Abstract
The characterization and synthesis of 3-chloro-2-{(2E)-2-[1-(4-chlorophenyl)ethylidene]hydrazinyl}pyridine (CCPEHP) was investigated in our study. Mass and UV-visible spectra were recorded in chloroform solvent. The CCPEHP molecule containing pyridine and chlorophenyl rings and hydrazone group crystallized in the triclinic system and P-1 space group. FTRaman and FTIR spectra were performed in the solid state. The optimized geometry of CCPEHP was computed by DFT/B3LYP method with 6–311 G (d, p) and 6–31 G (d, p) levels. The computed vibrational analysis, electronic absorption spectrum, electronic properties, molecular electrostatic potential, natural bond orbitals analysis and other calculated structural parameters were determined by using the DFT/B3LYP/6–31 G (d, p) basis set. The correlation of fundamental modes of the compound and the complete vibrational assignments analysis were studied. The strong and weak contacts were identified by using Hirshfeld surface analysis. The molecular modeling results showed that CCPEHP structure strongly binds to COVID-19 main protease by relative binding affinity of -6.4 kcal/mol.
Keywords: Vibrational spectroscopy, Density functional theory, COVID-19, X-ray diffraction
1. Introduction
Heterocyclic compounds are an important class of compounds in chemistry. As these compounds are associated with chemistry and biochemistry, they are used in the pharmaceutical design industry, veterinary products and agricultural chemicals. Small heterocyclic molecules or compounds with a heterocyclic structure have the highest proportion of drugs used in human medicine. Nine of the 15 best-selling prescription drugs in the world in 2009 were small molecules heterocyclic compounds [1]. Pyridine, showing antimicrobial pharmaceuticals, is the most important nitrogen-based compound in the heterocyclic ring system. It also plays a key role in catalyzing biological and chemical systems. Pyridine derivatives contain multi-functional groups and streptonigrone, steroidigrin, and lavendamycin of these groups have been reported as anti-cancer drugs and cerivastatin as HMG-CoA enzyme inhibitor drugs [2,3]. The importance of pyridine and phenol vibration spectra in biological and industrial systems attracts great attention from a spectroscopic point of view [4]. In addition, hydrazone-containing compounds show electrophilic or nucleophilic character thanks to the azomethine groups they contain, which makes them unique in drug development studies [5]. While forming the molecular system, atoms do not bond randomly, they interact using a certain order and rules according to the laws of nature. The best experimental technique used to analyze these seemingly complex structures is the X-ray diffraction method [6]. Ab inito methods are of great importance in examining the properties of molecules, which are very important for researchers [7]. Such calculations are based on quantum theory and all chemical properties of the material can be calculated. Again, thanks to this method, the properties of the drugs designed for the treatment of various fatal diseases are determined, especially in pharmacology, and the loss of money and time can be prevented by aiming to inhibit the target protein [8]. Computer-aided drug design has been a promising technique for designing new and powerful therapeutics. Many theoretical studies have been applied to identify inhibitors against COVID-19. Xu et al. (2020), created a three-dimensional SARS-CoV2 Mpro homology model based on SARS-CoV Mpro and screened it through protein modeling and virtualizing against 1903 drug inhibitors, highlighting nelfinavir as a potential drug inhibitor against SARS-CoV2 Mpro [9,10]. There is currently no defined treatment or antiviral drug for COVID-19 treatment. However, the US Food and Drug Administration (FDA) has approved both Chloroquine (CQ) and Hydroxychloroquine (HCQ) for COVID-19 control and treatment for emergency purposes [11]. Chloroquine and Hydroxychloroquine are notorious for toxicokinetic properties and can reveal dose-dependent toxicity. Recently, multi-centered global clinical trials are being conducted to evaluate the therapeutic potential of Chloroquine and its hydroxyl form, Hydroxychloroquine as a treatment for this infection [12], [13], [14]. The synthesized CCPEHP compound has a similar molecular structure with (CQ) and (HCQ) drugs. In our study, the behaviors of CCPEHP compound and (CQ) and (HCQ) drugs in molecular docking studies have been tried to be correlated. CCPEHP molecule was designed by different substituting (pyridine, hydrazone and heterocyclic) groups. These groups are considered for their potential to interact with amino acids in 4 different types (hydrophobic, hydrogen bonds, ionic and water bridges) [15,16]. More specifically, the characterization of the synthesized compound with X-ray diffraction, spectroscopic and packing analysis and with quantum chemistry calculations was evaluated in this paper. The crystalline reactive sites with MEP analyses, electronic properties with HOMO-LUMO gap energies, donor-acceptor stabilization energies E (2) with NBO approach, electronic transitions with UV–Vis spectrum analysis, the effects of the compound on crystal packaging and the definition of non-covalent forces with Hirshfeld surface analysis were investigated. Finally, the inhibitor-receptor 2D and 3D interactions of the CCPEHP compound were visualized. [17], [18], [19].
2. Materials and methods
2.1. Materials and physical measurements
Intensity data for CCPEHP were collected using the ω–θ2 scan technique on an Agilent SuperNova, (Single source at offset and Eos CCD detector) diffractometer with a SuperNova (Mo) X-ray Source (Mo-Kα, k = 0.71073 Å). The CRYSALISPRO software program was used for cell refinement, data collection and reduction [20]. The FTIR spectrum of the CCPEHP compound was obtained by the Perkin Elmer FTIR-Spectrometer Spectrum Two Model between 400 and 4000 cm−1. Mass spectrum (ESI) was recorded on TSQ Fortis™ Triple Quadrupole Mass Spectrometer LC-MS/MS model device. The FTRaman spectrum was obtained in the region 100–4000 cm−1 with a Thermo Fisher Scientific model DXR™ 3xi Raman Imaging Microscope with 1064 nm Nd: YAG laser source with a resolution of 3 cm−1. The melting point of the crystallized material was determined by the Stuart SMP10 device. The absorption spectrum was recorded using a Shimadzu UV-1800 spectrophotometer between 240 and 800 nm. 3‑chloro-2-hydrazinopyridine, 4′-chloroacetophenone, ethanol, acetonitrile were bought from Sigma Aldrich company.
2.2. Synthesis
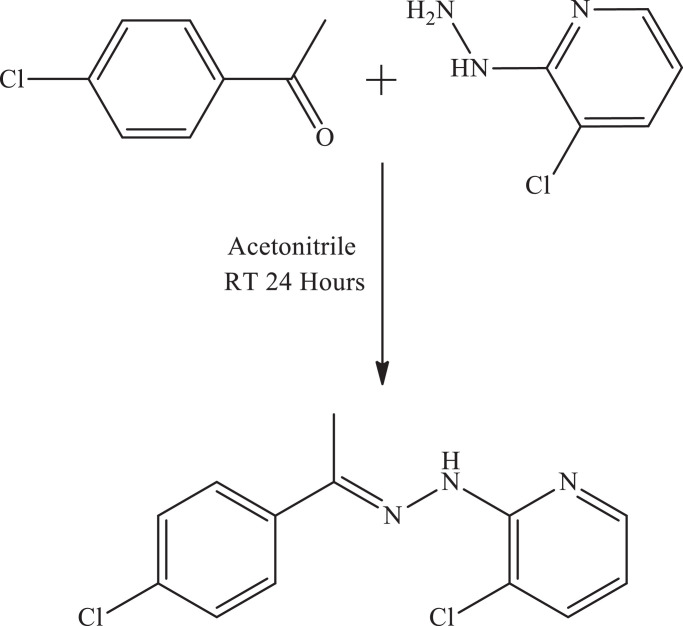
3-chloro-2-{(2E)−2-[1-(4-chlorophenyl)ethylidene]hydrazinyl}pyridine (CCPEHP) was prepared according to the literature [21]. The 4′-chloroacetophenone (1 mmol, 0.1545 g) in 15 ml acetonitrile was mixed with 3-chloro-2-hydrazinopyridine (1 mmol, 0.1435 g) in 10 ml acetonitrile (Fig. 1 ). The mixture was slowly stirred for 24 h at RT. After stirring, the solution was left at RT for 3 days and crem product was obtained and washed with water and recrystallized by acetonitrile, and dried over in a vacuum desicator. C13H11Cl2N3 [CCPEHP], Light Crem; Yield 79%. M.p.: 138 °C. δH (400 MHz, CDCl3-d6, ppm) 8.32 (1H, N—H), 8.29 (1H, Arpry-H), 7.80 (2H, Ar-H), 7.60 (1H, Arpry-H), 7.36 (2H, Ar-H), 6.80 (1H, Arpry-H), 2.33 (3H, -CH3), δC (100 MHz, CDCl3-d6, ppm) 150.32, 147.14, 146.41, 137.08, 136.88, 134.70, 128.51, 128.51, 127.62, 127.62, 116.20, 115.12, 12.75. MS (ESI+): m/z 280.15 [M]+ (100).
Fig. 1.
Synthesis of 3-chloro-2-{(2E)−2-[1-(4-chlorophenyl)ethylidene]hydrazinyl}pyridine.
