Abstract
Intervertebral disc degeneration (IDD) is a common chronic disease characterized by the loss of extracellular matrix (ECM) in the nucleus pulposus (NP). Accumulating evidence has revealed that abnormal expression of microRNAs (miRs) is closely associated with IDD development. The present study aimed to investigate the precise role and possible mechanism underlying the effects of miR-654-5p in the pathogenesis of IDD. NP cells were isolated from patients with IDD. Monodansylcadaverine staining was conducted to reveal cell autophagy, while western blotting was performed to detect the expression of ECM-related proteins in NP cells. Luciferase reporter and RNA immunoprecipitation assays were conducted to identify the binding between RNAs. The results demonstrated that miR-654-5p was significantly upregulated in degenerated NP tissues from patients with IDD and high miR-654-5p expression was positively associated with disc degeneration grade. Functional assays suggested that miR-654-5p facilitated ECM degradation by increasing the expression levels of MMP-3, MMP-9 and MMP-13, as well as decreasing collagen I, collagen II, SOX9 and aggrecan expression by inhibiting autophagy. Furthermore, autophagy-related gene 7 (ATG7) was verified as a direct downstream target gene of miR-654-5p. miR-654-5p could bind to the 3′ untranslated region of ATG7 to inhibit its mRNA expression and further reduce its translation. Notably, ATG7 knockdown abrogated the effects of the miR-654-5p inhibitor on ECM degradation and autophagy regulation. Furthermore, miR-654-5p inhibited autophagy in NP cells by increasing the protein expression levels of phosphorylated (p)-PI3K, p-AKT and p-mTOR in an ATG7-dependent manner. In conclusion, the results of the present study revealed that miR-654-5p may enhance ECM degradation via inhibition of autophagy by targeting ATG7 to activate the PI3K/AKT/mTOR signaling pathway. These findings may provide novel insights into the treatment of IDD.
Keywords: microRNA-654-5p, autophagy-related gene 7, intervertebral disc degeneration, extracellular matrix degradation, autophagy
Introduction
Low back pain (LBP) is a common public health problem; ~80% of the global population experiences LBP, which represents a significant economic and societal burden (1,2). Intervertebral disc degeneration (IDD) is a leading cause of LBP and is characterized by progressive degradation of extracellular matrix (ECM) (3). Current therapies mainly focus on alleviating the clinical symptoms of patients with IDD, and there are substantial barriers to targeting the potential pathological changes in the disc. Therefore, a deeper understanding of the pathogenesis of IDD may help to develop strategies for improving clinical outcomes.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs with a length of 18–22 nucleotides (4). The biological function of miRNAs has been reported in various diseases in recent years, including IDD (5,6). For example, miR-139 has been reported to promote the development of osteoarthritis by facilitating apoptosis of chondrocytes (7). miR-133 was shown to suppress cell proliferation and promote apoptosis in lupus nephritis (8), and it has been suggested that overexpression of miR-154 may contribute to the degradation of ECM in IDD (9). Moreover, miR-654-5p has been reported to be involved in several diseases, such as ovarian cancer, gastric cancer and oral squamous cell carcinoma (10–12). In addition, a recent study revealed that miR-654-5p is significantly upregulated in NP cells from patients with IDD (9). However, the specific role of miR-654-5p in IDD pathology remains to be further investigated.
miRNAs participate in the development of diverse diseases by targeting specific genes (13). For example, miR-128 has been reported to targes PPARγ to promote the progression of Alzheimer's disease (14). In addition, miR-373 has been reported to aggravate renal fibrosis by targeting Sirtuin 1 and regulating NF-κB/MMP-9 signaling (15). miR-129-5p inhibition may contribute to IDD development by accelerating NP cell apoptosis by targeting BMP2 (16). Moreover, increasing evidence has indicated that miR-654-5p is involved in multiple diseases in a similar manner. For example, miR-654-5p has been shown to target epithelial stromal interaction 1 to suppress cell growth and invasion in breast cancer development (17). miR-654-5p may promote cell proliferation and migration by modulating the GRAP/Ras/MAPK signaling pathway in oral squamous cell carcinoma (12). However, to the best of our knowledge, the mechanism underlying the effects of miR-654-5p on IDD development has not yet been revealed.
The present study aimed to investigate the key role of miR-654-5p in IDD development. The results revealed that miR-654-5p promoted ECM degradation by inhibiting autophagy via suppression of autophagy-related gene 7 (ATG7) and activation of the PI3K/AKT/mTOR signaling pathway. These findings may provide information that aids in identifying novel therapeutic targets for IDD.
Materials and methods
Sample collection
A total of 76 NP tissue samples were obtained from patients with IDD who underwent discectomy and 20 healthy control samples were donated by patients with lumbar vertebral fracture who underwent anterior discectomy in Zhongda Hospital of Southeast University between September 2016 and December 2019. Ages of the 76 patients with IDD (males 45; females 31) were between 31–53 years, and the mean age was 42.3 years. Ages of the 20 control participants (males 13; females 7) were between 24–45 years, and the mean age was 33.6 years. Participants who had IDD were excluded from the control group. The grade of IDD was estimated by T2-weighted images following the Pfirrmann classification system (18). The present study was approved by the Ethics Committee of The Affiliated Zhongda Hospital of Southeast University (Nanjing, China; approval no. 2015ZDKYSB014) and followed the guidelines of the Declaration of Helsinki (19). All patients provided written informed consent prior to the operation.
Isolation and culture of degenerated NP cells
After washing twice with PBS, the degenerated NP specimens from patients with IDD were cut into small fragments (1 mm3). Subsequently, the fragments were treated with 0.25% trypsin solution (Sigma-Aldrich; Merck KGaA) for 30 min at 37°C and centrifuged at 1,000 × g for 3 min at 4°C. After centrifugation, the supernatant was discarded, and precipitates were stimulated with 0.2% type II collagenase (Sigma-Aldrich; Merck KGaA) at 37°C for 4 h. After filtration through a 0.45-µm nylon filter membrane, the suspension was collected and the NP cells were resuspended in DMEM/F12 supplemented with 15% FBS (both Gibco; Thermo Fisher Scientific, Inc.) at 37°C in an atmosphere containing 5% CO2. After digestion with 0.25% trypsin solution, the cells were incubated. The culture medium was changed three times a week, and the second-generation cells were collected and used in subsequent experiments.
