Abstract
The bitter taste sensation is important to warn mammals of the ingestion of potentially toxic food compounds. For mammals, whose nutrition relies on highly specific food sources, such as blood in the case of vampire bats, it is unknown if bitter sensing is involved in prey selection. By contrast to other bat species, vampire bats exhibit numerous bitter taste receptor pseudogenes, which could point to a decreased importance of bitter taste. However, electrophysiological and behavioural studies suggest the existence of functional bitter taste transmission. To determine the agonist spectra of the three bitter taste receptors that are conserved in all three vampire bat species, we investigated the in vitro activation of Desmodus rotundus T2R1, T2R4 and T2R7. Using a set of 57 natural and synthetic bitter compounds, we were able to identify agonists for all three receptors. Hence, we confirmed a persisting functionality and, consequently, a putative biological role of bitter taste receptors in vampire bats. Furthermore, the activation of the human TAS2R7 by metal ions is shown to be conserved in D. rotundus.
Keywords: vampire bat, T2R, bitter taste, metal ions
1. Introduction
The mammalian sense of taste provides important information about the food constituents prior to consumption [1]. Typically, mammals are able to detect and distinguish the five basic taste qualities: salty, sour, sweet, umami and bitter [2]. Whereas salty and sour tastes are mediated via ion channels [3,4], the remaining three taste qualities are based on G protein-coupled receptors [5]. These receptors belong to two different gene families, named taste 1 receptors (note the use of species-specific gene symbols: TAS1R, human; Tas1r, mouse; T1R, common for other vertebrates) and taste 2 receptors (TAS2R, Tas2r, T2R). The group of TAS1Rs consists of the three members TAS1R1, TAS1R2 and TAS1R3, which are responsible for sweet and umami tasting, and thus mediate the sensing of energy-rich food items [6–9]. By contrast, bitter taste, which is facilitated by TAS2Rs, is proposed to prevent the ingestion of potentially poisonous compounds, although a general correlation between toxicity and bitterness is lacking [10–13]. Instead, a mild bitterness can be appreciated by humans and may further serve to guide a range of mammals to pharmacologically active food items for self-medication purposes [14–16]. Mostly bitter compounds are rather complex organic molecules with an extraordinary chemical diversity [17]. However, inorganic molecules such as magnesium sulfate, which is known as ‘bitter salt’ or ‘Epsom salt’, also activate the human TAS2R7 [18,19]. Magnesium ions can profoundly affect human physiology, as evidenced by their effects as a laxative [20], suppressant of premature labour (= tocolytic) [21] and by their interference with cardiac parameters [22]. Given these varied and drastic effects, magnesium sensation by bitter taste receptors, similar to the more ‘classical’ organic bitter compounds, might be broadly selected for as a mechanism to regulate ingestion.
The repertoires of functional bitter taste receptors differ widely between species (for a recent review see [23]) and might be related to the dietary habits [24]. However, the mere number of bitter taste receptors does not allow conclusions about their importance for individual species as their average tuning breadths can fluctuate considerably such that few receptors may enable the detection of a large array of bitter compounds [25]. With more than 1000 different species, bats (Chiroptera) represent one of the largest mammalian clades [26]. Bats exhibit a wide range of dietary habits with herbivorous, insectivorous, frugivorous, carnivorous, piscivorous and sanguivorous species [27]. The differential endowment of bat species with taste receptors reflects these very distinct feeding habits. The T1R1 gene is absent or nonfunctional in all investigated bat species, supposing the general loss of canonical umami taste in Chiroptera [28]. To date, rather little information about the T2R repertoires in bats exists. Based on the hypothesis that the taste receptor repertoire is evolutionary modified according to the feeding habits, it has been suggested that herbivorous and insectivorous bats might have more functional T2Rs compared to others [24,29,30]. While the insectivorous Myotis species indeed showed an increased number of T2Rs, the hypothesis could not be confirmed for all vertebrates including bats in general [24,31,32]. The three members of the Myotis genus, Myotis davidii, Myotis brandtii and Myotis lucifugus, are spread over largely diverse habitats [33,34]. Therefore, the high number of potentially functional T2Rs is considered important for the adaptation to different prey insect repertoires [32].
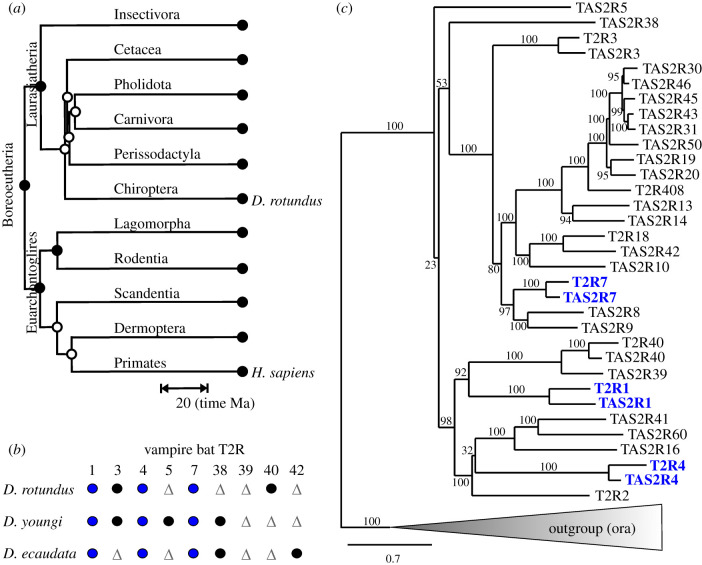
Vampire bats represent a special group among the Chiroptera, as their nutrition relies exclusively on blood [27]. There are only three species known, named Desmodus rotundus, Diaemus youngi and Diphylla ecaudatus. In vampire bats all three T1R genes including the already mentioned T1R1 are absent from the genomes or became pseudogenes, suggesting that not only umami, but also sweet taste perception has been lost [28,35,36]. The bitter taste sensation is also speculated to play a minor role in food selection, since blood is unlikely to contain a high quantity of toxic compounds and as it was shown that vampire bats primarily use their sense of odour, echolocation and infrared sensors for prey detection and the identification of capillary-rich regions [27,37,38]. These facts correlate with an evolutionary reduction of taste receptors in vampire bats. Indeed, the number of T2R pseudogenes in vampire bats is significantly higher than in non-blood-sucking bats. Whereas 13 of the 21 T2Rs of the common vampire bat D. rotundus are pseudogenes and only 8 T2Rs are considered functional, the pseudogene ratio for the above-mentioned Myotis species is below 25% [32]. However, three intact T2Rs, named T2R1, T2R4 and T2R7, are conserved in all three vampire bat species indicating a persisting function [24] (figure 1b). Moreover, functional bitter taste signal transduction components such as CALHM1 exist [36]. Interestingly, vampire bats avoid blood adulterated with bitter compounds, indicating a remaining role of T2Rs for taste perception [42]. Furthermore, anatomical and electrophysiological studies identified normal taste buds and functional taste receptor cells in vampire bats, although vallate papillae, one of three common types of taste papillae, are absent in D. rotundus [43–45]. The existence of these structures and molecules suggests an intact gustatory system in vampire bats. A recent study confirmed the activation of D. rotundus T2Rs by mainly synthetic bitter compounds in vitro [46].
Figure 1.
