Abstract
This study presents an approach for mining structured information from clinical narratives in Electronic Health Records (EHRs), by using Rich Text Formatted (RTF) records. RTF is adopted by many medical information management systems. There is rich structural information in these files which can be extracted and interpreted, yet such information is largely ignored. Analysts who focus on structured EHR data often do not treat EHR narratives as structured; NLP researchers often start from plain text instead of RTF files, casting structured data as narratives. We investigate multiple types of EHR narratives in the Enterprise Data Warehouse from a multisite large healthcare chain consisting of both the academic medical center and community hospitals. We focus on the RTF constructs related to tables and sections that are not available in plain text EHR narratives. We show how to parse these RTF constructs, analyze their prevalence and characteristics in the context of multiple types of EHR narratives. Our case study demonstrates the additional utility, of the features derived from RTF constructs, over plaintext oriented NLP.
Keywords: Electronic Health Records, Natural Language Processing, Rich Text Format
Introduction
Nationwide adoption of Electronic Health Records (EHRs) has given rise to a large amount of digital health data, which can be used for secondary analysis [1]. The data within these records are expected to improve efficiency and overall healthcare. In addition to structured data, a large amount of clinically relevant data remains present in an unstructured free-text format. These narratives are largely variable between institutions and even healthcare professionals within them making it difficult for data extraction. In order to optimize research to improve quality or clinical needs, a systematic representation of EHRs for both structured and unstructured data is necessary. For instance, computational phenotyping has been used to mine or predict both clinically and scientifically significant phenotypes from structured EHR data, unstructured clinical narratives, and their combinations [2]. Computational phenotyping calls for efficient approaches to mine a large volume of clinical narratives for structured information on patient pathophysiology. To this end, natural language processing (NLP) is widely utilized as an effective tool for turning unstructured narratives into structured information [3].
Applications of language processing methods to clinical data in many projects have achieved reasonable success in analyzing various types of biomedical data, including clinical narratives [4, 5]. However, when it comes to EHRs, there is a gap between what data can support and what the method asks for. NLP generally assumes plain text as inputs, but clinical narratives in EHR often possess rich-text format (RTF) such as tables, different fonts, and font sizes. The majority of studies ignore the informative RTF information in EHR narratives. To our knowledge, there are very few, if any, studies that process RTF EHR narratives and utilize the formatting for additional information. When evaluating numerous clinical note repositories for shared task challenges (e.g., challenges from BioNLP [6-8], i2b2 [9, 10], SemEval [11, 12], and BioCreative [13]), there are few RTF EHR narratives.
The task of processing RTF documents is relatively straightforward though non-trivial. Despite the existence of standard packages to parse RTF or convert it to other formats such as HTML, research in the field of biomedical NLP largely ignore RTF and still use plain-text as their raw input. Typical NLP processes include sentence breaking, word tokenization, part-of-speech tagging, constituency or dependency parsing, named entity detection, semantic role labeling, and event frame extraction, etc. These processes largely ignore the formatting information. As a result, EHR databases (e.g., Enterprise Data Warehouse) for medical institutions often additionally store the plain text format of EHR narratives in addition to their RTF counterparts, or simply store in plain text format instead of RTF. Furthermore, information loss can sometimes occur during the conversion of RTF to plain-text formats.
Interestingly, researchers who focus on studying structured data do not treat EHR narratives as structured and often rely on NLP to extract structured pieces out of EHR narratives from plain text. Many studies utilize generic medical NLP systems such as MedLEE [14], MetaMap [15], cTakes [16], or GATE [17]. Other studies take advantage of special purpose NLP systems such as MedEx [18] for medication detection or TEES [19] for medication event frame extraction. Finally, many authors developed their NLP components or even systems to serve specific data mining tasks such as computational phenotyping [20-27].
Our study addresses the gap between structured EHR and clinical NLP research regarding the usage of rich-text formatted EHR narratives. In the current study, we demonstrate how to extract the structured pieces (e.g., tables and section headings) from RTF EHR narratives and investigate the prevalence of such structures in EHR narratives. Our objective is to expose the need to start from rich-text formatted EHR narratives, making it suitable for NLP or structured EHR research, and demonstrate that this can be done without much hassle. We also perform a case study to show the efficacy of mined structured information in improving computational phenotyping tasks.
