Abstract
Bacillus subtilis develops genetic competence for the uptake of foreign DNA when cells enter stationary phase and a high cell density is reached. These signals are integrated by the competence transcription factor ComK, which is subject to transcriptional, post-transcriptional and post-translational regulation. Many proteins are involved in the development of competence, both to control ComK activity and to mediate DNA uptake. However, for many proteins, the precise function they play in competence development is unknown. In this study, we assessed whether proteins required for genetic transformation play a role in the activation of ComK or rather act downstream of competence gene expression. While these possibilities could be distinguished for most of the tested factors, we assume that two proteins, PNPase and the transcription factor YtrA, are required both for full ComK activity and for the downstream processes of DNA uptake and integration. Further analyses of the role of the transcription factor YtrA for the competence development revealed that the overexpression of the YtrBCDEF ABC transporter in the ytrA mutant causes the loss of genetic competence. Moreover, overexpression of this ABC transporter also affects biofilm formation. Since the ytrGABCDEF operon is naturally induced by cell wall-targeting antibiotics, we tested the cell wall properties upon overexpression of the ABC transporter and observed an increased thickness of the cell wall. The composition and properties of the cell wall are important for competence development and biofilm formation, suggesting that the observed phenotypes are the result of the increased cell wall thickness as an outcome of YtrBCDEF overexpression.
Keywords: genetic competence, biofilm formation, ABC transporter, cell wall homeostasis, Bacillus subtilis
Introduction
The Gram-positive model bacterium Bacillus subtilis has evolved many different ways to survive harsh environmental conditions, i.e., it can form highly resistant spores, secrete toxins to kill and cannibalize neighboring cells, form resistant macroscopic biofilms or become competent for transformation [reviewed in López and Kolter (2010)].
Development of genetic competence is a strategy, which allows bacterial cells to take up foreign DNA from the environment in order to increase the genetic variability of the population. Competence is developed during the transition from exponential to stationary phase of growth as a response to increased cell density and nutrient limitation. In B. subtilis, genetic competence is developed in a bistable manner, meaning that only about 10–20% of the cells of a population change their physiological characteristics and become competent for transformation, leaving the rest of the population non-competent (Haijema et al., 2001; Maamar and Dubnau, 2005). Whether a specific cell becomes competent or not depends on the level of the master regulator ComK (van Sinderen et al., 1995), whose cellular amount is tightly controlled by a complex network of regulators acting on the transcriptional, post-transcriptional as well as on post-translational levels [for a detailed overview see Maier (2020)].
Transcription of the comK gene is controlled by three repressor proteins, Rok, CodY, and AbrB (Serror and Sonenshein, 1996; Hoa et al., 2002; Hamoen et al., 2003), moreover, comK transcription is activated by the transcriptional regulator DegU (Hamoen et al., 2000). Another important player for regulation of comK expression is Spo0A-P, which controls the levels of the AbrB repressor and additionally supports activation of comK expression by antagonizing Rok (Hahn et al., 1995; Mirouze et al., 2012). The presence of phosphorylated Spo0A directly links competence to other lifestyles, since Spo0A-P is also involved in pathways leading to sporulation or biofilm formation (Aguilar et al., 2010). When ComK levels reach a certain threshold, it binds its own promoter region to further increase its own expression, thereby creating a positive feedback loop, which leads to full activation of competence (Maamar and Dubnau, 2005; Smits et al., 2005).
ComK levels are also controlled post-transcriptionally by the Kre protein, which destabilizes the comK mRNA (Gamba et al., 2015). Post-translational regulation is achieved through the adapter protein MecA, which sequesters ComK and directs it toward degradation by the ClpCP protease (Turgay et al., 1998). During competence, this degradation is prevented by a small protein, ComS, which is expressed in response to quorum sensing (Nakano et al., 1991).
ComK activates expression of more than 100 genes (Berka et al., 2002; Hamoen et al., 2002; Ogura et al., 2002; Boonstra et al., 2020). Whereas a clear role in competence development has been assigned to many of the ComK regulon members, the roles of some ComK-dependent genes remain unclear. Similarly, many single deletion mutant strains were identified as competence deficient, for which the reasons for this deficiency are known. However, there are still many single deletion mutants, in which the reason for the loss of competence remains unknown. Typical examples for this are mutants lacking various RNases, namely RNase Y, RNase J, or PNPase (Luttinger et al., 1996; Figaro et al., 2013). Recently, a library of B. subtilis single gene deletion mutants was screened for various phenotypes, including competence development (Koo et al., 2017). This screen revealed 21 mutants with completely abolished competence. Out of those, 16 are known to be involved in the control of the ComK master regulator, DNA uptake or genetic recombination. However, in case of the other five competence-deficient strains the logical link to competence remains elusive.
Here, we have focused on some of these factors to investigate their role in genetic competence in more detail. We took advantage of the fact that artificial overexpression of ComK and ComS significantly increases transformation efficiency independently of traditional ComK and ComS regulation (Rahmer et al., 2015). This enables the identification of genes that are involved in competence development due to a function in comK expression or for other specific reasons downstream of ComK activity. We identified the YtrBCDEF ABC transporter, which is encoded in the ytrGABCDEF operon as an important player for B. subtilis differentiation, since its overexpression does not only result in a complete loss of competence by a so far unknown mechanism, it also affects the proper development of other lifestyles of B. subtilis. We hypothesize that the production of a thicker cell wall upon overexpression of the proteins encoded by the ytrGABCDEF operon is likely the cause of the observed competence and biofilm defects.
Materials and Methods
Bacterial Strains and Growth Conditions
Bacillus subtilis strains used in this study are listed in Table 1. Lysogeny broth (LB) (Sambrook et al., 1989) was used to grow Escherichia coli and B. subtilis, unless otherwise stated. When required, media were supplemented with antibiotics at the following concentrations: ampicillin 100 μg ml–1 (for E. coli) and chloramphenicol 5 μg ml–1, kanamycin 10 μg ml–1, spectinomycin 250 μg ml–1, tetracycline 12.5 μg ml–1, and erythromycin 2 μg ml–1 plus lincomycin 25 μg ml–1 (for B. subtilis). For agar plates, 15 g l–1 Bacto agar (Difco) was added.
TABLE 1.
Bacillus subtilis strains used in this study.