The 3 extant T2Rs of vampire bats are direct one-to-one orthologues of human TAS2Rs. (a) The phylogenetic tree of Boreoeutheria demonstrates the evolutionary distant relationship of the vampire bat Desmodus rotundus with humans. The scale bar indicates the time of divergence in million years (computed with TimeTree (www.timetree.org) [39]). (b) Extant and pseudogenized T2R genes in the 3 vampire bat species. T2R pseudogenes (triangles, grey), extant T2R genes (circles, black) and T2R genes that are extant and conserved among vampire bats (circles, grey/blue online) are shown (taken from [32]). (c) Phylogenetic tree showing the D. rotundus T2Rs together with the 25 potentially functional human TAS2Rs. The one-to-one orthologues (confirmed with the software UPhO [40]) are highlighted in bold type/blue online. The six zebrafish pheromone receptors, ORA1 to 6, were included as an outgroup [41]. Branch support (%) is indicated by grey numbers, scale bar = amino acid substitutions per site. (Online version in colour.)
Beside the gustatory system, T2Rs are expressed in extra-oral tissues, indicating a role beyond taste sensation [47]. Specific agonists for taste receptors present in non-gustatory tissues are largely unknown. However, recent studies presented the activation of the human TAS2R7 by bi- and trivalent metal ions, which were discussed to have a putative biological function as endogenous agonists as they are released upon excitation of cells like Zn2+ in β-cells [18,19]. Interestingly, the three D. rotundus receptors, T2R1, T2R4 and T2R7 are one-to-one orthologues to human TAS2R1, TAS2R4 and TAS2R7 (figure 1c), raising the possibility for a functional conservation of these orthologues including metal ion responses.
In this work, we aimed to identify the spectrum of bitter compounds detected by vampire bats by functional analyses of the three highly conserved bitter taste receptors T2R1, T2R4 and T2R7 of D. rotundus. We hypothesize that they may play a role in feeding decisions and, perhaps, detect bitter components in the blood or other materials relevant to their behaviour, ecology and physiology.
2. Material and methods
(a). Bioinformatics
To highlight the great evolutionary distance between human and D. rotundus, we obtained a phylogenetic tree of Boreoeutheria using the TimeTree webserver (www.timetree.org, [39]). Amino acid sequences of the 25 human TAS2Rs were taken from Meyerhof et al. [48], the amino acid sequences of D. rotundus T2R1 (XP_024434157.1), T2R4 (XP_024410857.1) and T2R7 (XP_024431669.1) were retrieved through NCBI resources (www.ncbi.nlm.nih.gov). As an outgroup, the amino acid sequences of the small zebrafish pheromone receptor family ORA (olfactory receptor class A-related, [41], ORA1 (NP_001124140.1), ORA2 (NP_001091865.1), ORA3 (XP_009294044.1), ORA4 (XP_005168339.1), ORA5 (XP_002663506.1) and ORA6 (XP_003200835.2) were included and fetched from the NCBI database. The phylogenetic tree of the 25 human and three D. rotundus bitter taste receptors was established using the phylogeny.fr webserver (www.phylogeny.fr). Briefly, sequences were aligned with ClustalW (v. 2.1), the phylogenetic tree was reconstructed using the maximum likelihood method implemented in PhyML (v. 3.1) and reliability of the internal branch was assessed using aLRT (χ2-based parametric, v. 3.0). Tree rendering was done with TreeDyn (v. 198.3). For the determination of orthogroups the phylogenetic tree construction was done as before including now, in addition to the 25 human TAS2Rs, all 8 putatively functional D. rotundus T2Rs [36], all 35 mouse Tas2rs [49], the 12 cat and 15 dog T2Rs [50]. The resulting Tree file was then subjected to orthogroup detection using the software UPhO [40]. Estimation of the rates of non-synonymous/synonymous substitutions (dN/dS) for human and D. rotundus orthologous bitter taste receptor pairs was done using the program PAL2NAL (www.bork.embl.de/pal2nal [51]).
(b). Chemicals
For the functional screening of D. rotundus T2R1, T2R4 and T2R7, we used a set of 57 bitter compounds. The substances (see electronic supplementary material, table S1) were selected based on previous successful screenings covering a large range of vertebrate bitter taste receptors, including human [48,49,52], mouse [49], rat [53], chimpanzee [54], various bird and frog [25], zebrafish and coelacanth [55] receptors, using a wide range of diverse natural and synthetic bitter compounds. Stock solutions were prepared either in the assay buffer C1 (130 mM NaCl, 10 mM HEPES pH 7.4, 5 mM KCl, 2 mM CaCl2, 0.18% glucose) or in DMSO. The final DMSO concentration in the screening procedure was kept below 0.5% to prevent unspecific cellular responses. The highest concentrations of substances employed in the receptor screening were chosen based on previous experiments to avoid receptor-independent cellular responses or solubility issues.
The finding of rather strong negative selective pressure on D. rotundus T2R7 and its human orthologue TAS2R7 prompted us to screen the two receptors additionally with metal salts, a compound group recently found to elicit responses specifically on human TAS2R7 [18,19].
(c). Cloning
The predicted cDNAs covering the entire coding regions of D. rotundus bitter taste receptors T2R1 (XM_024578389), T2R4 (XM_024555089) and T2R7 (XM_024575901) (electronic supplementary material, appendix S1), elongated by 5′- and 3′-restriction sites, were synthesized (BioCat GmbH, Heidelberg, Germany) and cloned into the pcDNA 5 FRT T/O expression vector (Thermo Fisher Scientific, Waltham, USA). The open reading frames of the receptor genes were elongated by an N-terminal sst3-tag for effective cell surface expression and a C-terminal HSV-tag, which is necessary for immunocytochemical detection. The tags were already present in the expression vector or in case of T2R4 the synthetic cDNA was elongated by the sequence. The vector and the synthetic cDNAs were cleaved using the restriction enzymes EcoRI (Thermo Fisher Scientific) and NotI (Thermo Fisher Scientific) for T2R1 and T2R7 and BamHI (Thermo Fisher Scientific) and NotI for T2R4. Subsequently, linearized vector DNA was dephosphorylated using FastAP Thermosensitive Alkaline Phosphatase (Thermo Fisher Scientific) and, after purification, ligated using T4 DNA ligase (New England Biolabs, Frankfurt, Germany) with the receptor cDNAs. The integrity of the final constructs was confirmed by sequencing (Eurofins Genomics GmbH, Ebersberg, Germany).
(d). Cultivation of HEK293T–Gα16gust44 cells
HEK293T–Gα16gust44 cells [56] were grown in Dulbecco's modified eagle medium (DMEM) (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (Sigma Aldrich, St Louis, USA), 2 mM l-glutamine (Sigma Aldrich), 100 units ml−1 penicillin (Sigma Aldrich) and 100 µg ml−1 streptomycin (Sigma Aldrich) at 37°C and 5% CO2.
(e). Transient transfection
The day before transfection, HEK293T–Gα16gust44 cells were seeded onto 96-well plates coated with 10 µg ml−1 poly-d-lysine to reach a confluence of 40–60% the next day. Transient transfection was performed with 150 ng of the cloned plasmid DNA and 0.3 µl lipofectamine 2000 in serum-free DMEM per well. For negative controls empty vector DNA (mock) was transfected. Five hours post transfection, the media was changed to DMEM supplemented as described above.
