb04456.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Clary DO, Wolstenholme DR. 1985. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J. Mol. Evol. 22, 252-271. ( 10.1007/BF02099755) [DOI] [PubMed] [Google Scholar]
- 128.Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44, W54-W57. ( 10.1093/nar/gkw413) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol. Biol. 1962, 1-14. ( 10.1007/978-1-4939-9173-0_1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads—a baiting and iterative mapping approach. Nucleic Acids Res. 41, e129. ( 10.1093/nar/gkt371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, e18. ( 10.1093/nar/gkw1060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Al-Nakeeb K, Petersen TN, Sicheritz-Pontén T. 2017. Norgal: extraction and de novo assembly of mitochondrial DNA from whole-genome sequencing data. BMC Bioinf. 18, 510. ( 10.1186/s12859-017-1927-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47, e63. ( 10.1093/nar/gkz173) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wyman SK, Jansen RK, Boore JL. 2004. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20, 3252-3255. ( 10.1093/bioinformatics/bth352) [DOI] [PubMed] [Google Scholar]
- 135.Jung J, Kim JI, Jeong Y-S, Yi G. 2018. AGORA: organellar genome annotation from the amino acid and nucleotide references. Bioinformatics 34, 2661-2663. ( 10.1093/bioinformatics/bty196) [DOI] [PubMed] [Google Scholar]
- 136.Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 69, 313-319. ( 10.1016/j.ympev.2012.08.023) [DOI] [PubMed] [Google Scholar]
- 137.Fourdrilis S, de Frias Martins AM, Backeljau T. 2018. Relation between mitochondrial DNA hyperdiversity, mutation rate and mitochondrial genome evolution in Melarhaphe neritoides (Gastropoda: Littorinidae) and other Caenogastropoda. Sci. Rep. 8, 17964. ( 10.1038/s41598-018-36428-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Prada CF, Boore JL. 2019. Gene annotation errors are common in the mammalian mitochondrial genomes database. BMC Genomics 20, 73. ( 10.1186/s12864-019-5447-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Birky CW Jr. 2001. The inheritance of genes in mitochondria and chloroplasts: laws, mechanisms, and models. Annu. Rev. Genet. 35, 125-148. ( 10.1146/annurev.genet.35.102401.090231) [DOI] [PubMed] [Google Scholar]
- 140.Barr CM, Neiman M, Taylor DR. 2005. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 168, 39-50. ( 10.1111/j.1469-8137.2005.01492.x) [DOI] [PubMed] [Google Scholar]
- 141.Capt C, Bouvet K, Guerra D, Robicheau BM, Stewart DT, Pante E, Breton S. 2020. Unorthodox features in two venerid bivalves with doubly uniparental inheritance of mitochondria. Sci. Rep. 10, 1087. ( 10.1038/s41598-020-57975-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Breton S, Stewart DT. 2015. Atypical mitochondrial inheritance patterns in eukaryotes. Genome 58, 423-431. ( 10.1139/gen-2015-0090) [DOI] [PubMed] [Google Scholar]
- 143.Zouros E. 2013. Biparental inheritance through uniparental transmission: the doubly uniparental inheritance (DUI) of mitochondrial DNA. Evol. Biol. 40, 1-31. ( 10.1007/s11692-012-9195-2) [DOI] [Google Scholar]
- 144.Cao L, Kenchington E, Zouros E. 2004. Differential segregation patterns of sperm mitochondria in embryos of the blue mussel (Mytilus edulis). Genetics 166, 883-894. ( 10.1534/genetics.166.2.883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Cogswell AT, Kenchington ELR, Zouros E. 2006. Segregation of sperm mitochondria in two- and four-cell embryos of the blue mussel Mytilus edulis: implications for the mechanism of doubly uniparental inheritance of mitochondrial DNA. Genome 49, 799-807. ( 10.1139/g06-036) [DOI] [PubMed] [Google Scholar]
- 146.Sato K, Sato M. 2017. Multiple ways to prevent transmission of paternal mitochondrial DNA for maternal inheritance in animals. J. Biochem. 162, 247-253. ( 10.1093/jb/mvx052) [DOI] [PubMed] [Google Scholar]