Methods
A. Dataset
We constructed multiple rich-text formatted corpora consisting of different types of EHR narratives, by querying Northwestern Medicine Enterprise Data Warehouse (NMEDW). The NMEDW is a joint initiative between the Northwestern University Feinberg School of Medicine and Northwestern Memorial HealthCare [28]. The NMEDW is a multi-center data repository with heterogeneous types of data. We constructed multiple patient corpora over a specific period from breast cancer patients. In this study, the progress notes were queried, which are records of events during a patient’s office visit or hospitalization that can be used to communicate opinions, findings, and plans between healthcare professionals. A well-documented progress note is complete, accurate, and concise for the care delivered, including diagnosis and treatments [29]. We extracted progress notes that were dated between 07/01/2015 and 10/01/2015. In order to test performance evaluation, we extracted progress notes for breast cancer patients from a previous study [30] matching their plain text counterparts as a use case.
B. RTF processing
Many word processors support the reading and writing of rich-text format (RTF) files, as well as many of the medical information management systems. In this study, we found and identified the important RTF structural information from medical narratives, namely tables and section headings. A medical history table may contain several columns with headings stressed in bold, underlines, or inserted as the first line of the table. These tables often summarize important clinical information such as prescriptions, allergies, or medical histories. The format of RTF text is defined by groups, control-words, and delimiters. Groups are enclosed in braces, and control-words start with backslash (\) characters. For example, a 2-by-2 table and the corresponding RTF code is shown in Figure 1.
Figure 1:

A 2-by-2 table and the corresponding RTF code. (a) A 2-by-2 table defined by the RTF code (b) RTF code snippet that defines a 2-by-2 table.
As illustrated in Figure 1, the codes are within closed braces with control-words in the first line indicating the format and encoding used in this group. Each row in the table begins with a “\trowd” tag, and ends with a “\row” tag. Thus, lines 2-7 define the first row, and lines 8-13 defines the second row. The “\cellx###” tag declares the position of the right side of a cell, and the “\cell” tag denotes the end of a cell. The above code generates a 2-by-2 table, with cell widths equal to 1000. Searching for the “\trowd” tag in RTF files allow for table discovery. In addition to “\trowd” tags, many tables in medical notes are also defined by tab-delimited-tables, i.e., cells in a table row are separated by tab characters. To identify these tab characters, RTF uses the “\tab” tag.
Section headings in medical notes often describe the topic of that section. The information within a section is usually cohesive and can easily be interpreted by both NLP programs and manual curation. As a result, the Common Data Model of Observational Medical Outcomes Partnership specifies that sections be stored as annotations to facilitate portable computational phenotyping (https://github.com/OHDSI/CommonDataModel) [accessed on March 20, 2019]. Section headings tend to share some characteristics such as bold or underlined formatting font. The headings may or may not occupy a whole line, and may or may not be capitalized or be followed by a colon. Heading levels are often indicated by indentation. In summary, section headings provide important information, and the RTF format allows us to apply specific searching criteria to accurately and efficiently identify section headings and their levels.
However, the syntax of RTF is relatively more obscure and lacks powerful software packages for data parsing compared to more popular formats such as HTML, XML, and LaTex. On the other hand, HTML is a much more commonly used format with many existing tools for parsing and extraction of structured information, such as Python’s BeautifulSoup package (https://www.crummy.com/software/BeautifulSoup/bs4/doc/) [accessed on March 20, 2019]. To address this, we convert RTF to HTML using LibreOffice prior to parsing any data and then analyze the structures using python’s BeautifulSoup web scraping package. For regular tables, we searched for the HTML <table> tag, and summarized the rows by counting the row tag <tr>. For tab-delimited-tables, we matched them using the regular expression “\S+ *\t *\S+”, i.e. two non-space characters separated by a tab and zero or more spaces characters. The section headings in the documents are usually stressed in bold (HTML tag of <b>) or with an underline (HTML tag of <u>). The stressed phrase should occupy a whole line.