(f). Calcium imaging
Twenty-four hours after transfection HEK293T–Gα16gust44 cells were loaded with the fluorescence dye Fluo-4-AM (Invitrogen) in the presence of 2.5 mM probenecid (Sigma Aldrich) for 1 h as described previously [25,49]. Cells were washed with buffer C1 using a BioTek Cell Washer, incubated in the dark for half an hour and washed again. Measurement of fluorescence changes upon automated application of different agonist concentrations was done using a FLIPRTETRA system (Molecular Devices, San Jose, USA). Somatostatin 14 (final concentration 100 nM) (Bachem, Bubendorf, Switzerland) was used as cell viability control.
(g). Data analysis
For data export, the FLIPR software ScreenWorks was used. Data were negative control corrected using the data of mock-transfected cells and exported. Exported fluorescence intensities were standardized to basal fluorescence and normalized to the buffer-only control to obtain the relative changes in fluorescence (ΔF/F) using the Microsoft Excel software. Statistical evaluation was done using SigmaPlot. Statistical significance (p < 0.01) was determined using Student's t-test.
3. Results
(a). Screening of Desmodus rotundus T2Rs with a set of known bitter compounds
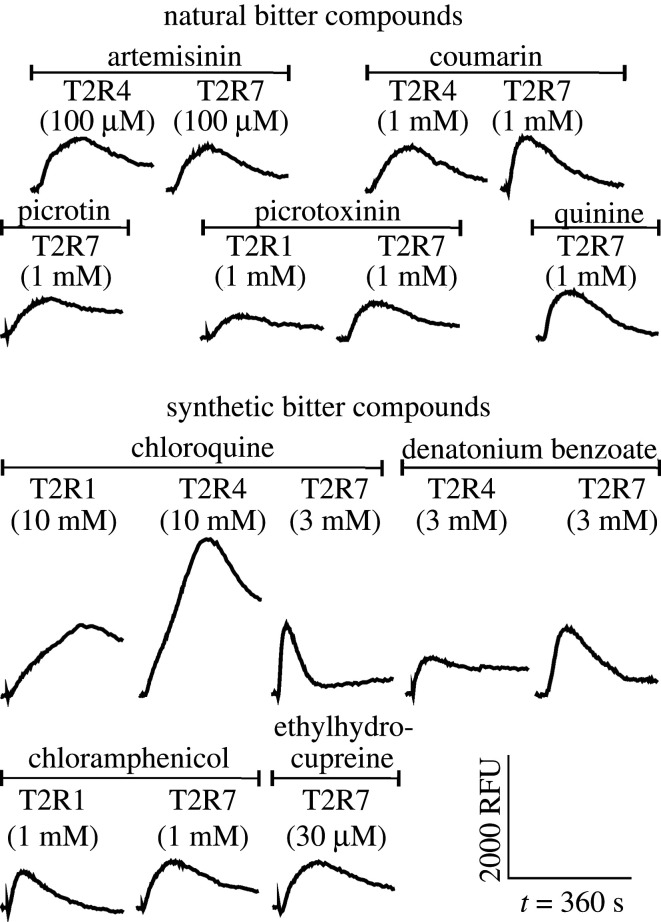
Prior to performing functional receptor assays with transiently transfected cells, the expression and membrane localization of the T2Rs was confirmed (electronic supplementary material, appendix S2). As described in the material and methods section, we initially used a set of 57 natural and synthetic bitter compounds with diverse chemical structures to cover a large chemical space for receptor characterization (electronic supplementary material, table S1). These compounds were tested for their activation of the D. rotundus bitter taste receptors T2R1, T2R4 and T2R7. In total, we detected 3 agonists for T2R1 (approx. 5%), 4 for T2R4 (approx. 7%) and 9 for T2R7 (approx. 16%) (table 1 and figure 2) indicating different tuning breadth. All of the receptors exhibited responses to substances of different compound classes. Among these, common structural motifs were not detected, similar to what has been observed for the human orthologous receptors TAS2R4 and TAS2R7 [48]. The group of agonists include natural, as well as synthetic compounds, with different toxicities among these substances. When comparing the agonists of the three receptors, we observed overlapping agonist profiles. Chloroquine was found to activate all three tested receptors. T2R1 and T2R7 responded to chloramphenicol and picrotoxinin and T2R4 and T2R7 to artemisinin, coumarin and denatonium benzoate. Only T2R7 was activated by ethylhydrocupreine, quinine and picrotin (table 1). For 48 of the 57 compounds (approx. 84%) we observed no responses (electronic supplementary material, table S1).
Table 1.
Identified agonists of the Desmodus rotundus bitter taste receptors T2R1, T2R4 and T2R7. Natural and synthetic bitter substances, which showed receptor activation in Ca2+-imaging experiments are presented. Multiple activators per receptor were identified, and their maximal applied concentrations (cmax) are presented. No activation is indicated by (—) and for agonists efficacies and the corresponding standard deviations are given as relative fluorescence (ΔF/F) (n = 3). Further, activation threshold concentrations are given in parenthesis for compounds, which activation threshold concentration is different from the maximal concentration.
| cmax | T2R1 | T2R4 | T2R7 | |
|---|---|---|---|---|
| artemisinin | 100 µM | — | 0.12 ± 0.01 | 0.13 ± 0.01 |
| chloramphenicol | 1 mM | 0.08 ± 0.02 | — | 0.09 ± 0.02 |
| chloroquine | 10 mM | 0.16 ± 0.03 | 0.29 ± 0.08 | 0.34 ± 0.10 (3 mM) |
| coumarin | 1 mM | — | 0.13 ± 0.03 | 0.16 ± 0.01 |
| denatonium benzoate | 10 mM | — | 0.32 ± 0.02 | 0.32 ± 0.04 (300 µM) |
| ethylhydrocupreine | 30 µM | — | — | 0.10 ± 0.02 |
| picrotin | 1 mM | — | — | 0.08 ± 0.02 |
| picrotoxinin | 1 mM | 0.08 ± 0.02 | — | 0.13 ± 0.03 |
| quinine sulfate | 30 µM | — | — | 0.11 ± 0.01 |
Figure 2.
Cellular responses to application of organic bitter agonists. HEK293T–Gα16gust44 cells transfected with D. rotundus T2R1, T2R4 and T2R7 were loaded with a Ca2+-indicator (Fluo-4-AM) and fluorescence emission after ligand application was measured. The panels show the FLIPR recordings after negative control (mock) correction. All identified agonists of D. rotundus T2Rs and their corresponding concentrations are presented. Relative light units (from −200 to 3000) are plotted against the time (6 min).
