Abstract
Tryptophan 2,3-dioxygenase (TDO2) is a key rate-limiting enzyme in the kynurenine pathway and promotes tumor growth and escape from immune surveillance in different types of cancer. The present study aimed to investigate whether TDO2 serves a role in the development of ovarian cancer. Reverse transcription-quantitative PCR and western blotting were used to detect the expression of TDO2 in different cell lines. The effects of TDO2 overexpression, TDO2 knockdown and TDO2 inhibitor on ovarian cancer cell proliferation, migration and invasion were determined by MTS, colony formation and Transwell assays. The expression of TDO2 in ovarian cancer tissues, normal ovarian tissues and fallopian tube tissues were analyzed using the gene expression data from The Cancer Genome Atlas and Genotype-Tissue Expression project. Immune cell infiltration in cancer tissues was evaluated using the single sample gene set enrichment analysis algorithm. The present study found that RasV12-mediated oncogenic transformation was accompanied by the upregulation of TDO2. In addition, it was demonstrated that TDO2 was upregulated in ovarian cancer tissues compared with normal ovarian tissues. TDO2 overexpression promoted proliferation, migration and invasion of ovarian cancer cells, whereas TDO2 knockdown repressed these phenotypes. Treatment with LM10, a TDO2 inhibitor, also repressed the proliferation, migration and invasion of ovarian cancer cells. The present study indicated that TDO2 can be used as a new target for the treatment of ovarian cancer.
Keywords: tryptophan 2,3-dioxygenase; ovarian cancer; proliferation; migration; invasion
Introduction
Ovarian cancer is the seventh commonest type of cancer in women and the eighth most common cause of cancer death worldwide, with a 5-year survival rate <45%. The incidence has increased with increased life expectancy, especially in developing countries. As the symptoms of early-stage ovarian cancer are mild and non-specific, most cases are diagnosed at an advanced stage and require aggressive cytoreductive surgery, followed by platinum and taxane-based chemotherapy (1). At present, ~85% of patients with advanced ovarian cancer who have achieved full remission following surgery and chemotherapy develop recurrent disease. The median survival time of patients with advanced ovarian cancer ranges only 12–24 months (2,3).
The kynurenine pathway is an important pathway for tryptophan metabolism; >95% of the tryptophan in the body is metabolized through the kynurenine pathway (4,5). In this pathway, tryptophan is first metabolized into the intermediate N-formyl kynurenine by one of three first-step enzymes, namely, indoleamine 2,3-dioxygenase 1 (IDO1), indoleamine 2,3-dioxygenase 2 (IDO2) and tryptophan 2,3-dioxygenase (TDO2) and then converted into kynurenine by arylformamidase (6). Kynurenine subsequently undergoes a series of enzymatic reactions and is converted into 3-hydroxykynurenine, 3-hydroxyanthranilic acid, 2-amino-3-carboxymuconatesemialdehyde and quinolinic acid, after which it is finally converted to nicotinamide adenine dinucleotide for metabolic use. The metabolites of the kynurenine pathway are involved in multiple physiological activities, mainly in the nervous and the immune systems (4,5). The first-step enzymes, namely IDO1, IDO2 and TDO2, are the most important rate-limiting enzymes in the kynurenine pathway (7). At the amino acid level, IDO1 and IDO2 have ~43% sequence similarity, whereas TDO2 has little similarity with IDO1 and IDO2 (8,9). Studies have shown that one or more of these enzymes are upregulated to different degrees in tumor tissues (10–16). Upregulation of these enzymes leads to tryptophan depletion and kynurenine accumulation, thereby impairing the anticancer immune response (17–23). The expression and roles of IDO1 in ovarian cancer have been extensively investigated (24–29). Inaba et al (30) reported that higher expression of IDO1 in ovarian cancer tissues is associated with shorter survival time. Furthermore, overexpression of IDO1 in the human ovarian carcinoma cell line SKOV3 promotes xenograft tumor growth (30), whereas knockdown of IDO1 in SKOV3 cells suppresses xenograft tumor growth (31). The tumor-promoting effect of IDO2 in SK-IDO-xenografted mice is blocked by oral administration of the IDO inhibitor 1-methyl-tryptophan (30). The present study aimed to investigate the expression and roles of TDO2 in ovarian cancer.
Materials and methods
Plasmid and reagents
The TDO2 overexpression plasmid was designed by Sino Biological Inc. Briefly, the coding sequence of pCMV3-TDO2 was amplified using 5′-TAATACGACTCACTATAGGG-3′ and 5′-TAGAAGGCACAGTCGAGG−3′ as primers and inserted into the KpnI/XbaI site of pCMV3-untagged vector (Sino Biological Inc.). LM10, which is an effective TDO inhibitor, was purchased from Selleck Chemicals. The cells were treated with LM10 at a final concentration of 500 µM (32).
Cell culture
The human ovarian epithelial cell lines T29 and T29H were provided by Dr Jinsong Liu (School & Hospital of Stomatology, Wenzhou Medical University, China) (33,34). Briefly, isolated human surface ovarian epithelium cells were infected sequentially by retroviruses containing SV40 T/t antigens and hTERT genes to generate T29 cells. The immortalized, but non-oncogenic T29 cells were further transformed by introducing an oncogenic H-RASV12 in a pLNCX retroviral vector to form the T29H cell. The SKOV3 cell line was provided by Dr Xueqiong Zhu (The Second Affiliated Hospital of Wenzhou Medical University, China). The OVSAHO (JCRB1046) cells were from JCRB Cell Bank. T29, T29H and OVSAHO cells were maintained in high-glucose Dulbecco's modified Eagle's medium (DMEM; HyClone; Cytiva) supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) and 1% penicillin-streptomycin (Gibco; Thermo Fisher Scientific, Inc.). Human ovarian cell line SKOV3 cells were maintained in Roswell Park Memorial Institute-1640 medium (RPMI-1640; HyClone; Cytiva) supplemented with 10% FBS and 1% penicillin-streptomycin. Cells were cultured in an incubator in a 5% CO2 humidified atmosphere at 37°C. All cell lines were mycoplasma free and cells passaged at the Digestive Cancer Center, The First Affiliated Hospital of Wenzhou Medical University for >6 months following receipt were authenticated by genetic profiling using polymorphic short-tandem repeat loci (35).
