Abstract
Protein–protein interaction (PPI) networks are key to nearly all aspects of cellular activity. Therefore, the identification of PPIs is important for understanding a specific biological process in an organism. Compared with conventional methods for probing PPIs, the recently described proximity labeling (PL) approach combined with mass spectrometry (MS)-based quantitative proteomics has emerged as a powerful approach for characterizing PPIs. However, the application of PL in planta remains in its infancy. Here, we summarize recent progress in PL and its potential utilization in plant biology. We specifically summarize advances in PL, including the development and comparison of different PL enzymes and the application of PL for deciphering various molecular interactions in different organisms with an emphasis on plant systems.
Keywords: protein interactions, proximity labeling, biotin ligase, plant, membrane contact sites, organelles
Proximity labeling (PL) has emerged as a powerful approach for the characterization of protein–protein interactions. This review summarizes advances in PL, including the development and comparison of different PL enzymes and the application of PL for dissecting molecular interactions in different organisms with an emphasis on plant systems.
Introduction
All proteins in cells act cooperatively by interacting with other cellular components, including proteins and nucleic acids. These protein complexes constitute the basic unit of life activities and participate in various life processes, including biological signaling, gene expression, cell metabolism, cell-cycle regulation, and others. Due to the central role of protein components in these diverse processes, the development of approaches for identifying proteins within various complexes is very important for their further functional characterization.
Several conventional methods have been used to screen for protein–protein interactions (PPIs), including co-immunoprecipitation (co-IP) combined with mass spectrometry (MS) and yeast two-hybrid (Y2H) assays. In most cases, co-IP can detect only high-affinity interactions but not weak or transient interactions. Co-IP also shows low efficiency for the identification of the interaction partners of insoluble proteins, in particular membrane-associated proteins. Y2H is another commonly used approach for identifying PPIs. However, it is often labor-intensive to construct cDNA libraries suitable for Y2H, and some proteins, such as transcription factors, are not suitable for the Y2H assay. In addition, both the traditional co-IP and Y2H assays are limited to the identification of PPIs under in vitro or non-physiological conditions.
The recently developed proximity labeling (PL) approach overcomes some inherent limitations of conventional methods such as affinity purification-coupled MS and Y2H. PL is generally conducted by fusing a catalytic enzyme to a protein of interest (bait protein) or anchoring it to a subcellular compartment. Supplementation with corresponding substrates will activate the covalent tagging of proteins that are proximal to the bait protein with reactive small molecules in living cells. Therefore, abundant soluble proteins as well as insoluble membrane proteins that are difficult to obtain by conventional approaches can be effectively enriched under stringent denaturing conditions, thus allowing subsequent MS identification of proteins proximal to the bait protein or an organelle (Figure 1A). PL can detect weak, transient, or hydrophobic PPIs in their native state, thereby revealing an unprecedented spatial and temporal protein interaction network to better understand a specific biological process. Hence, PL-based affinity purification coupled with MS has been widely used in the last few years to study protein interaction networks, topology, protein–DNA or protein–RNA interactions, and other phenomena in animal systems. However, the application of PL in planta remains in its infancy. In this review, we summarize the classification of different PL enzymes, the development of PL technology, and the use of PL to study different types of protein interactions; we also underscore its increasing use in plant systems.
Figure 1.
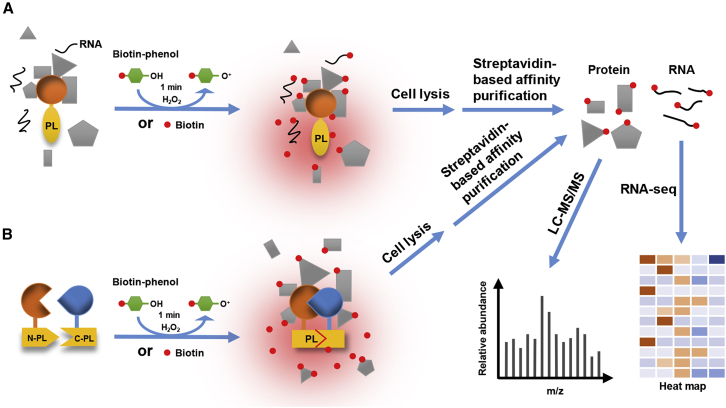
Schematic representation of proximity-labeling systems.
(A) Proximity labeling (PL) assay based on the intact PL enzyme. Biotin ligase or the APEX PL enzyme is fused to the target protein of interest and expressed in living cells. Upon the addition of substrates, such as biotin or biotin-phenol and hydrogen peroxide (H2O2), to the medium, proteins or RNAs (for APEX2-based PL) in the vicinity of the target protein can be tagged by biotin. By lysing the cells and incubating with streptavidin beads, biotin-labeled proteins or RNAs can be enriched for subsequent LC-MS/MS or high-throughput sequencing analysis.
(B) Split-PL system for identifying the composition of protein complexes. The N- and C-terminal parts of the PL enzyme are fused to a pair of known interacting proteins. As the pair interacts in cells, the two halves of split-PL are pulled into close proximity, leading to the reconstitution of the full PL enzyme and initiating the labeling of nearby partners of the protein complex.
PL enzymes: development, classification, and comparison
PL enzymes
The two most commonly used PL enzymes are the BirA mutant (also called BioID) and ascorbate peroxidase (APEX) (Table 1). The BioID-based PL technique was first used in mammalian cells to characterize the interactome of lamin-A protein, which is a component of the nuclear lamina (Roux et al., 2012). BioID is based on the Escherichia coli biotin ligase BioID (R118G), and non-toxic biotin serves as the substrate for BioID-based PL. Upon the addition of biotin and ATP, BioID catalyzes biotin to form reactive biotinol-5ʹ-AMP, which can promiscuously biotinylate the lysine residues of proteins that are in close proximity to the BioID enzyme (within a radius of 10 nm). The use of flexible linkers between the PL enzyme and the bait protein has been proposed to increase the labeling radius (Kim et al., 2014). The optimal labeling time for BioID is about 18–24 h (Roux et al., 2012). The improved biotin ligase BioID2 from Aquifex aeolicus has been adapted for PL and is 10 kDa smaller than BioID (Kim et al., 2016). BioID2 has the advantage of maintaining the proper localization of the fused protein and requires less biotin for labeling compared with BioID. In addition, an appropriate labeling radius can be achieved by adding linkers of different lengths between BioID2 and the bait protein (Kim et al., 2016).
Table 1.
Current proximity-labeling enzymes used for the identification of PPIs.
| Enzymes | Origin | Size (kDa) | Optimal temperature (°C) | Labeling time | Labeling radius (nm) | Substrate and cytotoxicity | Organisms | References |
|---|---|---|---|---|---|---|---|---|
| APEX | Pea | 28 | 37 | 1 min | <20 | Biotin-phenol + H2O2 (toxic) | Mammalian cells, flies | (Rhee et al., 2013) |
| APEX2 | Soybean | Mammalian cells, bacteria, Chlamydia, yeast | (Lam et al., 2015) | |||||
| BioID | E. coli | 35 | 37 | 18–24 h | ~10 | Biotin (atoxic) | Mammalian and plant cells, yeast, T. brucei, Dictyostelium | (Roux et al., 2012) |
| BioID2 | A. aeolicus | 25 | Mammalian and plant cells | (Kim et al., 2016) | ||||
| BASU | B. subtilis | 1 min | Mammalian cells | (Ramanathan et al., 2018) | ||||
| AirID | E. coli | 37 | 26 | 3–24 h | Mammalian cells and wheat cell-free systems | (Kido et al., 2020) | ||
| miniTurboID | 28 | 25 | ≥10 min | Mammalian and plant cells, flies, worms, and yeast | (Branon et al., 2018) | |||
| TurboID | 35 | |||||||
| HRP | Horseradish | 44 | 37 | 10 min to 2 h | 200–300 | Biotin-phenol + H2O2 (toxic) | Human and chicken cells | (Li et al., 2014) |
| EXCELL | Staph | 23 | 30 min | Not available | Biotin-LPETG (atoxic) | Mammalian cells | (Ge et al., 2019) | |
| PUP-IT | Mycobacteria | 54 | 24 h | Not available | Pup (atoxic) | Mammalian cells | (Liu et al., 2018) | |
| NEDDylation | Human | 20 | 24–36 h | Direct contact | NEDD8 (atoxic) | Mammalian cells | (Hill et al., 2016) |
APEX is a 27-kDa monomeric protein that promiscuously biotinylates proteins within a 20-nm radius of the target protein when supplied with ATP, H2O2, and biotin-phenol at 37°C (Rhee et al., 2013). All neighboring proteins that are rich in tyrosine residues have the potential to be biotinylated by the APEX-fused bait protein within just 1 min, compared with 18–24 h in the case of BioID. Because H2O2 and biotin-phenol are toxic to living cells, an improved variant with increased catalytic activity, APEX2, was developed. APEX2 efficiently biotinylates nearby proteins even at low levels of enzyme expression and with less H2O2 and biotin-phenol, thus reducing the toxic effect on living cells (Lam et al., 2015). Compared with APEX, BioID has the advantage because it is easier to introduce the non-toxic biotin substrate than H2O2 and biotin-phenol to a whole organism, thereby avoiding oxidative stress to cells or tissues. Hence, BioID has been widely used in the past few years (Uezu et al., 2016; Gu et al., 2017; Lin et al., 2017; Opitz et al., 2017). However, the labeling time of BioID (18–24 h) is significantly longer than that of APEX (1 min), restricting its application for the study of dynamic processes such as cell signaling and transient PPIs.
Recently, the new PL enzyme TurboID (35 kDa) and its truncated mutant miniTurboID (28 kDa) were developed using yeast display-based directed evolution of BirA (Branon et al., 2018). These two new PL enzymes combine the advantages of BioID and APEX: they exhibit fast kinetics and use a non-toxic biotin substrate. TurboID and miniTurboID are able to identify the interactions involved in fast and dynamic processes without causing damage to living cells (Smith et al., 2018; Mair et al., 2019). Moreover, a notable breakthrough of TurboID- and miniTurboID-based PL is that efficient biotinylation can be achieved at room temperature (25°C), compared with 37°C required for the BioID and APEX systems. In addition, although it has lower labeling efficiency than TurboID, miniTurboID has the advantage of reducing the labeling background and minimizing undesired effects of the tag fusion on target protein function. These improvements in TurboID and miniTurboID provided the basis for extending the PL technique beyond mammalian cells to other model systems, such as flies, worms, and plants, as described below (Branon et al., 2018; Zhang et al., 2019). Very recently, a new PL enzyme, AirID, was developed by Kido et al. (2020). This system uses an engineered ancestral BirA enzyme to label surrounding proteins in the presence of ATP and biotin. Compared with TurboID, the AirID system showed more specific tagging of the interaction partners and was less toxic to cells over long incubation periods, providing an additional useful PL tool for long-lasting experiments in live cells.
