Abstract
The presence of autoantibodies and autoreactive T cells to citrullinated proteins and citrullinating enzymes in patients with rheumatoid arthritis (RA), together with the accumulation of citrullinated proteins in rheumatoid joints, provides substantial evidence that dysregulated citrullination is a hallmark feature of RA. However, understanding mechanisms that dysregulate citrullination in RA has important challenges. Citrullination is a normal process in immune and non-immune cells which is likely activated by different conditions (e.g. inflammation) with no pathogenic consequences. In a complex inflammatory environment such as the RA joint, unique strategies are therefore required to dissect specific mechanisms involved in the abnormal production of citrullinated proteins. Here, we will review current models of citrullination in RA and discuss critical components that, in our view, are relevant to understanding the accumulation of citrullinated proteins in the RA joint, collectively referred to as the RA citrullinome. In particular, we will focus on potential caveats in the study of citrullination in RA, and will highlight methods to precisely detect citrullinated proteins in complex biological samples, which is a confirmatory approach to mechanistically link the RA citrullinome with unique pathogenic pathways in RA.
Keywords: Citrullination, rheumatoid arthritis, ACPA, peptidylarginine deiminase, NETs, mass spectrometry
1. INTRODUCTION
Major advances in science have resulted from serendipity. The discovery of the antibodies to citrullinated antigens is no exception. An important breakthrough in the field of cellular biology was the development of the immunofluorescence technique by Albert H. Coons in 1942.1 Using this clever method, antibodies chemically labelled with fluorescein isocyanate allowed the visual detection of antigens in tissues for the first time. After the discovery that patients with systemic lupus erythematosus (SLE) have antibodies to DNA and nuclear components, the immunofluorescence technique was adopted into the arsenal for the study and characterization of anti-nuclear antibodies (ANA).2 In 1961, John Swanson Beck found that different patterns of nuclear fluorescence are produced by serum from patients with autoimmune diseases,3 which sparked interest on the study of autoimmune sera using the Coons’s immunofluorescent technique. Although tissue sections from small mammals, such as liver and kidney from rats and guinea-pigs, were the usual antigens for ANA detection, the laboratory of Robert M. Kark in Chicago started using smears of epithelial cells from healthy human buccal mucosa as substrate for ANAs instead.4 In 1959, Enno Mandema, a Dutch physician, was visiting Dr. Kark for research on the antinuclear factors,5 were he studied ANAs using smears from buccal mucosa.6 Enno Mandema became head of the Department of Medicine in Groningen in 1960,5 and four years later, he and Bob Nienhuis discovered that sera from patients with rheumatoid arthritis (RA) had a factor that generates a unique pattern of staining on epithelial cells from human buccal mucosa, termed anti-perinuclear factor (APF). This pattern was undetectable when using other tissues from animals (i.e. kidney, liver, thyroid, stomach and adrenal sections) and humans (i.e. cartilage, synovia and vaginal smears) as antigens.7 They concluded that APF was a novel antibody targeting a compound in human buccal mucosa, which was either not present, or was present in a different form, in other tissues.7
Fifteen years later, in the late 1970’s, Terry J. Hamblin’s group was studying the prevalence of anti-keratin antibodies (AKA) using the Coons’s technique on rat esophagus tissue and sera from a wide variety of medical diseases. They noticed that a large subset of AKA positive sera were also positive for rheumatoid factor (RF), leading to the interest of studying AKA in RA.8 They found that 58% of patients with RA were AKA positive, while these antibodies were only positive in 1.9% of other non-RA autoimmune arthritides and absent in healthy controls. They also confirmed that AKAs were distinct to RF,8 suggesting that these were a novel type of autoantibody in RA. While neither APF nor AKA were broadly used for clinical diagnosis in RA, their casual discovery provided the framework that would later lead to the breakthrough by Guy Serre’s group that both antibodies target the same antigen,9, 10 and key observation by Walther J van Venrooij’s group that citrullinated residues are the major determinants for APF/AKA antigen recognition.11
During the last 20 years, important advances have been made regarding the clinical use, specificity, potential origin and molecular features of autoantibodies to citrullinated proteins (known as ACPAs) in RA.12, 13 In addition, more than 100 citrullinated proteins have been identified in the RA joint,14–17 collectively referred to as the RA citrullinome, providing clues about the pathophysiologic targets of ACPA and the potential sources of citrullinated autoantigens in RA. The large repertoire of citrullinated proteins in the RA citrullinome strongly suggests that citrullination is dysregulated in RA.18 This notion has focused interest on understanding the mechanisms that trigger citrullination in the RA joint, highlighting a key area for treatment and preventive interventions in RA. However, although some pathogenic models of citrullination have been suggested in RA, it is still uncertain how much each may contribute to generating the RA citrullinome. Often, citrullination assays are performed under non-physiological conditions and the magnitude or extent of protein citrullination is not considered relative to the RA citrullinome observed in patients. In some cases, the identification of citrullinated proteins is plagued by technical or analytical challenges, which may lead to misleading conclusions. In this review, we discuss potential caveats in the study of citrullination in RA and underscore precise methods to detect citrullinated proteins in complex biological samples. The corollary of this work is to provide an initial basis to better understand pathogenic pathways linked to the generation of the citrullinome in patients with RA.