2.3. Calculation/theory
The geometries of 3-chloro-2-{(2E)−2-[1-(4-chlorophenyl)ethylidene]hydrazinyl}pyridine was optimized with Gaussian 09 W package program using Beck's triply parameter hybrid Exchange function B3 along with Lee-Yang-Parr (LYP) functions with 6–311 G (d, p) and 6–31G(d, p) basis sets [22,23]. Diffuse functions on all atoms support these basis sets. Vibrational frequencies and IR intensity values of CCPEHP compound optimized with DFT/B3LYP/6–31 G (d, p) level were calculated. In addition, potential energy distributions of vibration modes were determined using the VEDA4 program [24]. UV spectrum of the CCPEHP compound in chloroform solvent was computed using the TD-DFT method via DFT/B3LYP/6–31 G (d, p) level [25]. The LUMO and HOMO gap energies, molecular electrostatic potential (MEP) and natural bonding orbitals (NBO) were calculated using the B3LYP method and 6–31 G (d, p) level [1,26,27]. The Gauss-view 6.0 visualization software and the Gaussian 09 w package program were used for all the computerized calculations [28], [29], [30]. Gamess US software was used as an auxiliary theoretical calculation program [31]. CrystalExplorer package program generated Hirsfeld surfaces and 2D fingerprint plots which were used to determine the analysis of the intermolecular interactions in the crystal case [32].
2.4. Protein and ligand preparation for docking
Targeting the 3CLpro protease of the corona virus consisting of structural and nonstructural polyproteins could constitute a valid approach for the treatment of COVID-19 potential drug design. One of the structures of 3CLpro like protease protein was downloaded (PDBID: 6LU7, 2.16 Å) from Protein Data Bank (PDB) (http://www.rcsb.org), in 3D format [33]. Preparation for simulation and simulation processes of COVID-19 3CLpro/Mpro structure and ligand were carried out by AutoDock Tools 1.5.6, MG Tools of AutoDock Vina program and Discovery Studio 2020 Client (Dassault Systemes BIOVIA) [34,35]. In protein preparation, the missing residues were first checked and there were no missing residues of the viral protein. Chain A, one of two chains of main protein, was selected and the binding affinity of this chain was performed with our potential drug candidate molecule. Water molecules and hetero atoms were deleted from the COVID-19 main protease, then Kollman charges and polar hydrogen atoms were added to the target protein (chain A) and saved with AutoDock Tools in pdbqt format [10,36]. Autodock vina algorithm with the most appropriate configurations (exhaustiveness= 8, binding modes= 9, energy difference= 3 kcal/mol, feasible with x, y, and z coordinate) parameters is used in simulation studies [34,37]. Default settings were used in the other parameters. The grid box value and intermolecular interactions of the ligand-protein complex were visualized with the Discovery studio 2020 client program. Hydrophobic gap coordinates of the binding site were used in the docking of N3 (as control) and fungal metabolites. The active site of the amino acid was calculated as sized (30 Å x 30 Å x 30 Å) and centered (−10.71 Å x 12.41 Å x 68.83 Å). (Default grid spacing= 0.375 Å) [38]. In order to prepare the ligand for docking, the CIF data obtained from the X-ray crystal structure [39] was converted to mol2 format. The partial charge and torsion angels were changed and saved in pdbqt format with AutoDock Tools. Before the simulation, the separate pdbqt formats and docking parameters of the drug candidate compound and COVID-19 main protease were combined into a single .txt file and the complexation process was completed through the AutoDock Vina software.
3. Results and discussion
3.1. X-ray data collection and structure refinement
The structure was solved using SHELXT [40] and then refined by full-matrix least-squares refinements on F2 using the SHELXL [41] in Olex2 Software Package [42]. Crystallographic data and refinement details of the data collection for CCPEHP are tabulated in Table 1 . Geometrical calculations and crystal structure validations were performed using Platon software [43]. Mercury software [44] was used for visualization of the cif file. Additional crystallographic data with CCDC reference number 2,068,456 for CCPEHP has been deposited within the Cambridge Crystallographic Data Center via www.ccdc.cam.ac.uk/deposit.
Table 1.
Crystal data and refinement parameters for CCPEHP.
| CCDC | 2,068,456 |
|---|---|
| Empirical formula | C13H11Cl2N3 |
| Formula weight/g. mol−1 | 280.15 |
| Temperature/K | 293(2) |
| Radiation, Wavelength (Å) | MoKα (λ = 0.71073) |
| Crystal system | Triclinic |
| Space group | P-1 |
| a/Å | 8.0121(5) |
| b/Å | 11.9756(8) |
| c/Å | 14.7888(10) |
| α/° | 78.321(6) |
| β/° | 78.139(6) |
| γ/° | 74.029(6) |
| Crystal size/mm3 | 0.15 × 0.13 × 0.12 |
| Volume/Å3 | 1318.94(16) |
| Z | 4 |
| ρcalcd (g. cm−3) | 1.411 |
| µ (mm−1) | 0.476 |
| F(000) | 576 |
| 2θ range for data collection (°) | 6.952 to 50 |
| h/k/l | −9 ≤ h ≤ 9, -14 ≤ k ≤ 14, |
| -11 ≤ l ≤ 17 | |
| Reflections collected | 7011 |
| Independent reflections | 4607 [Rint = 0.0187, Rsigma = 0.0514] |
| Data/restraints/parameters | 4607/0/327 |
| Goodness-of-fit on F2 (S) | 1.006 |
| Final R indices [I > 2σ(I)] | R1 = 0.0509, wR2 = 0.1117 |
| R indices (all data) | R1 = 0.0934, wR2 = 0.1353 |
| Largest diff. peak/hole / e Å−3 | 0.24/−0.27 |
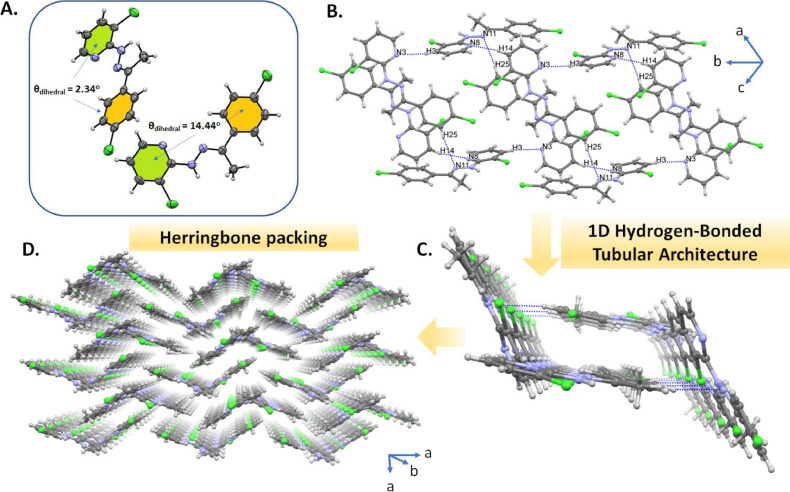
In order to get a better insight into the intermolecular interactions in the solid state, the clear colorless crystals of CCPEHP for single crystal X-ray diffraction were grown by slow evaporation, and structural analysis exhibits that CCPEHP crystallizes in the triclinic space group P-1, with the asymmetric unit containing two CCPEHP molecules. The structure exhibited chlorophenyl subunit with the chloropyridine moieties lying nearly in the same plane with the tilt angles 2.34° and 14.44°, which matches well with the computationally optimized geometries, for two molecules in the asymmetric unit, and these values are comparable to those observed in 2-{2-[1-(4-nitrophenyl)ethylidene]hydrazinyl}pyridine [CSD Ref Code: MEMQIH] [45]. In the solid state of CCPEHP, the weak hydrogen bonds (C11‧‧‧Cl27 = 3.636(4) Å as D‧‧‧A) between the CH of pyridine ring and chloride moiety connected two molecules in the asymmetric unit. As shown in Fig. 2 b, the major intermolecular interactions governing the crystal packing are identified as weak CH‧‧‧N hydrogen bonding contacts (C3‧‧‧N3 = 3.397(5) Å, C14‧‧‧N8 = 3.522(4) Å, C25‧‧‧N11 = 3.661(4) Å), and these interactions lead a 1D hydrogen-bonded tubular architecture (Fig. 2c) along the b-axis in the crystal packing. Furthermore, 1D hydrogen bonded tubulars are expanded into a 3D supramolecular network with herringbone crystal packing by the weak π‧‧‧π-stacking interactions (4.1639(18) and 4.305(2) Å) between the aromatic rings (Fig. 2d).
Fig. 2.
a) ORTEP drawings of the crystal structure of CCPEHP (30% probability level) showing the corresponding dihedral angles between the chlorophenyl and pyridine aromatic rings. b) and c) View of one-dimensional hydrogen bonded tubular architecture by CH‧‧‧N (blue dotted lines) contacts. d) Perspective view of the 3D supramolecular network showing herringbone packing fashion formed by the weak π‧‧‧π interactions.
3.2. Geometric optimization
The optimized structure of the CCPEHP compound, namely C13H11Cl2N3, was calculated with the help of quantum theory [30]. The selected DFT optimization and metrical parameters from X-ray diffraction are tabulated in Table 2 . It is understood from the results given in Table 2 that, experimental and theoretical data are in good agreement with each other. Although the results are compatible with each other, there are still slight differences. The reason for this difference should be kept in mind that the experimental results were obtained in the crystal phase, while the theoretical results obtained with 6–31 G (d, p) and 6–311 G (d, p) levels were obtained in the gas phase [46]. The bond length C(1)-N(8) in the molecule was observed as 1.323 Å and the same bond length was calculated as 1.335 and 1.331 Å for 6–31 G (d, p) and 6–311 G (d, p) basis sets [28].
Table 2.