Cell transfection and treatment
Isolated NP cells were seeded into 6-well plates at a density of 1×105 cells/well before transfection and 2 ml complete tissue culture medium was added to each well. Subsequently, small interfering RNA (siRNA) targeting ATG7 (si-ATG7) was designed and synthesized by Shanghai GenePharma Co., Ltd., and was used to knockdown ATG7. The target sequence for si-ATG7 was 5′-GGAGTCACAGCTCTTCCTT-3′. Scrambled siRNA-negative control (si-NC; Shanghai GenePharma Co., Ltd.) served as the NC for si-ATG7. The sequence for si-NC was 5′-GCACTGAGTAGCTCCTCTT-3′. miR-654-5p mimics and NC mimics, miR-654-5p inhibitor and NC inhibitor, and pcDNA3.1-ATG7 vector for overexpression of ATG7 and its negative control (empty pcDNA3.1 vector) were obtained from Shanghai GenePharma Co., Ltd. The sequence for miR-654-5p mimics was 5′-UGGUGGGCCGCAGAACAUGUGC-3′, and that for scrambled NC mimics was 5′-GAGUAGCCGUGGCUGCUAAGCG-3′. The sequence for miR-654-5p inhibitor was 5′-GCACAUGUUCUGCGGCCCACCA-3′, and that for scrambled NC inhibitor was 5′-CGGUCUCGACACACUCGAUCGC-3′.
miR-654-5p mimics (10 nM) and NC mimics (10 nM), miR-654-5p inhibitor (10 nM), NC inhibitor (10 nM), si-ATG7 (20 nM) and si-NC (20 nM) were transfected into NP cells (1×105) using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 48 h. The time between transfection and subsequent experimentation was 48 h. Subsequently, transfected cells were cultured in DMEM/F12 containing 10% FBS. Moreover, to interfere with autophagy, cells were treated with rapamycin (Rap; 10 nM; autophagy activator; Sigma-Aldrich; Merck KGaA) or 3-methyladenine (3-MA; 10 mM; autophagy inhibitor; Sigma-Aldrich; Merck KGaA) at 37°C for 6 h before transfection.
RNA isolation and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted from NP tissues or cells using TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. The RNA concentration was measured using a NanoDrop ND-1000 spectrophotometer (NanoDrop; Thermo Fisher Scientific, Inc.) and agarose gel electrophoresis was used to determine RNA integrity. Subsequently, RT was conducted using a miRcute miRNA RT kit (Tiangen Biotech Co., Ltd.) or a RevertAid RT kit (Thermo Fisher Scientific). The RT reaction conditions were as follows: 37°C for 60 min, followed by 85°C for 5 min and 4°C. All qPCR amplification reactions were performed in triplicate using the 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.) with a final volume of 20 µl 2X SYBR Green mix (Thermo Fisher Scientific). The thermocycling conditions were as follows: Initial denaturation at 95°C for 2 min, followed by 40 cycles of denaturation at 95°C for 15 sec, annealing at 59°C for 20 sec and elongation at 72°C for 20 sec; and a final extension at 72°C for 10 min. GAPDH served as an internal control for mRNA analysis and U6 served as an internal control for miR-654-5p analysis. The relative expression levels were calculated using the 2−ΔΔCq method (20). Primer sequences used for PCR were provided in Table I.
Table I.
Relative primer sequences.
| Targets | Sequences |
|---|---|
| miR-654-5p | F: 5′-UGGUGGGCCGCAGAACAUGU-3′ |
| R: 5′-CTCTACAGCTATATTGCCAGCCAC-3′ | |
| U6 | F: 5′-CTCGCTTCGGCAGCACA-3′ |
| R: 5′-AACGCTTCACGAATTTGCGT-3′ | |
| Collagen I | F: 5′-AAAGATGGAGAGGCTGGAG-3′ |
| R: 5′-ATCACCCTTAGCACCATCG-3′ | |
| Collagen II | F: 5′-CAAGGAGACAGAGGAGAAGC-3′ |
| R: 5′-CTTGAGGACCCTGGATTCC-3′ | |
| SOX9 | F: 5′-CTCTGGAGACTTCTGAACGA-3′ |
| R: 5′-ACTTGTAATCCGGGTGGTC-3′ | |
| MMP-3 | F: 5′-GGACAAATACTGGAGATTTGATGAG-3′ |
| R: 5′-CCCTGGAAAGTCTTCAGCT-3′ | |
| MMP-9 | F: 5′-TACTGTGCCTTTGAGTCCG-3′ |
| R: 5′-GAATCGCCAGTACTTCCCA-3′ | |
| MMP-13 | F: 5′-CTGGGCCAAATTATGGAGGA-3′ |
| R: 5′-GAAACAAGTTGTAGCCTTTGGA-3′ | |
| ATG3 | F: 5′-TCACAACACAGGTATTACAGGA-3′ |
| R: 5′-GCTGAGCAATCTTGAAGCC-3′ | |
| ATG5 | F: 5′-GGAAACTCATGGAATATCCTGC-3′ |
| R: 5′-GGTCTTTCAGTCGTTGTCTG-3′ | |
| ATG7 | F: 5′-GGAGUCACAGCUCUUCCUUdTdT-3′ |
| R: 5′-AAGGAAGAGCUGUGACUCCTdTd-3′ | |
| ATG16L1 | F: 5′-CCAATCGGCTTAATGCAGAG-3′ |
| R: 5′-TCATCATCCTGTTCGACTGG-3′ | |
| Beclin 1 | F: 5′-GAACTACAAACGCTGTTTGGA-3′ |
| R: 5′-AGCTCCTTTAGCTCCATCTG-3′ | |
| ULK1 | F: 5′-CCGAGAGGCTCATCTTCAG-3′ |
| R: 5′-CTGGAACATCTCGTCCAGG-3′ | |
| GAPDH | F: 5′-GCATCCTGGGCTACACTG-3′ |
| R: 5′-TGGTCGTTGAGGGCAAT-3′ |
miR, microRNA; MMP, matrix metalloproteinase; ATG, autophagy-related gene; ULK1, unc-51 like autophagy activating kinase 1. F, forward; R, reverse.
Luciferase reporter assay
The binding site of miR-654-5p on ATG7 was identified using starBase (http://starbase.sysu.edu.cn/index.php). A wild-type (Wt) or mutant (Mut) ATG7 fragment was subcloned into a pmirGLO vector (Promega Corporation), The mutation gene sequence was synthesized by Shanghai GenePharma Co., Ltd., and NP cells at a density of 1×105 cells/well were then cotransfected with miR-654-5p mimics (10 nM) or NC mimics (10 nM) and Wt or Mut vectors (1 µg) using Lipofectamine 2000 at 37°C for 48 h. After 24 h, the Dual Luciferase Reporter Assay system (Promega Corporation) was employed to detect luciferase activity. Relative luciferase activity was calculated as the ratio of firefly luciferase activity to Renilla luciferase activity.
