Abstract
Oral squamous cell carcinoma (OSCC) is a frequent malignant tumor worldwide. Long non-coding RNAs (lncRNAs) are known to play key roles in different types of cancer, including OSCC. It was previously reported that lncRNA deleted in lymphocytic leukemia 1 (DLEU1) is notably upregulated in OSCC; however, the role of DLEU1 in OSCC remains unclear. Gene and protein expression levels in OSCC cells were detected by reverse transcription-quantitative PCR and western blotting, respectively, in the present study. A Transwell assay was performed to measure cell migration and invasion. Flow cytometry was used to detect cell apoptosis, and the dual-luciferase reporter assay was applied to confirm the interaction between DLEU1, microRNA (miR)-149-5p and CDK6 in OSCC cells. DLEU1 expression was negatively associated with the survival rate of patients with OSCC. In addition, silencing of DLEU1 notably inhibited the proliferation of OSCC cells by inducing apoptosis. Meanwhile, DLEU1 directly bound to miR-149-5p, and CDK6 was found to be the direct target of miR-149-5p. Furthermore, DLEU1 knockdown-induced inhibition of OSCC cell proliferation was significantly reversed by the miR-149-5p antagomir. Knockdown of lncRNA DLEU1 reversed the proliferation of OSCC cells via regulation of the miR-149-5p/CDK6 axis. Thus, DLEU1 may serve as a novel target for treating OSCC.
Keywords: oral squamous cell carcinoma, deleted in lymphocytic leukemia 1, microRNA-149-5p, cyclin dependent kinase 6
Introduction
Oral squamous cell carcinoma (OSCC) is one of the most frequent malignant tumors with a poor prognosis (1,2), and the incidence of OSCC is ~300,000 new cases per year worldwide (3). Recent reports revealed that betel quid chewing, smoking and human papillomavirus infections are the three biggest risk factors that contribute to the tumorigenesis of OSCC (4,5). In addition, OSCC is known to exhibit a high propensity for metastasis (6). Currently, the primary treatment options for OSCC are surgery, chemotherapy and radiotherapy (7); however, these therapeutic strategies have limited effects, particularly for the patients with advanced stage OSCC (8). Therefore, it is necessary to investigate novel effective strategies for the treatment of OSCC.
Previous studies have reported that non-coding RNAs (ncRNAs) play important roles in multiple diseases (9,10). Among these ncRNAs are long ncRNAs (lncRNAs), which are >200 nucleotides in length (11). In addition, lncRNAs are involved in OSCC. For example, Ghapanchi et al (12) revealed that lncRNA H19 could increase the proliferation of OSCC cells. Wu et al (13) revealed that lncRNA RC3H2 was an oncogene via regulating microRNA (miRNA/miR)-101-3p in OSCC.
Meanwhile, the lncRNA deleted in lymphocytic leukemia 1 (DLEU1) has been confirmed to regulate the progression of multiple types of cancer (12,13), and elevated DLEU1 expression contributes to the development of OSCC, suggesting that DLEU1 may play an important role in OSCC (14); however, the detailed function of DLEU1 in OSCC remains unclear.
miRNAs are endogenic non-coding small RNAs that are found in abundance in the human body (15). In addition, dysregulation of miRNAs are known to be associated with the progression of OSCC (16). For example, it was previously reported that miR-770 could promote the migration and invasion of OSCC cells by regulating the NAD-dependent protein deacetylase sirtuin-7 (Sirt7)/Smad4 pathway (17). Moreover, miR-128 and miR-142 could regulate the tumorigenesis and epithelial-mesenchymal transition in OSCC through mediation of homeobox protein Hox-A10 (18). Meanwhile, Luo et al (19) found that miR-149-5p could regulate cisplatin chemosensitivity, cell growth, and metastasis of OSCC cells by targeting TGFβ2. However, the association between DLEU1 and miR-149-5p in OSCC is unclear.
The present study aimed to investigate the biological function of DLEU1 in OSCC, in order to identify novel potential therapeutic strategies for treating OSCC.
Materials and methods
Cell culture and cell transfection
OSCC cell lines (Cal-27 and SCC-9; American Type Culture Collection) were maintained in DMEM (Gibco; Thermo Fisher Scientific, Inc.), supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and antibiotics (100 U/ml penicillin and 100 µg/ml streptomycin) in an incubator at 37°C with 5% CO2.
Small interfering (si)RNAs targeted against DLEU1 (DLEU1 siRNA1, DLEU1 siRNA2 and DLEU1 siRNA3; 10 nM) and a negative control (NC) siRNA (siRNA-NC) were purchased from Guangzhou RiboBio Co., Ltd., and were transfected into OSCC cells (5×103) using Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.). Then, cells were incubated at 37°C for 6 h and the transfection efficiency was determined via reverse transcription-quantitative PCR (RT-qPCR). After 24 h of incubation, transfected cells were used for subsequent experiments. The siRNA sequences were as follows: siRNA-NC, 5′-UUCUCCGAACGUGUCACGUTT-3′; DLEU1 siRNA1, 5′-GGAAUGAAGCAACUGAGAUUU-3′; DLEU1 siRNA2, 5′-GGGTTACGATTGCCCAGAT-3′; and DLEU1 siRNA3, 5′-CGTTAAGGTTCCGGACGAC-3′.
The NC, miR-149-5p agomir and miR-149-5p antagomir were synthesized and obtained from Shanghai GenePharma Co., Ltd. Cells (5×103) were transfected with 10 nM control agomir (NC), miR-149-5p agomir or miR-149-5p antagomir for 24 h with Lipofectamine 2000 at 37°C. After 24 h of transfection, transfected cells were used for subsequent experiments. Meanwhile, the concentration of miR-149-5p agomir/antagomir (50 nM) was selected according to the instructions of the manufacturer. The sequences were as follows: miR-149-5p agomir, 5′-UCUGGCUCCGUGUCUUCACUCCC-3′; miR-149-5p antagomir, 5′-GGGAGUGAAGACACGGAGCCAGA-3′; and NC, 5′-UUCUCCGAACGUGUCACGUTT-3′.
Tissue collection
In total, 10 pairs of OSCC samples and adjacent normal tissues (~2 cm from tumor) were collected from the Affiliated Stomatological Hospital of Guizhou Medical University (Guiyang, China) between April 2018 and April 2019. The patients were informed of the purpose of the experiments and provided written informed consent. The clinical information of the patients is listed in Table I. Patients were diagnosed with OSCC following the American College of Rheumatology classification criteria for the diagnosis of OSCC (20). Meanwhile, the samples were used for investigation of DLEU1 and CDK6 levels. The present study was approved by the Institutional Ethical Committee of Affiliated Stomatological Hospital of Guizhou Medical University (approval no. ASHGMU20190523).
Table I.
