Abstract
Mollusca are the second largest and arguably most diverse phylum of the animal kingdom. This is in sharp contrast to our very limited knowledge concerning epigenetic mechanisms including DNA methylation in this invertebrate group. Here, we inferred DNA methylation patterns by analysing the normalized dinucleotide CG content in protein-coding sequences and identified DNA methyltransferases (DNMT1 and 3) in published transcriptomes and genomes of 140 species across all eight classes of molluscs. Given the evolutionary age and morphological diversity of molluscs, we expected to find evidence for diverse methylation patterns. Our inferences suggest that molluscs possess substantial levels of DNA methylation in gene bodies as a rule. Yet, we found deviations from this general picture with regard to (i) the CpG observed/expected distributions indicating a reduction in DNA methylation in certain groups and (ii) the completeness of the DNMT toolkit. Reductions were evident in Caudofoveata, Solenogastres, Polyplacophora, Monoplacophora, as well as Scaphopoda. Heterobranchia and Oegopsida were remarkable as they lacked DNMT3, usually responsible for de novo methylation, yet showed signs of DNA methylation. Our survey may serve as guidance for direct empirical analyses of DNA methylation in molluscs.
This article is part of the Theo Murphy meeting issue ‘Molluscan genomics: broad insights and future directions for a neglected phylum’.
Keywords: CpG observed/expected distribution, DNA methylation, DNA methyltransferase, epigenetics, molluscs
1. Introduction
Epigenetic modifications are reversible changes to the DNA/RNA or histones of a cell that, among others, affect gene function without entailing changes in DNA/RNA sequence. The most important mechanisms include DNA methylation, histone modifications, nucleosome remodelling and RNA-mediated modifications [1,2]. When mitotically or meiotically inherited, these modifications may play a fundamental role in the development and adaptation of organisms [3]. In animals, the most comprehensively investigated mechanism is DNA methylation, i.e. the covalent bonding of methyl groups to cytosine residues, mostly to CpG dinucleotides [4,5]. DNA methylation is functionally associated with the regulation of gene expression and the silencing of transposable element expression. It is relevant for development, adaptation (including phenotypic plasticity), stress compensation, chromosomal inactivation, and parental imprinting or memory formation, to mention just some of its functions [6–11].
Cytosine methylation in animals is mediated by two types of DNA methyltransferases (DNMTs; [12]). DNMT1 is traditionally known to perform maintenance methylation and DNMT3 is responsible for de novo methylation. However, current studies in mammalian systems show that both can be involved in de novo and maintenance methylation. [13,14]. By contrast, DNMT2, also known as tRNA aspartic acid methyltransferase 1, is not active on the DNA level [12,13,15,16]. By contrast with vertebrates, the DNMT toolkit of some invertebrate taxa, mostly insects, relies on DNMT1 homologues for both de novo and maintenance methylation [17–19]. In molluscs, the DNA methylation machinery has only been studied in a few species (Crassostrea gigas: [20]; Biomphalaria glabrata: [21]; Patinopecten yessoensis: [22]). Thus, the distribution of DNMTs across all eight classes of molluscs is unknown.
Our knowledge on the role of DNA methylation is largely restricted to model organisms, [5,23,24]. Comprehensive large-scale analyses, such as those made by Provataris et al. [18] for insects allowing for the inference of macro-evolutionary patterns, are still exceptional. Aliaga et al. [24] inferred the universality of what they call the DNA methylation codes across eukaryotes. However, since many taxa are only sparsely represented, e.g. molluscs by one snail and one mussel each, and the normalized CpG dinucleotide content among and within taxa varies considerably, the conclusion of universality may be premature.
In a recent review, Fallet et al. [25] collated our knowledge about DNA methylation (and histone modifications) in molluscs. DNA methylation has been directly investigated by bisulfite sequencing or immunoprecipitation in a total of only 16 species of molluscs: eight gastropods, seven bivalves and one cephalopod. The analysis of an eighth bivalve was published later [26]. Genome-wide analyses are restricted to the freshwater gastropod B. glabrata [27,28] and the congeneric marine bivalves C. gigas and C. virginica [20,26,29,30]. In addition, the normalized dinucleotide CG content has been analysed in the marine limpet Lottia gigantea [24]. These four species exhibit mosaic DNA methylation, i.e. stretches of hypermethylated DNA alternate with hypomethylated stretches. The former largely characterize housekeeping genes, while the latter have predominately inducible expression [31]. In invertebrates in general and in B. glabrata as well as C. gigas and C. virginica, methylation is mainly targeted to gene bodies, while intergenic regions are less methylated [20,21,26,28,29,32]. Evidence for regulatory consequences of differential promoter methylation is ambiguous (summarized in [25]). In molluscs, variation in DNA methylation is essential in development (e.g. [21,33]), memory formation (e.g. [11]) and adaptation (e.g. [34,35]) related to ageing (e.g. [36,37]); and induced by biotic (e.g. [38]) and abiotic stressors (e.g. [39,40]). For a comprehensive compilation, see Fallet et al. [25].
Considering that Mollusca is the second largest animal phylum with greater than 125 000 described species, many of them of economic and medical importance [41], we have hardly scratched the surface in this area of research: with gastropods, bivalves and one cephalopod, only three of the eight classes of molluscs are represented among the above studies. Molluscs, probably rooted in the late Precambrian, are notorious for their morphological disparity, which is linked to their ecological success, as they have colonized practically all types of marine, freshwater and terrestrial habitats [41–43]. Given the diversity on multiple levels, we are obviously well advised not to draw any general conclusions on DNA methylation and its implications in molluscs at this stage.
To provide a basis for future empirical methylation studies leading to a better understanding of the evolution and function of DNA methylation in molluscs, we used transcriptomic and genomic data of molluscan species from all classes to infer the status of DNA methylation by calculating the CpG observed/expected (o/e) ratio and to characterize the DNMT toolkit. The CpG o/e approach is based on the known hyper-mutability of methylated cytosines, which readily deaminate to thymine residues [44]. Regions that have been historically methylated in the germline of a species are thus depleted of CpG dinucleotides over evolutionary time. DNA methylation prediction by CpG o/e ratio is well established and correspondence to directly inferred methylation and gene expression has been demonstrated for a variety of organisms [32,45–50] including, among molluscs, B. glabrata as well as C. gigas and C. virginica [20,26–28,31,51]. Given the evolutionary age and morphological diversity of molluscs, we expected to find evidence for diverse methylation patterns and mechanisms similar to what has been reported for other invertebrate groups [17–19].