RNA immunoprecipitation (RIP) assay
The RIP assay was performed using an EZ-Magna RIP Kit (EMD Millipore) according to the manufacturer's instructions. NP cell lysates were lysed with RIPA buffer (Beyotime Institute of Biotechnology) for 5 min at 4°C. Antibodies including anti-Argonaute 2 (anti-Ago2; cat. no. ab186733; 1:50; Abcam) and anti-Immunoglobulin G (anti-IgG; cat. no. 12-370; 1:100; EMD Millipore) were incubated with protein A/G magnetic beads (Pierce; Thermo Fisher Scientific, Inc.) for 1 h at 4°C. Then, cell lysate was mixed with the beads to incubate for 4 h at 4°C. Beads were washed twice using PBS buffer (Sangon Biotech Co., Ltd.), and the mixture was centrifuged at 2,500 × g for 10 min at 4°C. RNA was purified with 150 µl proteinase K buffer (Roche Diagnostics) and extracted using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.), and the immunoprecipitated RNA was detected via RT-qPCR as aforementioned.
Western blot analysis
Total proteins were extracted from NP cells with radioimmunoprecipitation assay lysis buffer (Beyotime Institute of Biotechnology) containing phenylmethylsulfonyl fluoride. A bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology) was used to evaluate protein concentration. Subsequently, equal amounts (50 µg per lane) of protein were separated by SDS-PAGE on 10% gels. After electrophoresis, proteins were transferred onto PVDF membranes (EMD Millipore), which were blocked with 5% nonfat milk at room temperature for 2 h. Membranes were then incubated with primary antibodies at 4°C overnight, followed by further incubation with horseradish peroxidase-conjugated goat-anti rabbit secondary antibody (cat. no. ab205718; 1:2,000; Abcam) at room temperature for 1 h. The following primary antibodies provided by Abcam were used: β-actin (cat. no. ab8227; 1:1,000), collagen I (cat. no. ab34710; 1:1,000), collagen II (cat. no. ab188570; 1:1,000), aggrecan (cat. no. ab36861; 1:2,000), SOX9 (cat. no. ab185966; 1:1,000), MMP-3 (cat. no. ab53015; 1:500), MMP-9 (cat. no. ab38898; 1:1,000), MMP-13 (cat. no. ab39012; 1:3,000), Beclin-1 (cat. no. ab62557; 1:2,000), ATG7 (cat. no. ab53255; 1:2,000), ATG12 (cat. no. ab109491; 1:1,000), ATG13 (cat. no. ab105392; 1:2,000), LC3 (cat. no. ab192890; 1:2,000), phosphorylated (p)-PI3K (cat. no. ab182651; 1:500), p-AKT (cat. no. ab38449; 1:500), p-mTOR (cat. no. ab109268; 1:1,000), PI3K (cat. no. ab191606; 1:1,000), AKT (cat. no. ab182729; 1:5,000) and mTOR (cat. no. ab134903; 1:10,000). The protein expression levels were normalized to those of β-actin. Finally, the bands were visualized using an Enhanced Chemiluminescence Substrate kit (EMD Millipore) and analyzed using a Quantity One software (version 4.62; Bio-Rad Laboratories, Inc.).
Monodansylcadaverine (MDC) staining
Transfected NP cells at the concentration of 106/ml were exposed to MDC solution (0.1 mM; Cayman Chemical Company) in 60-mm dishes at 37°C for 1 h. After washing three times with PBS, cells were fixed with 4% paraformaldehyde at room temperature for 15 min. To evaluate autophagic vacuoles (acidic granular vacuoles), cells were observed using a fluorescence microscope (Olympus Corporation) under an ultraviolet filter.
Statistical analysis
All experiments were independently conducted three times. Data are presented as the mean ± standard deviation. All statistical analyses were conducted using SPSS 22.0 software (IBM Corp.) and graphs were drawn with GraphPad software (version 8; GraphPad Software, Inc.). All data were assessed for normal distribution (Shapiro-Wilk test) and homogeneity of variance (Bartlett's test). All results were corrected for multiple comparisons using the false discovery rate method (21). Independent Student's t-test was used to analyze the differences between two groups. One-way analysis of variance followed by post hoc Dunnett's test (for comparisons with one control) and Tukey's test (for comparisons among various groups) was employed to analyze the differences among more than two groups. Spearman's correlation analysis was applied to analyze the correlation between miR-654-5p expression levels and Pfirrmann scores of patients with IDD. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-654-5p is upregulated in IDD
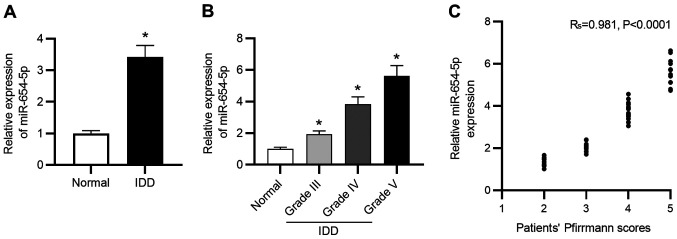
To explore the role of miR-654-5p in the progression of IDD, RT-qPCR was conducted to measure miR-654-5p expression in NP tissues. As shown in Fig. 1A, miR-654-5p expression levels were significantly elevated in degenerated NP tissues compared with those in healthy control tissues. Moreover, the expression levels of miR-654-5p were gradually elevated with an increased degree of disc degeneration (Fig. 1B). In addition, as shown in Fig. 1C, miR-654-5p expression was positively correlated with Pfirrmann scores in patients with IDD.