- 147.Milani L, Ghiselli F, Passamonti M. 2012. Sex-linked mitochondrial behavior during early embryo development in Ruditapes philippinarum (Bivalvia Veneridae) a species with the doubly uniparental inheritance (DUI) of mitochondria. J. Exp. Zool. B 318, 182-189. ( 10.1002/jez.b.22004) [DOI] [PubMed] [Google Scholar]
- 148.Ghiselli F, Milani L, Passamonti M. 2011. Strict sex-specific mtDNA segregation in the germ line of the DUI species Venerupis philippinarum (Bivalvia: Veneridae). Mol. Biol. Evol. 28, 949-961. ( 10.1093/molbev/msq271) [DOI] [PubMed] [Google Scholar]
- 149.Breton S, Bouvet K, Auclair G, Ghazal S, Sietman BE, Johnson N, Bettinazzi S, Stewart DT, Guerra D. 2017. The extremely divergent maternally- and paternally-transmitted mitochondrial genomes are co-expressed in somatic tissues of two freshwater mussel species with doubly uniparental inheritance of mtDNA. PLoS ONE 12, e0183529. ( 10.1371/journal.pone.0183529) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Obata M, Kamiya C, Kawamura K, Komaru A. 2006. Sperm mitochondrial DNA transmission to both male and female offspring in the blue mussel Mytilus galloprovincialis. Dev. Growth Differ. 48, 253-261. ( 10.1111/j.1440-169X.2006.00863.x) [DOI] [PubMed] [Google Scholar]
- 151.Kyriakou E, Zouros E, Rodakis GC. 2010. The atypical presence of the paternal mitochondrial DNA in somatic tissues of male and female individuals of the blue mussel species Mytilus galloprovincialis. BMC Res. Notes 3, 222. ( 10.1186/1756-0500-3-222) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ghiselli F, et al. 2019. Natural heteroplasmy and mitochondrial inheritance in bivalve molluscs. Integr. Comp. Biol. 59, 1016-1032. ( 10.1093/icb/icz061) [DOI] [PubMed] [Google Scholar]
- 153.Milani L, Ghiselli F, Passamonti M. 2016. Mitochondrial selfish elements and the evolution of biological novelties. Curr. Zool. 62, 687-697. ( 10.1093/cz/zow044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kenchington EL, Hamilton L, Cogswell A, Zouros E. 2009. Paternal mtDNA and maleness are co-inherited but not causally linked in mytilid mussels. PLoS ONE 4, e6976. ( 10.1371/journal.pone.0006976) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Birky CW Jr. 1995. Uniparental inheritance of mitochondrial and chloroplast genes: mechanisms and evolution. Proc. Natl Acad. Sci. USA 92, 11 331-11 338. ( 10.1073/pnas.92.25.11331) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kenchington E, MacDonald B, Cao L, Tsagkarakis D, Zouros E. 2002. Genetics of mother-dependent sex ratio in blue mussels (Mytilus spp.) and implications for doubly uniparental inheritance of mitochondrial DNA. Genetics 161, 1579-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ghiselli F, Milani L, Chang PL, Hedgecock D, Davis JP, Nuzhdin SV, Passamonti M. 2012. De novo assembly of the Manila clam Ruditapes philippinarum transcriptome provides new insights into expression bias, mitochondrial doubly uniparental inheritance and sex determination. Mol. Biol. Evol. 29, 771-786. ( 10.1093/molbev/msr248) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Milani L, Ghiselli F, Nuzhdin SV, Passamonti M. 2013. Nuclear genes with sex bias in Ruditapes philippinarum (Bivalvia, Veneridae): mitochondrial inheritance and sex determination in DUI species. J. Exp. Zool. B 320, 442-454. ( 10.1002/jez.b.22520) [DOI] [PubMed] [Google Scholar]
- 159.Diz AP, Dudley E, Cogswell A, MacDonald BW, Kenchington ELR, Zouros E, Skibinski DOF. 2013. Proteomic analysis of eggs from Mytilus edulis females differing in mitochondrial DNA transmission mode. Mol. Cell. Proteomics 12, 3068-3080. ( 10.1074/mcp.M113.031401) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Punzi E, Milani L, Ghiselli F, Passamonti M. 2018. Lose it or keep it: (how bivalves can provide) insights into mitochondrial inheritance mechanisms. J. Exp. Zool. B 330, 41-51. ( 10.1002/jez.b.22788) [DOI] [PubMed] [Google Scholar]