2. Methods
(a). Data acquisition
Transcriptomic and genomic (predicted coding sequences (CDS) and proteins of structural annotations) data publicly available at the end of the year 2019 were downloaded from various sources. During the course of analysis, we added a few important taxa whose genomes or transcriptomes became available only in 2020 (electronic supplementary material, appendix SA). Our analyses were mostly based on assembled and, in the case of genomic data, annotated sequences. However, in order to put together a taxonomically comprehensive dataset, we performed de novo assemblies of RNAseq data and structural annotation of genome assemblies to cover all mollusc classes. Publicly available RNAseq data of 14 mollusc species were downloaded and assembled (electronic supplementary material, appendix SB). The transcriptome assemblies were carried out using Trinity 2.11.0 [52]. To approximate the completeness of the transcriptome assemblies, we used BUSCO 4.1.4 [53] with the provided metazoa_odb10 dataset (for more details see electronic supplementary material, appendix SB). Additionally, we carried out structural annotation of publicly available genome assemblies from eight mollusc species using GeMoMa 1.6.4 [54,55]. The quality of the final annotations was evaluated by checking the annotation contiguity with a custom script and BUSCO 4.1.4 (see electronic supplementary material, appendix SB). In total, we analysed annotations of 31 genomes and 124 transcriptomes of a total of 140 species covering all extant mollusc classes. The presence of DNMTs and the CpG o/e ratios were analysed in 126 and 136 species, respectively. Taxonomic information of all investigated species, information on data quality and the type of data we used can be found in the electronic supplementary material, appendix SB.
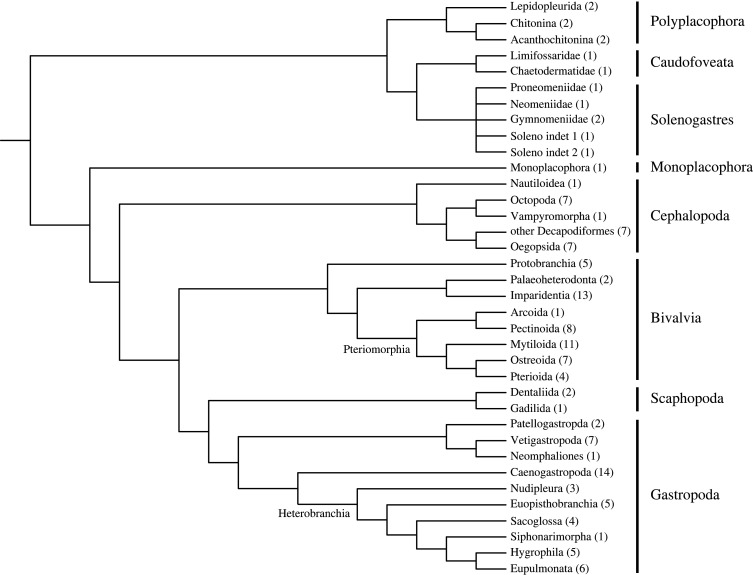
A cladogram reflecting actual insights into the phylogeny of molluscs is given in figure 1 in order to show the phylogenetic dispersion of the taxa analysed. The backbone of this cladogram was based on the most recent phylogenetic analysis of Mollusca by Kocot et al. [56]. For more details of tree construction, see electronic supplementary material, appendix SB. The tree only includes taxa represented in the data.
Figure 1.
Cladogram reflecting the phylogenetic dispersion of the analysed genomic data. The numbers in brackets indicate the species studied per taxonomic group.
(b). Identification of DNA methyltransferases
We identified genes that encode for DNMT1 and DNMT3. In our search, we also controlled for DNMT2. However, apart from reporting our findings, we do not go into further detail regarding DNMT2. As described in Provataris et al. [18], we generated Profile Hidden Markov Models (pHMMs) based on amino acid sequences of metazoan DNMT1 and DNMT3 obtained from OrthoDB v. 8 [57,58]. The search option for mollusc DNMTs produced too little output to create a suitable search pattern. We used MAFFT L-INS-i v. 7 [59] to align each group of orthologous sequences. Accordingly, we created pHMMs from each alignment using hmmbuild (package of HMMER 3.1b2; www.hmmer.org). Transcriptomic sequences were translated into their six reading frames with fastatranslate of Exonerate 2.4.0 [60]. We searched the predicted protein data (translated transcriptome or genome) of each species using the generated pHMMs of DNMT1 and DNMT3.
Subsequently, we scanned each candidate DNMT1 and DNMT3 sequence obtained in the previous step against the Pfam-A database version 32.0 using hmmscan (part of HMMER) to identify their protein domains. Only sequences that contained a DNA methylase domain (Pfam accession number: PF00145) and/or DNMT1/DNMT3 specific domains (DNMT1-RFD, Pfam accession number: PF12047, or ADD_DNMT3, Pfam accession number: PF17980) were used in downstream analyses. To distinguish between DNMT1 and DNMT3 candidate sequences, we searched the remaining candidate sequences against the official gene set of C. gigas [61] and excluded candidate sequences that did not have the corresponding C. gigas DNMT as their best match. Finally, we searched remaining candidate sequences against the non-redundant NCBI database using blastp 2.6.0+ [62] and only kept sequences whose best matches belonged to metazoan species.
In order to control for a relationship between data quality and detection of DNMTs, we constructed generalized linear models (GLMs) with binary response variable (presence/absence) and logit link function in PAST 4.01 [63]. Independent variables reflecting quality were BUSCO C (complete), 100 – BUSCO M (missing) = BUSCO present, number of contigs and total sequence length (electronic supplementary material, appendix SC).
(c). Calculation of normalized CpG dinucleotide content
In genes that have been historically methylated, CG dinucleotides tend to be underrepresented, while in genes that have been unaffected by DNA methylation CG dinucleotides are present at a relatively higher frequency [64]. Thus, if methylation is present in only a part of the gene repertoire of a species, two classes of genes will be present: one depleted of CG dinucleotides and another one unaffected by CpG depletion. By contrast, species with very little or no DNA methylation are expected to have only one class of genes that is unaffected by CpG depletion. CpG depletion of genes can be quantified by calculating the normalized CpG dinucleotide content provided by the following equation:
where PCpG, PC and PG are the frequencies of 5′-CpG-3 dinucleotides, C nucleotides and G nucleotides, respectively. In addition, the normalized GpC dinucleotide content was calculated to control for causative factors unrelated to cytosine DNA methylation, like GC content [65]. Sequences containing less than 200 nucleotides or greater than 5% ambiguous nucleotides (N) were excluded from the calculation of the normalized dinucleotide content.