In addition to the commonly used PL enzymes discussed above, other PL enzymes have been developed for proximity-dependent labeling, such as horseradish peroxidase (HRP) (Kotani et al., 2008; Li et al., 2014), Staphylococcus aureus transpeptidase sortase A variant (mgSrtA) (Ge et al., 2019), BASU (Ramanathan et al., 2018), proteasomal accessory factor A (PafA) (Liu et al., 2018), and NEDD8-conjugating enzyme (Hill et al., 2016) (Table 1). HRP can label nearby proteins within a range of 200–300 nm in the presence of H2O2 and biotin-phenol. By linking HRP to a specific antibody against a plasma membrane protein of interest and incubating it with bait cells in an H2O2- and biotin-phenol-containing labeling buffer, researchers were able to characterize several cell-type surface receptor clusters (Li et al., 2014). In most cases, HRP-based PL is used to map the protein interactome at the cell surface (Loh et al., 2016; Yamashita et al., 2011); it is not suitable for intracellular labeling because of the low activity of HRP in reductive environments. In addition to HRP, another enzyme-mediated proximity cell labeling (EXCELL) system was developed, which uses mgSrtA as the labeling enzyme and a biotin-tagged small peptide (biotin-LPETG) as the substrate. Displaying mgSrtA on the surface of the target cell leads to the labeling of neighboring cells upon the addition of biotin-LPETG, thus allowing the detection of cell–cell interactions (Ge et al., 2019). The labeling enzyme for the EXCELL system (23 kDa) is smaller than HRP (44 kDa, Table 1). Unlike the HRP system, EXCELL uses non-toxic substrates and does not require specific antibodies to the bait cell membrane. The BASU-based PL method depends on an engineered Bacillus subtilis-derived biotin ligase (28 kDa), which dramatically reduces the labeling time to as little as 1 min without toxicity to living cells (Ramanathan et al., 2018). The NEDDylation technique uses a genetically modified NEDD8 E2-conjugating enzyme, Ubc12, as the labeling enzyme and the His6-biotin-tagged NEDD8 protein (HB-NEDD8) as the substrate (Hill et al., 2016). The HB-NEDD8 protein tag can only be transferred to the prey protein by direct attack of the prey's surface lysine ϵ-amines upon the thioester in the active site of the bait-fused Ubc12. Therefore, direct bait–prey contact is essential for HB-NEDD8 tagging; this is a unique feature among PL methods and can be helpful for reducing false positives. However, the occurrence of NEDDylation requires the endogenous NEDD8 pathway of mammalian cells, potentially influencing their normal biological processes and limiting the use of NEDDylation in other organisms (Hill et al., 2016). For pupylation-based interaction tagging (PUP-IT), the 7-kDa prokaryotic ubiquitin-like protein (Pup) is tagged to proximal proteins using the PafA ligase from mycobacteria. Although the PafA enzyme has lower activity than APEX, the PUP-IT system has the distinct advantage that all of its components can be expressed in living cells without the addition of molecules in vitro, thus permitting its potential application in live animals. Moreover, compared with NEDDylation, the PUP-IT method takes advantage of the mycobacteria pupylation pathway that does not exist in mammalian cells, thereby avoiding interference with their normal biological processes. Nevertheless, the substrate of PafA ligase, Pup, is relatively large compared with those of BioID and APEX, limiting its utilization in probing proteomes within organelles (Liu et al., 2018). The characteristics of different PL enzymes are summarized and compared in Table 1.
Split-PL enzymes
Like the split fluorescent proteins used in the bimolecular fluorescence complementation assay, several PL enzymes, such as HRP, APEX2, BioID, and TurboID, can be split into two parts and reconstituted to a functional entity when they are brought into close proximity (Figure 1B). The split-PL system is particularly useful for studying additional interacting factors of spatiotemporally defined protein complexes and membrane contact sites (MCSs). HRP was the first PL enzyme used for the development of the split-PL system. Because HRP shows low activity in the reductive cytoplasmic environment, the split-HRP method is limited to mapping the protein interactome at the cell surface (Martell et al., 2016). To overcome this limitation, the split-APEX2 system was developed; this system enables the identification of PPIs in live cells with an improved signal-to-noise ratio (Han et al., 2019; Xue et al., 2017). BioID can also be split into two parts at different amino acid sites (Table 2). Comparison of the activity of two split-BioIDs cleaved at different amino acid sites, E140/Q141 and E256/G257, revealed that the E256/G257 version exhibited a stronger biotinylation signal (De Munter et al., 2017; Schopp et al., 2017). Using split-BioID, additional novel interactors of several spatiotemporally defined protein complexes, such as PP1-NIPP1 and Ago2/Dicer, were identified, demonstrating the feasibility of this method for detecting additional partners of conditional protein complexes. In addition, the split-BioID system effectively reduced background biotinylation compared with intact BioID (De Munter et al., 2017; Schopp et al., 2017). Recently, Kwak et al. (2020) developed the Contact-ID method, which splits BioID at amino acid site G78/G79 and shows higher biotinylation activity than the split-BioID pair cleaved at the E256/G257 site. Contact-ID was successfully used to decipher the kinetic composition of endoplasmic reticulum (ER)–mitochondria contact sites under normal and stress conditions and revealed topology information for integral membrane proteins. At the same time, a version of TurboID split at amino acid site L73/G74 was developed and used to profile the composition of ER–mitochondria contact sites (Cho et al., 2020). Contact-ID and split-TurboID identified similar numbers of proteins, but split-TurboID seemed to show lower biased labeling than Contact-ID, as demonstrated by a greater balance between the identified outer mitochondrial membrane (OMM) and ER membrane proteins. Contact-ID labeling requires 16 h, whereas split-TurboID labeling requires only 4 h, and this may account for the biased labeling displayed by these two systems. Considering the high biotin ligation activity of TurboID at room temperature, the split-TurboID system holds promise for the study of MCS or conditional protein complex interactomes in planta. The characteristics of different split-PL methods are summarized and compared in Table 2.
Table 2.
Current split-proximity labeling systems.
| Method | Holoenzyme | Split sites (amino acid) | Applications | Organisms | References |
|---|---|---|---|---|---|
| Split-HRP | Horseradish peroxidase | G213/N214 | Map intercellular PPIs at the synapses. Allows the visualization of interactions between specific sets of neurons by fluorescent labeling. | Mammalian cells | (Martell et al., 2016) |
| Split-APEX2 | Ascorbate peroxidase | G201/L202 | Identify key interacting regions of the STIM1 and Orai1 protein complex. | Mammalian cells | (Xue et al., 2017) |
| Split-APEX2 | E200/G201 | Split pairs can reconstitute at target nucleic acids and ER–mitochondria contact sites. | Mammalian cells | (Han et al., 2019) | |
| Split-BioID | Biotin ligase | E140/E141 | Identify transient substrates and interactors of a phosphatase holoenzyme. | Mammalian cells | (De Munter et al., 2017) |
| Split-BioID | E256/G257 | Map the Ago2-mediated silencing pathway. Identify additional interactors of the Cdc25C/14-3-3ϵ, Ago2/TNRC6C, and Ago2/Dicer protein–protein complexes. | Mammalian cells | (Schopp et al., 2017) | |
| Contact-ID | G78/G79 | Dissect the components of ER–mitochondria contact sites. | Mammalian cells | (Kwak et al., 2020) | |
| Split-TurboID | L73/G74 | Dissect the components of ER–mitochondria contact sites. | Mammalian cells | (Cho et al., 2020) |
Application of the PL technique for probing diverse molecular interactions
PL technology overcomes several limitations of traditional interaction detection approaches and has been widely used to decipher different molecular interactions in diverse biological contexts. Below, we discuss representative applications of the PL technique for addressing various biological questions (Figure 2).
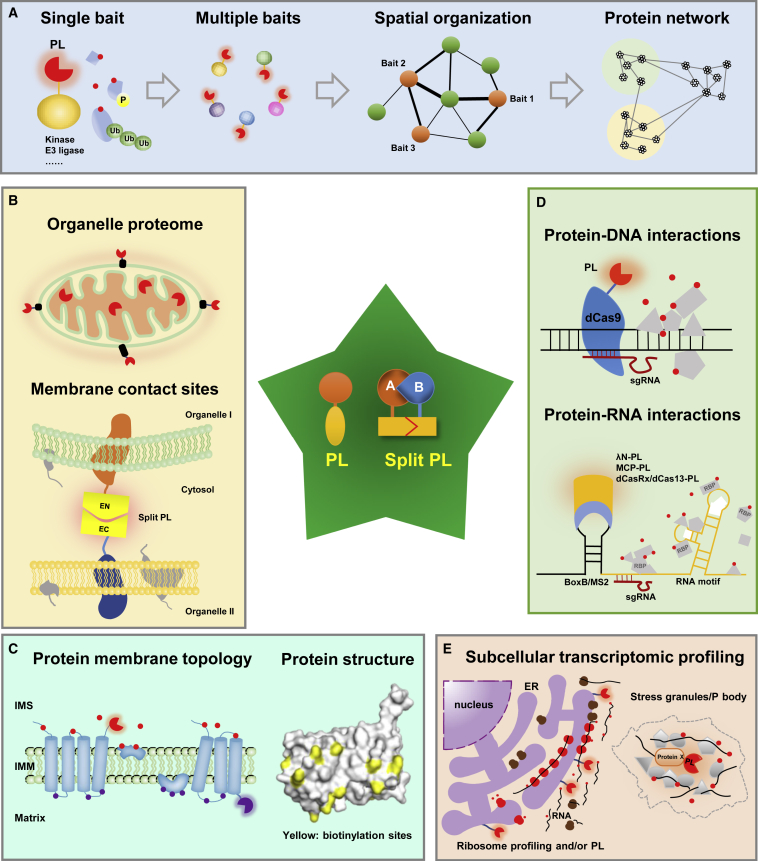
Figure 2.
Application of PL in probing diverse molecular interactions.
(A) Using PL for the identification of a single PPI, deciphering the spatial relationship of different proteins, and constructing protein interaction networks.
(B) Proteomic analysis of subcellular compartments and membrane contact sites by PL or split-PL.
(C) Application of PL to determine the topology of proteins and gain structural insights into the target protein. Due to the impermeability of the inner mitochondrial membrane (IMM) to small molecules, PL will occur exclusively at either the IMS (red) or the matrix side (brown).
(D) Identification of proteins bound to a specific genomic locus or RNA motif by combining PL with other existing techniques.