Figure 1.
miR-654-5p is upregulated in IDD. (A) Expression levels of miR-654-5p were detected in nucleus pulposus tissues from healthy controls and patients with IDD by reverse transcription-quantitative PCR. (B) niR-654-5p expression was compared between tissues from healthy controls and patients with IDD with various Pfirrmann grades. (C) Correlation of miR-654-5p expression with Pfirrmann score was analyzed. *P<0.05 vs. Normal. IDD, intervertebral disc degeneration; miR-654-5p, microRNA-654-5p.
miR-654-5p contributes to ECM degradation in NP cells
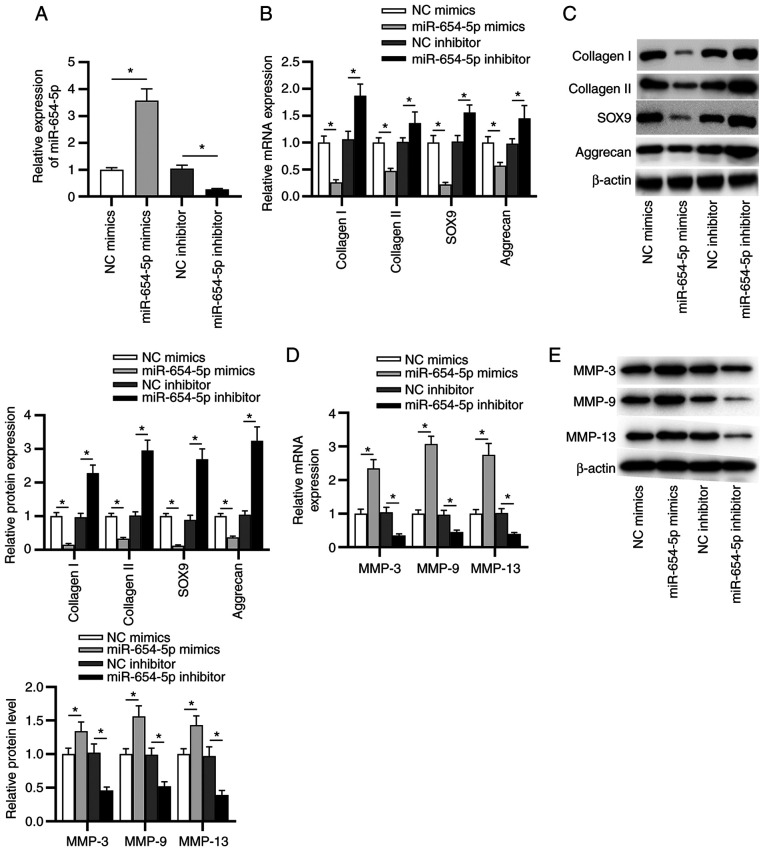
To further investigate the effects of miR-654-5p on the progression of IDD, the subsequent experiments were performed. As shown in Fig. 2A, the transfection efficiency of miR-654-5p mimics or the miR-654-5p inhibitor was confirmed by RT-qPCR. Next, the expression levels of type I/II collagen, SOX9 and aggrecan were detected in transfected NP cells. The results suggested that the mRNA and protein expression levels of collagen I, collagen II, SOX9 and aggrecan were significantly reduced by miR-654-5p mimics, and were elevated by the miR-654-5p inhibitor (Fig. 2B and C). In addition, the expression levels of MMP-3, MMP-9 and MMP-13 were detected in transfected NP cells. As shown in Fig. 2D and E, the mRNA and protein expression levels of MMP-3, MMP-9 and MMP-13 were significantly increased in NP cells transfected with miR-654-5p mimics, whereas an opposite trend was revealed in miR-654-5p inhibitor-transfected NP cells.
Figure 2.
miR-654-5p contributes to extracellular matrix degradation in NP cells. (A) RT-qPCR was used to determine the transfection efficiency of miR-654-5p mimics or an miR-654-5p inhibitor in NP cells. (B) RT-qPCR and (C) western blotting were carried out to measure the mRNA and protein expression levels of collagen I, collagen II, SOX9 and aggrecan, respectively. (D) mRNA and (E) protein expression levels of MMP-3, MMP-9 and MMP-13 were evaluated by RT-qPCR and western blot analysis, respectively. *P<0.05. miR-654-5p, microRNA-654-5p; NC, negative control; NP, nucleus pulposus; RT-qPCR, reverse transcription-quantitative PCR.
miR-654-5p suppresses autophagy in IDD
First, MDC staining was employed to detect autophagic vacuoles. As displayed in Fig. 3A, the accumulation of MDC-labeled vacuoles in the cytoplasm of NP cells was inhibited by miR-654-5p mimics and promoted by miR-654-5p inhibitor. Beclin-1, ATG12, ATG13 and LC3 (subtypes: LC3-I and LC3-II) have been widely identified as autophagy markers; therefore, western blot analysis was performed to examine the expression levels of these proteins in transfected NP cells. It was revealed that miR-654-5p mimics reduced the protein expression levels of Beclin-1, ATG12, ATG13 and the LC3-II/LC3-I ratio; however, silencing miR-654-5p exerted the opposite effects (Fig. 3B). These data indicated that miR-654-5p may inhibit autophagy in IDD.
Figure 3.
miR-654-5p suppresses autophagy in intervertebral disc degeneration. (A) Monodansylcadaverine staining was performed to observe autophagic vacuoles in transfected nucleus pulposus cells. Scale bar, 20 µm. (B) Protein expression levels of LC3-II/LC3-I, ATG12, ATG13 and Beclin-1 were assessed by western blotting. *P<0.05. ATG, autophagy-related gene; miR-654-5p, microRNA-654-5p; NC, negative control.
Autophagy is involved in miR-654-5p-induced ECM degradation in NP cells
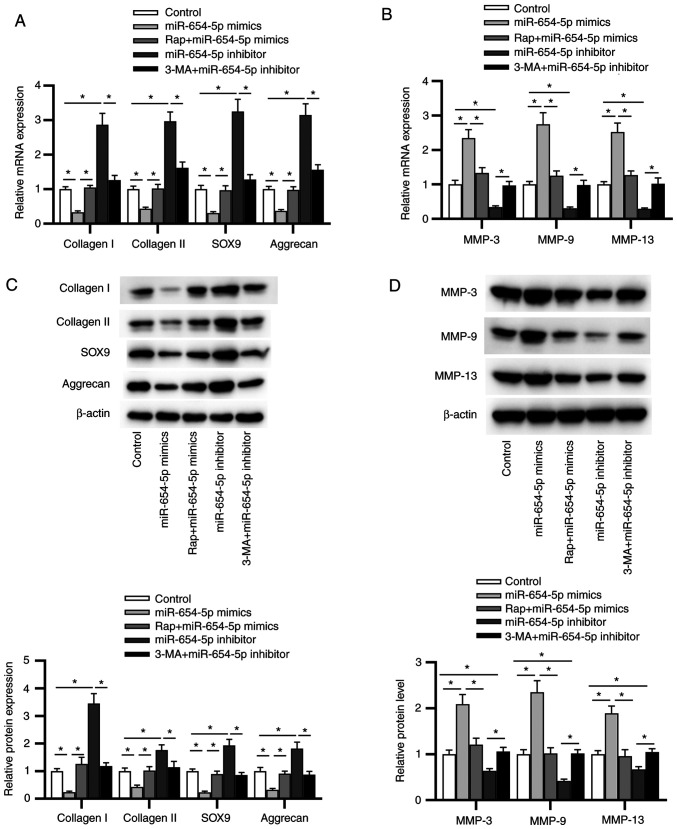
As autophagy is a crucial mechanism regulating ECM metabolism, it was hypothesized that miR-654-5p may enhance the degradation of ECM by inhibiting autophagy. To verify this hypothesis, NP cells were cultured with Rap or 3-MA to activate or antagonize autophagy, and were then transfected with miR-654-5p mimics or miR-654-5p inhibitor, respectively. RT-qPCR analysis revealed that the decreased expression levels of collagen I, collagen II, SOX9 and aggrecan, and the increased expression levels of MMP-3, MMP-9 and MMP-13 induced by miR-654-5p mimics transfection were reversed by Rap pretreatment. Moreover, 3-MA abrogated the increase in collagen I, collagen II, SOX9 and aggrecan expression, as well as the decline in MMP-3, MMP-9 and MMP-13 expression induced by the miR-654-5p inhibitor (Fig. 4A and B). Furthermore, similar results were obtained by western blot analysis (Fig. 4C and D). Overall, miR-654-5p-induced ECM degradation may be sustained by the inhibition of autophagy in IDD.