2. MASS SPECTROMETRY APPLIED TO THE STUDY OF CITRULLINATION IN COMPLEX SAMPLES
Citrullination (or deimination) is a posttranslational modification (PTM) that involves the enzymatic conversion of arginine residues to citrulline residues. This process is mediated by the peptidylarginine deiminases (PADs), a family of five calcium dependent enzymes designated PAD1 to PAD4 and PAD6.18 Among these enzymes, only PAD1-4 have citrullinating activity.19 PADs are highly specific for peptidylarginine and can only citrullinate arginine residues within polypeptide chains but not at their termini (i.e. they are endodeiminases).20, 21 Among PADs, PAD2 and PAD4 have gained prominence as potential candidates that drive citrullination of self-antigens in RA due to their increased expression in rheumatoid synovial tissue and fluid.22–24
Despite emerging biological studies, the physiological role of PAD enzymes and protein citrullination is far less well-studied compare to other protein PTMs. Query of the Uniprot knowledge database, under the category PTM, yielded 511876 results with only 219 (0.04%) belonging to citrullination. This, in part, is attributed to a lack of robust biochemical enrichment and detection methods. The commonly used approaches to study citrullination, such as immunodetection and rhodamine-phenylglyoxal based visualization,25–27 are convenient methods for assessing citrullination at the whole protein level, but have important limitations. Antibodies targeting specific citrullinated proteins (e.g. citrullinated histones or citrullinated fibrinogen) fail to recognize global citrullination, which is the hallmark in RA.14–17 Antibodies to peptidylcitrulline (e.g. F95 monoclonal antibody),28 antibodies to modified citrulline (Senshu method),25 and rhodamine-phenylglyoxal 26 can detect citrullinated proteins broadly, but provide no information regarding the identity of citrullinated proteins. Moreover, both the Senshu method and rhodamine-phenylglyoxal lack specificity to distinguish citrullination and carbamylation.26, 29 This is important since, like citrullinated proteins, carbamylated antigens can also be recognized by autoantibodies in patients with RA.30
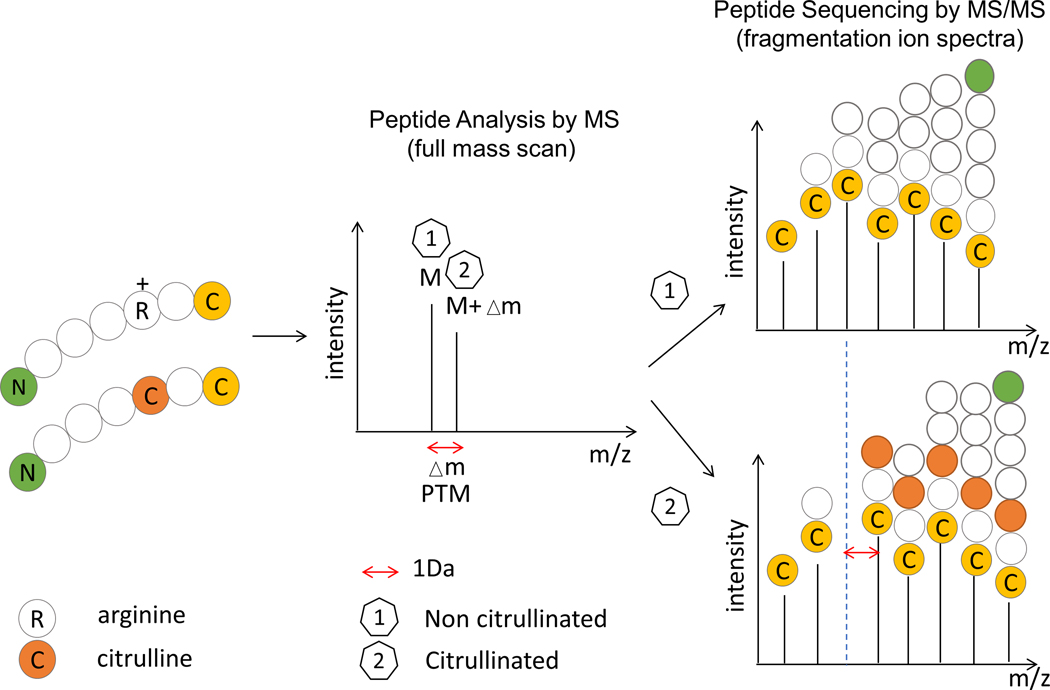
The ability to address this sample complexity is precisely why tandem mass spectrometry (MS/MS) now dominates proteomic analysis and has become the preferred method for studying protein citrullination. Liquid chromatography mass spectrometry (LC-MS/MS) instruments can analyze mixtures of thousands of peptides, acquiring the mass-to-charge ratio (m/z) of each peptide precursor ion (MS1) and fragmenting the most abundant precursors for a MS/MS analysis. The resulting fragment ions are recorded as MS/MS spectra, which are searched against a theoretical database. During this search, observed precursor and fragmentation product ion masses are compared with those of a theoretical protein digest, returning the amino acid sequence and statistical confidence of the sequence match.31 This precision makes MS/MS a key technique for peptide sequencing and PTM analysis as it facilitates mass determination and sequencing of peptides, thereby enabling the detection of site-specific PTMs. Most PTMs can be detected by the molecular weight shift of the modified amino acids relative to the unmodified form, and these mass increments or deficits can be detected by MS.32 Indeed, MS is currently the gold standard to confirm citrullination as well as to define the identity and sites of citrullination in proteins. Still, there are important features that need to be taken in consideration to avoid misleading interpretation of MS data.
2.1. Mass increment of 0.9840 Da is not citrullination specific
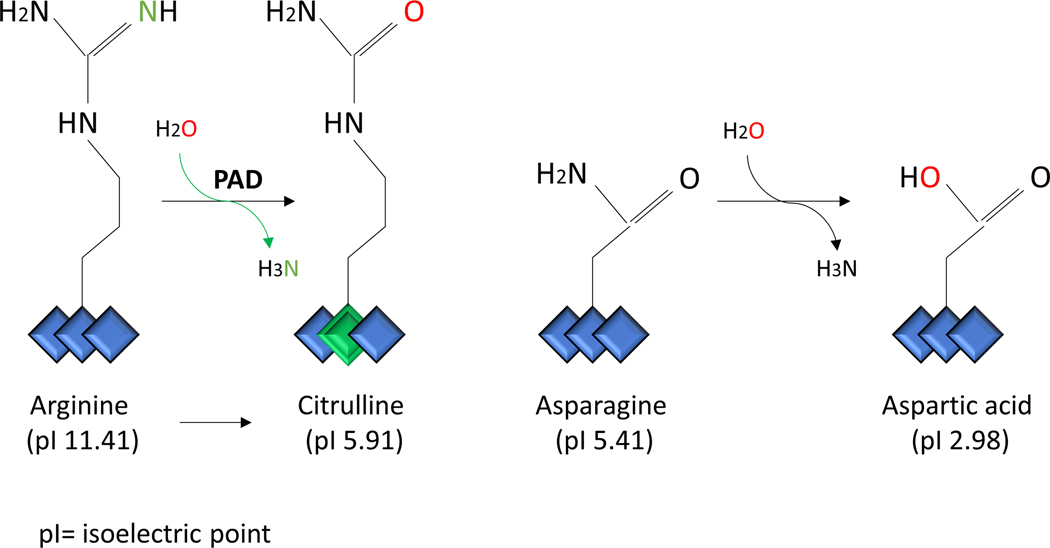
During citrullination, the guanidine group of the arginine side chain (-NH2 group) is converted to an ureido group with a hydroxyl -OH group via hydrolysis.33 This conversion results in a mass increment of 0.9840 Da in all the fragment ions that contain the citrullinated residue relative to the unmodified arginine-containing peptide 34 (Figure 1). These Δm values are very useful for annotation of PTM peptide MS/MS spectra, as they aid in identifying and assigning the modified residues. However, although these features demonstrate that citrullinated proteins can be characterized directly by MS/MS, the mass increment of 0.9840 Da is not specific for citrullination. This exact mass increment also occurs on one of the most frequent modifications, the deamidation of asparagine (Asn) or glutamine (Gln), which is a spontaneous non-enzymatic conversion of an uncharged side-chain residue (Figure 2). Occurrence of deamidated and citrullinated residues in the same peptide leads to interference during precursor ion isolation and can result in incorrect assignments of modified amino acids (i.e., co-isolation of other species), which leads to mixed or contaminated MS/MS spectra.35 In fact, deamidation of Asn/Gln occurs much more frequently than citrullination. Thus, this is the major problem leading to a high degree of ambiguity during data interpretation. When searching MS/MS data against a database of citrullinated peptides, it is therefore imperative that both modifications, citrullination and deamidation are included to prevent false positives corresponding to deamidated peptides. In addition, it is preferably to check if either the (potential) citrullination site or the (potential) deamidation site is covered by the fragment ions in the MS/MS spectrum.