| Strain | Genotype | Sourcea |
| 168 | trpC2 | Laboratory collection |
| BKE30420 | trpC2 ΔytrE::ermC | Koo et al., 2017 |
| BKE30430 | trpC2 ΔytrD::ermC | Koo et al., 2017 |
| BKE30440 | trpC2 ΔytrC::ermC | Koo et al., 2017 |
| BKE30450 | trpC2 ΔytrB::ermC | Koo et al., 2017 |
| PG389 | amyE::PcomG-lacZ-gfp-cat | Gamba et al., 2015 |
| PG10b | yvcA::(PmtlA-comKS) | Reuß et al., 2017 |
| DK1042 | comIQ12L | Konkol et al., 2013 |
| CCB434 | ΔrnjA::spc | Figaro et al., 2013 |
| CCB441 | Δrny::spc | Figaro et al., 2013 |
| GP811 | trpC2 ΔgudB::cat rocG::Tn10 spc amyE::(gltA-lacZ aphA3) ΔansR::tet | Flórez et al., 2011 |
| GP1152 | trpC2 ΔansR::tetR | GP811 → 168 |
| GP1748 | trpC2 ΔpnpA::aphA3 | Cascante-Estepa et al., 2016 |
| GP2155 | trpC2 ΔnrnA::aphA3 | See section “Materials and Methods” |
| GP2501 | trpC2 Δrny::spc | CCB441 → 168 |
| GP2506 | trpC2 ΔrnjA::spc | CCB434 → 168 |
| GP2559 | comIQ12L ΔymdB::cat | Kampf et al., 2018 |
| GP2612 | trpC2 ΔgreA::aphA3 | See section “Materials and Methods” |
| GP2618 | trpC2 yvcA-PmtlA-comKS-ermC-hisI | See section “Materials and Methods” |
| GP2620 | trpC2 yvcA-PmtlA-comKS-cat-hisI | See section “Materials and Methods” |
| GP2621 | trpC2 yvcA-PmtlA-comKS-ermC-hisI ΔpnpA::aphA3 | GP1748 → GP2618 |
| GP2624 | trpC2 yvcA-PmtlA-comKS-ermC-hisI Δrny::spc | GP2501 → GP2618 |
| GP2626 | trpC2 yvcA-PmtlA-comKS-ermC-hisI ΔrnjA::spc | GP2506 → GP2618 |
| GP2630 | trpC amyE::PcomG-lacZ-gfp-cat | PG389 → 168 |
| GP2640 | trpC2 ΔftsH::aphA3 | See section “Materials and Methods” |
| GP2641 | trpC2 ΔytrA::spc | See section “Materials and Methods” |
| GP2643 | trpC2 ΔcomEC::spc | See section “Materials and Methods” |
| GP2644 | trpC2 ΔdegU::aphA3 | See section “Materials and Methods” |
| GP2646 | trpC2 ΔytrGABCDEF::ermC | See section “Materials and Methods” |
| GP2647 | trpC2 ΔytrA::ermC | See section “Materials and Methods” |
| GP2652 | trpC2 yvcA-PmtlA-comKS-cat-hisI ΔftsH::aphA3 | GP2640 → GP2620 |
| GP2653 | trpC2 yvcA-PmtlA-comKS-cat-hisI ΔnrnA::aphA3 | GP2155 → GP2620 |
| GP2654 | trpC2 yvcA-PmtlA-comKS-cat-hisI ΔgreA::aphA3 | GP2612 → GP2620 |
| GP2655 | trpC2 yvcA-PmtlA-comKS-cat-hisI ΔytrA::spc | GP2641 → GP2620 |
| GP2659 | trpC2 yvcA-PmtlA-comKS-cat-hisI ΔcomEC::spc | GP2643 → GP2620 |
| GP2660 | trpC2 yvcA-PmtlA-comKS-cat-hisI ΔdegU::aphA3 | GP2644 → GP2620 |
| GP2664 | trpC2 amyE::PcomG-lacZ-gfp ΔftsH::aphA3 | GP2640 → GP2630 |
| GP2665 | trpC2 amyE::PcomG-lacZ-gfp ΔnrnA::aphA3 | GP2155 → GP2630 |
| GP2666 | trpC2 amyE::PcomG-lacZ-gfp ΔgreA::aphA3 | GP2612 → GP2630 |
| GP2667 | trpC2 amyE::PcomG-lacZ-gfp ΔytrA::spc | GP2641 → GP2630 |
| GP2671 | trpC2 amyE::PcomG-lacZ-gfp ΔcomEC::spc | GP2643 → GP2630 |
| GP2672 | trpC2 amyE::PcomG-lacZ-gfp ΔdegU::aphA3 | GP2644 → GP2630 |
| GP2700 | trpC2 ΔytrF::cat | See section “Materials and Methods” |
| GP3186 | trpC2 ΔytrGABCDE::ermC | See section “Materials and Methods” |
| GP3187 | trpC2 ΔytrF::cat ΔytrA::ermC | GP2647 → GP2700 |
| GP3188 | trpC2 ΔytrB | pDR244 → BKE30450 |
| GP3189 | trpC2 ΔytrC | pDR244 → BKE30440 |
| GP3190 | trpC2 ΔytrD | pDR244 → BKE30430 |
| GP3191 | trpC2 ΔytrE | pDR244 → BKE30420 |
| GP3193 | trpC2 ΔytrA::ermC ΔytrB | See section “Materials and Methods” |
| GP3194 | trpC2 ΔytrA::ermC ΔytrC | See section “Materials and Methods” |
| GP3195 | trpC2 ΔytrA::ermC ΔytrD | GP2647 → GP3190 |
| GP3196 | trpC2 ΔytrA::ermC ΔytrE | See section “Materials and Methods” |
| GP3197 | trpC2 ganA::PxylA-ytrF-aphA3 | pGP2184 → 168 |
| GP3200 | trpC2 amyE::PcomG-lacZ-gfp-cat ytrGABCDEF::ermC | GP2646 → GP2630 |
| GP3205 | trpC2 ΔytrCD::cat | See section “Materials and Methods” |
| GP3206 | trpC2 ΔytrA::ermC ΔytrB ΔytrE | See section “Materials and Methods” |
| GP3207 | comIQ12L ΔytrGABCDEF::ermC | GP2646 → DK1042 |
| GP3212 | comIQ12L ΔytrA::spc | GP2641 → DK1042 |
| BLMS2 | trpC2 ΔytrCD | See section “Materials and Methods” |
| BLMS3 | trpC2 ΔytrA::erm ΔytrCD | See section “Materials and Methods” |
aArrows indicate construction by transformation. bThis genome-reduced strain [see Reuß et al. (2017) for details] was used to amplify the PmtlA-comKS cassette.
DNA Manipulation and Strain Construction
S7 Fusion DNA polymerase (Mobidiag, Espoo, Finland) was used as recommended by the manufacturer. DNA fragments were purified using the QIAquick PCR Purification Kit (Qiagen, Hilden, Germany). DNA sequences were determined by the dideoxy chain termination method (Sambrook et al., 1989). Chromosomal DNA from B. subtilis was isolated using the peqGOLD Bacterial DNA Kit (Peqlab, Erlangen, Germany) and plasmids were purified from E. coli using the NucleoSpin Plasmid Kit (Macherey-Nagel, Düren, Germany). Oligonucleotides used in this study are listed in Supplementary Table 1. Deletion of the degU, comEC, ftsH, greA, ytrA, nrnA, and ytrF genes as well as ytrG-ytrE, and ytrGABCDEF regions was achieved by transformation with PCR products containing an antibiotic resistance cassette flanked by up- and downstream fragments of the target genes as described previously (Youngman, 1990; Guérout-Fleury et al., 1995; Wach, 1996). The identity of the modified genomic regions was verified by DNA sequencing. To construct strains GP2618 and GP2620 harboring the PmtlA-comKS cassette coupled to the antibiotic resistance gene, we first amplified PmtlA-comKS from strain PG10 (Reuß et al., 2017) as well as the resistance genes from pDG646 and pGEM-cat, respectively (Youngman, 1990; Guérout-Fleury et al., 1995) and the genes flanking the intended integration site, i.e., yvcA and hisI from B. subtilis 168. Subsequently, those DNA fragments were fused in another PCR reaction and the final product was used to transform B. subtilis 168. Correct insertion was verified by PCR amplification and sequencing. Markerless deletions of ytrB, ytrC, ytrD, and ytrE genes were generated using the plasmid pDR244 as previously described (Koo et al., 2017). In short, strains BKE30450, BKE30440, BKE30430, and BKE30420 were transformed with plasmid pDR244 and transformants were selected on LB agar plates supplemented with spectinomycin at 30°C. Transformants were then streaked on plain LB agar plates and incubated at 42°C to cure the plasmid, which contains a thermo-sensitive origin of replication. Single colonies were screened for spectinomycin and erythromycin/lincomycin sensitivity. The markerless deletion of ytrCD was achieved using the cre-lox system. First, Lox71 and Lox66 sites were attached to the kanamycin resistance gene using primers CZ168/169. The resulting PCR product was cut with EcoRI and XbaI and ligated with plasmid pBluescript II that had been cut with the same enzymes, yielding plasmid pGP2514. The kanamycin resistance cassette flanked by Lox71 and Lox66 sites was subsequently amplified from pGP2514 using primers CZ200/201. Next, 800 bp up- and downstream of ytrCD were amplified using primers MB198/LMS262 and MB201/LMS260, respectively. Primers LMS262 and LMS260 contained overhangs complementary to primers CZ200/201. The three fragments were subsequently fused using primers MB198/MB201, the resulting PCR product transformed into the B. subtilis wildtype strain 168 and transformants selected on SP medium containing kanamycin. The strain was cured from the kanamycin resistance cassette using plasmid pDR244 as described above, resulting in the construction of strain BLMS2. Markerless deletion was confirmed by PCR with primers flanking the deletion site. The resulting strains GP3188, GP3189, GP3190, GP3191, and BLMS2 were used for subsequent deletion of the ytrA gene. For the construction of GP3193, GP3194, and GP3196, the ytrA deletion cassette was amplified using primers MB66/69 and genomic DNA of strains GP3188, GP3189, and GP3191, respectively, as template. For the construction of BLMS3, the ΔytrA:erm region was amplified using primers MB173/70 and genomic DNA of strain GP2647. The corresponding ytrA deletion cassettes were subsequently transformed into GP3188, GP3189, GP3191, and BLMS2. Strain GP3195 was constructed by transformation with genomic DNA of the ytrA deletion strain. Deletion of the ytrA gene and preservation of selected markerless deletions were confirmed via PCR. To construct GP3206, the ΔytrA:erm ΔytrB region of strain GP3193 was amplified using primers MB66 and MB180 and transformed into GP3191.
Transformation of Bacillus subtilis Strains
Transformation experiments were conducted based on the two-step protocol as described previously (Kunst and Rapoport, 1995). Briefly, cells were grown at 37°C at 200 rpm in 10 ml MNGE medium containing 2% glucose, 0.2% potassium glutamate, 100 mM potassium phosphate buffer (pH 7), 3.4 mM trisodiumcitrate, 3 mM MgSO4, 42 μM ferric ammonium citrate, 0.24 mM L-tryptophan and 0.1% casein hydrolyzate. During the transition from exponential to stationary phase, the culture was diluted with another 10 ml of MNGE medium (without casein hydrolyzate) and incubated for 1 h at 37°C with shaking. For transformation experiments with strain GP3197, 0.5% xylose was added to both media. Afterward, 250 ng of chromosomal DNA was added to 400 μl of cells and incubated for 30 min at 37°C. One hundred microliter of Expression mix (2.5% yeast extract, 2.5% casein hydrolyzate, 1.22 mM tryptophan) was added and cells were grown for 1 h at 37°C, before spreading onto selective LB plates containing appropriate antibiotics.