Results
A. Parsing the tables
In our study, we extracted and used 158,948 breast cancer progress notes. Given that column names are often listed in the first row of a table, we also extracted the cells in the first rows to summarize the topic of the tables. The distribution of the number of tables, and the most common first rows of these tables are summarized in Figure 2. A Histogram indicating table frequency is shown on the upper panel of the figures. Note that the y-axis is in log-scale; the number of tables in a medical note approximately follows an exponential distribution – the probability decreases by a factor of 10 for roughly every 10 additional tables. The most common first rows are summarized on the lower panel of the figures. Adjacent cells are separated by a comma.
Figure 2:
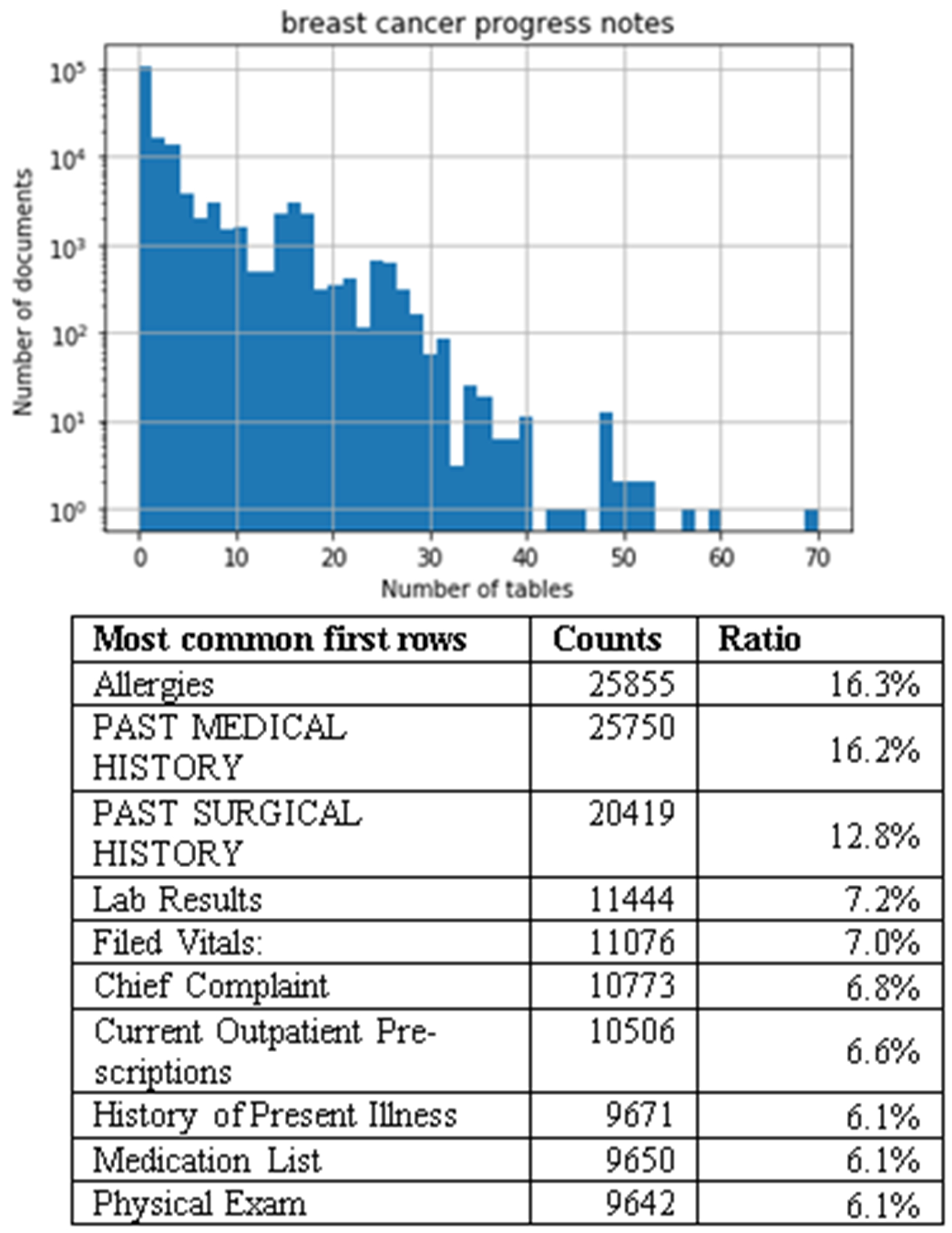
Histogram of the number of tables and the most common first rows in breast cancer progress notes. The “Ratio” column is the count of the first rows over the total number of notes of this type.
