Abstract
Background
ctDNA sequencing could be used for early cancer screening, prognosis prediction, and medication guidance. However, data of its application in gastric cancer are still lacking. In this study, using ctDNA sequencing, we aimed to screen the mutant genes closely associated with gastric cancer and to explore the impact of these genes on gastric cancer development.
Methods
ctDNA for high‐throughput sequencing was obtained from gastric cancer patients, and the high‐frequency mutant gene KMT2D was identified. Immunohistochemical examination was conducted to assess the expression of KMT2D in gastric cancer tissues. KMT2D knockdown was performed to establish the stably transfected gastric cancer cells. Then, CCK8, plate clone formation assay, and Transwell assay were conducted, and a subcutaneous tumor‐bearing model was induced in nude mice to investigate the changes in cell proliferation and invasion capability. Transcriptome sequencing was also performed to investigate the differences in cellular gene expression.
Results
Detection of ctDNA found 113 gastric cancer‐related mutations, 11 of which are the top 20 high‐frequency mutations of gastric cancer recorded by COSMIC (Catalogue of Somatic Mutations in Cancer, COSMIC). They are TP53, ARID1A, CDH1, PIK3CA, KMT2C, KMT2D, APC, SPEN, CTNNB1, SETBP1, and KMT2A. The gene closely related to the clinical characteristics of the patient is KMT2D. The high‐frequency mutant gene KMT2D was identified in gastric cancer tissues. The positive rate of KMT2D expression in cancer tissues was 74.3%, which was higher than that in para‐carcinoma tissues (56.8%). The knockdown of KMT2D inhibited the proliferation, invasion, and tumor formation capacity of the gastric cancer cells, causing differences in the gene expression profiles, and the expression of different functional gene clusters was up‐ or downregulated.
Conclusion
The findings of this study revealed that KMT2D could be an oncogene capable of promoting gastric cancer proliferation.
Keywords: ctDNA, gastric cancer, KMT2D
The knockdown of KMT2D inhibited the proliferation, invasion, and tumor formation capacity of the gastric cancer cells, causing differences in the gene expression profiles, and the expression of different functional gene clusters was up‐ or downregulated. This revealed that KMT2D could be an oncogene capable of promoting gastric cancer proliferation.
1. INTRODUCTION
Gastric cancer is one of the most common malignant tumors and the second leading cause of cancer‐related death throughout the world. 1 The incidence of gastric cancer is influenced by several categories of factors, including genetic, dietetic, and geographical. 1 The incidence and mortality of gastric cancer in China are both approximately 2‐fold higher than the world average levels. 2 This type of cancer represents a highly heterogeneous tumor; the early diagnosis of which is difficult while the risks of postoperative relapse and metastasis are relatively high. Polygenic predictor analysis could contribute to increasing the early diagnosis rate of gastric cancer and thus improving the prognosis of patients. 3 The application of genomic examinations in addition to conventional pathological staging could allow the further understanding of the pathological mechanisms and molecular classification of gastric cancer. Such insights are an essential precondition for improving the prognoses of patients through providing individualized treatments. 4
In comparison with obtaining tumor tissue samples, the collection of peripheral blood is convenient, non‐invasive, and suitable for dynamic observation. Circulating tumor DNA (ctDNA), the major component used for liquid biopsy examination, is an important biomarker for tracking and analyzing cancerous mutations. 5 An enzymatic on/off switch‐mediated assay can identify KRAS mutations in a normal background from an minimal amount ctDNA of peripheral blood. 6 Measuring the ctDNA of patients provides important information for the screening and diagnosis of a tumor and promotes the efficacy of monitoring and medication guidance.
Lysine (K)‐specific methyltransferase 2D (KMT2D) belongs to the mammalian histone H3 lysine 4 (H3 K4) methyltransferase family. 7 A high‐frequency mutation of KMT2D was previously identified in small‐cell lung cancer (SCLC) and pheochromocytoma. 8 , 9 KMT2D is expressed in various tumors and is critically involved in regulating the tumor development, differentiation, metabolism, and suppression. 10 The inhibition of KMT2D expression, therefore, could be an applicable strategy for tumor treatment. 7 Treatments targeting the methyltransferase KMT2D in pancreatic ductal adenocarcinoma exerted beneficial therapeutic effects. 11 However, KMT2D was also identified as a tumor suppressor gene, whose early loss promotes lymphomagenesis by remodeling of the epigenetic landscape of the cancer precursor cells. 12 Ablating the KMT2D gene in B cells enhanced the progression of lymphomas in mice. 13 However, more studies are still needed to uncover the exact mechanisms through which KMT2D is involved in gastric cancer development.
Therefore, in the present study, we aimed to screen the mutant genes closely associated with gastric cancer by next‐generation sequencing of gastric cancer ctDNA, investigate the influence of KMT2D on gastric cancer development, and further explore the targets for gastric cancer treatment and indicators of treatment efficacy monitoring.
2. SUBJECTS AND METHODS
2.1. Ethics statement
This study was approved by the Ethics Committee of the Fujian Medical University Union Hospital (Fuzhou, Fujian Province, China). All the patients or families signed informed consents before the collection of peripheral blood and tissue samples. The animal experiments in this investigation were approved by the Ethics Committee of Fujian Zoological Society and were performed in agreement with the Chinese Animal Management Regulations.