Clinical characteristics of patients with OSCC in the present study.
| Clinical characteristics | Number |
|---|---|
| Age, years | |
| ≥60 | 4 |
| <60 | 6 |
| Sex | |
| Male | 7 |
| Female | 3 |
| Tumor size, cm | |
| ≥3 | 6 |
| <3 | 4 |
| TNM stage | |
| I/II | 6 |
| III/IV | 4 |
OSCC, oral squamous cell carcinoma; TNM, tumor node metastasis.
RT-qPCR
Total RNA was extracted from OSCC cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA was reverse transcribed into cDNA using the PrimeScript RT reagent kit (Takara Bio, Inc.) according to the manufacturer's instructions. Then, RT-qPCR was performed using a SYBR® Premix Ex Taq™ II kit (Takara Bio, Inc.) on a 7900HT system (Applied Biosystems; Thermo Fisher Scientific, Inc.) according to the following conditions: 60°C for 1 min, 90°C for 15 min, followed by 40 cycles of application at 90°C for 15 sec and 55°C for 60 sec. The primers used were as follows: U6 forward, 5′-CGCTTCGGCAGCACATATAC-3′ and reverse, 5′-AAATATGGAACGCTTCACGA-3′; DLEU1 forward, 5′-GGTCCACGGCACGTTAACA-3′ and reverse, 5′-CCAATTGAAGGCCTTAAGG-3′; miR-149-5p forward, 5′-TGCGCTAGCAGCGGGAACAGTTC-3′ and reverse, 5′-CCAGTGCAGGGTCCGAGGTATT-3′; and β-actin forward, 5′-GTCCACCGCAAATGCTTCTA-3′ and reverse, 5′-TGCTGTCACCTTCACCGTTC-3′. The relative expression level was quantified by normalizing to β-actin or U6 using the 2−ΔΔCq method (21).
Bioinformatics analysis
The survival curve was calculated based on the data from The Cancer Genome Atlas (TCGA). The result shown here is in part based upon data generated by the TCGA Research Network (https://www.cancer.gov/tcga). In addition, the data from TCGA were analyzed using Gene Expression Profiling Interactive Analysis (GEPIA) as previously described (22).
Bioinformatics prediction
The potential downstream miRNA of DLEU1 was predicted using StarBase (http://starbase.sysu.edu.cn/index.php). In addition, the target mRNA of miR-149-5p was predicted using TargetScan (http://www.targetscan.org/vert_72/), miRDB (http://www.mirdb.org/) and miRWalk (http://mirwalk.umm.uni-heidelberg.de/).
Cell Counting Kit-8 (CCK-8) assay
SCC-9 or Cal-27 cells (5.0×103 cells/well) were transfected with siRNA-NC, DLEU1 siRNA2 or DLEU1 siRNA3 for 0, 24, 48 or 72 h. Then, 10 µl CCK-8 reagent (Beyotime Institute of Biotechnology) was added into each well, and the plate was incubated for 2 h at 37°C. The absorbance was detected at 450 nm using a microplate reader (Bio-Rad Laboratories, Inc.).
Immunofluorescence staining
OSCC cells were plated onto a 96-well plate at a density of 5.0×103 cells/well. Following incubation, cells were transfected with siRNA-NC or DLEU1 siRNA2 for 72 h. After that, cells were fixed with 4% paraformaldehyde for 20 min at room temperature and blocked with 2% bovine serum albumin (Beyotime Institute of Biotechnology) at room temperature for 30 min. Subsequently, cells were incubated with rabbit monoclonal antibody anti-Ki67 (1:100; cat. no. ab92742; Abcam) at 4°C overnight. Then, cells were incubated with anti-rabbit IgG secondary antibody (1:1,000; cat. no. ab150077; Abcam) for 1 h at room temperature. The results were observed under a fluorescence microscope (Olympus BX53; Olympus Corporation).
Transwell assay
For the cell migration assay, 2×105 OSCC cells were seeded into the upper chambers of the 24-well plates in 200 µl serum-free DMEM (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 0.2% bovine serum albumin (Beyotime Institute of Biotechnology). The lower chambers contained RPMI-1640 medium supplemented with 1% FBS. After incubation for 24 h at 37°C, the non-migrating cells were gently removed from the upper side of each chamber with a cotton swab, while the cells that had migrated were fixed with 95% alcohol for 10 min at room temperature and stained with 0.1% crystal violet (Sigma-Aldrich; Merck KGaA) for 5 min at room temperature. Finally, cells were counted under an inverted light microscope (Olympus Corporation) at ×400 magnification.
For the cell invasion assay, the upper chamber was pre-treated with 100 µl Matrigel (BD Biosciences) for 4 h at 37°C. Subsequently, transfected OSCC cells (2×105) were seeded into the upper chambers with serum-free DMEM, while the media in the lower chambers was supplemented with 1% FBS. After incubation for 24 h, the Transwell chambers were fixed with 95% alcohol for 10 min at room temperature and stained with 0.1% crystal violet for 5 min at room temperature. The Transwell chamber was observed and the invaded cells were counted under a microscope.
Flow cytometry assay
Cells were trypsinized and resuspended in binding buffer. Then, cells were stained with 5 µl Annexin V-FITC (BD Biosciences) and propidium iodide (PI; BD Biosciences) in the dark at 37°C for 30 min. Flow cytometry (FACScan™; BD Biosciences) was applied to analyze the apoptosis rate using CellQuest™ software (version 5.1; BD Biosciences).
Western blotting
OSCC cells were lysed in RIPA lysis buffer (Nanjing KeyGen Biotech Co., Ltd.), and the protein concentration was determined using a BCA Assay kit (Beijing Solarbio Science & Technology Co., Ltd.). Proteins (30 µg per lane) were separated via SDS-PAGE (10% gel), and separated proteins were then transferred onto PVDF membranes (Thermo Fisher Scientific, Inc.). The membranes were blocked in 5% non-fat dried milk in TBS with Tween-20 (10%) for 1 h at room temperature. Then, the PVDF membranes were incubated at 4°C overnight with the following primary antibodies: Anti-Bax (cat. no. ab182733; 1:1,000), anti-cleaved caspase-3 (cat. no. ab49822; 1:1,000), anti-Bcl-2 (cat. no. ab196495; 1:1,000), anti-CDK6 (cat. no. ab131469; 1:1,000), anti-CDK4 (cat. no. ab68266; 1:1,000), anti-cyclin-dependent kinase inhibitor p27 (p27 Kip1; cat. no. ab32034; 1:1,000) and anti-β-actin (cat. no. ab8226; 1:1,000). Then, the membrane was incubated with horseradish peroxidase-labeled goat anti-rabbit secondary antibody (cat. no. ab7090; 1:5,000) at room temperature for 1 h. All antibodies were obtained from Abcam. Enhanced chemiluminescence reagent (Thermo Fisher Scientific, Inc.) was used to visualize the protein bands. ImageJ software (version 2.0; National Institutes of Health) was used to quantify the intensity of the bands.