Figure 1.
MS/MS characteristic fragmentation of citrullinated peptides. During citrullination the guanidine group of the arginine side chain is converted to an ureido group. This conversion results in a mass increment (Δm) of 0.9840 Da compared with the unmodified arginine.
Figure 2.
Comparison between peptidyl-arginine deimidation vs. peptidyl-asparagine deamidation. Citrullination is the enzymatic conversion of arginine residues to citrulline residues, that results in a permanent change of the positive charge to electrically neutral. Deamidation is a spontaneous non-enzymatic reaction in which an amide functional group in the side chain of asparagine or glutamine residues is converted to another functional group. Usually, asparagine is converted to aspartic acid and glutamine to glutamic acid (not shown).
Citrullination can also be misidentify as 13C isotopes as the mass increment for 13C isotope is 1.0036 Da. Misidentification of 13C isotopes can be circumvented by searching the database with a small enough parent mass tolerance, ideally <5 ppm.36 There are typically two sets of mass tolerances that are inputs to a database search: peptide mass tolerance and fragment mass tolerance. The peptide mass tolerance setting is based on the mass accuracy of the analyzer used to measure the precursor ions masses in the MS scans. The fragment mass tolerance is based on the mass accuracy of the analyzer used to measure the fragment ions in the MS/MS scans. This parameter can be referred to as MS/MS tolerance, fragment mass error, or product ion tolerance. The ideal parent mass tolerances for citrullination should be <5pmm and the MS/MS tolerance to ~0.6 Da.
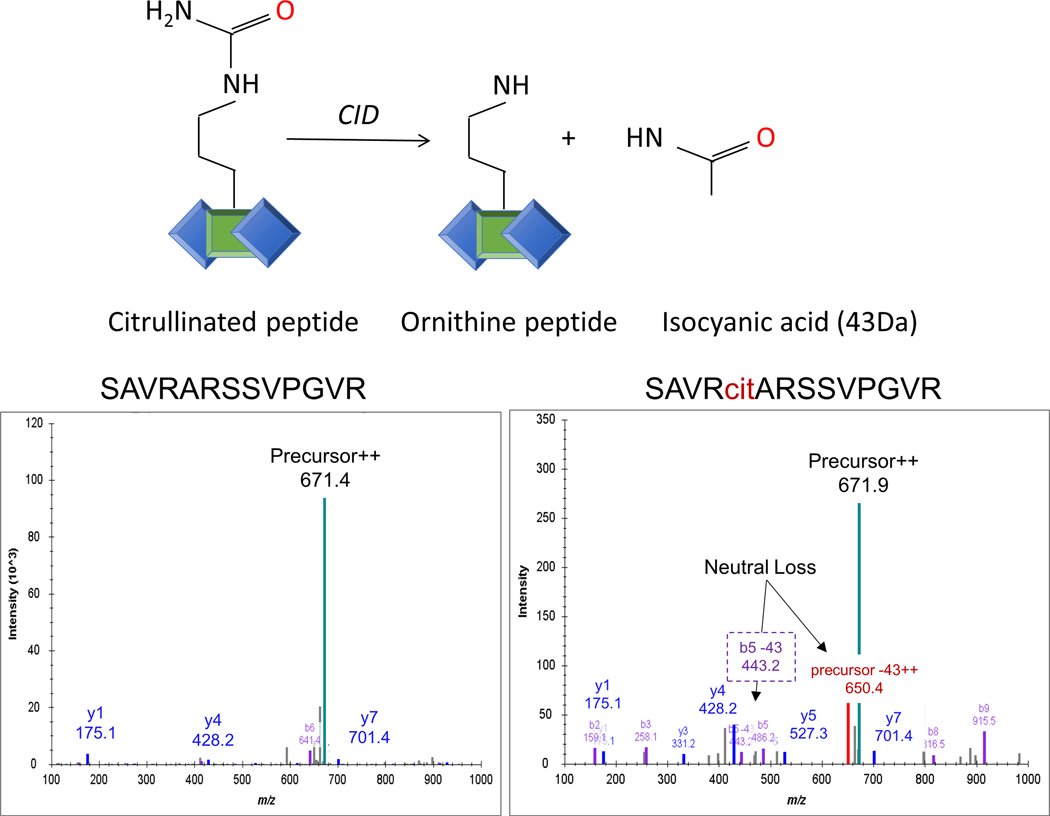
2.2. Neutral loss of isocyanic acid in peptide collision-induced dissociation is a unique fragmentation feature of citrullinated peptides.
Precise identification of any modification and the modification site can be very challenging regarding the stability of the modification and the gas phase dissociation behavior of the modified peptide precursor. Fortunately, citrullination is a stable PTM for which peptide backbone fragmentation has been observed due to the dominant neutral loss of isocyanic acid (HN=CO) of 43.0058 Da from the deiminated arginine amino acid side-chain (Figure 3). This unique fragmentation feature, which is reconciled by the release of an isocyanic acid moiety from the citrulline ureido group,37 can be observed with complementary fragmentation modes, like collision-induced dissociation (CID) MS/MS,37 electron transfer dissociation (ETD) and high-energy collision dissociation (HCD).38 In 2009, Hao et al. used CID, neutral loss (isocyanic acid) and ion series to identify citrullination sites and to distinguish citrullination from deamidation, which does not produce a neutral loss (Figure 3).37 The technical advance of CID type of fragmentation stretched the capabilities of mass spectrometry-based proteomics technology. However, it should accompany good practices for localization of citrullination sites in peptides, which requires a full ion series (all b and y ions of −43 Da) around modified residue.39
Figure 3.
Neutral loss of isocyanic acid in citrullinated peptide collision-induced dissociation (CID). The schematic representation of the of isocyanic acid (HN=CO) loss in collision-induced dissociation (CID) spectra. The signature neutral loss of 43 Da from citrullinated peptide precursor ions comes from HN=CO moiety (isocyanic acid) from the citrulline ureido group. The elimination occurs in multiple charge states of precursor ions and also in b and y ions.
Later, as an alternative approach, Creese and colleagues combined CID with ETD, as ETD cleaves randomly along the peptide backbone, while often leaving side chains and modifications intact.34 If neutral loss of isocyanic acid is observed in the CID spectrum, ETD of the parent ion is performed, which improved the identification and localization of the modification site.40 In 2013, the complementary fragmentation mode, also suitable for citrullination, was reported by Jin and colleagues.41 They used CID in combination with HCD. The fragmentation pattern of HCD is featured with higher activation energy and shorter activation time comparing the traditional ion trap CID, which allows for detection of lower m/z ions (b2, b3). CID-triggered HCD fragmentation can differentiate between citrullination and deamidation with great confidence.
