Abstract
Streptococcus sobrinus is an etiologic cause of dental caries (tooth decay) in humans. Our knowledge of S. sobrinus is scant despite the organism’s important role in oral health. It is widely believed that S. sobrinus lacks the natural competence pathways that are used by other streptococci to regulate growth, virulence, and quorum sensing. The lack of natural competence has also prevented genetic manipulation of S. sobrinus, limiting our knowledge of its pathogenicity. We discovered that most strains of S. sobrinus contain a new class of the ComRS competence system. Although S. sobrinus is typically placed among the mutans group streptococci, the S. sobrinus ComRS system is most similar to the competence pathways in the salivarius group. Unlike all other ComRS systems, the S. sobrinus pathway contains 2 copies of the transcriptional regulator ComR and has a peptide pheromone (XIP) that lacks any aromatic amino acids. Synthetic XIP enables transformation of S. sobrinus with plasmid or linear DNA, and we leverage this newfound genetic tractability to confirm that only 1 of the ComR homologs is required for induced competence while the other appears to suppress competence. Exogenous XIP increases the expression of bacteriocin gene clusters and produces an antimicrobial response that inhibits growth of S. mutans. We also identified 2 strains of S. sobrinus that appear to be “cheaters” by either not responding to or not producing XIP. We show how a recombination event in the nonresponsive strain could restore function of the ComRS pathway but delete the gene encoding XIP. Thus, the S. sobrinus ComRS pathway provides new tools for studying this pathogen and offers a lens into the evolution of ecological cheaters.
Keywords: dental caries, genetics, microbiology, streptococcus, pheromones, quorum sensing
Introduction
Dental caries results from acid fermentation by bacteria, most commonly the oral pathogens Streptococcus mutans and Streptococcus sobrinus. Although S. sobrinus is rarer thanS. mutans, studies show that S. sobrinus is more strongly associated with the development of caries, especially in children (Hirose et al. 1993; Nurelhuda et al. 2010; Gross et al. 2012; Singla et al. 2016). Coinfection with S. sobrinus and S. mutans is associated with greater incidence or severity of caries (Seki et al. 2006; Kanasi et al. 2010; Okada et al. 2012), suggesting that S. sobrinus and S. mutans may interact by an unknown mechanism to exacerbate the disease. Such an interaction is consistent with the ecological plaque hypothesis (Takahashi and Nyvad 2008; Philip et al. 2018), a viewpoint that caries results from complex interactions among microbes, the host, and the environment.
Streptococci use peptide pheromones for intra- and intercellular communication. The pheromones regulate competence pathways that innervate metabolism (Underhill et al. 2019), virulence (Koirala et al. 2018; Lin and Lau 2019), quorum sensing (Shanker and Federle 2017), and antibiotic tolerance (Slager et al. 2014). However, no peptide pheromones or functional competence pathways have been discovered in S. sobrinus. The apparent lack of competence pathways raises questions about how S. sobrinus interacts with S. mutans and other microbes during cariogenesis.
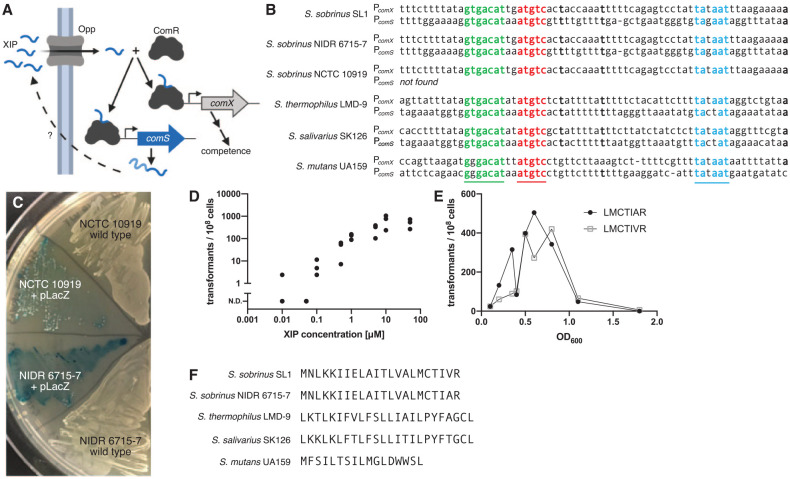
Avery et al. first discovered the natural competence of streptococci in 1944. Fifty years later, researchers showed that a peptide pheromone named CSP controlled competence through the ComCDE pathway (Håvarstein et al. 1995; Håvarstein et al. 1996). A second competence pathway, ComRS, was later discovered in many streptococci (Fontaine et al. 2010; Mashburn-Warren et al. 2010). ComRS systems contain a transcriptional regulator (ComR) that is activated by XIP, a small peptide derived from a precursor peptide ComS. The classic XIP pathway functions as an autocrine loop (Fig. 1A; Fontaine et al. 2015). The precursor peptide ComS is exported and cleaved to form active extracellular XIP. The processed XIP is imported by an unknown mechanism and binds to the transcriptional regulator ComR. The ComR/XIP complex initiates transcription of the competence-inducing sigma factor ComX (Talagas et al. 2016).