Dual-luciferase reporter assay
CDK6 3′-untranslated region (3′UTR) containing the putative binding site of miR-149-5p (CDK6 WT 3′UTR) and the CDK6 3′UTR with the mutation binding site (CDK6 MT 3′UTR) were synthesized by Shanghai GenePharma Co., Ltd. The mutation was generated using a site directed mutagenesis kit (Promega Corporation). The partial sequences of DLEU1 were synthesized by Shanghai GenePharma Co., Ltd. These were then cloned into the vectors (pmirGLO; Promega Corporation). Subsequently, DLEU1 (WT/MT) and CDK6 (WT/MT) were transfected into OSCC cells using Lipofectamine 2000. After 48 h of transfection, the relative luciferase activity was detected using the Dual-Luciferase Reporter kit (Promega Corporation). The data were normalized to Renilla luciferase activity.
RNA pull-down assay
Probe-control or probe-DLEU1 was transcribed and labeled with a Biotin RNA Labeling Mix (Roche Diagnostics). Cells were lysed with Poly-lysis buffer (ELK bioscience), washed with PBS and centrifuged at 1000 × g for 5 min at 4°C. Secondary structure formation in the biotin-labeled RNAs was induced with RNA structure buffer (Thermo Fisher Scientific, Inc.). Streptavidin beads (75 µl; Thermo Fisher Scientific, Inc.) were washed and incubated overnight. After that, streptavidin bead-RNA complexes were obtained by separating the mixture. Then, cell lysates (5×107 cells) were added to the complexes and incubated for 1 h. Following incubation, the mixture was separated again, and the supernatant of cell lysates was utilized to detect the enrichment of miR-149-5p. RT-qPCR was used to detect the enrichment of miR-149-5p.
Cell cycle detection
Cell cycle distribution was assessed as previously described (23). Briefly, OSCC cells (5×105) were harvested, fixed with 75% ethanol on ice for 20 min, permeabilized with 0.25% Triton X-100 and stained with Pharmingen PI/RNase (BD Biosciences). After incubation for 15 min at 4°C, cells were analyzed using a flow cytometer (BD FACSAria III; BD Biosciences) and ModFit (version 3.0; Verity Software House, Inc.). The data were quantified using FlowJo software (version 3.0; FlowJo, LLC).
Statistical analysis
Data are presented as the mean ± SD. The CCK-8 assay was performed five times. Immunofluorescence staining, RT-qPCR, western blotting, flow cytometry and Transwell invasion assays were repeated in triplicate. In addition, all other experiments were repeated three times. One-way analysis of variance and Tukey's post hoc tests were used for comparisons between ≥3 groups. A paired Student's t-test was used for comparisons between tumor tissues and adjacent normal tissues of the same patients, while an unpaired Student's t-test was used for comparisons between unpaired groups. P<0.05 was considered to indicate a statistically significant difference.
Results
DLEU1 expression is negatively associated with the survival rate of patients with OSCC
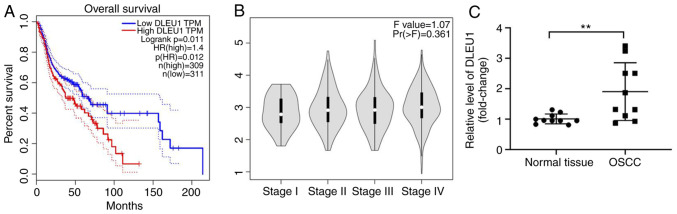
To detect the role of DLEU1 in OSCC, TCGA data were analyzed. As presented in Fig. 1A, DLEU1 level was negatively associated with the survival rate of patients with OSCC. In addition, DLEU1 was closely associated with patients with advanced stage OSCC (Fig. 1B). Furthermore, the expression of DLEU1 was significantly higher in OSCC tissues (Fig. 1C). Taken together, the expression of DLEU1 was upregulated in OSCC.
Figure 1.
DLEU1 expression is negatively associated with the survival rate of patients with OSCC. (A) TCGA dataset was used to explore the association between DLEU1 expression and the survival rate of patients with OSCC. (B) The relationship between DLEU1 and different stages of OSCC was analyzed using TCGA dataset. (C) The expression of DLEU1 in OSCC and adjacent normal tissues was assessed via reverse transcription-quantitative PCR. **P<0.01 vs. normal tissues. DLEU1, deleted in lymphocytic leukemia 1; OSCC, oral squamous cell carcinoma; TCGA, The Cancer Genome Atlas.
Silencing of DLEU1 significantly inhibits the proliferation of OSCC cells
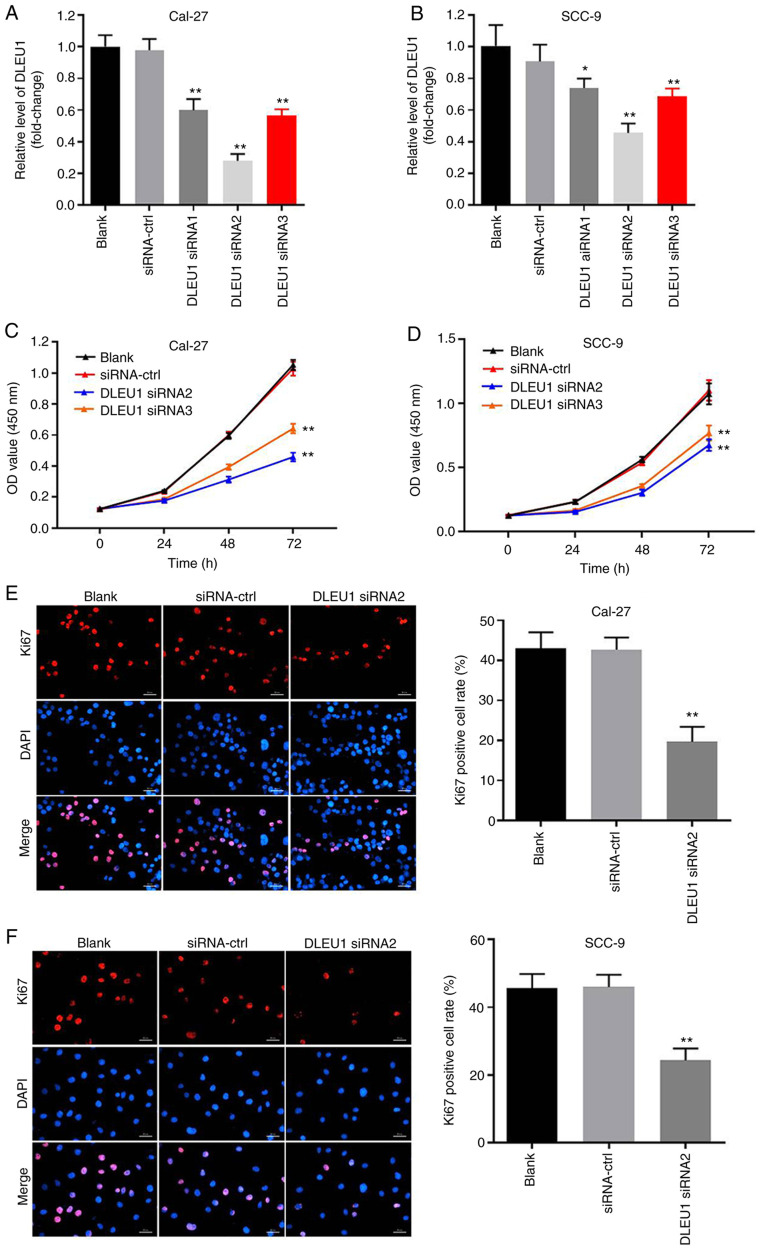
In order to evaluate the efficacy of cell transfection, RT-qPCR was performed. The data indicated that the expression of DLEU1 was significantly downregulated in OSCC cells when transfected with DLEU1 siRNAs (Fig. 2A and B). In addition, knockdown of DLEU1 significantly suppressed the cell viability of OSCC cells (Fig. 2C and D). DLEU1 siRNA2 had the most significant inhibitory effects on the expression of DLEU1, thus DLEU1 siRNA2 was used in subsequent investigations. Furthermore, the results of the Ki67 staining revealed that DLEU1 siRNA2 significantly inhibited the proliferation of OSCC cells (Fig. 2E and F), and it was revealed that Cal-27 cells were more susceptible to DLEU1 siRNA2 treatment. Thus, Cal-27 cells were selected for use in the subsequent analyses. Overall, silencing of DLEU1 significantly inhibited the proliferation of OSCC cells.