The single-crystal X-ray and optimized selected geometrical parameters of CCPEHP.
| Bond length (Å) | Exp. | 6–31 G (d, p) | 6–311 G (d, p) | Dihedral angles (o) | Exp. | 6–31G | 6–311 G |
|---|---|---|---|---|---|---|---|
| (d, p) | (d, p) | ||||||
| C(1)-C(2) | 1.395 | 1.415 | 1.413 | N(8)-C(1)-C(2)-C(3) | 0.8 | −0.074 | 0.083 |
| C(1)-N(8) | 1.323 | 1.335 | 1.331 | N(9)-C(1)-C(2)-C(3) | 179.7 | −179.632 | 179.735 |
| C(1)-N(9) | 1.381 | 1.382 | 1.382 | N(9)-C(1)-C(2)-Cl(29) | 0.0 | 0.371 | −0.269 |
| C(2)-C(3) | 1.363 | 1.384 | 1.380 | N(8)-C(1)-C(2)-Cl(29) | −178.9 | 179.929 | −179.9203 |
| C(2)-Cl(29) | 1.737 | 1.766 | 1.766 | C(5)-C(4)-C(3)-C(2) | 0.0 | 0.090 | −0.085 |
| C(3)-C(4) | 1.381 | 1.397 | 1.394 | N(8)-C(1)-N(9)-N(11) | −11.6 | 3.585 | −3.217 |
| C(4)-C(5) | 1.367 | 1.392 | 1.389 | C(1)-N(9)-N(11)-C(12) | 174.6 | −178.399 | 178.316 |
| C(5)-N(8) | 1.331 | 1.335 | 1.333 | N(9)-N(11)-C(12)-C(13) | −2.7 | 0.956 | −0.900 |
| N(9)-N(11) | 1.360 | 1.349 | 1.347 | C(13)-C(12)-C(17)-C(18) | −175.1 | 167.388 | 13.075 |
| N(11)-C(12) | 1.289 | 1.294 | 1.289 | C(18)-C(20)-C(24)-C(22) | −0.2 | −0.192 | −0.347 |
| C(12)-C(13) | 1.496 | 1.512 | 1.511 | C(19)-C(22)-C(24)-Cl(27) | −178.8 | −179.713 | −179.880 |
| C(12)-C(17) | 1.480 | 1.482 | 1.483 | C(18)-C(20)-C(24)-Cl(27) | 178.8 | 179.840 | 179.697 |
| C(24)-Cl(27) | 1.735 | 1.758 | 1.759 | N(9)-N(11)-C(12)-C(17) | 178.4 | −179.277 | 179.337 |
| Bond angles (o) | Bond angles (o) | ||||||
| C(1)-C(2)-Cl(29) | 119.1 | 120.4 | 120.4 | C(5)-C(4)-C(3) | 118.2 | 117.6 | 117.6 |
| N(8)-C(1)-N(9) | 118.9 | 119.1 | 119.2 | C(1)-N(9)-N(11) | 121.2 | 121.3 | 121.4 |
| C(3)-C(2)-Cl(29) | 120.6 | 119.7 | 119.7 | C(13)-C(12)-C(17) | 120.7 | 121.0 | 121.1 |
| C(22)-C(24)-Cl(27) | 120.1 | 119.6 | 119.5 | C(1)-N(8)-C(5) | 117.1 | 118.6 | 118.8 |
3.3. Vibrational analysis
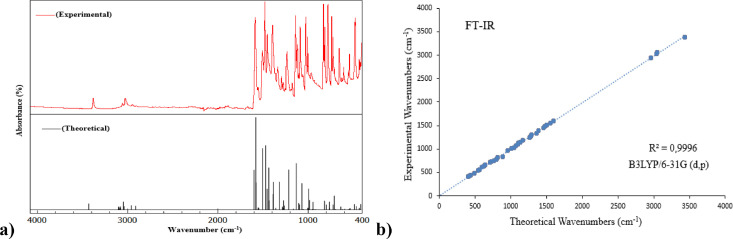
Vibrational frequencies calculations were obtained using optimized structure parameters. 81 fundamental vibrational modes of CCPEHP were calculated based on quantum chemical calculations. DFT potential symmetrically makes vibration wavenumbers higher than experimental data. Inconsistencies in these wavenumbers are corrected by calculating non-harmonic corrections or corrected by presenting a scaled area or by an appropriate factor directly calculated. In order to eliminate these systematic errors, the vibration wavenumbers were scaled with a 0.961 scaling factor [28]. (http://cccbdb.nist.gov/vsfx.asp). After scaling the wavenumbers, the observed and the theoretical vibration wavenumbers showed a very good agreement with each other. The basic vibration modes of CCPEHP compound which are characteristic N—H, C—Cl, C—H, C—C, C = N vibrations found in pyridine and chlorophenyl rings and hydrazone group were tried to be investigated. The scaled and unscaled calculated vibrational modes and intensities of FTIR with their PED and observed FTIR and FTRaman bands are reported in Table 3 . It is observed in Table 3 that the scaled FTIR bands of the CCPEHP have a close relationship between experimental frequencies [47]. The CCPEHP molecule has 29 atoms. All vibration modes are assigned thanks to the graphical interface GaussView 6.0 software program [29]. The pyridine ring of CCPEHP involves 1 N atom and 5 C atoms which generate 4 C—C and 2 C=N stretching vibrations [48]. Additionally, there are also C=N peaks belonging to the hydrazone group in this structure. The C=N vibration peaks are recorded at 1030–1590 cm−1 in FTRaman and 1030–1600 cm−1 in FTIR in the literature [49,50]. The stretching vibration of C=N groups were observed at 1030, 1239, 1589 cm−1 in FTIR and 1031, 1257, 1590 cm−1 in FTRaman and scaled calculated bands at 1045, 1216, 1264, 1319, 1437, 1504, 1554, 1601 cm−1 for CCPEHP molecule. The C—H stretching vibrations in compounds containing aromatic groups are generally in the range 3000–3100 cm−1 [51,52]. The absorption peaks at 3069 and 3025, 3055 cm−1 were assigned to the FTRaman and FTIR spectra. The bands attributed to 3039, 3046, 3079, 3086, 3088, 3096, 3100 and 3103 cm−1 were calculated as C—H stretching vibrations in chlorophenyl and pyridine rings. In literature, the C—H peaks in-plane bending vibrations of aromatic rings are observed at 1020–1300 cm−1 [28]. The C—H in-plane bending vibrations were calculated at 1057, 1069, 1096, 1107, 1165, 1216, 1278 cm−1, respectively based on PED results. Generally, the stretching vibrations of C—C bands occur in the range 1430–1625 cm−1 in the aromatic ring [53]. This band was observed at 1506, 1557 cm−1 in FTIR and 1509, 1560, 1582 cm−1 in FTRaman and that it was well coherent with the simulated data at 1504, 1548, 1554, 1577 and 1580 cm−1. The peaks calculated at 395, 392, 468, 620, 639, 711, 764, 886, 985, 995 cm−1 were attributed for C—C—C in-plane bending vibrations of CCPEHP and the peaks at 78, 211, 265, 349, 404, 488, 536, 697, 916, 955 cm−1 were attributed for C—C—C—C torsion vibrations [54]. Since compound CCPEHP has more than one chlorine atom, it can occur in symmetric and asymmetric modes [55]. The C—Cl peaks were observed in both FTRaman and FTIR at 427 and 440 cm−1, respectively [56]. The stretching vibrations of C—Cl mode were allotted at 392, 711 and 995 cm−1 with significant PED (>10%) [1]. Symmetrical stretching vibrations of methyl (-CH3) groups are generally found in between 2850 and 2940 cm−1 [57]. The vibrations of methyl group were observed at 2910 in FT-Raman and 2948 cm−1 in FTIR. Similarly, computed values of 2912 and 2961 cm−1 were attributed to C—H stretching vibrations in the CH3 group. The N—H stretching vibrations of aliphatic amines are found between 3300 and 3500 cm−1. In this study, this vibration was computed at 3431 cm−1 and observed at 3383 cm−1 in FTRaman and 3380 cm−1 in FTIR. PED column is contributing exactly 100% to this mode [3]. The combined spectra and correlation diagram of FTIR data between theoretical and experimental frequencies are given in Fig. 3 . The linear and the equality in the correlation diagram are y = 0.9922x + 6.9917 (R² = 0.9996).
Table 3.
Observed, computed vibrational assignments of CCPEHP at B3LYP level.