Transformation of strains harboring PmtlA-comKS, in which the expression of comK and comS is induced in the presence of mannitol, was performed as previously described (Rahmer et al., 2015). Briefly, an overnight culture was diluted in 5 ml LB to an initial OD600 of 0.1 and incubated at 37°C and 200 rpm. After 90 min of incubation, 5 ml of fresh LB containing 1% mannitol and 5 mM MgCl2 were added and the bacterial culture was incubated for an additional 90 min. Cells were then pelleted by centrifugation for 10 min at 2,000 × g and the pellet was re-suspended in the same amount of fresh LB medium. 1 ml aliquots were distributed into 1.5 ml reaction tubes and 250 ng of chromosomal DNA was added to each of them. The cell suspension was incubated for 1 h at 37°C and transformants were selected on LB plates as described above.
Plasmid Construction
All plasmids used in this study are listed in Table 2. E. coli DH5a (Sambrook et al., 1989) was used for plasmid constructions and transformation using standard techniques (Sambrook et al., 1989). To overproduce the YtrF protein, the ytrF gene was placed under the control of a xylose inducible promotor. For this purpose, we cloned the ytrF gene into the backbone of pGP888 via the XbaI and KpnI sites (Diethmaier et al., 2011).
TABLE 2.
Plasmids used in this study.
| Plasmid | Relevant characteristics | Primers | References |
| pDR244 | cre + Ts origin | – | Koo et al., 2017 |
| pGEM-cat | Amplification of the cat cassette | – | Youngman, 1990 |
| pDG646 | Amplification of the ermC cassette | – | Guérout-Fleury et al., 1995 |
| pDG780 | Amplification of the aphA3 cassette | – | Guérout-Fleury et al., 1995 |
| pDG1726 | Amplification of the spc cassette | – | Guérout-Fleury et al., 1995 |
| pGP888 | ganA:PxylA; aphA3 | – | Diethmaier et al., 2011 |
| pGP2184 | pGP888-ytrF | MB186/MB187 | This study |
| pGP2514 | Amplification of aphA3-lox for the cre-lox system | CZ200/CZ201 | This study |
Quantitative Real-Time PCR
For the isolation of RNA, a single colony was used to inoculate 4 ml LB medium containing the appropriate antibiotics. The cells were grown over the day at 37°C with agitation and used to inoculate 10 ml MNGE defined medium and incubated overnight. The next day, 100 ml MNGE medium were inoculated to an OD600 of 0.1 and the cells were incubated at 37°C until they reached an OD600 of 1.0. 25 ml of each culture were mixed with 15 ml frozen killing buffer (20 mM Tris, pH 7.5, 5 mM MgCl2, 20 mM NaN3), followed by a 5-min centrifugation step at 8,000 rpm and 4°C. Pellets were snap-frozen in liquid nitrogen and stored at −80°C. Disruption of cells and isolation of RNA was carried out as previously described (Meinken et al., 2003). 5 μg isolated RNA were digested with 5 μl DNase I (1 U/μl, Thermo Scientific) for 40 min at 37°C. The reaction was stopped by adding 2.5 μl 25 mM EDTA and incubating the samples at 65°C for 10 min. To verify that the isolated RNA is free of DNA, a check PCR was performed using primers KG44/KG45. Genomic DNA from B. subtilis 168 was used as control.
Quantitative real-time PCR (qRT-PCR) was carried out using the One-Step reverse transcription PCR kit, the Bio-Rad iCycler and the Bio-Rad iQ5 software (Bio-Rad, Munich, Germany). Three technical and three biological repeats were performed. Primers KG44/45 and KG42/43 were used to determine transcript amounts of the ribosomal genes rpsE and rpsJ, respectively, which were used as internal controls. Transcript amounts for ytrE and ytrF were monitored using primers MB219/220 and MB224/225, respectively. The average of the cycle threshold (CT) values of rpsE and rpsJ were used to normalize the CT-values obtained for ytrE and ytrF. For each strain, the fold changes of ytrE and ytrF expression were calculated using the ΔΔCT-method.
Biofilm Assay
To analyze biofilm formation, selected strains were grown in LB medium to an OD600 of about 0.5–0.8 and 10 μl of the culture were spotted onto MSgg agar plates (Branda et al., 2001). Plates were incubated for 3 days at 30°C.
Fluorescence Microscopy
For fluorescence microscopy imaging, B. subtilis cultures were grown in 10 ml MNGE medium until the transition from exponential to stationary phase and then diluted with another 10 ml of MNGE medium as described for the transformation experiments (see section “Transformation of Bacillus subtilis Strains”). 2 ml of the bacterial culture were harvested and resuspended in 500 μl PBS. The cell suspension was mixed with 25 μl of 100 μg/ml nile red solution to stain the bacterial membranes. 5 μl of cells were pipetted on microscope slides coated with a thin layer of 1% agarose and covered with a cover glass. Fluorescence images were obtained with the AxioImager M2 fluorescence microscope, equipped with the digital camera AxioCam MRm and an EC Plan-NEOFLUAR 100X/1.3 objective (Carl Zeiss, Göttingen, Germany). Filter sets 38 (EX BP 470/40, FT 495, EM BP 525/50; Carl Zeiss) and 43 (EX BP 545/25, FT 579, EM BP 605/70; Carl Zeiss) were applied for GFP and nile red detection, respectively. Images were processed with the AxioVision Rel 4.8 software. Ratio of GFP expressing cells to the total number of cells was determined by manual examination from at least six independent growth experiments. For each experiment, the ratio of GFP expressing wildtype cells was set to one and used to calculate the relative GFP fluorescence for each mutant of the same experiment.
Transmission Electron Microscopy
To examine cell wall thickness of B. subtilis strains, cells were prepared for Transmission Electron Microscopy (TEM) as previously described (Rismondo et al., 2021). An overnight culture was inoculated to an OD600 of 0.05 in 30 ml MNGE medium and grown to an OD600 of 0.6 ± 0.1 at 37°C and 200 rpm. Cells were centrifuged for 10 min at 4,000 rpm to obtain a 100 μl cell pellet, which was then washed twice in phosphate-buffered saline (PBS, 127 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) and fixed overnight in 2.5% (w/v) glutaraldehyde at 4°C. Cells were mixed with 1.5% (w/v, final concentration in PBS) molten Bacto-Agar, kept liquid at 55°C. After solidification, the resulting agar block was cut into 1 mm3 pieces. A dehydration series was performed (15% aqueous ethanol solution for 15 min, 30, 50, 70, and 95% for 30 min and 100% for 2 × 30 min) at 0°C, followed by an incubation step in 66% LR white resin mixture (v/v, in ethanol) (Plano, Wetzlar, Germany) for 2 h at room temperature and embedment in 100% LR-White solution overnight at 4°C. One agar piece was transferred to a gelatin capsule filled with fresh LR-white resin, which was subsequently polymerized at 55°C for 24 h. The gelatin capsule was shaped into a truncated pyramid using a milling tool (TM 60, Fa. Reichert and Jung, Vienna, Austria) An ultramicrotome (Reichert Ultracut E, Leica Microsystems, Wetzlar, Germany) and a diamond knife (Delaware Diamond Knives, Wilmington, DE, United States) were subsequently used to obtain ultrathin sections (80 nm) of the samples. The resulting sections were mounted onto mesh specimen grids (Plano, Wetzlar, Germany) and stained with 4% (w/v) uranyl acetate solution (pH 7.0) for 10 min. Microscopy images were taken on a Jeol JEM 1011 transmission electron microscope (Jeol Germany GmbH, Munich, Germany) at 80 kV, with a magnification of 30,000 and recorded with an Orius SC1000 CCD camera (Gatan Inc., Pleasanton, CA, United States). For each replicate, 20 cells were photographed and cell wall thickness was measured at three different locations using the software ImageJ (Rueden et al., 2017).