(d). Inferring the presence of DNA methylation based on CpG observed/expected distributions
To infer the presence of DNA methylation in mollusc species, we calculated the CpG o/e of protein-coding sequences. The modality of the CpG o/e distributions was tested using the Gaussian mixture modelling software package mclust (5.4.6 [66] in R 4.0.3 ([67]; see [48]) and fitted two Gaussian distributions to the CpG o/e and GpC o/e distributions of the respective species. To conclude on the presence of germline DNA methylation in the protein-coding sequences of a species, we followed the criteria established by Provataris et al. [18]:
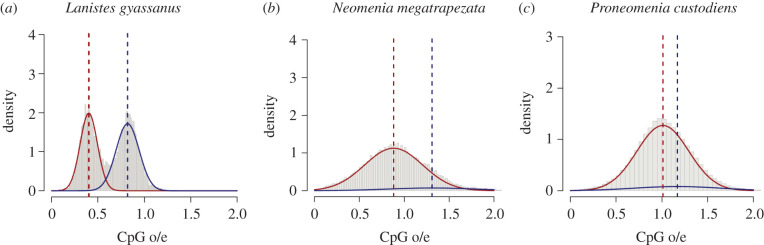
1. A CpG o/e distribution is bimodal if the absolute difference of the means of the two Gaussian distributions is at least 0.25 and one of the means less than 0.7. Also the proportion of data to the smallest of the fitted components should be greater than 0.1. These criteria of bimodality should not apply to the GpC o/e distribution, because GpC dinucleotides are not affected by DNA methylation. A CpG o/e distribution that meets these criteria is called ‘bimodal depleted' (figure 2a).
2. If clear bimodality is not observed, the criteria of bimodality do not apply. Based on the findings in insects that did not show a bimodal distribution but had experimentally verified cytosine methylation, a large part of the data belongs to the smaller of the two fitted distributions [32,47,50]. Therefore, the threshold for the proportion of the smaller of the fitted normal distributions was set to ≥0.36 (based on experimental data of Bombyx mori [68]). Again, this should not hold for the corresponding GpC o/e distribution. The CpG o/e distributions of such species are called ‘unimodal, indicative of DNA methylation' (figure 2b).
3. If the above criteria were not met, we considered the evidence as insufficient to conclude the presence of DNA methylation. Such CpG o/e distributions are described as ‘unimodal, not indicative of DNA methylation' (figure 2c).
Figure 2.
Examples of CpG o/e distribution patterns in protein-coding sequences in three mollusc species. The red and blue curves represent the two Gaussian distributions fitted to the data. The correspondingly coloured dashed lines represent the mean values of the fitted distributions. (a) Lanistes nyassanus shows a typical bimodal CpG o/e distribution with a portion that has low CpG o/e values (sequences mainly affected by CpG depletion) and one that has high CpG o/e values (sequences less affected by CpG depletion) (b) Neomenia megatrapezata displays a ‘unimodal, indicative of methylation' CpG o/e distribution. This distribution lacks clear bimodality, but their low CpG o/e component has a characteristically large tail containing a significant amount of data. (c) Proneomenia custodiens shows a ‘unimodal, not indicative of DNA methylation' CpG o/e distribution. The mean values of the two fitted distributions are almost identical and the proportion of data belonging to the smaller component is small. (Online version in colour.)
3. Results
(a). The distribution of DNA methyltransferases in molluscs
We found homologues of all three DNMTs in the transcriptomes and genomes of all eight mollusc classes (electronic supplementary material, appendix SC; in particular for DNMT2, which is otherwise not further mentioned). Only in nine of the 140 species investigated neither DNMT1 nor DNMT3 were found. These were single, distantly related representatives from different classes: (i) the cephalopods Sepia pharaonis and Sepioloidea lineolata; (ii) the bivalves Eurhomalea rufa, Mytilus galloprovincialis, Mytilus trossulus and Septifer virgata; and (iii) the gastropods Haliotis fulgens, Plakobranchus ocellatus as well as Siphonaria pectinata. In the following three species, DNMT1 was not found: (i) the cephalopd Sepiella maindroni, as well as in the two bivalves (ii) Margaritifera margaritifera and (iii) Placopecten magellanicus. In all other species, we did detect a DNMT1 homologue. By contrast, DMNT3 homologues were consistently not found in all investigated representatives of certain taxonomic groups: these were (i) within Cephalopoda, the order Oegopsida with seven species from five different families (Chiroteuthis calyx, Dosidicus gigas and Stenoteuthis oualaniensis, Octopoteuthis deletron, Onychoteuthis banksii, as well as Pterygioteuthis hoylei and Watasenia scintillans); and (ii) within Gastropoda, the sub-class Heterobranchia with 21 not closely related species (Melibe leonina, Phylliroe bucephala, Aplysia californica, Limacina antarctica, L. helicina, L. retroversa, Clione limacina, Elysia chlorotica, E. cornigera, E. timida, Plakobranchus ocellatus, Siphonaria pectinata, B. glabrata, B. pfeifferi, Lymnaea stagnalis, Physella acuta, Radix auricularia, Achatina fulica, Arion vulgaris, Bradybaena similaris and Cepaea nemoralis). Furthermore, DNMT3 was not detected in individual species of different taxonomic groups: one caudofoveate, three solenogastres, two more polyplacophorans, one additional decapodiform cephalopod, another twelve bivalves, two scaphopods, three vetigastropods, one patellogastropod and eight caenogastropods (for details see electronic supplementary material, appendix SC).
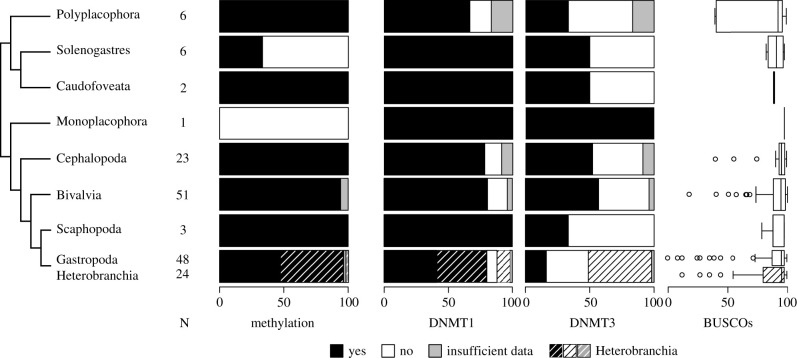
The GLMs predicting presence/absence of DNMTs based on quality scores for the transcriptomes and genomes clearly suggested a direct relationship. The detection of DNMT1 depended on complete BUSCOs, present BUSCOs and total sequence length (G = 14.476, p = 0.00014; G = 17.821, p < 0.00005; G = 6.718, p = 0.00954). The number of contigs was not decisive (G = 2.725, p = 0.09882). The presence of DNMT3 was predicted by complete BUSCOs and present BUSCOs (G = 11.47, p = 0.00071; G = 8.048, p = 0.00456). Number of contigs and total sequence length did not have an influence (G = 0.016, p = 0.90095; G = 1.297, p = 0.25477). The distribution of DNMTs and BUSCO completeness across classes is summarized in figure 3.