2.2. Samples
Pathologically confirmed gastric cancer patients, treated in the Fujian Medical University Union Hospital from November 2005 to December 2018, were included. Finally, 10 patients (5 males and 5 females) were included, of whom 5 were >50 years and 5 were <50 years old. Seven of the patients were with poorly differentiated gastric cancer, whereas the other three were with moderately or highly differentiated gastric cancer. Repeated blood samplings were conducted for the patients, and a total number of 26 blood samples were obtained for ctDNA analysis. In addition, 74 samples of gastric cancer and para‐carcinoma tissues (gastric tissues of ≥3 cm from the margin of the cancer tissues) were collected, which were subjected to paraffin embedding for further immunohistochemical examinations.
2.3. ctDNA sequencing
A volume of 10 mL of peripheral blood was obtained, followed by separation of its plasma and blood cells. Then, genomic DNA was extracted from the plasma and blood cells using DNeasy® Blood & Tissue Kit (Qiagen, Inc., Valencia, CA, USA). The samples were quantified by Nanodrop 8000 (Thermo Scientific, Wilmington, DE, USA) to ensure the DNA volume in each sample was ≥1 ug, while the DNA was free from evident degradation. CAN_panel_EZ_HX3 (1.7 M) target region capture chip (Roche, USA) and HiSeq 3000 ((Illumina, San Diego, CA, USA) were used for capture sequencing. The sequencing results of the leukocytes of the same sample were used as the control. The germline mutations were excluded, and the somatic cell variations were screened.
2.4. Cell lines
Human gastric cancer lines AGS, BGC823, HGC‐27, SNU‐16, and MGC803, as well as normal gastric epithelial cell line GES‐1, were purchased from the Shanghai Cell Bank of Chinese Academy of Sciences (Shanghai, China). The cells were cultured with DMEM culture medium (HyClone, USA) or 1640 culture medium (HyClone, USA) containing 10% fetal bovine serum (FBS), in an incubator at 37 °C, 100% humidity, and 5% CO2.
2.5. Immunohistochemistry
The paraffin‐embedded tissue slices were subjected to dewaxing, hydration, and rinsing with running water. Next, the antigen was repaired by the use of citrates at a high temperature and a high pressure, and peroxidase was blocked. Animal serum was used for blocking, and then, Anti‐MLL2 Antibody (1:200; Merck, USA), the primary antibody, was added, and the slices were incubated at 4 °C overnight. DAB development was then conducted, followed by counterstaining with hematoxylin, dehydration, and mounting with neutral balsam. The slices were independently reviewed by two pathologists. Ten visual fields were randomly selected and observed at 400× magnification. The expression of KMT2D in the cancer and para‐carcinoma tissues was scored independently. The staining results were classified as negative (score of 0), light yellow (score of 1), brown‐yellow (score of 2), and dark brown (score of 3). The percentage of positive cells was classified as follows: 0‐1/3 (1), 1/3‐2/3 (2), and >2/3 (3). The KMT2D score was calculated by adding these two scores and was classified as follows: negative (score of 0), mild positive (+, score of 2–3), moderate positive (++, score of 4), and strong positive (+++, score of 5–6). 14
2.6. Quantitative reverse transcription PCR (RT‐qPCR)
Gastric cancer cells were collected for total RNA extraction. Then, first‐strand cDNA synthesis was conducted using a reverse transcription kit (FastQuant RT Kit; Tiangen, China). The following primers were utilized for KMT2D gene: (F) 5’‐ATCGTCGCTGCTGCTATC‐3’, and (R) 5’‐GGAGAGGCGTCAGTGAAG‐3’. GAPDH was selected as the internal reference. The reaction system was prepared using Eastep RT‐qPCR Master Mix (Promega, China), according to the instructions of the manufacturers. ABI 7500 (ABI, USA) was used for the analysis of the samples.
2.7. Western blotting
The proteins were extracted from gastric cancer cells by RIPA lysis buffer (Beyotime, China), and then, the BCA quantification kit (Beyotime, China) was employed to measure the protein concentration. Protein sample (24 µg) was added to 10% SDS‐PAGE gel and electrophoresed, followed by electric transfer to a PVDF membrane (Pall Corporation, East Hills, NY, USA). After blocking, the membrane was incubated with anti‐KMT2D antibody (1:1000; Merck, USA) and anti‐GAPDH antibody (1:2000; Abcam, USA) at 4 °C overnight. Next, the membrane was rinsed with Tween‐20 three times, followed by incubation with horseradish peroxidase‐labeled goat‐anti‐rabbit IgG antibody (1:10,000; Abcam, USA) at 37 °C. ELC (Advanasta, USA) enhanced chemiluminescence was then conducted, and the protein strips were analyzed by a gel imaging analysis system (Gel Doc 1000; Bio‐Rad, Cambridge, MA, USA).