Figure 1.
The peptide XIP induces competence in Streptococcus sobrinus. (A) The ComRS competence pathway in Streptococcus mutans forms an autocrine signaling loop. An unknown protein cleaves the leader peptide from ComS and exports the XIP precursor. Activated XIP is imported, where it facilitates dimerization of the transcriptional regulator ComR. The ComR/XIP complex binds a DNA motif to promote transcription of comX and comS. (B) The ComR/XIP binding motif appears upstream of the sigma factor comX. Using this sequence, we identified a comS gene in 2 strains of S. sobrinus. The comS gene appears downstream of a homolog of the regulator comR. (C) S. sobrinus strains NIDR 6715-7 and NCTC 10919 can be transformed with exogenous XIP. Both strains were transformed with a plasmid expressing LacZ. When plated with X-gal, the plasmid-carrying strains produce a blue color, but the wild type strains do not. (D) The transformation efficiency of S. sobrinus strain NCTC 10919 increases with XIP concentration. Transformation assays used linear DNA with homology to regions flanking comR. No transformants were observed without XIP. (E) Transformation efficiency peaks in the midexponential phase. Transformation assays were performed with strain NCTC 10919 and the pLacZ plasmid (Appendix Fig. 1) by using the predicted XIP for strains SL1 (LMCTIVR) and NIDR 6715-7 (LMCTIAR). No XIP precursor gene (comS) is found in the NCTC 10919 genome. (F) The S. sobrinus ComS peptides differ from sequences in S. mutans, Streptococcus salivarius, and Streptococcus thermophilus. In particular, all previously known XIP sequences in streptococci contain 2 aromatic amino acids; the XIP in S. sobrinus has none.
S. mutans has functional ComCDE and ComRS pathways, but genomic studies predicted that both pathways are incomplete in S. sobrinus (Song et al. 2013; Conrads et al. 2014). The lack of natural competence has led to a widespread belief that S. sobrinus is genetically intractable. Researchers can genetically modify other streptococci using synthetic CSP or XIP (Morrison et al. 2015; Junges et al. 2017; Salvadori et al. 2017), but these methods do not work with S. sobrinus. The paucity of genetic tools for S. sobrinus creates gaps in our mechanistic understanding of how this pathogen affects oral health.
Here we report that S. sobrinus does, in fact, contain a functional ComRS competence pathway. Although S. sobrinus is commonly classified among the mutans streptococci, theS. sobrinus ComRS system is most similar to pathways in the salivarius group. Most strains of S. sobrinus contain 2 homologs of ComR, and we show that only 1 is essential for transformation with foreign DNA. Unlike all known XIP sequences, the XIPs from S. sobrinus contain no aromatic amino acids. The ComRS pathway regulates bacteriocin production in S. sobrinus, and we discovered that S. sobrinus inhibits the growth of S. mutans through a ComRS-dependent response. Finally, we report 2 strains of S. sobrinus that appear to be “cheating” by not sensing or not producing XIP. Overall, the discovery of a novel competence pathway suggests thatS. sobrinus can communicate extracellularly within the complex oral microbiome. The ComRS pathway also provides a robust method for genetic manipulation of S. sobrinus.
Methods
Strains, Reagents, and Growth Conditions
Appendix Data Set 3 lists the strains and plasmids used in this study. Complete genome sequences are now available for all of the strains used in this article (Sales et al. 2018), confirming that the strains are S. sobrinus.
Liquid cultures were grown anaerobically (5% H2, 10% CO2, and 85% N2) at 37 °C in chemically defined medium (CDM; Chang et al. 2011) containing 1% glucose. Solid agar plates were made with Todd-Hewitt broth plus 0.5% yeast extract, 1.5% agar, and antibiotic selection when needed: 1-mg/mL kanamycin, 400-µg/mL spectinomycin, or 4 µg/ml chloramphenicol. Plates were incubated aerobically under 5% CO2 at 37 °C.
All chemicals were purchased from Sigma Aldrich unless otherwise stated. Enzymes were purchased from New England Biolabs. Oligonucleotides were synthesized by Integrated DNA Technologies. Synthetic peptides were purchased from GenScript, Inc. at >90% purity.
DNA Manipulation and Strain Construction
Linear DNA fragments were constructed by Golden Gate assembly (Engler et al. 2008) following the procedures outlined in the Appendix Methods. A new Golden Gate–compatible backbone (pRW17) was constructed from the Escherichia coli/streptococcal shuttle vector pDL278 (LeBlanc et al. 1992). Appendix Table 4 lists all primers and genomic coordinates.
