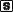Abstract
The putative NTPase/helicase protein from severe acute respiratory syndrome coronavirus (SARS-CoV) is postulated to play a number of crucial roles in the viral life cycle, making it an attractive target for anti-SARS therapy. We have cloned, expressed, and purified this protein as an N-terminal hexahistidine fusion in Escherichia coli and have characterized its helicase and NTPase activities. The enzyme unwinds double-stranded DNA, dependent on the presence of a 5′ single-stranded overhang, indicating a 5′o 3′ polarity of activity, a distinct characteristic of coronaviridae helicases. We provide the first quantitative analysis of the polynucleic acid binding and NTPase activities of a Nidovirus helicase, using a high throughput phosphate release assay that will be readily adaptable to the future testing of helicase inhibitors. All eight common NTPs and dNTPs were hydrolyzed by the SARS helicase in a magnesium-dependent reaction, stimulated by the presence of either single-stranded DNA or RNA. The enzyme exhibited a preference for ATP, dATP, and dCTP over the other NTP/dNTP substrates. Homopolynucleotides significantly stimulated the ATPase activity (15–25-fold) with the notable exception of poly(G) and poly(dG), which were non-stimulatory. We found a large variation in the apparent strength of binding of different homopolynucleotides, with dT24 binding over 10 times more strongly than dA24 as observed by the apparent Km .
The etiological agent of severe acute respiratory syndrome (SARS)1 in humans was recently identified as a novel coronavirus, SARS coronavirus (SARS-CoV), an enveloped, single-stranded RNA positive-strand virus with a genome of ∼30 kb (1, 2, 3, 4, 5). According to the World Health Organization, as of the July 9, 2003 there have been 8436 reported probable cases of SARS worldwide including 812 deaths (www.who.int/csr/sars/en). Although this initial global outbreak of SARS appears to have been successfully contained, SARS will remain a serious concern while there continues to be no suitable vaccine or effective drug treatment.
The coronaviruses and arteriviruses belong to the order Nidovirales, members of which share a similar polycistronic genomic composition and organization (6, 7). The 29.7-kb SARS-CoV genome is dominated by a 21.2-kb replicase gene (orf1ab) in the 5′-most region, which incorporates a ribosomal slippage site (3, 4, 7). Termination of translation without ribosomal frameshifting generates the Orf1a protein, while the (–1) frameshift results in translation of the extended Orf1ab polyprotein (pp1ab) (7). These two initial translation products are then autoproteolytically processed primarily by the main proteinase (Mpro, also called 3CLpro), encoded within the 5′-proximal region of the replicase gene (6), to release a number of non-structural proteins (nsPs), including the RNA-dependent RNA polymerase (RdRp, nsP9) and the NTPase/helicase (Hel, nsP10). Within the N-terminal region of the predicted SARS-CoV Hel protein is a cysteine-rich putative metal binding domain (MBD), which from mutational studies in the related equine arteritis virus (EAV) has been implicated in subgenomic mRNA synthesis, genome replication, and virion biogenesis (8, 9).
The main proteinase (Mpro), the RNA-dependent RNA polymerase (RdRp), and the NTPase/helicase (Hel) proteins are all thought to be essential for nidovirus viability (7, 8, 10) and are therefore potential targets for the development of anti-SARS drugs (11, 12). Promising NTPase/helicase inhibitors are currently being tested for the treatment of herpes simplex virus (13, 14, 15) and hepatitis C viral infections (16). As a foundation to the development of specific SARS-CoV NTPase/helicase inhibitors, we have cloned, expressed, purified, and characterized the biochemical properties of the SARS-CoV NTPase/helicase domain.
EXPERIMENTAL PROCEDURES
Source DNA and Cloning—Cultured Vero cells were infected with SARS coronavirus strain HKU 39487. Total RNA was isolated with TRIZOL (Invitrogen) as instructed by the manufacturer. Purified RNA was reverse-transcribed using Superscript II reverse transcriptase (Invitrogen) and random hexamers. The NTPase/helicase domain (nsP10) was amplified by PCR, using AmpliTaq Gold (Applied Biosystems) with the forward primer 5′-GGCGGATCCGCCATGGCTGTAGGTGCTTGTGTATTG-3′ and the reverse primer 5′-GGCCTCGAGTCATTGTAATGTAGCCACATTGCG-3′. The PCR product was gel purified, digested with BamHI/XhoI and ligated with a similarly digested pET28a(+) vector (Novagen) to make the plasmid pHelA12.
