The crystal structures of two benzoyl amides: 2-chloro-4-ethoxy-3,5-dimethoxy-N-(3-oxocyclohex-1-en-1-yl)benzamide and 2-chloro-N-(5,5-dimethyl-3-oxocyclohex-1-en-1-yl)-4-ethoxy-3,5-dimethoxybenzamide have been determined.
Keywords: crystal structure, enaminones, bioactivity
Abstract
The first title benzoyl amide, C17H20ClNO5 (3a), crystallizes in the monoclinic space group P21/c with Z = 4 and the second, C19H24ClNO5 (3b), also crystallizes in P21/c with Z = 8 (Z′ = 2), thus there are two independent molecules in the asymmetric unit. In 3a, the phenyl ring makes a dihedral angle of 50.8 (3)° with the amide moiety with the C=O group on the same side of the molecule as the C—Cl group. One methoxy group is almost in the plane of the benzene ring, while the ethoxy and other methoxy substituent are arranged on opposite sides of the ring with the ethoxy group occupying the same side of the ring as the C=O group in the amide moiety. For one of the two molecules in 3b, both the amide and 5,5-dimethyl-3-oxocyclohex-1-en-1-yl moieties are disordered over two sets of sites with occupancies of 0.551 (2)/0.449 (2) with the major difference between the two conformers being due to the conformation adopted by the cyclohex-2-en-1-one ring. The three molecules in 3b (i.e., the undisordered molecule and the two disorder components) differ in the arrangement of the subsituents on the phenyl ring and the conformation adopted by their 5,5-dimethyl-3-oxocyclohex-1-en-1-yl moieties. In the crystal of 3a, N—H⋯O hydrogen bonds link the molecules into a zigzag chain propagating in the [001] direction. For 3b a combination of C—H⋯O and N—H⋯O intermolecular interactions link the molecules into a zigzag ribbon propagating in the [001] direction.
Chemical context
Enaminones are compounds in which a nitrogen atom is conjugated through a carbon–carbon double bond to an ester (vinylogous urethane) or a ketone (vinylogous amide) functional group (see Scheme). Enaminones may be viewed as amides into which a vinyl fragment has been interpolated. Designations often used, such as enamino ketone or β-amino-α, β-unsaturated ketone, are misleading in that the compounds rarely exhibit the physical properties normally associated with ketones. Enaminones, compounds possessing the structural unit NH2—C=C—C=O, are versatile synthetic intermediates that combine the ambient nucleophilicity of enamines with the ambient electrophilicity of enones (Greenhill, 1976 ▸; Lue & Greenhill, 1996 ▸).
β-Enaminones may be used in the synthesis of many bioactive molecules with a heterocyclic unit. Enaminones as intermediates are responsible for a wide range of therapeutic agents from both natural and synthetic sources including taxol, anticonvulsants, anti-inflammatories, and duocarmycin, and consequently have been the subject of numerous structural bioactivity investigations in recent times (Misra et al., 2008 ▸; Greenhill, 1977 ▸; Boger et al., 1989 ▸; Eddington et al., 2003 ▸; Stoltz et al., 2016 ▸; Jerach & Elassar, 2015 ▸; Kalita et al., 2017 ▸). In spite of the breadth of research related to the biological properties of enaminones, recent research also indicates that enaminones, particularly the cyclic 3-(phenylamino)-2-cyclohexen-1-one (PACO), contain spectroscopic signatures of intramolecular charge transfer (ICT), making cyclic enaminones ideal components for molecules that mimic natural photosynthetic energy and electron transfer (Lue & Greenhill, 1996 ▸). A later study conducted in 2009 concluded that PACO has a low lying strongly polar singlet excited state with significant intramolecular charge transfer (Misra et al., 2009 ▸).
We herein describe the synthesis and structural characterization of the title benzoyl amides 2-chloro-4-ethoxy-3,5-dimethoxy-N-3-oxocyclohex-1-en-1-yl)benzamide, 3a and 2-chloro-N-(5,5-dimethyl-3-oxocyclohex-1-en-1-yl)-4-ethoxy-3,5-dimethoxybenzamide, 3b developed in connection with an ongoing research interest.
Structural commentary
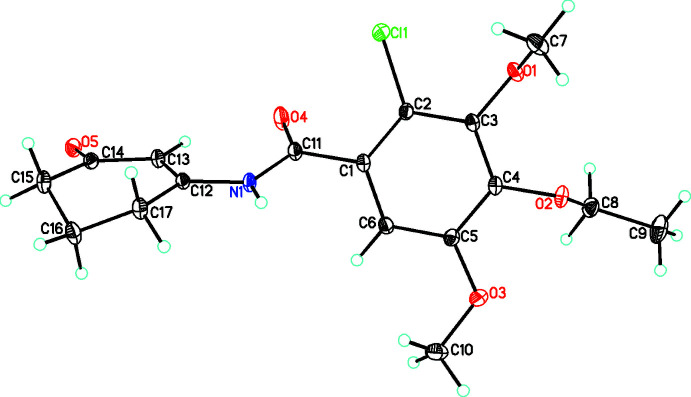
In view of the bioactivity of enaminones, the conformation adopted by a molecule is crucial to its activity. Thus an analysis of this for both molecules is appropriate. The benzoyl amide, C17H20ClNO5 (3a), crystallizes in the monoclinic space group P21/c with Z = 4. The compound is the result of the condensation of the enaminone 1a with the acid chloride 2. In the case of 3a (Fig. 1 ▸), the central phenyl ring makes a dihedral angle of 50.8 (3)° with the amide moiety; with the C=O group on the same side of the molecule as the C—Cl group; in the 3-oxocyclohex-1-en-1-yl group the C=O moiety is on the same side with respect to the phenyl ring [the pseudo torsion angle for O4—C11⋯C14—O5 = 21.8 (1)°]. One of the methoxy groups (O3—C10) attached to the C1–C6 benzene ring is close to the plane of the ring [torsion angle between the ring and C5—O3—C10 = 17.72 (2)°], while the ethoxy and the other methoxy substituent are arranged on opposite sides of the ring with the ethoxy group occupying the same side of the ring as the C=O group in the amide moiety [C8—O2⋯C11—O4 = −44.0 (1) and C7—O1⋯C11—O4 = 123.6 (1)°]. The extended conformation of the ethoxy group with respect to the ring is shown by a torsion angle of −170.8 (1)° for C4—O2—C8—C9.
Figure 1.
The molecular structure of 3a with atom labeling and with atomic displacement parameters shown at the 30% probability level.
The benzoyl amide, C19H24ClNO5 (3b), crystallizes in the monoclinic space group P21/c with Z = 8 (Z′ = 2), thus there are two independent molecules in the asymmetric unit. The compound is the result of the condensation of the enaminone 1b with the acid chloride 2. For one of the two molecules, both the amide and 5,5-dimethyl-3-oxocyclohex-1-en-1-yl moieties are disordered over two inequivalent conformations with occupancies of 0.551 (2)/0.449 (2). The major difference between the two conformers is due to the conformation adopted by the cyclohex-2-en-1-one ring (vide infra).
The conformations of both independent molecules will be discussed separately and then comparisons will be made between the conformation of 3a and the two molecules of 3b in which, due to disorder, one has adopted two different conformations. For simplicity, these will be called 3ba, 3bb and 3bc (where 3bb and 3bc are the major and minor components, respectively, of the disordered molecule). For 3ba (Fig. 2 ▸) the central phenyl ring makes a dihedral angle of 54.5 (3)° with the amide moiety with the C=O group on the opposite side of the molecule as the C—Cl group in contrast to the situation in 3a (this is illustrated by the respective C2—C1⋯C11—O4 torsion angles of 47.2 (2) and −129.5 (2) for 3a and 3ba, respectively). In both the amide moiety and the 3-oxocyclohex-1-en-1-yl group, the C=O moiety is on the same side [the torsion angle for O4A—C11A⋯C14A—O5A = −17.5 (1)°]. For the substituents on the phenyl ring, one methoxy group is almost coplanar with the ring [torsion angle between the ring and C5A—O3A—C10A = 3.5 (2)°] while in contrast to the situation in 3a, both the other methoxy and ethoxy substituents are on the same side of the ring [torsion angles for C7A—O1A⋯C11A—O4A and C8A—O2A⋯C11A—O4A = −32.4 (2) and −6.4 (2)°, respectively]. The conformation of the ethoxy substituent is different than that in 3a in that it has not adopted a fully extended aspect [C4A—O2A—C8A—C9A = −148.77 (16)].
Figure 2.
The molecular structure of 3ba with atom labeling and with atomic displacement parameters shown at the 30% probability level.
As indicated above, 3bb and 3bc are the major and minor components of the disordered 5,5-dimethyl-3-oxocyclohex-1-en-1-yl moieties with occupancies of 0.551 (2)/0.449 (2) (Fig. 3 ▸). The difference in the conformation of this group can be seen by the torsion angles for the C12—C17—C16—C15 grouping in 3a, 3ba, 3bb and 3bc of −48.67 (17), 50.11 (15), −51.7 (7) and 53.9 (10)°, respectively. From this it can be seen that for this moiety, 3a and 3bb have a similar conformation and 3ba and 3bc also have a similar conformation. For 3bb, the central phenyl ring makes a dihedral angle of 55.8 (9)° with the amide moiety with the C=O group on the opposite side of the molecule as the C—Cl group [torsion angle for C2B—C1B⋯C11B—O4B = −122.81 (13)]. In both the amide moiety and the 3-oxocyclohex-1-en-1-yl group, the C=O moiety is on the same side [O4B—C11B⋯C14B—O5B = 13.7 (2)°]. For the substituents on the phenyl ring, one methoxy group is almost coplanar with the ring [torsion angle between ring and methoxy group of 2.3 (2)] while the other methoxy group and ethoxy groups are on opposite sides of the ring [torsion angles for C7B—O1B⋯C11B—O4B and C8B—O2B⋯C11B—O4B = 165.4 (2) and −46.6 (2)°, respectively]. The conformation of the ethoxy substituent is different than that in 3a in that it has not adopted an extended aspect [C4B—O2B—C8B—C9B = 67.92 (16)°].
Figure 3.
The molecular structure of the disordered molecule in 3b showing both disorder components (3bb and 3bc) with atom labeling and with atomic displacement parameters shown at the 30% probability level.
Both 3bb and 3bc retain the same (undisordered) phenyl moiety and the only differences are in the conformation of the 5,5-dimethyl-3-oxocyclohex-1-en-1-yl moiety, thus in discussing this molecule we only have to consider the amide moiety and the 3-oxocyclohex-1-en-1-yl group where the C=O moiety is on the same side [O4B—C11B⋯C14C—O5C = −9.1 (2)°].
Supramolecular features
For 3a, N—H⋯O hydrogen bonds (Table 1 ▸) link the molecules into a zigzag chain propagating in the [001] direction as shown in Fig. 4 ▸. For 3b, a combination of C—H⋯O and N—H⋯O intermolecular interactions (Table 2 ▸) link the molecules into a zigzag ribbon propagating in the [001] direction (Fig. 5 ▸).
Table 1. Hydrogen-bond geometry (Å, °) for 3a .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1A⋯O5i | 0.844 (18) | 2.098 (18) | 2.9410 (15) | 177.1 (16) |
| C7—H7A⋯Cl1ii | 0.98 | 2.86 | 3.7022 (16) | 145 |
| C7—H7A⋯O4ii | 0.98 | 2.48 | 3.293 (2) | 140 |
| C9—H9A⋯O4iii | 0.98 | 2.53 | 3.470 (2) | 161 |
Symmetry codes: (i) x, -y+{\script{3\over 2}}, z+{\script{1\over 2}}; (ii) x, -y+{\script{1\over 2}}, z+{\script{1\over 2}}; (iii) -x, -y+1, -z+1.
Figure 4.