Figure 4.
Autophagy is involved in miR-654-5p-induced extracellular matrix degradation in NP cells. NP cells were treated with Rap or 3-MA, followed by transfection with miR-654-5p mimics or an miR-654-5p inhibitor. mRNA expression of levels of (A) collagen I, collagen II, SOX9 and aggrecan, and (B) MMP-3, MMP-9 and MMP-13 were detected by reverse transcription-quantitative PCR. Western blot analysis was used to measure the protein expression levels of (C) collagen I, collagen II, SOX9 and aggrecan, and (D) MMP-3, MMP-9 and MMP-13. *P<0.05. 3-MA, 3-methyladenine; miR-654-5p, microRNA-654-5p; NC, negative control; NP, nucleus pulposus; Rap, rapamycin.
ATG7 is a direct downstream target gene of miR-654-5p
The present study conducted RT-qPCR to assess the expression of autophagy-related genes in NP cells transfected with miR-654-5p mimics. The results revealed that the miR-654-5p mimics significantly reduced the expression levels of ATG7, ATG16L1 and unc-51 like autophagy activating kinase 1, and ATG7 expression exhibited the most obvious decline (Fig. 5A). Moreover, ATG7 was markedly downregulated in NP tissues obtained from patients with IDD compared with that in tissues obtained from the normal control group (Fig. 5B). Therefore, ATG7 was selected for further study and it was hypothesized that miR-654-5p might interact with ATG7 in IDD development. To test this hypothesis, starBase was used to predict the potential targets of miR-654-5p and it was revealed that a binding site exists between miR-654-5p and the ATG7 3′ untranslated region. Subsequently, a luciferase reporter assay revealed that miR-654-5p significantly attenuated the luciferase activity of pmirGLO-ATG7-Wt vectors but not pmirGLO-ATG7-Mut vectors (Fig. 5C). In addition, miR-654-5p and ATG7 were immunoprecipitated by anti-Ago2 antibodies but not anti-IgG antibodies (Fig. 5D). Furthermore, miR-654-5p mimics significantly reduced the mRNA and protein expression levels of ATG7 (Fig. 5E). Collectively, these data indicated that miR-654-5p may directly target ATG7.
Figure 5.
ATG7 is a direct downstream target gene of miR-654-5p. (A) mRNA expression levels of autophagy-related proteins in NP cells transfected with miR-654-5p mimics. *P<0.05 vs. NC mimics. (B) RT-qPCR was carried out to detect the expression levels of ATG7, ATG16L1 and ULK1. *P<0.05 vs. Normal. (C) A luciferase reporter assay was performed in NP cells cotransfected with miR-654-5p mimics and pmirGLO-ATG7-Wt vectors or pmirGLO-ATG7-Mut vectors. *P<0.05 vs. NC mimics. (D) RNA immunoprecipitation assays were used to further confirm the interaction between miR-654-5p and ATG7. *P<0.05 vs. IgG. (E) mRNA and protein expression levels of ATG7 were detected by RT-qPCR and western blotting in miR-654-5p mimic-transfected NP cells, respectively. *P<0.05 vs. NC mimics. ATG, autophagy-related gene; IDD, intervertebral disc degeneration; miR-654-5p, microRNA-654-5p; Mut, mutant; NC, negative control; NP, nucleus pulposus; RT-qPCR, reverse transcription-quantitative PCR; Wt, wild-type; ULK1, unc-51 like autophagy activating kinase 1.
miR-654-5p inhibits autophagy by targeting ATG7
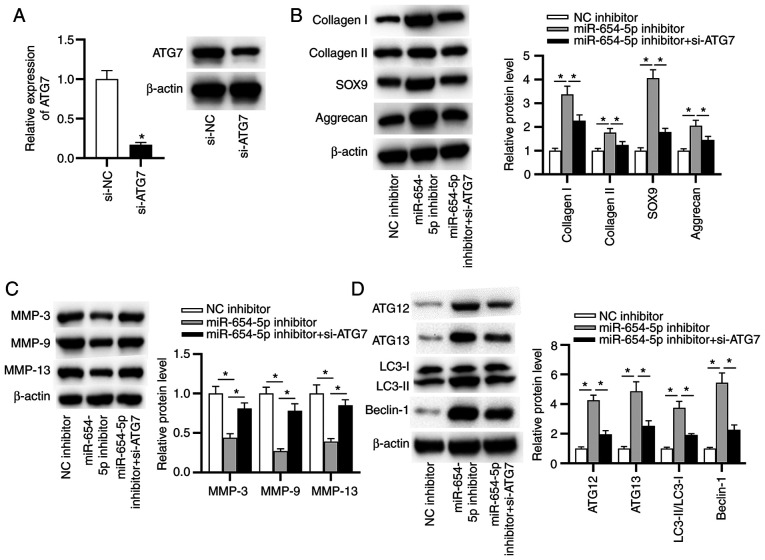
To determine whether miR-654-5p generates its effects on IDD by targeting ATG7, follow-up rescue assays were performed. First, the knockdown efficiency of ATG7 was assessed by RT-qPCR. The results indicated that the mRNA and protein expression levels of ATG7 were significantly decreased in NP cells transfected with si-ATG7 (Fig. 6A). Notably, ATG7 knockdown counteracted the inhibitory effects of the miR-654-5p inhibitor on ECM degradation, including the reduced protein expression levels of MMP-3, MMP-9 and MMP-13, as well as the elevated protein expression levels of collagen I, collagen II, SOX9 and aggrecan (Fig. 6B and C). Moreover, knockdown of ATG7 rescued the miR-654-5p inhibitor-mediated promotion of autophagy (Fig. 6D). In summary, miR-654-5p facilitated ECM degradation by inhibiting autophagy by modulating ATG7.