Protein Purification—4 liters of LB broth supplemented with kanamycin (50 μg/ml) were inoculated with saturated pHelA12/BL21 (DE3) culture (1/200 dilution) and grown at 37 °C until A 600 = 0.6. Protein expression was induced by addition of isopropyl-1-thio-β-d-galactopyranoside (0.5 mm), and cultures were incubated at 25 °C for 6 h. After cooling to 4 °C, the cells were harvested by centrifugation and resuspended in Buffer A (25 mm Tris-HCl, pH 6.8, 20 mm imidazole, 0.5 m NaCl, 0.1% Triton X-100, protease inhibitors (Complete, Roche Applied Science), 1 g of wet cell pellet/5 ml of buffer). Cells were lysed by sonication (Sonics VibraCell, microtip, 30% power for 3 min (4 s on, 10 s off) with ice cooling) and then centrifuged (30 min, 30,000 × g), and the supernatant was filtered (Corning syringe filter, 0.45 μm) and then applied to 2 × 5 ml nickel-charged HiTrap chelating columns directly connected in series (Amersham Biosciences). The 10-ml column was washed with 100 ml of Buffer A, then 50 ml Buffer B (25 mm Tris-HCl, pH 6.8, 50 mm imidazole, 0.5 m NaCl) before eluting the protein with a stepwise gradient over 100 ml versus Buffer C (25 mm Tris-HCl, pH 6.8, 250 mm imidazole, 0.5 m NaCl). Collected fractions (2 ml) were immediately diluted with 2 ml of Buffer D (20 mm Tris-HCl, pH 6.8, 5 mm β-mercaptoethanol (β-ME), 10% glycerol) to reduce protein precipitation. The purest fractions (determined by SDS-PAGE) were combined, dialyzed against 500 volumes of Buffer D, and then filtered (0.45 μm) before application to a column containing 0.5 ml of Red Sepharose CL-6B resin (Amersham Biosciences). The resin was washed with 5 ml of Buffer B (containing 5 mm β-ME), before elution with a stepwise gradient over 10 ml versus Buffer E (25 mm Tris-HCl, pH 6.8, 50 mm imidazole, 2 m NaCl, 5% glycerol, 5 mm β-ME, 2% Nonidet P-40). Pure fractions (by SDS-PAGE) were combined and stored at 4 °C for short term or frozen at –70 °C for long term storage.
NTPase Assays—NTPase assays were performed by measuring phosphate release using a colorimetric method based on complexation with malachite green and molybdate (AM/MG reagent) (17, 18). In a 50-μl volume, typically 0.2–0.7 pmol of protein, 50 mm Tris-HCl, pH 6.6, 5 mm MgCl2, 1 mm ATP, and 25 μg/ml poly(U) (Amersham Biosciences, s 20, w = 4.8) were incubated for 10 min at 25 °C. The reaction was stopped by the addition of 10 μl of 0.5 m EDTA, and then the color was developed for 5 min following the addition of 200 μl of AM/MG reagent (0.034% malachite green, 1.05% ammonium molybdate, 0.04% Tween 20, in 1 m HCl). 40 μl of 34% (w/v) trisodium citrate dihydrate was then added and the A 630 read, typically using a 96-well plate reader. For the assays stimulated by homopolynucleotides, the polynucleotide concentration was accurately measured using the A 260 and quoted as a molar concentration of 24-mer. The amount of phosphate released was quantified by comparison with a standard phosphate calibration curve. Data were fitted using Origin (Microcal Software).
Preparation of Duplex DNA Substrates—The synthetic oligonucleotide “released” (5′-GGTGCAGCCGCAGCGGTGCTCG-3′, Proligo (Singapore), de-salted) was labeled with [γ-32P]ATP (3000 Ci/mmol, Amersham Biosciences) using T4 polynucleotide kinase (Amersham Biosciences) according to the manufacturer's instructions. Labeled DNA was purified by desalting (AutoSeq G-50 spin column, Amersham Biosciences), phenol:chloroform extraction, and ethanol precipitation and was resuspended in deionized water. The labeled released oligo (100 pmol) was annealed to 150 pmol of oligos “Tblunt” (5′-CGAGCACCGCTGCGGCTGCACC-3′), “3T20” (5′-CGAGCACCGCTGCGGCTGCACCTTTTTTTTTTTTTTTTTTTT-3′), and “5T20” (5′-TTTTTTTTTTTTTTTTTTTTCGAGCACCGCTGCGGCTGCACC-3′) in 100 μl of Buffer F (25 mm Hepes, pH 7.4, 1 mm EDTA, 25 mm NaCl) by heating to 95 °C for 5 min, followed by gradual cooling to room temperature at 0.5–1 °C/min, to form substrate DNA duplexes dB, d3T, and d5T, respectively. DNA duplexes were not purified further.