In breast cancer progress notes, the most common first row contains a single cell with the most common terms being “Allergies” and “Past Medical History” making up 16.3% and 16.2%, of the notes, respectively. The 16.3% of the medical notes have an Allergies table. In addition to these terms, we also identified terms involving lab results, vital information, complaints, current prescriptions, illness history, medication list, and physical exam information (Figure 2). It is evident that the related tables belong to the pre-formatted template.
B. Parsing the section headings
Section headings often contain critical information that can be used to identify key features. We searched the section headings in progress notes of patients with breast cancer from the NMEDW. Among the 158,948 breast cancer progress notes, we found 46.6% files have first level headings, and we found 2.4% files have higher level headings. Histograms of the number of first-level section headings (section headings with the smallest indentation) in the notes are summarized in Figure 3. The number of headings in a note generally follows an exponential distribution. The lower panel of the figures shows the most common first-level section headings.
Figure 3:
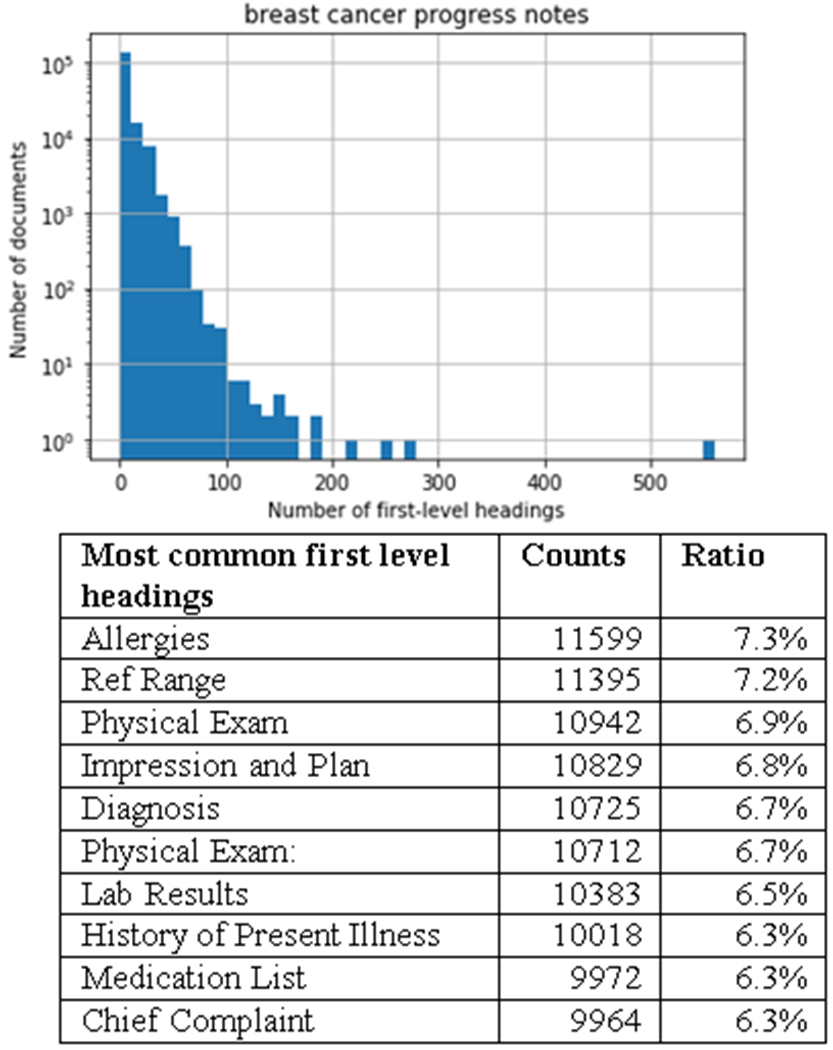
Histogram of the number of first-level section headings, and the most common first-level headings in the breast cancer progress notes. Note that the y-axis is in log-scale. The “Ratio” column is the count of the heading over the total number of notes of this type.