Figure 2.
Silencing of DLEU1 significantly inhibits the proliferation of OSCC cells. (A) Cal-27 or (B) SCC-9 cells were transfected with NC, DLEU1 siRNA1, DLEU1 siRNA2 or DLEU1 siRNA3 for 24 h. Then, the efficiency of DLEU1 siRNA transfection was detected via reverse transcription-quantitative PCR. (C) Cal-27 or (D) SCC-9 cells were treated with NC, DLEU1 siRNA2 or DLEU1 siRNA3 for 0, 24, 48 or 72 h. Then, OD values of OSCC cells were determined via a Cell Counting Kit-8 assay. (E and F) The proliferation of OSCC cells was measured by Ki67 staining. Red fluorescence shows Ki67 expression. Blue fluorescence shows DAPI staining. Scale bar, 100-µm. *P<0.05, **P<0.01 vs. control. DLEU1, deleted in lymphocytic leukemia 1; OSCC, oral squamous cell carcinoma; NC, negative control; siRNA, small interfering RNA.
DLEU1 sponges miR-149-5p in OSCC
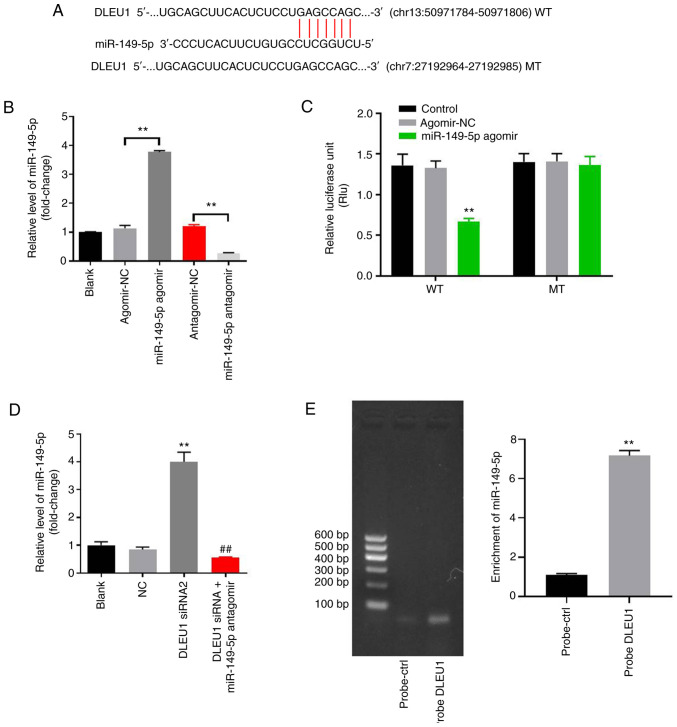
In order to investigate the underlying mechanism by which DLEU1 mediated the proliferation of OSCC cells, StarBase was used. As indicated in Fig. 3A, DLEU1 had a putative miR-149-5p targeting site. In addition, miR-149-5p is known to regulate the tumorigenesis of OSCC (19). Thus, miR-149-5p was selected for further analysis. The level of miR-149-5p in OSCC cells was upregulated by miR-149-5p agomir, but was downregulated by miR-149-5p antagomir (Fig. 3B). This result suggested that miR-149-5p agomir or antagomir was stably transfected into OSCC cells. Co-transfection of the WT-DLEU1 vector with miR-149-5p agomir resulted in a significant decrease in the luciferase activity when compared with the MT-DLEU1 vector (Fig. 3C). Furthermore, the expression of miR-149-5p in OSCC cells was significantly increased by the knockdown of DLEU1, while this was completely reversed by miR-149-5p antagomir (Fig. 3D). In addition, the results of the pull-down assay suggested that DLEU1 bound to miR-149-5p directly in OSCC cells (Fig. 3E). Overall, DLEU1 could sponge miR-149-5p in OSCC.
Figure 3.
DLEU1 can sponge miR-149-5p in OSCC. (A) Gene structure of DLEU1 indicated the binding site of miR-149-5p in its 3′UTR. (B) OSCC cells were transfected with antagomir/agomir NC, miR-149-5p agomir/antagomir for 24 h. Then, the expression of miR-149-5p in OSCC cells was detected via RT-qPCR. (C) The relative luciferase activity in Cal-27 cells after co-transfection with miR-149-5p and WT/MT DLEU1 3′UTR was measured using a dual-luciferase reporter assay. (D) Cal-27 cells were treated with NC, DLEU1 siRNA2 or DLEU1 siRNA2 + miR-149-5p. Then, the expression of miR-149-5p in Cal-27 cells was detected via RT-qPCR. (E) The association between DLEU1 and miR-149-5p was explored using an RNA pull-down assay. **P<0.01 vs. control; ##P<0.01 vs. DLEU1 siRNA2. DLEU1, deleted in lymphocytic leukemia 1; OSCC, oral squamous cell carcinoma; NC, negative control; siRNA, small interfering RNA; miR, microRNA; UTR, untranslated region; WT, wild-type; MT, mutant; RT-qPCR, reverse transcription-quantitative PCR.