(E) Application of PL to map the membrane-enclosed or membrane-less organelle RNAome together with other techniques, such as ribosome profiling. These RNAs can be either mRNAs for translation or noncoding RNAs.
PL techniques: from single PPI identification to protein interaction networks
As discussed above, the PL technique can capture interactions with rapid kinetics (Lobingier et al., 2017; Paek et al., 2017). A number of studies have identified interactors of proteins of interest using PL methods, and these have been summarized by other excellent reviews (Kim and Roux, 2016; Varnaitė and MacNeill, 2016; Béganton et al., 2019; Nguyen et al., 2020). Here, we would like to underscore the great potential of PL methods for probing weak or transient interactions, in particular enzyme–substrate interactions, which are often difficult to capture by conventional methods. For example, the mitogen-activated protein kinase (MAPK) signaling pathway is often dynamically involved in various physiological processes under different stress conditions, and it is difficult for traditional methods to simultaneously capture MAPK substrates in different states. Dumont et al. (2019) used APEX2-based PL to map the interactome of p38α and p38γ MAPKs under both steady-state and activated conditions and identified novel substrates of p38. Likewise, the identification of E3 ubiquitin ligase substrates is very challenging due to their transient interactions and the rapid degradation of their substrates by the 26S proteasome. The PL method also provides a powerful tool for addressing this issue. For example, Coyaud et al. (2015) used BioID-based PL to investigate the interaction partners of the Skp-Cullin-F-box (SCF)β−TrCP1/2 E3 ligase and identified many novel SCFβ−TrCP1/2 substrates. Koerver et al. (2019) fused APEX2 to the C terminus of the ubiquitin-conjugating enzyme UBE2QL1 and identified several potential ubiquitination targets. In addition, Carter-O’Connell et al. (2018) used BioID-based PL together with chemical genetics to identify the substrates of poly(ADP-ribose) polymerase 14 (PARP14) and uncovered 114 potential PARP14-specific protein substrates. These studies highlight the advantages of using the PL technique to identify the targets and reveal the dynamic interactomes of biological enzymes in vivo.
An increasing number of studies have extended the applications of the PL technique from mapping the proximal proteome of a single bait protein to building a global protein proximity network, thus providing a comprehensive view of the spatial relationships between groups of proteins (Gingras et al., 2019; Lundberg and Borner, 2019). By integrating the proximal proteomes from a number of baits within a subcellular compartment, a spatial protein proximity network can be constructed. Usually, PL experiments are designed by selecting specific known proteins localized to the same organelles as baits. The same prey may be tagged by several baits, which will indicate its spatial location in the proximity network. By integrating the data from baits in different positions and calculating the degree of overlap and crossover, the proximal relationship of core proteins can be determined and an overall proximity network generated (Gingras et al., 2019; Lundberg and Borner, 2019) (Figure 2A). For example, Gupta et al. (2015) mapped the composition and spatial organization of the centrosome–cilium interface with 58 bait proteins and produced a global protein interaction network comprising >7000 interactions. Anne-Claude Gingras's lab used 139 baits to systematically investigate the composition of mRNA-associated granules and bodies and generated a global network that consisted of 7424 unique proximal interactions among 1792 proteins (Youn et al., 2018). Recently, they developed a proximity atlas of human cells through BioID-based PL using 192 marker proteins from 32 different subcellular compartments (Go et al., 2019), providing a rich resource for the comprehensive understanding of various dynamic activities in human cells. Very recently, an elegant topological map of integrin adhesion complexes was also constructed using multiplexed BioID-based PL (Chastney et al., 2020). With the improvement of techniques, including high-throughput microscopy and PL, combined with machine learning and user-friendly data analysis software, we anticipate that the study of spatial proteome organization will be greatly advanced in the future.
Subcellular proteome mapping
Although biochemical fractionation allows the purification of most subcellular organelles for MS-based proteomics, the refined proteomic composition of specific regions of an organelle remains poorly understood due to the lack of techniques to purify these sub-organellar regions. For example, the human intact mitochondrial proteome has been well characterized in many previous studies (Calvo and Mootha, 2010); however, mitochondrial proteomes at the sub-organellar level remain largely unknown. To address these gaps, Alice Ting's group performed a systemic proteomic mapping of different sub-organellar regions of the mitochondrion, including the mitochondrial matrix (Rhee et al., 2013), the cytoplasm-facing OMM (Hung et al., 2017), and the intermembrane space (IMS) between the inner mitochondrial membrane (IMM) and the OMM (Hung et al., 2014) (Figure 2B). They identified many novel mitochondrial proteins and clarified and redefined several mitochondrial proteins whose subcellular localization was ambiguous or disputed in previous studies (Rhee et al., 2013; Hung et al., 2014, 2017). Likewise, numerous studies have been conducted to characterize the lipid droplet (LD) proteome via MS analysis of biochemically isolated LDs, which are often contaminated by other organelles or membrane fragments (Yang et al., 2012). Bersuker et al. (2018) exploited APEX2-based PL to re-map the proteomic composition of the LD and identified many known and novel LD proteins without the presence of common contaminating proteins. More importantly, they could dynamically monitor changes in the LD proteome and reveal novel signaling pathways involved in the regulation of LD metabolism. In addition, PL methods have also been used successfully to dissect the composition of several large membrane-associated protein complexes, such as the nuclear pore complex on the nuclear envelope (Kim et al., 2014) and G protein-coupled receptors (Lobingier et al., 2017; Paek et al., 2017) and CaV1.2 voltage-gated calcium channels (Liu et al., 2020) on the plasma membrane.
In addition to the proteomic mapping of membrane-bound organelles described above, PL-based methods can also be used to investigate non-membrane-enclosed organelles, which cannot be purified by traditional biochemical fractionation approaches. For example, mammalian primary cilia, important signaling organelles that are not enclosed by a membrane, were subjected to proteomic analysis by fusing APEX with ciliary-targeting signals, followed by in vivo biotinylation and MS analysis. Many known and several uncharacterized ciliary signaling proteins were identified (Mick et al., 2015). In recent years, PL-based techniques have also been used to intensively study the molecular composition of several other membrane-less compartments, such as stress granules and processing bodies (Markmiller et al., 2018; Youn et al., 2018), mitochondrial nucleoids (Han et al., 2017), the centrosome (Firat-Karalar et al., 2014), the human centrosome–cilium interface (Gupta et al., 2015), the chromocenter (Kochanova et al., 2020), and the replication fork (Srivastava et al., 2018). Taken together, all these studies highlight the versatility of the PL method for mapping the proteomes of subcellular compartments that were previously inaccessible.
MCSs
In the last decade, our understanding of organelles changed from separate compartments to interdependent cellular structures that communicate with each other through MCSs. Accumulating evidence shows that MCSs play an important role in maintaining cellular homeostasis, and their dysfunction can lead to a variety of diseases (Eisenberg-Bord et al., 2016; Prinz et al., 2020). Conventional methods have shown low efficiency for studying transient and hydrophobic PPIs, especially the tether proteins localized to the MCSs. However, the PL method has great advantages for the study of MCSs (Han et al., 2019; Cho et al., 2020; Kwak et al., 2020), as illustrated by the intensively studied ER–mitochondria contact sites. At the beginning, APEX2 was frequently anchored to the cytosol-facing organelle membranes by fusing with membrane-localized proteins such as STIM1 (Jing et al., 2015) or signal peptides of membrane proteins (Hung et al., 2017). In this way, proteins in the area of the MCSs could be labeled and identified by subsequent liquid chromatography-tandem MS (LC-MS/MS) analysis. Using PL, a number of potential proteins that localize to the ER–mitochondria or ER–plasma MCSs were identified and characterized (Jing et al., 2015; Hung et al., 2017).
Recently, split-PL enzymes, including split-APEX2 (Han et al., 2019), split-TurboID (Cho et al., 2020), and Contact-ID (Kwak et al., 2020), combined with chemically inducible FRB–FKBP dimerization systems were independently developed to map the composition of ER–mitochondria contact sites. The FRB–FKBP system is an inducible system consisting of a pair of proteins named FKBP12 (12 kDa FK506-binding protein) and FRB (FKBP–rapamycin-binding domain). These two proteins do not interact in the absence of rapamycin but form a tight FKBP–rapamycin–FRB ternary complex upon the addition of rapamycin (Banaszynski et al., 2005; Choi et al., 1996). The N- and C-terminal PL enzyme fragments were genetically conjugated with FKBP12 and FRB and targeted to different organellar membranes by fusing with signal peptides or membrane-localized proteins. Full enzyme activity can be reconstituted with the addition of rapamycin, leading to the labeling of proteins localized to the narrow gaps between two organellar membranes (Figure 2B). Using this system, many novel proteins that associate with ER–mitochondria contact sites were identified, and further analysis of the biotinylation sites also provided insight into the topologies of the identified integral membrane proteins (Han et al., 2019; Cho et al., 2020; Kwak et al., 2020). All these studies highlight the power and reliability of the PL technique to profile the molecular composition of organelle MCSs in living cells.
In addition to FRB and FKBP12, other protein pairs such as neurexin (NRX) and neuroligin (NLG) (Martell et al., 2016) or protein phosphatase 1 (PP1) and nuclear inhibitor of PP1 (NIPP1) (De Munter et al., 2017) have also been used for complementary fragment screening of PL enzymes. Under some conditions, the reconstitution of certain PL enzymes relies on the utilization of an interaction protein pair. For example, Han et al. (2019) reported that reconstitution of split APEX2 at mitochondria–ER contact sites was PPI dependent despite multiple days of coexpression of the sAPEX fragments. Martell et al. (2016) showed that split-HRP enzymes require a PPI for reconstitution at cell–cell contacts. Therefore, the fusion of interaction protein pairs to certain split PL enzyme fragments is necessary to reconstitute the active PL enzyme during the study of intracellular or intercellular contacts. However, this may raise concerns about whether the PPI-dependent reconstitution defines natural MCSs or whether the forced reconstitution of the split-PL mediated by PPI leads to an artifact that does not reflect natural events at the MCSs. For example, Martell et al. (2016) indicated that detection of authentic synapses required expression of NRX- and NLG-fused split-HRP fragments at low to moderate levels. When they used the strong CAG promoter instead of the weaker synapsin promoter, many of the split-HRP-labeled contact sites were oblong and much larger than physiological synapses, unlike those of endogenous synapses. To avoid artificial contacts, they lowered the expression level of the split-HRP constructs. Similarly, Han et al. (2019) also found that it was necessary to reduce overall protein expression by changing the promoter of OMM–FKBP–EX from CMV to human ubiquitin promoter (hUBC). These results imply that the fusion of interaction protein pairs like FRB–FKBP or NRX–NLG to the split-PL enzyme fragments may have an effect on the tested MCSs if their expression level is too high.