2.8. Constructing stable cells of gene knockdown by lentiviral transfection
A lentivirus expressing KMT2D‐siRNA (GenePharma, Shanghai, China) was used for the transfection (MOI, 100) of MGC803 gastric cancer cells. The sequence of KMT2D siRNA‐1, siRNA‐2, and siRNA‐3 was as follows: (S) 5'‐GATCCGCAGTGCCCTGAATGCAAAGTTTCAAGAGAACTTTGCATTCAGGGCACTGCTTTTTTG‐3’, (A) 5'‐AATTCAAAAAAGCAGTGCCCTGAATGCAAAGTTCTCTTGAAACTTTGCATTCAGGGCACTGCG‐3’; (S) 5'‐GATCCGGACAAATTTGCTGCAGAAGATTCAAGAGATCTTCTGCAGCAAATTTGTCCTTTTTTG‐3’, (A) 5'‐AATTCAAAAAAGGACAAATTTGCTGCAGAAGATCTCTTGAATCTTCTGCAGCAAATTTGTCCG‐3’; and (S) 5'‐GATCCGCATCTACATGTTCCGAATAATTCAAGAGATTATTCGGAACATGTAGATGCTTTTTTG‐3’, (A) 5'‐AATTCAAAAAAGCATCTACATGTTCCGAATAATCTCTTGAATTATTCGGAACATGTAGATGCG‐3'. RT‐qPCR and WB were employed to assess the inhibition of KMT2D. For co‐culture, 5 mg/mL of polybrene (MP, USA) was utilized to screen the stably transfected cells.
2.9. CCK8 assay
The cells were cultured in a 96‐well plate, with a density of 800 cells/well. After the cells were cultured for 6–8 h to allow adhesion, CCK‐8 was added, and that time point was referred to as 0 h. Afterward, CCK‐8 was also added at 24, 48, and 72 h, respectively, according to the instructions of the CCK‐8 kit (Dojindo, Japan). Multiskan GO microplate reader ((Thermo Scientific, Waltham, MA,, USA) was used to count the cells.
2.10. Cell cycle measurement
The cells were digested with trypsin and washed with PBS, and a cell suspension was prepared. Then, ethanol was added for fixation, and a PI cell cycle kit was used for staining (Keygen Biotech, China). The cell cycles were measured by flow cytometer (Accuri C6, BD, USA) and analyzed by Flow J software (Ashland, OR, USA).
2.11. Clone formation assay
The cells were cultured with a density of 400 cells/well, at 37 °7 s5% CO2 for 10–14 days. After the visible cell clone formed, the culturing was terminated. The culture medium was discarded, the cells were washed with PBS, fixed with methanol, stained with crystal violet, and the number of clones was counted.
2.12. Wound healing assay
The cells were cultured in a 6‐well plate with a density of 1 × 106 cells/well. After the cells adhered and reached full confluence, the tip of a sterilized 10‐μL pipette was used to create the scratch. The cells were washed with PBS to remove the detached cells. The culture medium was changed to FBS‐free medium, and the cells were cultured for 24 h. An inverted microscope was used to observe the migration of the cells, and the distance was measured by ImageJ software.
2.13. Transwell assay
Serum‐free medium was utilized to re‐suspend the cells. Further, 1 x 104 cells were added to the Matrigel‐coated upper chamber of Transwell (Corning, USA) while DMEM culture medium containing 10% FBS was added to the lower chamber. The cells were cultured at 37 °C, 5% CO2 for 24 h, followed by fixation with methanol, staining with crystal violet for 10 min, and washing by PBS. An inverted microscope (Otter BDS200, Chongqing, China) was employed for the subsequent observation and counting.
2.14. Animal experiment
Four‐week‐old BALB/C nude mice were purchased from SLACCAS Laboratory Animal Co., Ltd. (Shanghai, China). The MGC803 cells before and after KMT2D knockdown were harvested. The cells were subcutaneously injected into the midback of the mice, at a density of 1 × 107 cells/mouse. The mice were observed for a period of 18 continuous days after the injection, and their body weight (g), maximal diameter of the subcutaneous tumor (L), and the perpendicular minimal diameter (W) were measured every three days. The volume of the tumor was calculated according to the following equation: volume (mm3) =0.5 × L (mm) × W2 (mm2). 15
2.15. Transcriptome sequencing of the cells
The cells were collected, and the total RNA was extracted. The gene library was created by using NEBNext® UltraTM RNA Library Prep Kit (Illumina) kit. HiSeq 3000 (Illumina, USA) was employed for sequencing. Cluster Profiler R software was used for the GO enrichment analysis of the differentially expressed genes.
2.16. Statistical analysis
SPSS 24.0 software was used for the statistical analysis. Data were described as means ±standard deviations. A 2 × 2‐paired chi‐square test was performed to analyze the immunohistochemistry results. Independent t test was used for the comparisons of the data between groups. p < 0.05 was considered statistically significant.
3. RESULTS
3.1. ctDNA sequencing results
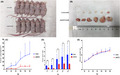
A total number of 113 gastric cancer‐related mutant genes were detected, of which 11 were among the leading 20 high‐frequency mutant genes of gastric cancer, documented in COSMIC. The sequencing results of each patient at different time mainly showed no evident patterns. Only the three samples from the patient 7 showed the same six mutant genes, which showed a pattern of decrease followed by increase with the change of disease conditions (Figure 1A). The detection frequency of KMT2D was 15.38% (4/26), which was the second most frequent gene following TP53 (30.8%, 8/26). The GO functional analysis of these six genes showed that the genes were associated with drug response, regulation of RNA polymerase II transcription, and cell adhesion (Figure 1B). KEGG pathway analysis revealed that the genes participated in various cancer signaling pathways, leukocyte migration, and regulation of the pluripotent pathway of stem cells (Figure 1C).