Knockdown of DLEU1 inhibits the tumorigenesis of OSCC via regulation of miR-149-5p
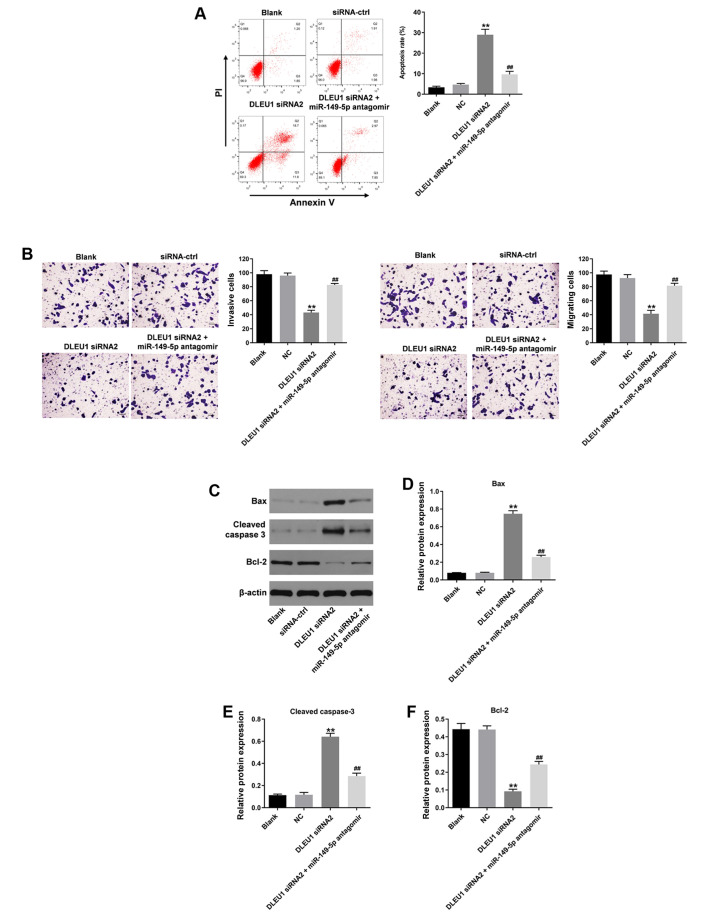
In order to further investigate the underlying mechanism by which DLEU1 regulated the progression of OSCC, flow cytometry was used. As indicated in Fig. 4A, DLEU1 siRNA2 significantly induced cell apoptosis, which was partially reversed by miR-149-5p antagomir (Fig. 4A). In addition, the migration and invasion of OSCC cells was significantly inhibited following knockdown of DLEU1, and this was also reversed by the miR-149-5p antagomir (Fig. 4B). Meanwhile, the expression levels of Bax and cleaved caspase-3 in OSCC cells were significantly increased by DLEU1 siRNA2 (Fig. 4C-E). By contrast, DLEU1 siRNA2 significantly inhibited the expression of Bcl-2 in OSCC cells (Fig. 4C and F). However, the effects of DLEU1 knockdown on these three proteins were significantly reversed by miR-149-5p antagomir (Fig. 4C-F). Knockdown of DLEU1 inhibited the tumorigenesis of OSCC via mediation of miR-149-5p.
Figure 4.
Silencing of DLEU1 inhibits the tumorigenesis of OSCC via sponging miR-149-5p. (A) The apoptosis of Cal-27 cells was detected via flow cytometry. (B) The migration and invasion of Cal-27 cells were determined using Transwell assays (magnification, ×400). (C) The protein expression levels of Bax, cleaved caspase-3 and Bcl-2 in Cal-27 cells were detected via western blotting. The relative expression levels of (D) Bax, (E) cleaved caspase-3 and (F) Bcl-2 were semi-quantified via normalization to β-actin. **P<0.01 vs. control; ##P<0.01 vs. DLEU1 siRNA2. DLEU1, deleted in lymphocytic leukemia 1; OSCC, oral squamous cell carcinoma; NC, negative control; siRNA, small interfering RNA; miR, microRNA.
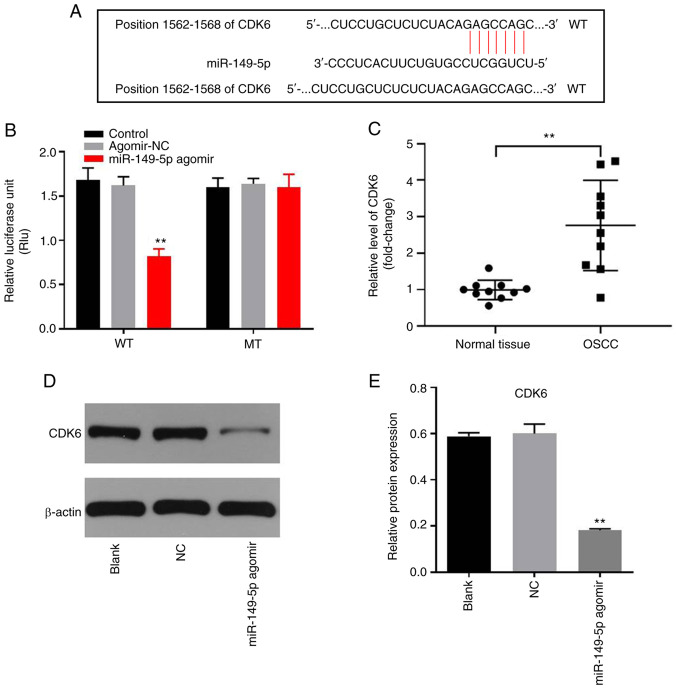
miR-149-5p directly targets CDK6 in OSCC cells
In order to identify the downstream target of miR-149-5p, TargetScan, miRDB and miRWalk were used in the present study. As presented in Fig. 5A, CDK6 was demonstrated to be the potential target of miR-149-5p using these three online tools. The co-transfection of the WT-CDK6 vector with miR-149-5p agomir significantly decreased the luciferase activity when compared with the MT-CDK6 vector (Fig. 5B). In addition, the level of CDK6 was significantly higher in OSCC tissues, compared with adjacent normal tissues (Fig. 5C). Furthermore, the protein expression of CDK6 in OSCC cells was significantly inhibited by miR-149-5p agomir (Fig. 5D and E). In summary, miR-149-5p directly targeted CDK6 in OSCC cells.
Figure 5.
miR-149-5p directly targets CDK6. (A) Gene structure of CDK6 at the position of 1562–1568 indicated the binding site of miR-149-5p in its 3′UTR. (B) The relative luciferase activity in Cal-27 cells after co-transfection with miR-149-5p and WT/MT CDK6 3′UTR was measured using a dual-luciferase reporter assay. (C) The expression of CDK6 in OSCC or adjacent normal tissues was detected via reverse transcription-quantitative PCR. (D and E) The protein expression of CDK6 in Cal-27 cells was measured via western blotting. **P<0.01 vs. control. UTR, untranslated region; WT, wild-type; MT, mutant; OSCC, oral squamous cell carcinoma; NC, negative control; miR, microRNA.