| S.No | Observed | FTIR |
Theory DFT/B3LYP 6–31G(d, p) |
Assignments(%PED)≥10% | ||
|---|---|---|---|---|---|---|
| FTRaman | Unscaled | Scaled | IR Intensity (km/mole) | |||
| (cm−1) | (cm−1) | (cm−1) | (cm−1) | |||
| 1 | 3383 | 3380 | 3571 | 3431 | 18,72 | νNH(100) |
| 2 | 3229 | 3103 | 2,48 | νCH(79)+ νCH(19) | ||
| 3 | 3225 | 3100 | 8,62 | νCH(97) | ||
| 4 | 3222 | 3096 | 8,48 | νCH(23)+ νCH(76) | ||
| 5 | 3213 | 3088 | 1,18 | νCH(−19)+ νCH(80) | ||
| 6 | 3211 | 3086 | 4,58 | νCH(97) | ||
| 7 | 3069 | 3204 | 3079 | 9,17 | νCH(74)+ νCH(−23) | |
| 8 | 3055 | 3170 | 3046 | 24,20 | νCH(97) | |
| 9 | 3025 | 3162 | 3039 | 9,88 | νCH(91) | |
| 10 | 2948 | 3081 | 2961 | 13,06 | νCH(99) | |
| 11 | 2910 | 3030 | 2912 | 11,29 | νCH(93) | |
| 12 | 1590 | 1589 | 1666 | 1601 | 124,88 | νNC(66) |
| 13 | 1582 | 1645 | 1580 | 358,58 | νCC(−41) | |
| 14 | 1641 | 1577 | 84,75 | νCC(51)+ δHCC(14) | ||
| 15 | 1560 | 1557 | 1617 | 1554 | 5,83 | νNC(17)+ νCC(12)+ νCC(11)+ δHNN(13) |
| 16 | 1611 | 1548 | 3,63 | νCC(−40) | ||
| 17 | 1509 | 1506 | 1565 | 1504 | 192,68 | νCC(−16)+ νNC(12)+ δHNN(25) |
| 18 | 1484 | 1483 | 1532 | 1473 | 201,63 | δHCC(48)+ δCCC(13) |
| 19 | 1453 | 1514 | 1455 | 64,91 | δHCH(62)+ τHCCC(19) | |
| 20 | 1495 | 1437 | 131,48 | νNC(−27)+ δHCC(14)+ δHCN(27) | ||
| 21 | 1494 | 1435 | 20,01 | δHCH(−68)+ τHCCC(16) | ||
| 22 | 1401 | 1395 | 1444 | 1388 | 48,52 | δHNN(10)+ δHCC(16)+ δHCC(10) |
| 23 | 1442 | 1385 | 86,90 | δHCC)−10)+ δHCC(12) | ||
| 24 | 1344 | 1338 | 1415 | 1360 | 4,67 | δHCH(82) |
| 25 | 1373 | 1319 | 87,24 | νNC(11)+ νNC(−17)+ δHCN(30) | ||
| 26 | 1297 | 1298 | 1339 | 1287 | 8,59 | νCC(69) |
| 27 | 1278 | 1278 | 1330 | 1278 | 4,39 | δHCC(75) |
| 28 | 1325 | 1273 | 28,46 | νCC(30) | ||
| 29 | 1257 | 1239 | 1316 | 1264 | 12,06 | νNC(61) |
| 30 | 1237 | 1265 | 1216 | 125,19 | νNC(15)+ δHCC(24)+ δCCC(10) | |
| 31 | 1178 | 1180 | 1212 | 1165 | 2,05 | νCC(20)+ δHCC(−70) |
| 32 | 1136 | 1141 | 1179 | 1133 | 145,17 | νNN(44) |
| 33 | 1122 | 1123 | 1151 | 1107 | 20,28 | νCC(−19)+ δHCC(41)+ δHCC(10) |
| 34 | 1092 | 1141 | 1096 | 15,51 | δHCC(−40) | |
| 35 | 1063 | 1062 | 1113 | 1069 | 82,63 | νCC(41)+ δHCC(22) |
| 36 | 1099 | 1057 | 1,41 | νCC(14)+ νCC(−10)+ δHCC(15) | ||
| 37 | 1031 | 1030 | 1087 | 1045 | 0,03 | νNC(−10)+ νCC(−16)+ τHCCC(−20) |
| 38 | 1008 | 1008 | 1046 | 1005 | 4,88 | δHCH(13)+ τHCCC (63)+ γCCNC(12) |
| 39 | 1036 | 995 | 65,43 | νNC(28)+ νClC(−18)+ δCCN(13)+ δCCC(10) | ||
| 40 | 1025 | 985 | 29,40 | δCCC(68) | ||
| 41 | 968 | 994 | 955 | 1,06 | τHCCC(80)+ τCCCC(−12) | |
| 42 | 985 | 947 | 23,40 | νCC(30)+ τHCCC(17) | ||
| 43 | 976 | 938 | 0,74 | τHCNC(72) | ||
| 44 | 954 | 917 | 0,81 | τHCCC(79)+ γClCCC(10) | ||
| 45 | 914 | 953 | 916 | 0,15 | τHCCC(−80)+ τCCCC(11) | |
| 46 | 834 | 922 | 886 | 0,12 | νNC(−12)+ δCCC(34) | |
| 47 | 820 | 854 | 821 | 27,11 | τHCCC(80)+ γCCCC(11) | |
| 48 | 783 | 783 | 831 | 799 | 15,33 | τHCCC(84) |
| 49 | 795 | 764 | 23,94 | τHCCN(−69)+ τHCCC(11) | ||
| 50 | 745 | 744 | 795 | 764 | 4,99 | νCC(−11)+ δCCC(−21) |
| 51 | 730 | 755 | 725 | 15,23 | τCNCC(−63) | |
| 52 | 711 | 740 | 711 | 42,97 | νCC(−13)+ νClC(14)+ δCCN(17)+ δCCC(−16) | |
| 53 | 725 | 697 | 0,76 | τCCCC(49)+ τCCNN(−11) | ||
| 54 | 660 | 659 | 665 | 639 | 8,86 | δCCN(−13)+ δCCC(23) |
| 55 | 629 | 646 | 620 | 0,81 | δCCC(−60)+ δCCC(15) | |
| 56 | 608 | 610 | 619 | 595 | 1,86 | νCC(−10)+ δNCN(42) |
| 57 | 556 | 557 | 581 | 558 | 0,86 | τHNNC(−10)+ γCCNC(−37) |
| 58 | 547 | 564 | 542 | 2,91 | νCC(12)+ δCNC(41) | |
| 59 | 558 | 536 | 3,61 | τHCNC(10)+ τCCCC(49) | ||
| 60 | 483 | 483 | 508 | 488 | 16,75 | τCCCC(11)+ γCCCC(29)+ γCCNC(10) |
| 61 | 487 | 468 | 10,27 | νCC(11)+ δCCC(−12)+ δCCC(13) | ||
| 62 | 427 | 440 | 453 | 435 | 4,53 | γClCCC(69) |
| 63 | 428 | 436 | 419 | 16,87 | τHNNC(−15) | |
| 64 | 408 | 410 | 420 | 404 | 1,13 | τCCCC(−80) |
| 65 | 415 | 399 | 30,78 | τHNNC-23 | ||
| 66 | 408 | 392 | 22,51 | νClC(−31)+ δCCC(−12)+ τHNNC(24) | ||
| 67 | 363 | 349 | 4,22 | τCCCC(13)+ τCCNN(56) | ||
| 68 | 307 | 329 | 316 | 1,33 | δCCN(54) | |
| 69 | 282 | 307 | 295 | 4,35 | δCCCl(34) | |
| 70 | 276 | 265 | 1,75 | τCCCC(46) | ||
| 71 | 274 | 263 | 6,72 | δCCCl(−46) | ||
| 72 | 220 | 211 | 0,27 | τCCCC(−13)+ τCCNN(−47) | ||
| 73 | 194 | 187 | 0,20 | δCCCl(−48) | ||
| 74 | 145 | 139 | 0,12 | τCNCN(49)+ τCNCC(−11) | ||
| 75 | 133 | 128 | 2,12 | δCNC(21)+ δCCCl(−16)+ τHCCC(11) | ||
| 76 | 105 | 101 | 2,50 | τHCCC(64) | ||
| 77 | 81 | 78 | 0,30 | τNCNN(14)+ τCCCC(36)+ γCCCC(−23) | ||
| 78 | 68 | 65 | 5,12 | τCNNC(−74) | ||
| 79 | 44 | 43 | 0,11 | δCNN(−76) | ||
| 80 | 33 | 32 | 0,17 | τCNNC(80) | ||
| 81 | 21 | 20 | 0,01 | τNCNN(58)+ τCCNN(−15) | ||
γ; out-of-plane bending, ν; stretching, δ; in-plane-bending, τ; torsional, PED; Potential Energy Distribution.
Fig. 3.
The a) combined spectra, b) correlation diagram for the observed and calculated FTIR wavenumbers (with scaling factor) of CCPEHP compound.
3.4. Ultraviolet (UV)-visible analysis
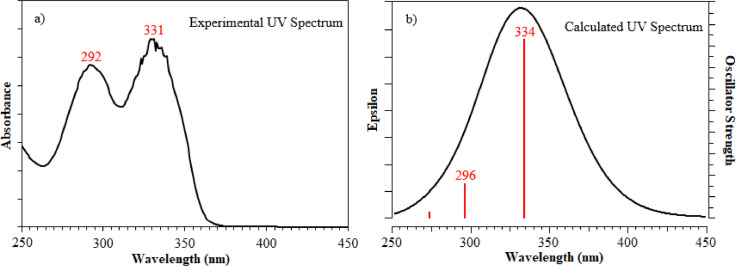
The electronic absorption spectrum of CCPEHP compound was computed by TD-DFT with B3LYP/6–31 G (d, p) level in chloroform solution. The oscillator strengths, excitation energies and wavelengths were estimated by using this method and basis set. The CCPEHP compound was studied at room temperature (RT) in 2 × 10−5 mol L−1 chloroform solution and in the wavelength range of 250–450 nm. Ultraviolet-visible spectroscopy of the compound was compared with the electron transition possibilities. Theoretical calculations of the CCPEHP and corresponding experimental UV–Vis spectra are shown in Fig. 4 . The observed spectrum demonstrated the maximum absorption (Amax) peaks at 292 and 331 nm which indicated that π_π* and n_π* transitions, respectively. According to the theoretical calculations, the bands at 296 and 334 nm were attributed to the π-π* and n-π* transitions. These transitions of oscillator strengths (f) 0.1551/0.9191 and excitation energies (eV) 4.1801/3.7101 were obtained from simulation results and these data are given in Table 4 . The π-π* transitions of the CCPEHP arise from the electrons of the pyridine ring, while the n-π* band is due to hydrazone groups of atoms [2,58].
Fig. 4.
a) Experimental, b) calculated UV-Visible spectra of CCPEHP molecule in chloroform.
Table 4.