Figure 1.
Results of the peripheral blood ctDNA sequencing and GO/KEGG functional analysis of the six genes of patient 7. A, The mutant gene cluster and mutation frequency changes of the peripheral blood ctDNA of the patient 7. B, GO analysis of the six mutant genes. The length of the bar indicates the number of genes enriched in the corresponding function. The P‐value is indicated by different colors: the darker color indicates a lower P‐value, suggesting more evident enrichment. C, Bubble diagram of the KEGG pathway enrichment analysis of the six genes. Each bubble in the bubble diagram indicates a signaling pathway. The horizontal axis of the bubble indicates the concentration factor of the pathway in the KEGG analysis, while the size of bubble indicates the number of genes enriched in this pathway.
3.2. Expression of KMT2D in gastric cancer tissues
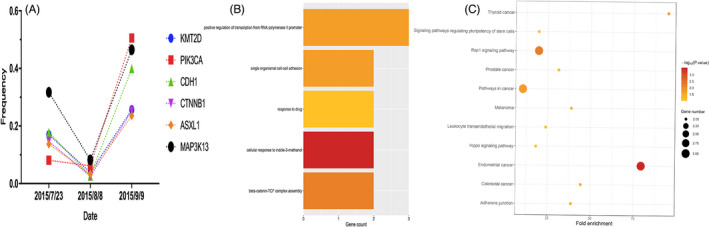
Immunohistochemistry analysis showed that KMT2D was positive in both the cytoplasm and the cellular nuclei, but mainly in the cellular nuclei. In addition, KMT2D was expressed in both cancer tissues and para‐carcinoma tissues (Figure 2A). The expression of KMT2D in cancer tissues was not significantly associated with the clinical pathological features (Table 1). However, the expression rate was significantly higher in the cancer tissues than in the para‐carcinoma tissues (p < 0.001) (Table 2).
Figure 2.
KMT2D expression in gastric cancer tissues and construction of stable KMT2D knockdown cells. A, KMT2D expression in the cancer tissues and para‐carcinoma tissues (400×); B, F, measuring KMT2D protein expression in different gastric cancer cell lines with Western blot, and the results of the gray‐scale analysis; C, G, KMT2D protein expression and gray‐scale analysis in MGC803 cells before and after KMT2D knockdown; (D) relative expression level of KMT2D mRNA in different gastric cancer cell lines; and (E) relative expression level of KMT2D mRNA in MGC803 cells before and after KMT2D knockdown.
Table 1.
Association between KMT2D expression and the clinical pathological features of gastric cancer.
| Clinical pathological feature | KMT2D staining n (%) | Total | Positive rate | p | |
|---|---|---|---|---|---|
| Negative (n = 19) | Positive (n = 65) | n | % | ||
| Age (years) | |||||
| <65 | 10 (52.6) | 26 (47.3) | 36 | 72.2 | 0.792 |
| ≥ 65 | 9 (47.4) | 29 (52.7) | 38 | 76.3 | |
| Gender | |||||
| M | 13 (68.4) | 41 (74.5) | 54 | 75.9 | 0.765 |
| F | 6 (31.6) | 14 (25.5) | 20 | 70 | |
| Differentiation degree | |||||
| Poor | 5 (26.3) | 28 (50.9) | 33 | 84.8 | 0.107 |
| Moderate to high | 14 (73.7) | 27 (49.1) | 41 | 65.9 | |
| Vessel carcinoma embolus | |||||
| Yes | 4 (21.1) | 21 (38.2) | 25 | 84 | 0.261 |
| No | 15 (78.9) | 34 (61.8) | 49 | 69.4 | |
| Nerve invasion | |||||
| Yes | 6 (31.6) | 19 (34.5) | 25 | 76 | 1.000 |
| No | 13 (68.4) | 36 (65.5) | 49 | 73.5 | |
| T stage | |||||
| T1‐2 | 3 (15.8) | 9 (16.4) | 12 | 75 | 1.0 |
| T3‐4 | 16 (84.2) | 46 (83.6) | 62 | 74.2 | |
| N stage | |||||
| N0‐1 | 8 (42.1) | 25 (45.5) | 33 | 75.8 | 1.0 |
| N2‐3 | 11 (57.9) | 30 (54.5) | 41 | 73.2 | |
| TNM stage | |||||
| Stages I–II | 7 (36.8) | 24 (43.6) | 31 | 77.4 | 0.788 |
| Stages III–IV | 12 (63.2) | 31 (56.4) | 43 | 72.1 | |
Table 2.
Expression of KMT2D in the cancer and para‐carcinoma tissues (n).
| Group | n | KMT2D | Positive rate (%) (+ to +++) | |||
|---|---|---|---|---|---|---|
| ‐ | + (2–3) | ++ (4) | +++ (5–6) | |||
| Para‐carcinoma tissues | 74 | 32 | 20 | 10 | 12 | 56.8 |
| Cancer tissues | 74 | 19 | 21 | 12 | 22 | 74.3*** |
p < 0.001.