Small interfering (si)RNA and transfection
siRNAs were purchased from Gema Gene. The siRNA sequences were: siTDO2, 5′-CGUUAAUCGCGUAUAAUACGCGUATT-3′ (sense), 5′-UACGCGUAUUAUACGCGAUUAACGTT-3′ (anti-sense); siNC (negative control siRNA), 5′-AAUUCUCCGAACGUGUCACGUTT-3′ (sense) and 5′-ACGUGACACGUUCGGAGAAUUTT-3′ (anti-sense).
Cells were seeded into 6-well plates at 3×105/well. The plasmids were transfected 2.5 µg/well and the siRNAs were transfected at a final concentration of 50 nM using Lipofectamine® 2000 Reagent (Invitrogen; Thermo Fisher Scientific, Inc.), following the manufacturer's instructions. The cells were then collected 48 h after transfection for subsequent analysis, including western blotting and reverse transcription-quantitative (RT-q) PCR.
Western blotting
Total protein was extracted from T29, T29H, OVSAHO and SKOV3 cells using RIPA lysis buffer (Beyotime Institute of Biotechnology). After mixing with SDS lysis buffer and boiling, the protein content in cell lysates were measured using BCA Protein Assay kit (Beyotime Institute of Biotechnology). Equal amounts of protein (30 µg) were loaded onto 10% SDS-PAGE and separated via electrophoresis, then separated proteins were transferred to PVDF membranes. After blocking in 5% skimmed milk at room temperature for 1 h, membranes were incubated overnight at 4°C with primary antibodies against TDO2 (cat. no. H00006999-A01; Abnova; 1:1,000) and anti-GAPDH (cat. no. 2118S; Cell Signaling Technology, Inc; 1:1,000). The membranes were washed with TBST, followed by incubation with goat anti-mouse IgG (cat. no. ab6789; Abcam; 1:10,000) or goat anti-rabbit IgG (cat. no. ab6721; Abcam; 1:10,000) antibodies at room temperature for 1 h. Subsequently, the bands were detected with ECL plus reagents (GE Healthcare). Densitometric analysis was performed using ImageJ software (version 1.8.0; National Institutes of Health). All experiments were repeated at least three times.
RT-qPCR
Total RNA was extracted using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), incubated with RNase-free DNase I (Promega Corporation) for 30 min and reverse transcribed using the M-MLV reverse transcription kit (Promega Corporation) according to the manufacturer's protocol. qPCR was subsequently performed using a SYBR Green PCR Master mix (Vazyme Biotech Co., Ltd.) on an ABI PRISM 7300 Sequence Detection system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used for qPCR: 95°C for 30 sec followed by 40 cycles of 95°C for 10 sec and 60°C for 30 sec. The following primer sequences were used: TDO2: 5′-TCCTCAGGCTATCACTACCTGC(forward) and 5′-ATCTTCGGTATCCAGTGTCGG−3′ (reverse) (36); GAPDH: 5′-GCAAATTCCATGGCACCGTC-3′ (forward) and 5′-CCTGGAAGATGGTGATGGGA-3′ (reverse). The ΔΔCt method was used to measure the relative expression levels of the subject genes. ΔCq was obtained by subtracting the Cq (threshold cycle) value of GAPDH from that of the subject gene and ΔΔCq was calculated by subtracting the ΔCq of the control sample from that of the subject sample. The fold change was calculated as 2−ΔΔCq and the relative expression level of the control sample was defined as 1 (37). All experiments were performed independently and repeated three times.
Cell proliferation assay
Following cell transfection for 24 h, the T29H, OVSAHO or SKOV3 cells were seeded in 96-well plates with 2×103 cells at each time point, CellTiter 96 Aqueous One Solution Cell Proliferation Assay (MTS) kit (Promega Corporation) was added to the corresponding plates and incubated for 3 h at 37°C according to the manufacturer's instructions. MTS is bioreduced by cells into a formazan product that is soluble in tissue culture medium. The absorbance of the formazan at 490 nm can be measured directly from the 96-well assay plates using a Multiskan Spectrum microplate reader (Thermo Fisher Scientific, Inc.) (38).
For cell number counting assays, cells were seeded into 96-well plates with 1×104/well. Cells were counted at 12, 24, 48 and 72 h after transfection or drug treatment using a Countess 3 Automated Cell Counter (Thermo Fisher Scientific, Inc.).
Colony formation assay
Following cell transfection for 24 h, the cells were digested with trypsin and resuspended with the cell medium and inoculated into a six-well plate with 200 cells/well and 2 ml of the complete medium was added. The six-well plate was placed in the incubator for further cultivation. After 18 days, the cells were fixed with absolute methanol for 20 min, stained with 0.5% crystal violet at room temperature for 20 min and the sample was rinsed and images captured for counting.
Cell migration and invasion assay
Transwell inserts of 8 µm-pore plain (to assess migration) or Matrigel-coated (to assess invasion; Costar; Corning, Inc.) were placed in the wells of 24-well culture plates and 500 µl of DMEM or RPMI-1640 containing 10% FBS was added to the lower chamber. The T29H, OVSAHO, or SKOV3 cells were washed once with Hanks' Balanced Salt Solution (Invitrogen; Thermo Fisher Scientific, Inc.) 12 h after transfection, resuspended in 100 µl serum-free medium (8×104 cells) and added to the upper chamber. After 12 h of incubation at 37°C with 5% CO2, the cells on the top side of the filter were manually removed with a cotton swab. The cells adherent to the bottom surface of the insert were fixed in cold absolute methanol for 10 min and then stained with 0.01% crystal violet in 20% ethanol at room temperature. After 10 min, the filters were washed thoroughly in water and images were captured under a DMI3000 M inverted manual microscope (Leica Microsystems GmbH). The number of migratory cells was recorded using an optical microscope at ×100 magnification. The average number of migrated cells was assessed by counting five randomly selected microscopic fields. The experiment was performed in triplicate.