Duplex DNA-unwinding Assays—Purified helicase protein (typically 6 pmol) was incubated with the labeled duplex DNA (30 fmol) in 60 μl of Buffer G (20 mm Hepes, pH 7.4, 5 mm magnesium acetate, 2.5 mm ATP, 2 mm dithiothreitol, 0.1 mg/ml bovine serum albumin, 10% glycerol) for 10 min at 30 °C. Reactions were halted by the addition of 15 μl of gel loading buffer (5% SDS, 100 mm EDTA, 15% w/v Ficoll, 0.1% xylene cyanol, 0.05% bromphenol blue), and the products (25 μl) were immediately resolved on 15% polyacrylamide gels (29:1 acrylamide: bisacrylamide in (×1) TBE (89 mm Tris base, 89 mm boric acid, 10 mm EDTA)). For the positive control, labeled duplex DNA (30 fmol) in 60 μl of Buffer F was denatured by heating to 95 °C for 5 min and then flash cooling in liquid nitrogen. Gels were analyzed on a Storm 860 PhosphorImager using ImageQuaNT software (Amersham Biosciences).
RESULTS
Cloning, Expression, and Purification of the Putative SARS Coronavirus NTPase/Helicase—A bacterial expression system was used to produce the SARS-CoV NTPase/helicase (Hel) in quantities suitable for enzymatic studies. SARS-CoV strain HKU 39487 was used to infect Vero cells, and total RNA was extracted and reverse-transcribed with random hexamers. PCR primers were designed to amplify the region coding for the full-length Hel domain (nsP10), as defined by the helicase Mpro proteolytic sites: Leu-Gln↓ (Ser, Ala, Gly) based on the work by Anand et al. (12). The PCR product of the expected length was ligated into a pET28a(+) expression vector, thereby introducing a small hexahistidine-T7 tag at the N terminus of the protein, to facilitate subsequent purification. Sequencing confirmed that the cloned PCR fragment was identical to that of the deposited sequence (NCBI accession number: NC_004718) apart from a silent CCC to CCG variation present at position 16,325. The His-tagged protein was purified from the soluble fraction using nickel affinity chromatography, followed by dye-affinity chromatography, resulting in a single band of the expected mass of ∼70 kDa, as observed by SDS-PAGE after staining with Coomassie Blue (Fig. 1 , panel A). Typical yields were 1–2 mg of purified protein/liter of bacterial culture.
Fig. 1.
Purification and DNA-unwinding activity of the SARS coronavirus NTPase/helicase.Panel A, purification of the SARS-CoV Hel as observed by 4–12% SDS-PAGE. Lane 1, marker, from bottom (in kilodaltons) 10, 15, 20, 25, 30, 40, 50 (strong), 60, 70, 80, 90, 100, 120, 160, 220. Lane 2, Whole cell extract from E. coli BL21 (DE3) expressing SARS-CoV Hel (5 μg). Lane 3, soluble extract from E. coli BL21 (DE3) expressing SARS-CoV Hel (10 μg). Lane 4, SARS-CoV Hel after purification by nickel affinity chromatography (1.5 μg). Lane 5, SARS-CoV Hel after purification on Red Sepharose CL-6B (0.4 μg). Panel B, schematic showing the fully double-stranded (dB) and partially double-stranded DNA substrates (d5T = 5′-oligo(dT)20 tail, d3T = 3′-oligo(dT)20 tail) used in the unwinding assay. An asterisk indicates the position of the 32P radiolabel. Panel C, the SARS-CoV Hel unwinds duplex DNA with a 5′ to 3′ polarity. The positions of the intact dsDNA duplexes (dB, d5T, and d3T), as well as the released (labeled) ssDNA strand on the polyacrylamide gel, are indicated with arrows. Lanes 1–5, reactions containing 30 fmol of DNA duplex; lanes 2–5, reactions containing 6 pmol of helicase; lanes 1–4, reactions containing 2.5 mm ATP; lane 1, the d5T DNA duplex was heat-denatured. Panel D, titration of helicase concentration. Lanes 1–4, reactions containing 150 fmol, 600 fmol, 1.5 pmol, and 3 pmol of helicase, respectively, in addition to 30 fmol of d5T DNA duplex and 2.5 mm ATP.