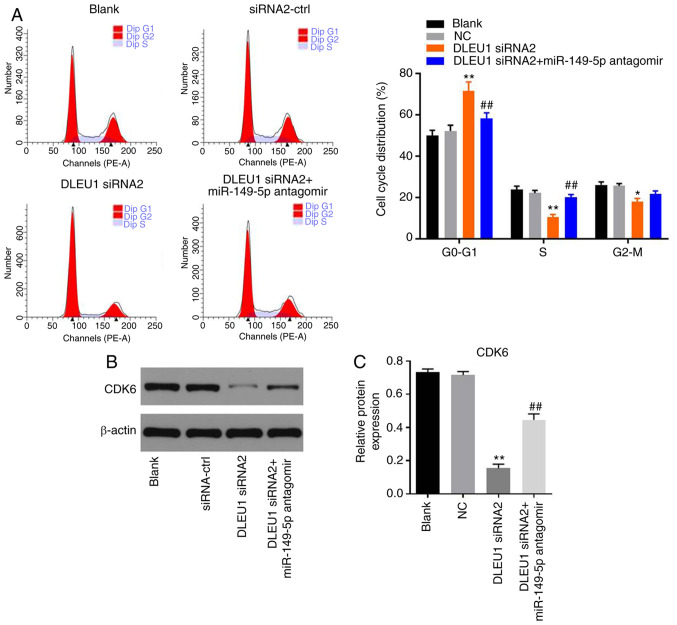
Knockdown of DLEU1 induces G1 arrest in OSCC cells via mediation of CDK6
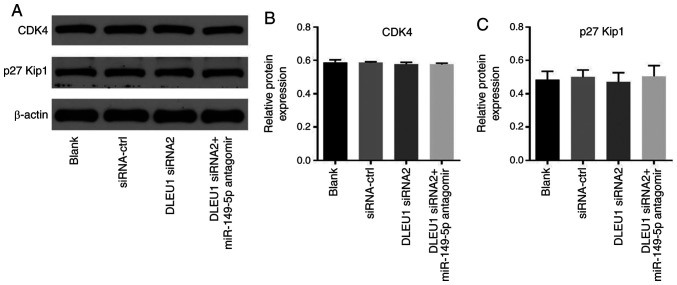
In order to determine cell cycle distribution, flow cytometry was performed in the present study. The data revealed that DLEU1 knockdown significantly induced G1 cell cycle arrest in OSCC cells, which was partially rescued by miR-149-5p antagomir (Fig. 6A). In addition, the expression of CDK6 in OSCC cells was significantly inhibited by DLEU1 siRNA2, while this phenomenon was partially reversed by miR-149-5p antagomir (Fig. 6B and C). Meanwhile, the expression levels of CDK4 and p27 Kip1 in OSCC cells were not affected by the knockdown of DLEU1 or the addition of miR-149-5p antagomir (Fig. 7A-C). In summary, knockdown of DLEU1 induced G1 cell cycle arrest in OSCC cells via mediation of CDK6.
Figure 6.
Silencing of DLEU1 induces G1 arrest in Cal-27 cells. (A) The cell cycle distribution was determined by flow cytometry. (B and C) The protein expression of CDK6 in Cal-27 cells was detected via western blotting. The relative expression was semi-quantified via normalization to β-actin. *P<0.05, **P<0.01 vs. control; ##P<0.01 vs. DLEU1 siRNA2. DLEU1, deleted in lymphocytic leukemia 1; NC, negative control; siRNA, small interfering RNA; miR, microRNA.
Figure 7.
Silencing of DLEU1 did not affect the expression of CDK4 and p27 Kip1 in OSCC cells. (A) The protein expressions levels of CDK4 and p27 Kip1 in OSCC cells were detected via western blotting. The relative expression levels of (B) CDK4 and (C) p27 Kip1 were semi-quantified via normalization to β-actin. OSCC, oral squamous cell carcinoma; DLEU1, deleted in lymphocytic leukemia 1; siRNA, small interfering RNA; miR, microRNA; p27 Kip1, cyclin-dependent kinase inhibitor p27.
Discussion
Several lncRNAs have been found to be up- or downregulated in OSCC, and dysregulated lncRNAs have been implicated to play an important role during the tumorigenesis of OSCC (24–26). A previous study indicated that the expression of DLEU1 was upregulated in OSCC tissues (14). The present study further investigated the function of DLEU1 in OSCC, confirming that DLEU1 could act as a key biomarker in OSCC. In addition, Lin et al (27) reported that upregulation of DLEU1 could promote the tumorigenesis of esophageal squamous cell carcinoma. Miao et al (24) revealed that DLEU1 promoted the progression of clear cell renal cell carcinoma. Based on these data, it can be suggested that DLEU1 could function as an oncogene in certain tumor types. According to Liu et al (28), Sp1 transcription factor (SP1) could bind to the promoter region of DLEU1 to activate DLEU1 transcription. In addition, SP1 could promote the tumorigenesis of OSCC (29). Thus, DLEU1 may be upregulated by SP1 in OSCC.
It was previously reported that miR-149-5p could act as a suppressor in certain types of malignancies (30,31). Luo et al (19) revealed that miR-149-5p could regulate cell proliferation and tumor metastasis in OSCC by targeting TGFβ2. Consistently, the results of the present study indicated that miR-149-5p antagomir could inhibit the effect of DLEU1 knockdown on the proliferation of OSCC cells, suggesting that DLEU1 could sponge miR-149-5p in OSCC. However, Chen et al (32) indicated that knockdown of DLEU1 inhibited the proliferation of osteosarcoma cells via sponging miR-671-5p. This difference may be due to the different tumor type.
miRNAs can mediate cancer tumorigenesis via the repression of target gene expression (33). In the present study, it was revealed that miR-149-5p could directly target CDK6 in OSCC cells. CDK6 is a regulator of the cell cycle that can prevent G0/G1 arrest in non-small cell lung, liver and gastric cancer cell lines (34–36). In addition, CDK6 has a specific oncogenic role in a variety of tumors (32). Li et al (33) revealed that Tanshinone IIA promoted cardiac differentiation and enhanced cell motility via mediation of CDK6; Liu et al (34) confirmed that CDK6 was directly targeted by miR-149-5p in lung cancer. Consistently, the results of the present study suggested that DLEU1 siRNA inhibited the tumorigenesis of OSCC via indirectly targeting CDK6. Meanwhile, a previous report revealed that knockdown of DLEU1 could induce G1 phase cell cycle arrest in gastric cancer cells, and DLEU1 could mediate the expression of KLF2 (37). The present study revealed that silencing of DLEU1 could increase G1 phase distribution in OSCC cells. KLF2 is known to act as a key regulator in G1 phase distribution (38,39). This similar function between CDK6 and KLF2 may contribute to the consistent data between the present study and the previous report. On the other hand, knockdown of DLEU1 could inhibit the invasion of OSCC cells, while miR-149-5p antagomir reversed this phenomenon. In addition, miR-149-5p directly targeted CDK6. Thus, DLEU1 knockdown may inhibit the invasion of OSCC cells via indirectly targeting CDK6. According to Chen et al (40), overexpression of CDK6 relieved the inhibitory action of miR-770-5p upregulation on cancer cell invasion. Thus, the present study was consistent with this previous research.