Results
ComK-Dependent and –Independent Functions of Proteins Are Required for the Development of Genetic Competence
Genetic work with B. subtilis is facilitated by the development of genetic competence, a process that depends on a large number of factors. While the specific contribution of many proteins to the development of competence is well understood, this requirement has not been studied for many other factors. In particular, several RNases (RNase Y, RNase J1, and PNPase) are required for competence, and the corresponding mutants have lost the ability to become naturally competent (Luttinger et al., 1996; Figaro et al., 2013). We are interested in the reasons for the loss of competence in these mutant strains, as well as in other single gene deletion mutants, in which the impairment in the development of natural competence is not understood (Koo et al., 2017). Therefore, we first tested the roles of the aforementioned RNases (encoded by the rny, rnjA, pnpA, and nrnA genes) as well as of the transcription elongation factor GreA, the metalloprotease FtsH and the transcription factor YtrA (Koo et al., 2017) for the development of genetic competence. For this purpose, we compared the transformation efficiencies of the corresponding mutant strains to that of a wild type strain. The comEC and degU mutants, which have completely lost their genetic competence for different reasons, were used as controls. The ComEC protein is responsible for the transport of the DNA molecule across the cytoplasmic membrane. Loss of ComEC blocks competence, but it should not affect the global regulation of competence development and expression of other competence factors (Draskovic and Dubnau, 2005). In contrast, DegU is a transcription factor required for the expression of the key regulator of competence, ComK, and thus indirectly also for the expression of all other competence genes (Hamoen et al., 2000; Shimane and Ogura, 2004). Our analysis confirmed the significant decrease in transformation efficiency for all tested strains (see Table 3). For five out of the seven strains, as well as the two control strains, competence was abolished completely, whereas transformation of strains GP2155 (ΔnrnA) and GP1748 (ΔpnpA) was possible, but severely impaired as compared to the wild type strain. These results confirm the implication of these genes in the development of genetic competence.
TABLE 3.
Effect of gene deletions on the development of genetic competence in dependence of the competence transcription factor ComKa.
| Wild type | PmtlA-comKS | |
| Mutant | Colonies per μg of DNA | |
| Wild type | 138,60017,006 | 47,9528,854 |
| ΔdegU | 00 | 60,85313,693 |
| ΔcomEC | 00 | 00 |
| ΔnrnA | 1,689316 | 34,9336,378 |
| ΔftsH | 00 | 00 |
| ΔgreA | 00 | 00 |
| Δrny | 00 | 00 |
| ΔrnjA | 00 | 00 |
| ΔpnpA | 176 | 29319 |
| ΔytrA | 00 | 467278 |
aCells were transformed with chromosomal DNA of strain GP1152 harboring a tetracycline resistance marker as described in section “Materials and Methods.”
The proteins that are required for genetic competence might play a more general role in the control of expression of the competence regulon (as known for the regulators that govern comK expression and ComK stability, e.g., the control protein DegU), or they may have a more specific role in competence development such as the control protein ComEC. To distinguish between these possibilities, we introduced the mutations into a strain that allows for the inducible overexpression of the comK and comS genes. The overexpression of comK and comS allows transformation in rich medium and hence facilitates the transformation of some competence mutants (Rahmer et al., 2015). For this purpose, we first constructed strains that contain mannitol inducible comK and comS genes fused to resistance cassettes (GP2618 and GP2620, for details see section “Materials and Methods”). Subsequently, we deleted our target genes in this genetic background and assayed transformation efficiency after induction of comKS expression (for details see section “Materials and Methods”). In the B. subtilis wild type strain 168, deletion of degU leads to a complete loss of competence, while the transformation efficiency of a degU mutant overexpressing comKS is comparable to its isogenic wild type strain. This suggests that DegU affects competence only by its role in comK expression and that DegU is no longer required in the strain with inducible comKS expression. In contrast, the competence of the comEC mutant could not be restored by the overexpression of comKS, reflecting the role of the ComEC protein on a process downstream of ComK, namely on DNA uptake (see Table 3). Of the tested strains, only the nrnA mutant showed a transformation efficiency similar to that of the isogenic control strain with inducible comKS expression. This observation suggests that nanoRNase A might be involved in the control of comK expression. In contrast, the ftsH, greA, rny, and rnjA mutants did not show any transformants even upon comKS overexpression, indicating that the corresponding proteins act downstream of ComK. Finally, we have observed a small but reproducible improvement of competence for the pnpA and ytrA mutants, indicating that PNPase and YtrA might affect comK expression. This finding is particularly striking in the case of the ytrA mutant, since this strain did not yield a single transformant in the 168 wild type background (see Table 3). However, the low number of transformants obtained with pnpA and ytrA mutants as compared to the isogenic wild type strain suggests that PNPase and the YtrA transcription factor play also a role downstream of ComK.
ComK activates transcription of many competence genes including comG (van Sinderen et al., 1995). Therefore, as a complementary approach to verify the results shown above, we decided to assess ComK activity using a fusion of the comG promoter to a promoterless GFP reporter gene (Gamba et al., 2015). For this purpose, we deleted the selected genes in strain GP2630 containing the PcomG-gfp construct. We grew the cells in competence inducing medium using the two-step protocol as we did in the previous transformation experiment. At the time point when DNA would be added to the cells during the transformation procedure, we assessed comG promoter activity in the cells using fluorescence microscopy. Since expression of ComK and thus also activation of competence takes place only in a subpopulation of cells (Smits et al., 2005), we determined the ratio of gfp expressing cells for each mutant strain compared to the wildtype strain as an indication of ComK activity (see Figure 1). Since RNase mutants tend to form chains, thus making it difficult to study fluorescence in individual cells, we did not include the RNase mutants for this analysis.
FIGURE 1.
Effect of gene deletions on the expression of a PcomG-gfp translational fusion as a readout for the activity of the competence transcription factor ComK. (A) Strains harboring the PcomG-gfp construct were grown in competence inducing medium as described in section “Materials and Methods.” Cells were analyzed by phase contrast and fluorescence microscopy and representative images are shown. Scale bar is 2 μm. (B) The percentage of GFP-expressing cells was determined for each strain. The ratio of GFP-expressing cells of each mutant compared to the wildtype strain was calculated and the result of at least six independent experiments plotted. For statistical analysis, a one-way ANOVA followed by a Dunnett’s multiple comparison test was used (*p ≤ 0.05, ***p ≤ 0.001, and ****p ≤ 0.0001).
In the wild type strain GP2630, about 40% of the cells expressed GFP, similar to previously published results (She et al., 2020), and slightly higher numbers were obtained for the control strain lacking ComEC, which is not impaired in comK and subsequent comG expression. In contrast, the control strain lacking DegU showed decreased amount of GFP expressing cells as compared to the wild type (Figure 1), which reflects the role of DegU on the activation of comK expression. In agreement with our previous finding that nanoRNase A affects comK expression or ComK activity, only about 4% of nrnA mutant cells showed expression from PcomG-gfp. Thus, the nanoRNase A encoded by nrnA seems to play a role in the regulation of comK expression, which has not been described so far. For the strain lacking GreA, we observed similar rates of GFP expressing cells as in the wild-type strain, indicating that ComK activation is not the problem, which causes loss of competence in the greA mutant. Surprisingly, we did not find any single cell expressing GFP for the ftsH mutant, where ComK expression does not seem to be the cause of competence deficiency as indicated by the previous transformation experiment. Additionally, we observed significantly decreased numbers of GFP producing cells for the ytrA deletion mutant, in which the competence deficiency could only be slightly restored by comKS overexpression. This observation suggests that FtsH and YtrA could potentially play a dual role in the development of genetic competence. On one hand, they both seem to be required for ComK activity but on the other hand, they also seem to have a ComK-independent function. However, it is also possible that the overexpression of comK and comS using the PmtlA promoter does not lead to the production of sufficient amounts of ComK to fully restore competence of the ftsH and ytrA mutants. The ytrA gene encodes a transcription factor, whose physiological function is poorly understood (Salzberg et al., 2011). Therefore, we focused our further work on understanding the role of YtrA on the development of genetic competence.
Overexpression of the YtrBCDEF ABC Transporter Inhibits Genetic Competence
The ytrA gene encodes a negative transcription regulator of the GntR family, which binds to the inverted repeat sequence AGTGTA-13bp-TACACT (Salzberg et al., 2011). In the B. subtilis genome, this sequence is present in front of two operons, its own operon ytrGABCDEF and ywoBCD. The deletion of ytrA leads to an overexpression of these two operons (Salzberg et al., 2011). It is tempting to speculate that overexpression of one of these operons is the cause for the loss of competence in the ytrA mutant. In case of the ytrA mutant used in this study, the expression of the downstream genes, namely ytrBCDEF, is controlled by the promoter of the ermC gene, which was inserted into the ytrA locus. This results in a 335- and 566-fold higher expression of ytrE and ytrF, respectively, in the ytrA mutant as compared to the B. subtilis wildtype strain (Supplementary Figure 1). To test whether the overexpression of ytrBCDEF leads to the competence deficiency of the ytrA mutant, we constructed strain GP2646, where the complete ytrGABCDEF operon is replaced by the ermC gene and assayed its genetic competence. This revealed that although deletion of ytrA fully blocks genetic competence, the transformation efficiency of the strain lacking the whole operon is comparable to the wild type strain 168 (Table 4). We conclude that overexpression of the ytrGABCDEF operon causes the loss of competence in the ytrA mutant strain. In addition, we assessed ComK activity in the mutant lacking the ytrGABCDEF operon, using the expression of the PcomG-gfp fusion as a readout. As observed for the wild type, about 40% of the mutant cells expressed comG, indicating that ComK is fully active in the mutant (Figure 1), and that the reduced activity in the ytrA mutant results from the overexpression of the operon. Initially we also aimed to delete the ywoBCD operon to assess a potential involvement in genetic competence, however several attempts to construct such a strain failed. As we were already able to show that the overexpression of the ytr operon causes the loss of competence in the ytrA mutant, we decided not to continue with this second YtrA-controlled operon.