DNA Duplex-unwinding Activity of the SARS Coronavirus NTPase/Helicase—Three radiolabeled DNA duplex and partial duplex substrates were prepared to probe the activity of the recombinant helicase protein in a fully defined in vitro system. The dB substrate was fully double-stranded (blunt ended), while d3T and d5T each contained a 20-residue 3′- or 5′-poly(dT) tail, respectively (Fig. 1, panel B), analogous to the system used by Seybert et al. (19). One strand in each of the three annealed duplexes, referred to as the released strand (10), was 5′-end-labeled with 32PO4, so that the substrate duplex and single-stranded DNA products could be resolved by polyacrylamide gel electrophoresis and visualized by phosphorimaging. When incubated with the helicase, only the d5T partial duplex with the 5′ single-stranded DNA tail was unwound to release the labeled strand (Fig. 1, panel C). This activity was entirely dependent upon the presence of ATP (Fig. 1, panel C) and Mg2+ (data not shown). Neither the fully double-stranded duplex (dB) nor a partial duplex with a 3′ single-stranded tail (d3T) could be unwound by the SARS-CoV helicase, indicating that it has activity with specific 5′ to 3′ polarity. We also show that the release of the labeled strand from the d5T duplex correlated with the quantity of helicase present in the reaction (Fig. 1, panel D).
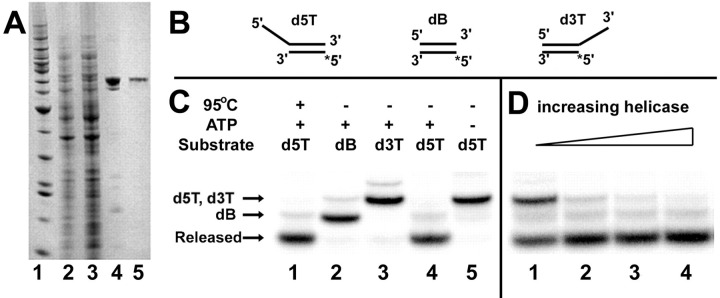
ATPase Activity of the SARS Coronavirus NTPase/Helicase— The ATPase activity was quantified using a sensitive colorimetric assay that measures the total amount of orthophosphate released, based on the highly colored complex of phosphomolybdate and malachite green (17). The assays were performed in parallel on a 50 μl scale, using a standard 96-well plate. In the absence of ssDNA or RNA, there was a basal rate of ATPase activity of ∼0.4 s–1. As found for all other RNA helicases studied to date (10), the ATPase activity of the SARS-CoV Hel was absolutely dependent on Mg2+, as no NTPase activity was observed when a molar excess of EDTA was added to sequester the Mg2+ (data not shown). Mn2+ (as a chloride salt) could substitute for Mg2+, but with reduced relative efficiency (∼40% at 2.5 mm, data not shown). The ATPase activity was stimulated significantly (15–25-fold) by the addition of most homopolyribo- and deoxyribonucleotides, in particular poly(U) (Fig. 2 ). However, the addition of either poly(G)24 or poly(dG)24 caused only a very minor increase in the ATPase activity of the helicase. Due to the significant stimulatory effect of poly(U), this was used as the standard polynucleotide for further analysis of the NTPase activities of the SARS-CoV Hel.
Fig. 2.
Comparative polynucleotide-induced stimulation of the ATPase activity of the SARS coronavirus NTPase/helicase. Each 24-mer polynucleotide was added to a concentration of 1 μm. The degree of stimulation was calculated as a factor, normalized to the rate of ATPase activity in the absence of polynucleotide. Experimental conditions were: 0.5 mm ATP, 5 mm MgCl2, 50 mm Tris-HCl, pH 6.6, 50-μl reaction volume, 0.5 pmol of helicase, 10-min reaction.