(b). Determination of activation threshold concentrations by Ca2+-imaging
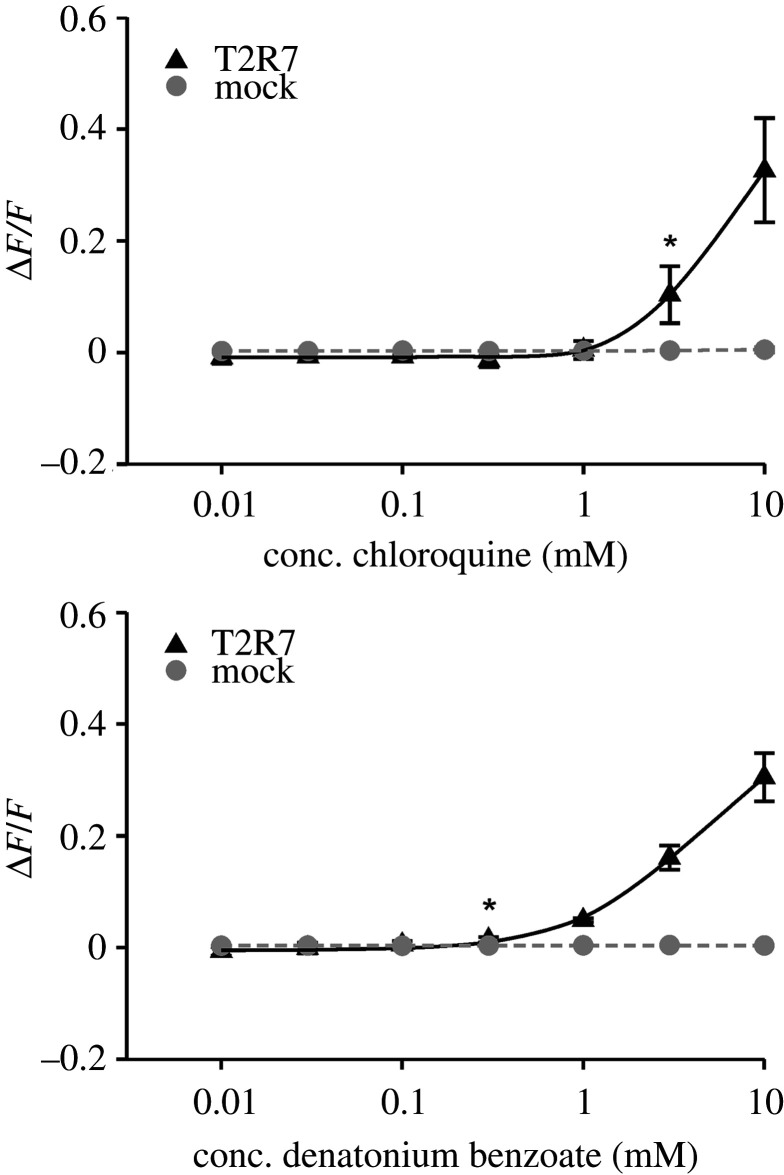
For the description of receptor–agonist relationships, potencies and efficacies are important parameters. The lowest agonist concentration, which resulted in a statistically significant fluorescence increase (p < 0.01), compared to the empty vector control, is defined as activation threshold concentration (table 1). For most of the agonists, activation was solely detectable at the maximal applied concentration. As T2R7 responded in a wider range to its agonists chloroquine and denatonium benzoate, we established dose–response relationships. However, the limited solubility of the test compounds and the occurrence of receptor-independent artefacts prevented us from using higher compound concentrations necessary for achieving signal saturation. Thus, the determination of EC50-concentrations was not possible (figure 3).
Figure 3.
Concentration–response relationships of the activation of Desmodus rotundus T2R7 by chloroquine and denatonium benzoate. HEK293T–Gα16gust44 cells were transiently transfected with D. rotundus T2R7 and the empty vector control (mock). Fluorescence intensities, according to the Ca2+-release upon receptor activation, were measured with an automated fluorometric imaging plate reader (FLIPRTetra). For dose–response curves seven different concentrations of chloroquine (upper panel) and denatonium benzoate (lower panel) (cmax = 10 mM) were used. The relative fluorescence intensities were mock subtracted and plotted against the agonist concentration in mM (n = 3). Statistical significance (p < 0.01) is presented by (*).
The efficacy of an agonist determines the extent of receptor activation. The efficacies observed for the activation of T2R7 by chloroquine (0.34 ± 0.10) and denatonium benzoate (0.32 ± 0.04) were almost identical (table 1). Similarly, we observed comparable efficacies for all other compounds that activated two receptors (table 1).
(c). The Desmodus rotundus bitter taste receptor T2R7 responds to metal ions
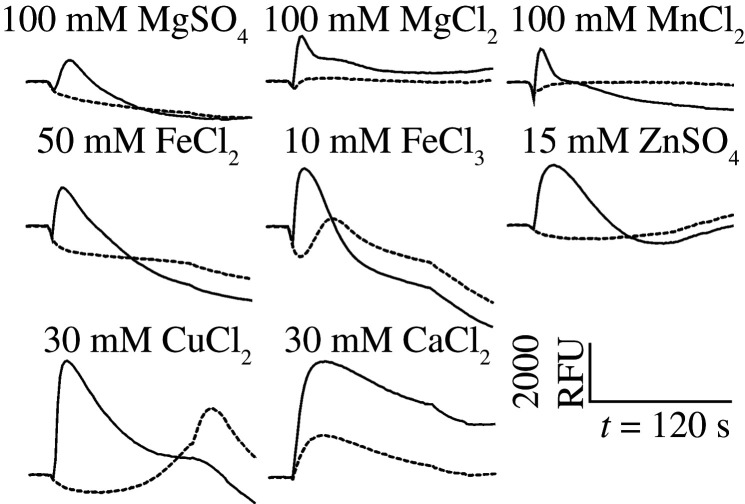
Comparing the rates of non-synonymous to synonymous substitutions among the investigated orthologous human and D. rotundus bitter taste receptors revealed that the strongest purifying selection exists for the TAS2R7/T2R7 pair of receptors with a dN/dS ratio of 0.52, whereas the TAS2R1/T2R1 (dN/dS = 0.73) and TAS2R4/T2R4 (dN/dS = 0.67) pairs exhibited lower ratios. Former works showed the activation of the human bitter taste receptor TAS2R7 by bitter salts [18,19]. In the light of the strong negative selective pressure identified for the D. rotundus orthologue T2R7, we further tested a set of bitter salts for their activation of the three D. rotundus receptors (table 2). While, as anticipated, no responses were evident for cells expressing T2R1 and T2R4 (data not shown), we observed responses for T2R7. All tested di- and trivalent salts showed an activation of the T2R7, whereas the monovalent salt potassium chloride did not. Because of the use of a Ca2+-imaging assay, the activation by CaCl2 cannot be fully confirmed, however, T2R7 transfected cells showed a higher response than mock-transfected cells suggesting the activation by this bitter salt as well (figure 4).
Table 2.