TABLE 4.
Effect of gene deletions in the ytrGABCDEF operon on the development of genetic competencea.
| Mutant | Colonies per μg of DNA |
| Wild type | 138,60017,006 |
| ΔytrGABCDEF | 114,73314,408 |
| ΔytrA | 00 |
| ΔytrAB | 00 |
| ΔytrAC | 00 |
| ΔytrAD | 242 |
| ΔytrAE | 13751 |
| ΔytrAF | 10,180549 |
| Pxyl-ytrF | 137,53326,595 |
| ΔytrGABCDE | 108,46714,836 |
| ΔytrABE | 30988 |
| Wild type | 142,60039,074 |
| ΔytrACD | 1.72.4 |
aCells were transformed with chromosomal DNA of strain GP1152 harboring a tetracycline resistance marker as described in section “Materials and Methods.”
The ytr operon consists of seven genes (see Figure 2A). Five proteins encoded by this operon (YtrB, YtrC, YtrD, YtrE, and YtrF) are components of a putative ABC transporter (see Figure 2B), which was suggested to play a role in acetoin utilization (Quentin et al., 1999; Yoshida et al., 2000). YtrB and YtrE are supposed to be nucleotide binding proteins, YtrC and YtrD membrane spanning proteins and YtrF the substrate binding protein. Finally, ytrG is another open reading frame, which is located upstream of ytrA and encodes a peptide of 45 amino acids, which is probably not part of the ABC transporter (Salzberg et al., 2011). The expression of the ytr operon is usually kept low due to transcriptional repression by YtrA. This repression is naturally relieved in response to several lipid II-binding antibiotics or during cold shock (Beckering et al., 2002; Salzberg et al., 2011; Wenzel et al., 2012).
FIGURE 2.
Genetic organization of the ytrGABCDEF operon and organization of the putative ABC transporter YtrBCDEF. (A) Reading frames are depicted as arrows with respective gene names. Green arrows indicate genes encoding proteins suggested to form the ABC transporter; the yellow arrow indicates the gene coding for the repressor YtrA and the gray arrow indicates the small open reading frame called ytrG. The map is based on information provided in Salzberg et al. (2011). (B) Organization of the putative ABC transporter YtrBCDEF as suggested by Yoshida et al. (2000). YtrB and YtrE are nucleotide binding proteins, YtrC and YtrD membrane spanning proteins and YtrF is a substrate binding protein. The role and localization of the YtrG peptide remain elusive.
To test the involvement of the individual components of the putative YtrBCDEF ABC transporter in the development of genetic competence, we constructed double mutants of ytrA together with each of the other genes of the operon, i.e., ytrB, ytrC, ytrD, ytrE, and ytrF, and assessed their genetic competence. The results revealed that most of the double mutants are deficient in genetic transformation, as observed for the single ytrA mutant GP2647 (Table 4). However, strain GP3187 with deletions of ytrA and ytrF but still overexpressing all the other parts of the transporter had partially restored competence. YtrF could thus be a major player for the loss of competence in the overexpressing strain.
To further test the role of YtrF overexpression for the loss of competence, we used two different approaches. First, we constructed a strain with artificial overexpression of ytrF from a xylose inducible promoter (GP3197) and second, we created a strain with deletion of all other components (ytrGABCDE) of the operon, leaving only overexpressed ytrF (GP3186). Overexpression of ytrF in strain GP3186 was confirmed by qRT-PCR analysis (Figure 2). In contrast to our expectations, competence was not blocked in any of the two strains, suggesting that increased levels of YtrF alone are not enough to block the competence and that YtrF might need assistance from other components of the putative transporter for its full action and/or proper localization. The ytr operon encodes two putative nucleotide binding proteins (YtrB and YtrE) and two putative membrane spanning proteins (YtrC and YtrD), whereas YtrF is the only substrate binding protein that interacts with the transmembrane proteins. Therefore, we hypothesized that YtrF overexpression might only block genetic competence if the protein is properly localized in the membrane via YtrC and YtrD. To check this possibility, we constructed strains GP3206 and BLMS3 lacking YtrA and the nucleotide binding proteins YtrB and YtrE or the membrane proteins YtrC and YtrD, respectively, and tested their transformability. qRT-PCR analysis was again used to confirm that ytrF is overexpressed in strains GP3206 and BLMS3 as compared to the wildtype strain 168 (Supplementary Figure 1). Strain GP3206 showed very few transformants, suggesting that the overexpression of ytrC, ytrD, and ytrF, encoding the remaining transporter components, are sufficient to cause the loss of competence. Surprisingly, similar results were obtained for a B. subtilis strain lacking YtrA, YtrC, and YtrD. We thus conclude that the observed loss of competence as a result of YtrF overexpression does not depend on the nucleotide binding proteins or the transmembrane proteins. Since we did not observe a loss of competence for B. subtilis strains, in which only YtrF is overexpressed, we hypothesize that YtrF might require assistance of another, unknown factor.
Overexpression of the ytrGABCDEF Operon Leads to Alterations in Colony Morphology During Biofilm Formation
Bacillus subtilis can employ various lifestyles which are tightly interconnected through regulatory proteins (Lopez et al., 2009). Therefore, we anticipated that the overexpression of the YtrBCDEF transporter might also affect other lifestyles of B. subtilis. Indeed, it was previously shown that the ytrA mutant has a reduced sporulation efficiency (Koo et al., 2017). We thus decided to examine the effect of ytrA deletion on biofilm formation. To that end, we deleted the ytrA gene or the whole ytrGABCDEF operon from the biofilm-forming strain DK1042 (Konkol et al., 2013). We then tested the biofilm formation of the resulting strains on biofilm inducing MSgg agar (Branda et al., 2001). As expected, the wild type strain DK1042 formed structured colonies that are indicative of biofilm formation. In contrast, the negative control GP2559, a ymdB mutant that is known to be defective in biofilm formation (Kampf et al., 2018), formed completely smooth colonies. The biofilm formed by the ytrA mutant GP3212 was less structured, more translucent and with only some tiny wrinkles on its surface, indicating that biofilm formation was slightly inhibited but not fully abolished upon loss of YtrA. In contrast, strain GP3207 lacking the complete ytrGABCDEF operon formed a biofilm indistinguishable from the one of the parental strain DK1042 (see Figure 3). This observation suggests that overexpression of components of the YtrBCDEF ABC transporter could interfere with biofilm formation in B. subtilis.
FIGURE 3.
Biofilm formation is affected by ytrA deletion. Biofilm formation was examined in the wild type strain DK1042 and respective deletion mutants of ymdB (GP2559), ytrA (GP3212), and ytrGABCDEF (GP3207). The biofilm assay was performed on MSgg agar plates as described in section “Materials and Methods.” The plates were incubated for 3 days at 30°C. All images were taken at the same magnification.
Overexpression of the ytr Operon Increases Cell Wall Thickness
In previous experiments, we have shown that the overexpression of the ytr operon interferes with the development of genetic competence and, to some extent, with biofilm formation. However, it remains unclear why competence and biofilm formation are affected by the overexpression of the ytr operon. The ytr operon is repressed under standard laboratory conditions by the YtrA transcription regulator and this repression is naturally relieved upon exposure to very specific stress conditions such as cell wall targeting antibiotics and cold shock (Beckering et al., 2002; Cao et al., 2002; Mascher et al., 2003; Salzberg et al., 2011; Nicolas et al., 2012; Wenzel et al., 2012). The possible link between antibiotic resistance, genetic competence, and biofilm formation is not apparent, however, cell wall properties might provide an answer. Indeed, it has been shown that wall teichoic acids, the uppermost layer of the cell wall, are important for biofilm formation and for DNA binding during transformation (Bucher et al., 2015; Mirouze et al., 2018; Zhu et al., 2018).