Measurement of kynurenine
Supernatants from the control group and knockdown TDO2 group cells were collected at 500 × g for 15 min at 4°C. Levels of kynurenine in the supernatant were determined using the human kynurenine ELISA kit (Cusabio Biotech Co., Ltd.; cat. no. CSB-E13659h).
Activities of caspase-3/7
The activities of caspase-3/7 were measured using a caspase-3/7 activity apoptosis assay kit (Sangon Biotech Co., Ltd.), according to the manufacturer's instructions.
The Cancer Genome Atlas (TCGA) and The Genotype-Tissue Expression (GTEx) dataset analysis
RNA sequencing data of ovarian serous cystadenocarcinoma were obtained from TCGA database (https://tcga-data.nci.nih.gov/tcga/). RNA sequencing data of normal ovarian tissues and fallopian tube tissues were obtained from GTEx project (http://gtexportal.org). The expression data were log2 (TPM+1) transformed. Bindea et al (39) examined the spatio-temporal dynamics of 24 different tumor-infiltrating immune cells. The relative quantities of these 24 immune cell types in ovarian cancer were evaluated by using the R software (www.R-project.org; version 3.6.2) GSVA (version 1.34.0) package (40). Correlation between the expression of TDO2 and the relative quantity of immune cells was calculated by the signature gene sets of the immune cells.
Statistical analysis
Data from three independent experiments are presented as the mean ± standard error of the mean. Differences were analyzed using a two-tailed unpaired Student's t-test. Univariate hazard ratios with 95% confidence intervals were calculated using the Cox proportional hazards regression and significance was calculated using Wald's test. Statistical analysis was performed using SPSS software (version 20.0; IBM Corp.). P<0.05 was considered to indicate a statistically significant difference.
Results
Upregulation of tryptophan 2,3-dioxygenase in ovarian cancer
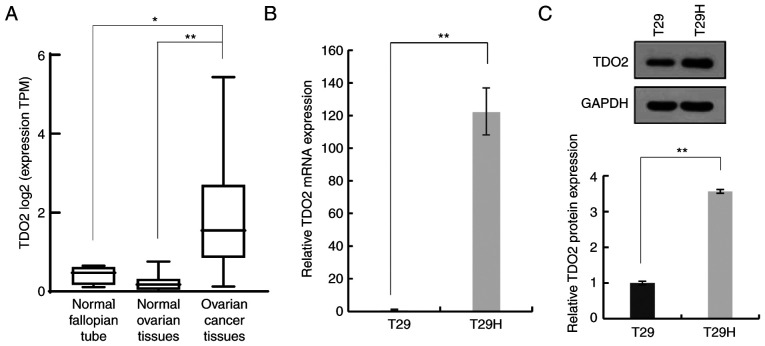
To investigate whether TDO2 serves a role in ovarian cancer development, the expression of TDO2 in ovarian cancer tissues, normal ovarian tissues and normal fallopian tube tissues was compared. As shown in Fig. 1A, the TDO2 mRNA level was significantly higher in ovarian cancer tissues compared with normal ovarian tissues and fallopian tube tissues. It was then evaluated whether TDO2 was upregulated in a genetically defined model of human ovarian cancer. The T29 cells were derived from primary human ovarian surface epithelial cells by stable transfection with the SV40 T/t antigens and hTERT. The immortalized but non-oncogenic T29 cells were further transformed by introducing oncogenic HRasV12 to generate the T29H cell line, which resembles natural ovarian cancer in several aspects. It was found that the mRNA level of TDO2 was >100-fold higher in T29H cells compared with T29 cells (Fig. 1B). Western blotting revealed that the TDO2 protein was ~3-fold higher in T29H cells compared with T29 cells (Fig. 1C). Taken together, these data indicated that TDO2 is upregulated in ovarian cancer cells.
Figure 1.
Upregulation of TDO2 in ovarian cancer cells; comparison of TDO2 mRNA expression. (A) Comparison of TDO2 mRNA expression in 374 cases of ovarian cancer, 88 normal ovarian tissues and 5 normal fallopian tube tissues. (B) Quantification of TDO2 levels in T29 and T29H cells using real-time polymerase chain reaction. (C) Representative western blotting of the TDO2 protein in T29 and T29H cells. *P<0.05; **P<0.01 by Student's t-test. TDO2, tryptophan 2,3-dioxygenase.