Recently, Chien-Yun Lee et al. used a straightforward method to look at the MS-based data of 30 human tissues to identify citrullination sites on endogenous proteins.42 This data mining process allows for the discovery of patterns in large data sets. The group identified ~13,000 citrullinated candidate spectra in 26 tissues based on database searching of ∼70 million tandem mass spectra. The spectra were manually inspected for the presence of neutral loss of 43.0058 Da, and putative citrulline immonium ion at 130.0975 m/z. The immonium ion is an internal fragment formed by specific cleavage; in the case of arginine, the ion mass is 129 m/z, which becomes 130 m/z after citrullination. Unfortunately, this ion does not give a strong MS signal and it is not very specific for the presence of a particular amino acid residue within a peptide.
In order to resolve the aforementioned ambiguity, Chien-Yun Lee’s group synthetized ∼2,200 citrullinated and 1,300 deamidated peptides to build a library of reference spectra.42 Conceptually, the premise of this approach is that the MS/MS fragmentation pattern of a peptide under some fixed conditions is a reproducible fingerprint of that peptide, such that spectral matching can identify unknown spectra acquired under the same conditions. The validation confirmed that loss of isocyanic acid was not entirely specific for citrulline and loss from fragment ions should always be included in the analysis if relying only on MS spectra interpretation. Nevertheless, this is the largest review of protein citrullination in human tissues, including 375 citrullination sites on 209 human proteins.42
2.3. Hydrophobicity and peptide elusion in the analysis of citrullination.
The concept of hydrophobicity has been a topic of much study in all aspects of science, particularly in the fields of biology and chemistry.43 The loss of a positive charge coming with citrullination affects the acidity of the amino acid residue, changing the isoelectric point from 11.41 for arginine to 5.91 for citrulline.44 The isoelectric point determines a protein’s minimum solubility level and its hydrophobicity that effects peptide elusion from the MS column. Hermansson et al.45 was the first to present the practicality of the hydrophilicity/hydrophobicity of arginine/citrulline peptides that allows for chromatographic separation and could enable their specific identification and quantitation. Using this concept, they were able to identify and quantify citrullinated fibrinogen in synovial tissue from rheumatoid arthritis patients.45 The limitation of this application comes from the necessity of including synthetic peptides for already known candidates, so cannot be used for other potentially unknown citrullinated peptides detected during acquisition.
The concept was taken and revised with the development of sequential window acquisition of all theoretical fragment ion spectra (SWATH) or data-independent acquisition (DIA) mass spectrometry (MS).46 The first advantage of the approach is a more comprehensive fragmentation, including low abundance sub-proteomes, such as citrullination. The second advantage is the reference assay library that should contain all the prior knowledge of the peptide components to be extracted from the SWATH data.47 Thus, assay library generation is one key challenge and limitation of this approach.
To overcome this problem, Fert-Bober et al. came up with the idea of hypercitrullination library creation.48 The hypercitrullinated library allows for enrichment of the lower abundance citrullinated peptides to be detected by the DIA MS approach. In contrast to previous enrichment methods, chemical derivatization of the citrulline residue with 2,3-butanedione and antipyrine,49 4-bromophenylglyoxal,50 or 4-hydroxyphenylglyoxal,51 the group used active PAD cocktail to citrullinate as many sites as possible in study samples. The hypercitrullinated enrichment results dramatically increased the sensitivity for detection of low abundant citrullinated peptides ex vivo. Since increased pooling may affect the quality of the citrullinated peptides, Fert-Bober et al. applied several validation steps that were incorporated into a publicly available search engine, at https://github.com/Citrullinome/CitFinder. Importantly, to avoid false matches after trypsin cleavage on the C-terminal arginine as the citrullination site (C-term Cit),52 the group used endoproteinase Lys-C that cleaves after the C-terminal side of lysine to obtain peptides with untouched arginine/citrullinate sites.
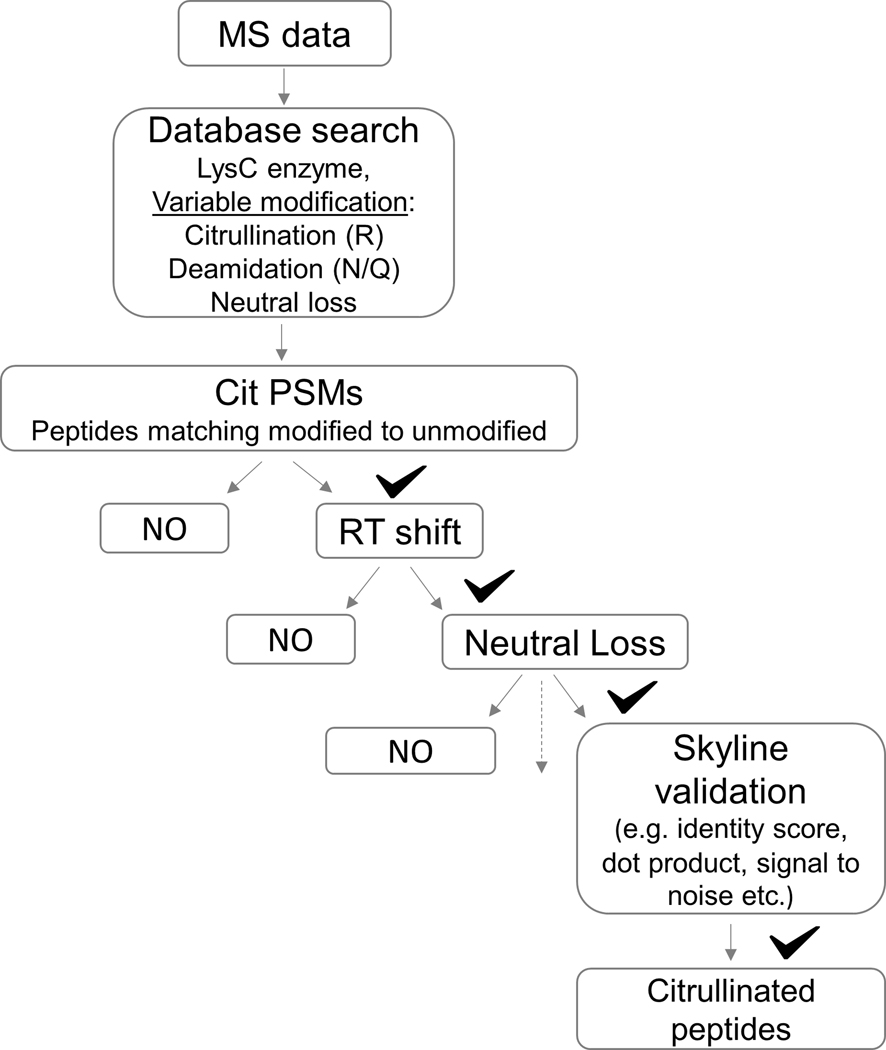
The large data sets that result from the multi-step identification of citrullinated sites require bioinformatics tools to efficiently process and validate these data to obtaining meaningful and reliable results. A decision tree, such as the Cit Finder Algorithm 52 (Figure 4), can be used to automate the correct assignment of protein citrullination in large data sets. Briefly, it is mandatory to specify the retention time (RT) shift between modified and unmodified peptide pairs (ΔRT). As presented, the ΔRT holds true for citrullinated peptides only, distinguishing it clearly from deamidated peptides. Second, neutral loss (43Da) and fragment ions should be specified to enhance confidence in the assignment of a given PTM. Third, false discovery rate (FDR) scores should be close to 1 with a peptide probability cutoff of ≥0.99 to increase probability that the peptide-spectrum matching is correct. Forth, Skyline validation should be made based on the features of peptide detection across acquisition. This series of analytical steps was shown to improve the specificity for identifying citrullinated residues, and has been incorporated under one algorithm, citFinder, that is a robust tool for the correct assignment of protein citrullination in large data sets.48 Consistent with the report by Chien-Yun et al,42 the largest number of citrullinated proteins in this study was found in the brain and lung.48 In addition, unique cellular diversity of the citrullinated proteins suggests a broad biological role for protein citrullination. In summary, to pinpoint true-positive citrullination annotations, the citrullinated residue must be distinguished from deaminated residue. In this regard, RT shift and/or neutral loss of isocyanic acid with fragment ion monitoring represent specific markers to identify citrullinated proteins.