However, it should be noted that although ectopic expression of the PPI-dependent split-PL enzymes may produce artificial MCSs, this does not mean that the strong binding affinity of the interaction protein pairs themselves will play a role in the formation of the MCSs. For example, Martell et al. (2016) addressed this concern by showing that NRX–NLG constructs did not perturb synapse size, number, location, or specificity in transgenic mouse brain. These studies indicate that the high binding affinity possessed by NRX–NLG or other protein pairs has little or marginal effect on the formation of MCSs.
In fact, the interaction protein pair is not absolutely necessary for split-PL systems during the study of certain MCSs. For example, split HRP can reconstitute in the ER in a PPI-independent manner, in contrast to its behavior at the synapse (Martell et al., 2016). Moreover, Alice Ting's and Hyun-Woo Rhee's labs identified the localized proteome at the ER–mitochondrial contact site in living cells using split-TurboID and Contact-ID, respectively (Cho et al., 2020; Kwak et al., 2020). Ting's group investigated the ER–mitochondrial contact site by fusing FRB–FKBP to split-TurboID fragments. With or without rapamycin, they captured proteins that indeed function at the ER–mitochondrial contact site, suggesting a PPI-independent reconstitution of the split-TurboID. Nevertheless, more proteins with ER, mitochondria, and MAM annotations were identified in the presence of rapamycin (Cho et al., 2020), suggesting that the introduction of PPIs was somewhat conducive to local proteome analysis at the MCS. By contrast to Ting's group, Rhee's lab used a BioID split-pair system, Contact-ID, to study the ER–mitochondrial contact site without the fusion of interaction protein pairs. They also identified a set of true proteins that function in the tethering of ER and mitochondrial membranes; their list of candidate proteins largely overlapped with that of Ting’s group (Cho et al., 2020; Kwak et al., 2020). These results further revealed the PPI-independence of some split-PL enzymes under certain conditions and also indicated that fusion of the interaction protein pair to the split-PL enzyme fragments can indeed generate true proteins that function at the MCS. Although PPI may have some potential or subtle effect on the MCS, this is outweighed by its ability to capture the true molecular composition of the MCS.
Protein membrane topology and surface-exposed protein subunit identification
Understanding the topology of membrane-localized proteins is critical for the in-depth study of their functional roles in signal transduction, molecule trafficking, and other cellular processes. However, conventional approaches that involve in vitro reactions often lead to unreliable results and do not accurately reflect membrane topology in living cells (Yoo and Rhee, 2020). The APEX-based method provides an effective solution for unraveling membrane topological information under physiological conditions (Lee et al., 2016; Yoo and Rhee, 2020). APEX is fused to the N or C terminus of target membrane proteins to observe their “restricted or diffusive” biotin-labeling patterns by electron or confocal microscopy (Figure 2C, left). To date, the membrane topology of many ER- or mitochondria-localized proteins has been intensively studied and redefined using APEX- or Contact-ID-based PL systems (Kwak et al., 2020; Lee et al., 2017; Yoo and Rhee, 2020). Most of these studies come from Rhee's lab. If the APEX-fused proteins generate a diffusive biotin-labeling pattern, this means that APEX is at the cytosolic face or within the IMS of the mitochondria. By contrast, APEX in the mitochondrial matrix will display a restricted biotin-labeling pattern due to the impermeability of the IMM to small molecules such as active biotin-phenoxyl radicals (Rhee et al., 2013). In addition, APEX in the ER lumen also displays a very restricted biotin-labeling pattern (Lee et al., 2016). Using this APEX-based biotinylation pattern method, the membrane topologies of several ER and mitochondria proteins, such as the mitochondrial calcium uniporter (Martell et al., 2012), heme oxygenase 1 (Lee et al., 2016), and sigma 1-receptor (Mavylutov et al., 2018), have been successfully characterized. Furthermore, Lee et al. (2017) developed an APEX-derived method, Spot-ID, to improve the efficiency of membrane topology determination from the single protein to the proteome level. Due to the impermeability of the IMM to small molecules and short-lived phenoxyl radicals, the solvent-accessible tyrosine residues at both the matrix and IMS sides will be exclusively labeled using the Spot-ID approach. With the Spot-ID system, membrane topology mapping of the submitochondrial proteome can be achieved by MS analysis of desthiobiotin-phenol-labeled peptides in a high-throughput manner (Lee et al., 2017; Yoo and Rhee, 2020). The Spot-ID method can also be expanded to the analysis of other subcellular compartments, such as the ER luminal proteins (Lee et al., 2017; Yoo and Rhee, 2020). Very recently, Rhee's group also revealed the membrane topologies of 85 ER- and mitochondria-associated membrane proteins using the Contact-ID system (Kwak et al., 2020). In addition, the exposure level of the Tyr or Lys on a protein is often positively correlated with its labeling intensity by PL enzymes, which can be assessed by quantitative proteomic analysis (Lee et al., 2017). Therefore, analysis of the biotinylation intensity of the peptides can provide insight into the dynamic structure of the proteins under a naive cellular environment (Figure 2C, right). This has been well documented by correlation analysis of the structures of either the cytochrome BC1 or mitochondrial ATP synthase complexes and the MS1 signal intensity of the labeled peptides (Lee et al., 2017). A recent reanalysis of four proximity proteomic datasets indicated that PL can be used to identify intrinsically disordered regions, as biotinylation occurs much faster in the unfolded and exposed regions of a protein than in the structured and less-accessible regions (Minde et al., 2020). Together, these studies indicate that the PL method is an effective tool for high-throughput topological analysis of membrane proteins; it complements existing structural biology tools by offering additional information on the dynamic structure of a target protein.
Protein–nucleic acid interactions
PL technology can be used to characterize both DNA–protein and RNA–protein interactions. The fine-tuning regulation of gene expression by interactions between transcription factors and specific genomic loci is crucial for the genomic stability of living cells (Ahmad et al., 2008; Kinney et al., 2007). However, mapping the proteins associated with a particular genomic locus in living cells is still challenging because of the limitations of traditional techniques, which are low throughput and time consuming. Recently, a novel method has been developed to identify genomic loci-associated proteins by combining the PL technique with catalytic dead RNA-guided nuclease Cas9 (dCas9) (Figure 2D). dCas9 loses its ability to cleave DNA but retains the ability to target a specific genomic locus when directed by a corresponding single-guide RNA (sgRNA) (Qi et al., 2013). Fusion with dCas9 enables the targeting of the PL enzyme to specific genomic loci and the subsequent labeling of neighborhood proteins. These tagged proteins can be further enriched and analyzed by LC-MS/MS, resulting in the identification of proteins associated with distinct genomic loci (Gao et al., 2019). Several labs have harnessed PL enzymes and the CRISPR-dCas9 system to develop systems for mapping DNA-associated proteins, including CasID (Schmidtmann et al., 2016), GLoPro (Myers et al., 2018), C-BERST (Gao et al., 2018), and CAPLOCUS (Qiu et al., 2019).
RNA–protein interactions play an important role in the regulation of gene expression and the processing, transport, translation, and stability of mRNAs (Bolognani and Perrone-Bizzozero, 2008). However, current methods for the identification of RNA-associated proteins depend on the purification of RNA–protein complexes and require in vitro RNA manipulation; they cannot reflect RNA–protein interactions in native cellular environments. Recently developed PL technology, together with other existing techniques, offers new solutions for studying RNA-associated proteins (Figure 2D). For example, Ramanathan et al. (2018) developed an RNA–protein interaction detection (RaPID) system by fusing BioID to λ bacteriophage antiterminator protein N (λN22) peptides, which can bind to the boxB stem loop structure with high affinity. The λN22-fused BioID can be recruited to the proximity of an RNA motif of interest by engineering boxB stem loops at both termini of the RNA motif, thereby biotinylating proteins bound to or near the RNA motif of interest. RaPID was successfully used to identify novel proteins bound to 3′ UTRs of Zika virus. The RaPID system was further improved by substituting BioID with BASU, and this modification led to a higher signal-to-noise ratio than the λN-BioID system (Ramanathan et al., 2018). Lu and Wei (2019) subsequently demonstrated that λN-BASU is more competent than λN-APEX2 for the detection of RNA–protein interactions in living cells. However, because the labeling radius in this method is approximately 20 nm, RaPID is suitable for studying short RNA motifs (≤132 nt) (Ramanathan et al., 2018). Recently, PL enzymes such as APEX2, BioID2, BASU, TurboID, or PafA were integrated with MS2-MCP, CRISPR-dCas13, or CRISPR-dCasRx systems to study RNA-binding proteins on specific RNAs. A number of novel proteins associating with RNAs were discovered (Mukherjee et al., 2019; Zhang et al., 2020b; Han et al., 2020; Li et al., 2020; Yi et al., 2020), highlighting the robustness of these newly developed PL methods for the investigation of dynamic RNA–protein interactions in living cells.
Subcellular transcriptomic profiling
In eukaryotic cells, RNAs are often translocated to specific subcellular compartments for local translation, which is crucial for many biological processes (Buxbaum et al., 2015; Jung et al., 2014). Conventional methods to study subcellular RNA include fluorescence in situ hybridization (FISH) for investigating RNA localization (Chen et al., 2015; Femino et al., 1998) and immunoprecipitation (Gilbert et al., 2004; Ule et al., 2003) coupled with microarray or deep sequencing for local transcriptome analysis. However, these methods have some drawbacks, such as limited throughput for FISH imaging and complicated biochemical manipulations that are difficult for most labs to master. Recently, PL methods have opened a new door for mapping the local transcriptome of subcellular compartments (Figure 2E). In 2014, two studies from the Weissman lab used BirA to promiscuously label ribosomes around an organelle in living cells; biotinylated ribosomes were then enriched, and the RNAs bound by the ribosomes were characterized by deep sequencing (i.e., ribosome profiling). Therefore, PL and ribosome profiling can be used to characterize the actively translated mRNAs near specific organelles such as the ER (Jan et al., 2014) and the mitochondria (Williams et al., 2014) in living cells. However, proximity-specific ribosome profiling cannot detect noncoding RNAs or non-translated mRNAs. To overcome these limitations, two new systems, APEX-RIP (Kaewsapsak et al., 2017) and Proximity-CLIP (Daniel et al., 2018), were developed by combining APEX-based PL with formaldehyde-mediated or UV-induced protein–RNA crosslinking methods. These two systems have been used to profile the subcellular transcriptome of various compartments, such as the mitochondrial matrix, nucleus, cytosol, ER membrane, and cell–cell interface (Daniel et al., 2018; Kaewsapsak et al., 2017). However, both APEX-RIP and Proximity-CLIP require crosslinking treatment, which may reduce the spatial resolution to some degree. To bypass the crosslinking step, another two methods, APEX-seq (Padrón et al., 2019) and CAP-seq (Wang et al., 2019), were developed to directly biotinylate RNAs near or within subcellular compartments such as the ER or stress granules (Padrón et al., 2019) (Figure 2E). Fazal et al. (2019) used the APEX-seq method to map RNAs near nine distinct subcellular locales and generated a global spatial map of the human transcriptome. Zhou et al. (2019) further optimized the APEX-seq method by substituting the biotin-phenol substrate with a novel biotin-aniline probe, which significantly improved RNA labeling efficiency and accuracy in the cellular environment. As well as expanding APEX2 substrates for RNA labeling, they also developed a miniSOG-mediated RNA labeling technique called CAP-seq to investigate local RNAs proximal to the ER and mitochondria (Wang et al., 2019). Because CAP-seq and APEX-seq use different enzymes, they may generate results with different spatial resolutions and can complement each other in mapping the subcellular transcriptome. In addition, APEX-RIP, Proximity-CLIP, and APEX-seq methods can provide information on both the local transcriptome and RNA-interacting proteins (Kaewsapsak et al., 2017; Daniel et al., 2018; Padrón et al., 2019), whereas CAP-seq can only map organelle-associated RNAs (Wang et al., 2019). However, because APEX-derived systems use H2O2, which may induce stress or perturb cellular functions, the light-activated CAP-seq systems may be applicable to more biological systems, such as plants. Overall, the PL-derived RNA identification system serves as a powerful tool for mapping spatial transcriptomes in a broad range of biological contexts.