3.3. Cell screening and construction of stable KMT2D knockdown cells
GES‐1 was used as the reference. The RT‐qPCR and Western blot results showed that KMT2D was overexpressed in all the five cell lines, but its relative expression in MGC803 cells was the highest (Figure 2B, D and F). Therefore, MGC803 cells were selected for the KMT2D knockdown. Cells transfected with lentiviral empty vector was used as the control, and the RT‐qPCR results showed siRNA‐1 (sh1KMT2D) had the best knockdown effects, which inhibited 45.4% of the KMT2D expression (p < 0.01) (Figure 2E). In addition, WB results also confirmed that the KMT2D protein level significantly decreased (Figure 2C and G). MGC803 cells stably transfected with siRNA‐1 (sh1KMT2D) was chosen for the consequent experiments (shKMT2D).
3.4. KMT2D knockdown inhibited cell proliferation and invasion
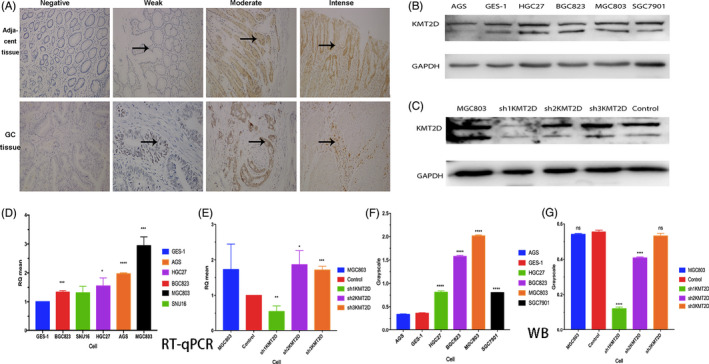
Compared with the control group, the cells in shKMT2D group showed significantly decreased proliferation in 24 h (Figure 3A). The relative proliferation rate of the cells also decreased, especially at 48 and 72 h (Figure 3B). The number of cells in G1 phase increased after the KMT2D knockdown; in addition, the percentage of cells in the S phase also increased, while percentage of cells in G2 phase declined (Figure 3C). These findings demonstrated that KMT2D knockdown promoted the cell entry from the G1 phase into the S phase, inhibiting the proliferation of gastric cancer cells. The plate clone formation assay revealed that the rate of clone formation was 49.78 ± 3.46 and 24.89 ± 1.73 before and after KMT2D knockdown, respectively, and the difference was statistically significant (p < 0.0001) (Figure 3D–F). The wound healing assay showed a decrease in the migration area of the cells in the shKMT2D group after the scratch; the migration capability was inhibited by approximately 30% (p < 0.001) (Figure 3G). Similarly, the Transwell assay also showed that the number of invasive cells decreased significantly in the shKMT2D group, and the invasion capability was inhibited by about 50% (p < 0.001) (Figure 3H). These findings demonstrated that KMT2D promotes gastric cancer proliferation through multiple pathways.
Figure 3.
Changes in the biological behaviors of MGC803 gastric cancer cells before and after the KMT2D knockdown. A, KMT2D knockdown inhibited MGC803 cell proliferation; (B) KMT2D knockdown decreased the relative proliferation rate of the cells; (C) cell cycle analysis before and after KMT2D knockdown; (D, E, and F) KMT2D knockdown reduced the clone formation capability of the cells; (G) KMT2D knockdown inhibited the migration capability of gastric cancer cells; and (H) KMT2D knockdown impeded the invasion capability of the cells. (**p < 0.01, ***p < 0.001, and ****p < 0.0001).
3.5. KMT2D knockdown inhibited subcutaneous tumor formation
After the administration of the subcutaneous injection with cells into the nude mice, visible xenografts were found on the 3rd day in both groups, yielding a tumor formation rate of 100% (Figure 4A and B). The volumes of the tumors were measured to plot the tumor growth curve, which showed significantly higher tumor volume and tumor growth rate in the control group than in the shKMT2D group since the 6th day (Figure 4C and D). The body weight of the mice in the two groups was not significantly different (Figure 4E).
Figure 4.
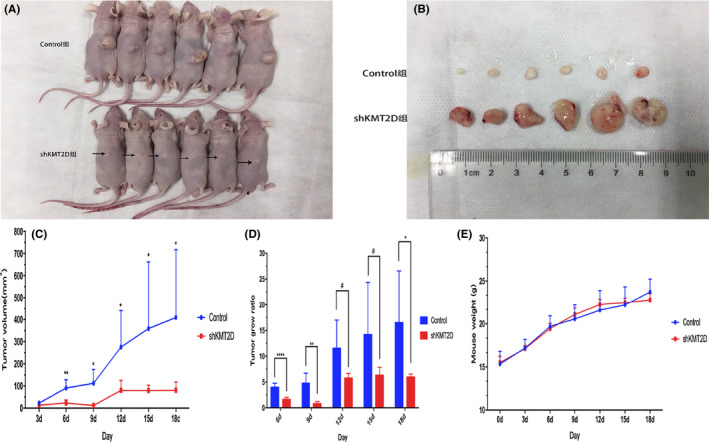
KMT2D knockdown inhibited the growth of subcutaneous xenograft in the nude mice. A, Tumor‐bearing mice. The arrows show the positions of the subcutaneous xenografts; (B) the xenografts obtained from the mice; (C) volume of the xenografts in the nude mice; (D) xenograft growth rate in the nude mice; (E) body weight changes of the nude mice. (*p < 0.05,**p < 0.01, and ****p < 0.0001).