Packing diagram for 3a viewed along the b axis showing the molecules linked by N—H⋯O hydrogen bonds (shown by dashed bonds) into chains propagating in the [001] direction
Table 2. Hydrogen-bond geometry (Å, °) for 3b .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1A—H1AA⋯O5B | 0.853 (16) | 2.040 (18) | 2.849 (8) | 158.1 (15) |
| N1A—H1AA⋯O5C | 0.853 (16) | 2.081 (19) | 2.909 (9) | 163.5 (15) |
| C7A—H7AA⋯O2B i | 0.98 | 2.44 | 3.3451 (17) | 153 |
| C8A—H8AB⋯Cl1A ii | 0.99 | 2.92 | 3.703 (2) | 137 |
| C17A—H17A⋯O5B | 0.99 | 2.42 | 3.285 (7) | 146 |
| C17A—H17A⋯O5C | 0.99 | 2.63 | 3.481 (8) | 144 |
| C10B—H10F⋯O4A iii | 0.98 | 2.46 | 3.4013 (15) | 161 |
| N1B—H1BA⋯O5A iv | 0.77 (5) | 2.31 (5) | 2.985 (10) | 147 (5) |
| N1C—H1CA⋯O5A iv | 0.88 (5) | 1.96 (4) | 2.795 (12) | 159 (4) |
| C17C—H17E⋯O5A iv | 0.99 | 2.54 | 3.364 (3) | 141 |
Symmetry codes: (i) -x+1, y+{\script{1\over 2}}, -z+{\script{1\over 2}}; (ii) -x+2, -y+2, -z+1; (iii) -x+1, -y+2, -z+1; (iv) x, -y+{\script{3\over 2}}, z-{\script{1\over 2}}.
Figure 5.
Packing diagram for 3b viewed along the a axis showing the molecules linked by both C—H⋯O and C—H⋯Cl interactions as well as N—H⋯O hydrogen bonds (all shown by dashed bonds) into chains propagating in the [001] direction
Database survey
A survey of the Cambridge Structural Database for similar compounds did not provide any hits. Even if the molecules are broken up into two components, one based on the trisubstituted phenyl ring and the other on the cyclohexene ring no hits for the former and only one hit for the latter fragment is obtained [Cambridge Structural Database refcode MOLPUA (Meng et al., 2014 ▸)]. Even in this structure the only similar chromophore is the cyclohex-2-ene-1-one fragment, but with the double bond in a different position in the ring. For similar structures to this fragment but containing a cyclohexane ring there are DOSDOE, DOSBUK (Romney et al., 2014 ▸) and KAVDAP (Alford et al., 2016 ▸).
Synthesis and crystallization
The methodology involves N-deprotonation of the commercially available enaminones 1a,b with sodium hydride followed by benzoylation of 2 to give the title benzoyl amides 3a,b in 54% and 51% yield, respectively, from a method previously reported (see Scheme 1; Anderson et al., 2004 ▸). Benzoyl chloride 2 was prepared via chlorination of commercially available 4 under previously reported conditions (Zheng et al., 2011 ▸).
Preparation of 2-chloro-4-ethoxy-3,5-dimethoxybenzoyl chloride (2)
A solution of commercially available 2-chloro-4-ethoxy-3,5-dimethoxybenzoic acid, 4 (2.07 g, 7.7 mmol), and a catalytic amount of DMF in thionyl chloride (5 ml) was stirred at 353–363 K for 3 h to give the crude acid chloride 2. The mixture was concentrated under reduced pressure and used without any further purification. 1H NMR: (400 MHz, DMSO): δ 1.40–1.45 (3H, t, CH3), δ 3.03 (3H, s, CH3), δ 3.19 (3H, s, CH3), 4.21–4.28 (2H, q, CH2), 7.48 (H, s, aromatic H).
Preparation of 2-chloro-4-ethoxy-3,5-dimethoxy- N -(3-oxocyclohex-1-en-1-yl)benzamide (3a)
The enaminone, 1a (0.799 g, 7.2 mmol), under an inert atmosphere, was stirred in a solution of NaH (0.391 g, 17.2 mmol) in dry THF (40 ml) maintaining the temperature below 293 K. The reaction was refluxed for 20 minutes, cooled to room temperature and stirred on an ice-bath for 5 minutes before a solution of benzoyl chloride 2 (2.09 g, 7.5 mmol) in dry THF (10 ml) was added dropwise over 5 minutes. After stirring at room temperature for a further 10 minutes, the mixture was quenched with concentrated hydrochloric acid (∼5 ml) and diluted with dichloromethane (25 ml). The mixture was transferred to a separatory funnel and washed successively with water (25 ml), 10% NaHCO3 and with water again. The organic layer was dried over sodium sulfate and concentrated in vacuo. The crude residue was purified by column chromatography (silica gel, EtOAc:hexanes = 5:5) to give compound 3a (1.37 g, 54%) as a faint yellow solid. (m.p. = 417–418 K) R f (EtOAc:hexanes 7:3) 1H NMR: (400 MHz, DMSO): 1.01 (6H, s, 2 × CH3), δ 1.27–1.32 (3H, t, CH3), 2.16 (2H, m, CH2), 2.43 (2H, t, CH2), 3.83 (6H, s, 2 × CH3), δ4.01–4.07 (2H, quart, CH2), δ 6.70 (H, s, CH), 7.04 (H, s, aromatic H), 10.25 (H, s, NH) ppm; 13C NMR (DMSO) δ 198.60, 165.69, 154.02, 152.27, 149.54, 142.88, 131.24, 115.82, 110.15, 107.81, 68.80, 60.83, 56.32, 49.96, 40.85, 32.13, 27.73, 15.35 ppm.
Preparation of 2-chloro- N -(5,5-dimethyl-3-oxocyclohex-1-en-1-yl)-4-ethoxy-3,5-δimethoxybenzamide (3b)
The same synthesis and purification method as for 3a was used to prepare 3b except that 1.00 g (7.2 mmol) of the enaminone 1b replaced 1a: this gave compound 3b (1.40 g, 51%) as a light white solid. (m.p. = 331–332 K) R f (EtOAc:hexanes 7:3) 1H NMR: (400 MHz, DMSO): δ 1.27–1.32 (3H, t, CH3), δ 1.8.7–1.95 (2H, quintet, CH2), 2.22–2.29 (2H, t, CH2), 3.32 (6, s, 2 × CH3, slight long-range coupling noticed), δ 4.00–4.06 (2H, quart, CH2), δ 6.71 (H, s, CH), 7.04 (H, s, aromatic H), 10.30 (H, s, NH) ppm; 13C NMR (DMSO) δ 198.62, 165.56, 156.14, 152.27, 149.55, 142.85, 131.27, 115.77, 110.76, 107.76, 68.80, 60.83, 56.32, 49.96, 40.85, 32.13, 27.73, 21.11, 15.35 ppm.
For both 3a and 3b crystals were grown from a 2:1 ethanol:water mixed solvent system.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 3 ▸. The N-bound H atoms were located in difference maps and their positions were freely refined. A riding model was used for the H atoms attached to C with C—H distances ranging from 0.95 to 0.99 Å and U iso(H) = 1.2U eq(C) [1.5U eq(CH3)]. For 3b there are two independent molecules in the asymmetric unit, in one of which the 5,5-dimethyl-3-oxocyclohex-1-en-1-yl moiety is disordered and was treated with similar metrical parameters with refined occupancies of 0.551 (2)/0.449 (2).
Table 3. Experimental details.
| 3a | 3b | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C17H20ClNO5 | C19H24ClNO5 |
| M r | 353.79 | 381.84 |
| Crystal system, space group | Monoclinic, P21/c | Monoclinic, P21/c |
| Temperature (K) | 150 | 150 |
| a, b, c (Å) | 14.654 (3), 8.9148 (17), 13.045 (2) | 14.6986 (13), 10.6309 (10), 25.131 (2) |
| β (°) | 102.581 (3) | 90.1851 (14) |
| V (Å3) | 1663.2 (5) | 3926.9 (6) |
| Z | 4 | 8 |
| Radiation type | Mo Kα | Mo Kα |
| μ (mm−1) | 0.26 | 0.22 |
| Crystal size (mm) | 0.34 × 0.32 × 0.10 | 0.48 × 0.44 × 0.21 |
| Data collection | ||
| Diffractometer | Bruker SMART APEXII CCD | Bruker SMART APEXII CCD |
| Absorption correction | Multi-scan (SADABS; Sheldrick, 2008 ▸) | Multi-scan (SADABS; Sheldrick, 2008 ▸) |
| T min, T max | 0.885, 0.975 | 0.841, 0.954 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 21434, 5291, 4023 | 67292, 12630, 10320 |
| R int | 0.041 | 0.026 |
| (sin θ/λ)max (Å−1) | 0.726 | 0.727 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.040, 0.111, 1.05 | 0.039, 0.115, 1.03 |
| No. of reflections | 5291 | 12630 |
| No. of parameters | 224 | 584 |
| No. of restraints | 0 | 399 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.41, −0.25 | 0.64, −0.35 |
Supplementary Material
Crystal structure: contains datablock(s) 3a, 3b, global. DOI: 10.1107/S2056989021001778/hb7962sup1.cif
Structure factors: contains datablock(s) 3a. DOI: 10.1107/S2056989021001778/hb79623asup2.hkl
Structure factors: contains datablock(s) 3b. DOI: 10.1107/S2056989021001778/hb79623bsup3.hkl
Supporting information file. DOI: 10.1107/S2056989021001778/hb79623asup4.cml
Supporting information file. DOI: 10.1107/S2056989021001778/hb79623bsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors wish to acknowledge the assistance of Dr Peter Zavalij at the University of Maryland for collecting the X-ray data.