Application of PL in planta remains in its infancy
In contrast to the wide application of PL in animal systems, its utilization in plants remains to be fully explored. In 2017, the Lin lab used BioID-based PL to identify interactors of the OsFD2 transcription factor in rice protoplasts (Lin et al., 2017). This was the first attempt to establish the PL method in planta. In 2018, two membrane-localized effectors of Pseudomonas syringae, AvrPto and HopF2, were fused with BioID to map their interactome in Nicotiana benthamiana and Arabidopsis thaliana, respectively (Conlan et al., 2018; Khan et al., 2018). In 2019, Wong's group used BioID-based PL to identify the interaction partners of autophagy-related protein 8 (ATG8) and the 126 kDa replicase of tobacco mosaic virus (TMV) (Das et al., 2019; Macharia et al., 2019). Very recently, Yangnan Gu's group used BioID2-based PL to profile the plant nuclear envelope and inner nuclear membrane proteomes in Arabidopsis (Huang et al., 2020). They identified a novel component of the nuclear pore complex and unraveled critical regulators of inner nuclear membrane protein degradation, all of which enhance our understanding of the composition of subnuclear structures and the underlying functions of nuclear membrane-associated proteins (Huang et al., 2020; Tang et al., 2020). These studies reinforced the robustness of PL methods for identifying membrane-associated interaction partners, which are inaccessible by traditional approaches, and indicated that PL methods could also be used to investigate global sub-organelle proteomes in plant systems as in mammalian cells (Kim et al., 2016; Cho et al., 2020). It should be noted that the optimal temperature for the catalytic activity of both BioID and BioID2 is 37°C, which may cause heat stress in plant cells, given that the optimal plant growth temperature is 22°C–25°C. However, despite the different temperatures required for plant incubation (25°C) and optimal BioID or BioID2 ligase activity (37°C), several in planta PL studies have revealed that both BioID- and BioID2-based PL work at room temperature, as demonstrated by the identification of some novel interaction partners (Khan et al., 2018; Das et al., 2019; Huang et al., 2020). Also, for experiments such as sub-organellar proteome profiling that do not require fast labeling kinetics, the use of TurboID is not necessarily imperative, although it may further improve labeling efficiency and result in more comprehensive profiling. In addition, adding biotin-phenol and H2O2 during APEX-based PL is toxic to plant cells and can lead to oxidative stress, which also limits its application in plant systems. The TurboID enzyme developed by Ting's group (Branon et al., 2018) shows fast kinetics at room temperature and uses a non-toxic biotin substrate; it has therefore paved the way for better application of PL to plant biology research.
To date, four labs have independently established the TurboID-based PL technique in plant systems, including Arabidopsis, tomato root cultures, and N. benthamiana (Kim et al., 2019; Mair et al., 2019; Zhang et al., 2019; Arora et al., 2020). Three labs compared the activity of BioID, BioID2, TurboID, and miniTurboID in different plant systems. All these studies demonstrated that TurboID has a higher efficiency in biotinylating proximal proteins in planta than BioID and BioID2. The nucleotide-binding leucine-rich repeat (NLR) immune receptors play a key role in mediating plant immune responses to diverse pathogens. Usually, NLRs are expressed at a very low level to avoid autoimmunity and detrimental effects on plant fitness (Tian et al., 2003). This low expression of the NLR proteins, together with the dynamic properties of the NLR regulatory network, makes it difficult to capture the proteins that are directly involved in the NLR signaling pathway by conventional methods. In 2019, we used TurboID-based PL to map the interactome of the N NLR immune receptor that confers resistance to TMV. We identified a novel regulator, UBR7, that can directly interact with the N NLR and mediate its turnover during N-mediated resistance to TMV. It has long been observed that N protein expression is elevated upon the perception of TMV’s entry into cells (Mestre and Baulcombe, 2006; Padmanabhan et al., 2013). Although the mechanistic basis for this process remains elusive, the identification of UBR7 by PL methods provides mechanistic insight into the intricate regulation of N NLR protein homeostasis (Zhang et al., 2019). This was also the first application of TurboID-based PL to plant systems, and it substantiated the robustness of TurboID in mapping the dynamic signaling network in planta (Zhang et al., 2019). A detailed step-by-step protocol for the identification of PPIs in N. benthamiana has recently been published (Zhang et al., 2020a). In addition to N. benthamiana, Mair et al. (2019) established TurboID-based PL in another model plant, Arabidopsis, and demonstrated the applicability of TurboID in dissecting the interacting proteome of low abundance proteins and mapping the interactome of cell-type-specific nuclear proteins such as FAMA. Furthermore, Arora et al. (2020) verified the possibility of using TurboID to capture membrane protein interactomes in N. benthamiana and provided topological information on membrane proteins. Kim et al. (2019) demonstrated the reliability of TurboID for unraveling highly dynamic PPIs by screening the protein substrates of a transient kinase.
Taken together, these excellent recent studies pave the way for further application of PL techniques in studying PPIs in planta. We anticipate that the PL-derived methods discussed above, which have been well developed in mammalian systems, can be easily transferred to plant systems, substantially complementing current approaches for identifying the interactomes of DNA, RNA, and proteins and accelerating the discovery of more novel interaction proteins in plants.
Some considerations for PL methods
Although PL methods have emerged as powerful and versatile tools to identify diverse protein interactions, the use of PL for a specific experimental design should be considered carefully in advance in order to obtain desirable potential candidate interaction partners for subsequent validation. Below are some suggestions to consider when implementing the PL method. First, researchers should choose a suitable PL enzyme for their studies. Based on published reports comparing different biotin ligases, TurboID and miniTurboID show higher biotinylation efficiency than BioID and BioID2 in plant systems (Zhang et al., 2019; Arora et al., 2020). However, whether the candidate protein interactors detected by TurboID-based PL are better than those detected with other PL enzymes such as BioID and BioID2 remains to be tested; such a comparison would be useful for the selection of suitable PL enzymes for individual experimental designs. miniTurboID can minimize the deleterious effect of tag fusion on the function of the target protein, but it has a lower labeling efficiency than TurboID (Branon et al., 2018). Second, researchers should make sure that the fusion of the PL enzyme to the bait protein does not interfere with its localization or functions. It is recommended to fuse the PL enzyme and a fluorescent protein to the N or C terminus of the protein of interest and perform confocal microscopy or a complementation test to confirm the correct targeting and functional maintenance of the tagged bait protein. In addition, to better mimic the physiological state of the bait protein in vivo, engineering of the tagged bait protein under an inducible or native promoter is also recommended (Mair et al., 2019). Third, as PL studies can generate large numbers of candidate interacting proteins, it is important to include appropriate controls and perform biological and technical replicates to minimize false positives or false negatives. The control protein should have a similar or at least overlapping localization pattern and a similar expression level to that of the bait protein. Inadequate expression of the control protein may cause false positives. Conversely, increased expression of the control protein may filter positive signals and cause false negatives (Zhang et al., 2019, 2020a). The PL enzyme alone or a fluorescent protein-tagged PL enzyme was often used as the control in previous PL-related studies. Alternatively, a functional mutant that disrupts the native localization or PPIs of the cognate bait protein can also be used as a control to minimize the false positives or negatives. Fourth, biotin ligase may not be applicable to some organelles, such as peroxisomes and vacuoles, because of their oxidative or acidic environments. In addition, some biotin ligases such as TurboID (Branon et al., 2018) utilize endogenous biotin, and caution should therefore be exercised in the use of TurboID for experiments that require strictly inducible biotinylation. Fifth, the labeling of PL enzymes requires negatively charged amino acid residues on the surface. If these residues are buried inside the bait protein, labeling will not occur, and this may lead to false negatives. This may partially explain the absence of some known interaction partners from candidate protein lists generated by PL assays, which has often been observed in previous studies (Khan et al., 2018; Zhang et al., 2019). Some PL enzyme-mediated covalent modifications occur on lysine and may affect subsequent digestion by trypsin or lysine C. However, not all lysines are modified during PL analysis if they are not surface-exposed. Also, other digestive enzymes, such as α-chymotrypsin, can be used together with trypsin or lysine C to achieve better proteolysis of the enriched bait protein. Sixth, because the optimal temperature of existing PL enzymes ranges from 25°C to 37°C, current PL methods may not be suitable for the characterization of PPIs under cold stress. Seventh, the labeling time should be optimized by conducting preliminary experiments. A longer labeling time has been reported to cause biotin depletion in cells, which may trigger stress responses (Branon et al., 2018). The extensive modifications of the proteins by biotin are irreversible and may also interfere with the normal cellular machinery. Also, a longer labeling time will inevitably increase false positives or false negatives. Eighth, for the interpretation of the MS data after PL analysis, a cutoff value of 1.5-fold enrichment compared with the control group during isobaric tandem mass tag (TMT) labeling-based quantitative proteomics is often used to select candidate interaction proteins for subsequent validation (Zhang et al., 2019). Because label-free MS-based quantitative proteome profiling exhibits a greater fold change than TMT labeling-based MS analysis, determination of the cutoff value depends largely on experimental design, and there is no universal cutoff standard. Known interactors identified in the MS dataset can be helpful for selecting the cutoff value. Last but not least, it is important to keep in mind that the PL method can identify proteins that enter the labeling radius of the PL enzyme-fused bait protein; whether these biotin-labeled proteins really interact with the target protein remains to be tested by additional experiments such as co-IP and bimolecular fluorescence complementation assays (Kim and Roux, 2016). Proteins involved in bait protein translation (e.g., ribosome proteins), folding (e.g., molecular chaperones), or trafficking (such as importin α protein for nuclear localization signal-containing cargo) may also be labeled by PL enzymes. Although they may not be false-positive candidates, these types of proteins should be considered carefully to avoid misleading interpretations during MS data analysis. It is also important to note that candidate PPIs identified by PL methods may not be consistently verified by conventional approaches because of their limitations in analyzing transient, weak, or hydrophobic PPIs. Thus, further genetic assays are required to confirm these PPIs. Also, although PL methods have many advantages, they cannot completely replace traditional approaches such as co-IP or Y2H.