Regulation of proliferation, migration and invasion in ovarian cancer cells by tryptophan 2,3-dioxygenase
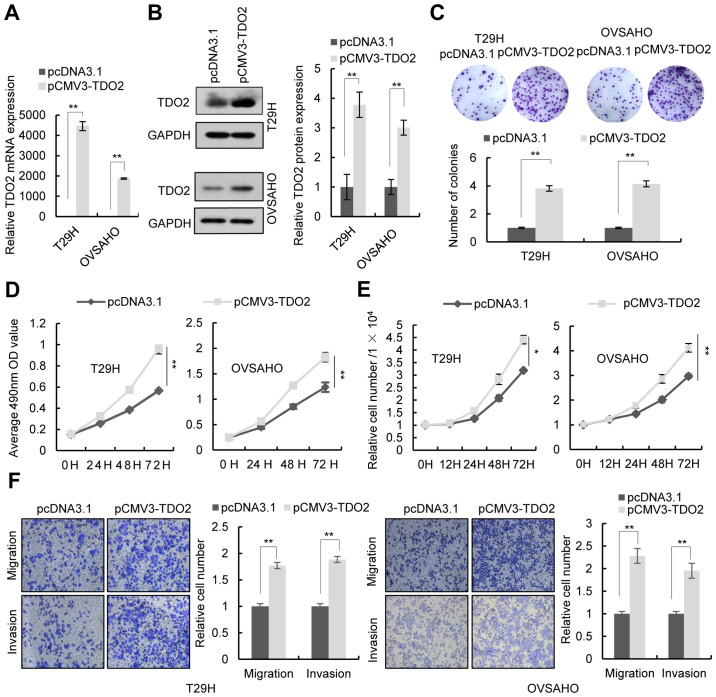
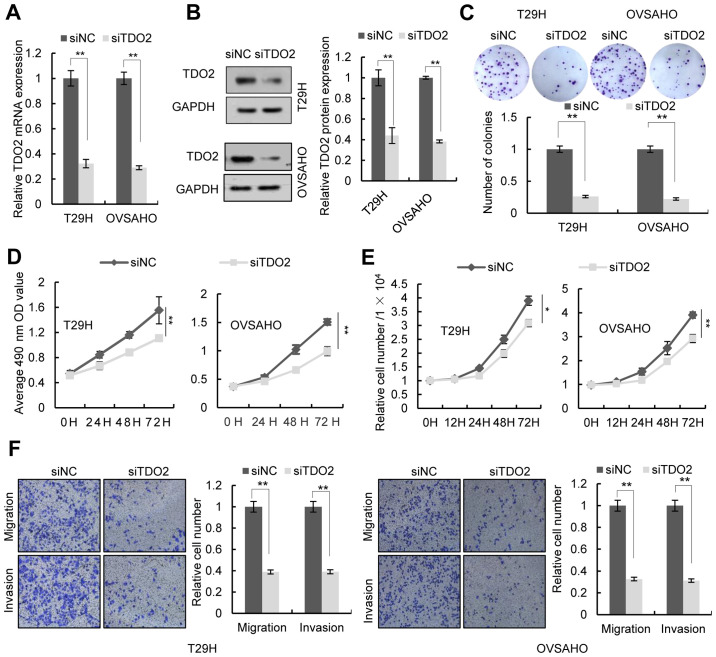
To investigate the function of TDO2 in ovarian cancer cells, the TDO2 overexpression plasmid was transfected into the human ovarian cancer cell lines T29H, OVSAHO and SKOV3. RT-qPCR and western blotting confirmed the overexpression of TDO2 in these cell lines (Figs. 2A and B and S1A and B). Colony formation assays, MTS assays and cell number counts revealed that TDO2 overexpression in T29H, OVSAHO and SKOV3 promoted cell proliferation (Figs. 2C-E and S1C-E). Transwell assays indicated that TDO2 overexpression promoted the migration and invasion of ovarian cells (Figs. 2F and S1F). It was then investigated whether TDO2 knockdown affected the proliferation, migration and invasion of ovarian cancer cells. Transfection of a TDO2-specific siRNA led to decreased expression of TDO2 in T29H, OVSAHO and SKOV3 cells (Figs. 3A and B and S1A and B). Colony formation assays, MTS assays, and cell number counts showed that knockdown of TDO2 decreased cell proliferation (Figs. 3C-E and S1C-E). Transwell assays indicated that TDO2 knockdown reduced cell migration and invasion (Figs. 3F and S1F). TDO2 knockdown did not increase the activity of caspase-3/7, indicating that the decrease of MTS signal and cell numbers in TDO2 knockdown cells was not a result of cell apoptosis (Fig. S2).
Figure 2.
Effect of TDO2 overexpression on proliferation, migration and invasion of ovarian cancer cells. T29H and OVSAHO cells transfected with pCMV3-TDO2-untagged (pCMV3-TDO2) or pcDNA3.1. (A) TDO2 mRNA levels quantified 48 h post-transfection using real-time polymerase chain reaction. (B) Representative western blotting of TDO2 protein levels 48 h post-transfection. (C) Representative images of colony formation assay (top) and the number of colonies (bottom). (D) Cell proliferation assays were performed at different time points following transfection; (E) Cell counts 12, 24, 48 and 72 h after transfection; (F) Representative images of cell migration and invasion assays (left) and quantification (right) from three independent experiments. Magnification, ×100. Data are shown as mean ± standard deviation from three independent experiments. *P<0.05; **P<0.01 by Student's t-test. TDO2, tryptophan 2,3-dioxygenase.
Figure 3.
Effect of TDO2 knockdown on proliferation, migration and invasion of ovarian cancer cells. T29H and OVSAHO cells transfected with siTDO2 or siNC. (A) The expression of TDO2 determined using reverse transcription-quantitative PCR and (B) western blotting 48 h after transfection. (C) Representative images of colony formation assay (top) and the number of colonies (bottom). (D) Cell proliferation assays were performed at different time points after transfection. (E) Cell counts 12, 24, 48 and 72 h after transfection. (F) Representative images of cell migration and invasion assays (left) and the quantification (right) from three independent experiments. Magnification, ×100. Data are shown as mean ± standard deviation from three independent experiments. *P<0.05; **P<0.01 by Student's t-test. TDO2, tryptophan 2,3-dioxygenase; si, short interfering; NC, negative control.
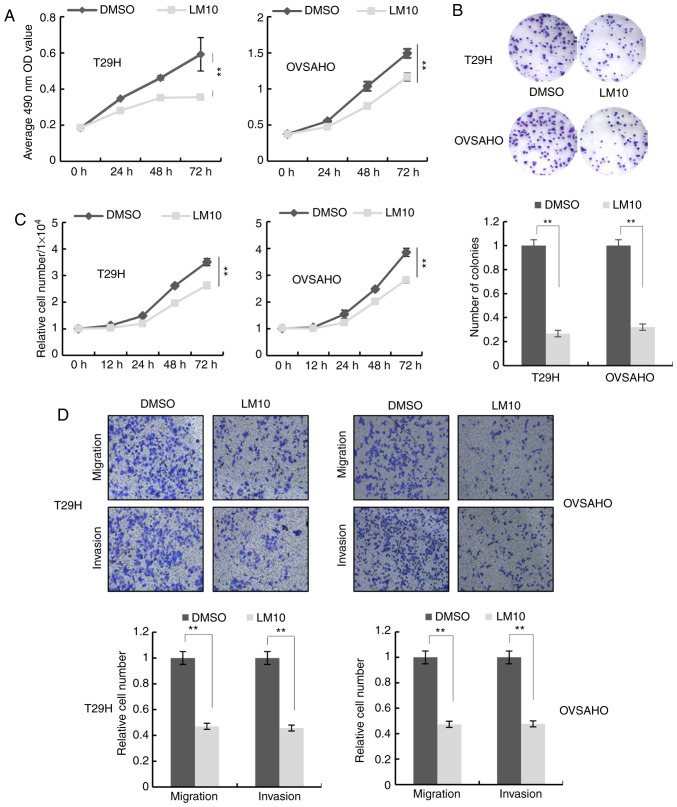
Inhibition of proliferation, migration and invasion of ovarian cancer cells by the tryptophan 2,3-dioxygenase inhibitor, LM10
LM10, a TDO2 inhibitor, reportedly exhibits anti-cancer activity by reversing the tumoral immune resistance caused by TDO2 overexpression (15). It was therefore examined whether LM10 could inhibit the proliferation, migration and invasion of ovarian cancer cells. MTS assays, colony formation assays and cell number counting revealed that treatment with LM10 inhibited cell proliferation (Figs. 4A-C and S1C-E). Transwell assays indicated that LM10 significantly repressed cell migration and invasion (Figs. 4D and S1F).