Figure 3.
Methylation in mollusc classes. From left to right: Topology displaying the relationships between the eight mollusc classes and the sub-class Heterobranchia; number of analysed species; percentage of species showing methylation derived from CpG o/e analyses; percentage of species showing DNMT1 and DNMT3 presence; boxplot of present BUSCOs percentages (present = 100 - missing; Nmetazoa_odb10 = 954). Heterobranchia are displayed in stacked barplots as shaded subgroups of Gastropoda. In the boxplots, the upper boxplot of Gastropoda reflects the distribution across all analysed gastropods and the lower shaded one the distribution within Heterobranchia.
(b). CpG observed/expected distributions
To infer the presence of DNA methylation in molluscs, we calculated the CpG o/e ratio of protein-coding sequences in 140 species covering all mollusc classes. Our results suggest that DNA methylation is widespread in the phylum as it was present in seven out of eight classes. Only in the class Monoplacophora, where we could examine just one representative (Laevipilina hyalina), did we find no evidence of DNA methylation. In addition, four out of six species in the Solenogastres showed no indication of DNA methylation: Greenland neomeniomorph, Neomeniomorpha sp1, Proneomenia custodiens and Wirenia argentea (figure 3; electronic supplementary material, appendix SC).
We found both ‘bimodal depleted' and ‘unimodal, indicative of methylation' CpG o/e distribution patterns in molluscs. While in Cephalopoda and Bivalvia, a bimodal CpG o/e distribution pattern predominated, aplacophoran species (Caudofoveata and Solenogastres) only showed unimodal CpG o/e distributions, except for Chaetoderma nitidulum. In gastropods, Littorina saxatilis and Potamopyrgus antipodarum showed ‘unimodal, indicative of methylation' CpG o/e distribution patterns. By contrast, all other gastropods had a bimodal CpG o/e distribution. In Polyplacophora and Scaphopda, we found both patterns. For more details, see electronic supplementary material, appendices SC, SD.
Solenogastres exhibited higher overall mean CpG o/e values (greater than 0.9), i.e. lower mean germline DNA methylation, in protein-coding sequences compared to the other molluscan classes except for Monoplacophora (mean CpG o/e value of almost 0.9), which showed no signature of cytosine DNA methylation. Within Caudofoveata the mean CpG o/e value was 0.66 and in Polyplacophora 0.7. In most cephalopods and bivalves, mean CpG o/e values were between 0.58 and 0.62. But there were two outliers to the bottom: (i) the cephalopod Nautilus pompilius appeared to be extremely CpG-depleted (mean CpG o/e value 0.45) and (ii) the two bivalve species Margaritifera margaritifera and Mytilus edulis displayed mean CpG o/e values of 0.41 and 0.43, respectively, indicating high CpG depletion. Furthermore, two bivalve species had a rather high mean CpG o/e value of 0.74 and 0.8, respectively (Dreissena rostriformes, D. polymorpha). In Scaphopoda, the mean CpG o/e value was 0.73 and in gastropods approximately 0.57. One upward outlier in gastropods was Potamopyrgus antipodarum with a mean CpG o/e value of 0.78 indicating less cytosine DNA methylation, whereas the gastropod species Siphonaria pectinata and B. glabrata appeared to be extremely CpG-depleted with mean CpG o/e values of 0.39 and 0.45, respectively. Mean CpG o/e values for classes are shown in the electronic supplementary material, appendix SB.
4. Discussion
Among invertebrates, DNA methylation has largely been studied in model organisms [24]. Comprehensive comparative analyses across higher, more inclusive taxa are scarce and arthropods are the notable exception among invertebrates [17,18,69]. Such large-scale analyses are partially based on indirect inference through the detection of DNMTs and evaluation of CpG o/e distributions. In the second largest phylum Mollusca, methylation has been experimentally studied in only 17 species [25,26] and Aliaga et al.'s [24] account included merely two. To our knowledge, this is the first study to comprehensively investigate methylation patterns in 140 species across all eight mollusc classes based on the normalized CpG dinucleotide content distributions and the presence of DNMTs.
(a). The distribution of DNA methyltransferases in molluscs
DNMTs were found in the majority of species and in all eight classes of molluscs. Therefore, the last common ancestor of molluscs likely possessed the enzymatic toolkit necessary for DNA methylation, which is in line with Aliaga et al. [24]. This general conclusion was also backed by the GLMs suggesting that the detection of DNMTs highly depended on the quality of the sequencing data. In particular, BUSCO values were good predictors for the presence of DNA methyltransferases. This means that the lack of DNMTs in species with low BUSCO completeness cannot be interpreted as evidence for their absence. By contrast, the presence of a DNA methyltransferase does not leave room for ambiguity. This also means that in species with high BUSCO completeness the absence of DNMTs is probably due to an evolutionary loss. This interpretation gained more weight in cases where the findings were similar among closely related species, suggesting an ancestral loss of DNMTs in these clades.
DNMT1 homologues could not be found in twelve species. By contrast, DNMT3 was not detected in 69 species from all eight classes, particularly gastropods. In most cases, the absence of DNMT1 can probably be explained by poor data quality. Only five of these species showed a BUSCO completeness greater than 80% and nine also lacked DNMT3. In all these cases, we only examined transcriptomes, which only reflect a snapshot of the genes that were currently transcribed when the respective species was fixed. Furthermore, due to the rapid degeneration of mRNA, this snapshot may comprise only a subsample of the genes expressed [70–72]. As the CpG o/e distributions of the twelve species lacking DNMT1 did indicate the presence of DNA methylation (electronic supplementary material, appendix SD), it is very likely that these species do possess the necessary DNMT toolkit.