Figure 4.
Decision tree summarizing the Cit Finder Algorithm aiding the identification and validation of citrullinated peptides. To exploit and increase the correct assignment of protein citrullination rates, a decision tree algorithm to automate the identification of protein citrullination in large data sets was developed 48. The steps include using LysC instead of trypsin for digestion, specifying deamidation as a variable modification, matching an unmodified-modified pair to specify the RT shift, and specifying the neutral loss of 43 Da in the search engine. These steps are automated under the Cit Finder algorithm and are required for the correct assignment of protein citrullination in large data sets.
3. THE NORMAL AND RA CITRULLINOMES
Citrullination is a normal process across multiple tissues in humans.42 MS analysis has identified more than 200 citrullinated proteins in different healthy human tissues.42 Together, these sets of proteins constitute the normal citrullinome. The effect of citrullination on the function of most of these proteins is unknown. In colon mucosa, patterns of citrullination in RA and healthy controls are almost identical among 223 citrullinated peptides from 121 proteins, with the exception of 3 citrullinated peptides that were specific for some patients with RA.53 Indeed, the majority of citrullinated autoantigen candidates targeted in RA 14, 54–65 are found across normal tissues,42 demonstrating that citrullination of these antigens is not an exclusive feature of RA. Nevertheless, although the targets and magnitude of citrullination may not differ among controls and RA patients in some tissues,53 it is interesting that the rheumatoid joint is enriched in citrullination.14–17
Proteomic analysis of the cellular and fluid components of RA synovial fluid has identified large amounts of citrullinated proteins in both compartments, suggesting that dysregulated citrullination occurs both in cells and extracellularly.14–17 To date, more than 100 citrullinated proteins have been identified in the RA joint,14–17 which represent the current RA citrullinome, but this list is likely to grow. The targets of citrullination in RA do not appear to be selected based on unique structural or functional features. Instead, members of the intra- and extracellular citrullinomes in RA contain a broad range of proteins including enzymes, ribonucleoproteins, histones, structural proteins, and members of the complement and coagulation pathways, among others.14–17 This unique pattern of citrullination that includes proteins spanning the range of molecular weights is termed hypercitrullination.16 Understanding the immune components in RA joint that are responsible for driving hypercitrullination and generating the RA citrullinome is thought to provide critical insights into mechanisms of autoantigen and ACPA production in RA.
4. DISCOVERING THE ORIGIN OF THE INTRACELLULAR CITRULLINOME IN RA
Defining the mechanisms responsible for generating the cellular citrullinome in RA has important challenges. Although animal models of autoimmune arthritis express synovial citrullinated proteins and recapitulate several features of the human disease,66, 67 they do not produce antibodies specific for citrullinated proteins as observed in RA.66, 68–71 The study of citrullination in RA therefore requires unique strategies to identify mechanisms that dysregulate citrullination using the human disease model. In particular, studies have been centered on identifying immune-effector pathways and target cells in the RA joint that can experimentally replicate the cellular RA citrullinome.
Although any PAD-expressing cell present in the RA joint could contribute to the RA citrullinome, including immune cells and resident fibroblast-like synoviocytes (FLSs),23, 24 neutrophils are considered the major source of citrullination in RA.16, 56, 72, 73 Neutrophils are the most abundant inflammatory cells in RA synovial fluid,74 are dominant expressors of PAD enzymes and have a unique capacity to hypercitrullinated.56 In regard to mechanisms that activate citrullination in neutrophils, it is noteworthy that although large amounts of citrullinated proteins are found in the cellular compartment in RA synovial fluid, intracellular citrullinated proteins are also detected extracellularly,17 providing initial clues that the citrullinating cells are suffering lytic cell death. Together, these findings are considered strong evidence to suggest that dying neutrophils are a major source of citrullinated autoantigens in RA and have focused interest on identifying mechanisms of cell death responsible for this process.
The conundrum in the study of citrullination in RA is that multiple inflammatory and death pathways are activated in the RA joint,16, 75–77 some of which could be erroneously linked to the RA citrullinome. Indeed, citrullination is a non-specific feature associated with inflammation.78, 79 Therefore, the search for unique forms of neutrophil death that can explain citrullination in the RA joint requires stringent criteria to dissect those that may be directly responsible for generating the RA citrullinome. Some important points to consider in this regard include: 1) the potential killing-effector pathway needs to be present in the RA joint; 2) this process should experimentally reproduce patterns of hypercitrullination found in the RA joint; and 3) the resulting citrullinome, determined by precise MS analysis, should mimic the RA citrullinome.