supplementary crystallographic information
2-Chloro-4-ethoxy-3,5-dimethoxy-N-(3-oxocyclohex-1-en-1-yl)benzamide (3a) . Crystal data
| C17H20ClNO5 | F(000) = 744 |
| Mr = 353.79 | Dx = 1.413 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 14.654 (3) Å | Cell parameters from 5173 reflections |
| b = 8.9148 (17) Å | θ = 2.7–31.0° |
| c = 13.045 (2) Å | µ = 0.26 mm−1 |
| β = 102.581 (3)° | T = 150 K |
| V = 1663.2 (5) Å3 | Prism, yellow |
| Z = 4 | 0.34 × 0.32 × 0.10 mm |
2-Chloro-4-ethoxy-3,5-dimethoxy-N-(3-oxocyclohex-1-en-1-yl)benzamide (3a) . Data collection
| Bruker SMART APEXII CCD diffractometer | 5291 independent reflections |
| Radiation source: sealed tube | 4023 reflections with I > 2σ(I) |
| Detector resolution: 8.333 pixels mm-1 | Rint = 0.041 |
| φ and ω scans | θmax = 31.1°, θmin = 2.7° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2008) | h = −21→21 |
| Tmin = 0.885, Tmax = 0.975 | k = −12→12 |
| 21434 measured reflections | l = −18→18 |
2-Chloro-4-ethoxy-3,5-dimethoxy-N-(3-oxocyclohex-1-en-1-yl)benzamide (3a) . Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.040 | Hydrogen site location: mixed |
| wR(F2) = 0.111 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.05 | w = 1/[σ2(Fo2) + (0.0464P)2 + 0.4383P] where P = (Fo2 + 2Fc2)/3 |
| 5291 reflections | (Δ/σ)max < 0.001 |
| 224 parameters | Δρmax = 0.41 e Å−3 |
| 0 restraints | Δρmin = −0.25 e Å−3 |
2-Chloro-4-ethoxy-3,5-dimethoxy-N-(3-oxocyclohex-1-en-1-yl)benzamide (3a) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Compound #4 |
2-Chloro-4-ethoxy-3,5-dimethoxy-N-(3-oxocyclohex-1-en-1-yl)benzamide (3a) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Cl1 | 0.29234 (3) | 0.32109 (4) | 0.48615 (3) | 0.02725 (10) | |
| O1 | 0.18273 (7) | 0.38773 (11) | 0.64208 (7) | 0.0241 (2) | |
| O2 | 0.10261 (6) | 0.65866 (12) | 0.67309 (7) | 0.0239 (2) | |
| O3 | 0.10408 (7) | 0.88836 (11) | 0.54071 (9) | 0.0302 (2) | |
| O4 | 0.25869 (8) | 0.47397 (12) | 0.27145 (8) | 0.0320 (3) | |
| O5 | 0.38442 (7) | 0.56929 (12) | −0.02661 (7) | 0.0249 (2) | |
| N1 | 0.33695 (8) | 0.69571 (13) | 0.31234 (8) | 0.0191 (2) | |
| H1A | 0.3526 (12) | 0.762 (2) | 0.3588 (13) | 0.026 (4)* | |
| C1 | 0.23398 (9) | 0.60558 (15) | 0.42223 (10) | 0.0193 (2) | |
| C2 | 0.23353 (9) | 0.48844 (14) | 0.49310 (10) | 0.0189 (2) | |
| C3 | 0.18745 (9) | 0.50552 (14) | 0.57581 (10) | 0.0186 (2) | |
| C4 | 0.14420 (9) | 0.64091 (15) | 0.58915 (10) | 0.0194 (2) | |
| C5 | 0.14667 (9) | 0.76025 (15) | 0.51996 (10) | 0.0204 (2) | |
| C6 | 0.19046 (9) | 0.74103 (15) | 0.43585 (10) | 0.0207 (3) | |
| H6A | 0.190600 | 0.820783 | 0.387583 | 0.025* | |
| C7 | 0.25364 (12) | 0.39211 (18) | 0.73685 (12) | 0.0323 (3) | |
| H7A | 0.247923 | 0.303626 | 0.779583 | 0.048* | |
| H7B | 0.246122 | 0.483071 | 0.776384 | 0.048* | |
| H7C | 0.315408 | 0.392639 | 0.719441 | 0.048* | |
| C8 | 0.00347 (10) | 0.62546 (17) | 0.64724 (12) | 0.0260 (3) | |
| H8A | −0.006469 | 0.516730 | 0.633878 | 0.031* | |
| H8B | −0.027186 | 0.680643 | 0.583095 | 0.031* | |
| C9 | −0.03754 (11) | 0.6722 (2) | 0.73783 (14) | 0.0410 (4) | |
| H9A | −0.104576 | 0.649464 | 0.722087 | 0.061* | |
| H9B | −0.028357 | 0.780238 | 0.749697 | 0.061* | |
| H9C | −0.006531 | 0.617541 | 0.801030 | 0.061* | |
| C10 | 0.12623 (12) | 1.02402 (17) | 0.49314 (14) | 0.0327 (3) | |
| H10A | 0.109255 | 1.110196 | 0.531760 | 0.049* | |
| H10B | 0.091199 | 1.028074 | 0.420061 | 0.049* | |
| H10C | 0.193419 | 1.026858 | 0.494974 | 0.049* | |
| C11 | 0.27614 (9) | 0.58307 (16) | 0.32791 (10) | 0.0213 (3) | |
| C12 | 0.38339 (9) | 0.70472 (14) | 0.22986 (10) | 0.0180 (2) | |
| C13 | 0.36265 (9) | 0.61821 (15) | 0.14259 (10) | 0.0204 (3) | |
| H13A | 0.315554 | 0.543565 | 0.137043 | 0.024* | |
| C14 | 0.41092 (9) | 0.63713 (15) | 0.05714 (10) | 0.0201 (2) | |
| C15 | 0.49279 (10) | 0.74267 (18) | 0.07335 (11) | 0.0262 (3) | |
| H15A | 0.499687 | 0.781566 | 0.004357 | 0.031* | |
| H15B | 0.550511 | 0.686924 | 0.104906 | 0.031* | |
| C16 | 0.48180 (11) | 0.87402 (17) | 0.14418 (11) | 0.0275 (3) | |
| H16A | 0.430304 | 0.939526 | 0.107746 | 0.033* | |
| H16B | 0.540060 | 0.934001 | 0.158934 | 0.033* | |
| C17 | 0.46077 (10) | 0.81930 (16) | 0.24729 (11) | 0.0234 (3) | |
| H17A | 0.518006 | 0.774546 | 0.291063 | 0.028* | |
| H17B | 0.442654 | 0.905888 | 0.286032 | 0.028* |
2-Chloro-4-ethoxy-3,5-dimethoxy-N-(3-oxocyclohex-1-en-1-yl)benzamide (3a) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.03470 (19) | 0.02092 (16) | 0.02974 (18) | 0.00364 (13) | 0.01494 (14) | −0.00058 (13) |
| O1 | 0.0298 (5) | 0.0226 (5) | 0.0213 (5) | −0.0047 (4) | 0.0089 (4) | 0.0046 (4) |
| O2 | 0.0204 (5) | 0.0342 (5) | 0.0200 (5) | −0.0020 (4) | 0.0107 (4) | −0.0029 (4) |
| O3 | 0.0330 (6) | 0.0225 (5) | 0.0407 (6) | 0.0067 (4) | 0.0200 (5) | 0.0031 (4) |
| O4 | 0.0448 (6) | 0.0290 (5) | 0.0281 (5) | −0.0144 (5) | 0.0210 (5) | −0.0079 (4) |
| O5 | 0.0320 (5) | 0.0273 (5) | 0.0173 (4) | 0.0001 (4) | 0.0092 (4) | −0.0014 (4) |
| N1 | 0.0220 (5) | 0.0222 (5) | 0.0153 (5) | −0.0033 (4) | 0.0087 (4) | −0.0023 (4) |
| C1 | 0.0192 (6) | 0.0227 (6) | 0.0177 (6) | −0.0027 (5) | 0.0076 (4) | 0.0000 (5) |
| C2 | 0.0194 (6) | 0.0191 (6) | 0.0202 (6) | −0.0008 (5) | 0.0085 (4) | −0.0009 (5) |
| C3 | 0.0189 (6) | 0.0203 (6) | 0.0176 (6) | −0.0035 (5) | 0.0065 (4) | 0.0012 (5) |
| C4 | 0.0187 (6) | 0.0239 (6) | 0.0173 (6) | −0.0023 (5) | 0.0075 (4) | −0.0007 (5) |
| C5 | 0.0181 (6) | 0.0215 (6) | 0.0227 (6) | 0.0005 (5) | 0.0071 (5) | 0.0002 (5) |
| C6 | 0.0205 (6) | 0.0222 (6) | 0.0206 (6) | 0.0003 (5) | 0.0072 (5) | 0.0039 (5) |
| C7 | 0.0401 (9) | 0.0305 (8) | 0.0242 (7) | −0.0003 (6) | 0.0026 (6) | 0.0077 (6) |
| C8 | 0.0210 (6) | 0.0297 (7) | 0.0301 (7) | −0.0005 (5) | 0.0114 (5) | −0.0007 (6) |
| C9 | 0.0256 (8) | 0.0645 (12) | 0.0379 (9) | 0.0013 (8) | 0.0178 (7) | −0.0059 (8) |
| C10 | 0.0356 (8) | 0.0216 (7) | 0.0420 (9) | 0.0061 (6) | 0.0107 (7) | 0.0059 (6) |
| C11 | 0.0230 (6) | 0.0242 (6) | 0.0190 (6) | −0.0021 (5) | 0.0094 (5) | 0.0011 (5) |
| C12 | 0.0183 (6) | 0.0206 (6) | 0.0165 (5) | 0.0011 (4) | 0.0065 (4) | 0.0028 (4) |
| C13 | 0.0217 (6) | 0.0245 (6) | 0.0165 (6) | −0.0025 (5) | 0.0072 (4) | 0.0009 (5) |
| C14 | 0.0220 (6) | 0.0229 (6) | 0.0165 (6) | 0.0032 (5) | 0.0065 (4) | 0.0027 (5) |
| C15 | 0.0258 (7) | 0.0357 (8) | 0.0202 (6) | −0.0046 (6) | 0.0116 (5) | −0.0005 (6) |
| C16 | 0.0305 (7) | 0.0290 (7) | 0.0263 (7) | −0.0101 (6) | 0.0132 (6) | −0.0001 (6) |
| C17 | 0.0240 (6) | 0.0274 (7) | 0.0208 (6) | −0.0066 (5) | 0.0096 (5) | −0.0030 (5) |
2-Chloro-4-ethoxy-3,5-dimethoxy-N-(3-oxocyclohex-1-en-1-yl)benzamide (3a) . Geometric parameters (Å, º)
| Cl1—C2 | 1.7352 (13) | C8—C9 | 1.497 (2) |
| O1—C3 | 1.3714 (15) | C8—H8A | 0.9900 |
| O1—C7 | 1.4317 (18) | C8—H8B | 0.9900 |
| O2—C4 | 1.3734 (14) | C9—H9A | 0.9800 |
| O2—C8 | 1.4486 (16) | C9—H9B | 0.9800 |
| O3—C5 | 1.3567 (16) | C9—H9C | 0.9800 |
| O3—C10 | 1.4285 (18) | C10—H10A | 0.9800 |
| O4—C11 | 1.2132 (17) | C10—H10B | 0.9800 |
| O5—C14 | 1.2350 (16) | C10—H10C | 0.9800 |
| N1—C11 | 1.3865 (17) | C12—C13 | 1.3536 (18) |
| N1—C12 | 1.3950 (15) | C12—C17 | 1.5061 (18) |