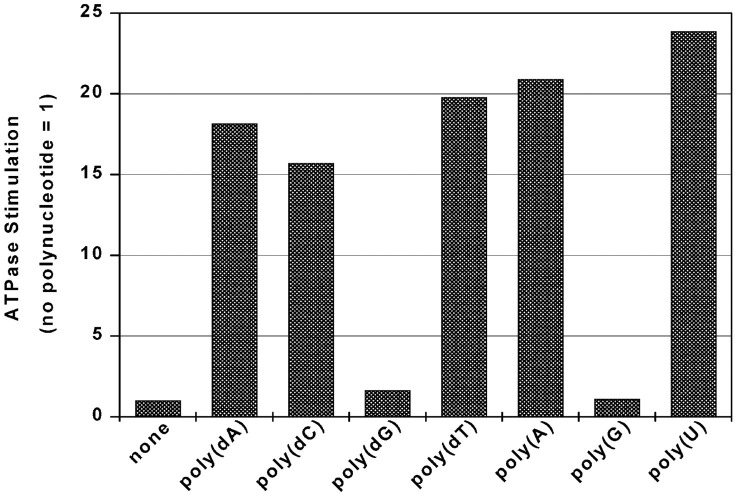
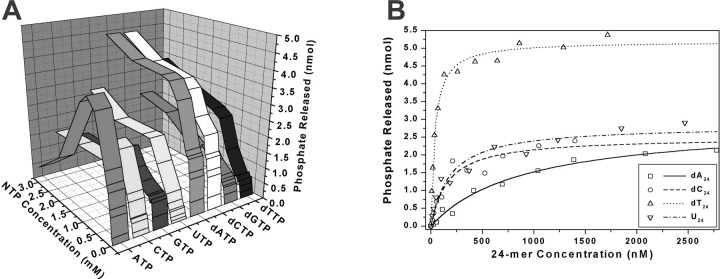
Comparison of NTP Substrates in the NTPase Reaction—The SARS-CoV helicase was able to hydrolyze all eight common NTPs and dNTPs in a magnesium-dependent reaction that was highly stimulated by certain polynucleotides (Fig. 3 , panel A). At lower concentrations of NTP (<1 mm), hydrolysis followed simple single-site Michaelis-Menten behavior, with the values of Km, kcat , and the specificity constant kcat /Km shown in Table I . Judging by the specificity constant, ATP, dATP, and dCTP were the “best” substrates of those tested in the NTPase assay. At higher concentrations (>1 mm) of ATP (and to a lesser extent for GTP and CTP) there was a marked reduction in NTPase activity, indicative of substrate inhibition, whereas the other NTPs continued to follow single-site Michaelis-Menten behavior.
Fig. 3.
Effect of NTP and polynucleotide concentration on rate of NTP hydrolysis catalyzed by the SARS coronavirus NTPase/helicase.Panel A, hydrolysis of NTP by the SARS coronavirus NTPase/helicase versus NTP concentration versus NTP identity for eight different nucleotides. Data for the NTPase activity for each NTP over a range of NTP concentrations (0–3 mm) were fitted to a simple Michaelis-Menten model; the results are shown in Table I. Experimental conditions were: 25 μg/ml poly(U), 5 mm MgCl2, 50 mm Tris-HCl, pH 6.6, 50-μl reaction volume, 0.4 pmol of helicase, 10-min reaction. Panel B, hydrolysis of dATP by the SARS coronavirus NTPase/helicase versus polynucleotide concentration for four different polynucleotides. Data are shown after subtraction of basal dATPase phosphate release. Data for the dATPase activity in the presence of each polynucleotide were fitted to a simple Michaelis-Menten model, with the fit shown by the four lines. Experimental conditions were: 1 mm dATP, 5 mm MgCl2, 50 mm Tris-HCl, pH 6.6, 50-μl reaction volume, 0.7 pmol of SARS-CoV helicase, 10-min reaction.
Table I.
Rate constants for the NTPase activity of the SARS coronavirus NTPase/helicase
Km and kcat values were calculated by least squares regression on a simple Michaelis-Menten fitting of the NTPase phosphate-release data shown in Fig. 3, panel A, as measured by the malachite green/molybdate assay. Experimental conditions were the same as for Fig. 3, panel A.
| Km | Kcat | kcat/Km | |
|---|---|---|---|
| mm | s-1 | mm-1 s-1 | |
| ATP | 0.33 | 19.1 | 57.9 |
| CTP | 0.47 | 13.9 | 29.6 |
| GTP | 0.20 | 5.4 | 27.0 |
| UTP | 0.55 | 10.5 | 19.1 |
| dATP | 0.20 | 14.8 | 74.0 |
| dCTP | 0.22 | 14.3 | 65.0 |
| dGTP | 0.35 | 7.5 | 21.4 |
|
dUTP |
0.93 |
14.8 |
15.9 |
Determination of the Strengths of Binding of Polynucleotides to the SARS Coronavirus NTPase/Helicase—The concentration that gave half-maximal stimulation of dATPase activity was measured for a selection of polynucleotides (24-mers), to obtain a set of apparent Km values. dATP was used in place of ATP as it was impossible to use ATP at a non-rate-limiting concentration due to substrate inhibition at higher ATP concentrations. When the concentration of dATP was non-rate-limiting, the dATPase activity of the enzyme displayed simple single-site behavior with respect to the concentration of the single-stranded polynucleotide present (Fig. 3, panel B). Apparent half-maximal stimulation values ( ) for the binding of poly(dA)24, poly(dC)24, poly(dT)24, and poly(U)24 to the SARS-CoV Hel were determined by fitting a simple Michaelis-Menten model to the ATPase data. The values obtained were 900, 165, 37, and 150 nm, respectively. The maximal rate of stimulation by (dT)24 appeared higher relative to the other polynucleotides in the presence of dATP (Fig. 3, panel B) as opposed to ATP (as in Fig. 2), but otherwise similar experiments using 1 mm ATP gave similar values for (results not shown). The values for poly(G)24 and poly(dG)24 could not be determined, due to the extremely low level of NTPase stimulation that their addition elicited. Poly(dT)24 stimulated the dATPase activity at the lowest concentrations: the most simple model would therefore suggest that poly(dT)24 binds most strongly to Hel.