Concluding remarks and future perspectives
PL methods are increasingly applied to identify PPIs across diverse research fields. Compared with conventional techniques, PL methods are more accessible, simpler, and, most importantly, more powerful in probing various molecular interactions, especially transient or weak PPIs and low-abundance and membrane-localized proteins. In fact, owing to its flexibility, easy implementation, and efficiency, the combination of PL methods with existing techniques will no doubt be broadly used to address more biological questions, which will be conducive to the development of different research fields. Indeed, our understanding of PPIs was largely fragmented and restricted to static events based on conventional approaches. The emerging development of PL techniques provides an unprecedented opportunity to profile dynamic interaction networks under different conditions, thus offering a comprehensive view of global and complete cellular functions with a high spatial and temporal resolution, which will greatly advance our understanding of various biological processes.
There is no one “perfect” method for all situations, and the PL technique also has its own strengths and caveats. In the future, we believe the development of new PL enzymes and the optimization of the currently available PL methods will continue. This will further enhance the applicability of PL methods by extending their use to special cellular environments such as vacuoles or peroxisomes. Moreover, the application of PL methods to species other than the model plants (Zhang et al., 2019; Arora et al., 2020), such as rice, maize, and wheat, needs to be demonstrated. Due to the large amount of data generated by LC-MS/MS analyses, the development of more standard and convenient bioinformatic tools for MS data interpretation and processing is also an urgent task that should be carried out in parallel with advances in PL techniques. Together, there is every reason to believe that PL methods will play increasing roles in addressing a variety of biological questions in plant science.
Funding
This work was supported by grants from the National Natural Science Foundation of China (31872637 to Y.Z. and 31830106 to D.L.); and NSF-IOS-1354434, NSF-IOS-1339185, and NIH-GM132582 to S.P.D.-K.
Author contributions
All authors contributed to the writing of the manuscript and the preparation of the figures.
Acknowledgments
We apologize to all colleagues whose original publications are not cited due to space limitations. No conflict of interest declared.
Published: December 15, 2020
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Contributor Information
Savithramma P. Dinesh-Kumar, Email: spdineshkumar@ucdavis.edu.
Yongliang Zhang, Email: cauzhangyl@cau.edu.cn.
References
- Ahmad S., Keskin O., Sarai A., Nussinov R. Protein-DNA interactions: structural, thermodynamic and clustering patterns of conserved residues in DNA-binding proteins. Nucleic Acids Res. 2008;36:5922–5932. doi: 10.1093/nar/gkn573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora D., Abel N.B., Liu C., Van Damme P., Yperman K., Eeckhout D., Vu L.D., Wang J., Tornkvist A., Impens F. Establishment of proximity-dependent biotinylation approaches in different plant model systems. Plant Cell. 2020;32:3388–3407. doi: 10.1105/tpc.20.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banaszynski L.A., Liu C.W., Wandless T.J. Characterization of the FKBP·rapamycin·FRB ternary complex. J. Am. Chem. Soc. 2005;127:4715–4721. doi: 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- Béganton B., Solassol I., Mangé A., Solassol J. Protein interactions study through proximity-labeling. Expert Rev. Proteomics. 2019;16:717–726. doi: 10.1080/14789450.2019.1638769. [DOI] [PubMed] [Google Scholar]
- Bersuker K., Peterson C.W.H., To M., Sahl S.J., Savikhin V., Grossman E.A., Nomura D.K., Olzmann J.A. A proximity labeling strategy provides insights into the composition and dynamics of lipid droplet proteomes. Dev. Cell. 2018;44:97–112.e7. doi: 10.1016/j.devcel.2017.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolognani F., Perrone-Bizzozero N.I. RNA–protein interactions and control of mRNA stability in neurons. J. Neurosci. Res. 2008;86:481–489. doi: 10.1002/jnr.21473. [DOI] [PubMed] [Google Scholar]
- Branon T.C., Bosch J.A., Sanchez A.D., Udeshi N.D., Svinkina T., Carr S.A., Feldman J.L., Perrimon N., Ting A.Y. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018;36:880–887. doi: 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum A.R., Haimovich G., Singer R.H. In the right place at the right time: visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol. 2015;16:95–109. doi: 10.1038/nrm3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo S.E., Mootha V.K. The mitochondrial proteome and human disease. Annu. Rev. Genomics Hum. Genet. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter-O’Connell I., Vermehren-Schmaedick A., Jin H., Morgan R.K., David L.L., Cohen M.S. Combining chemical genetics with proximity-dependent labeling reveals cellular targets of poly(ADP-ribose) polymerase 14 (PARP14) ACS Chem. Biol. 2018;13:2841–2848. doi: 10.1021/acschembio.8b00567. [DOI] [PubMed] [Google Scholar]
- Chastney M.R., Lawless C., Humphries J.D., Warwood S., Jones M.C., Knight D., Jorgensen C., Humphries M.J. Topological features of integrin adhesion complexes revealed by multiplexed proximity biotinylation. J. Cell Biol. 2020;219:e202003038. doi: 10.1083/jcb.202003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.H., Boettiger A.N., Moffitt J.R., Wang S., Zhuang X. RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K.F., Branon T.C., Rajeev S., Svinkina T., Udeshi N.D., Thoudam T., Kwak C., Rhee H.W., Lee I.K., Carr S.A. Split-TurboID enables contact-dependent proximity labeling in cells. Proc. Natl. Acad. Sci. U S A. 2020;117:12143–12154. doi: 10.1073/pnas.1919528117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Chen J., Schreiber S.L., Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- Conlan B., Stoll T., Gorman J.J., Saur I., Rathjen J.P. Development of a rapid in planta bioid system as a probe for plasma membrane-associated immunity proteins. Front Plant Sci. 2018;9:1882. doi: 10.3389/fpls.2018.01882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyaud E., Mis M., Laurent E.M.N., Dunham W.H., Couzens A.L., Robitaille M., Gingras A.-C., Angers S., Raught B. BioID-based identification of Skp Cullin F-box (SCF)β-TrCP1/2 E3 ligase substrates. Mol. Cell. Proteomics. 2015;14:1781–1795. doi: 10.1074/mcp.M114.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel, Benhalevy D., Anastasakis, Markus H. Proximity-CLIP provides a snapshot of protein-occupied RNA elements in subcellular compartments. Nat. Methods. 2018;15:1074–1082. doi: 10.1038/s41592-018-0220-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das P.P., Macharia M.W., Lin Q., Wong S.M. In planta proximity-dependent biotin identification (BioID) identifies a TMV replication co-chaperone NbSGT1 in the vicinity of 126 kDa replicase. J. Proteomics. 2019;204:103402. doi: 10.1016/j.jprot.2019.103402. [DOI] [PubMed] [Google Scholar]
- De Munter S., Gornemann J., Derua R., Lesage B., Qian J., Heroes E., Waelkens E., Van Eynde A., Beullens M., Bollen M. Split-BioID: a proximity biotinylation assay for dimerization-dependent protein interactions. FEBS Lett. 2017;591:415–424. doi: 10.1002/1873-3468.12548. [DOI] [PubMed] [Google Scholar]
- Dumont A.-A., Dumont L., Berthiaume J., Auger-Messier M. p38α MAPK proximity assay reveals a regulatory mechanism of alternative splicing in cardiomyocytes. Biochim. Biophys. Acta. 2019;1866:118557. doi: 10.1016/j.bbamcr.2019.118557. [DOI] [PubMed] [Google Scholar]
- Eisenberg-Bord M., Shai N., Schuldiner M., Bohnert M. A tether is a tether is a tether: tethering at membrane contact sites. Dev. Cell. 2016;39:395–409. doi: 10.1016/j.devcel.2016.10.022. [DOI] [PubMed] [Google Scholar]
- Fazal F.M., Han S., Parker K.R., Kaewsapsak P., Xu J., Boettiger A.N., Chang H.Y., Ting A.Y. Atlas of subcellular RNA localization revealed by APEX-seq. Cell. 2019;178:473–490.e26. doi: 10.1016/j.cell.2019.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino A.M., Fay F.S., Fogarty K., Singer R.H. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Firat-Karalar, Elif N., Rauniyar N., Yates J.R., Stearns T. Proximity interactions among centrosome components identify regulators of centriole duplication. Curr. Biol. 2014;24:664–670. doi: 10.1016/j.cub.2014.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X.D., Rodriguez T.C., Sontheimer E.J. Adapting dCas9-APEX2 for subnuclear proteomic profiling. Methods Enzymol. 2019;616:365–383. doi: 10.1016/bs.mie.2018.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X.D., Tu L.-C., Mir A., Rodriguez T., Ding Y., Leszyk J., Dekker J., Shaffer S.A., Zhu L.J., Wolfe S.A. C-BERST: defining subnuclear proteomic landscapes at genomic elements with dCas9-APEX2. Nat. Methods. 2018;15:433–436. doi: 10.1038/s41592-018-0006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Chen L., Liu S., Zhao J., Zhang H., Chen P.R. Enzyme-mediated intercellular proximity labeling for detecting cell-cell interactions. J. Am. Chem. Soc. 2019;141:1833–1837. doi: 10.1021/jacs.8b10286. [DOI] [PubMed] [Google Scholar]
- Gilbert C., Kristjuhan A., Winkler G.S., Svejstrup J.Q. Elongator interactions with nascent mRNA revealed by RNA immunoprecipitation. Mol. Cell. 2004;14:457–464. doi: 10.1016/s1097-2765(04)00239-4. [DOI] [PubMed] [Google Scholar]
- Gingras A.C., Abe K.T., Raught B. Getting to know the neighborhood: using proximity-dependent biotinylation to characterize protein complexes and map organelles. Curr. Opin. Chem. Biol. 2019;48:44–54. doi: 10.1016/j.cbpa.2018.10.017. [DOI] [PubMed] [Google Scholar]
- Go C.D., Knight J.D.R., Rajasekharan A., Rathod B., Hesketh G.G., Abe K.T., Youn J.-Y., Samavarchi-Tehrani P., Zhang H., Zhu L.Y. A proximity biotinylation map of a human cell. bioRxiv. 2019:796391. doi: 10.1101/796391. [DOI] [Google Scholar]