| N1—H1A | 0.844 (18) | C13—C14 | 1.4543 (17) |
| C1—C6 | 1.3949 (18) | C13—H13A | 0.9500 |
| C1—C2 | 1.3956 (18) | C14—C15 | 1.5029 (19) |
| C1—C11 | 1.5056 (17) | C15—C16 | 1.522 (2) |
| C2—C3 | 1.4001 (16) | C15—H15A | 0.9900 |
| C3—C4 | 1.3917 (18) | C15—H15B | 0.9900 |
| C4—C5 | 1.4008 (19) | C16—C17 | 1.5241 (19) |
| C5—C6 | 1.3969 (17) | C16—H16A | 0.9900 |
| C6—H6A | 0.9500 | C16—H16B | 0.9900 |
| C7—H7A | 0.9800 | C17—H17A | 0.9900 |
| C7—H7B | 0.9800 | C17—H17B | 0.9900 |
| C7—H7C | 0.9800 | ||
| C3—O1—C7 | 113.37 (11) | C8—C9—H9C | 109.5 |
| C4—O2—C8 | 112.80 (10) | H9A—C9—H9C | 109.5 |
| C5—O3—C10 | 117.91 (11) | H9B—C9—H9C | 109.5 |
| C11—N1—C12 | 126.28 (11) | O3—C10—H10A | 109.5 |
| C11—N1—H1A | 119.2 (12) | O3—C10—H10B | 109.5 |
| C12—N1—H1A | 114.3 (12) | H10A—C10—H10B | 109.5 |
| C6—C1—C2 | 119.68 (11) | O3—C10—H10C | 109.5 |
| C6—C1—C11 | 119.99 (11) | H10A—C10—H10C | 109.5 |
| C2—C1—C11 | 120.24 (12) | H10B—C10—H10C | 109.5 |
| C1—C2—C3 | 120.11 (12) | O4—C11—N1 | 123.29 (11) |
| C1—C2—Cl1 | 122.32 (9) | O4—C11—C1 | 122.22 (12) |
| C3—C2—Cl1 | 117.53 (10) | N1—C11—C1 | 114.48 (11) |
| O1—C3—C4 | 119.88 (11) | C13—C12—N1 | 123.91 (12) |
| O1—C3—C2 | 120.10 (11) | C13—C12—C17 | 122.49 (11) |
| C4—C3—C2 | 119.99 (11) | N1—C12—C17 | 113.59 (11) |
| O2—C4—C3 | 119.54 (11) | C12—C13—C14 | 121.39 (12) |
| O2—C4—C5 | 120.30 (12) | C12—C13—H13A | 119.3 |
| C3—C4—C5 | 120.11 (11) | C14—C13—H13A | 119.3 |
| O3—C5—C6 | 124.74 (12) | O5—C14—C13 | 120.63 (12) |
| O3—C5—C4 | 115.67 (11) | O5—C14—C15 | 121.24 (11) |
| C6—C5—C4 | 119.59 (12) | C13—C14—C15 | 118.12 (11) |
| C1—C6—C5 | 120.47 (12) | C14—C15—C16 | 112.36 (11) |
| C1—C6—H6A | 119.8 | C14—C15—H15A | 109.1 |
| C5—C6—H6A | 119.8 | C16—C15—H15A | 109.1 |
| O1—C7—H7A | 109.5 | C14—C15—H15B | 109.1 |
| O1—C7—H7B | 109.5 | C16—C15—H15B | 109.1 |
| H7A—C7—H7B | 109.5 | H15A—C15—H15B | 107.9 |
| O1—C7—H7C | 109.5 | C15—C16—C17 | 110.98 (12) |
| H7A—C7—H7C | 109.5 | C15—C16—H16A | 109.4 |
| H7B—C7—H7C | 109.5 | C17—C16—H16A | 109.4 |
| O2—C8—C9 | 108.30 (12) | C15—C16—H16B | 109.4 |
| O2—C8—H8A | 110.0 | C17—C16—H16B | 109.4 |
| C9—C8—H8A | 110.0 | H16A—C16—H16B | 108.0 |
| O2—C8—H8B | 110.0 | C12—C17—C16 | 111.97 (11) |
| C9—C8—H8B | 110.0 | C12—C17—H17A | 109.2 |
| H8A—C8—H8B | 108.4 | C16—C17—H17A | 109.2 |
| C8—C9—H9A | 109.5 | C12—C17—H17B | 109.2 |
| C8—C9—H9B | 109.5 | C16—C17—H17B | 109.2 |
| H9A—C9—H9B | 109.5 | H17A—C17—H17B | 107.9 |
| C6—C1—C2—C3 | 1.71 (19) | C11—C1—C6—C5 | 176.51 (12) |
| C11—C1—C2—C3 | −174.66 (12) | O3—C5—C6—C1 | 178.77 (13) |
| C6—C1—C2—Cl1 | −175.87 (10) | C4—C5—C6—C1 | −1.9 (2) |
| C11—C1—C2—Cl1 | 7.76 (18) | C4—O2—C8—C9 | −170.82 (13) |
| C7—O1—C3—C4 | −85.34 (15) | C12—N1—C11—O4 | 2.7 (2) |
| C7—O1—C3—C2 | 96.38 (15) | C12—N1—C11—C1 | −178.18 (12) |
| C1—C2—C3—O1 | 176.51 (12) | C6—C1—C11—O4 | −129.20 (15) |
| Cl1—C2—C3—O1 | −5.80 (17) | C2—C1—C11—O4 | 47.2 (2) |
| C1—C2—C3—C4 | −1.77 (19) | C6—C1—C11—N1 | 51.64 (17) |
| Cl1—C2—C3—C4 | 175.92 (10) | C2—C1—C11—N1 | −132.00 (13) |
| C8—O2—C4—C3 | −94.55 (14) | C11—N1—C12—C13 | 12.4 (2) |
| C8—O2—C4—C5 | 88.21 (15) | C11—N1—C12—C17 | −166.70 (13) |
| O1—C3—C4—O2 | 4.46 (18) | N1—C12—C13—C14 | 177.36 (12) |
| C2—C3—C4—O2 | −177.25 (11) | C17—C12—C13—C14 | −3.6 (2) |
| O1—C3—C4—C5 | −178.29 (12) | C12—C13—C14—O5 | −171.94 (13) |
| C2—C3—C4—C5 | −0.01 (19) | C12—C13—C14—C15 | 7.3 (2) |
| C10—O3—C5—C6 | −18.0 (2) | O5—C14—C15—C16 | 146.86 (13) |
| C10—O3—C5—C4 | 162.68 (13) | C13—C14—C15—C16 | −32.36 (18) |
| O2—C4—C5—O3 | −1.55 (19) | C14—C15—C16—C17 | 52.88 (17) |
| C3—C4—C5—O3 | −178.78 (12) | C13—C12—C17—C16 | 24.94 (19) |
| O2—C4—C5—C6 | 179.06 (12) | N1—C12—C17—C16 | −155.90 (12) |
| C3—C4—C5—C6 | 1.8 (2) | C15—C16—C17—C12 | −48.67 (17) |
| C2—C1—C6—C5 | 0.1 (2) |
2-Chloro-4-ethoxy-3,5-dimethoxy-N-(3-oxocyclohex-1-en-1-yl)benzamide (3a) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1A···O5i | 0.844 (18) | 2.098 (18) | 2.9410 (15) | 177.1 (16) |
| C7—H7A···Cl1ii | 0.98 | 2.86 | 3.7022 (16) | 145 |
| C7—H7A···O4ii | 0.98 | 2.48 | 3.293 (2) | 140 |
| C9—H9A···O4iii | 0.98 | 2.53 | 3.470 (2) | 161 |
Symmetry codes: (i) x, −y+3/2, z+1/2; (ii) x, −y+1/2, z+1/2; (iii) −x, −y+1, −z+1.
2-Chloro-N-(5,5-dimethyl-3-oxocyclohex-1-en-1-yl)-4-ethoxy-3,5-dimethoxybenzamide (3b) . Crystal data
| C19H24ClNO5 | F(000) = 1616 |
| Mr = 381.84 | Dx = 1.292 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 14.6986 (13) Å | Cell parameters from 23388 reflections |
| b = 10.6309 (10) Å | θ = 2.4–31.0° |
| c = 25.131 (2) Å | µ = 0.22 mm−1 |
| β = 90.1851 (14)° | T = 150 K |
| V = 3926.9 (6) Å3 | Prism, colourless |
| Z = 8 | 0.48 × 0.44 × 0.21 mm |
2-Chloro-N-(5,5-dimethyl-3-oxocyclohex-1-en-1-yl)-4-ethoxy-3,5-dimethoxybenzamide (3b) . Data collection
| Bruker SMART APEXII CCD diffractometer | 12630 independent reflections |
| Radiation source: sealed tube | 10320 reflections with I > 2σ(I) |
| Detector resolution: 8.333 pixels mm-1 | Rint = 0.026 |
| φ and ω scans | θmax = 31.1°, θmin = 2.1° |
| Absorption correction: multi-scan (SADABS; Sheldrick, 2008) | h = −21→21 |
| Tmin = 0.841, Tmax = 0.954 | k = −15→15 |
| 67292 measured reflections | l = −36→36 |
2-Chloro-N-(5,5-dimethyl-3-oxocyclohex-1-en-1-yl)-4-ethoxy-3,5-dimethoxybenzamide (3b) . Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.039 | Hydrogen site location: mixed |
| wR(F2) = 0.115 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.0599P)2 + 1.0458P] where P = (Fo2 + 2Fc2)/3 |
| 12630 reflections | (Δ/σ)max = 0.006 |
| 584 parameters | Δρmax = 0.64 e Å−3 |
| 399 restraints | Δρmin = −0.35 e Å−3 |
2-Chloro-N-(5,5-dimethyl-3-oxocyclohex-1-en-1-yl)-4-ethoxy-3,5-dimethoxybenzamide (3b) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Compound #6 |
2-Chloro-N-(5,5-dimethyl-3-oxocyclohex-1-en-1-yl)-4-ethoxy-3,5-dimethoxybenzamide (3b) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Cl1A | 0.74167 (2) | 1.06636 (3) | 0.50622 (2) | 0.03341 (7) | |
| O1A | 0.85300 (6) | 0.92470 (9) | 0.43031 (3) | 0.03358 (19) | |
| O2A | 0.96808 (6) | 0.73233 (10) | 0.45218 (4) | 0.0371 (2) | |
| O3A | 0.98422 (6) | 0.64044 (10) | 0.55396 (4) | 0.0411 (2) | |
| O4A | 0.79762 (5) | 0.97694 (9) | 0.66166 (3) | 0.03064 (18) | |
| O5A | 0.54514 (6) | 1.01715 (10) | 0.78213 (3) | 0.0376 (2) | |
| N1A | 0.67015 (6) | 0.94345 (9) | 0.61067 (4) | 0.02155 (17) | |
| H1AA | 0.6506 (11) | 0.9153 (15) | 0.5810 (7) | 0.034 (4)* | |
| C1A | 0.81762 (7) | 0.89610 (10) | 0.57423 (4) | 0.02277 (19) | |
| C2A | 0.81118 (7) | 0.94022 (11) | 0.52235 (4) | 0.0245 (2) | |
| C3A | 0.86370 (7) | 0.88575 (12) | 0.48185 (4) | 0.0272 (2) | |
| C4A | 0.92222 (7) | 0.78611 (12) | 0.49363 (5) | 0.0293 (2) | |
| C5A | 0.92801 (7) | 0.74122 (12) | 0.54613 (5) | 0.0293 (2) | |
| C6A | 0.87719 (7) | 0.79758 (11) | 0.58605 (4) | 0.0263 (2) | |
| H6AA | 0.882994 | 0.768878 | 0.621695 | 0.032* | |
| C7A | 0.91558 (11) | 1.01997 (17) | 0.41412 (5) | 0.0474 (4) | |
| H7AA | 0.903236 | 1.043380 | 0.377078 | 0.071* | |
| H7AB | 0.977879 | 0.987895 | 0.417198 | 0.071* | |
| H7AC | 0.908551 | 1.093994 | 0.436981 | 0.071* | |
| C8A | 1.06559 (9) | 0.7271 (2) | 0.45645 (6) | 0.0624 (5) | |