Figure 4.
Effect of LM10 on proliferation, migration and invasion of ovarian cancer cells. T29H and OVSAHO cells were treated with 500 µM LM10 or an equivalent volume of Dulbecco's modified Eagle's medium. (A) Cell proliferation assays performed at different time points. (B) Representative images of colony formation assay (top) and the number of colonies (bottom). (C) Cell counts 12, 24, 48 and 72 h after treatment. (D) Representative images of cell migration and invasion assays (top) and quantification (bottom) from three independent experiments. Magnification, ×100. Data are shown as mean ± standard deviation from three independent experiments. **P<0.01 by Student's t-test. TDO2, tryptophan 2,3-dioxygenase.
Association of tryptophan 2,3-dioxygenase expression with pathological characteristics in patients with ovarian cancer
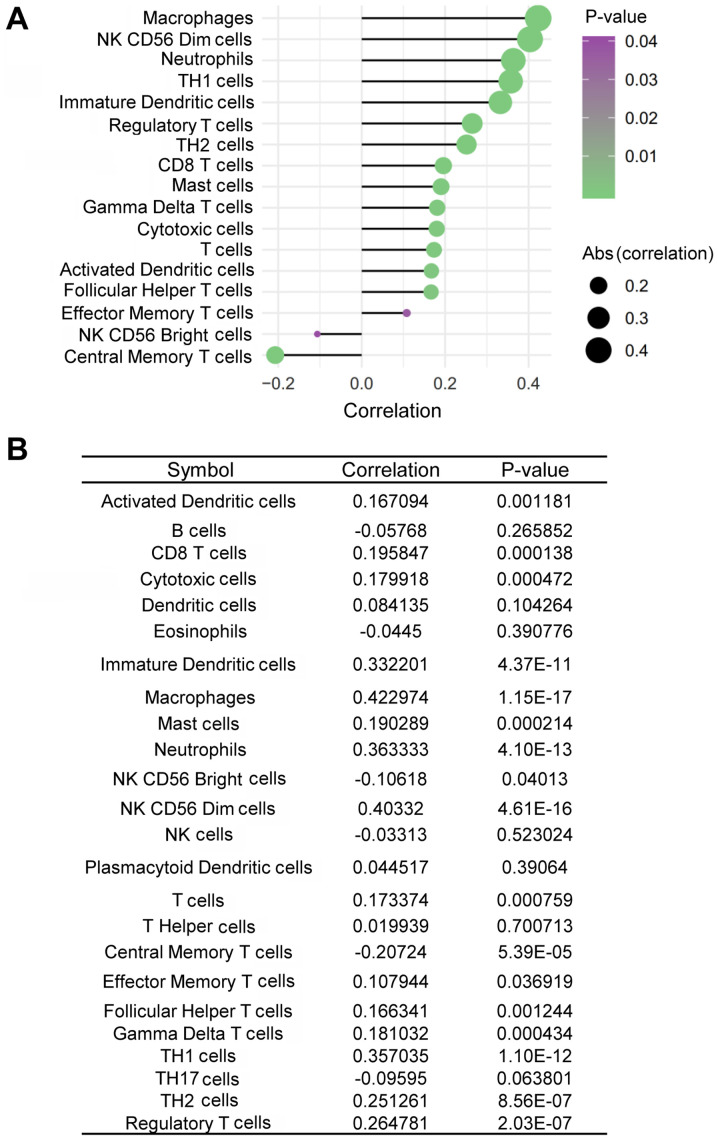
TDO2 is capable of inhibiting anti-CD3-driven T-cell proliferation and inducing CD8+ T-cell death (41,42). It was therefore assessed whether the expression of TDO2 was inversely associated with immune cell infiltration in ovarian cancer tissues. TDO2 expression was positively, rather than negatively, associated with the infiltration of CD8+ T cells in ovarian cancer tissues (Fig. 5A and B). In addition, TDO2 expression was also positively associated with the infiltration of other immune cells, including macrophages, CD56dim natural killer (NK) cells, neutrophils, T-helper type 1 (Th1) cells, immature dendritic cells (iDCs), regulatory T cells (Tregs), T-helper type 2 (Th2) cells, mast cells, gamma delta T (γδT) cells, cytotoxic T cells, activated dendritic cells (aDCs), follicular helper T (TFH) cells and effector helper T cells in ovarian cancer tissues, whereas it was negatively associated with the infiltration of CD56bright NK cells and central memory T (TCM) cells (Fig. 5A and B).
Figure 5.
Association of TDO2 expression with the infiltration of different immune cells of patients with ovarian cancer. (A and B) Association of TDO2 expression with the infiltration of different immune cells in ovarian cancer tissues. TDO2, tryptophan 2,3-dioxygenase.