3.6. KMT2D knockdown influenced the cellular gene expression
Clustering analysis following transcriptome sequencing of the genes showed significant differences between the control and shKMT2D groups (Figure 5A). The analysis of the differential genes showed 2062 of the differential genes were upregulated (red), and 575 of the differential genes were downregulated (green) (Figure 5B). GO functional analysis showed that after KMT2D knockdown, the genes related to the negative regulation of immune responses, regulation of inflammatory reactions, regulation of interleukins, and activities of signal receptors (such as MAPK13, CCL5, FOXP3, and JAK3) were upregulated (Figure 5C, up), whereas the expression of the genes associated with the termination of gene translation, reassembly of the cytoskeleton, and the release of cytochrome C was downregulated (Figure 5D, down).
Figure 5.
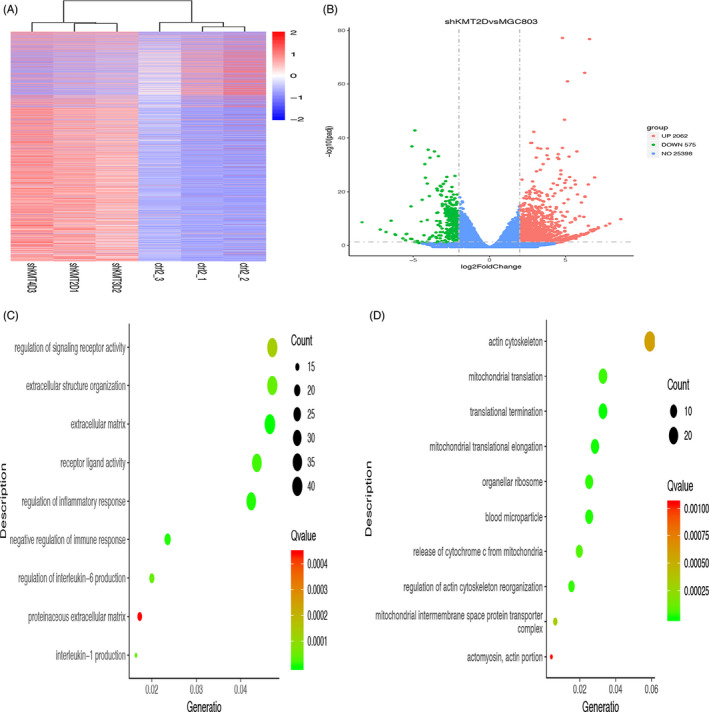
Effects of KMT2D knockdown on the expression of genes in MGC803 gastric cancer cells. A, Gene clustering analysis after the KMT2D knockdown; (B) analysis of the differential genes in the cells after the KMT2D knockdown; (C) GO functional enrichment analysis of the upregulated genes after the KMT2D knockdown; (D) GO functional enrichment analysis of the downregulated genes after the KMT2D knockdown (each bubble in the bubble diagram indicates a function. The horizontal axis of the bubble indicates the concentration factor of the function in the GO analysis, while the size of bubble indicates the number of genes enriched in this function).
4. DISCUSSION
In this study, we conducted ctDNA sequencing to investigate the high‐frequency gastric cancer‐related mutant gene KMT2D. The positive rate of KMT2D expression was higher in the gastric cancer tissues than para‐carcinoma tissues. We further constructed stable cell lines of KMT2D knockdown, and the in vitro and in vivo studies showed that KMT2D knockdown suppressed the proliferation, invasion, clone formation, and in vitro tumor formation capabilities of gastric cancer cells.
Previous studies of breast cancers revealed that the higher expression of KMT2D is associated with poorer prognosis of the patients, which maintained the tumor initiation capability of cancer stem cell‐like cells. 16 , 17 After binding to KMT2D, AKT induced phosphorylation of KMT2D, which consequently reduced the activities of methyltransferase and functions of ER. However, the suppression of PI3Kα enhanced the activities of KMT2D. These findings provided theoretic basis for epigenetic therapy of ER‐positive breast cancer. 18 , 19 Here, we detected a high‐frequency KMT2D mutation in the ctDNA of gastric cancer patients, the frequency of which was associated with changes in the disease development. KMT2D gene ranks the 4th of the TOP20 genes listed by COSMIC. Gastric cancer tissue examinations confirmed that the positive rate of KMT2D was higher in the cancer tissues than in the para‐carcinoma tissues. Our findings are in agreement with the results reported by Xiong et al., 20 who found that KMT2D was overexpressed in gastric cancer tissues and was closely associated with poorer prognosis of the patients. These findings suggest that KMT2D could be a representative of a new type of oncogenes.