DISCUSSION
On the basis of sequence homology, helicases are divided into five major groups: the superfamilies SF-1, SF-2, and SF-3, a fourth family consisting of helicases related to the Escherichia coli DnaB protein, and a fifth family typified by the transcription termination factor Rho (20). The majority of the viral helicases that have been characterized to date belong to SF-2, as typified by the hepatitis C virus ns3 helicase domain, for which a crystal structure was solved 6 years ago (21). The primary sequence of the predicted SARS-CoV Hel, however, places it in SF-1, a less well characterized family with regards to its viral helicase members. Initial studies on virus-encoded helicases from SF-1 showed ATPase activity but no detectable helicase activity (10). Subsequently, the HCoV 229E NTPase/helicase was shown to have DNA and RNA duplex-unwinding activity with a 5′ to 3′ polarity (19), as was found for the EAV helicase from the closely related arterivirus family (22). Studies on a helicase from a further arterivirus, porcine reproductive and respiratory syndrome virus confirmed a strict 5′ to 3′ polarity (23). Our results show that the SARS-CoV Hel has very similar properties to these three helicases: a 5′ to 3′ polarity of unwinding and a stimulation of ATPase activity by single stranded polynucleotides. This stands in stark contrast to the SF-2 viral helicases, which display a 3′ to 5′ polarity of unwinding (18). Although it was previously suggested that a 5′ to 3′ unwinding was a signature of all SF-1 viral helicases (19), it has recently been reported that SF-1 helicases from two hordeiviruses and a potexvirus (both plant (+) ssRNA viruses) unwind RNA duplexes in both directions (24). This would suggest that helicases of the Nidovirales (including coronaviruses and arteriviruses), possibly together with SF1 helicases from other animal (+) ssRNA viruses (including alphaviruses, rubiviruses, and hepatitis E viruses) form a distinct class of 5′ to 3′ RNA viral helicases within SF-1 (see Supplementary Material).
The SARS-CoV Hel hydrolyzed all eight common NTPs and dNTPs, exhibiting a marked preference for ATP, dATP, and dCTP (Fig. 3, panel A, and Table I). The ability of Hel to hydrolyze a wide variety of NTPs and dNTPs is consistent with the triphosphate moiety forming the basis of the recognition factor for binding and hydrolysis, as has been suggested for the hepatitis C virus ns3 helicase (18, 25). It is possible that this lack of selectivity in NTP hydrolysis also enables the Hel to act as a broad RNA 5′-triphosphatase in viral 5′ capping, as reported for a related potexvirus helicase (26). The ATP hydrolysis rate decreased at higher ATP concentrations, suggesting that ATP may be able to bind to a weaker set of ATP-binding sites in a putative homomultimeric enzyme complex, thereby inhibiting the net ATPase activity of the enzyme (16, 27).
The basal ATPase activity of Hel was stimulated by most polynucleotides 15–25-fold, with the exception of poly(G) and poly(dG) (Fig. 2). Variation in strength of homopolynucleotide binding was particularly significant (Fig. 3, panel B), suggesting that the SARS-CoV Hel may possess elements of secondary structure/sequence recognition capability. We speculate that this specificity may play a role in the localization of a putative RdRp-helicase complex to selected genomic locations during subgenomic RNA synthesis, augmenting proposed base-pairing factors (28). The 5′ to 3′ direction of unwinding suggests to us that the helicase may aid the replicative process by disrupting secondary structure or displacing bound proteins and annealed RNA fragments, putative roles that will require experimental validation.