There are some limitations in this research, which are as follows: i) The underlying mechanism by which DLEU1 is upregulated in OSCC is unknown; ii) the detailed effect of CDK6 on cell invasion of OSCC is not clear; iii) the association between DLEU1 and three risk factors (betel quid chewing, smoking and human papillomavirus infections) for OSCC needs to be further investigated; iv) more tissue samples need to be collected in further study; and v) DLEU1 overexpression experiments need to be supplemented. Thus, in future studies, we will improve these limitations. In addition, more underlying mechanisms by which DLEU1 mediates the tumorigenesis of OSCC will be investigated.
In conclusion, silencing of DLEU1 suppressed the tumorigenesis of OSCC via mediation of the miR-149-5p/CDK6 axis in the present study. Therefore, DLEU1 may serve as a target for the treatment of OSCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
TL, HL and WH conceived and supervised the study. TL and WH confirm the authenticity of all the raw data. TL and HL designed the study. TL, HL and YW performed the experiments and analyzed the data. All authors reviewed the results, and read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Institutional Ethical Committee of Affiliated Stomatological Hospital of Guizhou Medical University (Guiyang, China). Written informed consent was obtained from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Andisheh-Tadbir A, Goharian AS, Ranjbar MA. Glypican-3 expression in patients with oral squamous cell carcinoma. J Dent (Shiraz) 2020;21:141–146. doi: 10.30476/DENTJODS.2019.84541.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ueda S, Kanda M, Sato Y, Baba H, Nakamura S, Sawaki K, Shimizu D, Motoyama S, Fujii T, Kodera Y, et al. Chromobox 2 expression predicts prognosis after curative resection of oesophageal squamous cell carcinoma. Cancer Genomics Proteomics. 2020;17:391–400. doi: 10.21873/cgp.20198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsao YC, Chang YJ, Wang CH, Chen L. Discovery of isoplumbagin as a novel NQO1 substrate and anti-cancer quinone. Int J Mol Sci. 2020;21:21. doi: 10.3390/ijms21124378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y, Ci HS, Mao YL, Li JW, Zuo JH. CircRNA_002178 promotes the proliferation and migration of oral squamous cell carcinoma cells by activating the Akt/mTOR pathway. Eur Rev Med Pharmacol Sci. 2020;24:6122–6130. doi: 10.26355/eurrev_202006_21507. [DOI] [PubMed] [Google Scholar]
- 5.Zhu L, Yan D, Chen Y, Chen S, Chen N, Han J. The identification of autophagy-related genes in the prognosis of oral squamous cell carcinoma. Oral Dis. 2020;26:1659–1667. doi: 10.1111/odi.13492. [DOI] [PubMed] [Google Scholar]
- 6.Deng W, Fu J, Wang T, Chen JX, Fu LB, Peng W. Hsa_circRNA_101036 acts as tumor-suppressor in oral squamous cell carcinoma cells via inducing endoplasmic reticulum stress. Eur Rev Med Pharmacol Sci. 2020;24:6111–6121. doi: 10.26355/eurrev_202006_21506. [DOI] [PubMed] [Google Scholar]
- 7.Zhou YM, Yao YL, Liu W, Shen XM, Shi LJ, Wu L. MicroRNA-134 inhibits tumor stem cell migration and invasion in oral squamous cell carcinomas via downregulation of PI3K-Akt signaling pathway by inhibiting LAMC2 expression. Cancer Biomark. 2020;29:51–67. doi: 10.3233/CBM-191362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gissi DB, Gabusi A, Tarsitano A, Asioli S, Rossi R, Marchetti C, Montebugnoli L, Foschini MP, Morandi L. Application of a non-invasive oral brushing procedure based on bisulfite sequencing of a 13-gene panel to study high-risk OSCC patients. Cancer Biomark. 2020;28:499–510. doi: 10.3233/CBM-190422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma YS, Chu KJ, Ling CC, Wu TM, Zhu XC, Liu JB, Yu F, Li ZZ, Wang JH, Gao QX, et al. Long Noncoding RNA OIP5-AS1 promotes the progression of liver hepatocellular carcinoma via regulating the hsa-miR-26a-3p/EPHA2 axis. Mol Ther Nucleic Acids. 2020;21:229–241. doi: 10.1016/j.omtn.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Li A, Mallik S, Luo H, Jia P, Lee DF, Zhao Z. H19, a Long Non-coding RNA, mediates transcription factors and target genes through interference of MicroRNAs in pan-cancer. Mol Ther Nucleic Acids. 2020;21:180–191. doi: 10.1016/j.omtn.2020.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan C, Zhang Z, Bao S, Hou P, Zhou M, Xu C, Sun J. Computational methods and applications for identifying disease-associated lncRNAs as potential biomarkers and therapeutic targets. Mol Ther Nucleic Acids. 2020;21:156–171. doi: 10.1016/j.omtn.2020.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghapanchi J, Ranjbar Z, Mokhtari MJ, Koohpeima F, Derakhshan M, Khademi B, Ghaderi H, Sheikhbahaei S, Aliabadi E. The LncRNA H19 rs217727 polymorphism is associated with oral squamous cell carcinoma susceptibility in iranian population. BioMed Res Int. 2020;2020:1634252. doi: 10.1155/2020/1634252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu K, Jiang Y, Zhou W, Zhang B, Li Y, Xie F, Zhang J, Wang X, Yan M, Xu Q, et al. Long noncoding RNA RC3H2 facilitates cell proliferation and invasion by targeting MicroRNA-101-3p/EZH2 axis in OSCC. Mol Ther Nucleic Acids. 2020;20:97–110. doi: 10.1016/j.omtn.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishiyama K, Maruyama R, Niinuma T, Kai M, Kitajima H, Toyota M, Hatanaka Y, Igarashi T, Kobayashi JI, Ogi K, et al. Screening for long noncoding RNAs associated with oral squamous cell carcinoma reveals the potentially oncogenic actions of DLEU1. Cell Death Dis. 2018;9:826. doi: 10.1038/s41419-018-0893-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saikishore R, Velmurugan P, Ranjithkumar D, Latha R, Sathiamoorthi T, Arun A, Ravi AV, Sivakumar S. The circular RNA-miRNA Axis: A special RNA signature regulatory transcriptome as a potential biomarker for OSCC. Mol Ther Nucleic Acids. 2020;22:352–361. doi: 10.1016/j.omtn.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Padmavathi G, Ramkumar KM. MicroRNA mediated regulation of the major redox homeostasis switch, Nrf2, and its impact on oxidative stress-induced ischemic/reperfusion injury. Arch Biochem Biophys. 2021;698:108725. doi: 10.1016/j.abb.2020.108725. [DOI] [PubMed] [Google Scholar]