The experimental and computed UV-visible spectral parameters of CCPEHP.
| Experimental | Calculated B3LYP/6–31 G(d, p) basis set | Transition | ||
|---|---|---|---|---|
| λ (nm) | λ (nm) | Excitation energy (eV) | Oscillator strenght (f) | |
| 292 | 296 | 4.1801 | 0.1551 | π-π* |
| 331 | 334 | 3.7101 | 0.9191 | n-π* |
3.5. Hirshfeld surface analysis
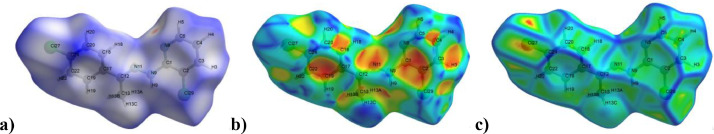
Hirshfeld surface analysis emerged from an attempt to define the area covered by the molecule forming the crystal lattice. It is an effective tool that provides more information about the crystal structure by color-coding the short or long contacts in the molecule system [59]. Crystal packing differences occur between the particular types of intermolecular interactions due to molecular abilities. The formation of intermolecular interactions between the donor and acceptor affects biological activity [60]. The Hirshfeld surface analysis and 2D fingerprint plots studies were carried out using CIF data [61]. CCPEHP compound mapped over dnorm, shape index and curvedness forms and surfaces are shown in Fig. 5 . The volumetric and area surface properties of mapped CCPEHP were found as 325.08 Å3 and 311.95 Å2, respectively [58]. Short and strong contacts on the dnorm surface are attributed to red spot color, while long and weak contacts are identified with blue areas in Fig. 5a. According to the dnorm surface analysis, it is understood that weak intermolecular interactions are dominant in the CCPEHP molecule. The regions observed in shape index (the orange-red colored) and in curvedness (the large green straight surface, which is separated by a blue outline) represent π-π stacking interactions of the CCPEHP molecule (Figs. 5b and 5c). Furthermore, these defined regions indicated that intermolecular interactions with neighboring molecules [62] .
Fig. 5.
Views of the a)dnorm, b) shape index and c) curvedness forms of Hirshfeld surfaces of CCPEHP.
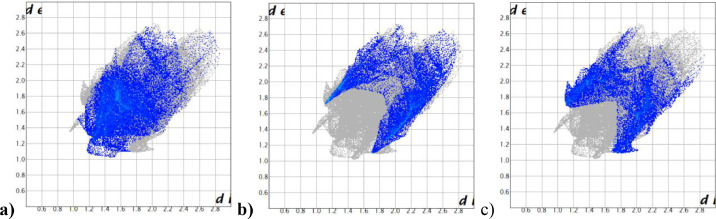
The 2D fingerprints plots are associated with Hirshfeld Surface area and the interactions of H‧‧‧H, Cl‧‧‧H/H‧‧‧Cl and C‧‧‧H/H‧‧‧C were the most contributing close contact interactions to the total surface and are illustrated in Fig. 6 . The intermolecular interaction areas are shown in blue, and areas outside of this interaction are shown in gray; H‧‧‧H 31.2%, Cl‧‧‧H/H‧‧‧Cl 20.7%, C‧‧‧H/H‧‧‧C 20.3%, N‧‧‧H/H‧‧‧N 10.4%, Cl‧‧‧Cl 7.1%, C‧‧‧Cl/Cl‧‧‧C 4.0%, C‧‧‧C 3.8%, C‧‧‧N/N‧‧‧C 1.8%, N‧‧‧Cl/Cl‧‧‧N 0.6% were obtained from the 2D fingerprint plot study [17]. When the results of the surface analysis are evaluated, the stability of the CCPEHP compound is due to the presence of hydrogen bonds in the structure. Additionally, examining the physical and chemical properties of potential biologically active compounds in simulation studies allows us to predict their behavior in in-vitro studies.
Fig. 6.
Decomposed 2D fingerprint plots of the a) H‧‧‧H, b) Cl‧‧‧H/H‧‧‧Cl and c) C‧‧‧H/H‧‧‧C interactions for CCPEHP crystal.
3.6. Natural bond orbital analysis
Natural bonding orbital gives very important theoretical information about conjugation and hyperconjugation and intra and intermolecular charge transfer of the molecular orbital system [2]. Donor–acceptor interaction indicates that delocalization of electron density between formally unoccupied non-Lewis type natural bonding orbitals and occupied Lewis type natural bonding orbitals. The more intense the interaction between the electron acceptor and donor, the higher the E (2) value [63]. Decolalizations effects of electrons have been obtained from the second order perturbation results performed by the NBO 3.1 package [27]. DFT/B3LYP method with 6–31 G (d, p) basis set was used to understand and interpret different intermolecular interactions of atoms in molecules. For CCPEHP, the some strongest π to π* interactions were calculated as π(C1-N8)→π*(C4-C5) 28.20 kcal/mol, π(C2-C3)→π*(C1-N8) 24.70 kcal/mol, π(C2-C3)→π*(C4-C5) 13.10 kcal/mol, π(C4-C5)→π*(C2-C3) 27.10 kcal/mol, π(C17-C19)→π*(C18-C20) 18.80 kcal/mol, π(C17-C19)→π*(C22-C24) 21.90 kcal/mol, π(C18-C20)→π*(C17-C19) 18.60 kcal/mol, π(C18-C20)→π*(C22-C24) 21.00 kcal/mol, π(C22-C24)→π*(C17-C19) 18.10 kcal/mol, π(C22-C24)→π*(C18-C20) 17.80 kcal/mol. Regarding the lone pairs of electrons delocalization, LP1 (N9) to π*(C1-N8) and LP1 (N9) to π*(N11-C12) with the hyperconjugative stabilization energy data were obtained as 49.10 kcal/mol and 30.80 kcal/mol, respectively. The other strongest hyperconjugative interactions were calculated as LP1 (N8)→π*(C1-C2) 11.15 kcal/mol and LP1 (N11)→π*(C12-C13) 11.60 kcal/mol [64]. According to the results of NBO analysis, stabilization energy values were also obtained between the antibonding donor orbitals and the antibonding receptor orbitals.
3.7. HOMO–LUMO analysis
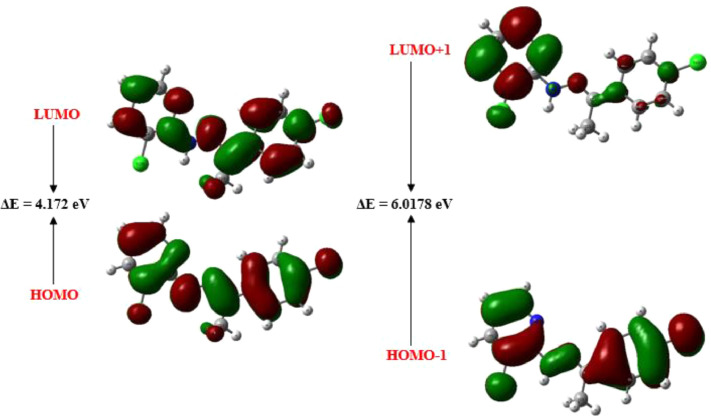
Determining the electronic properties of the molecule is a very important tool in interpreting the results of biological activity [65]. In the molecular system, frontier molecular orbitals (FMOs) are of great importance for drug designing. The highest occupied molecular orbital (HOMO) corresponded to ionization potential, while the lowest unoccupied molecular orbital (LUMO) corresponded to electron affinity [66]. Thanks to the FMOs energy gap, important information about the chemical stability and reactivity of molecules can be obtained. The chemical stability of the compound plays a crucial role in drug design investigations. A large energy gap brings about to the molecule less reactive and more stable while smaller energy gab brings about to less stable and more reactive in the system [2]. The electronic properties of the CCPEHP and the theoretical energies of (EHOMO) and (ELUMO) orbitals of the CCPEHP compound were investigated (Fig. 7 ).
Fig. 7.
Frontier molecular orbitals of CCPEHP.
The energy difference (ΔE) was found as 4.1720 eV in (LUMO—HOMO) and 6.0178 eV in (LUMO+1)-(HOMO-1). According to the result obtained from the energy gap values, the CCPEHP molecule has high kinetic stability. The reactivity descriptors parameters are calculated from HOMO and LUMO orbitals where electron affinity, chemical softness, chemical hardness, electronic chemical potential, ionization potential are tabulated in Table 5 [67,68]. The chemical stability of the molecule was supported by chemical hardness and softness in addition to the energy gap [18]. In Fig. 7, while the red color symbolizes the positive lobes, the green color symbolizes the negative lobes of the molecule. Generally, HOMO and LUMO charges are localized over the whole molecule [1].
Table 5.
The calculated FMO and molecular description of CCPEHP molecule.
| Frontier molecular orbital parameter | Reactivity descriptions | ||
|---|---|---|---|
| ELUMO (eV) | −1.3600 | Ionization potential (I) | 5.5320 |
| EHOMO (eV) | −5.5320 | Electron affinity (A) | 1.3600 |
| EHOMO-LUMO ΔE (eV) | 4.1720 | Chemical hardness (η) | 2.0860 |
| ELUMO+1 (eV) | −0.7831 | Chemical softness (S) | 0.2396 |
| EHOMO-1(eV) | −6.8009 | Electronegativity (χ) | 3.4460 |
| EHOMO-1-LUMO+1 (eV) | 6.0178 | Chemical potential (µ) | −3.4460 |
ΔE = ELUMO- EHOMO, I = –EHOMO, η = (I–A)/2, A = –ELUMO, S = 1/2η, µ = –(I + A)/2, χ = (I + A)/2.