To further investigate the effects of KMT2D on gastric cancer development, we constructed KMT2D‐knockdown cell lines. The proliferation capability of cancer cells is a key factor affecting tumor growth. The findings of this study showed that the KMT2D knockdown decreased the proliferation and clone formation capabilities of the gastric cancer cells. Previous studies in gastric and prostate cancers showed that KMT2D knockdown inhibited the proliferation, invasion, and migration of the cells. 20 , 21 Our findings confirmed that KMT2D knockdown altered the cell cycle, affecting the proliferation of gastric cancer cells. Earlier investigations found that inhibiting the cells in the S stage suppressed the proliferation of gastric cancer cells. 22 In addition, arresting cell cycle also increased the sensitivity of gastric cancer cells to some chemotherapeutic drugs. 23 Furthermore, identifying the genetic profiles closely associated with the cell cycle facilitated the assessment of the survival rate of gastric cancer patients. 24 Invasion and metastases are the biological characteristics of malignant tumors that endanger the health and cause death of patients. Guo et al. 7 found that the absence of KMT2D led to reduced cell proliferation and cell migration. Our findings of the wound healing assay showed that KMT2D knockdown inhibited the cell migration capability by approximately 30%. Similarly, the invasion of the cells was suppressed by about 50% after KMT2D knockdown, as revealed by the Transwell assay results. We further induced xenograft models in nude mice and obtained results that were in agreement with the aforementioned in vitro findings. Therefore, KMT2D knockdown inhibits the tumor formation capability of the cancer cells in vivo. In agreement with the results of previous studies, our findings also showed that the absence of KMT2D effectively inhibited the growth of tumors. 21
The interactions among multiple genes are involved in the pathogenesis of gastric cancer. The gastric cancer‐related genes participate in various signaling pathways, such as MAPK and PIK pathways. 25 Deletion or absence of the genes could affect the copy numbers of the genes and thus consequently affect the activities of various signal transduction pathways. 26 Previous studies in breast cancer have demonstrated the interactions in the expressions of KMT2D, AKT, and PI3Ka. 18 The transcriptome sequencing of the cells in this study showed that the expression levels of the genes in gastric cancer cells changed after KMT2D knockdown. The GO functional analysis showed that the genes related to the activities of signal receptors and negative regulation of immune responses were upregulated. Immune is closely associated with the development and progression of tumors. Immune checkpoint therapy is the key of the individualized immunotherapy of gastric cancer, while the application of inhibitor of immune checkpoint could improve the prognosis of gastric cancer patients. 27 , 28 , 29 While on the other hand, the genes related to the termination of gene translation and release of cytochrome C were downregulated. However, more studies are needed to elucidate the exact mechanisms involved in the effects of KMT2D knockdown on down‐ and upregulation of gene expression.
In summary, the findings of this study confirmed that KMT2D is closely associated with gastric cancer. This gene promoted the proliferation and invasion of gastric cancer cells altered the expression of the genes in the cells and thus enhanced the progression of gastric cancer.
5. SUMMARY
In this study, we used ctDNA sequencing to screen out the gastric cancer‐related high‐frequency mutant gene KMT2D, which was highly expressed in gastric cancer tissues. KMT2D knockdown altered the cell cycle of the gastric cancer cells and inhibited the proliferation, invasion, migration, clone formation, and in vivo tumor formation capabilities of gastric cancer cells. In addition, KMT2D knockdown also led to down‐ or upregulation of other genes in the cells. The results of this study indicate that KMT2D could be an oncogene that can be used for gastric cancer treatment. However, these findings need to be further verified in KMT2D overexpression cell lines. In addition, more studies are required to investigate the downstream pathways of KMT2D and further explore the mechanisms involved in the effects of this gene.
CONFLICT OF INTEREST
None.
ACKNOWLEDGEMENTS
This study was supported by the Specialized Research Fund of Fujian Provincial Department of Science and Technology (2013YZ0002‐2).