- 17.Jia B, Zhang S, Wu S, Zhu Q, Li W. MiR-770 promotes oral squamous cell carcinoma migration and invasion by regulating the Sirt7/Smad4 pathway. IUBMB Life. 2021;73:264–272. doi: 10.1002/iub.2426. [DOI] [PubMed] [Google Scholar]
- 18.Yao Y, Xu Q, Yan L, Jiao Y, Su Q, Li X, Liu C, Zhao F. MiRNA-128 and MiRNA-142 regulate tumorigenesis and EMT in oral squamous cell carcinoma through HOXA10. Cancer Manag Res. 2020;12:9987–9997. doi: 10.2147/CMAR.S250093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo K, He J, Yu D, Açil Y. MiR-149-5p regulates cisplatin chemosensitivity, cell growth, and metastasis of oral squamous cell carcinoma cells by targeting TGFβ2. Int J Clin Exp Pathol. 2019;12:3728–3739. [PMC free article] [PubMed] [Google Scholar]
- 20.Jain A, Kotimoole CN, Ghoshal S, Bakshi J, Chatterjee A, Prasad TS, Pal A. Identification of potential salivary biomarker panels for oral squamous cell carcinoma. Sci Rep. 2021;11:3365. doi: 10.1038/s41598-021-82635-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roudnicky F, Poyet C, Buser L, Saba K, Wild P, Otto VI, Detmar M. Characterization of tumor blood vasculature expression of human invasive baldder cancer by laser capture microdissection and transcriptional profiling. Am J Pathol. 2020;190:1960–1970. doi: 10.1016/j.ajpath.2020.05.020. [DOI] [PubMed] [Google Scholar]
- 24.Miao T, Si Q, Wei Y, Fan R, Wang J, An X. Identification and validation of seven prognostic long non-coding RNAs in oral squamous cell carcinoma. Oncol Lett. 2020;20:939–946. doi: 10.3892/ol.2020.11603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghafouri-Fard S, Dashti S, Taheri M. The role of long non-coding RNA CASC2 in the carcinogenesis process. Biomed Pharmacother. 2020;127:110202. doi: 10.1016/j.biopha.2020.110202. [DOI] [PubMed] [Google Scholar]
- 26.Yin Y, Tan Y, Yao Y, Lu N, Zhang F. SNHG12/miR-326/E2F1 feedback loop facilitates the progression of oral squamous cell carcinoma. Oral Dis. 2020;26:1631–1639. doi: 10.1111/odi.13458. [DOI] [PubMed] [Google Scholar]
- 27.Lin P, Li Q, Lv X, Qu J, Wang D, Li A, Jiang G. HMGA1 promotes the development of esophageal squamous cell carcinoma by mediating miR-671-5p/lncRNA DLEU1. Panminerva Med. 2020 Jan 30; doi: 10.23736/S0031-0808.19.03843-6. (Epub ahead of print). doi: 10.23736/S0031-0808.19.03843-6. [DOI] [PubMed] [Google Scholar]
- 28.Liu X, Chen R, Liu L. SP1-DLEU1-miR-4429 feedback loop promotes cell proliferative and anti-apoptotic abilities in human glioblastoma. Biosci Rep. 2019;39:39. doi: 10.1042/BSR20190994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu XB, Wang J, Li K, Fan XN. Sp1 promotes cell migration and invasion in oral squamous cell carcinoma by upregulating Annexin A2 transcription. Mol Cell Probes. 2019;46:101417. doi: 10.1016/j.mcp.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 30.Zhou C, Wang P, Tu M, Huang Y, Xiong F, Wu Y. Long non-coding RNA PART1 promotes proliferation, migration and invasion of hepatocellular carcinoma cells via miR-149-5p/MAP2K1 axis. Cancer Manag Res. 2020;12:3771–3782. doi: 10.2147/CMAR.S246311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian D, Tian M, Ma ZM, Zhang LL, Cui YF, Li JL. Anesthetic propofol epigenetically regulates breast cancer trastuzumab resistance through IL-6/miR-149-5p axis. Sci Rep. 2020;10:8858. doi: 10.1038/s41598-020-65649-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, Zhang C, Wang X. Long noncoding RNA DLEU1 aggravates osteosarcoma carcinogenesis via regulating the miR-671-5p/DDX5 axis. Artif Cells Nanomed Biotechnol. 2019;47:3322–3328. doi: 10.1080/21691401.2019.1648285. [DOI] [PubMed] [Google Scholar]
- 33.Li Y, Shi B, Dong F, Zhu X, Liu B, Liu Y. Long bon-coding RNA DLEU1 promotes cell proliferation, invasion, and confers cisplatin resistance in bladder cancer by regulating the miR-99b/HS3ST3B1 axis. Front Genet. 2019;10:280. doi: 10.3389/fgene.2019.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu L, Chen Y, Li Q, Duan P. lncRNA HNF1A-AS1 modulates non-small cell lung cancer progression by targeting miR-149-5p/Cdk6. J Cell Biochem. 2019;120:18736–18750. doi: 10.1002/jcb.29186. [DOI] [PubMed] [Google Scholar]
- 35.Li K, Wang X, Fan C, Wu C, Li S, Liu H. Tanshinone IIA promotes cardiac differentiation and improves cell motility by modulating the Wnt/β-catenin signaling pathway. Mol Med Rep. 2020;22:1839–1846. doi: 10.3892/mmr.2020.11272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang D, Sun Y, Li W, Ye F, Zhang Y, Guo Y, Zhang DY, Suo J. Antiproliferative effects of the CDK6 inhibitor PD0332991 and its effect on signaling networks in gastric cancer cells. Int J Mol Med. 2018;41:2473–2484. doi: 10.3892/ijmm.2018.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X, Li Z, Liu Z, Xiao J, Yu S, Song Y. Long non-coding RNA DLEU1 predicts poor prognosis of gastric cancer and contributes to cell proliferation by epigenetically suppressing KLF2. Cancer Gene Ther. 2018;25:58–67. doi: 10.1038/s41417-017-0007-9. [DOI] [PubMed] [Google Scholar]
- 38.Zheng S, Qian Z, Jiang F, Ge D, Tang J, Chen H, Yang J, Yao Y, Yan J, Zhao L, et al. CircRNA LRP6 promotes the development of osteosarcoma via negatively regulating KLF2 and APC levels. Am J Transl Res. 2019;11:4126–4138. [PMC free article] [PubMed] [Google Scholar]
- 39.Hu CC, Liang YW, Hu JL, Liu LF, Liang JW, Wang R. LncRNA RUSC1-AS1 promotes the proliferation of breast cancer cells by epigenetic silence of KLF2 and CDKN1A. Eur Rev Med Pharmacol Sci. 2019;23:6602–6611. doi: 10.26355/eurrev_201908_18548. [DOI] [PubMed] [Google Scholar]
- 40.Chen B, Ji F, Wen X, Jin Z. Circular RNA circ_ASAP2 promotes cell viability, migration, and invasion of gastric cancer cells by regulating the miR-770-5p/CDK6 axis. Int J Clin Exp Pathol. 2020;13:2806–2819. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.