Figure 6.
miR-654-5p inhibits autophagy by targeting ATG7. (A) Knockdown efficiency of ATG7 was determined by reverse transcription-quantitative PCR. *P<0.05 vs. si-NC. Western blot analysis was used to measure the protein expression levels of (B) collagen I, collagen II, SOX9 and aggrecan, (C) MMP-3, MMP-9 and MMP-13, and (D) LC3-II/LC3-I, ATG12, ATG13 and Beclin-1 in transfected nucleus pulposus cells. *P<0.05. ATG, autophagy-related gene; miR-654-5p, microRNA-654-5p; NC, negative control; si, small interfering.
miR-654-5p activates the PI3K/AKT/mTOR signaling pathway via ATG7 in IDD
As shown in Fig. 7A, the protein expression levels of p-PI3K, p-AKT and p-mTOR were significantly elevated by miR-654-5p mimics, and significantly reduced by the miR-654-5p inhibitor. However, no significant difference was detected in the total amount of PI3K, AKT and mTOR protein among the control group and other groups. Moreover, transfection with a pcDNA3.1-ATG7 vector effectively increased ATG7 expression in NP cells (Fig. 7B). Upregulation of ATG7 reversed the effects of miR-654-5p mimics on the P13K/AKT/mTOR signaling pathway (Fig. 7C). These findings suggested that miR-654-5p may suppress autophagy by activation of the P13K/AKT/mTOR signaling pathway.
Figure 7.
miR-654-5p regulates autophagy via the PI3K/AKT/mTOR signaling pathway in IDD. (A) Effects of miR-654-5p mimics and an miR-654-5p inhibitor on the protein expression levels of p-PI3K, p-AKT and p-mTOR were measured by western blotting. *P<0.05. (B) mRNA expression levels of ATG7 in NP cells transfected with pcDNA3.1-ATG7 were evaluated by reverse transcription-quantitative PCR. *P<0.05 vs. Empty pcDNA3.1. (C) Transfection with the pcDNA3.1-ATG7 vector rescued the effects of miR-654-5p on the PI3K/AKT/mTOR pathway, as detected by western blotting. *P<0.05. ATG, autophagy-related gene; miR-654-5p, microRNA-654-5p; NC, negative control; p, phosphorylated.
Discussion
IDD is a common cause of LBP (22). It has been demonstrated that the etiology of IDD involves several complex mechanisms, including genetic, developmental and biochemical aspects (23). Although IDD therapy has been improved in recent decades, there is still much work to be done to achieve effective treatment.
It has been reported that miR-654-5p may have an important role in several diseases, including ovarian cancer, gastric cancer and oral squamous cell carcinoma (11,12,24). In particular, miR-654-5p has been identified to be upregulated in NP cells from patients with IDD (9). Consistent with a previous study, the present study revealed that miR-654-5p exhibited high expression levels in degenerated NP tissues and that elevated miR-654-5p levels were closely associated with exacerbation of IDD.
Increasing evidence has indicated that early degenerative changes suggestive of IDD appear in NP cells, and the main characteristic of IDD is progressive degradation of ECM macromolecules, of which collagen II and aggrecan show significantly decreased expression levels (25). MMPs are the main enzymes that promote the cleavage of collagen II and aggrecan (3). MMP-3, MMP-9 and MMP-13 are members of the MMP family proteins and are well known to be highly expressed in IDD (26). Moreover, reduced MMP-3, MMP-9 and MMP-13 expression can facilitate ECM repair and disc regeneration (26). Therefore, a deeper understanding of ECM homeostasis is helpful for developing novel therapeutic approaches for IDD. The present study revealed that miR-654-5p contributed to ECM degradation by decreasing collagen II and aggrecan levels, as well as increasing the expression levels of MMP-3, MMP-9 and MMP-13 in NP cells.
Autophagy is a homeostatic process that participates in the degradation and digestion of intracellular components by lysosomes (27). Compared with that in healthy controls, the number of autophagosomes has been shown to be significantly decreased in human degenerative NP cells (28). In addition, the activation of autophagy has been found to suppress MMP-3 expression in NP cells treated with TNF-α, suggesting that autophagy may promote disc ECM anabolism (29,30). Furthermore, previous studies have reported that miRNAs may function as regulators of autophagy in various diseases, including IDD (31). For example, inhibition of miR-20 promoted chondrocyte proliferation and autophagy in osteoarthritis (32). In addition, miR-889 maintained mycobacterial survival by suppressing autophagy in patients with latent tuberculosis infection (33). miR-21 may contribute to ECM degradation by inhibiting autophagy via the PTEN/AKT/mTOR signaling pathway in IDD (34). Similarly, the present findings indicated that autophagy is involved in miR-654-5p-induced ECM degradation in IDD.
ATG7, in conjunction with various signaling pathways, serves an important role in autophagosome formation and vesicle progression (35). Recently, a great number of miRNAs have been shown to suppress the autophagic process by targeting ATG7. For example, miR-96-5p suppressed autophagy to inhibit hepatic stellate cell activation by targeting ATG7 (36). Furthermore, miR-210 inhibited autophagy by targeting ATG7 in IDD (37). Notably, in the present study, ATG7 was found to be directly targeted by miR-654-5p. Moreover, the present findings revealed that ATG7 knockdown significantly reversed the effects of the miR-654-5p inhibitor on ECM degradation and autophagy.
The PI3K/AKT/mTOR signaling pathway has been reported to play a crucial role in regulating autophagy in various diseases. For example, miR-129-5p inhibited autophagy by targeting ATG14 and activated the PI3K/AKT/mTOR signaling pathway in ischemic heart disease (38). miR-20 suppressed chondrocyte proliferation and autophagy by regulating the PI3K/AKT/mTOR pathway in osteoarthritis (39). In addition, PTEN has been considered a negative regulator of the PI3K/AKT/mTOR pathway (40). It has been reported that silencing ATG7 may promote AKT phosphorylation via the c-JUN/PTEN axis to promote the AKT pathway (41). Moreover, direct targets of miR-654-5p are associated with the AKT pathway in ovarian cancer (11). The results of the present study indicated that miR-654-5p promoted the activation of the PI3K/AKT/mTOR signaling pathway by downregulating ATG7 in NP cells.