In contrast with DNMT1, the lack of DNMT3 showed a systematic distribution. All 21 fairly distantly related representatives of the Heterobranchia (Gastropoda), all seven Oegopsida (Cephalopoda) and both species of Dreissena (Bivalvia) as well as Littorina (Gastropoda) lacked DNMT3 homologues. The largely high BUSCO completeness—only six heterobranchs and one oegopsid had values less than 85%—suggests that in these taxa the respective most recent common ancestors already lacked DNMT3. In seven Heterobranchia, this was backed by genome data. The loss of DNMT3 in these four groups apparently occurred independently given their distant relationships. The lack of DNMT3 in the heterobranchs B. glabrata and A. californica despite experimentally proven DNA methylation has already been reported [21,28]. Also, in insects, the frequent loss of DNMT3 has been documented [17,18]. It was hypothesized that the function of the DNMT3 may be compensated and taken over by DNMT1 or another enzyme. Likewise, the methylomes of these four groups of molluscs may depend on DNMT1 for both maintenance and de novo methylation. Thus, DNMT3 is evolutionarily dispensable for many invertebrate taxa and its absence does not seem to negatively impact functional methylation systems.
(b). CpG observed/expected patterns
Our inferences on the occurrence of DNA methylation were based on CpG o/e distributions of gene bodies and in particular exons/exonic sequences. This is justified through the finding that DNA methylation predominates in gene bodies throughout invertebrates. Although it is not yet well understood how gene regulation through methylation of gene bodies functions, experimental studies have established a clear relationship between gene expression and gene body methylation [73–76] including evidence from B. glabrata as well as C. gigas and C. virginica [21,26,51,77]. Given this tight correspondence, inferring DNA methylation patterns through CpG o/e distributions allows for a meaningful, phylogenetically broad survey based on existing genome and transcriptome data revealing general patterns, hence, providing a basis for future experimental research.
The results of our CpG o/e analyses showed that a bimodal CpG o/e distribution pattern was dominant in all mollusc classes except for Solenogastres and Monoplacophora, suggesting that DNA methylation is widespread in molluscs. According to this, genes in these groups appear to be divided into highly and lowly methylated genes. Unimodal CpG o/e distributions were frequently observed in Caudofoveata, Monoplacophora, Polyplacophora, Scaphopoda and Solenogastres. Especially remarkable were the Solenogastres, in which all six investigated species showed a unimodal CpG o/e distribution, with four representatives entirely lacking a methylation signature despite the presence of DNMTs. The only monoplacophoran species examined also showed no sign of methylation in its CpG o/e distribution. Thus, species belonging to these two groups likely possess a sparse or even no-DNA methylation pattern in protein-coding sequences. A similar pattern has been reported for holometabolous insects [18]. Experimental investigations on the patterns of Holometabola have shown that DNA methylation is sparsely targeted to exons of protein-coding genes (reviewed in [78]). Thus, we suggest that many representatives belonging to Caudofoveata, Monoplacophora, Polyplacophora, Scaphopoda and Solenogastres likely possess a pattern of DNA methylation that is similar to Holometabola.
In comparison, almost all representatives of the Cephalopoda, Bivalvia and Gastropoda showed a bimodal CpG o/e distribution with lower mean CpG o/e values. These results indicated high levels of DNA methylation in these groups, which was also detected experimentally in the bivalves Chlamys farreri, Crassostrea gigas, C. virginica, Pinctada fucata and Saccostrea glomerata, the cephalopod Octopus vulgaris, and the gastropods Haliotis discus hannai, A. californica, B. glabrata and Lymnaea sp. (see review of [25] and [26]). Similarly low mean CpG o/e values have been detected in many hemimetabolous insects species [17,18]. This observation, also backed by experimental evidence, leads to the hypotheses that DNA methylation levels were high in the last common ancestor of insects and were subsequently reduced in holometabolous insects [18]. However, inferring macro-evolutionary trends in molluscs is probably premature at this stage as the classes Solenogastres, Caudofoveata, Polyplacophora, Monoplacophora and Scaphopoda were represented only by low numbers of species. Ideally, such inference would be based on comprehensive experimental evidence, though, which is still a far way to go.
Interestingly, all Heterobranchia (Gastropoda) showed a bimodal CpG o/e distribution pattern indicating the presence of DNA methylation, despite all of them lacking DNMT3. This further supports the idea that the function of DNMT3 is compensated by another enzyme, likely DNMT1.
5. Conclusion
Our inferences suggest that molluscs possess substantial levels of DNA methylation in gene bodies as a rule. Yet, we did detect deviations from this general pattern both regarding CpG o/e distributions indicating reduced methylation as well as the completeness of the DNMT toolkit. These deviations were scattered across the mollusc tree indicating convergent evolution. On the other hand, we have to concede that the data basis in particular for the smaller classes is unsatisfying so that it is premature to infer general evolutionary trends of DNA methylation among Mollusca. In comparison to other groups of animals such as vertebrates and arthropods, methylomic data of molluscs are still rather scarce [25,79]. One reason for this may lie at the very beginning of such analyses: extraction of suitable DNA. The polysaccharides contained in the mucus of molluscs are known to inhibit enzymes used in DNA extractions, the preparation of sequencing libraries and even in the sequencing processes itself [80,81]. Protocols working for one species may be inappropriate for others. Such difficulties may have been responsible also for the lower quality in several of the datasets available to us. Regardless of technical problems, direct analyses of DNA methylation are the principal route of research we want to encourage. These data will be invaluable for a more comprehensive understanding of evolution and development in molluscs. Our indirect inferences provide a solid basis for targeted empirical investigations and testing of the hypotheses we presented.
Acknowledgements
We thank Andreas Wanninger (University of Vienna, Austria) for providing data and Bernhard Misof (Director of the Zoological Research Museum Alexander Koenig, Bonn, Germany) for his encouragement. We further would like to thank Angus Davison (University of Nottingham, UK) and Maurine Neimann (University of Iowa, USA) for organizing the workshop ‘Pearls of wisdom: synergising leadership and expertise in molluscan genomics' in 2019. Two anonymous referees are acknowledged for their constructive comments.
Data accessibility
Data are available as electronic supplementary material.
Authors' contributions
C.G. conceived the project, downloaded data, contributed to the CpG o/e ratio calculation and wrote the manuscript. M.H. downloaded data, performed the GLM analyses and wrote the manuscript. L.M. downloaded data, contributed to the DNMT search and wrote the manuscript. P.P. designed and contributed to analyses on the identification of DNMTs and the occurrence of DNA methylation. T.S. performed de novo transcriptome assemblies, structural annotation, BUSCO searches, contributed to the DNMT searches and contributed to the CpG o/e ratio calculation. All authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
L.M. and M.H. were involved in the Research Training Group 2010 RESPONSE funded by the Deutsche Forschungsgemeinschaft.