Transformation Assays
An overnight culture of S. sobrinus in CDM with antibiotic selection was diluted 150× into fresh CDM without antibiotics. The culture was grown to the midexponential phase (OD600 between 0.55 and 0.75). XIP dissolved in DMSO was added to a final concentration of 10 µM. The culture was mixed well by flicking the tube, and 200 µL was transferred to a microcentrifuge tube where 400 ng of DNA was added. After another 2 h of anaerobic incubation, 150 µL was plated on solid agar and incubated for 24 h aerobically. Transformation efficiency was calculated by comparing colony counts after 24 h on selective and nonselective plates. A detailed transformation protocol is given in the Appendix Methods.
Inhibition Assays
An inhibition assay protocol was adapted from Mignolet et al. (2018) and Van de Rijn and Kessler (1980). Assay plates (60 mm petri dish) were made with 2 layers: a 3-mL bottom layer of 1% agarose and CDM, supplemented with XIP, and a 0.6-mL top layer of 0.4% agarose and CDM mixed with 12 µL of overnight culture of S. mutans UA159. A 3-µL spot of S. sobrinus (OD 0.6 to 0.85, grown in CDM) was added to the top layer. Plates were imaged after 24 h of incubation on an Axygen Gel Documentation System.
Growth Assays
Overnight CDM cultures of S. sobrinus were diluted 100× into fresh CDM and grown to near OD 0.8. A 96-well plate was prepared with 200 µL of fresh CDM and either 10µM XIP (dissolved in DMSO) or an equal volume of DMSO and prewarmed in the incubator. A 50-µL inoculum was added to the prewarmed 96-well plate, and growth was monitored every 30 min with a BioTek Epoch2 spectrophotometer.
Gene Expression Profiling
Changes in gene expression were measured by quantitative reverse transcription polymerase chain reaction via biological triplicates at each time point. Protocols (including RNA isolation) and primer information are available in the Appendix Methods and Appendix Table 5.
Statistical Analysis
We used a linear statistical model to quantify the overall effects of deleting or complementing comR1 and comR2. Deleting comR2 reduces transformation efficiency below our limit of detection (P < 1 × 10−16). On average, strains of NIDR 6715-7 with extra copies of comR2 in trans are transformable with 10.5-fold higher efficiency (P < 8 × 10−15). Deleting comR1 increases transformation efficiency by 2.7-fold (P < 2 × 10−4), and strains carrying extra copies of comR1 have a transformation efficiency that is barely detectable (110-fold decrease, P < 2 × 10−16).
Bioinformatic Searches
Targeted queries for ComRS homologs were performed via BLAST with blastp or tblastn through the NCBI web interface. Searches for homologs of ComR in all S. sobrinus strains was performed offline through custom R scripts and the blastp toolkit (Camacho et al. 2009).
Results
Previous studies have searched unsuccessfully for ComCDE and ComRS systems in S. sobrinus (Song et al. 2013; Conrads et al. 2014). Searches for ComS (XIP) and ComC (CSP) have also failed, although these small genes are difficult to find. We used a hybrid strategy to search the recently completed genomes of 3 S. sobrinus strains (Sales et al. 2018) for both ComR homologs and the promoter sequence recognized by the ComR/XIP complex. The ComR/XIP motif is usually located upstream of the sigma factor comX and the comS gene (Mashburn-Warren et al. 2010). The ComR/XIP motif from S. mutans UA159 (GGGACATNNATGTC) was not present in S. sobrinus; however, the ComR/XIP motif from Streptococcus thermophilus LMD-9 and Streptococcus salivarius SK126 (GTGACAT NNATGTC) was found upstream of comX in S. sobrinus (Fig. 1B, Appendix Data Set 1). In 2 S. sobrinus strains (SL1 and NIDR 6715-7), the ComR/XIP motif appeared between a homolog of the S. thermophilus comR gene and a short ORF that we believed to be comS (Fig. 1B). The third strain (NCTC 10919) contained a homolog of comR but no comS gene.
Based on ComS cleavage patterns in S. thermophilus andS. mutans, we purchased peptides containing the last 7 amino acids of ComS for strains NIDR 6715-7 and SL1. Both synthetic peptides induced competence in strain NIDR 6715-7, allowing us to transform the strain with either linear or plasmid DNA (Fig. 1C). Both peptides also induced competence in NCTC 10919 (the strain lacking a comS gene), but neither peptide induced competence in strain SL1. Synthetic peptides with only the last 9 amino acids of ComS also induced competence in NIDR 6715-7 and NCTC 10919, but peptides with only the last 5 amino acids did not. We used the 7–amino acid peptide for all subsequent experiments and refer to this peptide as XIP; however, we do not know the exact location of the ComS cleavage site that produces active XIP in vivo.