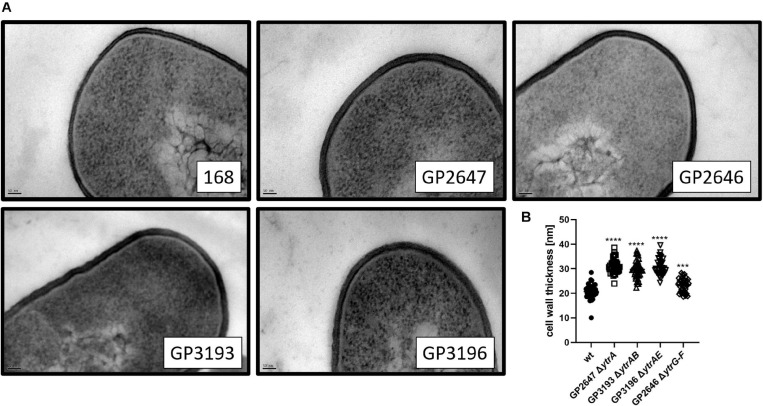
To test the hypothesis that overexpression of the putative ABC transporter encoded by the ytrGABCDEF operon affects cell wall properties of B. subtilis cells, we compared the cell morphology of the wild type, the non-competent ytrA, ytrAB, ytrAE mutants as well as the competent ytrGABCDEF mutant lacking the complete operon by TEM. While the wild type strain showed an average cell wall thickness of 21 nm, which is in agreement with previous studies (Beveridge and Murray, 1979), the ytrA (GP2647) mutant showed a significant increase in cell wall thickness with an average of 31 nm. B. subtilis strains lacking one of the nucleotide binding proteins of the transporter, YtrB or YtrE, in addition to YtrA also produced a thicker cell wall with an average thickness of 30 nm and 31 nm, respectively. In contrast, such an increase was not observed for the whole operon mutant (GP2646) that had an average cell wall thickness of 23 nm (see Figure 4). These observations are in agreement with the hypothesis that the overexpression of the YtrBCDEF ABC transporter affects cell wall properties and could potentially explain the loss of genetic competence and the minor biofilm formation defect of the ytrA mutant.
FIGURE 4.
Cell wall thickness is increased in ytrA, ytrAB, and ytrAE mutants. (A) Shown are representative transmission electron microscopy images of the wild type strain 168, the ytrA mutant (GP2647), the ytrAB (GP3193), and the ytrAE (GP3196) double mutants and the ytrGABCDEF mutant (GP2646). Scale bar is 50 nm. (B) Cell wall thickness of B. subtilis wild type and ytr mutants. The cell wall thickness of 40 individual cells per strain was measured as described in section “Materials and Methods”and plotted. For statistical analysis, a one-way ANOVA followed by a Dunnett’s comparison test was used (***p ≤ 0.001 and ****p ≤ 0.0001).
Discussion
Genetic competence is a multifactorial process in the Gram-positive organism B. subtilis. In a genome-wide study, several single deletion mutants were identified, which are significantly impaired in competence development (Koo et al., 2017). However, it remains to be determined why the deletion of the corresponding genes affect this process. Here, we aimed to investigate the role of the transcription elongation factor GreA, the metalloprotease FtsH, the transcriptional regulator YtrA and the RNases RNase Y, RNase J1, PNPase, and nanoRNase A on competence development in B. subtilis. In accordance with previous studies, deletion of greA, ftsH, ytrA, rny, and rnjA resulted in a complete loss of competence and the competence of strains lacking the PNPase or nanoRNase A was severely reduced (Figaro et al., 2013; Koo et al., 2017). The overexpression of comKS, coding for the master regulator of competence ComK and the small antiadapter protein ComS, could not restore the competence of the rny, rnjA, and greA mutants, suggesting that RNase Y, RNase J1, and GreA act downstream of the control of comK expression. In contrast, overexpression of comKS restored the competence efficiency of the nrnA mutant to a level comparable to the isogenic wild type strain and we hypothesize that nanoRNase A directly or indirectly affects comK expression. Lack of the metalloprotease FtsH led to a complete loss of genetic competence that could not be restored by the overexpression of comKS suggesting that FtsH acts downstream of ComK. However, we could not detect any expression from the comG promoter, which contradicts the first result. We hypothesize that FtsH might have a dual role: on one hand, it could influence the expression of comK and on the other hand, it could have a ComK-independent role. However, it is also possible that ComK is not properly expressed in the ftsH mutant or that FtsH is involved in the turnover of MecA, Rok or AbrB leading to a reduced expression of comK or an enhanced degradation of ComK. Further studies are required to understand the precise role of FtsH on competence development in B. subtilis. For the pnpA and ytrA mutants, we observed a slight improvement of competence, suggesting that PNPase and YtrA could play a role in comK expression control. However, the genetic competence could not be fully restored by comKS overexpression, indicating that ComK might not be the only factor contributing to the competence deficiency of the pnpA and ytrA mutants. Again, elevated ComK levels might not be sufficient to completely suppress the competence deficiency of these two mutants.
The transcriptional repressor YtrA is encoded in the ytrGABCDEF operon, whose expression is induced in the presence of cell wall-acting antibiotics and cold shock (Beckering et al., 2002; Salzberg et al., 2011; Wenzel et al., 2012). The ytrGABCDEF operon further codes for a putative ABC transporter, YtrBCDEF, which was suggested to be involved in acetoin utilization in B. subtilis (Quentin et al., 1999; Yoshida et al., 2000). Previous work already revealed that lack of YtrA leads to a competence defect and a decreased sporulation efficiency (Koo et al., 2017). In this work we could show that loss of genetic competence of the ytrA mutant is caused by the overexpression of the ytrGABCDEF operon. Furthermore, this phenotype seems to partially depend on the presence of the substrate binding protein YtrF. Surprisingly, transformations of a ytrA mutant also lacking both ATP-binding proteins, YtrB and YtrE, or both transmembrane proteins, YtrC and YtrD, only leads to a small number of colonies, suggesting that the loss of genetic competence of a ytrA mutant does not require the activity of the YtrBCDEF transporter.
Based on the partial complementation of genetic competence of the ytrA mutant upon comKS overexpression, one might expect that the loss of YtrA and the concomitant overexpression of the ABC transporter somehow interfere with a process upstream of ComK activation. However, competence is developed in an all or nothing scenario, and cells in which the ComK levels reach a certain threshold should become competent (Haijema et al., 2001; Maamar and Dubnau, 2005). Our observation that comKS overexpression only partially restores competence of the ytrA mutant suggests that ComK levels are not the only factor that limit competence of the ytrA mutant. If the ytrA deletion would only interfere with ComK activation, one would expect wild type-like competence upon overexpression of comKS, which was not the case. However, it is also possible that ComK levels in the ytrA mutant background are not sufficient to restore genetic competence, even after mannitol-induced overexpression of comKS. Another explanation could be that the activity of MecA is enhanced in the absence of YtrA leading to a faster degradation of ComK by the ClpCP complex. In addition to the loss of competence, the ytrA mutant produces a thicker cell wall as compared to the B. subtilis wild type, which could also have an impact on genetic competence. The DNA uptake apparatus must be adapted to cell wall thickness in order to ensure that the extracellular DNA can reach the ComG/ComE DNA transport complex. Due to the increased cell wall thickness upon overexpression of the YtrBCDEF ABC transporter, the DNA is probably unable to get in contact with the ComG pili. Overexpression of ComK will then result in the increased production of DNA-binding ComG on the cell surface of all cells of the population (compared to about 10% in the wild-type strain transformed with the classical two-step protocol). This would simply increase the probability that foreign DNA reaches the DNA uptake machinery in some cells, which then leads to the appearance of a few transformants as observed in our study. On the other hand, the results obtained by fluorescence microscopy revealed a decreased transcription from the ComK dependent comG promoter in the ytrA mutant. However, this expression is expected to be wild type-like if the action of the YtrBCDEF ABC transporter would not interfere with ComK activity and only block DNA uptake as a result of the thicker cell wall as suggested above. Again, the thicker cell wall might be responsible, since ComK expression is induced by the detection of extracellular quorum-sensing signals (both ComXPA and Rap-Phr systems) and this induction depends on the accessibility of the sensor domains for the pheromones [reviewed in Maier (2020)], which might be impaired in the strain with altered cell wall thickness.
In addition to the loss of genetic competence, it was previously shown that ytrA deletion leads to decreased sporulation efficiency (Koo et al., 2017) and we have shown that it also has a minor effect on biofilm formation. Considering the changed cell wall properties, this is in agreement with previous studies, which showed hampered biofilm formation upon disruption of cell wall biosynthesis (Bucher et al., 2015; Zhu et al., 2018). Taken together, we conclude that the overexpression of the YtrBCDEF ABC transporter upon deletion of ytrA leads to pleiotropic effects on alternative lifestyles of B. subtilis and to increased cell wall thickness, however, at the current stage it is unknown whether the proteins encoded in the ytr operon have a direct impact on competence, sporulation and biofilm formation or whether these pleiotropic effects are indirect consequences of the increased cell wall thickness.