References
- 1.Ho DH, Burggren WW. 2010. Epigenetics and transgenerational transfer: a physiological perspective. J. Exp. Biol. 213, 3-16. ( 10.1242/jeb.019752) [DOI] [PubMed] [Google Scholar]
- 2.Becker PB, Workman JL. 2013. Nucleosome remodeling and epigenetics. Cold Spring Harbor Perspect. Biol. 5, a017905. ( 10.1101/cshperspect.a017905) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu C-T, Morris JR. 2001. Genes, genetics, and epigenetics: a correspondence. Science 293, 1103-1105. ( 10.1126/science.293.5532.1103) [DOI] [PubMed] [Google Scholar]
- 4.Mandrioli M. 2007. A new synthesis in epigenetics: towards a unified function of DNA methylation from invertebrates to vertebrates. Cell. Mol. Life Sci. 64, 2522-2524. ( 10.1007/s00018-007-7231-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts BS, Gavery MR. 2012. Is there a relationship between DNA methylation and phenotypic plasticity in invertebrates? Front. Physiol. 2, 116. ( 10.3389/fphys.2011.00116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. 2004. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900-903. ( 10.1038/nature02633) [DOI] [PubMed] [Google Scholar]
- 7.Kucharski R, Maleszka J, Foret S, Maleszka R. 2008. Nutritional control of reproductive status in honeybees via DNA methylation. Science 319, 1827-1830. ( 10.1126/science.1153069) [DOI] [PubMed] [Google Scholar]
- 8.Angers B, Castonguay E, Massicotte R. 2010. Environmentally induced phenotypes and DNA methylation: how to deal with unpredictable conditions until the next generation and after. Mol. Ecol. 19, 1283-1295. ( 10.1111/j.1365-294X.2010.04580.x) [DOI] [PubMed] [Google Scholar]
- 9.Moore TL, Le T, Fan G. 2012. DNA methylation and its basic function. Neuropsychopharmacol. 28, 23-38. ( 10.1038/npp.2012.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schübeler D. 2015. Function and information content of DNA methylation. Nature 517, 321-326. ( 10.1038/nature14192) [DOI] [PubMed] [Google Scholar]
- 11.Forest J, Sunada H, Dodd S, Lukowiak K. 2016. Training Lymnaea in the presence of a predator scent results in a long-lasting ability to form enhanced long-term memory. J. Comp. Physiol. A 202, 399-409. ( 10.1007/s00359-016-1086-z) [DOI] [PubMed] [Google Scholar]
- 12.Goll MG, Bestor TH. 2005. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481-514. ( 10.1146/annurev.biochem.74.010904.153721) [DOI] [PubMed] [Google Scholar]
- 13.Jeltsch A, Jurkowska RZ. 2014. New concepts in DNA methylation. Trends Biochem. Sci. 39, 310-318. ( 10.1016/j.tibs.2014.05.002) [DOI] [PubMed] [Google Scholar]
- 14.Jeltsch A, Jurkowska RZ. 2016. Allosteric control of mammalian DNA methyltransferases—a new regulatory paradigm. Nucleic Acids Res. 44, 8556-8575. ( 10.1093/nar/gkw723) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh C-L, Zhang X, Golic KG, Jacobsen SE, Bestor TH. 2006. Methylation of tRNA. Science 311, 395-398. ( 10.1126/science.1120976) [DOI] [PubMed] [Google Scholar]
- 16.Lyko F. 2017. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 19, 81-92. ( 10.1038/nrg.2017.80) [DOI] [PubMed] [Google Scholar]
- 17.Bewick AJ, Vogel KJ, Moore AJ, Schmitz RJ. 2017. Evolution of DNA methylation across insects. Mol. Biol. Evol. 34, 654-665. ( 10.1093/molbev/msw264) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Provataris P, Meusemann K, Niehuis O, Grath S, Misof B. 2018. Signatures of DNA methylation across insects suggest reduced DNA methylation levels in Holometabola. Genome Biol. Evol. 10, 1185-1197. ( 10.1093/gbe/evy066) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosic S, et al. 2018. Evolutionary analysis indicates that DNA alkylation damage is a byproduct of cytosine DNA methyltransferase activity. Nat. Genet. 50, 452-459. ( 10.1038/s41588-018-0061-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, et al. 2014. Genome-wide and single-base resolution DNA methylomes of the Pacific oyster Crassostrea gigas provide insight into the evolution of invertebrate CpG methylation. BMC Genomics 15, 1119. ( 10.1186/1471-2164-15-1119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geyer KK, et al. 2017. The Biomphalaria glabrata DNA methylation machinery displays spatial tissue expression, is differentially active in distinct snail populations and is modulated by interactions with Schistosoma mansoni. PLoS Negl. Trop. Dis. 11, e0005246. ( 10.1371/journal.pntd.0005246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Zhang L, Li Y, Li W, Guo Z, Li R, Hu X, Bao Z, Wang S. 2019. Dynamics of DNA methylation and DNMT expression during gametogenesis and early development of scallop Patinopecten yessoensis. Mar. Biotechnol. 21, 196-205. ( 10.1007/s10126-018-09871-w) [DOI] [PubMed] [Google Scholar]
- 23.Law JA, Jacobsen SE. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204-220. ( 10.1038/nrg2719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aliaga B, Bulla I, Mouahid G, Duval D, Grunau C. 2019. Universality of the DNA methylation codes in Eucaryotes. Sci. Rep. 9, 173. ( 10.1038/s41598-018-37407-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fallet M, Luquet E, David P, Cosseau C. 2020. Epigenetic inheritance and intergenerational effects in mollusks. Gene 729, 144166. ( 10.1016/j.gene.2019.144166) [DOI] [PubMed] [Google Scholar]
- 26.Venkataraman YR, Downey-Wall AM, Ries J, Westfield I, White SJ, Roberts SB, Lotterhos KE. 2020. General DNA methylation patterns and environmentally-induced differential methylation in the eastern oyster (Crassostrea virginica). Front. Mar. Sci. 7, 225. ( 10.3389/fmars.2020.00225) [DOI] [Google Scholar]