- Gu B., Lambert J.P., Cockburn K., Gingras A.C., Rossant J. AIRE is a critical spindle-associated protein in embryonic stem cells. eLife. 2017;6:e28131. doi: 10.7554/eLife.28131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G.D., Coyaud É., Gonçalves J., Mojarad B.A., Liu Y., Wu Q., Gheiratmand L., Comartin D., Tkach J.M., Cheung S.W.T. A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell. 2015;163:1484–1499. doi: 10.1016/j.cell.2015.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Udeshi N.D., Deerinck T.J., Svinkina T., Ellisman M.H., Carr S.A., Ting A.Y. Proximity biotinylation as a method for mapping proteins associated with mtDNA in living cells. Cell Chem. Biol. 2017;24:404–414. doi: 10.1016/j.chembiol.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Zhao B.S., Myers S.A., Carr S.A., He C., Ting A.Y. RNA-protein interaction mapping via MS2 or Cas13-based APEX targeting. bioRxiv. 2020 doi: 10.1073/pnas.2006617117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Branon T.C., Martell J.D., Boassa D., Shechner D., Ellisman M.H., Ting A. Directed evolution of split APEX2 peroxidase. ACS Chem. Biol. 2019;14:619–635. doi: 10.1021/acschembio.8b00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill Z.B., Pollock S.B., Zhuang M., Wells J.A. Direct proximity tagging of small molecule protein targets using an engineered NEDD8 ligase. J. Am. Chem. Soc. 2016;138:13123–13126. doi: 10.1021/jacs.6b06828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A., Tang Y., Shi X., Jia M., Zhu J., Yan X., Chen H., Gu Y. Proximity labeling proteomics reveals critical regulators for inner nuclear membrane protein degradation in plants. Nat. Commun. 2020;11:3284. doi: 10.1038/s41467-020-16744-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V., Lam S.S., Udeshi N.D., Svinkina T., Guzman G., Mootha V.K., Carr S.A., Ting A.Y. Proteomic mapping of cytosol-facing outer mitochondrial and ER membranes in living human cells by proximity biotinylation. eLife. 2017;6:e24463. doi: 10.7554/eLife.24463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung V., Zou P., Rhee H.W., Udeshi N.D., Cracan V., Svinkina T., Carr S.A., Mootha V.K., Ting A.Y. Proteomic mapping of the human mitochondrial intermembrane space in live cells via ratiometric APEX tagging. Mol. Cell. 2014;55:332–341. doi: 10.1016/j.molcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan C.H., Williams C.C., Weissman J.S. Principles of ER cotranslational translocation revealed by proximity-specific ribosome profiling. Science. 2014;346:1257521. doi: 10.1126/science.1257521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing J., He L., Sun A., Quintana A., Ding Y., Ma G., Tan P., Liang X., Zheng X., Chen L. Proteomic mapping of ER-PM junctions identifies STIMATE as a regulator of Ca2+ influx. Nat. Cell Biol. 2015;17:1339–1347. doi: 10.1038/ncb3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H., Gkogkas C.G., Sonenberg N., Holt C.E. Remote control of gene function by local translation. Cell. 2014;157:26–40. doi: 10.1016/j.cell.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewsapsak P., Shechner D.M., Mallard W., Rinn J.L., Ting A.Y. Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking. eLife. 2017;6:e29224. doi: 10.7554/eLife.29224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Youn J.Y., Gingras A.C., Subramaniam R., Desveaux D. In planta proximity dependent biotin identification (BioID) Sci. Rep. 2018;8:9212. doi: 10.1038/s41598-018-27500-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido K., Yamanaka S., Nakano S., Motani K., Shinohara S., Nozawa A., Kosako H., Ito S., Sawasaki T. AirID, a novel proximity biotinylation enzyme, for analysis of protein-protein interactions. eLife. 2020;9:e54983. doi: 10.7554/eLife.54983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.I., Birendra K.C., Zhu W., Motamedchaboki K., Doye V., Roux K.J. Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc. Natl. Acad. Sci. U S A. 2014;111:E2453–E2461. doi: 10.1073/pnas.1406459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.I., Jensen S.C., Noble K.A., Kc B., Roux K.H., Motamedchaboki K., Roux K.J. An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell. 2016;27:1188–1196. doi: 10.1091/mbc.E15-12-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.I., Roux K.J. Filling the void: proximity-based labeling of proteins in living cells. Trends Cell Biol. 2016;26:804–817. doi: 10.1016/j.tcb.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.-W., Park C.H., Hsu C.-C., Zhu J.-Y., Hsiao Y., Branon T., Xu S.-L., Ting A.Y., Wang Z.-Y. Application of TurboID-mediated proximity labeling for mapping a GSK3 kinase signaling network in Arabidopsis. bioRxiv. 2019:636324. [Google Scholar]
- Kinney J.B., Tkacik G., Callan C.G., Jr. Precise physical models of protein-DNA interaction from high-throughput data. Proc. Natl. Acad. Sci. U S A. 2007;104:501–506. doi: 10.1073/pnas.0609908104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochanova N.Y., Schauer T., Mathias G.P., Lukacs A., Schmidt A., Flatley A., Schepers A., Thomae A.W., Imhof A. A multi-layered structure of the interphase chromocenter revealed by proximity-based biotinylation. Nucleic Acids Res. 2020;48:4161–4178. doi: 10.1093/nar/gkaa145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koerver L., Papadopoulos C., Liu B., Kravic B., Rota G., Brecht L., Veenendaal T., Polajnar M., Bluemke A., Ehrmann M. The ubiquitin-conjugating enzyme UBE2QL1 coordinates lysophagy in response to endolysosomal damage. EMBO Rep. 2019;20:e48014. doi: 10.15252/embr.201948014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani N., Gu J., Isaji T., Udaka K., Taniguchi N., Honke K. Biochemical visualization of cell surface molecular clustering in living cells. Proc. Natl. Acad. Sci. U S A. 2008;105:7405–7409. doi: 10.1073/pnas.0710346105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak C., Shin S., Park J.S., Jung M., Nhung T.T.M., Kang M.G., Lee C., Kwon T.H., Park S.K., Mun J.Y. Contact-ID, a tool for profiling organelle contact sites, reveals regulatory proteins of mitochondrial-associated membrane formation. Proc. Natl. Acad. Sci. U S A. 2020;117:12109–12120. doi: 10.1073/pnas.1916584117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S.S., Martell J.D., Kamer K.J., Deerinck T.J., Ellisman M.H., Mootha V.K., Ting A.Y. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods. 2015;12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.-Y., Kang M.-G., Park J.-S., Lee G., Ting A.Y., Rhee H.-W. APEX fingerprinting reveals the subcellular localization of proteins of interest. Cell Rep. 2016;15:1837–1847. doi: 10.1016/j.celrep.2016.04.064. [DOI] [PubMed] [Google Scholar]
- Lee S.Y., Kang M.G., Shin S., Kwak C., Kwon T., Seo J.K., Kim J.S., Rhee H.W. Architecture mapping of the inner mitochondrial membrane proteome by chemical tools in live cells. J. Am. Chem. Soc. 2017;139:3651–3662. doi: 10.1021/jacs.6b10418. [DOI] [PubMed] [Google Scholar]
- Li X.W., Rees J.S., Xue P., Zhang H., Hamaia S.W., Sanderson B., Funk P.E., Farndale R.W., Lilley K.S., Perrett S. New insights into the DT40 B cell receptor cluster using a proteomic proximity labeling assay. J. Biol. Chem. 2014;289:14434–14447. doi: 10.1074/jbc.M113.529578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Liu S., Cao L., Luo Y., Du H., Li S., You F. CBRPP: a new RNA-centric method to study RNA-protein interactions. bioRxiv. 2020 doi: 10.1101/2020.04.09.033290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q., Zhou Z., Luo W., Fang M., Li M., Li H. Screening of proximal and interacting proteins in rice protoplasts by proximity-dependent biotinylation. Front. Plant Sci. 2017;8:749. doi: 10.3389/fpls.2017.00749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Papa A., Katchman A.N., Zakharov S.I., Roybal D., Hennessey J.A., Kushner J., Yang L., Chen B.X., Kushnir A. Mechanism of adrenergic CaV1.2 stimulation revealed by proximity proteomics. Nature. 2020;577:695–700. doi: 10.1038/s41586-020-1947-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Zheng J., Sun W., Huo Y., Zhang L., Hao P., Wang H., Zhuang M. A proximity-tagging system to identify membrane protein-protein interactions. Nat. Methods. 2018;15:715–722. doi: 10.1038/s41592-018-0100-5. [DOI] [PubMed] [Google Scholar]
- Lobingier B.T., Hüttenhain R., Eichel K., Miller K.B., Ting A.Y., von Zastrow M., Krogan N.J. An approach to spatiotemporally resolve protein interaction networks in living cells. Cell. 2017;169:350–360.e12. doi: 10.1016/j.cell.2017.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K.H., Stawski P.S., Draycott A.S., Udeshi N.D., Lehrman E.K., Wilton D.K., Svinkina T., Deerinck T.J., Ellisman M.H., Stevens B. Proteomic analysis of unbounded cellular compartments: synaptic clefts. Cell. 2016;166:1295–1307.e21. doi: 10.1016/j.cell.2016.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M., Wei W. Proximity labeling to detect RNA-protein interactions in live cells. FEBS Open Bio. 2019;9:1860–1868. doi: 10.1002/2211-5463.12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg E., Borner G.H.H. Spatial proteomics: a powerful discovery tool for cell biology. Nat. Rev. Mol. Cell Biol. 2019;20:285–302. doi: 10.1038/s41580-018-0094-y. [DOI] [PubMed] [Google Scholar]