| H8AA | 1.083977 | 0.647258 | 0.473668 | 0.075* | |
| H8AB | 1.087544 | 0.797509 | 0.478864 | 0.075* | |
| C9A | 1.10703 (11) | 0.7356 (2) | 0.40297 (7) | 0.0628 (5) | |
| H9AA | 1.172751 | 0.721418 | 0.405797 | 0.094* | |
| H9AB | 1.095707 | 0.819284 | 0.388025 | 0.094* | |
| H9AC | 1.080069 | 0.671610 | 0.379685 | 0.094* | |
| C10A | 0.99347 (11) | 0.59578 (18) | 0.60744 (7) | 0.0550 (4) | |
| H10A | 1.033598 | 0.522173 | 0.607983 | 0.082* | |
| H10B | 0.933524 | 0.572283 | 0.621209 | 0.082* | |
| H10C | 1.019610 | 0.662303 | 0.629735 | 0.082* | |
| C11A | 0.76218 (7) | 0.94515 (10) | 0.61997 (4) | 0.02202 (19) | |
| C12A | 0.60074 (6) | 0.97178 (10) | 0.64589 (4) | 0.02102 (18) | |
| C13A | 0.61233 (7) | 0.98962 (10) | 0.69867 (4) | 0.02312 (19) | |
| H13A | 0.671961 | 0.987378 | 0.713341 | 0.028* | |
| C14A | 0.53549 (7) | 1.01213 (11) | 0.73334 (4) | 0.0258 (2) | |
| C15A | 0.44417 (8) | 1.03239 (13) | 0.70805 (5) | 0.0322 (2) | |
| H15A | 0.437492 | 1.122477 | 0.698850 | 0.039* | |
| H15C | 0.396169 | 1.011049 | 0.734077 | 0.039* | |
| C16A | 0.43022 (7) | 0.95307 (15) | 0.65763 (5) | 0.0363 (3) | |
| C17A | 0.50923 (7) | 0.98156 (14) | 0.61962 (4) | 0.0327 (3) | |
| H17A | 0.506711 | 0.922034 | 0.589352 | 0.039* | |
| H17B | 0.501634 | 1.067588 | 0.605191 | 0.039* | |
| C18A | 0.34099 (9) | 0.9921 (3) | 0.63114 (6) | 0.0782 (8) | |
| H18A | 0.290687 | 0.979665 | 0.656099 | 0.117* | |
| H18B | 0.330783 | 0.940598 | 0.599323 | 0.117* | |
| H18C | 0.344203 | 1.080952 | 0.620991 | 0.117* | |
| C19A | 0.42926 (11) | 0.81411 (18) | 0.67166 (7) | 0.0577 (5) | |
| H19A | 0.380973 | 0.797814 | 0.697561 | 0.087* | |
| H19B | 0.488156 | 0.790343 | 0.687029 | 0.087* | |
| H19C | 0.418020 | 0.764497 | 0.639432 | 0.087* | |
| Cl1B | 0.33068 (2) | 0.36791 (3) | 0.37042 (2) | 0.03251 (7) | |
| O1B | 0.19443 (6) | 0.32113 (8) | 0.28774 (3) | 0.03058 (17) | |
| O2B | 0.12498 (6) | 0.49472 (9) | 0.21650 (3) | 0.03228 (18) | |
| O3B | 0.17637 (6) | 0.73524 (8) | 0.22015 (3) | 0.03071 (18) | |
| O4B | 0.35516 (6) | 0.74079 (10) | 0.40209 (4) | 0.0415 (2) | |
| C1B | 0.31411 (6) | 0.60638 (10) | 0.33084 (4) | 0.02011 (18) | |
| C2B | 0.28768 (7) | 0.48116 (10) | 0.32756 (4) | 0.02208 (19) | |
| C3B | 0.22268 (7) | 0.44399 (10) | 0.28987 (4) | 0.02362 (19) | |
| C4B | 0.18562 (7) | 0.53194 (11) | 0.25503 (4) | 0.0240 (2) | |
| C5B | 0.21397 (7) | 0.65796 (10) | 0.25735 (4) | 0.02301 (19) | |
| C6B | 0.27647 (7) | 0.69485 (10) | 0.29595 (4) | 0.02189 (18) | |
| H6BA | 0.293725 | 0.780733 | 0.298645 | 0.026* | |
| C7B | 0.23634 (11) | 0.25212 (13) | 0.24515 (7) | 0.0436 (3) | |
| H7BA | 0.215324 | 0.164658 | 0.245936 | 0.065* | |
| H7BB | 0.302605 | 0.254294 | 0.249483 | 0.065* | |
| H7BC | 0.219649 | 0.290379 | 0.211010 | 0.065* | |
| C8B | 0.03306 (9) | 0.47519 (14) | 0.23569 (6) | 0.0416 (3) | |
| H8BA | 0.034665 | 0.416637 | 0.266315 | 0.050* | |
| H8BB | −0.003821 | 0.435696 | 0.207205 | 0.050* | |
| C9B | −0.01135 (10) | 0.59598 (18) | 0.25249 (10) | 0.0675 (6) | |
| H9BA | −0.072982 | 0.578521 | 0.265278 | 0.101* | |
| H9BB | −0.014447 | 0.653490 | 0.222061 | 0.101* | |
| H9BC | 0.024414 | 0.634758 | 0.281091 | 0.101* | |
| C10B | 0.20442 (9) | 0.86380 (11) | 0.22096 (5) | 0.0310 (2) | |
| H10D | 0.173044 | 0.909934 | 0.192567 | 0.047* | |
| H10E | 0.270299 | 0.868615 | 0.215424 | 0.047* | |
| H10F | 0.189185 | 0.901108 | 0.255464 | 0.047* | |
| C11B | 0.37827 (7) | 0.65588 (10) | 0.37276 (4) | 0.02269 (19) | |
| O5B | 0.5926 (5) | 0.7923 (8) | 0.5286 (3) | 0.0383 (12) | 0.551 (2) |
| N1B | 0.4614 (6) | 0.6071 (10) | 0.3766 (4) | 0.0274 (15) | 0.551 (2) |
| H1BA | 0.471 (3) | 0.552 (4) | 0.3576 (18) | 0.026 (13)* | 0.551 (2) |
| C12B | 0.5307 (6) | 0.6264 (11) | 0.4131 (5) | 0.0259 (10) | 0.551 (2) |
| C13B | 0.5224 (5) | 0.7057 (8) | 0.4537 (3) | 0.0210 (9) | 0.551 (2) |
| H13B | 0.466093 | 0.746700 | 0.460177 | 0.025* | 0.551 (2) |
| C14B | 0.6004 (6) | 0.7288 (8) | 0.4881 (4) | 0.0293 (11) | 0.551 (2) |
| C15B | 0.69268 (14) | 0.6766 (2) | 0.47298 (9) | 0.0304 (5) | 0.551 (2) |
| H15B | 0.729500 | 0.665206 | 0.505684 | 0.036* | 0.551 (2) |
| H15D | 0.724367 | 0.738554 | 0.450190 | 0.036* | 0.551 (2) |
| C16B | 0.6870 (4) | 0.5507 (7) | 0.4434 (3) | 0.0262 (9) | 0.551 (2) |
| C17B | 0.62225 (13) | 0.5682 (2) | 0.39563 (8) | 0.0261 (4) | 0.551 (2) |
| H17C | 0.610886 | 0.485622 | 0.378636 | 0.031* | 0.551 (2) |
| H17D | 0.651346 | 0.623624 | 0.368993 | 0.031* | 0.551 (2) |
| C18B | 0.65099 (17) | 0.4478 (2) | 0.48043 (10) | 0.0384 (5) | 0.551 (2) |
| H18D | 0.692949 | 0.437144 | 0.510480 | 0.058* | 0.551 (2) |
| H18E | 0.646148 | 0.368375 | 0.460797 | 0.058* | 0.551 (2) |
| H18F | 0.590875 | 0.471931 | 0.493723 | 0.058* | 0.551 (2) |
| C19B | 0.78134 (15) | 0.5136 (3) | 0.42324 (10) | 0.0436 (7) | 0.551 (2) |
| H19D | 0.823551 | 0.507798 | 0.453410 | 0.065* | 0.551 (2) |
| H19E | 0.803155 | 0.577384 | 0.398177 | 0.065* | 0.551 (2) |
| H19F | 0.777791 | 0.431964 | 0.405263 | 0.065* | 0.551 (2) |
| O5C | 0.6059 (7) | 0.7977 (10) | 0.5201 (4) | 0.0413 (15) | 0.449 (2) |
| N1C | 0.4643 (5) | 0.5949 (11) | 0.3708 (4) | 0.0156 (9) | 0.449 (2) |
| H1CA | 0.476 (3) | 0.549 (4) | 0.3423 (18) | 0.019 (12)* | 0.449 (2) |
| C12C | 0.5324 (8) | 0.6138 (14) | 0.4091 (6) | 0.0262 (12) | 0.449 (2) |
| C13C | 0.5340 (6) | 0.6981 (11) | 0.4488 (4) | 0.0268 (13) | 0.449 (2) |
| H13C | 0.485165 | 0.756476 | 0.450818 | 0.032* | 0.449 (2) |
| C14C | 0.6048 (7) | 0.7067 (10) | 0.4889 (5) | 0.0310 (13) | 0.449 (2) |
| C15C | 0.67207 (18) | 0.6008 (3) | 0.49113 (11) | 0.0341 (6) | 0.449 (2) |
| H15E | 0.649682 | 0.536354 | 0.516327 | 0.041* | 0.449 (2) |
| H15F | 0.730553 | 0.633284 | 0.505144 | 0.041* | 0.449 (2) |
| C16C | 0.6890 (6) | 0.5385 (9) | 0.4371 (4) | 0.0293 (12) | 0.449 (2) |
| C17C | 0.59763 (16) | 0.5023 (2) | 0.41227 (10) | 0.0244 (5) | 0.449 (2) |
| H17E | 0.607993 | 0.468961 | 0.376009 | 0.029* | 0.449 (2) |
| H17F | 0.569501 | 0.434700 | 0.433730 | 0.029* | 0.449 (2) |
| C18C | 0.7464 (2) | 0.4183 (3) | 0.44515 (14) | 0.0434 (8) | 0.449 (2) |
| H18G | 0.806403 | 0.441080 | 0.459204 | 0.065* | 0.449 (2) |
| H18H | 0.753484 | 0.375020 | 0.410976 | 0.065* | 0.449 (2) |
| H18I | 0.715614 | 0.362431 | 0.470371 | 0.065* | 0.449 (2) |
| C19C | 0.74033 (19) | 0.6285 (3) | 0.39994 (12) | 0.0373 (7) | 0.449 (2) |
| H19G | 0.706920 | 0.708007 | 0.397146 | 0.056* | 0.449 (2) |
| H19H | 0.745573 | 0.590307 | 0.364584 | 0.056* | 0.449 (2) |
| H19I | 0.801241 | 0.644531 | 0.414378 | 0.056* | 0.449 (2) |
2-Chloro-N-(5,5-dimethyl-3-oxocyclohex-1-en-1-yl)-4-ethoxy-3,5-dimethoxybenzamide (3b) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1A | 0.03295 (13) | 0.04046 (16) | 0.02684 (13) | 0.00661 (11) | 0.00220 (10) | 0.00619 (11) |
| O1A | 0.0289 (4) | 0.0525 (5) | 0.0193 (4) | −0.0066 (4) | 0.0016 (3) | −0.0010 (4) |
| O2A | 0.0223 (4) | 0.0560 (6) | 0.0330 (4) | −0.0005 (4) | 0.0088 (3) | −0.0150 (4) |
| O3A | 0.0336 (4) | 0.0458 (5) | 0.0439 (5) | 0.0154 (4) | 0.0105 (4) | 0.0032 (4) |
| O4A | 0.0233 (3) | 0.0448 (5) | 0.0238 (4) | −0.0036 (3) | 0.0004 (3) | −0.0060 (3) |
| O5A | 0.0335 (4) | 0.0616 (6) | 0.0178 (4) | −0.0051 (4) | 0.0043 (3) | 0.0003 (4) |