The transformation efficiency of S. sobrinus increases with XIP concentration, peaking at 10−5 transformants per cell with 10µM XIP for strain NIDR 6715-7 (Fig. 1D). The transformation efficiency also depends on the growth phase of the cells. The efficiency is highest in the midexponential phase (Fig. 1E), similar to other streptococci transformed with XIP (Junges et al. 2017; Salvadori et al. 2017).
The S. sobrinus XIP sequence is unlike those of other streptococci (Fig. 1F). ComRS systems are classified by the identity and location of 2 aromatic amino acids in the XIP peptide (Fontaine et al. 2010; Talagas et al. 2016). These aromatic residues are believed to be essential for binding between XIP and ComR (Talagas et al. 2016). We were surprised that the S. sobrinus ComS sequence contains no aromatic amino acids and therefore cannot be grouped into any of the 3 established classes of ComRS systems.
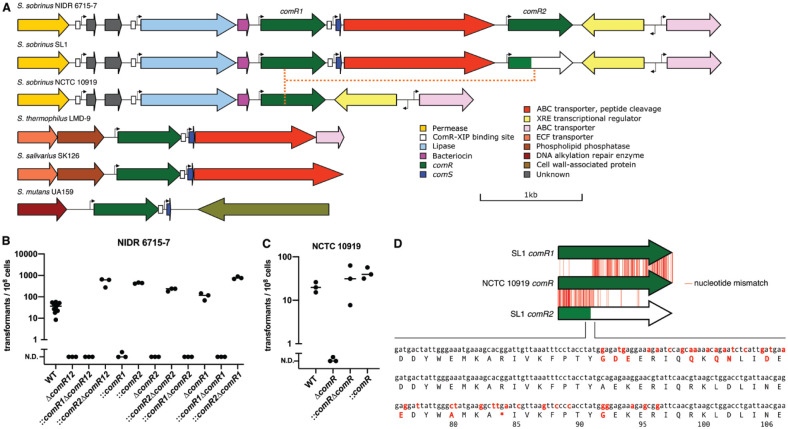
To our knowledge, all ComRS gene clusters contain a single copy of the transcriptional regulator gene comR. Two S. sobrinus strains (SL1 and NIDR 6715-7) contain 2 homologs of comR in the same gene cluster (Fig. 2A, Appendix Data Set 2). These genes (comR1 and comR2) flank the comS gene and the gene encoding the putative ComS exporter. We used strain NIDR 6715-7 to test if comR1, comR2, or both genes are required for competence (Fig. 2B). Deleting comR1, comR2, and the 2 genes in between (comS and its putative exporter) abolished transformation. Adding a second copy of comR2 in trans restored transformability, but adding back comR1 did not. In fact, adding extra copies of comR1 to the wild type strain reduced the transformation efficiency by 111-fold. We verified our results by deleting and complementing comR2 or comR1 alone. In all cases, transformation required at least 1 copy of comR2, and the transformation efficiency was inversely correlated with the number of copies of comR1.
Figure 2.
The Streptococcus sobrinus ComRS gene cluster is distinct from other streptococci. (A) The ComRS gene cluster in strains NIDR 6715-7 and SL1 contain 2 homologs of comR that we call comR1 and comR2. The type strain SL1 contains a truncated comR2 gene (green/white) and cannot be transformed. Strain NCTC 10919 has a single homolog of comR and no comS gene or a ComS export/cleavage gene. (B) Strain NIDR 6715-7 cannot be transformed if comR2 or the region from comR1 to comR2 is deleted. An additional copy of comR1 on a plasmid does not rescue transformation, but strains complemented with extra comR2 can be transformed. The horizontal bars represent the mean of the biological replicates (black dots). (C) The single comR homolog in strain NCTC 10919 is required for transformation. (D) The comR gene in strain NCTC 10919 appears to be a fusion of comR1 and comR2. A recombination event that produced comR would have removed the premature stop codon found in the comR2 gene of strain SL1.
Strain SL1 is unresponsive to XIP, which we believe is explained by a premature stop codon in its comR2 gene. We predict that the truncated ComR2 in SL1 is nonfunctional on the basis of structural studies in other streptococci (Talagas et al. 2016). The strain NCTC 10919 contains only 1 comR gene, and this gene is essential for induced competence (Fig. 2C). It appears that the single copy of comR is the result of a recombination event between comR1 and comR2, since the 5′ end of the gene is more similar to comR1 and the 3′ end is more similar to comR2 (Fig. 2D). Such a recombination event would have deleted the comS gene and removed the premature stop codon in the comR2 of strain SL1. It is possible that strain NCTC 10919 lost its ability to produce XIP in exchange for repairing the broken ComRS pathway in a common ancestor with SL1.