Activation of human TAS2R7 and Desmodus rotundus T2R7 by metal salts. The tested metal salts are presented including their maximal applied concentration (cmax). No activation is indicated by (x). As a Ca2+-imaging assay was used, the activation of the TAS2R7 by CaCl2 cannot be confirmed with certainty. Dose–response relationships were established for the metal salts CuCl2, FeCl3, MgSO4, MgCl2, FeCl2, MnCl2 and ZnSO4, which activated the human TAS2R7 and the D. rotundus T2R7. For comparison of both receptors, the activation threshold concentrations, the EC50-concentrations in mM and the efficacies in relative light units (RLU) are presented. EC50-concentrations, that were not determinable, are indicated by (—). The compound concentrations triggering the strongest receptor responses are given in parenthesis.
| cmax | threshold |
EC50 |
efficacy |
||||
|---|---|---|---|---|---|---|---|
| T2R7 | TAS2R7 | T2R7 | TAS2R7 | T2R7 | TAS2R7 | ||
| CaCl2 | 30 mM | ? | ? | ? | ? | ? | ? |
| KCl | 100 mM | x | x | x | x | x | x |
| CuCl2 | 30 mM | 1.3 | 1.3 | 1.62 ± 0.11 | 2.03 ± 0.03 | 0.81 ± 0.15 (10 mM) | 0.35 ± 0.02 (10 mM) |
| FeCl3 | 30 mM | 0.75 | 0.9 | 0.77 ± 0.09 | — | 0.82 ± 0.17 (3 mM) | 0.40 ± 0.04 (1 mM) |
| MgSO4 | 300 mM | 30 | 10 | — | 10.45 ± 4.02 | 0.19 ± 0.05 (100 mM) | 0.37 ± 0.02 (100 mM) |
| MgCl2 | 100 mM | 10 | 30 | 12.74 ± 8.66 | — | 0.17 ± 0.06 | 0.21 ± 0.04 |
| FeCl2 | 50 mM | 3 | 1 | 4.51 ± 0.46 | — | 0.21 ± 0.05 | 0.14 ± 0.04 (10 mM) |
| MnCl2 | 100 mM | 10 | 3 | — | 3.94 ± 0.32 | 0.14 ± 0.04 (30 mM) | 0.36 ± 0.09 (30 mM) |
| ZnSO4 | 15 mM | 3 | 0.3 | — | 0.38 ± 0.02 | 0.23 ± 0.01 | 0.42 ± 0.05 |
Figure 4.
Identification of bitter salts as agonists for the Desmodus rotundus bitter taste receptor T2R7. HEK293T–Gα16gust44 cells were transiently transfected with the D. rotundus bitter taste receptor T2R7 and the empty vector control (mock). Fluorescence intensities, according to the Ca2+-release upon receptor activation, were measured with an automated fluorometric imaging plate reader (FLIPRTetra). Responses of receptor (solid lines) and mock-transfected (dashed lines) cells to the exposure of MgSO4, MgCl2, MnCl2, FeCl2, FeCl3, ZnSO4, CuCl2 and CaCl2 are presented.
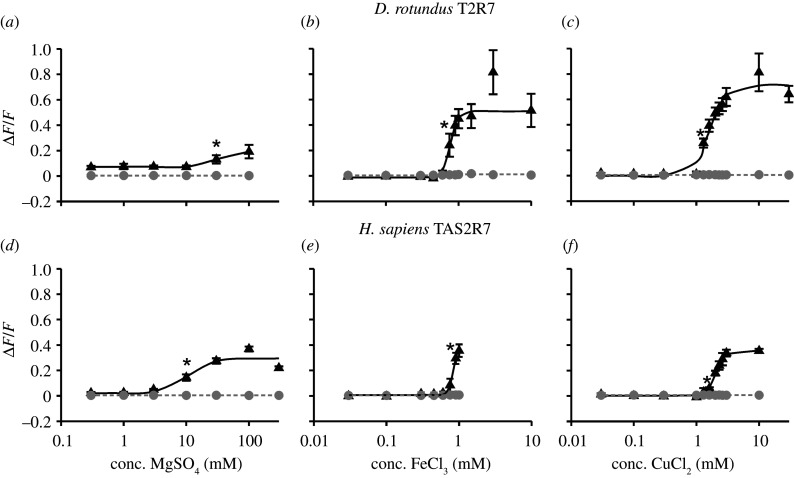
To compare the human and the D. rotundus receptor orthologues in potency and efficacy, we monitored dose–response relationships for the tested bitter salts (figure 5; electronic supplementary material, figures S1 and S2). Considering the determined activation threshold values, human TAS2R7 and D. rotundus T2R7 showed similar sensitivities with some minor deviations (table 2). By contrast, the receptors showed different efficacies for the metal ions, except for MgCl2 and FeCl2. We were not able to determine most of the EC50 values, because of lacking receptor signal saturation in the range of applicable agonist concentrations. Only for CuCl2 an EC50 value for both receptors was calculated. With 1.62 ± 0.11 mM for the D. rotundus T2R7 and 2.03 ± 0.03 mM for the human TAS2R7, these values were in a comparable range, indicating a similar potency of CuCl2 to activate both receptors (table 2).
Figure 5.
Concentration–response relationships of the activation of human TAS2R7 and Desmodus rotundus T2R7 by selected bitter salts. HEK293T–Gα16gust44 cells were transiently transfected with human TAS2R7 (d–f) and D. rotundus T2R7 (a–c) and the empty vector control (mock). Fluorescence intensities, according to the Ca2+-release upon receptor activation, were measured with an automated fluorometric imaging plate reader (FLIPRTetra). Different concentrations of MgSO4 (a,d), FeCl3 (b,e) and CuCl2 (c,f) were used. The relative fluorescence intensities (black) were mock (grey) subtracted and plotted against the ligand concentration in mM (n = 3). Statistical significance (p < 0.01) is presented by (*).
4. Discussion
Vampire bats' nutrition is solely based on blood and their prey selection is suggested to be primarily facilitated through smell, echolocation and infrared sensors [27,37,38]. Therefore, the functionality of T2Rs and their involvement in prey selection and sensing of bitter substances has been debated in former works [32]. We found that 84% of the tested natural and synthetic bitter compounds did not activate the investigated D. rotundus receptors, which may indeed hint at a more restricted perceptual space of D. rotundus. For comparison, human receptors did not respond to approximately 12%, frog T2Rs to 25%, chicken T2Rs to 32%, mouse Tas2rs to 34%, turkey T2Rs to 43%, domestic cat T2Rs to 50% and zebra finch T2Rs to 75% of a similar set of natural and synthetic bitter substances [25,48–50]. It has to be considered that in the case of frog, zebra finch and D. rotundus not all of the putatively functional T2Rs were included in the studies and that the bitter compound sets used in the different studies are slightly varying. Consequently, the real percentage of undetected bitter compounds by these T2Rs may differ from those indicated. Moreover, it is important to point out that the bitter compound libraries used by us and others are biased towards humans, and thus species tailored libraries may result in different fractions of activating bitter substances. It is further hypothesized that the bitter taste sensation abilities of mammals are correlated to nutritional requirements [57]. Vertebrates with limited nutrition spectra, like blood for vampire bats, might have a decreased or more specialized ability to taste bitter substances [24,31]. Hence, important agonists for vampire bat T2Rs may not have been included in our set of bitter compounds.
However, genetic data showed evolutionary conserved T2Rs in vampire bats and some electrophysiological and behavioural studies already confirmed their functionality [32,42,44]. The characterization of the three D. rotundus T2Rs in our work as well as in the recent report by Lu et al. [46], suggests the persistence of function for these receptors, which may, therefore, be involved in taste sensation by responding to bitter compounds in blood. Except for the synthetic compounds ethylhydrocupreine, chloroquine and denatonium benzoate, a substance known as a very strong bitter agent for humans, all of the identified agonists are natural. Whether these or other yet to be discovered agonists appear in the blood in concentration ranges able to activate bitter taste receptors of vampire bats remains elusive.