In conclusion, the results of the present study revealed that miR-654-5p may participate in IDD development by directly targeting ATG7 and promoting activation of the PI3K/AKT/mTOR signaling pathway. The present study may provide valuable information for future explorations into the beneficial effects of targeting miR-654-5p as a potential therapeutic strategy for IDD. However, the sample size in the present study was limited and further studies are required to investigate the role of miR-654-5p in animal models of intervertebral disc NP.
Acknowledgements
Not applicable.
Funding Statement
This work was supported by the National Natural Science Foundation of China (grant no. 81572188).
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 81572188).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
SW designed the study. SW, YG, XZ and CW performed the experiments. SW and CW wrote the paper. SW and CW contributed to data analysis. SW, YG, XZ and CW confirm the authenticity of all the raw data. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The Ethics Committee of The Affiliated Zhongda Hospital of Southeast University (Nanjing, China) approved the present study (approval no. 2015ZDKYSB014). All patients provided written informed consent prior to the operation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Clouet J, Fusellier M, Camus A, Le Visage C, Guicheux J. Intervertebral disc regeneration: From cell therapy to the development of novel bioinspired endogenous repair strategies. Adv Drug Deliv Rev. 2019;146:306–324. doi: 10.1016/j.addr.2018.04.017. [DOI] [PubMed] [Google Scholar]
- 2.Karran EL, McAuley JH, Traeger AC, Hillier SL, Grabherr L, Russek LN, Moseley GL. Can screening instruments accurately determine poor outcome risk in adults with recent onset low back pain? A systematic review and meta-analysis. BMC Med. 2017;15:13. doi: 10.1186/s12916-016-0774-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang WJ, Yu XH, Wang C, Yang W, He WS, Zhang SJ, Yan YG, Zhang J. MMPs and ADAMTSs in intervertebral disc degeneration. Clin Chim Acta. 2015;448:238–246. doi: 10.1016/j.cca.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 4.Evangelatos G, Fragoulis GE, Koulouri V, Lambrou GI. MicroRNAs in rheumatoid arthritis: From pathogenesis to clinical impact. Autoimmun Rev. 2019;18:102391. doi: 10.1016/j.autrev.2019.102391. [DOI] [PubMed] [Google Scholar]
- 5.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: Background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 6.Zhou X, Chen L, Grad S, Alini M, Pan H, Yang D, Zhen W, Li Z, Huang S, Peng S. The roles and perspectives of microRNAs as biomarkers for intervertebral disc degeneration. J Tissue Eng Regen Med. 2017;11:3481–3487. doi: 10.1002/term.2261. [DOI] [PubMed] [Google Scholar]
- 7.Makki MS, Haqqi TM. MiR-139 modulates MCPIP1/IL-6 expression and induces apoptosis in human OA chondrocytes. Exp Mol Med. 2015;47:e189. doi: 10.1038/emm.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Z, Pang G, Huang YG, Li C. MiR-133 inhibits proliferation and promotes apoptosis by targeting LASP1 in lupus nephritis. Exp Mol Pathol. 2020;114:104384. doi: 10.1016/j.yexmp.2020.104384. [DOI] [PubMed] [Google Scholar]
- 9.Wang J, Liu X, Sun B, Du W, Zheng Y, Sun Y. Upregulated miR-154 promotes ECM degradation in intervertebral disc degeneration. J Cell Biochem. 2019 doi: 10.1002/jcb.28471. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 10.Zhu W, Li L, Li D. Rs11655237 polymorphism of LINC00673 affects the prognosis of cervical cancer by interfering with the interaction between LINC00673 and microRNA-1231. J Cell Physiol. 2020;235:8155–8166. doi: 10.1002/jcp.29470. [DOI] [PubMed] [Google Scholar]
- 11.Majem B, Parrilla A, Jiménez C, Suárez-Cabrera L, Barber M, Marín A, Castellví J, Tamayo G, Moreno-Bueno G, Ponce J, et al. MicroRNA-654-5p suppresses ovarian cancer development impacting on MYC, WNT and AKT pathways. Oncogene. 2019;38:6035–6050. doi: 10.1038/s41388-019-0860-0. [DOI] [PubMed] [Google Scholar]
- 12.Lu M, Wang C, Chen W, Mao C, Wang J. MiR-654-5p Targets GRAP to promote proliferation, metastasis, and chemoresistance of oral squamous cell carcinoma through Ras/MAPK Signaling. DNA Cell Biol. 2018;37:381–388. doi: 10.1089/dna.2017.4095. [DOI] [PubMed] [Google Scholar]
- 13.Pu M, Chen J, Tao Z, Miao L, Qi X, Wang Y, Ren J. Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci. 2019;76:441–451. doi: 10.1007/s00018-018-2940-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Zhang Y, Liu P, Bai H, Li X, Xiao J, Yuan Q, Geng S, Yin H, Zhang H, et al. MicroRNA-128 knockout inhibits the development of Alzheimer's disease by targeting PPARγ in mouse models. Eur J Pharmacol. 2019;843:134–144. doi: 10.1016/j.ejphar.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Yang H, Liao D, Tong L, Zhong L, Wu K. MiR-373 exacerbates renal injury and fibrosis via NF-κB/MatrixMetalloproteinase-9 signaling by targeting Sirtuin1. Genomics. 2019;111:786–792. doi: 10.1016/j.ygeno.2018.04.017. [DOI] [PubMed] [Google Scholar]
- 16.Yang W, Sun P. Downregulation of microRNA-129-5p increases the risk of intervertebral disc degeneration by promoting the apoptosis of nucleus pulposus cells via targeting BMP2. J Cell Biochem. 2019;120:19684–19690. doi: 10.1002/jcb.29274. [DOI] [PubMed] [Google Scholar]
- 17.Tan YY, Xu XY, Wang JF, Zhang CW, Zhang SC. MiR-654-5p attenuates breast cancer progression by targeting EPSTI1. Am J Cancer Res. 2016;6:522–532. [PMC free article] [PubMed] [Google Scholar]
- 18.Pfirrmann CW, Metzdorf A, Zanetti M, Hodler J, Boos N. Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine (Phila Pa 1976) 2001;26:1873–1878. doi: 10.1097/00007632-200109010-00011. [DOI] [PubMed] [Google Scholar]