4.1. Neutrophil extracellular traps (NETs) as source of the RA citrullinome
NETs, an antimicrobial form of neutrophil death, was the first mechanism proposed to be associated with the production of citrullinated autoantigens in RA.76 This form of cell death is present in the RA joint,76 in vitro evidence suggests that neutrophils from patients with RA have increased susceptibility to form NETs,76 and in vivo evidence suggests that the formation of NETs is increased in RA.80 An association between a NETs and citrullination was initially proposed by the finding that PAD4-mediated citrullination of histones was necessary for NET formation in mice.81 Later, this finding was erroneously taken as direct evidence that citrullination, particularly of histones, is a hallmark and exclusive marker of NETs.82 Thus, in the absence of other mechanisms that may explain citrullination in RA, the co-existence of NETs and citrullination in the RA joint led to the conclusion that the production of citrullinated proteins in RA is the consequence of dysregulated formation of NETs.76 Although further studies have questioned the role of citrullination in NET formation in humans,83, 84 potential discrepancies in the study of NETs appear to be explained from the use of distinct artificial stimuli, such as phorbol 12-myristate 13-acetate (PMA) and calcium ionophores, which activate different pathways to induce NETs or neutrophil lysis.83, 84 In this regard, although there is a large volume of publications linking NETs, citrullination and RA, only few studies have explored autoantigen citrullination in NETs induced by stimuli relevant for RA pathogenesis.76, 85
Using NETs induced by RF, ACPA-positive RA-IgG or tumor necrosis factor (TNF)-α, Khandpur et al. analyzed the NET protein cargo by LC/ESI MS/MS.76 However, while several proteins were identified in NETs, it is noteworthy that the proteomic analysis did not search for citrullinated peptides. Instead, the possible presence of citrullination in NETs was determined by the indirect analysis of a single protein (i.e. vimentin) using antibodies.76 More recently, Carmona-Rivera et al. used MS to confirm the presence of citrullination in NETs induced by RF.85 Using trypsin to digest the NET cargo, the study found 28 citrullinated peptides from 20 proteins in NETs. However, it is remarkable that 25 of the 28 peptides were reported as C-terminally citrullinated,85 questioning whether interpretation of MS data was affected by the previously discuss caveats in the analysis of protein citrullination (discussed in the “MASS SPECTROMETRY APPLIED TO THE STUDY OF CITRULLINATION IN COMPLEX SAMPLES: section). In particular, since trypsin is a serine protease that cleaves peptide bonds after the basic amino acids arginine and lysine, trypsin does not efficiently cleave after neutrally charged citrulline residues.60, 86–88 In addition, the majority of the C-terminally citrullinated peptides identified also contained asparagine and/or glutamine residues, which introduces additional caveats to the data interpretation. Therefore, tryptic peptides that report C-terminal citrullination should be taken with caution and subjected to additional analysis to confirm the presence of citrullination and distinguish it from deamination. While NET formation appears to be increased in RA, rigorous MS analysis of NET cargo is warranted to determine if NETs represent a major source of citrullinated autoantigens in RA or are capable of recapitulating the RA citrullinome.
4.2. Immune-mediated membranolytic pathways and the citrullinome in RA
During the analysis of potential death mechanisms associated with cellular hypercitrullination in the RA joint, it was noticed that apoptotic cell death was increased in synovial fluid cells from approximately half of patients with RA, while the other half had evidence of complement pathway activation.16 In the apoptotic group, cleavage of BH3-interacting domain death agonist (BID, a marker of extrinsic apoptosis) was enriched, providing initial evidence that cytotoxic-cell death was involved in this process.16 Consistent with this finding, further studies demonstrated that among multiple forms of neutrophil death, cytotoxic-mediated killing (via perforin) was the only mechanism able to reproduce patterns of hypercitrullination found in the RA joint.16 In the other half of patients with RA who have no evidence of cytotoxic death activity, synovial fluid cells showed deposition of the membrane attack complex (MAC), suggesting complement-induced cell damage. Similar to perforin, complement-mediated neutrophil killing was associated with prominent hypercitrullination as observed in the RA joint.16 MS analysis of neutrophils killed with perforin or MAC further confirmed the presence of protein citrullination and their protein identity overlapped substantially with the RA citrullinome.16 providing direct evidence that these pathways can experimentally reproduce citrullination found in the RA joint.
Pore-forming proteins, such as perforin and MAC, induce membrane destabilization, influx of extracellular calcium, and ultimately osmotic lysis.89–92 During this process, cytotoxic loss of membrane integrity results in calcium-dependent hyperactivation of PADs and hypercitrullination of cellular substrates, which are subsequently released into the extracellular space likely contributing to the extracellular citrullinome.16 This form of neutrophil death induced by these pore-forming mechanisms has been termed leukotoxic hypercitrullination (LTH).84 Interestingly, the induction of LTH is not limited to perforin and MAC. Some bacterial pore-forming toxins are also potent inducers of LTH,72, 84 which has been suggested as a mechanism by which some bacteria may contribute to RA pathogenesis.72, 84 In this regard, it is important to underscore that perforin and MAC are the only mechanisms active in the RA joint that have been able to experimentally replicate the RA citrullinome.16
5. THE GLITCH OF EXTRACELLULAR CITRULLINATION IN RA
The discovery of citrullinated fibrin(ogen) in the RA joint identified the first relevant substrate for ACPAs,93 and provided initial evidence that citrullination in RA can occur extracellularly. Further MS analysis of synovial fluid supernatants confirmed that fibrin(ogen) is citrullinated in RA, and additionally identified a large number of extracellular citrullinated proteins, including components of the complement and coagulation pathways, among many others.17 Two potential sources of extracellular PADs have been suggested to be responsible for this process. These include PADs secreted from activated neutrophils, and PADs released during neutrophil death.73, 94, 95 Nevertheless, while there is convincing evidence that soluble PADs are found in RA synovial fluid,95, 96 the mechanisms involved in extracellular PAD activation are not fully understood.
PAD activity requires a reducing environment to maintain the active site free thiol cysteine necessary for catalysis.95 The oxidizing extracellular environment, which contrasts with the reducing milieu inside cells,97 is therefore hostile for citrullination. Indeed, neither endogenous nor recombinant PADs have catalytic activity in synovial fluid unless reducing agents, such as dithiothreitol or reduced glutathione, are exogenously added.95, 96 Similarly, PADs secreted from activated neutrophils, during PMA-induced NET formation, or from necrotic neutrophils are only active in the presence of high amounts of dithiothreitol.73, 94, 95 In some cases, supraphysiologic amounts of calcium (>1.5mM, with 1–1.5mM normally present in extracellular compartments) are also added exogenously to increase extracellular PAD activity.73, 94, 96 Importantly, however, these conditions do not co-exist in vivo during physiologic conditions and whether these conditions are altered in the rheumatoid joint is unknown. Nevertheless, while extracellular citrullination is likely a very inefficient process, the presence of extracellular PAD substrates supports the existence of mechanisms that may facilitate extracellular citrullination in RA.
5.1. PAD4 agonistic autoantibodies
In addition to citrullinated proteins, citrullinating enzymes themselves (i.e. PAD2 and PAD4) are also targets of autoantibodies in RA.98–100 A subset of antibodies that enhance the catalytic activity of PAD4 were recently described in RA,100 and discovered because of their cross-reactivity with PAD3 (so called PAD3/4 autoantibodies). These antibodies identify a unique subgroup of patients with the most erosive disease and pulmonary involvement,100–102 supporting their pathogenic potential in RA. Interestingly, these antibodies decrease the amount of calcium required for PAD4 to catalyze citrullination and enhance PAD4 activity in the absence of exogenous reducing agents.100 Since PADs released from activated and dying neutrophils are transiently active,95 it is possible that activating antibodies in synovial fluid may accelerate PAD4 activity before its inactivation, thus enhancing dysregulated extracellular citrullination.