An independent and concurrent study recently reported the ligands of some D. rotundus T2Rs [46]. Lu and colleagues screened a set of 19 mostly synthetic bitter compounds selected from past research conducted by members of the present authorship at a single concentration and identified agonists for 5 of the 8 investigated D. rotundus T2Rs. Surprisingly, some differences in the activation of the tested T2R1, T2R4 and T2R7 were evident. Considering these 3 T2Rs, Lu and colleagues showed activation of all receptors by denatonium benzoate and activation of T2R1 and T2R4 by chloramphenicol, whereas we could not confirm T2R1 responses to denatonium benzoate and found T2R1 and T2R7 responding to chloramphenicol instead. One additional compound, the natural bitter substance yohimbine was reported to activate T2R7, although our screening efforts could not confirm a T2R7 response to this substance. Despite intrinsic similarities in the two screening approaches (both performed calcium-imaging analyses), we speculate that the observed differences reflect the numerous variations in the experimental setups including the use of different cell lines, different screening strategies (single compound concentrations versus high and low concentrations) and different sensitivities of the employed fluorometric imaging devices. By contrast to Lu et al. [46], we identified quinine, which was shown to be avoided by D. rotundus when added to blood samples, as an agonists of D. rotundus T2R7 [42]. The behavioural experiments with quinine were performed in a concentration range between 0.1 and 30 mM, with a statistically significant avoidance at 30 mM. In our screening, the T2R7 response to quinine was already detectable at a concentration of 30 µM, which is one thousandth of the published data. As the bitter substances in our assay are dissolved in a simple buffer system, whereas blood is very complex and rich in carrier proteins for rather hydrophobic substances such as quinine, we speculate that this has prevented a more sensitive detection in the in vivo experiment [58]. Assuming that this is also true for other bitter compounds potentially present in blood, vampire bats may generally require T2Rs whose threshold concentrations are able to detect their agonists despite quenching by blood components. Further influences, like the composition of the vertebrate's saliva, can manipulate the sensitivity of bitter-tasting in vivo [59–61].
Artemisinin and quinine represent typical members of sesquiterpene lactone and alkaloid bitter compounds and even chloroquine, albeit a synthetic compound, shows chemical similarities with quinine, hence, these activators may represent prototypical plant bitter substances. Therefore, in light of the assumption that the common ancestor of phyllostomid bats, including Desmodontinae, exhibited an insectivorous lifestyle with slight frugivory prior to the massive adaptive radiation giving rise to multiple nutritionally specialized bat clades [62], the responses to plant-derived bitter substances may represent retained functionality inherited from a insectivorous/frugivorous ancestor.
The alignment of D. rotundus T2R7 and human TAS2R7 (figure 1; electronic supplementary material, appendix S3) revealed an amino acid sequence identity of approximately 77%. It was reported previously that the human TAS2R7 is responding to bitter-tasting salts, with H943.37 and E2647.32 as crucial residues for the interaction with metal ions [18,19]. These residues are conserved in D. rotundus T2R7 (electronic supplementary material, appendix S3). Interestingly, we demonstrated that the D. rotundus T2R7 is indeed also activated by these substances and activation threshold concentrations of human TAS2R7 and D. rotundus T2R7 are in similar ranges (table 2). However, these concentrations are in a high µM to low mM range and bitter salts are, therefore, unlikely to appear in taste relevant concentrations in the blood of prey animals of D. rotundus (table 2) [19]. Occasionally, vampire bats do drink water [63] and some springs (e.g. hot volcanic springs in Cauca valley, Colombia, a habitat of D. rotundus [64]) contain water highly enriched in bitter metal salts with magnesium ion concentrations of up to 15.5 mM [65]. This concentration is above the half-maximal activating (EC50-) concentration for D. rotundus T2R7. As metal salts were shown to interact with heparin [66], the digestibility of blood meals might become changed by water from bitter springs, providing a possible explanation for this T2R function.
Mammalian bitter taste receptors are also present in various extra-gustatory tissues, where they might play important roles in fulfilling significant endogenous biological functions [47]. A putative extra-oral function of the activation of the D. rotundus T2R7 by bitter salts, as was proposed for the human orthologue [19], may also be possible and is a promising avenue for future research.
In conclusion, we were able to characterize the three T2Rs common to all extant vampire bat species, with T2R7 as the most broadly tuned receptor. In combination with previously published data, our results confirm a persisting functionality of D. rotundus T2Rs [36,42,44,46]. In particular, D. rotundus T2R7 shares di- and trivalent metal ions as common agonists with human TAS2R7. To fully understand the role of D. rotundus T2Rs in taste sensation and signalling in extra-gustatory tissues, further research concerning the functionality and activation mechanisms of these receptors in vivo is necessary.
Supplementary Material
Acknowledgement
We thank Catherine Delaporte and Eva Boden for excellent technical assistance.
Data accessibility
All data of this study are provided in the main text and electronic supplementary material.
Authors' contributions
M.B. and F.Z. designed the work. F.Z. performed the experiments. F.Z. and M.B. analysed the data. M.B. and F.Z. wrote the article. All authors have read and agreed to the published version of the manuscript.
Competing interests
We declare we have no competing interests.
Funding
We received no specific funding for this study.
References
- 1.Lindemann B. 1996. Taste reception. Physiol. Rev. 76, 719-766. ( 10.1152/physrev.1996.76.3.719) [DOI] [PubMed] [Google Scholar]
- 2.Yarmolinsky DA, Zuker CS, Ryba NJ. 2009. Common sense about taste: from mammals to insects. Cell 139, 234-244. ( 10.1016/j.cell.2009.10.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tu YH, et al. 2018. An evolutionarily conserved gene family encodes proton-selective ion channels. Science 359, 1047-1050. ( 10.1126/science.aao3264) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chandrashekar J, Kuhn C, Oka Y, Yarmolinsky DA, Hummler E, Ryba NJ, Zuker CS. 2010. The cells and peripheral representation of sodium taste in mice. Nature 464, 297-301. ( 10.1038/nature08783) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrens M, Meyerhof W. 2016. G protein-coupled taste receptors. In Chemosensory transduction (eds Zufall F, Munger SD), pp. 227-244. New York, NY: Academic Press. [Google Scholar]
- 6.Hoon MA, Adler E, Lindemeier J, Battey JF, Ryba NJ, Zuker CS. 1999. Putative mammalian taste receptors: a class of taste-specific GPCRs with distinct topographic selectivity. Cell 96, 541-551. ( 10.1016/S0092-8674(00)80658-3) [DOI] [PubMed] [Google Scholar]
- 7.Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. 2001. Mammalian sweet taste receptors. Cell 106, 381-390. ( 10.1016/S0092-8674(01)00451-2) [DOI] [PubMed] [Google Scholar]
- 8.Li X, Staszewski L, Xu H, Durick K, Zoller M, Adler E. 2002. Human receptors for sweet and umami taste. Proc. Natl Acad. Sci. USA 99, 4692-4696. ( 10.1073/pnas.072090199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson G, Chandrashekar J, Hoon MA, Feng L, Zhao G, Ryba NJP, Zuker CS. 2002. An amino-acid taste receptor. Nature 416, 199-202. ( 10.1038/nature726) [DOI] [PubMed] [Google Scholar]
- 10.Glendinning JI. 1994. Is the bitter rejection response always adaptive? Physiol. Behav. 56, 1217-1227. ( 10.1016/0031-9384(94)90369-7) [DOI] [PubMed] [Google Scholar]
- 11.Garcia J, Hankins W. 1975. The Evolution of Bitter and the Acquisition of Toxiphobia. In Olfaction and Taste V. Proceedings of the 5th International Symposium in Melbourne (eds DA Denton, JP Coghlan), pp. 39–45. Australia, NY: Academic Press.