| N1A | 0.0193 (4) | 0.0277 (4) | 0.0176 (4) | −0.0009 (3) | 0.0024 (3) | −0.0040 (3) |
| C1A | 0.0181 (4) | 0.0292 (5) | 0.0211 (4) | −0.0022 (4) | 0.0033 (3) | −0.0018 (4) |
| C2A | 0.0195 (4) | 0.0320 (5) | 0.0220 (5) | −0.0011 (4) | 0.0017 (3) | 0.0000 (4) |
| C3A | 0.0208 (4) | 0.0409 (6) | 0.0200 (5) | −0.0046 (4) | 0.0029 (4) | −0.0032 (4) |
| C4A | 0.0189 (4) | 0.0416 (6) | 0.0274 (5) | −0.0031 (4) | 0.0070 (4) | −0.0080 (5) |
| C5A | 0.0200 (4) | 0.0353 (6) | 0.0326 (6) | 0.0019 (4) | 0.0053 (4) | −0.0016 (5) |
| C6A | 0.0196 (4) | 0.0340 (6) | 0.0252 (5) | 0.0002 (4) | 0.0042 (4) | 0.0016 (4) |
| C7A | 0.0488 (8) | 0.0673 (10) | 0.0262 (6) | −0.0183 (7) | 0.0043 (5) | 0.0060 (6) |
| C8A | 0.0219 (6) | 0.1254 (17) | 0.0400 (8) | 0.0095 (8) | 0.0090 (5) | −0.0176 (9) |
| C9A | 0.0377 (7) | 0.0939 (14) | 0.0570 (10) | −0.0074 (8) | 0.0225 (7) | −0.0248 (10) |
| C10A | 0.0494 (8) | 0.0604 (10) | 0.0552 (9) | 0.0266 (8) | 0.0120 (7) | 0.0177 (8) |
| C11A | 0.0197 (4) | 0.0250 (5) | 0.0213 (4) | −0.0013 (3) | 0.0031 (3) | −0.0002 (4) |
| C12A | 0.0201 (4) | 0.0228 (5) | 0.0202 (4) | 0.0008 (3) | 0.0031 (3) | −0.0002 (4) |
| C13A | 0.0214 (4) | 0.0287 (5) | 0.0193 (4) | −0.0013 (4) | 0.0018 (3) | 0.0004 (4) |
| C14A | 0.0266 (5) | 0.0311 (5) | 0.0199 (5) | −0.0002 (4) | 0.0039 (4) | 0.0008 (4) |
| C15A | 0.0273 (5) | 0.0462 (7) | 0.0232 (5) | 0.0103 (5) | 0.0048 (4) | −0.0002 (5) |
| C16A | 0.0187 (4) | 0.0670 (9) | 0.0232 (5) | 0.0029 (5) | 0.0020 (4) | −0.0056 (5) |
| C17A | 0.0219 (5) | 0.0560 (8) | 0.0201 (5) | 0.0068 (5) | 0.0006 (4) | −0.0016 (5) |
| C18A | 0.0225 (6) | 0.180 (2) | 0.0325 (7) | 0.0184 (10) | −0.0011 (5) | −0.0117 (11) |
| C19A | 0.0454 (8) | 0.0640 (11) | 0.0639 (10) | −0.0278 (7) | 0.0217 (7) | −0.0218 (8) |
| Cl1B | 0.03134 (13) | 0.03067 (14) | 0.03546 (15) | −0.00338 (10) | −0.00739 (11) | 0.01257 (11) |
| O1B | 0.0322 (4) | 0.0260 (4) | 0.0335 (4) | −0.0104 (3) | −0.0015 (3) | 0.0034 (3) |
| O2B | 0.0319 (4) | 0.0379 (5) | 0.0270 (4) | −0.0106 (3) | −0.0110 (3) | 0.0017 (3) |
| O3B | 0.0340 (4) | 0.0294 (4) | 0.0287 (4) | −0.0042 (3) | −0.0122 (3) | 0.0075 (3) |
| O4B | 0.0345 (4) | 0.0486 (6) | 0.0412 (5) | 0.0168 (4) | −0.0129 (4) | −0.0232 (4) |
| C1B | 0.0169 (4) | 0.0262 (5) | 0.0172 (4) | 0.0002 (3) | 0.0008 (3) | −0.0015 (4) |
| C2B | 0.0195 (4) | 0.0258 (5) | 0.0209 (4) | −0.0007 (3) | 0.0004 (3) | 0.0043 (4) |
| C3B | 0.0222 (4) | 0.0248 (5) | 0.0239 (5) | −0.0057 (4) | 0.0008 (4) | 0.0015 (4) |
| C4B | 0.0214 (4) | 0.0296 (5) | 0.0209 (5) | −0.0053 (4) | −0.0030 (3) | 0.0004 (4) |
| C5B | 0.0223 (4) | 0.0262 (5) | 0.0205 (4) | −0.0012 (4) | −0.0023 (3) | 0.0029 (4) |
| C6B | 0.0211 (4) | 0.0229 (5) | 0.0217 (4) | −0.0012 (3) | −0.0013 (3) | −0.0001 (4) |
| C7B | 0.0510 (8) | 0.0262 (6) | 0.0538 (9) | −0.0048 (5) | 0.0071 (6) | −0.0044 (6) |
| C8B | 0.0284 (6) | 0.0426 (7) | 0.0536 (8) | −0.0126 (5) | −0.0136 (5) | 0.0074 (6) |
| C9B | 0.0263 (6) | 0.0535 (10) | 0.1226 (18) | −0.0024 (6) | −0.0072 (8) | 0.0030 (11) |
| C10B | 0.0358 (6) | 0.0266 (5) | 0.0306 (6) | 0.0012 (4) | −0.0068 (4) | 0.0051 (4) |
| C11B | 0.0206 (4) | 0.0276 (5) | 0.0199 (4) | 0.0015 (4) | −0.0020 (3) | −0.0019 (4) |
| O5B | 0.044 (3) | 0.0460 (17) | 0.025 (2) | 0.0018 (18) | −0.0094 (18) | −0.0140 (14) |
| N1B | 0.0313 (19) | 0.034 (2) | 0.0166 (19) | 0.0050 (12) | 0.0028 (10) | −0.0165 (13) |
| C12B | 0.0168 (14) | 0.039 (3) | 0.022 (2) | 0.0082 (14) | −0.0033 (13) | −0.0089 (15) |
| C13B | 0.0186 (16) | 0.0277 (16) | 0.0168 (14) | 0.0033 (13) | −0.0052 (14) | −0.0116 (11) |
| C14B | 0.0250 (14) | 0.036 (2) | 0.0273 (17) | 0.0022 (14) | −0.0035 (11) | −0.0064 (16) |
| C15B | 0.0246 (9) | 0.0413 (12) | 0.0254 (9) | −0.0051 (8) | −0.0078 (7) | −0.0024 (9) |
| C16B | 0.0154 (13) | 0.0386 (19) | 0.0245 (18) | 0.0026 (12) | −0.0049 (11) | 0.0001 (14) |
| C17B | 0.0198 (8) | 0.0370 (11) | 0.0216 (9) | 0.0018 (7) | −0.0014 (7) | −0.0058 (8) |
| C18B | 0.0415 (12) | 0.0370 (12) | 0.0367 (12) | 0.0060 (9) | 0.0004 (9) | 0.0069 (9) |
| C19B | 0.0224 (9) | 0.0731 (19) | 0.0351 (12) | 0.0124 (10) | −0.0029 (8) | −0.0055 (12) |
| O5C | 0.039 (2) | 0.059 (2) | 0.026 (3) | −0.0141 (15) | 0.0007 (16) | −0.0192 (19) |
| N1C | 0.0102 (13) | 0.025 (2) | 0.012 (2) | 0.0063 (12) | −0.0053 (14) | −0.0123 (18) |
| C12C | 0.028 (2) | 0.035 (2) | 0.0154 (18) | 0.0042 (16) | −0.0047 (15) | −0.0139 (15) |
| C13C | 0.024 (2) | 0.029 (2) | 0.028 (2) | 0.0077 (15) | 0.0039 (15) | −0.0020 (16) |
| C14C | 0.034 (2) | 0.039 (3) | 0.0197 (18) | −0.0018 (18) | −0.0065 (15) | −0.009 (2) |
| C15C | 0.0299 (12) | 0.0471 (16) | 0.0252 (12) | 0.0027 (11) | −0.0088 (9) | −0.0059 (11) |
| C16C | 0.029 (2) | 0.035 (2) | 0.024 (2) | 0.0004 (15) | −0.0035 (15) | −0.0040 (16) |
| C17C | 0.0234 (10) | 0.0253 (11) | 0.0246 (11) | 0.0024 (9) | −0.0036 (8) | −0.0028 (9) |
| C18C | 0.0343 (14) | 0.0524 (18) | 0.0434 (17) | 0.0172 (13) | −0.0090 (12) | −0.0012 (14) |
| C19C | 0.0297 (12) | 0.0467 (16) | 0.0356 (14) | −0.0080 (11) | 0.0034 (10) | −0.0072 (12) |
2-Chloro-N-(5,5-dimethyl-3-oxocyclohex-1-en-1-yl)-4-ethoxy-3,5-dimethoxybenzamide (3b) . Geometric parameters (Å, º)
| Cl1A—C2A | 1.7331 (12) | C3B—C4B | 1.3908 (15) |
| O1A—C3A | 1.3684 (14) | C4B—C5B | 1.4042 (15) |
| O1A—C7A | 1.4281 (17) | C5B—C6B | 1.3903 (14) |
| O2A—C4A | 1.3678 (13) | C6B—H6BA | 0.9500 |
| O2A—C8A | 1.4381 (16) | C7B—H7BA | 0.9800 |
| O3A—C5A | 1.3667 (15) | C7B—H7BB | 0.9800 |
| O3A—C10A | 1.4313 (19) | C7B—H7BC | 0.9800 |
| O4A—C11A | 1.2163 (13) | C8B—C9B | 1.502 (2) |
| O5A—C14A | 1.2349 (13) | C8B—H8BA | 0.9900 |
| N1A—C11A | 1.3722 (13) | C8B—H8BB | 0.9900 |
| N1A—C12A | 1.3859 (12) | C9B—H9BA | 0.9800 |
| N1A—H1AA | 0.853 (16) | C9B—H9BB | 0.9800 |
| C1A—C2A | 1.3885 (15) | C9B—H9BC | 0.9800 |
| C1A—C6A | 1.3964 (15) | C10B—H10D | 0.9800 |
| C1A—C11A | 1.5047 (14) | C10B—H10E | 0.9800 |
| C2A—C3A | 1.4044 (15) | C10B—H10F | 0.9800 |
| C3A—C4A | 1.3956 (17) | C11B—N1B | 1.330 (8) |
| C4A—C5A | 1.4053 (17) | C11B—N1C | 1.421 (8) |
| C5A—C6A | 1.3886 (15) | O5B—C14B | 1.229 (8) |
| C6A—H6AA | 0.9500 | N1B—C12B | 1.384 (8) |
| C7A—H7AA | 0.9800 | N1B—H1BA | 0.77 (5) |
| C7A—H7AB | 0.9800 | C12B—C13B | 1.330 (7) |
| C7A—H7AC | 0.9800 | C12B—C17B | 1.545 (8) |
| C8A—C9A | 1.480 (2) | C13B—C14B | 1.455 (7) |
| C8A—H8AA | 0.9900 | C13B—H13B | 0.9500 |
| C8A—H8AB | 0.9900 | C14B—C15B | 1.515 (8) |
| C9A—H9AA | 0.9800 | C15B—C16B | 1.535 (7) |
| C9A—H9AB | 0.9800 | C15B—H15B | 0.9900 |
| C9A—H9AC | 0.9800 | C15B—H15D | 0.9900 |
| C10A—H10A | 0.9800 | C16B—C19B | 1.530 (7) |
| C10A—H10B | 0.9800 | C16B—C18B | 1.532 (8) |
| C10A—H10C | 0.9800 | C16B—C17B | 1.540 (6) |
| C12A—C13A | 1.3501 (14) | C17B—H17C | 0.9900 |
| C12A—C17A | 1.5001 (15) | C17B—H17D | 0.9900 |
| C13A—C14A | 1.4486 (14) | C18B—H18D | 0.9800 |
| C13A—H13A | 0.9500 | C18B—H18E | 0.9800 |
| C14A—C15A | 1.4989 (16) | C18B—H18F | 0.9800 |
| C15A—C16A | 1.5351 (18) | C19B—H19D | 0.9800 |
| C15A—H15A | 0.9900 | C19B—H19E | 0.9800 |
| C15A—H15C | 0.9900 | C19B—H19F | 0.9800 |
| C16A—C19A | 1.519 (2) | O5C—C14C | 1.244 (9) |
| C16A—C18A | 1.5263 (19) | N1C—C12C | 1.401 (9) |