Our results demonstrate that the YtrBCDEF ABC transporter is involved in the control of cell wall homeostasis, but it is not yet clear how this is achieved. An easy explanation would be that the system exports molecules necessary for cell wall synthesis, however, based on the presence of the substrate binding protein YtrF and on the critical role of this protein in preventing genetic competence, it can be assumed that the ABC transporter rather acts as an importer. YtrBCDEF could therefore be involved in the import of components required for the synthesis of peptidoglycan precursors or an unknown signal, which is involved in the regulation of peptidoglycan precursor production. In the latter case, overproduction of the ABC transporter YtrBCDEF would lead to an increased import of this unknown signal molecule and thus, to an enhanced peptidoglycan precursor synthesis leading to the production of a thicker cell wall. It is also possible that YtrBCDEF may not act as a transporter at all and simply modulate the activity of other enzymes that participate in cell wall metabolism. This could potentially explain why the B. subtilis strain lacking YtrA as well as the ATP-binding proteins, latter of which are usually essential for the activity of ABC transporters, still produce a thicker cell wall and remain non-competent. Strikingly, the C-terminus of YtrF contains a FtsX-like domain. In B. subtilis, the ABC transporter FtsEX activates the cell wall hydrolase CwlO via direct protein-protein interaction thereby affecting cell elongation (Meisner et al., 2013). We speculate that the FtsX-like domain of YtrF could thus be required for the interaction with other proteins.
Based on its expression pattern, the ytr operon was described as a reporter for glycopeptide antibiotics, such as vancomycin or ristocetin (Hutter et al., 2004) and antibiotics that interfere with the lipid II cycle, such as nisin (Wenzel et al., 2012). Whether this induction of ytrGABCDEF expression leads to an increased resistance toward those antibiotics is not clear, but recent results indicate that it has indeed an impact on nisin resistance (Senges et al., 2021). Interestingly, the substrate binding lipoprotein YtrF contains a MacB-like periplasmic core domain in its N-terminus. MacB proteins are usually involved in resistance toward antibiotics and peptide toxins (Greene et al., 2018), suggesting that YtrF could potentially bind antibiotics. It is tempting to speculate that cell wall-acting antibiotics serve as an exogenous signal to activate the expression of the ABC transporter YtrBCDEF, which leads to the production of a thicker cell wall and with this, to an increased resistance to these antibiotics. However, how this could still be achieved in the absence of the ATP-binding proteins YtrB and YtrE remains elusive. Future work will need to address the precise mechanism by which the YtrBCDEF ABC transporter affects cell wall synthesis in B. subtilis.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Author Contributions
MB, JS, and JR: design of the study and writing the paper. MB and LS: experimental work. MB, LS, JR, and JS: data analysis. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Julia Busse, Melin Güzel, Christopher Patrick Zschiedrich, and Leon Daniau for the help with some experiments. We are grateful to Josef Altenbuchner, Jan Gundlach, Leendert Hamoen, Daniel Kearns, Daniel Reuss, Sarah Wilcken, and the Bacillus Genetic Stock Center for providing B. subtilis strains. We would also like to thank Michael Hoppert for providing access to the Transmission Electron Microscope.
Footnotes
Funding. This research received funding from the Deutsche Forschungsgemeinschaft via SFB860.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.587035/full#supplementary-material
References
- Aguilar C., Vlamakis H., Guzman A., Losick R., Kolter R. (2010). KinD is a checkpoint protein linking spore formation to extracellular-matrix production in Bacillus subtilis biofilms. mBio 1:e00035-10. 10.1128/mBio.00035-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckering C. L., Steil L., Weber M. H. W., Völker U., Marahiel M. A. (2002). Genomewide transcriptional analysis of the cold shock response in Bacillus subtilis. J. Bacteriol. 184 6395–6402. 10.1128/JB.184.22.6395-6402.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berka R. M., Hahn J., Albano M., Draskovic I., Persuh M., Cui X., et al. (2002). Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 43 1331–1345. 10.1046/j.1365-2958.2002.02833.x [DOI] [PubMed] [Google Scholar]
- Beveridge T. J., Murray R. G. E. (1979). How thick is the Bacillus subtilis cell wall? Curr. Microbiol. 2 1–4. 10.1007/BF02601723 [DOI] [Google Scholar]
- Boonstra M., Schaffer M., Sousa J., Morawska L., Holsappel S., Hildebrandt P., et al. (2020). Analyses of competent and non-competent subpopulations of Bacillus subtilis reveal yhfW, yhxC and ncRNAs as novel players in competence. Environ. Microbiol. 22 2312–2328. 10.1111/1462-2920.15005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda S. S., González-Pastor J. E., Ben-Yehuda S., Losick R., Kolter R. (2001). Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 98 11621–11626. 10.1073/pnas.191384198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher T., Oppenheimer-Shaanan Y., Savidor A., Bloom-Ackermann Z., Kolodkin-Gal I. (2015). Disturbance of the bacterial cell wall specifically interferes with biofilm formation. Environ. Microbiol. Rep. 7 990–1004. 10.1111/1758-2229.12346 [DOI] [PubMed] [Google Scholar]
- Cao M., Wang T., Ye R., Helmann J. D. (2002). Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis σW and σM regulons. Mol. Microbiol. 45 1267–1276. 10.1046/j.1365-2958.2002.03050.x [DOI] [PubMed] [Google Scholar]
- Cascante-Estepa N., Gunka K., Stülke J. (2016). Localization of components of the RNA-degrading machine in Bacillus subtilis. Front. Microbiol. 7:1492. 10.3389/fmicb.2016.01492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diethmaier C., Pietack N., Gunka K., Wrede C., Lehnik-Habrink M., Herzberg C., et al. (2011). A novel factor controlling bistability in Bacillus subtilis: the Ymdb protein affects flagellin expression and biofilm formation. J. Bacteriol. 193 5997–6007. 10.1128/JB.05360-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draskovic I., Dubnau D. (2005). Biogenesis of a putative channel protein, ComEC, required for DNA uptake: membrane topology, oligomerization and formation of disulphide bonds. Mol. Microbiol. 55 881–896. 10.1111/j.1365-2958.2004.04430.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figaro S., Durand S., Gilet L., Cayet N., Sachse M., Condon C. (2013). Bacillus subtilis mutants with knockouts of the genes encoding ribonucleases RNase Y and RNase J1 are viable, with major defects in cell morphology, sporulation, and competence. J. Bacteriol. 195 2340–2348. 10.1128/JB.00164-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flórez L. A., Gunka K., Polanía R., Tholen S., Stülke J. (2011). SPABBATS: a pathway-discovery method based on Boolean satisfiability that facilitates the characterization of suppressor mutants. BMC Syst. Biol. 5:5. 10.1186/1752-0509-5-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamba P., Jonker M. J., Hamoen L. W. (2015). A novel feedback loop that controls bimodal expression of genetic competence. PLoS Genet. 11:e1005047. 10.1371/journal.pgen.1005047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene N. P., Kaplan E., Crow A., Koronakis V. (2018). Antibiotic resistance mediated by the MacB ABC transporter family: a structural and functional perspective. Front. Microbiol. 9:950. 10.3389/fmicb.2018.00950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérout-Fleury A. M., Shazand K., Frandsen N., Stragier P. (1995). Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167 335–336. 10.1016/0378-1119(95)00652-4 [DOI] [PubMed] [Google Scholar]
- Hahn J., Roggiani M., Dubnau D. (1995). The major role of Spo0A in genetic competence is to downregulate abrB, an essential competence gene. J. Bacteriol. 177 3601–3605. 10.1128/JB.177.12.3601-3605.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haijema B.-J., Hahn J., Haynes J., Dubnau D. (2001). A ComGA-dependent checkpoint limits growth during the escape from competence. Mol. Microbiol. 40 52–64. 10.1046/j.1365-2958.2001.02363.x [DOI] [PubMed] [Google Scholar]
- Hamoen L. W., Kausche D., Marahiel M. A., Sinderen D., Venema G., Serror P. (2003). The Bacillus subtilis transition state regulator AbrB binds to the -35 promoter region of comK. FEMS Microbiol. Lett. 218 299–304. 10.1111/j.1574-6968.2003.tb11532.x [DOI] [PubMed] [Google Scholar]
- Hamoen L. W., Smits W. K., de Jong A., Holsappel S., Kuipers O. P. (2002). Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res. 30 5517–5528. 10.1093/nar/gkf698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamoen L. W., Van Werkhoven A. F., Venema G., Dubnau D. (2000). The pleiotropic response regulator DegU functions as a priming protein in competence development in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 97 9246–9251. 10.1073/pnas.160010597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoa T. T., Tortosa P., Albano M., Dubnau D. (2002). Rok (YkuW) regulates genetic competence in Bacillus subtilis by directly repressing comK. Mol. Microbiol. 43 15–26. 10.1046/j.1365-2958.2002.02727.x [DOI] [PubMed] [Google Scholar]