- 12.Holloway GJ, de Jong PW, Brakefield PM, de Vos H. 1991. Chemical defence in ladybird beetles (Coccinellidae). I. Distribution of coccinelline and individual variation in defence in 7-spot ladybirds (Coccinella septempunctata). Chemoecology 2, 7-14. ( 10.1007/BF01240660) [DOI] [Google Scholar]
- 13.Nissim I, Dagan-Wiener A, Niv MY. 2017. The taste of toxicity: a quantitative analysis of bitter and toxic molecules. IUBMB Life 69, 938-946. ( 10.1002/iub.1694) [DOI] [PubMed] [Google Scholar]
- 14.Koshimizu K, Ohigashi H, Huffman MA. 1994. Use of Vernonia amygdalina by wild chimpanzee: possible roles of its bitter and related constituents. Physiol. Behav. 56, 1209-1216. ( 10.1016/0031-9384(94)90368-9) [DOI] [PubMed] [Google Scholar]
- 15.Vitazkova S, Long E, Paul A, Glendinning J. 2001. Mice suppress malaria infection by sampling a ‘bitter’ chemotherapy agent. Anim. Behav. 61, 887-894. ( 10.1006/anbe.2000.1677) [DOI] [Google Scholar]
- 16.Villalba JJ, Miller J, Ungar ED, Landau SY, Glendinning J. 2014. Ruminant self-medication against gastrointestinal nematodes: evidence, mechanism, and origins. Parasite 21, 31. ( 10.1051/parasite/2014032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behrens M. 2020. Chapter 3.11 – Bitter Taste. In The Senses: A Comprehensive Reference (eds B Fritzsch), pp. 231–246. Oxford, NY: Academic Press. [Google Scholar]
- 18.Behrens M, Redel U, Blank K, Meyerhof W. 2019. The human bitter taste receptor TAS2R7 facilitates the detection of bitter salts. Biochem. Biophys. Res. Commun. 512, 877-881. ( 10.1016/j.bbrc.2019.03.139) [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, et al. 2019. Metal ions activate the human taste receptor TAS2R7. Chem. Senses 44, 339-347. ( 10.1093/chemse/bjz024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bokhari SR, Siriki R, Teran FJ, Batuman V. 2018. Fatal hypermagnesemia due to laxative use. Am. J. Med. Sci. 355, 390-395. ( 10.1016/j.amjms.2017.08.013) [DOI] [PubMed] [Google Scholar]
- 21.Zheng K, et al. 2017. Bitter taste receptors as targets for tocolytics in preterm labor therapy. FASEB J. 31, 4037-4052. ( 10.1096/fj.201601323RR) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolte D, Vijayaraghavan K, Khera S, Sica D, Frishman WH. 2007. Role of magnesium in cardiovascular diseases. Cardiol. Rev. 22, 182-192. ( 10.1097/CRD.0000000000000003) [DOI] [PubMed] [Google Scholar]
- 23.Behrens M, Meyerhof W. 2018. Vertebrate bitter taste receptors: keys for survival in changing environments. J. Agric. Food Chem. 66, 2204-2213. ( 10.1021/acs.jafc.6b04835) [DOI] [PubMed] [Google Scholar]
- 24.Li D, Zhang J. 2014. Diet shapes the evolution of the vertebrate bitter taste receptor gene repertoire. Mol. Biol. Evol. 31, 303-309. ( 10.1093/molbev/mst219) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Behrens M, Korsching SI, Meyerhof W. 2014. Tuning properties of avian and frog bitter taste receptors dynamically fit gene repertoire sizes. Mol. Biol. Evol. 31, 3216-3227. ( 10.1093/molbev/msu254) [DOI] [PubMed] [Google Scholar]
- 26.Wilson DE, Reeder DM. 2005. Mammal species of the world: a taxonomic and geographic reference, 3rd edn. Baltimore, MD: Johns Hopkins University Press. [Google Scholar]
- 27.Altringham JD. 1996. Bats: biology and behavior. Oxford, UK: Oxford University Press. [Google Scholar]
- 28.Zhao H, Xu D, Zhang S, Zhang J. 2012. Genomic and genetic evidence for the loss of umami taste in bats. Genome Biol. Evol. 4, 73-79. ( 10.1093/gbe/evr126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang K, Zhao H. 2015. Birds generally carry a small repertoire of bitter taste receptor genes. Genome Biol. Evol. 7, 2705-2715. ( 10.1093/gbe/evv180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang P, Josue J, Li X, Glaser D, Li W, Brand JG, Margolskee RF, Reed DR, Beauchamp GK. 2012. Major taste loss in carnivorous mammals. Proc. Natl Acad. Sci. USA 109, 4956-4961. ( 10.1073/pnas.1118360109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao H, Zhang J. 2012. Mismatches between feeding ecology and taste receptor evolution: an inconvenient truth. Proc. Natl Acad. Sci. USA 109, E1464, author reply E5. ( 10.1073/pnas.1205205109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiao H, Wang Y, Zhang L, Jiang P, Zhao H. 2018. Lineage-specific duplication and adaptive evolution of bitter taste receptor genes in bats. Mol. Ecol. 27, 4475-4488. ( 10.1111/mec.14873) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruedi M, Mayer F. 2001. Molecular systematics of bats of the genus Myotis (Vespertilionidae) suggests deterministic ecomorphological convergences. Mol. Phylogenet. Evol. 21, 436-448. ( 10.1006/mpev.2001.1017) [DOI] [PubMed] [Google Scholar]
- 34.Ruedi M, Stadelmann B, Gager Y, Douzery EJP, Francis CM, Lin L-K, Guillén-Servent A, Cibois A. 2013. Molecular phylogenetic reconstructions identify East Asia as the cradle for the evolution of the cosmopolitan genus Myotis (Mammalia, Chiroptera). Mol. Phylogenet. Evol. 69, 437-449. ( 10.1016/j.ympev.2013.08.011) [DOI] [PubMed] [Google Scholar]
- 35.Zhao H, Zhou Y, Pinto CM, Charles-Dominique P, Galindo-Gonzalez J, Zhang S, Zhang J. 2010. Evolution of the sweet taste receptor gene Tas1r2 in bats. Mol. Biol. Evol. 27, 2642-2650. ( 10.1093/molbev/msq152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong W, Zhao H. 2014. Vampire bats exhibit evolutionary reduction of bitter taste receptor genes common to other bats. Proc. Biol. Sci. 281, 20141079. ( 10.1098/rspb.2014.1079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kürten L, Schmidt U. 1982. Thermoperception in the common vampire bat (Desmodus rotundus). J. Comp. Physiol. A 146, 223-228. ( 10.1007/BF00610241) [DOI] [Google Scholar]
- 38.Bahlman JW, Kelt DA. 2007. Use of olfaction during prey location by the common vampire bat (Desmodus rotundus). Biotropica 39, 147-149. ( 10.1111/j.1744-7429.2006.00218.x) [DOI] [Google Scholar]
- 39.Kumar S, Stecher G, Suleski M, Hedges SB. 2017. TimeTree: a resource for timelines, timetrees, and divergence times. Mol. Biol. Evol. 34, 1812-1819. ( 10.1093/molbev/msx116) [DOI] [PubMed] [Google Scholar]