Discussion
TDO2 has been found to be upregulated in different types of cancers including Merkel cell carcinoma, breast carcinoma, bladder carcinoma, colorectal carcinoma, hepatocarcinoma and melanoma (15,43,44). However, its expression and role in ovarian cancer remain to be elucidated. The present study found that TDO2 was upregulated in ovarian cancer tissues compared with that in the normal ovarian tissues. Overexpression of TDO2 promoted the proliferation, migration and invasion of ovarian cancer cells, whereas knockdown of TDO2 suppressed these phenotypes. In addition, inhibiting the activity of TDO2 with LM10 suppressed the proliferation of ovarian cancer cells. Kynurenine is reported to promote proliferation, migration and invasion of cancer cells (6,13). The results of the present study showed that knockdown of TDO2 by ~70% did not significantly reduce the levels of kynurenine (Fig. S3), suggesting that TDO2 may promote cancer cell proliferation, migration and invasion in a kynurenine- independent mechanism. Taken together, these data suggested that TDO2 functions as an oncogene in ovarian cancer and that LM10 may be a candidate drug for ovarian cancer treatment.
TDO2 has been shown to inhibit the proliferation and activation of T cells in the tumor microenvironment, thereby allowing tumors to escape immune surveillance (9,45). Pilotte et al (15) reported that cancer cells with increased expression of TDO2 exhibited increased immune tolerance in mice and grew rapidly and the oral TDO2 inhibitor LM10 reversed this immune tolerance and inhibited tumor growth by activating anti-tumoral immunity. Consistent with these studies, the present study found that TDO2 expression was positively associated with Treg infiltration in ovarian cancer tissues. By contrast, the expression levels of TDO2 were also positively associated with CD8+ T-cell infiltration. These conflicting observations suggested that further studies are needed to determine whether TDO2 can negatively regulate the functions of CD8+ T cells. In addition, TDO2 expression was also positively associated with the infiltration of other immune cells, including macrophages, CD56dim NK cells, neutrophils, Th1 cells, iDCs, Th2 cells, mast cells, γδT cells, aDCs and TFH cells but negatively associated with the infiltration of CD56bright NK and TCM cells in ovarian cancer tissues. Future efforts should be directed to address the effects of TDO2 on these immune cells.
In summary, the findings of the present study indicated that TDO2 can promote proliferation, migration and invasion of ovarian cancer cells and that TDO2 expression is associated with immune cell infiltration in ovarian cancer tissues. Future studies should construct different animal models to investigate the in vivo role of TDO2 in ovarian cancer and to elucidate the underlying molecular mechanisms.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- TDO2
tryptophan 2,3-dioxygenase
- IDO1
indoleamine 2,3-dioxygenase 1
- IDO2
indoleamine 2, 3-dioxygenase 2
- RT-qPCR
reverse transcription-quantitative PCR
- NK
natural killer cells
- Th1
T-helper type 1 cells
- iDCs
immature dendritic cells
- Tregs
regulatory T cells
- Th2
T-helper type 2 cells
- γδT
gamma delta T cells
- aDCs
activated dendritic cells
- TFH
follicular helper T cells
- TCM
central memory T cell
Funding Statement
This work was supported by the National Natural Science Foundation of China (grant nos. 81572780, 81773011 and 81972648) and the Zhejiang Provincial Natural Sciences Foundation (grant nos. LZ16H160004, LY16C050004 and LY18H030008).
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 81572780, 81773011 and 81972648) and the Zhejiang Provincial Natural Sciences Foundation (grant nos. LZ16H160004, LY16C050004 and LY18H030008).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
KFT conceived and devised the study. KFT, YX, RO, WC, XW and FT designed the experiments and performed the analysis. YZ drafted the manuscript and revised the manuscript critically for important content. FT drafted the original manuscript. YZ, FT, JJ, LC, JD, XC, QH and SZ performed the experiments and analyzed the data. KFT, YX, RO, WC and XW contributed reagents and materials. KFT, YX and YZ confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests
References
- 1.Orr B, Edwards RP. Diagnosis and treatment of ovarian cancer. Hematol Oncol Clin North Am. 2018;32:943–964. doi: 10.1016/j.hoc.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 2.Gunderson CC, Moore KN. Olaparib: An oral PARP-1 and PARP-2 inhibitor with promising activity in ovarian cancer. Future Oncol. 2015;11:747–757. doi: 10.2217/fon.14.313. [DOI] [PubMed] [Google Scholar]
- 3.Corrado G, Salutari V, Palluzzi E, Distefano MG, Scambia G, Ferrandina G. Optimizing treatment in recurrent epithelial ovarian cancer. Expert Rev Anticancer Ther. 2017;17:1147–1158. doi: 10.1080/14737140.2017.1398088. [DOI] [PubMed] [Google Scholar]
- 4.Platten M, Nollen EAA, Röhrig UF, Fallarino F, Opitz CA. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat Rev Drug Discov. 2019;18:379–401. doi: 10.1038/s41573-019-0016-5. [DOI] [PubMed] [Google Scholar]