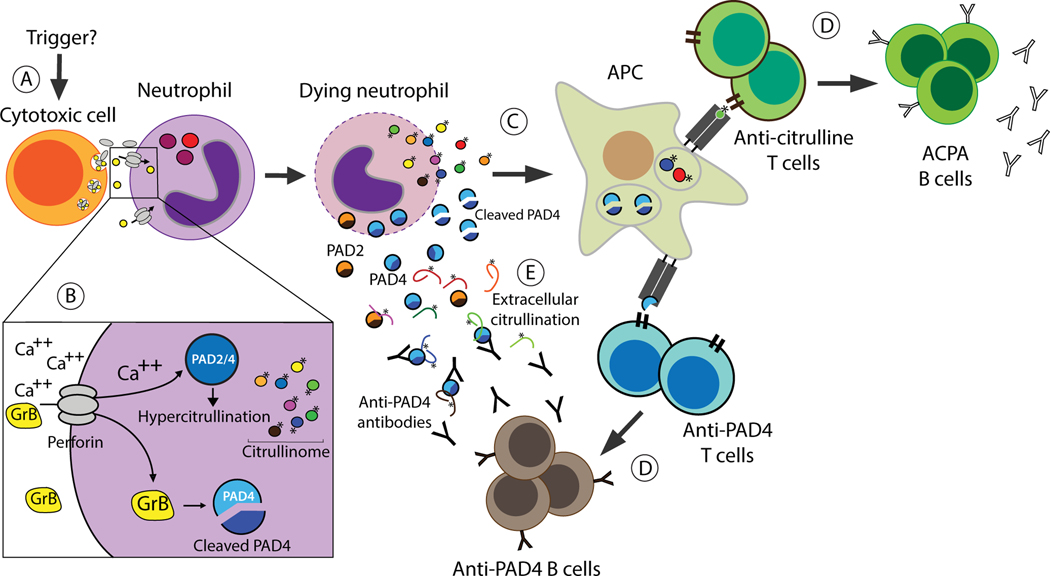
The presence of PAD4 agonistic autoantibodies is not only relevant for their contribution to citrullination, but their potential origin also provides important insights into disease mechanisms in RA. Analysis of recombinant antibodies cloned from single PAD4-specific memory B cells from patients with RA has revealed these antibodies arise from germline PAD4-reactive precursors, which likely bypassed early tolerance checkpoints.103 However, affinity maturation was shown to be essential for the antibodies to gain efficient agonistic activity against PAD4, strongly suggesting that these antibodies are driven by some immunogenic form of PAD4. In this regard, recent evidence has shown that granzyme B-mediated cleavage of PAD4 increases its recognition by CD4+ T cells from patients with RA.104 Granzyme B is a cytotoxic protease that is released together with perforin by killer cells. Perforin facilitates the delivery of granzyme B into the cytosol, which cleaves intracellular substrates and induces target cell death.105, 106 Thus, depending on genetic and environmental risk factors, PAD4-autoreactive naïve B cells may have a selective advantage for developing into high affinity autoreactive memory B cells through somatic mutations and affinity maturation, which may be driven by T cells raised against granzyme B-cleaved PAD4. In the context of RA pathogenesis, these findings, together with the role of cytotoxic cells in the induction of hypercitrullination, suggest a model connecting cytotoxic-effector pathways (i.e. perforin and granzyme B) to the targeting of citrullinated antigens and PAD4 in RA (Figure 5).
Figure 5.
A pivotal role of cytotoxic cells in the immune response to citrullinated antigens and PAD4. (A) The rheumatoid joint is enriched with numerous types of cytotoxic cells 145–150, although their trigger is still unknown. (B) Neutrophil death induced by perforin and granzyme B may have a dual effect on autoantigen production in RA. While perforin drives hyperactivation of PAD enzymes (via calcium influx) and hypercitrullination, cleavage of PAD4 by granzyme B changes its immunogenicity. (C) Dying neutrophils loaded with citrullinated antigens and cleaved PAD4 provide a source of autoantigens to antigen presenting cells, which can further activate antigen-specific autoreactive T cells. (D) In pre-clinical RA, this process may promote the expansion and maturation of autoreactive naïve B cells, generating high affinity autoreactive ACPA and anti-PAD4 memory B cells, respectively. In established RA, this process may provide a continued source of autoantigens to propagate and sustain the autoimmune response. (E) Newly released PADs from dying neutrophils may maintain transient but steady citrullination in the RA joint, which may be enhanced by agonistic antibodies to PAD4. Citrullinated antigens are indicated (*). GrB, granzyme B; APC, antigen presenting cell.
While agonistic antibodies to PAD4 provide a rational paradigm to explain the presence of extracellular citrullination despite the non-permissive conditions in synovial fluid, it is important to highlight that these antibodies are only present in ~20% of patients with RA.100 Therefore, it is likely that other mechanisms also exist, which increase PAD activity in the RA joint. Alternatively, extracellular citrullination in anti-PAD3/4 antibody negative individuals may depend on newly released PADs from large amounts of dying cells, which may maintain a transient but steady source of active citrullinating enzyme (Figure 5).
6. THE RA CITRULLINOME IN EXTRAARTICULAR SITES
Since ACPAs are detected years before the onset of joint symptoms in RA,107 it is uncertain whether the immune response to citrullinated proteins is initiated in the joints. Instead, there is growing support for the hypothesis that breach of immunologic tolerance to citrullinated antigens in pre-clinical RA initially occurs at mucosal sites such as the oral mucosa, lung, and gastrointestinal tract.108–110 In this model, environmental risk factors associated with RA may trigger aberrant protein citrullination at specific mucosal sites, driving the initial production of autoantibodies. Similar to citrullination in the RA joint, it is important to identify factors associated with pathogenic citrullination at extraarticular sites of disease initiation, to distinguish between non-specific citrullination associated with inflammation that can occur in barrier tissues and dysregulated citrullination that may predispose to RA. Since the pre-clinical production of ACPAs is a slow process that appears to be driven by epitope spreading targeting multiple citrullinated antigens,111 pathways that transiently citrullinate one or few proteins may not be relevant for RA pathogenesis. Instead, pathogenic citrullination at mucosal sites is expected to be robust, chronic and generate multiple citrullinated proteins, potentially mimicking patterns of citrullination in the rheumatoid joint.
6.1. The RA citrullinome in periodontitis
A tantalizing association between periodontitis and RA has been suggested for decades,112 which sparked the search for potential pathogenic mechanisms that may link both diseases. One such mechanism is citrullination. Proteomic analysis of gingival crevicular fluid (GCF) and periodontal tissue from patients with periodontitis with and without RA has shown abundant citrullination that overlaps with the RA citrullinome,72, 113 supporting the notion that periodontitis is a potential source of citrullinated autoantigens. Mechanisms that dysregulate citrullination in periodontitis have been mainly attributed to periodontal pathogens, in particular Porphyromonas gingivalis (P. gingivalis) and Aggregatibacter actinomycetemcomitans (Aa).112 While P. gingivalis is a keystone pathogen in periodontitis and produces an enzyme that citrullinates C-terminal arginine residues (i.e. an exodeiminase known as PPAD),114–116 the contribution of this pathogen in generating the citrullinome in periodontitis is unclear.112 Furthermore, peptide spectra of citrullinated proteins in periodontitis are consistent with endocitrullination,72 suggesting that an abnormal activation of host PADs, rather than PPAD, is responsible for this process. Nevertheless, although the contribution of PPAD to the periodontal citrullinome is unclear, it has been proposed that C-terminal citrullination may have some immunogenic advantage to break tolerance to citrullinated proteins.109 Further study of the contribution of PPAD to the RA and periodontal citrullinome is needed to support this interesting hypothesis.