- 161.Passamonti M, Ricci A, Milani L, Ghiselli F. 2011. Mitochondrial genomes and doubly uniparental inheritance: new insights from Musculista senhousia sex-linked mitochondrial DNAs (Bivalvia Mytilidae). BMC Genomics 12, 442. ( 10.1186/1471-2164-12-442) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Guerra D, Ghiselli F, Passamonti M. 2014. The largest unassigned regions of the male- and female-transmitted mitochondrial DNAs in Musculista senhousia (Bivalvia Mytilidae). Gene 536, 316-325. ( 10.1016/j.gene.2013.12.005) [DOI] [PubMed] [Google Scholar]
- 163.Ghiselli F, Milani L, Iannello M, Procopio E, Chang PL, Nuzhdin SV, Passamonti M. 2017. The complete mitochondrial genome of the grooved carpet shell, Ruditapes decussatus (Bivalvia, Veneridae). PeerJ 5, e3692. ( 10.7717/peerj.3692) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Robicheau BM, Breton S, Stewart DT. 2017. Sequence motifs associated with paternal transmission of mitochondrial DNA in the horse mussel, Modiolus modiolus (Bivalvia: Mytilidae). Gene 605, 32-42. ( 10.1016/j.gene.2016.12.025) [DOI] [PubMed] [Google Scholar]
- 165.Ghiselli F, Milani L, Guerra D, Chang PL, Breton S, Nuzhdin SV, Passamonti M. 2013. Structure, transcription, and variability of metazoan mitochondrial genome: perspectives from an unusual mitochondrial inheritance system. Genome Biol. Evol. 5, 1535-1554. ( 10.1093/gbe/evt112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kyriakou E, Kravariti L, Vasilopoulos T, Zouros E, Rodakis GC. 2015. A protein binding site in the M mitochondrial genome of Mytilus galloprovincialis may be responsible for its paternal transmission. Gene 562, 83-94. ( 10.1016/j.gene.2015.02.047) [DOI] [PubMed] [Google Scholar]
- 167.Breton S, Beaupré HD, Stewart DT, Piontkivska H, Karmakar M, Bogan AE, Blier PU, Hoeh WR. 2009. Comparative mitochondrial genomics of freshwater mussels (Bivalvia: Unionoida) with doubly uniparental inheritance of mtDNA: gender-specific open reading frames and putative origins of replication. Genetics 183, 1575-1589. ( 10.1534/genetics.109.110700) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Milani L, Ghiselli F, Guerra D, Breton S, Passamonti M. 2013. A comparative analysis of mitochondrial ORFans: new clues on their origin and role in species with doubly uniparental inheritance of mitochondria. Genome Biol. Evol. 5, 1408-1434. ( 10.1093/gbe/evt101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mitchell A, Guerra D, Stewart D, Breton S. 2016. In silico analyses of mitochondrial ORFans in freshwater mussels (Bivalvia: Unionoida) provide a framework for future studies of their origin and function. BMC Genomics 17, 597. ( 10.1186/s12864-016-2986-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Guerra D, et al. 2019. Variability of mitochondrial ORFans hints at possible differences in the system of doubly uniparental inheritance of mitochondria among families of freshwater mussels (Bivalvia: Unionida). BMC Evol. Biol. 19, 229. ( 10.1186/s12862-019-1554-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Breton S, Stewart DT, Shepardson S, Trdan RJ, Bogan AE, Chapman EG, Ruminas AJ, Piontkivska H, Hoeh WR. 2011. Novel protein genes in animal mtDNA: a new sex determination system in freshwater mussels (Bivalvia: Unionoida)? Mol. Biol. Evol. 28, 1645-1659. ( 10.1093/molbev/msq345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pozzi A, Plazzi F, Milani L, Ghiselli F, Passamonti M. 2017. SmithRNAs: could mitochondria ‘bend’ nuclear regulation? Mol. Biol. Evol. 34, 1960-1973. ( 10.1093/molbev/msx140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Passamonti M, Calderone M, Delpero M, Plazzi F. 2020. Clues of in vivo nuclear gene regulation by mitochondrial short non-coding RNAs. Sci. Rep. 10, 8219. ( 10.1038/s41598-020-65084-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pozzi A, Dowling DK. 2019. The genomic origins of small mitochondrial RNAs: are they transcribed by the mitochondrial DNA or by mitochondrial pseudogenes within the nucleus (NUMTs)? Genome Biol. Evol. 11, 1883-1896. ( 10.1093/gbe/evz132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Breton S, Beaupré HD, Stewart DT, Hoeh WR, Blier PU. 2007. The unusual system of doubly uniparental inheritance of mtDNA: isn't one enough? Trends Genet. 23, 465-474. ( 10.1016/j.tig.2007.05.011) [DOI] [PubMed] [Google Scholar]