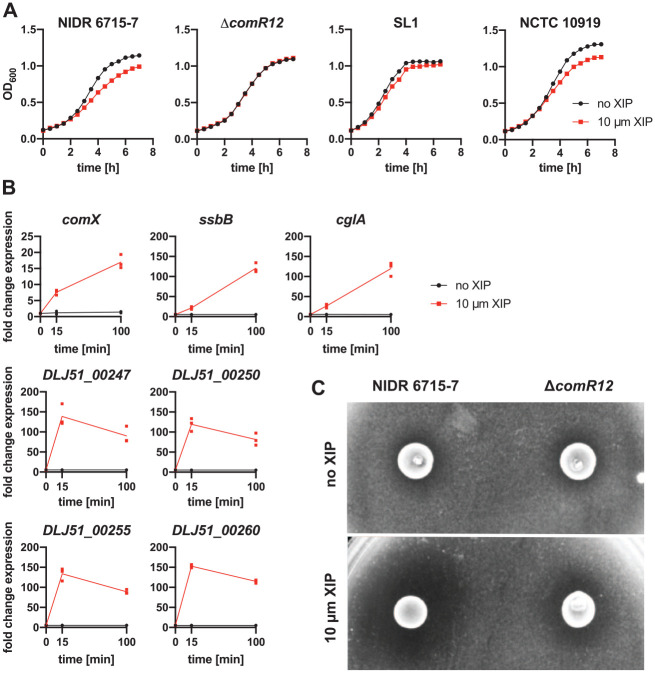
XIP can decrease the growth rate of some streptococci (Desai et al. 2012). Adding exogenous XIP slows the growth of S. sobrinus strain NIDR 6715-7, but no defect is observed in a ComRS deletion strain (Fig. 3A). The growth of strain SL1, which is not transformable with XIP, is unaffected by XIP. A XIP-induced growth defect is observed in the competent strain NCTC 10919.
Figure 3.
The ComRS pathway controls phenotypic changes in Streptococcus sobrinus. (A) Adding XIP to cultures of strain NIDR 6715-7 causes a growth defect. An NIDR 6715-7 deletion strain lacking the ComRS pathway (comR1, comS, the ComS exporter, and comR2) does not show a growth defect after XIP is added. The SL1 wild type strain cannot be transformed with XIP and does not show a growth defect, but the NCTC 10919 strain is transformable and grows slower in the presence of XIP. (B) XIP increases the expression of the comX, ssbB, and cglA genes in strain NIDR 6715-7. The ComR/XIP binding motif appears upstream of 4 genes in a bacteriocin gene cluster (DLJ51_00247, DLJ51_00250, DLJ51_00255, and DLJ51_00260). All 4 genes are upregulated after XIP is added. Each point is a biological replicate, and fold change is relative to the expression at time zero. (C) S. sobrinus inhibits the growth of S. mutans when XIP is added to cultures on solid agarose. A base layer of agarose containing XIP is topped with a second layer of agarose with embedded S. mutans UA159. Spotting S. sobrinus 6715-7 on top of the agarose inhibits the growth of Streptococcus mutans on plates containing XIP. The inhibition is reduced when the ComRS pathway is deleted from S. sobrinus.
It has been hypothesized that bacteria with a ComRS system could “cheat” by either not producing or not responding to XIP (Fontaine et al. 2010). Instead, the cheating strains would rely on other bacteria to perform the community-level processes controlled by the competence pathway. Two of our S. sobrinus strains appear to cheat. Strain SL1 does not respond to XIP due to the loss of a functional comR2 gene. Strains with this mutation would enjoy a selective advantage by avoiding the growth defect in the presence of XIP. Strain NCTC 10919 does not appear to produce XIP, although it can still respond to XIP produced by other strains.
XIP increases the expression of the competence-related genes comX, ssbB, and cglA. Transcript levels of these genes are increased by 15 min and continue to increase for up to 100 min (Fig. 3B). The sustained activation of competence genes is consistent with the ComRS system in S. mutans (Morrison et al. 2015).
We identified additional ComR/XIP binding sites in S. sobrinus that are upstream of putative bacteriocins (Fig. 2A). These genes are also upregulated upon stimulation by XIP (Fig. 3B). Surprisingly, we discovered that S. sobrinus inhibits the growth of S. mutans when the former is stimulated with XIP (Fig. 3C). The inhibition is attenuated when the ComRS pathway is deleted from S. sobrinus.