- 19.World Medical Association. World medical association declaration of Helsinki: Ethical principles for medical research involving human subjects. JAMA. 2013;310:2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 22.Feng C, Liu H, Yang Y, Huang B, Zhou Y. Growth and differentiation factor-5 contributes to the structural and functional maintenance of the intervertebral disc. Cell Physiol Biochem. 2015;35:1–16. doi: 10.1159/000369670. [DOI] [PubMed] [Google Scholar]
- 23.Feng C, Liu H, Yang M, Zhang Y, Huang B, Zhou Y. Disc cell senescence in intervertebral disc degeneration: Causes and molecular pathways. Cell Cycle. 2016;15:1674–1684. doi: 10.1080/15384101.2016.1152433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li ZY, Wang XL, Dang Y, Zhu XZ, Zhang YH, Cai BX, Zheng L. Long non-coding RNA UCA1 promotes the progression of paclitaxel resistance in ovarian cancer by regulating the miR-654-5p/SIK2 axis. Eur Rev Med Pharmacol Sci. 2020;24:591–603. doi: 10.26355/eurrev_202001_20035. [DOI] [PubMed] [Google Scholar]
- 25.Le Maitre CL, Pockert A, Buttle DJ, Freemont AJ, Hoyland JA. Matrix synthesis and degradation in human intervertebral disc degeneration. Biochem Soc Trans. 2007;35:652–655. doi: 10.1042/BST0350652. [DOI] [PubMed] [Google Scholar]
- 26.Vo NV, Hartman RA, Yurube T, Jacobs LJ, Sowa GA, Kang JD. Expression and regulation of metalloproteinases and their inhibitors in intervertebral disc aging and degeneration. Spine J. 2013;13:331–341. doi: 10.1016/j.spinee.2012.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lippai M, Szatmari Z. Autophagy-from molecular mechanisms to clinical relevance. Cell Biol Toxicol. 2017;33:145–168. doi: 10.1007/s10565-016-9374-5. [DOI] [PubMed] [Google Scholar]
- 28.Jiang W, Zhang X, Hao J, Shen J, Fang J, Dong W, Wang D, Zhang X, Shui W, Luo Y, et al. SIRT1 protects against apoptosis by promoting autophagy in degenerative human disc nucleus pulposus cells. Sci Rep. 2014;4:7456. doi: 10.1038/srep07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang XH, Zhu L, Hong X, Wang YT, Wang F, Bao JP, Xie XH, Liu L, Wu XT. Resveratrol attenuated TNF-α-induced MMP-3 expression in human nucleus pulposus cells by activating autophagy via AMPK/SIRT1 signaling pathway. Exp Biol Med (Maywood) 2016;241:848–853. doi: 10.1177/1535370216637940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu K, Chen W, Wang X, Peng Y, Liang A, Huang D, Li C, Ye W. Autophagy attenuates the catabolic effect during inflammatory conditions in nucleus pulposus cells, as sustained by NF-κB and JNK inhibition. Int J Mol Med. 2015;36:661–668. doi: 10.3892/ijmm.2015.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akkoc Y, Gozuacik D. MicroRNAs as major regulators of the autophagy pathway. Biochim Biophys Acta Mol Cell Res. 2020;1867:118662. doi: 10.1016/j.bbamcr.2020.118662. [DOI] [PubMed] [Google Scholar]
- 32.Gaviraghi M, Vivori C, Pareja Sanchez Y, Invernizzi F, Cattaneo A, Santoliquido BM, Frenquelli M, Segalla S, Bachi A, Doglioni C, et al. Tumor suppressor PNRC1 blocks rRNA maturation by recruiting the decapping complex to the nucleolus. EMBO J. 2018;37:e99179. doi: 10.15252/embj.201899179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen DY, Chen YM, Lin CF, Lo CM, Liu HJ, Liao TL. MicroRNA-889 inhibits autophagy to maintain mycobacterial survival in patients with latent tuberculosis infection by targeting TWEAK. mBio. 2020;11:e03045–19. doi: 10.1128/mBio.03045-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang WJ, Yang W, Ouyang ZH, Xue JB, Li XL, Zhang J, He WS, Chen WK, Yan YG, Wang C. MiR-21 promotes ECM degradation through inhibiting autophagy via the PTEN/akt/mTOR signaling pathway in human degenerated NP cells. Biomed Pharmacother. 2018;99:725–734. doi: 10.1016/j.biopha.2018.01.154. [DOI] [PubMed] [Google Scholar]
- 35.Xiong J. Atg7 in development and disease: Panacea or Pandora's Box? Protein Cell. 2015;6:722–734. doi: 10.1007/s13238-015-0195-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu K, Li N, Cheng Q, Zheng J, Zhu M, Bao S, Chen M, Shi G. MiR-96-5p prevents hepatic stellate cell activation by inhibiting autophagy via ATG7. J Mol Med (Berl) 2018;96:65–74. doi: 10.1007/s00109-017-1593-6. [DOI] [PubMed] [Google Scholar]
- 37.Wang C, Zhang ZZ, Yang W, Ouyang ZH, Xue JB, Li XL, Zhang J, Chen WK, Yan YG, Wang WJ. MiR-210 facilitates ECM degradation by suppressing autophagy via silencing of ATG7 in human degenerated NP cells. Biomed Pharmacother. 2017;93:470–479. doi: 10.1016/j.biopha.2017.06.048. [DOI] [PubMed] [Google Scholar]
- 38.Zhang H, Zhang X, Zhang J. MiR-129-5p inhibits autophagy and apoptosis of H9c2 cells induced by hydrogen peroxide via the PI3K/AKT/mTOR signaling pathway by targeting ATG14. Biochem Biophys Res Commun. 2018;506:272–277. doi: 10.1016/j.bbrc.2018.10.085. [DOI] [PubMed] [Google Scholar]
- 39.He W, Cheng Y. Inhibition of miR-20 promotes proliferation and autophagy in articular chondrocytes by PI3K/AKT/mTOR signaling pathway. Biomed Pharmacother. 2018;97:607–615. doi: 10.1016/j.biopha.2017.10.152. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y, Kwok-Shing Ng P, Kucherlapati M, Chen F, Liu Y, Tsang YH, de Velasco G, Jeong KJ, Akbani R, Hadjipanayis A, et al. A pan-cancer proteogenomic atlas of PI3K/AKT/mTOR pathway alterations. Cancer Cell. 2017;31:820–832.e3. doi: 10.1016/j.ccell.2017.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao D, Zhang S, Wang X, Gao D, Liu J, Cao K, Chen L, Liu R, Liu J, Long J. ATG7 regulates hepatic Akt phosphorylation through the c-JUN/PTEN pathway in high fat diet-induced metabolic disorder. FASEB J. 2019;33:14296–14306. doi: 10.1096/fj.201901414RR. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.