DATA AVAILABILITY STATEMENT
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
REFERENCES
- 1. Kim J, Cho YA, Choi WJ, Jeong SH. Gene‐diet interactions in gastric cancer risk: a systematic review. World J Gastroenterol. 2014;20(28):9600‐9610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA: A Cancer J Clin. 2016;66(2):115‐132. [DOI] [PubMed] [Google Scholar]
- 3. Ai K, Jia Y, Li J, Wang C, Wang Y. Systematic analysis of multigene predictors in gastric cancer exploiting gene expression signature. J Cell Biochem. 2019;120(5):8069‐8077. [DOI] [PubMed] [Google Scholar]
- 4. Kidd JM, Cooper GM, Donahue WF, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453(7191):56‐64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Khatami F, Tavangar SM. Circulating tumor DNA (ctDNA) in the era of personalized cancer therapy. J Diabet Metab Disord. 2018;17(1):19‐30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang QLZC, Yin YF, Xiao L, Wang Y, Li K. An enzymatic on/off switch‐mediated assay for KRAS hotspot point mutation detection of circulating tumor DNA. J Clin Lab Anal. 2020;34(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guo C, Chen LH, Huang Y, et al. KMT2D maintains neoplastic cell proliferation and global histone H3 lysine 4 monomethylation. Oncotarget. 2013;4(11):2144‐2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Augert A, Zhang Q, Bates B, et al. Small cell lung cancer exhibits frequent inactivating mutations in the histone methyltransferase KMT2D/MLL2: CALGB 151111 (Alliance). J Thorac Oncol. 2017;12(4):704‐713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Juhlin CC, Stenman A, Haglund F, et al. Whole‐exome sequencing defines the mutational landscape of pheochromocytoma and identifies KMT2D as a recurrently mutated gene. Genes Chromosomes Cancer. 2015;54(9):542‐554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Froimchuk E, Jang Y, Ge K. Histone H3 lysine 4 methyltransferase KMT2D. Gene. 2017;627:337‐342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dawkins JB, Wang J, Maniati E, et al. Reduced expression of histone methyltransferases KMT2C and KMT2D correlates with improved outcome in pancreatic ductal adenocarcinoma. Cancer Res. 2016;76(16):4861‐4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang J, Dominguez‐Sola D, Hussein S, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med. 2015;21(10):1190‐1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ortega‐Molina A, Boss IW, Canela A, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med. 2015;21(10):1199‐1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sun P, Wu T, Sun X, et al. KMT2D inhibits the growth and metastasis of bladder Cancer cells by maintaining the tumor suppressor genes. Biomed Pharmacother. 2019;115:108924. [DOI] [PubMed] [Google Scholar]
- 15. Tomayko MM, Reynolds CP. Determination of subcutaneous tumor size in athymic (nude) mice. Cancer Chemother Pharmacol. 1989;24(3):148‐154. [DOI] [PubMed] [Google Scholar]
- 16. Yasuda K, Hirohashi Y, Kuroda T, et al. MAPK13 is preferentially expressed in gynecological cancer stem cells and has a role in the tumor‐initiation. Biochem Biophys Res Commun. 2016;472(4):643‐647. [DOI] [PubMed] [Google Scholar]
- 17. Han H, Chen Y, Cheng L, Prochownik EV, Li Y. microRNA‐206 impairs c‐Myc‐driven cancer in a synthetic lethal manner by directly inhibiting MAP3K13. Oncotarget. 2016;7(13):16409‐16419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Toska E, Osmanbeyoglu HU, Castel P, et al. PI3K pathway regulates ER‐dependent transcription in breast cancer through the epigenetic regulator KMT2D. Science. 2017;355(6331):1324‐1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koren S, Bentires‐Alj M. Tackling resistance to PI3K inhibition by targeting the epigenome. Cancer Cell. 2017;31(5):616‐618. [DOI] [PubMed] [Google Scholar]
- 20. Xiong WDZ, Tang Y, Deng Z, Li M. Downregulation of KMT2D suppresses proliferation and induces apoptosis of gastric cancer. Biochem Biphys Res Commun. 2018;504(1):pp. 129–136, 129–36. [DOI] [PubMed] [Google Scholar]
- 21. Lv S, Ji L, Chen B, et al. Histone methyltransferase KMT2D sustains prostate carcinogenesis and metastasis via epigenetically activating LIFR and KLF4. Oncogene. 2017;37(10):1354‐1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xie H, Li X, Chen Y, Lang M, Shen Z, Shi L. Ethanolic extract of Cordyceps cicadae exerts antitumor effect on human gastric cancer SGC‐7901 cells by inducing apoptosis, cell cycle arrest and endoplasmic reticulum stress. J Ethnopharmacol. 2019;231:230‐240. [DOI] [PubMed] [Google Scholar]
- 23. Du H, Liu Y, Chen X, et al. DT‐13 synergistically potentiates the sensitivity of gastric cancer cells to topotecan via cell cycle arrest in vitro and in vivo. Eur J Pharmacol. 2018;818:124‐131. [DOI] [PubMed] [Google Scholar]
- 24. Zhao L, Jiang L, He L, et al. Identification of a novel cell cycle‐related gene signature predicting survival in patients with gastric cancer. J Cell Physiol. 2019;234(5):6350‐6360. [DOI] [PubMed] [Google Scholar]
- 25. Lin Y, Wu Z, Guo W, Li J. Gene mutations in gastric cancer: a review of recent next‐generation sequencing studies. Tum Biol. 2015;36(10):7385‐7394. [DOI] [PubMed] [Google Scholar]
- 26. Liang L, Fang JY, Xu J. Gastric cancer and gene copy number variation: emerging cancer drivers for targeted therapy. Oncogene. 2016;35(12):1475‐1482. [DOI] [PubMed] [Google Scholar]
- 27. Lazar DC, Avram MF, Romosan I, Cornianu M, Taban S, Goldis A. Prognostic significance of tumor immune microenvironment and immunotherapy: Novel insights and future perspectives in gastric cancer. World J Gastroenterol. 2018;24(32):3583‐3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Togasaki K, Sukawa Y, Kanai T, Takaishi H. Clinical efficacy of immune checkpoint inhibitors in the treatment of unresectable advanced or recurrent gastric cancer: an evidence‐based review of therapies. OncoTargets Ther. 2018;11:8239‐8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alsina M, Moehler M, Hierro C, Guardeno R, Tabernero J. Immunotherapy for Gastric Cancer: A Focus on Immune Checkpoints. Target Oncol. 2016;11(4):469‐477. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.