The ComRS pathway is widespread among strains of S. sobrinus. A BLAST search revealed that all 54 publicly available genomes for S. sobrinus contain a homolog of the comR gene. In 83% of these genomes, the ComR amino acid sequence has >97% amino acid identity with ComR2 in S. sobrinus NIDR 6715-7. The conservation of ComR suggests that XIP may be useful for transforming a wide range of S. sobrinus strains. While some strains of S. sobrinus, like SL1, remain genetically intractable, strains with a functional ComRS system can be manipulated to reveal differences between S. sobrinus and other cariogenic species. Thus, the discovery of this novel competence pathway will begin a new phase of S. sobrinus research where the mechanisms of cariogenicity can be unraveled with forward genetics.
Discussion
The S. sobrinus ComRS system is unique on the basis of its 2 comR genes and a XIP sequence that lacks any aromatic amino acids; however, the arrangement and sequence of the ORFs in the ComRS gene cluster is most similar to ComRS clusters in the salivarius group (S. salivarius and S. thermophilus; Fig. 2A). S. sobrinus was originally a subspecies of S. mutans and is frequently grouped with the mutans streptococci based on genomic phylogenies (Song et al. 2013). Recent phylogenetic trees place S. sobrinus closer to S. salivarius and S. thermophilus in a separate sobrinus clade (Patel and Gupta 2018). Our functional data on ComRS would agree with S. sobrinus’s placement in or near the salivarius group. However, neitherS. salivarius nor S. thermophilus is cariogenic, raising questions about why S. sobrinus is associated with aggressive dental caries.
S. sobrinus, like the noncariogenic species S. salivarius, has direct links between competence and bacteriocin production. Such a direct link has not been observed in cariogenic streptococci (Mignolet et al. 2018). The ComRS-activated bacteriocins in S. sobrinus can kill S. mutans, a surprising finding given that coinfection by these 2 species leads to worse oral health outcomes (Seki et al. 2006; Kanasi et al. 2010; Okada et al. 2012). Our scant knowledge of S. sobrinus and its behavior has been drawn through the presumed similarity of S. sobrinus to S. mutans. However, the structural, functional, and phylogenetic differences between S. sobrinus and S. mutans suggest that the association between S. sobrinus and the mutans streptococci needs to be reconsidered.
Streptococci use the ComRS system for intercellular communication, and we now know that S. sobrinus can participate in these community-level conversations. Competence affects individual bacteria and the community as a whole, and it has been hypothesized that bacteria with a ComRS system could “cheat” by either not producing or not responding to XIP (Fontaine et al. 2010). Two of our S. sobrinus strains appear to cheat. The type strain SL1 does not respond to XIP, most likely because its comR2 gene is truncated. Strains of S. sobrinus that do not respond to XIP would avoid the XIP-induced growth defect (Fig. 3A) and the metabolic costs of producing and exporting bacteriocins; however, they can still benefit from the bacteriocin defenses produced by neighboring cells (Matsumoto-Nakano and Kuramitsu 2006; Shanker and Federle 2017; Mignolet et al. 2018). Even worse, SL1 appears to have functional genes for producing and exporting XIP, so it could activate the ComRS systems in neighboring cells without taking part in the community response.
Strain NCTC 10919 has the potential to cheat by not producing or exporting XIP. NCTC 10919 can sense XIP produced by neighboring cells and participate in the community-wide competence state. This maximizes NCTC 10919’s chances of taking up beneficial foreign DNA while avoiding the metabolic costs of producing and exporting XIP. While strain SL1 and NCTC 10919 have the potential to cheat, the mechanisms and benefits of cheating are different. It is interesting that a single recombination event could have switched an SL1-type cheater to an NCTC 10919-type cheater, thereby swapping one type of cheating for another.
The ability to transform S. sobrinus clears the way for forward genetic studies into the bacterium’s pathogenicity. As compared with S. mutans, S. sobrinus produces more acid (de Soet et al. 1989) and better tolerates acid stress (Nascimento et al. 2004). The S. sobrinus acid tolerance response is poorly characterized, but we know that it is mechanistically distinct from the acid tolerance response of S. mutans (Nascimento et al. 2004; Conrads et al. 2014). Other studies have observed that S. sobrinus is capable of immune suppression (Veiga-Malta et al. 2004), hydrogen peroxide production (García-Mendoza et al. 1993; Conrads et al. 2014), and enhanced glucan synthesis (Conrads et al. 2014). Our new genetic system for S. sobrinus will allow us to study the molecular details of these phenotypes and identify new targets for reducing cariogenesis by S. sobrinus.
Author Contributions
J.W. Li, R.M. Wyllie, P.A. Jensen, contributed to conception, design, and data analysis, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, sj-pdf-1-jdr-10.1177_0022034520979150 for A Novel Competence Pathway in the Oral Pathogen Streptococcus sobrinus by J.W. Li, R.M. Wyllie and P.A. Jensen in Journal of Dental Research
Acknowledgments
We thank Bill Metcalf, Jim Slauch, and the members of the Jensen Lab for insightful discussions. We also thank Will Herbert and Emma Lee for assisting with the bioinformatic searches.