3.8. Molecular electrostatic potential (MEP)
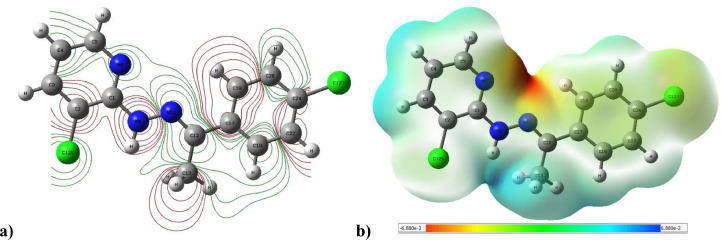
Molecular electrostatic potential gives information about the partial charges and electronegativities of different atoms in the molecule [26]. The color scale for (MEP) map surface: red zones are for negative potential, electron rich, strong repulsion; blue zones for positive potential, electron lacking, strong attraction; green zones for neutral. The map of 2D contour and electrostatic potential for CCPEHP are shown in Fig. 8 . The MEP values of the CCPEHP molecule were predicted in the range of −6.880 e−2 and 6.880 e−2. In CCPEHP crystal, the red surfaces of the negative potentials are spread over the pyridine ring and hydrazone group of nitrogen atoms. As a result of this, red zones (N8, N9 and N11) which have negative electrostatic potential leads to electrophilic attack and this indicates the strongest repulsion. In Fig. 8a, the 2D contour map of the molecule's reactants clearly confirms the proton transfer [18,28,69].
Fig. 8.
a) 2D contour map and b) 3D map for CCPEHP molecule.
3.9. Molecular docking
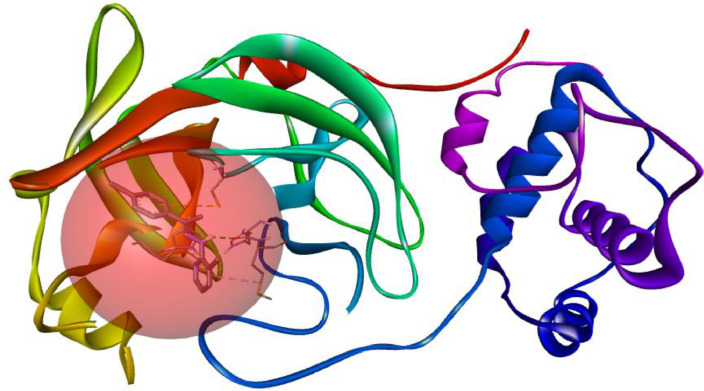
One of the best ways to understand ligand and protein binding interactions in the pharmaceutical design industry is molecular docking studies. Molecular docking is one of the best simulation methods applied to estimate drug-target interactions by calculating energy minimization and binding energy. It identifies the best position against the target protein. Autodock vina algorithm determines the most suitable ligand conformation with minimized energy with the help of some scoring functions [37,70]. After the simulation process, The lowest binding energy and the most hydrogen binding pose were selected from the ViewDock window. Ligand_1–6lu7 was chosen as the best position is most associated with hydrogen binding. Hydrogen binding and other interactions between the ligand and the 6lu7 protein were examined in the Biovia Discovery Studio (DS) visualizer (Fig. 9 ). The highest binding affinity was determined with COVID-19 Mpro, which exhibits covalent bond interaction thanks to molecular dynamic simulation [71]. The residues from the N3 binding site are Ser46, His41, Met49, Phe140, Tyr54, Leu141, Gly143, Asn142, Cys145, His164, His163, Met165, Leu167, Glu166, Pro168, Phe185, Asp187, His172, Gln189, Thr190, Ala191 and Gln192 [72]. When the molecular docking result applied in our study was analyzed, it was observed that the ligand binds to similar enzyme sites.
Fig. 9.
Main protease (6LU7) with CCPEHP inhibitor.
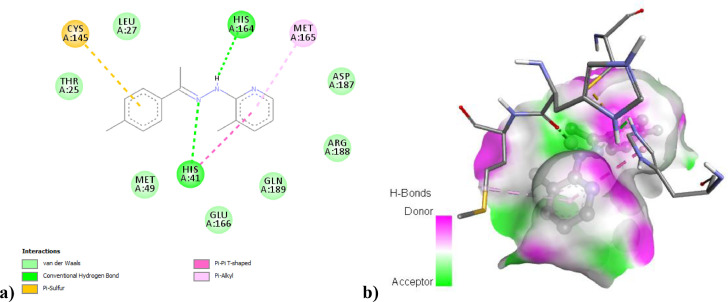
CCPEHP molecule has shown five interactions with COVID-19 Mpro (Fig. 10a) [38] . The CCPEHP ligand was observed to have interacted with two conventional hydrogen bonds His164 (2.31 Å) and His41 (2.92 Å), a Pi-Pi T-Shaped type with His41 (4.65 Å), a Pi-Sulfur with Cys145 (5.76 Å) and a Pi-alkyl with Met165 (4.91 Å). Pi-Pi T-Shaped and Pi-alkyl type interactions are included in the hydrophobic category. The binding affinity of this complex was found to be −6.4 kcal/mol. When the simulated binding interactions of the molecule were evaluated, the binding generally occurred in regions suitable for electrophilic attacks. While Pi interactions occurred on aromatic pyridine and chlorophenyl rings, hydrogen bond interactions occurred in the hydrazone region. In biological processes, the presence of hydrogen bonds is of great importance in the interaction of the inhibitor with the receptor. As shown in Fig. 10b, the green areas represent the electron acceptor, while the pink areas represent the electron donor. It was revealed that the theoretically calculated inhibitor-receptor complex interaction gave a very successful result and in this sense, the CCPEHP compound we designed has a good inhibitory capacity [18,19]. The behavior of molecular docking studies of FDA-approved drugs in the literature is examined. In a study by Dossary et al. (2021), the docking pose analysis showed that the chloroquine (CQ) bonded to Leu141, Asn142, His163, Pro168, Cys145 and Glu166 binding residues in the 6LU7 protein [73]. In another study on the main SARS-CoV-2 protease carried out by Nobre Júnior et al. (2020), the CCPEHP compound was showing better docking scores (−6.4 kcal/mol) as well as better ligand-receptor interactions in comparison to hydroxychloroquine and chloroquine (−5.0, and −4.7 respectively) [66].
Fig. 10.
(a) and (b) visualize the 2D and 3D interactions between CCPEHP and COVID-19 Mpro.
4. Conclusion
3-chloro-2-{(2E)−2-[1-(4-chlorophenyl)ethylidene]hydrazinyl}pyridine was synthesized and characterized the experimental technique by single-crystal X-ray crystallography and on the other hand, quantum theory to support the experimental data. The geometrical parameters, vibrational frequencies and UV-visible spectrum were obtained from the optimized structure of the CCPEHP compound and compared with the experimental results. The experimental values of vibrational frequencies were in good agreement with the scaled calculated values. Hirshfeld Surface analysis proved that important contacts were the H‧‧‧H 31.2%, Cl‧‧‧H/H‧‧‧Cl 20.7% and C‧‧‧H/H‧‧‧C 20.3% interactions. Stabilization energy values between donor-acceptor orbitals and hyperconjugation interactions were determined by NBO orbital analysis. The reactive zones of CCPEHP were determined by molecular electrostatic potential (MEP) results in the nitrogens N8, N9 and N11 were nucleophilic zones. Frontier molecular orbitals (FMOs) and properties related to molecular interaction ability were predicted. By examining almost all the properties of the CCPEHP molecule, its capacity in in-silico studies has been tried to be revealed. The results of molecular docking, spectroscopic and theoretical techniques provide better information about inhibitor-receptor interactions. The rational design of drugs developed against dangerous diseases and understanding the mechanisms of these drugs at the molecular level provide valuable information on drug-receptor interactions. From molecular docking analysis, CCPEHP compound was fitted snuggly into the sites of COVID-19 Mpro and exhibited Pi-Pi T-Shaped, Pi-alkyl type and hydrogen bond interactions. CCPEHP successfully docked with the amino acid molecule of the main protease of SARS-CoV2 with a high negative binding affinity. In conclusion, we hope that the successful in-silico results we obtained in the CCPEHP molecule will be confirmed by in vitro and in vivo studies.
Authors' contribution
T. Topal: designed, did the in silico and theoretical calculation studies, Writing, supervised the project and edited the manuscript. N. Karapınar: did the experimental work and spectroscopic analysis. Y. Zorlu: Methodology, Visualization, Writing, did the X-ray analysis and refinement.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
I thank lecturer Süleyman Çelik for his contributions. The calculations reported in this article were completely/partially carried out at TUBITAK ULAKBIM, Grid Computing Center and High Performance (TRUBA resources).