In the breast cancer progress notes, we found the most common heading to be “Allergies” similarly to the most common first row found in these notes. Interestingly, we also find headings involving Physical Exams, Lab Results, and Medication Lists, which are also the most common first rows of tables in these notes (Figure 3). These results suggest that section headings are important too. In addition, the structure of a progress note can be better understood by combining data from tables and section headings. Note that the vital signs (which is the first row of a table) are also deemed as the section headings because they are usually emphasized in bold with occupying a whole line.
B. Case study on breast cancer phenotyping
The structured data extracted from the medical notes can be used as features in machine learning models to improve their performances. As a case study, we added the structure information extracted to identify breast cancer patients with contralateral events. The problem was investigated in our previous study [30], with a cohort of 1063 patients who were diagnosed with breast cancer. In the cohort, 33 patients were identified with contralateral events (the case group). We previously identified the breast cancer contralateral events from the cohort using progress notes and counts of pathology reports. In our previous study, the progress notes were treated as plain text; structured information was ignored. This previous study was used as a baseline because it utilized an 8-step relatively sophisticated NLP system, and applied extensive feature engineering (e.g., filtered powerset method) to build the classification model. Thus, our previous system provides competitive plain-text oriented NLP baselines. In this case study, we pulled out the corresponding RTF versions of the same progress notes for the 1063 patients. We extracted the past medical history, past surgical history, and lab test results from the tables in the progress notes, and included them as features in the classification model. For lab test results, we used the reference ranges presented in the test result tables to determine whether the measured results are higher or lower than the ranges. We chose common features that occured in at least 10 patients’ progress notes, resulting in 37 medical history entries, 33 surgical history entries, and 10 abnormal lab test features. The most common medical/surgical history entries, and abnormal lab tests are summarized in Table 1.
Table 1:
Most common medical/surgical history entries, and abnormal lab tests.
| Common medical history entries (# of occurrences) | Common surgical history entries (# of occurrences) | Common abnormal lab test (# of occurrences) |
|---|---|---|
| Breast cancer (313) | Lumpectomy (213) | MCHC low (112) |
| Hypertension (213) | Right breast (81) | Blood Urea Nitrogen high (32) |
| Hyperlipidemia (134) | Colonoscopy (78) | Hemoglobin low (23) |
| Disorder of refraction and accommodation (77) | Remove of tonsils and adenoids (78) | Albumin low (21) |
| Hypothyroidism (72) | Left breast (73) | MCH low (20) |
We encoded the structured features as whether a patient had a condition in the past medical/surgical history, and whether a lab test is below or above the reference range. We adopted the same train-test split (7:3) and the same evaluation metric (AUC) as in our previous study [30]. We used logistic regression (LR) model, and 5-fold cross-validation to tune the regularization factor of LR, and obtained an AUC of 0.89 on the held-out test set. By building an ensemble model that averages the predicted probabilities from the structured-feature model and the NLP-feature model, an AUC of 0.92 was obtained. The model performance was compared with previous results in Table 2. Using much fewer structured features, we can achieve a comparable classification performance using unstructured MetaMap CUI features (83 vs. 1285). Combining the two models further improves the classification performance. Note that the pathology report count is essentially a type of structured feature though not derived from RTF tables [30]. For a fair comparison, the top 3 models in Table 2 (including the 3rd model that was reported as the best model in our previous study [30]) all use the pathology report count. In this case study, including the structured features improved the performance from 0.89 to 0.92. The overall results suggest that structured features extracted from RTF tables can be effective features in computational phenotyping tasks, and may work even better when combined with unstructured features extracted by NLP.
Table 2:
Comparison of classification performance on the held-out test set using different features. Shades indicate the results from our previous study[30] that treated clinical narratives as plain text.
| Model | Feature Number | Test AUC |
|---|---|---|
| Combined MetaMap + Structured Features + Pathology Report Count | 1365 | 0.92 |
| Structured Features + Pathology Report Count | 83 | 0.89 |
| Combined MetaMap + Pathology Report Count | 1285 | 0.89 |
| Combined MetaMap | 1282 | 0.68 |
| Pathology Report Count | 3 | 0.67 |
| Full MetaMap without Combination | 42 | 0.30 |
| Bag of Words | 55192 | 0.70 |
Discussion
This study is a preliminary work that tries to introduce the informatics community to the rich-text formatted EHR narratives, and it comes with limitations. First and foremost, we note that different institutions may have different clinical documentation systems, templates, and styles, especially across different healthcare chains on a national scale. Although our study uses multi-site data (a mixture of academic medical center and community hospitals) from regional healthcare chains, we expect challenges may arise when generalizing our study on a national scale. In addition, different healthcare chains may use different EHR systems, with possible variations in the prevalence of the rich-text structures. Thus, we plan to expand our study in the future to include more types of EHR narratives, from different EHR systems, and systematically study their variations to better inform the data mining practice over EHR. We will also study the potential impact of rich-text format on improving the interoperability in health information exchange and portable computational phenotyping, as it brings more regularity to the otherwise “free” text.