Mutational studies on the EAV MBD have suggested that this region is involved in virion biogenesis, subgenomic mRNA synthesis, genome replication, and possibly other processes crucial to the viral life cycle (8, 9, 22). Although there are differences in the primary sequence between the EAV MBD and those from HCoV and SARS-CoV, these MBDs are most likely zinc finger domains (29) sharing a common role (8). It remains unclear how the putative zinc finger and helicase domains of the SARS-CoV nsP10 are functionally related to one another and to the general viral life cycle.
We report here the expression, purification, and biochemical characterization of the predicted NTPase/helicase domain (nsP10) of the replicase polyprotein (pp1ab) from SARS-CoV. We demonstrate that the nsp10 protein is a functional helicase, which specifically unwinds polynucleotide duplexes with a 5′ to 3′ polarity, placing it in a distinct class of 5′ to 3′ RNA viral helicases within SF-1. The sensitive, high throughput phosphate-release assay that we have used here may be easily adapted to screen libraries of known antiviral compounds for their SARS-CoV helicase/NTPase inhibitory properties. Taken together, our experiments provide the first quantitative analysis of NTPase activity and selectivity in polynucleotide binding for a nidovirus helicase, providing invaluable insight into the biochemical properties of this key SARS-CoV protein. This study presents both an opportunity and a challenge for the future development of helicase-targeted anti-SARS drugs.
Addendum—During the process of publication, work in another group also showed the helicase activity of the SARS coronavirus NTPase/helicase (30). The protein was expressed as a maltose-binding protein fusion and shown to have both ATPase activity and DNA duplex-unwinding activity; both activities were abolished when the conserved lysine residue in the Walker A box was replaced with alanine.
Footnotes
The amino acid sequence of this protein can be accessed through NCBI Protein Database under NCBI accession number NC_828870.
The abbreviations used are: SARS, severe acute respiratory syndrome; CoV, coronavirus; MBD, metal binding domain; EAV, equine arteritis virus; β-ME, β-mercaptoethanol; ss, single-stranded.
Supplementary Material
References
- 1.Peiris J.S., Lai S.T., Poon L.L., Guan Y., Yam L.Y., Lim W., Nicholls J., Yee W.K., Yan W.W., Cheung M.T., Cheng V.C., Chan K.H., Tsang D.N., Yung R.W., Ng T.K., Yuen K.Y. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. N. Engl. J. Med. 2003;348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 3.Rota P.A., Oberste M.S., Monroe S.S., Nix W.A., Campagnoli R., Icenogle J.P., Penaranda S., Bankamp B., Maher K., Chen M.H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J.L., Chen Q., Wang D., Erdman D.D., Peret T.C., Burns C., Ksiazek T.G., Rollin P.E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Gunther S., Osterhaus A.D., Drosten C., Pallansch M.A., Anderson L.J., Bellini W.J. Science. 2003;300:1394–1399. doi: 10.1126/science.1085952. [DOI] [PubMed] [Google Scholar]
- 4.Marra M.A., Jones S.J., Astell C.R., Holt R.A., Brooks-Wilson A., Butterfield Y.S., Khattra J., Asano J.K., Barber S.A., Chan S.Y., Cloutier A., Coughlin S.M., Freeman D., Girn N., Griffith O.L., Leach S.R., Mayo M., McDonald H., Montgomery S.B., Pandoh P.K., Petrescu A.S., Robertson A.G., Schein J.E., Siddiqui A., Smailus D.E., Stott J.M., Yang G.S., Plummer F., Andonov A., Artsob H., Bastien N., Bernard K., Booth T.F., Bowness D., Czub M., Drebot M., Fernando L., Flick R., Garbutt M., Gray M., Grolla A., Jones S., Feldmann H., Meyers A., Kabani A., Li Y., Normand S., Stroher U., Tipples G.A., Tyler S., Vogrig R., Ward D., Watson B., Brunham R.C., Krajden M., Petric M., Skowronski D.M., Upton C., Roper R.L. Science. 2003;300:1399–1404. doi: 10.1126/science.1085953. [DOI] [PubMed] [Google Scholar]