- 40.Ballesteros JA, Hormiga G. 2016. A new orthology assessment method for phylogenomic data: unrooted phylogenetic orthology. Mol. Biol. Evol. 33, 2117-2134. ( 10.1093/molbev/msw069) [DOI] [PubMed] [Google Scholar]
- 41.Saraiva LR, Korsching SI. 2007. A novel olfactory receptor gene family in teleost fish. Genome Res. 17, 1448-1457. ( 10.1101/gr.6553207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson RD, Elias DJ, Shumake SA, Gaddis SE. 1982. Taste preferences of the common vampire bat (Desmodus rotundus). J. Chem. Ecol. 8, 715-721. ( 10.1007/BF00988313) [DOI] [PubMed] [Google Scholar]
- 43.Park H, Hall ER. 1951. The gross anatomy of the tongues and stomachs of eight new world bats. Trans. Kans. Acad. Sci. 54, 64-72. ( 10.2307/3626255) [DOI] [Google Scholar]
- 44.Suthers RA. 1970. Vision, olfaction taste. In Biology of bats (ed. WA Wimsatt), pp. 265–304. New York, NY: Academic Press. [Google Scholar]
- 45.Brightman V (ed.). 1976. Vallate-foliate papilla complex and suckling behavior. Anatomical record. New York, NY: Wiley-Liss. [Google Scholar]
- 46.Lu Q, Jiao H, Wang Y, Norbu N, Zhao H. In press. Molecular evolution and deorphanization of bitter taste receptors in the vampire bat. Integr. Zool. ( 10.1111/1749-4877.12509) [DOI] [PubMed] [Google Scholar]
- 47.Behrens M, Meyerhof W. 2011. Gustatory and extragustatory functions of mammalian taste receptors. Physiol. Behav. 105, 4-13. ( 10.1016/j.physbeh.2011.02.010) [DOI] [PubMed] [Google Scholar]
- 48.Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, Bufe B, Appendino G, Behrens M. 2010. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem. Senses 35, 157-170. ( 10.1093/chemse/bjp092) [DOI] [PubMed] [Google Scholar]
- 49.Lossow K, Hubner S, Roudnitzky N, Slack JP, Pollastro F, Behrens M, Meyerhof W. 2016. Comprehensive analysis of mouse bitter taste receptors reveals different molecular receptive ranges for orthologous receptors in mice and humans. J. Biol. Chem. 291, 15358-15377. ( 10.1074/jbc.M116.718544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lei W, Ravoninjohary A, Li X, Margolskee RF, Reed DR, Beauchamp GK, Jiang P. 2015. Functional analyses of bitter taste receptors in domestic cats (Felis catus). PLoS ONE 10, e0139670. ( 10.1371/journal.pone.0139670) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suyama M, Torrents D, Bork P. 2006. PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609-W612. ( 10.1093/nar/gkl315) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thalmann S, Behrens M, Meyerhof W. 2013. Major haplotypes of the human bitter taste receptor TAS2R41 encode functional receptors for chloramphenicol. Biochem. Biophys. Res. Commun. 435, 267-273. ( 10.1016/j.bbrc.2013.04.066) [DOI] [PubMed] [Google Scholar]
- 53.Foster SR, Blank K, See Hoe LE, Behrens M, Meyerhof W, Peart JN, Thomas WG. 2014. Bitter taste receptor agonists elicit G-protein-dependent negative inotropy in the murine heart. FASEB J. 28, 4497-4508. ( 10.1096/fj.14-256305) [DOI] [PubMed] [Google Scholar]
- 54.Risso D, Behrens M, Sainz E, Meyerhof W, Drayna D. 2017. Probing the evolutionary history of human bitter taste receptor pseudogenes by restoring their function. Mol. Biol. Evol. 34, 1587-1595. ( 10.1093/molbev/msx097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Behrens M, Di Pizio A, Redel U, Meyerhof W, Korsching SI. 2020. At the root of T2R gene evolution: recognition profiles of coelacanth and zebrafish bitter receptors. Genome Biol. Evol. 13, evaa264. ( 10.1093/gbe/evaa264) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bufe B, Breslin PA, Kuhn C, Reed DR, Tharp CD, Slack JP, Kim UK, Drayna D, Meyerhof W. 2005. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr. Biol. 15, 322-327. ( 10.1016/j.cub.2005.01.047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi P, Zhang J, Yang H, Zhang YP. 2003. Adaptive diversification of bitter taste receptor genes in mammalian evolution. Mol. Biol. Evol. 20, 805-814. ( 10.1093/molbev/msg083) [DOI] [PubMed] [Google Scholar]
- 58.Wanwimolruk S, Denton JR. 1992. Plasma protein binding of quinine: binding to human serum albumin, α1-acid glycoprotein and plasma from patients with malaria. J. Pharm. Pharmacol. 44, 806-811. ( 10.1111/j.2042-7158.1992.tb03210.x) [DOI] [PubMed] [Google Scholar]
- 59.Matsuo R. 2000. Role of saliva in the maintenance of taste sensitivity. Crit. Rev. Oral Biol. Med. 11, 216-229. ( 10.1177/10454411000110020501) [DOI] [PubMed] [Google Scholar]
- 60.Sainz E, Cavenagh MM, Gutierrez J, Battey JF, Northup JK, Sullivan SL. 2007. Functional characterization of human bitter taste receptors. Biochem. J. 403, 537-543. ( 10.1042/BJ20061744) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheled-Shoval S, Behrens M, Korb A, Di Pizio A, Meyerhof W, Uni Z, Niv MY. 2017. From cell to beak: in-vitro and in-vivo characterization of chicken bitter taste thresholds. Molecules 22, 821. ( 10.3390/molecules22050821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Freeman P. 2000. Macroevolution in microchiroptera: recoupling morphology and ecology with phylogeny. Evol. Ecol. Res. 2, 317-335. [Google Scholar]
- 63.Busch C. 1988. Consumption of blood, renal function and utilization of free water by the vampire bat, Desmodus rotundus. Comp. Biochem. Physiol. A 90, 141-146. ( 10.1016/0300-9629(88)91019-5) [DOI] [PubMed] [Google Scholar]
- 64.Castro Castro F, Muñoz J, Uieda W. 2016. Análisis filogenético del murciélago hematófago Desmodus rotundus en el Valle del Cauca Colombia. Acta Agronómica 65, 65-71. ( 10.15446/acag.v65n1.47496) [DOI] [Google Scholar]
- 65.Torres-Ceron DA, Acosta-Medina CD, Restrepo-Parra E. 2019. Geothermal and mineralogic analysis of hot springs in the Puracé-La Mina sector in Cauca. Geofluids 2019, 3191454. ( 10.1155/2019/3191454) [DOI] [Google Scholar]
- 66.Stevic I, Parmar N, Paredes N, Berry LR, Chan AK. 2011. Binding of heparin to metals. Cell Biochem. Biophys. 59, 171-178. ( 10.1007/s12013-010-9129-5) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data of this study are provided in the main text and electronic supplementary material.