- Hutter B., Fischer C., Jacobi A., Schaab C., Loferer H. (2004). Panel of Bacillus subtilis reporter strains indicative of various modes of action. Antimicrob. Agents Chemother. 48 2588–2594. 10.1128/AAC.48.7.2588-2594.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampf J., Gerwig J., Kruse K., Cleverley R., Dormeyer M., Grünberger A., et al. (2018). Selective pressure for biofilm formation in Bacillus subtilis: differential effect of mutations in the master regulator SinR on bistability. mBio 9:e01464-18. 10.1128/mBio.01464-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkol M. A., Blair K. M., Kearns D. B. (2013). Plasmid-encoded ComI inhibits competence in the ancestral 3610 strain of Bacillus subtilis. J. Bacteriol. 195 4085–4093. 10.1128/JB.00696-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo B. M., Kritikos G., Farelli J. D., Todor H., Tong K., Kimsey H., et al. (2017). Construction and analysis of two genome-scale deletion libraries for Bacillus subtilis. Cell Syst. 4 291–305. 10.1016/j.cels.2016.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst F., Rapoport G. (1995). Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177 2403–2407. 10.1128/jb.177.9.2403-2407.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López D., Kolter R. (2010). Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol. Rev. 34 134–149. 10.1111/j.1574-6976.2009.00199.x [DOI] [PubMed] [Google Scholar]
- Lopez D., Vlamakis H., Kolter R. (2009). Generation of multiple cell types in Bacillus subtilis. FEMS Microbiol. Rev. 33 152–163. 10.1111/j.1574-6976.2008.00148.x [DOI] [PubMed] [Google Scholar]
- Luttinger A., Hahn J., Dubnau D. (1996). Polynucleotide phosphorylase is necessary for competence development in Bacillus subtilis. Mol. Microbiol. 19 343–356. 10.1046/j.1365-2958.1996.380907.x [DOI] [PubMed] [Google Scholar]
- Maamar H., Dubnau D. (2005). Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol. Microbiol. 56 615–624. 10.1111/j.1365-2958.2005.04592.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B. (2020). Competence and transformation in Bacillus subtilis. Curr. Issues Mol. Biol. 37 57–76. 10.21775/cimb.037.057 [DOI] [PubMed] [Google Scholar]
- Mascher T., Margulis N. G., Wang T., Ye R. W., Helmann J. D. (2003). Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50 1591–1604. 10.1046/j.1365-2958.2003.03786.x [DOI] [PubMed] [Google Scholar]
- Meinken C., Blencke H.-M., Ludwig H., Stülke J. (2003). Expression of the glycolytic gapA operon in Bacillus subtilis: differential syntheses of proteins encoded by the operon. Microbiology 149 751–761. 10.1099/mic.0.26078-0 [DOI] [PubMed] [Google Scholar]
- Meisner J., Montero Llopis P., Sham L. T., Garner E., Bernhardt T. G., Rudner D. Z. (2013). FtsEX is required for CwlO peptidoglycan hydrolase activity during cell wall elongation in Bacillus subtilis. Mol. Microbiol. 89 1069–1083. 10.1111/mmi.12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze N., Desai Y., Raj A., Dubnau D. (2012). Spo0A~P imposes a temporal gate for the bimodal expression of competence in Bacillus subtilis. PLoS Genet. 8:e1002586. 10.1371/journal.pgen.1002586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirouze N., Ferret C., Cornilleau C., Carballido-López R. (2018). Antibiotic sensitivity reveals that wall teichoic acids mediate DNA binding during competence in Bacillus subtilis. Nat. Commun. 9:5072. 10.1038/s41467-018-07553-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano M. M., Xia L. A., Zuber P. (1991). Transcription initiation region of the srfA operon, which is controlled by the comP-comA signal transduction system in Bacillus subtilis. J. Bacteriol. 173 5487–5493. 10.1128/JB.173.17.5487-5493.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas P., Mäder U., Dervyn E., Rochat T., Leduc A., Pigeonneau N., et al. (2012). Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335 1103–1106. 10.1126/science.1206848 [DOI] [PubMed] [Google Scholar]
- Ogura M., Yamaguchi H., Kobayashi K., Ogasawara N., Fujita Y., Tanaka T. (2002). Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J. Bacteriol. 184 2344–2351. 10.1128/JB.184.9.2344-2351.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quentin Y., Fichant G., Denizot F. (1999). Inventory, assembly and analysis of Bacillus subtilis ABC transport systems. J. Mol. Biol. 287 467–484. 10.1006/jmbi.1999.2624 [DOI] [PubMed] [Google Scholar]
- Rahmer R., Morabbi Heravi K., Altenbuchner J. (2015). Construction of a super-competent Bacillus subtilis 168 using the PmtlA-comKS inducible cassette. Front. Microbiol. 6:1431. 10.3389/fmicb.2015.01431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuß D. R., Altenbuchner J., Mäder U., Rath H., Ischebeck T., Sappa P. K., et al. (2017). Large-scale reduction of the Bacillus subtilis genome: consequences for the transcriptional network, resource allocation, and metabolism. Genome Res. 27 289–299. 10.1101/gr.215293.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rismondo J., Schulz L. M., Yacoub M., Wadhawan A., Hoppert M., Dionne M. S., et al. (2021). EslB is required for cell wall biosynthesis and modification in Listeria monocytogenes. J. Bacteriol. 203:e00553-20. 10.1128/JB.00553-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueden C. T., Schindelin J., Hiner M. C., DeZonia B. E., Walter A. E., Arena E. T., et al. (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18:529. 10.1186/s12859-017-1934-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg L. I., Luo Y., Hachmann A.-B., Mascher T., Helmann J. D. (2011). The Bacillus subtilis GntR family repressor YtrA responds to cell wall antibiotics. J. Bacteriol. 193 5793–5801. 10.1128/JB.05862-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, 2nd Edn. New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Senges C. H. R., Stepanek J. J., Wenzel M., Raatschen N., Ay Ü, Märtens Y., et al. (2021). Comparison of proteomic responses as global approach to antibiotic mechanism of action elucidation. Antimicrob. Agents Chemother. 65:e01373-20. 10.1128/AAC.01373-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serror P., Sonenshein A. L. (1996). CodY is required for nutritional repression of Bacillus subtilis genetic competence. J. Bacteriol. 178 5910–5915. 10.1128/jb.178.20.5910-5915.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Q., Hunter E., Qin Y., Nicolau S., Zalis E. A., Wang H., et al. (2020). Negative interplay between biofilm formation and competence in the environmental strains of Bacillus subtilis. mSystems 5:e00539-20. 10.1128/mSystems.00539-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimane K., Ogura M. (2004). Mutational analysis of the helix-turn-helix region of Bacillus subtilis response regulator DegU, and identification of cis-acting sequences for DegU in the aprE and comK promoters. J. Biochem. 136 387–397. 10.1093/jb/mvh127 [DOI] [PubMed] [Google Scholar]
- Smits W. K., Eschevins C. C., Susanna K. A., Bron S., Kuipers O. P., Hamoen L. W. (2005). Stripping Bacillus: comK auto-stimulation is responsible for the bistable response in competence development. Mol. Microbiol. 56 604–614. 10.1111/j.1365-2958.2005.04488.x [DOI] [PubMed] [Google Scholar]
- Turgay K., Hahn J., Burghoorn J., Dubnau D. (1998). Competence in Bacillus subtilis is controlled by regulated proteolysis of a transcription factor. EMBO J. 17 6730–6738. 10.1093/emboj/17.22.6730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Sinderen D., Luttinger A., Kong L., Dubnau D., Venema G., Hamoen L. (1995). comK encodes the competence transcription factor, the key regulatory protein for competence development in Bacillus subtilis. Mol. Microbiol. 15 455–462. 10.1111/j.1365-2958.1995.tb02259.x [DOI] [PubMed] [Google Scholar]
- Wach A. (1996). PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12 259–265. [DOI] [PubMed] [Google Scholar]
- Wenzel M., Kohl B., Münch D., Raatschen N., Albada H. B., Hamoen L., et al. (2012). Proteomic response of Bacillus subtilis to lantibiotics reflects differences in interaction with the cytoplasmic membrane. Antimicrob. Agents Chemother. 56 5749–5757. 10.1128/AAC.01380-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K.-I., Fujita Y., Ehrlich S. D. (2000). An operon for a putative ATP-binding cassette transport system involved in acetoin utilization of Bacillus subtilis. J. Bacteriol. 182 5454–5461. 10.1128/JB.182.19.5454-5461.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P. (1990). “Use of transposons and integrational vectors for mutagenesis and construction of gene fusions in Bacillus species,” in Molecular Biological Methods for Bacillus, eds C. R. Harwood and S. M. Cutting (Chichester: John Wiley & Sons Ltd.), 221–266. [Google Scholar]
- Zhu X., Liu D., Singh A. K., Drolia R., Bai X., Tenguria S., et al. (2018). Tunicamycin mediated inhibition of wall teichoic acid affects Staphylococcus aureus and Listeria monocytogenes cell morphology, biofilm formation and virulence. Front. Microbiol. 9:1352. 10.3389/fmicb.2018.01352 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.