- 5.Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A.E., Humphrey C.D., Shieh W.J., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J.Y., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. N. Engl. J. Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 6.Ziebuhr J., Snijder E.J., Gorbalenya A.E. J. Gen. Virol. 2000;81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 7.Lai M.M., Cavanagh D. Adv. Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Dinten L.C., van Tol H., Gorbalenya A.E., Snijder E.J. J. Virol. 2000;74:5213–5223. doi: 10.1128/jvi.74.11.5213-5223.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Marle G., van Dinten L.C., Spaan W.J., Luytjes W., Snijder E.J. J. Virol. 1999;73:5274–5281. doi: 10.1128/jvi.73.7.5274-5281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kadare G., Haenni A.L. J. Virol. 1997;71:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmes K.V. J. Clin. Invest. 2003;111:1605–1609. doi: 10.1172/JCI18819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anand K., Ziebuhr J., Wadhwani P., Mesters J.R., Hilgenfeld R. Science. 2003;300:1763–1767. doi: 10.1126/science.1085658. [DOI] [PubMed] [Google Scholar]
- 13.Kleymann G., Fischer R., Betz U.A., Hendrix M., Bender W., Schneider U., Handke G., Eckenberg P., Hewlett G., Pevzner V., Baumeister J., Weber O., Henninger K., Keldenich J., Jensen A., Kolb J., Bach U., Popp A., Maben J., Frappa I., Haebich D., Lockhoff O., Rubsamen-Waigmann H. Nat. Med. 2002;8:392–398. doi: 10.1038/nm0402-392. [DOI] [PubMed] [Google Scholar]
- 14.Kleymann G. Expert Opin. Investig. Drugs. 2003;12:165–183. doi: 10.1517/13543784.12.2.165. [DOI] [PubMed] [Google Scholar]
- 15.Crute J.J., Grygon C.A., Hargrave K.D., Simoneau B., Faucher A.M., Bolger G., Kibler P., Liuzzi M., Cordingley M.G. Nat. Med. 2002;8:386–391. doi: 10.1038/nm0402-386. [DOI] [PubMed] [Google Scholar]
- 16.Borowski P., Schalinski S., Schmitz H. Antiviral Res. 2002;55:397–412. doi: 10.1016/s0166-3542(02)00096-7. [DOI] [PubMed] [Google Scholar]
- 17.Baykov A.A., Evtushenko O.A., Avaeva S.M. Anal. Biochem. 1988;171:266–270. doi: 10.1016/0003-2697(88)90484-8. [DOI] [PubMed] [Google Scholar]
- 18.Wardell A.D., Errington W., Ciaramella G., Merson J., McGarvey M.J. J. Gen. Virol. 1999;80:701–709. doi: 10.1099/0022-1317-80-3-701. [DOI] [PubMed] [Google Scholar]
- 19.Seybert A., Hegyi A., Siddell S.G., Ziebuhr J. RNA (N. Y.) 2000;6:1056–1068. doi: 10.1017/s1355838200000728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caruthers J.M., McKay D.B. Curr. Opin. Struct. Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- 21.Yao N., Hesson T., Cable M., Hong Z., Kwong A.D., Le H.V., Weber P.C. Nat. Struct. Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- 22.Seybert A., van Dinten L.C., Snijder E.J., Ziebuhr J. J. Virol. 2000;74:9586–9593. doi: 10.1128/jvi.74.20.9586-9593.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bautista E.M., Faaberg K.S., Mickelson D., McGruder E.D. Virology. 2002;298:258–270. doi: 10.1006/viro.2002.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalinina N.O., Rakitina D.V., Solovyev A.G., Schiemann J., Morozov S.Y. Virology. 2002;296:321–329. doi: 10.1006/viro.2001.1328. [DOI] [PubMed] [Google Scholar]
- 25.Kim J.L., Morgenstern K.A., Griffith J.P., Dwyer M.D., Thomson J.A., Murcko M.A., Lin C., Caron P.R. Structure (Lond.) 1998;6:89–100. doi: 10.1016/s0969-2126(98)00010-0. [DOI] [PubMed] [Google Scholar]
- 26.Li Y.I., Shih T.W., Hsu Y.H., Han Y.T., Huang Y.L., Meng M. J. Virol. 2001;75:12114–12120. doi: 10.1128/JVI.75.24.12114-12120.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borowski P., Niebuhr A., Mueller O., Bretner M., Felczak K., Kulikowski T., Schmitz H. J. Virol. 2001;75:3220–3229. doi: 10.1128/JVI.75.7.3220-3229.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasternak A.O., van den Born E., Spaan W.J., Snijder E.J. EMBO J. 2001;20:7220–7228. doi: 10.1093/emboj/20.24.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laity J.H., Lee B.M., Wright P.E. Curr. Opin. Struct. Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 30.Thiel V., Ivanov K.A., Putics A., Hertzig T., Schelle B., Bayer S., Weissbrich B., Snijder E.J., Rabenau H., Doerr H.W., Gorbalenya A.E., Ziebuhr J. J. Gen. Virol. 2003;84:2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.