| C16A—C17A | 1.5364 (16) | N1C—H1CA | 0.88 (5) |
| C17A—H17A | 0.9900 | C12C—C13C | 1.342 (9) |
| C17A—H17B | 0.9900 | C12C—C17C | 1.527 (10) |
| C18A—H18A | 0.9800 | C13C—C14C | 1.450 (9) |
| C18A—H18B | 0.9800 | C13C—H13C | 0.9500 |
| C18A—H18C | 0.9800 | C14C—C15C | 1.499 (8) |
| C19A—H19A | 0.9800 | C15C—C16C | 1.532 (9) |
| C19A—H19B | 0.9800 | C15C—H15E | 0.9900 |
| C19A—H19C | 0.9800 | C15C—H15F | 0.9900 |
| Cl1B—C2B | 1.7334 (11) | C16C—C17C | 1.528 (9) |
| O1B—C3B | 1.3715 (13) | C16C—C19C | 1.537 (10) |
| O1B—C7B | 1.4379 (17) | C16C—C18C | 1.545 (9) |
| O2B—C4B | 1.3724 (12) | C17C—H17E | 0.9900 |
| O2B—C8B | 1.4510 (16) | C17C—H17F | 0.9900 |
| O3B—C5B | 1.3605 (13) | C18C—H18G | 0.9800 |
| O3B—C10B | 1.4277 (14) | C18C—H18H | 0.9800 |
| O4B—C11B | 1.2147 (13) | C18C—H18I | 0.9800 |
| C1B—C2B | 1.3891 (15) | C19C—H19G | 0.9800 |
| C1B—C6B | 1.3986 (14) | C19C—H19H | 0.9800 |
| C1B—C11B | 1.5065 (14) | C19C—H19I | 0.9800 |
| C2B—C3B | 1.4000 (14) | ||
| C3A—O1A—C7A | 114.34 (10) | O1B—C7B—H7BB | 109.5 |
| C4A—O2A—C8A | 116.92 (10) | H7BA—C7B—H7BB | 109.5 |
| C5A—O3A—C10A | 116.80 (10) | O1B—C7B—H7BC | 109.5 |
| C11A—N1A—C12A | 127.99 (9) | H7BA—C7B—H7BC | 109.5 |
| C11A—N1A—H1AA | 118.9 (11) | H7BB—C7B—H7BC | 109.5 |
| C12A—N1A—H1AA | 112.8 (11) | O2B—C8B—C9B | 112.15 (12) |
| C2A—C1A—C6A | 119.60 (10) | O2B—C8B—H8BA | 109.2 |
| C2A—C1A—C11A | 124.39 (10) | C9B—C8B—H8BA | 109.2 |
| C6A—C1A—C11A | 115.99 (9) | O2B—C8B—H8BB | 109.2 |
| C1A—C2A—C3A | 120.34 (10) | C9B—C8B—H8BB | 109.2 |
| C1A—C2A—Cl1A | 121.28 (8) | H8BA—C8B—H8BB | 107.9 |
| C3A—C2A—Cl1A | 118.34 (9) | C8B—C9B—H9BA | 109.5 |
| O1A—C3A—C4A | 119.96 (10) | C8B—C9B—H9BB | 109.5 |
| O1A—C3A—C2A | 119.96 (11) | H9BA—C9B—H9BB | 109.5 |
| C4A—C3A—C2A | 119.94 (10) | C8B—C9B—H9BC | 109.5 |
| O2A—C4A—C3A | 117.42 (11) | H9BA—C9B—H9BC | 109.5 |
| O2A—C4A—C5A | 122.99 (11) | H9BB—C9B—H9BC | 109.5 |
| C3A—C4A—C5A | 119.50 (10) | O3B—C10B—H10D | 109.5 |
| O3A—C5A—C6A | 124.09 (11) | O3B—C10B—H10E | 109.5 |
| O3A—C5A—C4A | 115.87 (10) | H10D—C10B—H10E | 109.5 |
| C6A—C5A—C4A | 120.03 (11) | O3B—C10B—H10F | 109.5 |
| C5A—C6A—C1A | 120.56 (11) | H10D—C10B—H10F | 109.5 |
| C5A—C6A—H6AA | 119.7 | H10E—C10B—H10F | 109.5 |
| C1A—C6A—H6AA | 119.7 | O4B—C11B—N1B | 120.3 (4) |
| O1A—C7A—H7AA | 109.5 | O4B—C11B—N1C | 127.6 (4) |
| O1A—C7A—H7AB | 109.5 | O4B—C11B—C1B | 120.55 (9) |
| H7AA—C7A—H7AB | 109.5 | N1B—C11B—C1B | 119.1 (4) |
| O1A—C7A—H7AC | 109.5 | N1C—C11B—C1B | 111.8 (4) |
| H7AA—C7A—H7AC | 109.5 | C11B—N1B—C12B | 131.6 (8) |
| H7AB—C7A—H7AC | 109.5 | C11B—N1B—H1BA | 115 (4) |
| O2A—C8A—C9A | 110.05 (14) | C12B—N1B—H1BA | 112 (4) |
| O2A—C8A—H8AA | 109.7 | C13B—C12B—N1B | 122.2 (7) |
| C9A—C8A—H8AA | 109.7 | C13B—C12B—C17B | 123.6 (6) |
| O2A—C8A—H8AB | 109.7 | N1B—C12B—C17B | 113.1 (6) |
| C9A—C8A—H8AB | 109.7 | C12B—C13B—C14B | 119.3 (6) |
| H8AA—C8A—H8AB | 108.2 | C12B—C13B—H13B | 120.4 |
| C8A—C9A—H9AA | 109.5 | C14B—C13B—H13B | 120.4 |
| C8A—C9A—H9AB | 109.5 | O5B—C14B—C13B | 120.7 (7) |
| H9AA—C9A—H9AB | 109.5 | O5B—C14B—C15B | 119.7 (7) |
| C8A—C9A—H9AC | 109.5 | C13B—C14B—C15B | 119.6 (6) |
| H9AA—C9A—H9AC | 109.5 | C14B—C15B—C16B | 113.2 (4) |
| H9AB—C9A—H9AC | 109.5 | C14B—C15B—H15B | 108.9 |
| O3A—C10A—H10A | 109.5 | C16B—C15B—H15B | 108.9 |
| O3A—C10A—H10B | 109.5 | C14B—C15B—H15D | 108.9 |
| H10A—C10A—H10B | 109.5 | C16B—C15B—H15D | 108.9 |
| O3A—C10A—H10C | 109.5 | H15B—C15B—H15D | 107.8 |
| H10A—C10A—H10C | 109.5 | C19B—C16B—C18B | 109.4 (4) |
| H10B—C10A—H10C | 109.5 | C19B—C16B—C15B | 109.7 (4) |
| O4A—C11A—N1A | 124.73 (9) | C18B—C16B—C15B | 110.3 (4) |
| O4A—C11A—C1A | 121.52 (9) | C19B—C16B—C17B | 109.4 (4) |
| N1A—C11A—C1A | 113.66 (9) | C18B—C16B—C17B | 110.3 (4) |
| C13A—C12A—N1A | 124.56 (9) | C15B—C16B—C17B | 107.8 (4) |
| C13A—C12A—C17A | 122.20 (9) | C16B—C17B—C12B | 111.3 (4) |
| N1A—C12A—C17A | 113.25 (9) | C16B—C17B—H17C | 109.4 |
| C12A—C13A—C14A | 121.19 (10) | C12B—C17B—H17C | 109.4 |
| C12A—C13A—H13A | 119.4 | C16B—C17B—H17D | 109.4 |
| C14A—C13A—H13A | 119.4 | C12B—C17B—H17D | 109.4 |
| O5A—C14A—C13A | 121.13 (10) | H17C—C17B—H17D | 108.0 |
| O5A—C14A—C15A | 120.98 (10) | C16B—C18B—H18D | 109.5 |
| C13A—C14A—C15A | 117.88 (9) | C16B—C18B—H18E | 109.5 |
| C14A—C15A—C16A | 112.84 (9) | H18D—C18B—H18E | 109.5 |
| C14A—C15A—H15A | 109.0 | C16B—C18B—H18F | 109.5 |
| C16A—C15A—H15A | 109.0 | H18D—C18B—H18F | 109.5 |
| C14A—C15A—H15C | 109.0 | H18E—C18B—H18F | 109.5 |
| C16A—C15A—H15C | 109.0 | C16B—C19B—H19D | 109.5 |
| H15A—C15A—H15C | 107.8 | C16B—C19B—H19E | 109.5 |
| C19A—C16A—C18A | 110.90 (16) | H19D—C19B—H19E | 109.5 |
| C19A—C16A—C15A | 110.13 (12) | C16B—C19B—H19F | 109.5 |
| C18A—C16A—C15A | 108.85 (12) | H19D—C19B—H19F | 109.5 |
| C19A—C16A—C17A | 110.10 (11) | H19E—C19B—H19F | 109.5 |
| C18A—C16A—C17A | 108.99 (12) | C12C—N1C—C11B | 123.0 (9) |
| C15A—C16A—C17A | 107.80 (11) | C12C—N1C—H1CA | 119 (3) |
| C12A—C17A—C16A | 113.03 (9) | C11B—N1C—H1CA | 117 (3) |
| C12A—C17A—H17A | 109.0 | C13C—C12C—N1C | 128.2 (9) |
| C16A—C17A—H17A | 109.0 | C13C—C12C—C17C | 118.0 (8) |
| C12A—C17A—H17B | 109.0 | N1C—C12C—C17C | 111.9 (7) |
| C16A—C17A—H17B | 109.0 | C12C—C13C—C14C | 124.8 (8) |
| H17A—C17A—H17B | 107.8 | C12C—C13C—H13C | 117.6 |
| C16A—C18A—H18A | 109.5 | C14C—C13C—H13C | 117.6 |
| C16A—C18A—H18B | 109.5 | O5C—C14C—C13C | 119.6 (9) |
| H18A—C18A—H18B | 109.5 | O5C—C14C—C15C | 123.6 (8) |
| C16A—C18A—H18C | 109.5 | C13C—C14C—C15C | 116.8 (7) |
| H18A—C18A—H18C | 109.5 | C14C—C15C—C16C | 113.6 (5) |
| H18B—C18A—H18C | 109.5 | C14C—C15C—H15E | 108.8 |
| C16A—C19A—H19A | 109.5 | C16C—C15C—H15E | 108.8 |
| C16A—C19A—H19B | 109.5 | C14C—C15C—H15F | 108.8 |
| H19A—C19A—H19B | 109.5 | C16C—C15C—H15F | 108.8 |
| C16A—C19A—H19C | 109.5 | H15E—C15C—H15F | 107.7 |
| H19A—C19A—H19C | 109.5 | C17C—C16C—C15C | 109.0 (6) |
| H19B—C19A—H19C | 109.5 | C17C—C16C—C19C | 110.0 (6) |
| C3B—O1B—C7B | 112.62 (9) | C15C—C16C—C19C | 110.6 (6) |
| C4B—O2B—C8B | 114.21 (10) | C17C—C16C—C18C | 108.9 (6) |
| C5B—O3B—C10B | 116.84 (9) | C15C—C16C—C18C | 109.4 (6) |
| C2B—C1B—C6B | 119.81 (9) | C19C—C16C—C18C | 109.0 (6) |
| C2B—C1B—C11B | 123.40 (9) | C12C—C17C—C16C | 112.2 (6) |
| C6B—C1B—C11B | 116.68 (9) | C12C—C17C—H17E | 109.2 |
| C1B—C2B—C3B | 120.05 (9) | C16C—C17C—H17E | 109.2 |
| C1B—C2B—Cl1B | 121.83 (8) | C12C—C17C—H17F | 109.2 |
| C3B—C2B—Cl1B | 118.10 (8) | C16C—C17C—H17F | 109.2 |
| O1B—C3B—C4B | 119.85 (9) | H17E—C17C—H17F | 107.9 |
| O1B—C3B—C2B | 120.07 (10) | C16C—C18C—H18G | 109.5 |
| C4B—C3B—C2B | 120.08 (10) | C16C—C18C—H18H | 109.5 |
| O2B—C4B—C3B | 120.17 (10) | H18G—C18C—H18H | 109.5 |
| O2B—C4B—C5B | 119.75 (10) | C16C—C18C—H18I | 109.5 |
| C3B—C4B—C5B | 119.98 (9) | H18G—C18C—H18I | 109.5 |
| O3B—C5B—C6B | 125.11 (10) | H18H—C18C—H18I | 109.5 |
| O3B—C5B—C4B | 115.33 (9) | C16C—C19C—H19G | 109.5 |
| C6B—C5B—C4B | 119.56 (10) | C16C—C19C—H19H | 109.5 |
| C5B—C6B—C1B | 120.46 (10) | H19G—C19C—H19H | 109.5 |
| C5B—C6B—H6BA | 119.8 | C16C—C19C—H19I | 109.5 |
| C1B—C6B—H6BA | 119.8 | H19G—C19C—H19I | 109.5 |
| O1B—C7B—H7BA | 109.5 | H19H—C19C—H19I | 109.5 |
| C6A—C1A—C2A—C3A | 0.43 (16) | C8B—O2B—C4B—C3B | 79.16 (14) |