Footnotes
A supplemental appendix to this article is available online.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Institutes of Health grants DE0 26817 and GM138210.
ORCID iD: P.A. Jensen  https://orcid.org/0000-0002-1257-9836
https://orcid.org/0000-0002-1257-9836
References
- Avery OT, Macleod CM, McCarty M. 1944. Studies on the chemical nature of the substance inducing transformation of pneumococcal types : induction of transformation by a desoxyribonucleic acid fraction isolated from Pneumococcus type III. J Exp Med. 79(2):137–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinform. 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JC, LaSarre B, Jimenez JC, Aggarwal C, Federle MJ. 2011. Two group A streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development. PLoS Pathog. 7(8):e1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrads G, de Soet JJ, Song L, Henne K, Sztajer H, Wagner-Döbler I, Zeng AP. 2014. Comparing the cariogenic species Streptococcus sobrinus and S. mutans on whole genome level. J Oral Microbiol. 6:26189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai K, Mashburn-Warren L, Federle MJ, Morrison DA. 2012. Development of competence for genetic transformation of Streptococcus mutans in a chemically defined medium. J Bacteriol. 194(15):3774–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Soet JJ, Toors FA, de Graaff J. 1989. Acidogenesis by oral streptococci at different pH values. Caries Res. 23(1):14–17. [DOI] [PubMed] [Google Scholar]
- Engler C, Kandzia R, Marillonnet S. 2008. A one pot, one step, precision cloning method with high throughput capability. PloS One. 3(11):e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine L, Boutry C, de Frahan MH, Delplace B, Fremaux C, Horvath P, Boyaval P, Hols P. 2010. A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J Bacteriol. 192(5):1444–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine L, Wahl A, Fléchard M, Mignolet J, Hols P. 2015. Regulation of competence for natural transformation in streptococci. Infect Genet Evol. 33:343–360. [DOI] [PubMed] [Google Scholar]
- García-Mendoza A, Liébana J, Castillo AM, de la Higuera A, Piédrola G. 1993. Evaluation of the capacity of oral streptococci to produce hydrogen peroxide. J Med Microbiol. 39(6):434–439. [DOI] [PubMed] [Google Scholar]
- Gross EL, Beall CJ, Kutsch SR, Firestone ND, Leys EJ, Griffen AL. 2012. Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS One. 7(10):e47722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håvarstein LS, Coomaraswamy G, Morrison DA. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci U S A. 92(24):11140–11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Håvarstein LS, Gaustad P, Nes IF, Morrison DA. 1996. Identification of the streptococcal competence-pheromone receptor. Mol Microbiol. 21(4):863–869. [DOI] [PubMed] [Google Scholar]
- Hirose H, Hirose K, Isogai E, Miura H, Ueda I. 1993. Close association between Streptococcus sobrinus in the saliva of young children and smooth-surface caries increment. Caries Res. 27(4):292–297. [DOI] [PubMed] [Google Scholar]
- Junges R, Khan R, Tovpeko Y, Åmdal HA, Petersen FC, Morrison DA. 2017. Markerless genome editing in competent streptococci. Methods Mol Biol. 1537:233–247. [DOI] [PubMed] [Google Scholar]
- Kanasi E, Johansson I, Lu SC, Kressin NR, Nunn ME, Kent R, Tanner ACR. 2010. Microbial risk markers for childhood caries in pediatricians’ offices. J Dent Res. 89(4):378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koirala B, Lin J, Lau GW, Tal-Gan Y. 2018. Development of a dominant negative competence-stimulating peptide (dnCSP) that attenuates Streptococcus pneumoniae infectivity in a mouse model of acute pneumonia. Chembiochem. 19(22):2380–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc DJ, Lee LN, Abu-Al-Jaibat A. 1992. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid. 28(2):130–145. [DOI] [PubMed] [Google Scholar]
- Lin J, Lau GW. 2019. DprA-dependent exit from the competent state regulates multifaceted Streptococcus pneumoniae virulence. Infect Immun. 87(11):e00349-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashburn-Warren L, Morrison DA, Federle MJ. 2010. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg Regulator. Mol Microbiol. 78(3):589–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto-Nakano M, Kuramitsu HK. 2006. Role of bacteriocin immunity proteins in the antimicrobial sensitivity of Streptococcus mutans.J Bacteriol. 188(23):8095–8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignolet J, Fontaine L, Sass A, Nannan C, Mahillon J, Coenye T, Hols P. 2018. Circuitry rewiring directly couples competence to predation in the gut dweller Streptococcus salivarius. Cell Rep. 22(7):1627–1638. [DOI] [PubMed] [Google Scholar]