Conclusions
Electronic Health Records (EHR) narratives are not equivalent to plain text but contain important structural information. One common medical note file format for archived EHR narratives is rich text format (RTF). We have utilized the low-level syntax tags in RTF files to mine the structured information of multiple types of EHR narratives. We ran our parsing systems across RTF clinical narratives from multiple community hospitals and an academic medical center, and successfully extracted tables and section headings from them. The tables contain patient allergies, medical histories, lab test results, prescriptions, vital signs, and even follow-up appointment information. A long EHR narrative may be divided into several sections, with each section containing cohesive information of a perspective and context of a patient’s pathophysiologic profile. Properly recognizing the tables and sections is useful to extract structured information from EHR, and prevents unnecessary input chunks of text to NLP systems. In addition, targeted NLP algorithms can be further applied to individual sections to extract information more precisely. Our case study on breast cancer phenotyping shows that tasks such as computational phenotyping can benefit from properly extracted structural information (e.g., tables) and more accurate NLP, thanks to the usage of RTF structural information – a hidden and often ignored trove in EHR.
Acknowledgements
This project is supported in part by NIH grant R21 LM012618.
References
- [1].Blumenthal D and Tavenner M, “The “meaningful use” regulation for electronic health records,” N Engl J Med, vol. 2010, no. 363, pp. 501–504, 2010. [DOI] [PubMed] [Google Scholar]
- [2].Shivade C et al. , “A review of approaches to identifying patient phenotype cohorts using electronic health records,” Journal of the American Medical Informatics Association, vol. 21, no. 2, pp. 221–230, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Luo Y et al. , “Natural Language Processing for EHR-Based Pharmacovigilance: A Structured Review,” (in eng), Drug Saf, vol. 40, no. 11, pp. 1075–1089, Nov 2017. [DOI] [PubMed] [Google Scholar]
- [4].Nadkarni PM, Ohno-Machado L, and Chapman WW, “Natural language processing: an introduction,” Journal of the American Medical Informatics Association, vol. 18, no. 5, pp. 544–551, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zeng Z, Deng Y, Li X, Naumann T, and Luo Y, “Natural Language Processing for EHR-based computational phenotyping,” IEEE/ACM Transactions on Computational Biology and Bioinformatics, pp. 1–1, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Nédellec C et al. , “Overview of BioNLP shared task 2013,” in Proceedings of the BioNLP Shared Task 2013 Workshop, 2013, pp. 1–7: Association for Computational Linguistics. [Google Scholar]
- [7].Kim J-D, Wang Y, Takagi T, and Yonezawa A, “Overview of genia event task in bionlp shared task 2011,” in Proceedings of the BioNLP Shared Task 2011 Workshop, 2011, pp. 7–15: Association for Computational Linguistics. [Google Scholar]
- [8].Kim J-D, Ohta T, Pyysalo S, Kano Y, and Tsujii J. i, “Overview of BioNLP’09 shared task on event extraction,” in Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing: Shared Task, 2009, pp. 1–9: Association for Computational Linguistics. [Google Scholar]
- [9].Uzuner Ö, Luo Y, and Szolovits P, “Evaluating the state-of-the-art in automatic de-identification,” Journal of the American Medical Informatics Association, vol. 14, no. 5, pp. 550–563, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Uzuner Ö, South BR, Shen S, and DuVall SL, “2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text,” Journal of the American Medical Informatics Association, vol. 18, no. 5, pp. 552–556, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Elhadad N, Pradhan S, Chapman W, Manandhar S, and Savova G, “SemEval-2015 task 14: Analysis of clinical text,” in Proc of Workshop on Semantic Evaluation. Association for Computational Linguistics, 2015, pp. 303–10. [Google Scholar]
- [12].Segura Bedmar I, Martínez P, and Herrero Zazo M, “Semeval-2013 task 9: Extraction of drug-drug interactions from biomedical texts (ddiextraction 2013),” 2013: Association for Computational Linguistics. [Google Scholar]