| C11A—C1A—C2A—C3A | −177.91 (10) | C8B—O2B—C4B—C5B | −104.32 (12) |
| C6A—C1A—C2A—Cl1A | −177.38 (8) | O1B—C3B—C4B—O2B | −3.90 (16) |
| C11A—C1A—C2A—Cl1A | 4.27 (15) | C2B—C3B—C4B—O2B | 177.06 (9) |
| C7A—O1A—C3A—C4A | −89.70 (15) | O1B—C3B—C4B—C5B | 179.58 (9) |
| C7A—O1A—C3A—C2A | 94.67 (14) | C2B—C3B—C4B—C5B | 0.54 (16) |
| C1A—C2A—C3A—O1A | 176.11 (10) | C10B—O3B—C5B—C6B | 0.54 (16) |
| Cl1A—C2A—C3A—O1A | −6.00 (14) | C10B—O3B—C5B—C4B | −179.34 (10) |
| C1A—C2A—C3A—C4A | 0.48 (16) | O2B—C4B—C5B—O3B | 0.86 (15) |
| Cl1A—C2A—C3A—C4A | 178.36 (9) | C3B—C4B—C5B—O3B | 177.39 (10) |
| C8A—O2A—C4A—C3A | 124.66 (15) | O2B—C4B—C5B—C6B | −179.03 (10) |
| C8A—O2A—C4A—C5A | −58.83 (18) | C3B—C4B—C5B—C6B | −2.50 (16) |
| O1A—C3A—C4A—O2A | 0.97 (16) | O3B—C5B—C6B—C1B | −177.06 (10) |
| C2A—C3A—C4A—O2A | 176.61 (10) | C4B—C5B—C6B—C1B | 2.82 (15) |
| O1A—C3A—C4A—C5A | −175.67 (10) | C2B—C1B—C6B—C5B | −1.17 (15) |
| C2A—C3A—C4A—C5A | −0.03 (17) | C11B—C1B—C6B—C5B | −177.57 (9) |
| C10A—O3A—C5A—C6A | −3.11 (19) | C4B—O2B—C8B—C9B | 67.92 (16) |
| C10A—O3A—C5A—C4A | 178.00 (13) | C2B—C1B—C11B—O4B | −122.81 (13) |
| O2A—C4A—C5A—O3A | 1.15 (17) | C6B—C1B—C11B—O4B | 53.44 (14) |
| C3A—C4A—C5A—O3A | 177.60 (10) | C2B—C1B—C11B—N1B | 58.5 (6) |
| O2A—C4A—C5A—C6A | −177.78 (10) | C6B—C1B—C11B—N1B | −125.2 (6) |
| C3A—C4A—C5A—C6A | −1.34 (17) | C2B—C1B—C11B—N1C | 59.4 (5) |
| O3A—C5A—C6A—C1A | −176.57 (11) | C6B—C1B—C11B—N1C | −124.3 (5) |
| C4A—C5A—C6A—C1A | 2.27 (17) | O4B—C11B—N1B—C12B | 7.2 (16) |
| C2A—C1A—C6A—C5A | −1.81 (16) | C1B—C11B—N1B—C12B | −174.2 (11) |
| C11A—C1A—C6A—C5A | 176.67 (10) | C11B—N1B—C12B—C13B | 0 (2) |
| C4A—O2A—C8A—C9A | −148.77 (16) | C11B—N1B—C12B—C17B | −169.0 (10) |
| C12A—N1A—C11A—O4A | −2.59 (18) | N1B—C12B—C13B—C14B | −175.2 (12) |
| C12A—N1A—C11A—C1A | 174.03 (10) | C17B—C12B—C13B—C14B | −7.6 (18) |
| C2A—C1A—C11A—O4A | −129.47 (12) | C12B—C13B—C14B—O5B | −173.3 (12) |
| C6A—C1A—C11A—O4A | 52.13 (15) | C12B—C13B—C14B—C15B | 8.2 (15) |
| C2A—C1A—C11A—N1A | 53.79 (14) | O5B—C14B—C15B—C16B | 148.7 (9) |
| C6A—C1A—C11A—N1A | −124.61 (10) | C13B—C14B—C15B—C16B | −32.8 (11) |
| C11A—N1A—C12A—C13A | −9.78 (18) | C14B—C15B—C16B—C19B | 172.5 (5) |
| C11A—N1A—C12A—C17A | 170.04 (11) | C14B—C15B—C16B—C18B | −66.9 (6) |
| N1A—C12A—C13A—C14A | −176.55 (10) | C14B—C15B—C16B—C17B | 53.5 (7) |
| C17A—C12A—C13A—C14A | 3.65 (17) | C19B—C16B—C17B—C12B | −170.7 (7) |
| C12A—C13A—C14A—O5A | 173.40 (12) | C18B—C16B—C17B—C12B | 68.9 (7) |
| C12A—C13A—C14A—C15A | −8.11 (17) | C15B—C16B—C17B—C12B | −51.5 (7) |
| O5A—C14A—C15A—C16A | −146.37 (12) | C13B—C12B—C17B—C16B | 31.0 (15) |
| C13A—C14A—C15A—C16A | 35.13 (16) | N1B—C12B—C17B—C16B | −160.4 (9) |
| C14A—C15A—C16A—C19A | 65.22 (14) | O4B—C11B—N1C—C12C | 9.7 (15) |
| C14A—C15A—C16A—C18A | −173.01 (13) | C1B—C11B—N1C—C12C | −172.7 (11) |
| C14A—C15A—C16A—C17A | −54.92 (14) | C11B—N1C—C12C—C13C | −8 (3) |
| C13A—C12A—C17A—C16A | −26.27 (17) | C11B—N1C—C12C—C17C | 155.4 (10) |
| N1A—C12A—C17A—C16A | 153.90 (11) | N1C—C12C—C13C—C14C | 176.1 (15) |
| C19A—C16A—C17A—C12A | −70.04 (15) | C17C—C12C—C13C—C14C | 13 (2) |
| C18A—C16A—C17A—C12A | 168.10 (14) | C12C—C13C—C14C—O5C | 171.1 (16) |
| C15A—C16A—C17A—C12A | 50.11 (15) | C12C—C13C—C14C—C15C | −11 (2) |
| C6B—C1B—C2B—C3B | −0.81 (14) | O5C—C14C—C15C—C16C | −151.4 (13) |
| C11B—C1B—C2B—C3B | 175.34 (9) | C13C—C14C—C15C—C16C | 30.3 (14) |
| C6B—C1B—C2B—Cl1B | −178.96 (8) | C14C—C15C—C16C—C17C | −51.9 (9) |
| C11B—C1B—C2B—Cl1B | −2.82 (14) | C14C—C15C—C16C—C19C | 69.1 (9) |
| C7B—O1B—C3B—C4B | 79.27 (13) | C14C—C15C—C16C—C18C | −170.8 (7) |
| C7B—O1B—C3B—C2B | −101.69 (13) | C13C—C12C—C17C—C16C | −35.8 (17) |
| C1B—C2B—C3B—O1B | −177.93 (9) | N1C—C12C—C17C—C16C | 158.7 (11) |
| Cl1B—C2B—C3B—O1B | 0.30 (14) | C15C—C16C—C17C—C12C | 53.9 (10) |
| C1B—C2B—C3B—C4B | 1.12 (15) | C19C—C16C—C17C—C12C | −67.5 (9) |
| Cl1B—C2B—C3B—C4B | 179.34 (8) | C18C—C16C—C17C—C12C | 173.2 (8) |
2-Chloro-N-(5,5-dimethyl-3-oxocyclohex-1-en-1-yl)-4-ethoxy-3,5-dimethoxybenzamide (3b) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1A—H1AA···O5B | 0.853 (16) | 2.040 (18) | 2.849 (8) | 158.1 (15) |
| N1A—H1AA···O5C | 0.853 (16) | 2.081 (19) | 2.909 (9) | 163.5 (15) |
| C7A—H7AA···O2Bi | 0.98 | 2.44 | 3.3451 (17) | 153 |
| C8A—H8AB···Cl1Aii | 0.99 | 2.92 | 3.703 (2) | 137 |
| C17A—H17A···O5B | 0.99 | 2.42 | 3.285 (7) | 146 |
| C17A—H17A···O5C | 0.99 | 2.63 | 3.481 (8) | 144 |
| C10B—H10F···O4Aiii | 0.98 | 2.46 | 3.4013 (15) | 161 |
| N1B—H1BA···O5Aiv | 0.77 (5) | 2.31 (5) | 2.985 (10) | 147 (5) |
| N1C—H1CA···O5Aiv | 0.88 (5) | 1.96 (4) | 2.795 (12) | 159 (4) |
| C17C—H17E···O5Aiv | 0.99 | 2.54 | 3.364 (3) | 141 |
Symmetry codes: (i) −x+1, y+1/2, −z+1/2; (ii) −x+2, −y+2, −z+1; (iii) −x+1, −y+2, −z+1; (iv) x, −y+3/2, z−1/2.
References
- Alford, J. S., Abascal, N. C., Shugrue, C. R., Colvin, S. M., Romney, D. K. & Miller, S. J. (2016). ACS Cent. Sci. 2, 733–739. [DOI] [PMC free article] [PubMed]
- Anderson, A. J., Nicholson, J. M., Bakare, O., Butcher, R. J. & Scott, K. R. (2004). J. Comb. Chem. 6, 950–954. [DOI] [PubMed]
- Boger, D. L., Ishizaki, T., Wysocki, J. R., Munk, S. A., Kitos, P. A. & Suntornwat, O. (1989). J. Am. Chem. Soc. 111, 6461–6463.
- Bruker (2010). APEX2 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Eddington, N. D., Cox, D. S., Khurana, M., Salama, N. N., Stables, J. P., Harrison, S. J., Negussie, A., Taylor, R. S., Tran, U. Q., Moore, J. A., Barrow, J. C. & Scott, K. R. (2003). Eur. J. Med. Chem. 38, 49–64. [DOI] [PubMed]
- Greenhill, J. V. (1976). J. Chem. Soc. Perkin Trans. 1, pp. 2207–2210.
- Greenhill, J. V. (1977). Chem. Soc. Rev. 6, 277–294.
- Jerach, B. & Elassar, A.-Z. A. (2015). Chemical Science Transactions, 4, 113–120.
- Kalita, U., Kaping, S., Nongkynrih, R., Boiss, I., Singha, L. I. & Vishwakarma, J. N. (2017). Monatsh. Chem. 148, 2155–2171.
- Lue, P. & Greenhill, J. V. (1996). Adv. Heterocycl. Chem. 67, 207–343.
- Meng, L.-H., Li, X.-M., Lv, C.-T., Huang, C.-G. & Wang, B. G. (2014). J. Nat. Prod. 77, 1921–1927. [DOI] [PubMed]
- Misra, R., Bhattacharyya, S. & Maity, D. K. (2008). Chem. Phys. Lett. 458, 54–57.
- Misra, R., Mandal, A., Mukhopadhyay, M., Maity, D. K. & Bhattacharyya, S. P. (2009). J. Phys. Chem. B, 113, 10779–10791. [DOI] [PubMed]
- Romney, D. K., Colvin, S. M. & Miller, S. J. (2014). J. Am. Chem. Soc. 136, 14019–14022. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Stoltz, B. M., Dougherty, D. A., Duquette, D. & Duffy, N. (2016). US Patent US 9,518,034 B2.
- Zheng, F. L., Ban, S. R., Feng, X. E., Zhao, C. X., Lin, W. & Li, Q. S. (2011). Molecules, 16, 4897–4911. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) 3a, 3b, global. DOI: 10.1107/S2056989021001778/hb7962sup1.cif
Structure factors: contains datablock(s) 3a. DOI: 10.1107/S2056989021001778/hb79623asup2.hkl
Structure factors: contains datablock(s) 3b. DOI: 10.1107/S2056989021001778/hb79623bsup3.hkl
Supporting information file. DOI: 10.1107/S2056989021001778/hb79623asup4.cml
Supporting information file. DOI: 10.1107/S2056989021001778/hb79623bsup5.cml
Additional supporting information: crystallographic information; 3D view; checkCIF report