References
- 1.Sangeetha M., Mathammal R. A complete synergy on the experimental and theoretical study of the pyridine derivatives - 2-hydroxy-5-nitropyridine and 2-chloro-5-nitropyridine. J. Mol. Struct. 2016;1117:121–134. [Google Scholar]
- 2.Sivaprakash S., Prakash S., Mohan S., Jose S.P. Quantum chemical studies and spectroscopic investigations on 2-amino-3-methyl-5-nitropyridine by density functional theory. Heliyon. 2019;5 doi: 10.1016/j.heliyon.2019.e02149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kose E., Atac A., Karabacak M., Karaca C., Eskici M., Karanfil A. Synthesis, spectroscopic characterization and quantum chemical computational studies of (S)-N-benzyl-1-phenyl-5-(pyridin-2-yl)-pent-4-yn-2- amine. Spectrochim. Acta - Part A Mol. Biomol. Spectrosc. 2012;97:435–448. doi: 10.1016/j.saa.2012.06.041. [DOI] [PubMed] [Google Scholar]
- 4.Boopathi M., Udhayakala P., Devi T.S.R., Rajendiran T.V. Vibrational spectroscopic (FT-IR, FT-Raman and NMR) and DFT analysis of 2-methoxy-3- (trifluoromethyl) pyridine. J. Chem. Pharm. Res. 2015;7:1172–1183. [Google Scholar]
- 5.Rollas S., Küçükgüzel Ş.G. Biological activities of hydrazone derivatives. Molecules. 2007;12:1910–1939. doi: 10.3390/12081910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.ATALAY Ş. 4-((4- (tert-bütil) benziliden) amino)-1,5-dimetil-2-fenil-1,2-dihidro-3H-pirazol-3-on'un X-ışını ve Teorik Çalışmaları. Süleyman Demirel Üniversitesi Fen Edeb. Fakültesi Fen Derg. 2018;13:108–120. [Google Scholar]
- 7.Muthu S., Elamurugu Porchelvi E., FTIR FT-RAMAN. NMR, spectra, normal co-ordinate analysis, NBO, NLO and DFT calculation of N, N-diethyl-4-methylpiperazine-1-carboxamide molecule. Spectrochim. Acta - Part A Mol. Biomol. Spectrosc. 2013;115:275–286. doi: 10.1016/j.saa.2013.06.011. [DOI] [PubMed] [Google Scholar]
- 8.Uzun S., Demircioğlu Z. Bis[2-(metilamino)troponato]bakır(II) Molekülünün Yapısal ve Elektronik Özelliklerinin Deneysel ve Kuramsal Analizleri. Süleyman Demirel Üniversitesi Fen Edeb. Fakültesi Fen Derg. 2020;15:9–22. [Google Scholar]
- 9.Xu Z., Peng C., Shi Y., Zhu Z., Mu K., Wang X., Zhu W. Nelfinavir was predicted to be a potential inhibitor of 2019-nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation. BioRxiv. 2020;1201:0–2. [Google Scholar]
- 10.Bello M. Prediction of potential inhibitors of the dimeric SARS-CoV2 main proteinase through the MM/GBSA approach. J. Mol. Graph. Model. 2020;101 doi: 10.1016/j.jmgm.2020.107762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derwand R., Scholz M. Does zinc supplementation enhance the clinical efficacy of chloroquine/hydroxychloroquine to win todays battle against COVID-19? Med. Hypotheses. 2020;142:1–9. doi: 10.1016/j.mehy.2020.109815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2 doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nimgampalle M., Devanathan V., Saxena A. Screening of chloroquine, hydroxychloroquine and its derivatives for their binding affinity to multiple SARS-CoV-2 protein drug targets. J. Biomol. Struct. Dyn. 2020:1102. doi: 10.1080/07391102.2020.1782265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zavodszky M.I. Side-chain flexibility in protein-ligand binding: the minimal rotation hypothesis. Protein Sci. 2005;14:1104–1114. doi: 10.1110/ps.041153605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beura S., Chetti P. In-silico strategies for probing chloroquine based inhibitors against SARS-CoV-2. J. Biomol. Struct. Dyn. 2020:1102. doi: 10.1080/07391102.2020.1772111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai Y.-.Y., Xu L.-.Y., Chai L.-.Q., Li Y.-.X. Synthesis, crystal structure, TD/DFT calculations and Hirshfeld surface analysis of 1-(4-((Benzo)dioxol-5-ylmethyleneamino)phenyl)ethanone oxime. J. Mol. Struct. 2020;1204 [Google Scholar]
- 18.Gatfaoui S., Sagaama A., Issaoui N., Roisnel T., Marouani H. Synthesis, experimental, theoretical study and molecular docking of 1-ethylpiperazine-1,4-diium bis(nitrate) Solid State Sci. 2020;106 [Google Scholar]
- 19.Venkateshan M., Muthu M., Suresh J., Kumar R.R. Azafluorene derivatives as inhibitors of SARS CoV-2 RdRp: synthesis, physicochemical, quantum chemical, modeling and molecular docking analysis. J. Mol. Struct. 2020:1220. doi: 10.1016/j.molstruc.2020.128741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Diffraction Rigaku Oxford. Rigaku Corporation; Oxford, UK: 2015. CrysAlisPro Software System, Version 1.171. [Google Scholar]
- 21.Topal T. Synthesis, crystallographic structure, Hirshfeld Surface analysis, drug-likeness properties and molecular docking studies of new oxime-pyridine compounds. Acta Chim. Slov. 2021;68:88–101. [PubMed] [Google Scholar]
- 22.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R. Vol. 201. Gaussian, Inc.; Wallingford CT: 2009. (etc., Gaussian 09, Revision d. 01). [Google Scholar]
- 23.Kohn W., Sham L.J. Self-consistent equations including exchange and correlation effects. Phys. Rev. 1965;140:A1133–A1138. [Google Scholar]
- 24.Jamróz M.H. Vibrational energy distribution analysis (VEDA): scopes and limitations. Spectrochim. Acta. A. Mol. Biomol. Spectrosc. 2013;114:220–230. doi: 10.1016/j.saa.2013.05.096. [DOI] [PubMed] [Google Scholar]
- 25.Casida M.E., Casida K.C., Salahub D.R. Excited-state potential energy curves from time-dependent density-functional theory: a cross section of Formaldehyde’s 1A1 manifold. Int. J. Quantum Chem. 1998;70:919–924. [Google Scholar]
- 26.Murray J.S., Sen K. 1st ed. Elsevier; 1996. Molecular Electrostatic potentials: Concepts and Applications. [Google Scholar]
- 27.E.D. Glendening, A.E. Reed, J.E. Carpenter, F. and Weinhold, NBO Version 3.1.
- 28.Sas E.B., Kurt M., Can M., Horzum N., Atac A. Spectroscopic studies on 9H-carbazole-9-(4-phenyl) boronic acid pinacol ester by DFT method. J. Mol. Struct. 2016;1118:124–138. [Google Scholar]
- 29.GaussView, Version 6, Dennington, Roy; Keith, Todd A.; Millam, John M. Semichem Inc., Shawnee Mission, KS, (2016).
- 30.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.a., Cheeseman J.R., Scalmani G., Barone V., Petersson G.a., Nakatsuji H., Li X., Caricato M., Marenich a.V., Bloino J., Janesko B.G., Gomperts R., Mennucci B., Hratchian H.P., Ortiz J.V., Izmaylov a.F., Sonnenberg J.L., Williams, Ding F., Lipparini F., Egidi F., Goings J., Peng B., Petrone A., Henderson T., Ranasinghe D., Zakrzewski V.G., Gao J., Rega N., Zheng G., Liang W., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Throssell K., Montgomery Jr. J.a., Peralta J.E., Ogliaro F., Bearpark M.J., Heyd J.J., Brothers E.N., Kudin K.N., Staroverov V.N., Keith T.a., Kobayashi R., Normand J., Raghavachari K., Rendell a.P., Burant J.C., Iyengar S.S., Tomasi J., Cossi M., Millam J.M., Klene M., Adamo C., Cammi R., Ochterski J.W., Martin R.L., Morokuma K., Farkas O., Foresman J.B., Fox D.J. Gaussian, Inc.; Wallingford CT: 2016. Gaussian 09, Revision A.02. [Google Scholar]
- 31.Schmidt M.W., Baldridge K.K., Boatz J.A., Elbert S.T., Gordon M.S., Jensen J.H., Koseki S., Matsunaga N., Nguyen K.A., Su S., Windus T.L., Dupuis M., Montgomery Jr J.A. General atomic and molecular electronic structure system. J. Comput. Chem. 1993;14:1347–1363. [Google Scholar]
- 32.Wolff S.K., Grimwood D.J., McKinnon J.J., Turner M.J., Jayatilaka D., Spackman M.A. Univ. West. Aust. Crawley, Aust.; 2012. Crystal Explorer. [Google Scholar]
- 33.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 34.Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009:31. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dassault Systemes BIOVIA. Dassault Systemes; San Diego: 2017. Discovery Studio Modeling Environment, Release. [Google Scholar]
- 36.Das N., Jena P.K., Pradhan S.K. Arabinosyltransferase C enzyme of Mycobacterium tuberculosis, a potential drug target: an insight from molecular docking study. Heliyon. 2020;6:e02693. doi: 10.1016/j.heliyon.2019.e02693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chowdhury P., Pathak P. Neuroprotective immunity by essential nutrient “Choline” for the prevention of SARS CoV2 infections: an in silico study by molecular dynamics approach. Chem. Phys. Lett. 2020;761 doi: 10.1016/j.cplett.2020.138057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.S. Khaerunnisa, H. Kurniawan, R. Awaluddin, S. Suhartati, Potential inhibitor of COVID-19 main protease (M pro) from several medicinal plant compounds by molecular docking study, Preprints. (2020) 1–14.