- 176.Passamonti M, Ghiselli F. 2009. Doubly uniparental inheritance: two mitochondrial genomes, one precious model for organelle DNA inheritance and evolution. DNA Cell Biol. 28, 79-89. ( 10.1089/dna.2008.0807) [DOI] [PubMed] [Google Scholar]
- 177.Yusa Y, Breton S, Hoeh WR. 2013. Population genetics of sex determination in Mytilus mussels: reanalyses and a model. J. Hered. 104, 380-385. ( 10.1093/jhered/est014) [DOI] [PubMed] [Google Scholar]
- 178.Capt C, Renaut S, Ghiselli F, Milani L, Johnson NA, Sietman BE, Stewart DT, Breton S. 2018. Deciphering the link between doubly uniparental inheritance of mtDNA and sex determination in bivalves: clues from comparative transcriptomics. Genome Biol. Evol. 10, 577-590. ( 10.1093/gbe/evy019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Stewart DT, Kenchington ER, Singh RK, Zouros E. 1996. Degree of selective constraint as an explanation of the different rates of evolution of gender-specific mitochondrial DNA lineages in the mussel Mytilus. Genetics 143, 1349-1357. ( 10.1093/genetics/143.3.1349) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Milani L, Ghiselli F, Iannello M, Passamonti M. 2014. Evidence for somatic transcription of male-transmitted mitochondrial genome in the DUI species Ruditapes philippinarum (Bivalvia: Veneridae). Curr. Genet. 60, 163-173. ( 10.1007/s00294-014-0420-7) [DOI] [PubMed] [Google Scholar]
- 181.Dégletagne C, Abele D, Held C. 2016. A distinct mitochondrial genome with DUI-like inheritance in the ocean quahog Arctica islandica. Mol. Biol. Evol. 33, 375-383. ( 10.1093/molbev/msv224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bettinazzi S, Rodríguez E, Milani L, Blier PU, Breton S. 2019. Metabolic remodelling associated with mtDNA: insights into the adaptive value of doubly uniparental inheritance of mitochondria. Proc. R. Soc. B 286, 20182708. ( 10.1098/rspb.2018.2708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bettinazzi S, Nadarajah S, Dalpé A, Milani L, Blier PU, Breton S. 2020. Linking paternally inherited mtDNA variants and sperm performance. Phil. Trans. R. Soc. B 375, 20190177. ( 10.1098/rstb.2019.0177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gusman A, Azuelos C, Breton S. 2017. No evidence of sex-linked heteroplasmy or doubly-uniparental inheritance of mtDNA in five gastropod species. J. Mollusc. Stud. 83, 119-122. ( 10.1093/mollus/eyw034) [DOI] [Google Scholar]
- 185.Gissi C, Iannelli F, Pesole G. 2008. Evolution of the mitochondrial genome of Metazoa as exemplified by comparison of congeneric species. Heredity 101, 301-320. ( 10.1038/hdy.2008.62) [DOI] [PubMed] [Google Scholar]
- 186.Stöger I, Schrödl M. 2013. Mitogenomics does not resolve deep molluscan relationships (yet?). Mol. Phylogenet. Evol. 69, 376-392. ( 10.1016/j.ympev.2012.11.017) [DOI] [PubMed] [Google Scholar]
- 187.Kern EMA, Kim T, Park J-K. 2020. The mitochondrial genome in nematode phylogenetics. Front. Ecol. Evol. 8, 250. ( 10.3389/fevo.2020.00250) [DOI] [Google Scholar]
- 188.Danovaro R, Dell'Anno A, Pusceddu A, Gambi C, Heiner I, Kristensen RM. 2010. The first metazoa living in permanently anoxic conditions. BMC Biol. 8, 30. ( 10.1186/1741-7007-8-30) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Elson JL, Lightowlers RN. 2006. Mitochondrial DNA clonality in the dock: can surveillance swing the case? Trends Genet. 22, 603-607. ( 10.1016/j.tig.2006.09.004) [DOI] [PubMed] [Google Scholar]
- 190.Masta SE, Longhorn SJ, Boore JL. 2009. Arachnid relationships based on mitochondrial genomes: asymmetric nucleotide and amino acid bias affects phylogenetic analyses. Mol. Phylogenet. Evol. 50, 117-128. ( 10.1016/j.ympev.2008.10.010) [DOI] [PubMed] [Google Scholar]