- [13].Krallinger M et al. , “Evaluation of text-mining systems for biology: overview of the Second BioCreative community challenge,” Genome biology, vol. 9, no. 2, p. S1, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Friedman C, Shagina L, Lussier Y, and Hripcsak G, “Automated encoding of clinical documents based on natural language processing,” Journal of the American Medical Informatics Association, vol. 11, no. 5, pp. 392–402, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Aronson AR, “Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program,” in Proceedings of the AMIA Symposium, 2001, p. 17: American Medical Informatics Association. [PMC free article] [PubMed] [Google Scholar]
- [16].Savova GK et al. , “Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications,” Journal of the American Medical Informatics Association, vol. 17, no. 5, pp. 507–513, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cunningham H, “GATE, a general architecture for text engineering,” Computers and the Humanities, vol. 36, no. 2, pp. 223–254, 2002. [Google Scholar]
- [18].Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, and Denny JC, “MedEx: a medication information extraction system for clinical narratives,” Journal of the American Medical Informatics Association, vol. 17, no. 1, pp. 19–24, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Björne J and Salakoski T, “TEES 2.1: Automated annotation scheme learning in the BioNLP 2013 Shared Task,” in Proceedings of the BioNLP Shared Task 2013 Workshop, 2013, pp. 16–25: Association for Computational Linguistics. [Google Scholar]
- [20].Richesson RL et al. , “Electronic health records based phenotyping in next-generation clinical trials: a perspective from the NIH Health Care Systems Collaboratory,” Journal of the American Medical Informatics Association, vol. 20, no. e2, pp. e226–e231, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hripcsak G and Albers DJ, “Next-generation phenotyping of electronic health records,” Journal of the American Medical Informatics Association, vol. 20, no. 1, pp. 117–121, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pathak J, Kho AN, and Denny JC, “Electronic health records-driven phenotyping: challenges, recent advances, and perspectives,” ed: The Oxford University Press, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zexian Z et al. , “Using Clinical Narratives and Structured Data to Identify Distant Recurrences in Breast Cancer,” in 2018 IEEE International Conference on Healthcare Informatics (ICHI), 2018, pp. 44–52: IEEE. [Google Scholar]
- [24].Zeng Z et al. , “Using natural language processing and machine learning to identify breast cancer local recurrence,” (in eng), BMC Bioinformatics, vol. 19, no. Suppl 17, p. 498, December 28 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Luo Y, Sohani AR, Hochberg EP, and Szolovits P, “Automatic lymphoma classification with sentence subgraph mining from pathology reports,” Journal of the American Medical Informatics Association, vol. 21, no. 5, pp. 824–832, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Luo Y, Xin Y, Hochberg E, Joshi R, Uzuner O, and Szolovits P, “Subgraph augmented non-negative tensor factorization (SANTF) for modeling clinical narrative text,” Journal of the American Medical Informatics Association, vol. 22, no. 5, pp. 1009–1019, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Luo Y, Xin Y, Hochberg E, Joshi R, Uzuner O, and Szolovits P, “Subgraph augmented non-negative tensor factorization (SANTF) for modeling clinical narrative text,” Journal of the American Medical Informatics Association, p. ocv016, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Starren JB, Winter AQ, and Lloyd-Jones DM, “Enabling a learning health system through a unified enterprise data warehouse: the experience of the Northwestern University Clinical and Translational Sciences (NUCATS) Institute,” Clinical and translational science, vol. 8, no. 4, pp. 269–271, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dick R and Steen E, “The computer-based patient record. Institute of Medicine,” ed: Washington, DC: National Academy Press, 1991. [Google Scholar]
- [30].Zeng Z et al. , “Contralateral breast cancer event detection using Nature Language Processing,” in AMIA Annual Symposium Proceedings, 2017, vol. 2017, pp. 1885–1892: American Medical Informatics Association. [PMC free article] [PubMed] [Google Scholar]