- 27.Fneich S, Dheilly N, Adema C, Tognon A, Reichelt M, Bulla J, Grunau C, Cosseau C. 2013. 5-methyl-cytosine and 5-hydroxy-methyl-cytosine in the genome of Biomphalaria glabrata, a snail intermediate host of Schistosoma mansoni. Parasit. Vectors 6, 167. ( 10.1186/1756-3305-6-167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adema CM, et al. 2017. Whole genome analysis of a schistosomiasis-transmitting freshwater snail. Nat. Commun. 8, 15451. ( 10.1038/ncomms15451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olson CE, Roberts SB. 2014. Genome-wide profiling of DNA methylation and gene expression in Crassostrea gigas male gametes. Front. Physiol. 5, 224. ( 10.3389/fphys.2014.00224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Riviere G, He Y, Tecchio S, Crowell E, Gras M, Sourdaine P, Guo X, Favrel P. 2017. Dynamics of DNA methylomes underlie oyster development. PLoS Genet. 13, e1006807. ( 10.1371/journal.pgen.1006807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gavery MR, Roberts SB. 2010. DNA methylation patterns provide insight into epigenetic regulation in the Pacific oyster (Crassostrea gigas). BMC Genomics 11, 1-9. ( 10.1186/1471-2164-11-483) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarda S, Zeng J, Hunt BG, Yi SV. 2012. The evolution of invertebrate gene body methylation. Mol. Biol. Evol. 29, 1907-1916. ( 10.1093/molbev/mss062) [DOI] [PubMed] [Google Scholar]
- 33.Riviere G, Wu GC, Fellous A, Goux D, Sourdaine P, Favrel P. 2013. DNA methylation is crucial for the early development in the oyster C. gigas. Mar. Biotechnol. 15, 739-753. ( 10.1007/s10126-013-9523-2) [DOI] [PubMed] [Google Scholar]
- 34.Kong N, Liu X, Li J, Mu W, Lian J, Xue Y, Li Q. 2017. Effects of temperature and salinity on survival, growth and DNA methylation of juvenile Pacific abalone, Haliotis discus hannai Ino. Chin. J. Oceanol. Limnol. 35, 1248-1258. ( 10.1007/s00343-016-5185-z) [DOI] [Google Scholar]
- 35.Suarez-Ulloa V, Rivera-Casas C, Michel M. 2019. Seasonal DNA methylation variation in the flat tree oyster Isognomon alatus from a mangrove ecosystem in North Biscayne Bay Florida. J. Shellfish Res. 38, 79-88. ( 10.2983/035.038.0108) [DOI] [Google Scholar]
- 36.Müller R, Charaf S, Scherer C, Oppold A, Oehlmann J, Wagner M. 2016. Phenotypic and epigenetic effects of vinclozolin in the gastropod Physella acuta. J. Molluscan Stud. 82, 320-327. ( 10.1093/mollus/eyv069) [DOI] [Google Scholar]
- 37.García-Fernández P, García-Souto D, Almansa E, Morán P, Gestal C. 2017. Epigenetic DNA methylation mediating Octopus vulgaris early development: effect of essential fatty acids enriched diet. Front. Physiol 8, 292. ( 10.3389/fphys.2017.00292) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farias ND, de Oliveira NFP, da Silva PM. 2017. Perkinsus infection is associated with alterations in the level of global DNA methylation of gills and gastrointestinal tract of the oyster Crassostrea gasar. J. Invertebr. Pathol. 149, 76-81. ( 10.1016/j.jip.2017.08.007) [DOI] [PubMed] [Google Scholar]
- 39.Bal N, Kumar A, Nugegoda D. 2017. Assessing multigenerational effects of prednisolone to the freshwater snail, Physa acuta (Gastropoda: Physidae). J. Hazard. Mater. 339, 281-291. ( 10.1016/j.jhazmat.2017.06.024) [DOI] [PubMed] [Google Scholar]
- 40.Nica D, Popescu C, Draghici G, Privistirescu I, Suciu M, Stöger R. 2017. Effect of cadmium on cytosine hydroxymethylation in gastropod hepatopancreas. Environ. Sci. Pollut. Res. 24, 15 187-15 195. ( 10.1007/s11356-017-9104-4) [DOI] [PubMed] [Google Scholar]
- 41.Haszprunar G, Wanninger A. 2012. Molluscs. Curr. Biol. 22, R510-R514. ( 10.1016/j.cub.2012.05.039) [DOI] [PubMed] [Google Scholar]
- 42.Parkhaev PY. 2017. Origin and the early evolution of the phylum Mollusca. Paleontol. J. 51, 663-686. ( 10.1134/S003103011706003X) [DOI] [Google Scholar]
- 43.Wanninger A, Wollesen T. 2019. The evolution of molluscs. Biol. Rev. 94, 102-115. ( 10.1111/brv.12439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. 1978. Molecular basis of base substitution hotspots in Escherichia coli. Nature 274, 775-780. ( 10.1038/274775a0) [DOI] [PubMed] [Google Scholar]
- 45.Elango N, Hung BG, Goodisman MAD, Yi SV. 2009. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc. Natl Acad. Sci. USA 106, 11 206-11 211. ( 10.1073/pnas.0900301106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiang H, et al. 2010. Single base-resolution methylome of the silkworm reveals a sparse epigenomic map. Nat. Biotechnol. 28, 516-520. ( 10.1038/nbt.1626) [DOI] [PubMed] [Google Scholar]
- 47.Glastad KM, Hunt BG, Yi SV, Goodisman MAD. 2011. DNA methylation in insects: on the brink of the epigenomic era. Insect Mol. Biol. 20, 553-565. ( 10.1111/j.1365-2583.2011.01092.x) [DOI] [PubMed] [Google Scholar]
- 48.Park J, Peng Z, Zeng J, Elango N, Park T, Wheeler D, Werren JH, Yi SV. 2011. Comparative analyses of DNA methylation and sequence evolution using Nasonia genomes. Mol. Biol. Evol. 28, 3345-3354. ( 10.1093/molbev/msr168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dixon GB, Bay LK, Matz MV. 2014. Bimodal signatures of germline methylation are linked with gene expression plasticity in the coral Acropora millepora. BMC Genomics 15, 1109. ( 10.1186/1471-2164-15-1109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cunningham CB, et al. 2015. The genome and methylome of a beetle with complex social behavior, Nicrophorus vespilloides (Coleoptera: Silphidae). Genome Biol. Evol. 7, 3383-3396. ( 10.1093/gbe/evv194) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gavery MR, Roberts SB. 2013. Predominant intragenic methylation is associated with gene expression characteristics in a bivalve mollusc. PeerJ 1, e215. ( 10.771/peerj.215) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grabherr MG, et al. 2011. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 29, 644-652. ( 10.1038/nbt.1883) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210-3212. ( 10.1093/bioinformatics/btv351) [DOI] [PubMed] [Google Scholar]