- 39.Yılmaz Z.T., Odabaşoğlu H.Y., Şenel P., Adımcılar V., Erdoğan T., Özdemir A.D., Gölcü A., Odabaşoğlu M. A novel 3-((5-methylpyridin-2-yl)amino)isobenzofuran-1(3H)-one: molecular structure describe, X-ray diffractions and DFT calculations, antioxidant activity, DNA binding and molecular docking studies. J. Mol. Struct. 2020:1205. [Google Scholar]
- 40.Sheldrick G.M. SHELXT - integrated space-group and crystal-structure determination. Acta Crystallogr. Sect. A. 2015;71:3–8. doi: 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheldrick G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. Sect. C Struct. Chem. 2015;71:3–8. doi: 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dolomanov O.V., Bourhis L.J., Gildea R.J., Howard J.A.K., Puschmann H. OLEX2 : a complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009;42:339–341. [Google Scholar]
- 43.Spek A.L. Structure validation in chemical crystallography. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009;65:148–155. doi: 10.1107/S090744490804362X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Macrae C.F., Edgington P.R., McCabe P., Pidcock E., Shields G.P., Taylor R., Towler M., Van De Streek J. Mercury: visualization and analysis of crystal structures. J. Appl. Crystallogr. 2006;39:453–457. [Google Scholar]
- 45.Orrego-Hernández J., Portilla J. Synthesis of dicyanovinyl-substituted 1-(2-pyridyl)pyrazoles: design of a fluorescent chemosensor for selective recognition of cyanide. J. Org. Chem. 2017;82:13376–13385. doi: 10.1021/acs.joc.7b02460. [DOI] [PubMed] [Google Scholar]
- 46.Albayati M.R., Kansız S., Dege N., Kaya S., Marzouki R., Lgaz H., Salghi R., Ali I.H., Alghamdi M.M., Chung I.M. Synthesis, crystal structure, Hirshfeld surface analysis and DFT calculations of 2-[(2,3-dimethylphenyl)amino]-N’-[(E)-thiophen-2-ylmethylidene]benzohydrazide. J. Mol. Struct. 2020:1205. [Google Scholar]
- 47.Agarwal P., Choudhary N., Gupta A., Tandon P. Density functional theory studies on the structure, spectra (FT-IR, FT-Raman, and UV) and first order molecular hyperpolarizability of 2-hydroxy-3-methoxy-N-(2-chloro-benzyl)-benzaldehyde-imine: comparison to experimental data. Vib. Spectrosc. 2013;64:134–147. [Google Scholar]
- 48.Konakanchi R., Jyothi P., Kotha L.R. Investigation of structures, FTIR, FT-Raman, in vivo anti-inflammatory, molecular docking and molecular characteristics of 2-amino-3-pyridine carboxaldehyde and its copper(ii) complex using experimental and theoretical approach. Polycycl. Aromat. Compd. 2020;0:1–23. [Google Scholar]
- 49.Topal T., Kart H.H., Tunay Taşlı P., Karapınar E. Synthesis and structural study on (1E,2E,1′E,2′E)-3,3′-bis[(4-bromophenyl)-3,3′-(4-methy-1,2-phenylene diimine)] acetaldehyde dioxime: a combined experimental and theoretical study. Opt. Spectrosc. 2015;118:897–912. [Google Scholar]
- 50.Karabacak M., Calisir Z., Kurt M., Kose E., Atac A. The spectroscopic (FT-IR, FT-Raman, dispersive Raman and NMR) study of ethyl-6-chloronicotinate molecule by combined density functional theory. Spectrochim. Acta - Part A Mol. Biomol. Spectrosc. 2016;153:754–770. doi: 10.1016/j.saa.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 51.Dayı B., Onac C., Kaya A., Akdogan H.A., Rodriguez-Couto S. New type biomembrane: transport and biodegradation of reactive textile dye. ACS Omega. 2020;5:9813–9819. doi: 10.1021/acsomega.9b04433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swarnalatha N., Gunasekaran S., Muthu S., Nagarajan M. Molecular structure analysis and spectroscopic characterization of 9-methoxy-2H-furo[3,2-g]chromen-2-one with experimental (FT-IR and FT-Raman) techniques and quantum chemical calculations. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2015;137:721–729. doi: 10.1016/j.saa.2014.08.125. [DOI] [PubMed] [Google Scholar]
- 53.Sebastian S., Sylvestre S., Sundaraganesan N., Amalanathan M., Ayyapan S., Oudayakumar K., Karthikeyan B. Vibrational spectra, molecular structure, natural bond orbital, first order hyperpolarizability, TD-DFT and thermodynamic analysis of 4-amino-3-hydroxy-1- naphthalenesulfonic acid by DFT approach. Spectrochim. Acta - Part A Mol. Biomol. Spectrosc. 2013;107:167–178. doi: 10.1016/j.saa.2013.01.041. [DOI] [PubMed] [Google Scholar]
- 54.Wilmshurst J., Bernstein H. The vibrational spectra of pyridine, pyridine-4-d, pyridine-2,6-d2, and pyridine-3,5-d2. Can. J. Chem. 2011;35:1183–1194. [Google Scholar]
- 55.Higuchi S., Tsuyama H., Tanaka S., Kamada H. Some considerations on the out-of-plane vibration bands of PhnX type molecules in the 800-670 cm−1 region in relation to the estimation of the twist angle θ of benzente rings from their intensities. Spectrochim. Acta Part A Mol. Spectrosc. 1974;30:463–477. [Google Scholar]
- 56.Socrates G. 3rd ed. Wiley; Chichester: 2001. Infrared and Raman Characteristic Group frequencies : Tables and Charts. [Google Scholar]
- 57.Karabacak M., Sinha L., Prasad O., Asiri A.M., Cinar M., Shukla V.K., FT-IR FT-Raman. NMR, UV and quantum chemical studies on monomeric and dimeric conformations of 3,5-dimethyl-4-methoxybenzoic acid. Spectrochim. Acta - Part A Mol. Biomol. Spectrosc. 2014;123:352–362. doi: 10.1016/j.saa.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 58.Topal T. Synthesis and characterization of zinc(II) complexes with new pyridine-based ligands: crystal structure, Hirshfeld surface analysis, and molecular docking study of lung cancer cell. J. Coord. Chem. 2020;73:3203–3222. [Google Scholar]
- 59.Hirshfeld F.L. Bonded-atom fragments for describing molecular charge densities. Theor. Chim. Acta. 1977;44:129–138. [Google Scholar]
- 60.Ivachtchenko A.V., Mitkin O.D., Kravchenko D.V., Kovalenko S.M., Shishkina S.V., Bunyatyan N.D., Konovalova I.S., Dmitrieva I.G., Ivanov V.V., Langer T. Synthesis, X-ray crystal structure, Hirshfeld surface analysis, and molecular docking study of novel inhibitor of hepatitis B: methyl 4-fluoro-3-(morpholinosulfonyl)benzo[b]thiophene-2-carboxylate. Heliyon. 2019;5:e02738. doi: 10.1016/j.heliyon.2019.e02738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spackman M.A., Jayatilaka D. Hirshfeld surface analysis. Cryst. Eng. Comm. 2009;11:19–32. [Google Scholar]
- 62.Wu Q., Tang Y., Zi Q. Synthesis, crystallographic structure, Hirshfeld surface analysis and DFT calculations of two salen-type halogenated Schiff-base Ni(II) complexes. Polyhedron. 2019;166:123–129. [Google Scholar]
- 63.Sebastian S., Sundaraganesan N. The spectroscopic (FT-IR, FT-IR gas phase, FT-Raman and UV) and NBO analysis of 4-Hydroxypiperidine by density functional method. Spectrochim. Acta - Part A Mol. Biomol. Spectrosc. 2010;75:941–952. doi: 10.1016/j.saa.2009.11.030. [DOI] [PubMed] [Google Scholar]
- 64.Kavitha T., Velraj G. Structural, spectroscopic (FT-IR, FT-Raman, NMR) and computational analysis (DOS, NBO, Fukui) of 3,5-dimethylisoxazole and 4-(chloromethyl)-3,5-dimethylisoxazole: a DFT study. J. Theor. Comput. Chem. 2016:15. [Google Scholar]
- 65.Kaddouri Y., Abrigach F., Yousfi E.B., El Kodadi M., Touzani R. New thiazole, pyridine and pyrazole derivatives as antioxidant candidates: synthesis, DFT calculations and molecular docking study. Heliyon. 2020;6 doi: 10.1016/j.heliyon.2020.e03185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abraham C.S., Prasana J.C., Muthu S. Quantum mechanical, spectroscopic and docking studies of 2-Amino-3-bromo-5-nitropyridine by density functional method. Spectrochim. Acta - Part A Mol. Biomol. Spectrosc. 2017;181:153–163. doi: 10.1016/j.saa.2017.03.045. [DOI] [PubMed] [Google Scholar]
- 67.Domingo L.R., Ríos-Gutiérrez M., Pérez P. Applications of the conceptual density functional theory indices to organic chemistry reactivity. Molecules. 2016;21 doi: 10.3390/molecules21060748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pearson R.G. Absolute electronegativity and hardness: applications to organic chemistry. J. Org. Chem. 1989;54:1423–1430. [Google Scholar]
- 69.Madhankumar S., Muthuraja P., Dhandapani M. Structural characterization, quantum chemical calculations and Hirshfeld surface analysis of a new third order harmonic organic crystal: 2-Amino-4-methylpyridinium benzilate. J. Mol. Struct. 2020;1201 [Google Scholar]
- 70.Yuriev E., Agostino M., Ramsland P.A. Challenges and advances in computational docking: 2009 in review. J. Mol. Recognit. 2011;24:149–164. doi: 10.1002/jmr.1077. [DOI] [PubMed] [Google Scholar]
- 71.Rao P., Shukla A., Parmar P., Rawal R.M., Patel B., Saraf M., Goswami D. Reckoning a fungal metabolite, Pyranonigrin A as a potential Main protease (Mpro) inhibitor of novel SARS-CoV-2 virus identified using docking and molecular dynamics simulation. Biophys. Chem. 2020;264 doi: 10.1016/j.bpc.2020.106425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marinho E.M., Batista de Andrade Neto J., Silva J., Rocha da Silva C., Cavalcanti B.C., Marinho E.S., Nobre Júnior H.V. Virtual screening based on molecular docking of possible inhibitors of Covid-19 main protease. Microb. Pathog. 2020;148:1–6. doi: 10.1016/j.micpath.2020.104365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Noureddine O., Issaoui N., Al-Dossary O. DFT and molecular docking study of chloroquine derivatives as antiviral to coronavirus COVID-19. J. King Saud Univ. - Sci. 2021;33 doi: 10.1016/j.jksus.2020.101248. [DOI] [PMC free article] [PubMed] [Google Scholar]