- 5.Savitz J. The kynurenine pathway: A finger in every pie. Mol Psychiatry. 2019;25:131–147. doi: 10.1038/s41380-019-0414-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Venkateswaran N, Lafita-Navarro MC, Hao YH, Kilgore JA, Perez-Castro L, Braverman J, Borenstein-Auerbach N, Kim M, Lesner NP, Mishra P, et al. MYC promotes tryptophan uptake and metabolism by the kynurenine pathway in colon cancer. Genes Dev. 2019;33:1236–1251. doi: 10.1101/gad.327056.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Puccetti P, Fallarino F, Italiano A, Soubeyran I, MacGrogan G, Debled M, Velasco V, Bodet D, Eimer S, Veldhoen M, et al. Accumulation of an endogenous tryptophan- derived metabolite in colorectal and breast cancers. PLoS One. 2015;10:e0122046. doi: 10.1371/journal.pone.0122046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ball HJ, Yuasa HJ, Austin CJ, Weiser S, Hunt NH. Indoleamine 2,3-dioxygenase-2; a new enzyme in the kynurenine pathway. Int J Biochem Cell Biol. 2009;41:467–471. doi: 10.1016/j.biocel.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Cheong JE, Sun L. Targeting the IDO1/TDO2-KYN-AhR pathway for cancer immunotherapy-challenges and opportunities. Trends Pharmacol Sci. 2018;39:307–325. doi: 10.1016/j.tips.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 10.Pham QT, Oue N, Sekino Y, Yamamoto Y, Shigematsu Y, Sakamoto N, Sentani K, Uraoka N, Yasui W. TDO2 overexpression is associated with cancer stem cells and poor prognosis in esophageal squamous cell carcinoma. Oncology. 2018;95:297–308. doi: 10.1159/000490725. [DOI] [PubMed] [Google Scholar]
- 11.Liu M, Wang X, Wang L, Ma X, Gong Z, Zhang S, Li Y. Targeting the IDO1 pathway in cancer: From bench to bedside. J Hematol Oncol. 2018;11:100. doi: 10.1186/s13045-018-0644-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D'Amato NC, Rogers TJ, Gordon MA, Greene LI, Cochrane DR, Spoelstra NS, Nemkov TG, D'Alessandro A, Hansen KC, Richer JK. A TDO2-AhR signaling axis facilitates anoikis resistance and metastasis in triple-negative breast cancer. Cancer Res. 2015;75:4651–4664. doi: 10.1158/0008-5472.CAN-15-2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bishnupuri KS, Alvarado DM, Khouri AN, Shabsovich M, Chen B, Dieckgraefe BK, Ciorba MA. IDO1 and kynurenine pathway metabolites activate PI3K-Akt signaling in the neoplastic colon epithelium to promote cancer cell proliferation and inhibit apoptosis. Cancer Res. 2019;79:1138–1150. doi: 10.1158/0008-5472.CAN-18-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith C, Chang MY, Parker KH, Beury DW, DuHadaway JB, Flick HE, Boulden J, Sutanto-Ward E, Soler AP, Laury-Kleintop LD, et al. IDO is a nodal pathogenic driver of lung cancer and metastasis development. Cancer Discov. 2012;2:722–735. doi: 10.1158/2159-8290.CD-12-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pilotte L, Larrieu P, Stroobant V, Colau D, Dolusic E, Frédérick R, De Plaen E, Uyttenhove C, Wouters J, Masereel B, Van den Eynde BJ. Reversal of tumoral immune resistance by inhibition of tryptophan 2,3-dioxygenase. Proc Natl Acad Sci USA. 2012;109:2497–2502. doi: 10.1073/pnas.1113873109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fatokun AA, Hunt NH, Ball HJ. Indoleamine 2,3- dioxygenase 2 (IDO2) and the kynurenine pathway: Characteristics and potential roles in health and disease. Amino Acids. 2013;45:1319–1329. doi: 10.1007/s00726-013-1602-1. [DOI] [PubMed] [Google Scholar]
- 17.Brenk M, Scheler M, Koch S, Neumann J, Takikawa O, Häcker G, Bieber T, von Bubnoff D. Tryptophan deprivation induces inhibitory receptors ILT3 and ILT4 on dendritic cells favoring the induction of human CD4+CD25+ Foxp3+ T regulatory cells. J Immuno. 2009;183:145–154. doi: 10.4049/jimmunol.0803277. [DOI] [PubMed] [Google Scholar]
- 18.Chen W, Liang X, Peterson AJ, Munn DH, Blazar BR. The indoleamine 2,3-dioxygenase pathway is essential for human plasmacytoid dendritic cell-induced adaptive T regulatory cell generation. J Immunol. 2008;181:5396–5404. doi: 10.4049/jimmunol.181.8.5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung DJ, Rossi M, Romano E, Ghith J, Yuan J, Munn DH, Young JW. Indoleamine 2,3-dioxygenase-expressing mature human monocyte-derived dendritic cells expand potent autologous regulatory T cells. Blood. 2009;114:555–563. doi: 10.1182/blood-2008-11-191197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curti A, Pandolfi S, Valzasina B, Aluigi M, Isidori A, Ferri E, Salvestrini V, Bonanno G, Rutella S, Durelli I, et al. Modulation of tryptophan catabolism by human leukemic cells results in the conversion of CD25- into CD25+ T regulatory cells. Blood. 2007;109:2871–2877. doi: 10.1182/blood-2006-07-036863. [DOI] [PubMed] [Google Scholar]
- 21.Fallarino F, Grohmann U, You S, McGrath BC, Cavener DR, Vacca C, Orabona C, Bianchi R, Belladonna ML, Volpi C. The combined effects of tryptophan starvation and tryptophan catabolites down-regulate T cell receptor zeta-chain and induce a regulatory phenotype in naive T cells. J Immunol. 2006;176:6752–6761. doi: 10.4049/jimmunol.176.11.6752. [DOI] [PubMed] [Google Scholar]
- 22.Hippen KL, O'Connor RS, Lemire AM, Saha A, Hanse EA, Tennis NC, Merkel SC, Kelekar A, Riley JL, Levine BL, et al. In vitro induction of human regulatory T cells using conditions of low tryptophan plus kynurenines. Am J Transplant. 2017;17:3098–3113. doi: 10.1111/ajt.14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma MD, Baban B, Chandler P, Hou DY, Singh N, Yagita H, Azuma M, Blazar BR, Mellor AL, Munn DH. Plasmacytoid dendritic cells from mouse tumor-draining lymph nodes directly activate mature Tregs via indoleamine 2,3-dioxygenase. J Clin Invest. 2007;117:2570–2582. doi: 10.1172/JCI31911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kristeleit R, Davidenko I, Shirinkin V, El-Khouly F, Bondarenko I, Goodheart MJ, Gorbunova V, Penning CA, Shi JG, Liu X, et al. A randomised, open-label, phase 2 study of the IDO1 inhibitor epacadostat (INCB024360) versus tamoxifen as therapy for biochemically recurrent (CA-125 relapse)-only epithelial ovarian cancer, primary peritoneal carcinoma, or fallopian tube cancer. Gynecol Oncol. 2017;146:484–490. doi: 10.1016/j.ygyno.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 25.Zhang GN, Zhu Y, Huang JM. Understanding of targeting MyD88, IDO1 and AHR at the heart of immunosuppressive signaling pathway for immunotherapy of epithelial ovarian cancer. Zhonghua fu chan ke za zhi. 2018;53:448–451. doi: 10.3760/cma.j.issn.0529-567x.2018.07.002. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 26.Awuah SG, Zheng YR, Bruno PM, Hemann MT, Lippard SJ. A Pt(IV) pro-drug preferentially targets indoleamine-2,3-dioxygenase, providing enhanced ovarian cancer immuno-chemotherapy. J Am Chem Soc. 2015;137:14854–14857. doi: 10.1021/jacs.5b10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qian F, Villella J, Wallace PK, Mhawech-Fauceglia P, Tario JD, Jr, Andrews C, Matsuzaki J, Valmori D, Ayyoub M, Frederick PJ, et al. Efficacy of levo-1-methyl tryptophan and dextro-1-methyl tryptophan in reversing indoleamine-2, 3-dioxygenase-mediated arrest of T-cell proliferation in human epithelial ovarian cancer. Cancer Res. 2009;69:5498–5504. doi: 10.1158/0008-5472.CAN-08-2106. [DOI] [PubMed] [Google Scholar]
- 28.Tanizaki Y, Kobayashi A, Toujima S, Shiro M, Mizoguchi M, Mabuchi Y, Yagi S, Minami S, Takikawa O, Ino K. Indoleamine 2,3-dioxygenase promotes peritoneal metastasis of ovarian cancer by inducing an immunosuppressive environment. Cancer Sci. 2014;105:966–973. doi: 10.1111/cas.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uyttenhove C, Pilotte L, Theate I, Stroobant V, Colau D, Parmentier N, Boon T, Van den Eynde BJ. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat Med. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 30.Inaba T, Ino K, Kajiyama H, Yamamoto E, Shibata K, Nawa A, Nagasaka T, Akimoto H, Takikawa O, Kikkawa F. Role of the immunosuppressive enzyme indoleamine 2,3-dioxygenase in the progression of ovarian carcinoma. Gynecol Oncol. 2009;115:185–192. doi: 10.1016/j.ygyno.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 31.Wang D, Saga Y, Mizukami H, Sato N, Nonaka H, Fujiwara H, Takei Y, Machida S, Takikawa O, Ozawa K, Suzuki M. Indoleamine-2,3-dioxygenase, an immunosuppressive enzyme that inhibits natural killer cell function, as a useful target for ovarian cancer therapy. Int J Oncol. 2012;40:929–934. doi: 10.3892/ijo.2011.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dolusić E, Larrieu P, Moineaux L, Stroobant V, Pilotte L, Colau D, Pochet L, Van den Eynde B, Masereel B, Wouters J, Frédérick R. Tryptophan 2,3-dioxygenase (TDO) inhibitors. 3-(2-(pyridyl)ethenyl)indoles as potential anticancer immunomodulators. J Med Chem. 2011;54:5320–5334. doi: 10.1021/jm2006782. [DOI] [PubMed] [Google Scholar]
- 33.Young T, Mei F, Liu J, Bast RC, Jr, Kurosky A, Cheng X. Proteomics analysis of H-RAS-mediated oncogenic transformation in a genetically defined human ovarian cancer model. Oncogene. 2005;24:6174–6184. doi: 10.1038/sj.onc.1208753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu J, Yang G, Thompson-Lanza JA, Glassman A, Hayes K, Patterson A, Marquez RT, Auersperg N, Yu Y, Hahn WC, et al. A genetically defined model for human ovarian cancer. Cancer Res. 2004;64:1655–1663. doi: 10.1158/0008-5472.CAN-03-3380. [DOI] [PubMed] [Google Scholar]
- 35.Metzgar D, Liu L, Hansen C, Dybvig K, Wills C. Domain-level differences in microsatellite distribution and content result from different relative rates of insertion and deletion mutations. Genome Res. 2002;12:408–413. doi: 10.1101/gr.198602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li R, Quan Y, Xia W. SIRT3 inhibits prostate cancer metastasis through regulation of FOXO3A by suppressing Wnt/β-catenin pathway. Exp Cell Res. 2018;364:143–151. doi: 10.1016/j.yexcr.2018.01.036. [DOI] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Cory AH, Owen TC, Barltrop JA, Cory JG. Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun. 1991;3:207–212. doi: 10.3727/095535491820873191. [DOI] [PubMed] [Google Scholar]
- 39.Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf AC, Angell H, Fredriksen T, Lafontaine L, Berger A, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. doi: 10.1016/j.immuni.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 40.Li L, Feng Q, Wang X. PreMSIm: An R package for predicting microsatellite instability from the expression profiling of a gene panel in cancer. Comput Struct Biotechnol J. 2020;18:668–675. doi: 10.1016/j.csbj.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt SK, Muller A, Heseler K, Woite C, Spekker K, MacKenzie CR, Däubener W. Antimicrobial and immunoregulatory properties of human tryptophan 2,3-dioxygenase. Eur J Immunol. 2009;39:2755–2764. doi: 10.1002/eji.200939535. [DOI] [PubMed] [Google Scholar]
- 42.Greene LI, Bruno TC, Christenson JL, D'Alessandro A, Culp-Hill R, Torkko K, Borges VF, Slansky JE, Richer JK. A role for tryptophan-2,3-dioxygenase in CD8 T-cell suppression and evidence of tryptophan catabolism in breast cancer patient plasma. Mol Cancer Re. 2019;17:131–139. doi: 10.1158/1541-7786.MCR-18-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wardhani LO, Matsushita M, Iwasaki T, Kuwamoto S, Nonaka D, Nagata K, Kato M, Kitamura Y, Hayashi K. Expression of the IDO1/TDO2-AhR pathway in tumor cells or the tumor microenvironment is associated with Merkel cell polyomavirus status and prognosis in Merkel cell carcinoma. Human Pathol. 2019;84:52–61. doi: 10.1016/j.humpath.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 44.Rogers TJ, Christenson JL, Greene LI, O'Neill KI, Williams MM, Gordon MA, Nemkov T, D'Alessandro A, Degala GD, Shin J, et al. Reversal of triple-negative breast cancer EMT by miR-200c decreases tryptophan catabolism and a program of immunosuppression. Mol Cancer Res. 2019;17:30–41. doi: 10.1158/1541-7786.MCR-18-0246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Badawy AA. Targeting tryptophan availability to tumors: The answer to immune escape? Immunol Cell Biol. 2018;96:1026–1034. doi: 10.1111/imcb.12168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.