- 191.Froufe E, Sobral C, Teixeira A, Sousa R, Varandas S, Aldridge D C, Lopes-Lima M. 2014. Genetic diversity of the pan-European freshwater mussel Anodonta anatina (Bivalvia: Unionoida) based on CO1: new phylogenetic insights and implications for conservation. Aquat. Conserv. 24, 561-574. ( 10.1002/aqc.2456) [DOI] [Google Scholar]
- 192.Bleidorn C, Podsiadlowski L, Bartolomaeus T. 2006. The complete mitochondrial genome of the orbiniid polychaete Orbinia latreillii (Annelida, Orbiniidae): a novel gene order for Annelida and implications for annelid phylogeny. Gene 370, 96-103. ( 10.1016/j.gene.2005.11.018) [DOI] [PubMed] [Google Scholar]
- 193.Park J-K, Kim K-H, Kang S, Kim W, Eom KS, Littlewood DTJ. 2007. A common origin of complex life cycles in parasitic flatworms: evidence from the complete mitochondrial genome of Microcotyle sebastis (Monogenea: Platyhelminthes). BMC Evol. Biol. 7, 11. ( 10.1186/1471-2148-7-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Min G-S, Park J-K. 2009. Eurotatorian paraphyly: revisiting phylogenetic relationships based on the complete mitochondrial genome sequence of Rotaria rotatoria (Bdelloidea: Rotifera: Syndermata). BMC Genomics 10, 533. ( 10.1186/1471-2164-10-533) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Yokobori S-I, Iseto T, Asakawa S, Sasaki T, Shimizu N, Yamagishi A, Oshima T, Hirose E. 2008. Complete nucleotide sequences of mitochondrial genomes of two solitary entoprocts, Loxocorone allax and Loxosomella aloxiata: implications for lophotrochozoan phylogeny. Mol. Phylogenet. Evol. 47, 612-628. ( 10.1016/j.ympev.2008.02.013) [DOI] [PubMed] [Google Scholar]
- 196.Schrödl M, Stöger I. 2014. A review on deep molluscan phylogeny: old markers, integrative approaches, persistent problems. J. Nat. Hist. 48, 2773-2804. ( 10.1080/00222933.2014.963184) [DOI] [Google Scholar]
- 197.Kocot KM, et al. 2011. Phylogenomics reveals deep molluscan relationships. Nature 477, 452-456. ( 10.1038/nature10382) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Smith SA, Wilson NG, Goetz FE, Feehery C, Andrade SCS, Rouse GW, Giribet G, Dunn CW. 2011. Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature 480, 364-367. ( 10.1038/nature10526) [DOI] [PubMed] [Google Scholar]
- 199.Vinther J, Sperling EA, Briggs DEG, Peterson KJ. 2012. A molecular palaeobiological hypothesis for the origin of aplacophoran molluscs and their derivation from chiton-like ancestors. Proc. R. Soc. B 279, 1259-1268. ( 10.1098/rspb.2011.1773) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Wanninger A, Wollesen T. 2018. The evolution of molluscs. Biol. Rev. Camb. Phil. Soc. 94, 102-115. ( 10.1111/brv.12439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Kocot KM, Poustka AJ, Stöger I, Halanych KM, Schrödl M. 2020. New data from Monoplacophora and a carefully-curated dataset resolve molluscan relationships. Sci. Rep. 10, 1-8. ( 10.1038/s41598-019-56728-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Gomes-dos-Santos A, Lopes-Lima M, Castro LFC, Froufe E. 2019. Molluscan genomics: the road so far and the way forward. Hydrobiologia 847, 1705-1726. ( 10.1007/s10750-019-04111-1) [DOI] [Google Scholar]
- 203.Uribe JE, Irisarri I, Templado J, Zardoya R. 2019. New patellogastropod mitogenomes help counteracting long-branch attraction in the deep phylogeny of gastropod mollusks. Mol. Phylogenet. Evol. 133, 12-23. ( 10.1016/j.ympev.2018.12.019) [DOI] [PubMed] [Google Scholar]
- 204.Irisarri I, Uribe JE, Eernisse DJ, Zardoya R. 2020. A mitogenomic phylogeny of chitons (Mollusca: Polyplacophora). BMC Evol. Biol. 20, 22. ( 10.1186/s12862-019-1573-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kocot KM, Wollesen T, Varney RM, Schwartz ML, Steiner G, Wanninger A. 2019. Complete mitochondrial genomes of two scaphopod molluscs. Mitochondrial DNA B 4, 3161-3162. ( 10.1080/23802359.2019.1666689) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Combosch DJ, et al. 2017. A family-level tree of life for bivalves based on a Sanger-sequencing approach. Mol. Phylogenet. Evol. 107, 191-208. ( 10.1016/j.ympev.2016.11.003) [DOI] [PubMed] [Google Scholar]