- Morrison DA, Khan R, Junges R, Åmdal HA, Petersen FC. 2015. Genome editing by natural genetic transformation in Streptococcus mutans. J Microbiol Methods. 119:134–141. [DOI] [PubMed] [Google Scholar]
- Nascimento MM, Lemos JAC, Abranches J, Gonçalves RB, Burne RA. 2004. Adaptive acid tolerance response of Streptococcus sobrinus. J Bacteriol. 186(19):6383–6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurelhuda NM, Al-Haroni M, Trovik TA, Bakken V. 2010. Caries experience and quantification of Streptococcus mutans and Streptococcus sobrinus in saliva of Sudanese school children. Caries Res. 44(4):402–407. [DOI] [PubMed] [Google Scholar]
- Okada M, Kawamura M, Oda Y, Yasuda R, Kojima T, Kurihara H. 2012. Caries prevalence associated with Streptococcus mutans and Streptococcus sobrinus in Japanese school children. Int J Paediatr Dent. 22(5):342–348. [DOI] [PubMed] [Google Scholar]
- Patel S, Gupta RS. 2018. Robust demarcation of fourteen different species groups within the genus Streptococcus based on genome-based phylogenies and molecular signatures. Infect Genet Evol. 66:130–151. [DOI] [PubMed] [Google Scholar]
- Philip N, Suneja B, Walsh L. 2018. Beyond Streptococcus mutans: clinical implications of the evolving dental caries aetiological paradigms and its associated microbiome. Br Dent J. 224(4):219–225. [DOI] [PubMed] [Google Scholar]
- Sales MJ, Herbert WG, Du Y, Sandur AS, Stanley NM, Jensen PA. 2018. Complete genome sequences of Streptococcus sobrinus SL1 (ATCC 33478 = DSM 20742), NIDR 6715-7 (ATCC 27351), NIDR 6715-15 (ATCC 27352), and NCTC 10919 (ATCC 33402). Microbiol Resour Announc. 7(3):e00804-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvadori G, Junges R, Khan R, Åmdal HA, Morrison DA, Petersen FC. 2017. Natural transformation of oral streptococci by use of synthetic pheromones. Methods Mol Biol. 1537:219–232. [DOI] [PubMed] [Google Scholar]
- Seki M, Yamashita Y, Shibata Y, Torigoe H, Tsuda H, Maeno M. 2006. Effect of mixed mutans streptococci colonization on caries development. Oral Microbiol Immunol. 21(1):47–52. [DOI] [PubMed] [Google Scholar]
- Shanker E, Federle MJ. 2017. Quorum sensing regulation of competence and bacteriocins in Streptococcus pneumoniae and mutans. Genes. 8(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singla D, Sharma A, Sachdev V, Chopra R. 2016. Distribution of Streptococcus mutans and Streptococcus sobrinus in dental plaque of Indian pre-school children using PCR and SB-20M agar medium. J Clin Diagn Res. 10(11):ZC60–ZC63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slager J, Kjos M, Attaiech L, Veening JW. 2014. Antibiotic-induced replication stress triggers bacterial competence by increasing gene dosage near the origin. Cell. 157(2):395–406. [DOI] [PubMed] [Google Scholar]
- Song L, Wang W, Conrads G, Rheinberg A, Sztajer H, Reck M, Wagner-Döbler I, Zeng AP. 2013. Genetic variability of mutans streptococci revealed by wide whole-genome sequencing. BMC Genomics. 14:430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Nyvad B. 2008. Caries ecology revisited: microbial dynamics and the caries process. Caries Res. 42(6):409–418. [DOI] [PubMed] [Google Scholar]
- Talagas A, Fontaine L, Ledesma-Garca L, Mignolet J, Li de la Sierra-Gallay I, Lazar N, Aumont-Nicaise M, Federle MJ, Prehna G, Hols P, et al. 2016. Structural insights into streptococcal competence regulation by the cell-to-cell communication system ComRS. PLOS Pathog. 12(12):e1005980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill SAM, Shields RC, Burne RA, Hagen SJ. 2019. Carbohydrate and PepO control bimodality in competence development by Streptococcus mutans. Mol Microbiol. 112(5):1388–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Rijn I, Kessler RE. 1980. Growth characteristics of group A streptococci in a new chemically defined medium. Infect Immun. 27(2):444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga-Malta I, Duarte M, Dinis M, Tavares D, Videira A, Ferreira P. 2004. Enolase from Streptococcus sobrinus is an immunosuppressive protein. Cell Microbiol. 6(1):79–88. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jdr-10.1177_0022034520979150 for A Novel Competence Pathway in the Oral Pathogen Streptococcus sobrinus by J.W. Li, R.M. Wyllie and P.A. Jensen in Journal of Dental Research