- Macharia M.W., Tan W.Y.Z., Das P.P., Naqvi N.I., Wong S.M. Proximity-dependent biotinylation screening identifies NbHYPK as a novel interacting partner of ATG8 in plants. BMC Plant Biol. 2019;19:326. doi: 10.1186/s12870-019-1930-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair A., Xu S.L., Branon T.C., Ting A.Y., Bergmann D.C. Proximity labeling of protein complexes and cell-type-specific organellar proteomes in Arabidopsis enabled by TurboID. eLife. 2019;8:e47864. doi: 10.7554/eLife.47864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markmiller S., Soltanieh S., Server K.L., Mak R., Jin W., Fang M.Y., Luo E.-C., Krach F., Yang D., Sen A. Context-dependent and disease-specific diversity in protein interactions within stress granules. Cell. 2018;172:590–604.e13. doi: 10.1016/j.cell.2017.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell J.D., Deerinck T.J., Sancak Y., Poulos T.L., Mootha V.K., Sosinsky G.E., Ellisman M.H., Ting A.Y. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell J.D., Yamagata M., Deerinck T.J., Phan S., Kwa C.G., Ellisman M.H., Sanes J.R., Ting A.Y. A split horseradish peroxidase for the detection of intercellular protein-protein interactions and sensitive visualization of synapses. Nat. Biotechnol. 2016;34:774–780. doi: 10.1038/nbt.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavylutov T., Chen X., Guo L., Yang J. APEX2-tagging of Sigma 1-receptor indicates subcellular protein topology with cytosolic N-terminus and ER luminal C-terminus. Protein Cell. 2018;9:733–737. doi: 10.1007/s13238-017-0468-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mestre P., Baulcombe D.C. Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell. 2006;18:491–501. doi: 10.1105/tpc.105.037234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mick D.U., Rodrigues R.B., Leib R.D., Adams C.M., Chien A.S., Gygi S.P., Nachury M.V. Proteomics of primary cilia by proximity labeling. Dev. Cell. 2015;35:497–512. doi: 10.1016/j.devcel.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minde D.P., Ramakrishna M., Lilley K.S. Biotin proximity tagging favours unfolded proteins and enables the study of intrinsically disordered regions. Commun. Biol. 2020;3:38. doi: 10.1038/s42003-020-0758-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee J., Hermesh O., Eliscovich C., Nalpas N., Franz-Wachtel M., Maček B., Jansen R.-P. β-Actin mRNA interactome mapping by proximity biotinylation. Proc. Natl. Acad. Sci. U S A. 2019;116:12863–12872. doi: 10.1073/pnas.1820737116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S.A., Wright J., Peckner R., Kalish B.T., Zhang F., Carr S.A. Discovery of proteins associated with a predefined genomic locus via dCas9-APEX-mediated proximity labeling. Nat. Methods. 2018;15:437–439. doi: 10.1038/s41592-018-0007-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.M.T., Kim J., Doan T.T., Lee M.-W., Lee M. APEX proximity labeling as a versatile tool for biological research. Biochemistry. 2020;59:260–269. doi: 10.1021/acs.biochem.9b00791. [DOI] [PubMed] [Google Scholar]
- Opitz N., Schmitt K., Hofer-Pretz V., Neumann B., Krebber H., Braus G.H., Valerius O. Capturing the Asc1p/receptor for activated C kinase 1 (RACK1) microenvironment at the head region of the 40S ribosome with quantitative BioID in yeast. Mol. Cell. Proteomics. 2017;16:2199–2218. doi: 10.1074/mcp.M116.066654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan M.S., Ma S., Burch-Smith T.M., Czymmek K., Huijser P., Dinesh-Kumar S.P. Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog. 2013;9:e1003235. doi: 10.1371/journal.ppat.1003235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrón A., Iwasaki S., Ingolia N.T. Proximity RNA labeling by APEX-seq reveals the organization of translation initiation complexes and repressive RNA granules. Mol. Cell. 2019;75:875–887.e75. doi: 10.1016/j.molcel.2019.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek J., Kalocsay M., Staus D.P., Wingler L., Pascolutti R., Paulo J.A., Gygi S.P., Kruse A.C. Multidimensional tracking of GPCR signaling via peroxidase-catalyzed proximity labeling. Cell. 2017;169:338–349.e11. doi: 10.1016/j.cell.2017.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz W.A., Toulmay A., Balla T. The functional universe of membrane contact sites. Nat. Rev. Mol. Cell Biol. 2020;21:7–24. doi: 10.1038/s41580-019-0180-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W., Xu Z., Zhang M., Zhang D., Fan H., Li T., Wang Q., Liu P., Zhu Z., Du D. Determination of local chromatin interactions using a combined CRISPR and peroxidase APEX2 system. Nucleic Acids Res. 2019;47:e52. doi: 10.1093/nar/gkz134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan M., Majzoub K., Rao D.S., Neela P.H., Zarnegar B.J., Mondal S., Roth J.G., Gai H., Kovalski J.R., Siprashvili Z. RNA-protein interaction detection in living cells. Nat. Methods. 2018;15:207–212. doi: 10.1038/nmeth.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee H.W., Zou P., Udeshi N.D., Martell J.D., Mootha V.K., Carr S.A., Ting A.Y. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux K.J., Kim D.I., Raida M., Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012;196:801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidtmann E., Anton T., Rombaut P., Herzog F., Leonhardt H. Determination of local chromatin composition by CasID. Nucleus. 2016;7:476–484. doi: 10.1080/19491034.2016.1239000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schopp I.M., Amaya Ramirez C.C., Debeljak J., Kreibich E., Skribbe M., Wild K., Bethune J. Split-BioID a conditional proteomics approach to monitor the composition of spatiotemporally defined protein complexes. Nat. Commun. 2017;8:15690. doi: 10.1038/ncomms15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith F.D., Omar M.H., Nygren P.J., Soughayer J., Hoshi N., Lau H.T., Snyder C.G., Branon T.C., Ghosh D., Langeberg L.K. Single nucleotide polymorphisms alter kinase anchoring and the subcellular targeting of A-kinase anchoring proteins. Proc. Natl. Acad. Sci. U S A. 2018;115:E11465–E11474. doi: 10.1073/pnas.1816614115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M., Chen Z., Zhang H., Tang M., Wang C., Jung S.Y., Chen J. Replisome dynamics and their functional relevance upon DNA damage through the PCNA interactome. Cell Rep. 2018;25:3869–3883.e4. doi: 10.1016/j.celrep.2018.11.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Huang A., Gu Y. Global profiling of plant nuclear membrane proteome in Arabidopsis. Nat. Plants. 2020;6:838–847. doi: 10.1038/s41477-020-0700-9. [DOI] [PubMed] [Google Scholar]
- Tian D., Traw M., Chen J., Kreitman M., Bergelson J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature. 2003;423:74. doi: 10.1038/nature01588. [DOI] [PubMed] [Google Scholar]
- Uezu A., Kanak D.J., Bradshaw T.W., Soderblom E.J., Catavero C.M., Burette A.C., Weinberg R.J., Soderling S.H. Identification of an elaborate complex mediating postsynaptic inhibition. Science. 2016;353:1123–1129. doi: 10.1126/science.aag0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J., Jensen K.B., Ruggiu M., Mele A., Ule A., Darnell R.B. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Varnaitė R., MacNeill S.A. Meet the neighbors: mapping local protein interactomes by proximity-dependent labeling with BioID. Proteomics. 2016;16:2503–2518. doi: 10.1002/pmic.201600123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Tang W., Li Z., Zou Z., Zhou Y., Li R., Xiong T., Wang J., Zou P. Mapping spatial transcriptome with light-activated proximity-dependent RNA labeling. Nat. Chem. Biol. 2019;15:1110–1119. doi: 10.1038/s41589-019-0368-5. [DOI] [PubMed] [Google Scholar]
- Williams C.C., Jan C.H., Weissman J.S. Targeting and plasticity of mitochondrial proteins revealed by proximity-specific ribosome profiling. Science. 2014;346:748–751. doi: 10.1126/science.1257522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M., Hou J., Wang L., Cheng D., Lu J., Zheng L., Xu T. Optimizing the fragment complementation of APEX2 for detection of specific protein-protein interactions in live cells. Sci. Rep. 2017;7:12039. doi: 10.1038/s41598-017-12365-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita R., Kotani N., Ishiura Y., Higashiyama S., Honke K. Spatiotemporally-regulated interaction between beta1 integrin and ErbB4 that is involved in fibronectin-dependent cell migration. J. Biochem. 2011;149:347–355. doi: 10.1093/jb/mvq148. [DOI] [PubMed] [Google Scholar]
- Yang L., Ding Y., Chen Y., Zhang S., Huo C., Wang Y., Yu J., Zhang P., Na H., Zhang H. The proteomics of lipid droplets: structure, dynamics, and functions of the organelle conserved from bacteria to humans. J. Lipid Res. 2012;53:1245–1253. doi: 10.1194/jlr.R024117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W., Li J., Zhu X., Wang X., Fan L., Sun W., Liao L., Zhang J., Li X., Ye J. CRISPR-assisted detection of RNA-protein interactions in living cells. Nat. Methods. 2020;17:685–688. doi: 10.1038/s41592-020-0866-0. [DOI] [PubMed] [Google Scholar]
- Yoo C.-M., Rhee H.-W. APEX, a master key to resolve membrane topology in live cells. Biochemistry. 2020;59:250–259. doi: 10.1021/acs.biochem.9b00785. [DOI] [PubMed] [Google Scholar]
- Youn J.-Y., Dunham W.H., Hong S.J., Knight J.D.R., Bashkurov M., Chen G.I., Bagci H., Rathod B., MacLeod G., Eng S.W.M. High-density proximity mapping reveals the subcellular organization of mRNA-associated granules and bodies. Mol. Cell. 2018;69:517–532.e11. doi: 10.1016/j.molcel.2017.12.020. [DOI] [PubMed] [Google Scholar]
- Zhang Y.L., Song G.Y., Lai N.K., Nagalakshmi U., Li Y.Y., Zheng W.J., Huang P.J., Branon T.C., Ting A.Y., Walley J.W. TurboID-based proximity labeling reveals that UBR7 is a regulator of N NLR immune receptor-mediated immunity. Nat. Commun. 2019;10:3252. doi: 10.1038/s41467-019-11202-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li Y., Yang X., Wen Z., Nagalakshmi U., Dinesh-Kumar S.P. TurboID-based proximity labeling for in planta identification of protein-protein interaction networks. J. Vis. Exp. 2020:e60728. doi: 10.3791/60728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Sun W., Shi T., Lu P., Zhuang M., Liu J.L. Capturing RNA-protein interaction via CRUIS. Nucleic Acids Res. 2020;48:e52. doi: 10.1093/nar/gkaa143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Wang G., Wang P., Li Z., Yue T., Wang J., Zou P. Expanding APEX2 substrates for proximity-dependent labeling of nucleic acids and proteins in living cells. Angew. Chem. Int. Ed. 2019;131:11889–11893. doi: 10.1002/anie.201905949. [DOI] [PubMed] [Google Scholar]