- 207.Zapata F, Wilson NG, Howison M, Andrade SCS, Jörger KM, Schrödl M, Goetz FE, Giribet G, Dunn CW. 2014. Phylogenomic analyses of deep gastropod relationships reject Orthogastropoda. Proc. R. Soc. B 281, 20141739. ( 10.1098/rspb.2014.1739) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.González VL, Andrade SCS, Bieler R, Collins TM, Dunn CW, Mikkelsen PM, Taylor JD, Giribet G. 2015. A phylogenetic backbone for Bivalvia: an RNA-seq approach. Proc. R. Soc. B 282, 20142332. ( 10.1098/rspb.2014.2332) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Cunha TJ, Giribet G. 2019. A congruent topology for deep gastropod relationships. Proc. R. Soc. B 286, 20182776. ( 10.1098/rspb.2018.2776) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Cong H, Lei Y, Kong L. 2020. The mitochondrial genome of the toothed top shell snail Monodonta labio (Gastropoda: Trochidae): the first complete sequence in the subfamily Monodontinae. Mitochondrial DNA B 5, 621-622. ( 10.1080/23802359.2019.1711221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Froufe E, et al. 2020. Mesozoic mitogenome rearrangements and freshwater mussel (Bivalvia: Unionoidea) macroevolution. Heredity 124, 182-196. ( 10.1038/s41437-019-0242-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Ye YY, Wu CW, Li JJ. 2015. Genetic population structure of Macridiscus multifarius (Mollusca: Bivalvia) on the basis of mitochondrial markers: strong population structure in a species with a short planktonic larval stage. PLoS ONE 10, e0146260. ( 10.1371/journal.pone.0146260) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Froufe E, et al. 2016. Phylogeny, phylogeography, and evolution in the Mediterranean region: news from a freshwater mussel (Potomida, Unionida). Mol. Phylogenet. Evol. 100, 322-332. ( 10.1016/j.ympev.2016.04.030) [DOI] [PubMed] [Google Scholar]
- 214.Fernández-Pérez J, Froufe E, Nantón A, Gaspar MB, Méndez J. 2017. Genetic diversity and population genetic analysis of Donax vittatus (Mollusca: Bivalvia) and phylogeny of the genus with mitochondrial and nuclear markers. Estuar. Coast. Shelf Sci. 197, 126-135. ( 10.1016/j.ecss.2017.08.032) [DOI] [Google Scholar]
- 215.Fonseca MM, Lopes-Lima M, Eackles MS, King TL, Froufe E. 2016. The female and male mitochondrial genomes of Unio delphinus and the phylogeny of freshwater mussels (Bivalvia: Unionida). Mitochondrial DNA B 1, 954-957. ( 10.1080/23802359.2016.1241677) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Abalde S, Tenorio MJ, Afonso CML, Uribe JE, Echeverry AM, Zardoya R. 2017. Phylogenetic relationships of cone snails endemic to Cabo Verde based on mitochondrial genomes. BMC Evol. Biol. 17, 231. ( 10.1186/s12862-017-1069-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Tang Y, Zheng X, Ma Y, Cheng R, Li Q. 2018. The complete mitochondrial genome of Amphioctopus marginatus (Cephalopoda: Octopodidae) and the exploration for the optimal DNA barcoding in Octopodidae. Conserv. Genet. Resour. 10, 115-118. ( 10.1007/s12686-017-0777-2) [DOI] [Google Scholar]
- 218.Shen X, Meng XP, Chu KH, Zhao NN, Tian M, Liang M, Hao J. 2014. Comparative mitogenomic analysis reveals cryptic species: a case study in Mactridae (Mollusca: Bivalvia). Comp. Biochem. Physiol. D 12, 1-9. ( 10.1016/j.cbd.2014.08.002) [DOI] [PubMed] [Google Scholar]
- 219.Morin PA, et al. 2010. Complete mitochondrial genome phylogeographic analysis of killer whales (Orcinus orca) indicates multiple species. Genome Res. 20, 908-916. ( 10.1101/gr.102954.109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Teacher AG, André C, Merilä J, Wheat CW. 2012. Whole mitochondrial genome scan for population structure and selection in the Atlantic herring. BMC Evol. Biol. 12, 248. ( 10.1186/1471-2148-12-248) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.