- 54.Keilwagen J, Wenk M, Erickson JL, Schattat MH, Grau J, Hartung F. 2016. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res. 44, e89. ( 10.1093/nar/gkw092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keilwagen J, Hartung F, Paulini M, Twardziok SO, Grau J. 2018. Combining RNA-seq data and homology-based gene prediction for plants, animals and fungi. BMC Bioinf. 19, 189. ( 10.1186/s12859-018-2203-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kocot KM, Poustka AJ, Stöger I, Halanych KM, Schrödl M. 2020. New data from Monoplacophora and a carefully-curated dataset resolve molluscan relationships. Sci. Rep. 10, 101. ( 10.1038/s41598-019-56728-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kriventseva EV, Tegenfeldt F, Petty TJ, Waterhouse RM, Simao FA, Pozdnyakov IA, Ioannidis P, Zdobnov EM. 2015. OrthoDB v8: update of the hierarchical catalog of orthologs and the underlying free software. Nucleic Acids Res. 43, D250-D256. ( 10.1093/nar/gku1220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zdobnov EM, Tegenfeldt F, Kuznetsov D, Waterhouse RM, Simão FA, Ioannidis P, Seppey M, Loetscher A, Kriventseva EV. 2017. OrthoDB v9.1: cataloging evolutionary and functional annotations for animal, fungal, plant, archaeal, bacterial and viral orthologs. Nucleic Acids Res. 45, D744-D749. ( 10.1093/nar/gkw1119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772-780. ( 10.1093/molbev/mst010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Slater GSC, Birney E. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinf. 6, 31. ( 10.1186/1471-2105-6-31) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Wang Z, Yan X, Yu R, Kong J, Liu J, Li X, Li Y, Guo X. 2012. Laboratory hybridization between two oysters: Crassostrea gigas and Crassostrea hongkongensis. J. Shellfish Res. 31, 619-625. ( 10.2983/035.031.0304) [DOI] [Google Scholar]
- 62.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST +: architecture and applications. BMC Bioinf. 10, 421. ( 10.1186/1471-2105-10-421) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hammer Ø, Harper DAT, Ryan PD. 2001. Past: paleontological statistics software package for education and data analysis. Palaeontol. Electronica 4, 1-9. [Google Scholar]
- 64.Bird AP. 1980. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 8, 1499-1504. ( 10.1093/nar/8.7.1499) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fryxell KJ, Moon WJ. 2005. CpG mutation rates in the human genome are highly dependent on local GC content. Mol. Biol. Evol. 22, 650-658. ( 10.1093/molbev/msi043) [DOI] [PubMed] [Google Scholar]
- 66.Scrucca L, Fop M, Murphy TB, Raftery AE. 2016. mclust 5: clustering, classification and density estimation using Gaussian finite mixture models. R J. 8, 205-233. [PMC free article] [PubMed] [Google Scholar]
- 67.R Core Team. 2020. R: a language and environment for statistical computing. http://www.R-project.org/.
- 68.Zemach A, McDaniel IE, Silva P, Zilberman D. 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328, 916-919. ( 10.1126/science.1186366) [DOI] [PubMed] [Google Scholar]
- 69.Lewis SH, et al. 2020. Widespread conservation and lineage-specific diversification of genome-wide DNA methylation patterns across arthropods. PLoS Genet. 16, e1008864. ( 10.1371/journal.pgen.1008864) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gygi SP, Rochon Y, Franza BR, Aebersold R. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720-1730. ( 10.1128/MCB.19.3.1720) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pascal LE, True LD, Campbell DS, Deutsch EW, Risk M, Coleman IM, Eichner LJ, Nelson PS, Liu AY. 2008. Correlation of mRNA and protein levels: cell type-specific gene expression of cluster designation antigens in the prostate. BMC Genomics 9, 246. ( 10.1186/1471-2164-9-246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yeung KY, Dombek KM, Lo K, Mittler JE, Zhu J, Schadt EE, Bumgarner RE, Raftery AE. 2011. Construction of regulatory networks using expression time-series data of a genotyped population. Proc. Natl Acad. Sci. USA 108, 19 436-19 441. ( 10.1073/pnas.1116442108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang X, Wheeler D, Avery A, Rago A, Choi J-H, Colburne JK, Clark AG, Werren JH. 2013. Function and evolution of DNA methylation in Nasonia vitripennis. PLoS Genet. 9, e1003872. ( 10.1371/journal.pgen.1003872) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Glastad KM, Gokhale K, Liebig J, Goddisman MAD. 2016. The caste- and sex-specific DNA methylome of the termite Zootermopsis nevadensis. Sci. Rep. 6, 37110. ( 10.1038/srep37110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gatzmann F, Falckenhayn C, Gutekunst J, Hanna K, Raddatz G, Coutinho Carneiro V, Lyko F. 2018. The methylome of the marbled crayfish links gene body methylation to stable expression of poorly accessible genes. Epigenet. Chromatin 11, 57. ( 10.1186/s13072-018-0229-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li Y, Liew YJ, Cui G, Cziesielski MJ, Zahran N, Michell CT, Voolstra CR, Aranda M. 2018. DNA methylation regulates transcriptional homeostasis of algal endosymbiosis in the coral model Aiptasia. Sci. Adv. 4, eaat2142. ( 10.1126/sciadv.aat2142) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Song K, Li L, Zhang G. 2017. The association between DNA methylation and exon expression in the Pacific oyster Crassostrea gigas. PLoS ONE 12, e0185224. ( 10.1371/journal.pone.0185224) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Glastad KM, Hunt BG, Goodisman MAD. 2019. Epigenetics in insects: genome regulation and the generation of phenotypic diversity. Annu. Rev. Entomol. 64, 185-203. ( 10.1146/annurev-ento-011118-111914) [DOI] [PubMed] [Google Scholar]
- 79.Gavery MR, Roberts SB. 2014. A context dependent role for DNA methylation in bivalves. Briefings Funct. Genomics 13, 217-222. ( 10.1093/bfgp/elt054) [DOI] [PubMed] [Google Scholar]
- 80.Aoki Y, Koshihara H. 1972. Inhibitory effects of acid polysaccharides from sea urchin embryos on RNA polymerase activity. Biochim. Biophys. Acta Nucleic Acids Protein Synth. 272, 33-43. ( 10.1016/0005-2787(72)90030-5) [DOI] [PubMed] [Google Scholar]
- 81.Sokolov EP. 2000. An improved method for DNA isolation from mucopolysaccharide-rich molluscan tissues. J. Molluscan Stud. 66, 573-575. ( 10.1093/mollus/66.4.573) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available as electronic supplementary material.