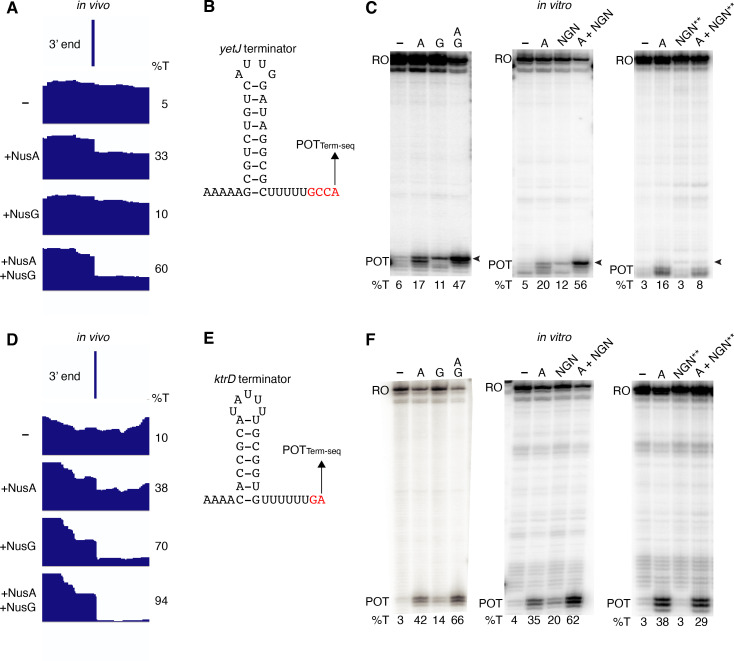
Figure 2. NusG stimulates intrinsic termination via its NGN domain.
(A) IGV screenshot of a genomic window centered around the yetJ terminator. Top track is the 3’ end identified by Term-seq. Bottom tracks are the RNA-seq coverage data for the nusAdep ΔnusG (-), ΔnusG (+NusA), nusAdep (+NusG), and wild-type (WT) (+NusA +NusG) strains. %T in each strain is shown on the right of each track. Transcription proceeds from left to right. (B) YetJ terminator showing the point of termination identified by Term-seq in vivo (POTTerm-seq). Disruptions in the U-rich tract are shown in red. The upstream A tract is also shown. (C) Single-round in vitro termination assay with the yetJ terminator. Experiments were performed in the absence (–) or presence of NusA (A), NusG (G), WT NGN domain, and/or mutant NGN domain as indicated (mutant NGN domain is signified as NGN**). Positions of terminated (POT) and run-off (RO) transcripts are marked. The arrowhead marks the most distal POT. %T is shown below each lane. (D–F) Identical to panels (A–C) except that it is the ktrD terminator.