Aa is a periodontal pathogen associated with severe forms of periodontitis,117–121 both localized aggressive periodontitis (LAP) and chronic periodontitis,117, 118, 122–131 In contrast to other oral bacteria, Aa has a unique capacity to induce neutrophil hypercitrullination, suggesting a potential role of this bacterium in generating the periodontal citrullinome.72 Similar to host-immune pore-forming pathways, Aa hyperactivates PADs by inducing membranolytic damage on neutrophils. The bacterium secretes a pore-forming toxin, leukotoxin A (LtxA), which bind CD18 initiating membrane destabilization, influx of extracellular calcium, PAD activation and neutrophil death by osmotic lysis. The citrullinome induced by LtxA is prominent, spanning more than 80 proteins with more than half overlapping with the RA citrullinome.72 These findings have focused interest on membranolytic damage induced by host and bacterial pore-forming proteins as a unifying mechanism to induce neutrophil hypercitrullination and reproduce the RA citrullinome.18, 84 Moreover, this work identified Aa-associated periodontitis as a potential risk factor linked to the lack of tolerance to citrullinated autoantigens in RA.
6.2. Citrullination in the lung
The finding that smoking (a major risk factor in RA) is associated with HLA-DRB1 shared epitope alleles and ACPA in patients with RA provided initial evidence to suggest a potential mechanistic relation between smoking and citrullination in the lung.132 This idea was further supported by the finding that non-RA smokers have increased citrullination in alveolar macrophages from bronchoalveolar lavage (BAL) compared to healthy non-smokers,132 suggesting the possibility that alveolar macrophages may increase citrullination in response to smoking or other environmental stimuli.132 The hypothesis that chronic inflammatory damage increases citrullination in the lung has been further explored using multiple strategies, with little distinction regarding specific lung pathologies or cellular sources of citrullinating enzymes (e.g. immune vs. epithelial cells). This has led to conflation of the results supporting the hypothesis that inflammation drives citrullination in the lung, despite important differences in the underlying pathology and potential contribution of distinct cellular sources of citrullination in the different disease states.
Several tissue sources have been studied to examine this hypothesis including: bronchoalveolar lavage (BAL), bronchial mucosa, lung tissue and/or sputum from a variety of individuals including healthy non-smokers and smokers, patients with RA-associated interstitial lung disease (RA-ILD), RA-associated interstitial pneumonia (RA-IP), idiopathic IP, non-RA patients with chronic obstructive pulmonary disease (non-RA COPD), RA patients without pulmonary symptoms and RA-free subjects at risk for RA.57, 133–142 Among these studies, only two addressed citrullination in the lung by MS. In lung tissue from non-RA COPD, MS was only applied to study citrullination of α-enolase and vimentin, but only confirmed citrullination in vimentin.139 In the second study, MS analysis of bronchial tissue from newly diagnosed RA only identified 7 citrullinated proteins.136 In the majority of the studies, however, the analysis of citrullination in lung samples (i.e. BAL and tissue) was done indirectly by immunodetection of citrullinated proteins,133, 135, 137, 138 rather than MS, which provides no information regarding the protein identity, extent of citrullination, or identify of citrullination sites. In addition, cellular expression of PAD enzymes, detection of ACPAs and/or the presence of markers of neutrophil lysis (denoted as NETs) in lung samples (i.e. BAL, tissue and/or sputum) have been used as indirect evidence to support dysregulated citrullination in the lung,133, 134, 137, 140–142 suggesting that detailed MS analysis of the lung citrullinome is needed.
It is important to underscore that the lung is the organ with the second largest number of citrullinated proteins in healthy humans.42 Understanding the cellular origin and magnitude of potential pathogenic citrullinomes in the lung therefore has important mechanistic implications for RA. First, MS analysis will aid in distinguishing the independent contribution of immune vs. epithelial cells in the production of citrullinated proteins in the lung. In this regard, it is noteworthy that while alveolar macrophages in BAL from non-RA smokers were initially shown to contain citrullinated proteins by immunostaining,132, 138 the presence of citrullination has not been addressed in inflammatory lung cells from patients with or at risk for RA. Moreover, although alveolar macrophages constitute up to 94% of the BAL cells in non-RA smokers,132 the proportion of lymphocytes is importantly increased in subsets of RA patients with and without lung involvement.143, 144 This highlights that differences in the composition of immune cells can be observed in distinct pathologic states and suggests that the cellular source of citrullination may not be uniform across lung pathologies. In addition, different to RA synovial fluid, neutrophils are among the least abundant cells in BAL.132, 143, 144 It is therefore important to define the independent contribution of PAD-expressing cells (e.g. neutrophils, macrophages, others) to the lung citrullinome using precise MS analysis. Second, defining the magnitude and protein identity of the citrullinome in the lung will help to determine whether patterns of citrullination are consistent with chronic inflammation or as result of PAD dysregulation (e.g. hypercitrullination). Third, defining the lung citrullinomes present during different pathologic states may reveal conditions or environmental stimuli, which recapitulate the RA citrullinome. These are critical steps in the search for mechanisms relevant for RA pathogenesis.
7. CONCLUSION
The precise definition of normal and pathogenic citrullinomes in human tissues have important implications for understanding disease mechanisms associated with dysregulated PAD activation. Under normal conditions, distinct patterns and levels of citrullination are detected in multiple tissues,42 suggesting that different cell types (i.e. both immune and non-immune) and stimuli (e.g. inflammation, hormones, and others) modulate PAD activity with no pathogenic consequences. The definition of pathogenic citrullinomes therefore requires precise methods to quantify citrullinated proteins in complex samples, such as in cells and tissues, and to determine citrullination sites in proteins. Otherwise, in the absence of a clear distinction between normal and dysregulated citrullination, any experimental condition that may activate PADs has the risk to be misclassified as pathogenic.
Recent advances in MS instrumentation and analytical methods have revolutionized the study of citrullination, but with the qualification that comprehensive analyses must be properly applied. In order to define true-positive citrullination events, citrullinated residues must be confidently distinguished from deaminated residues based on the RT shift and/or neutral loss of isocyanic acid with fragment ion monitoring for identification. Use of these two features can provide a specific strategy for the identification of citrullinated proteins, suggesting that this approach should become standard practice for the study of protein citrullination.
In RA, MS analysis of synovial fluid has been used to determine the identity and magnitude of the RA citrullinome, which represents the only available probe to identify mechanisms of autoantigen production in the rheumatoid joint. In this context, studies have been centered on pinpointing immune-effector pathways in the RA joint that can experimentally replicate the RA citrullinome. Yet, despite important advances in MS analysis and a vast body of literature regarding the role of citrullination in RA, there are only four studies that have applied MS analysis to address the citrullinomes generated by stimuli relevant for RA pathogenesis.16, 72, 76, 85 While these studies are highly cited, none of these findings have been attempted to be replicated by others. Furthermore, membranolytic damage induced by pore-forming proteins is the only mechanism that has been shown to dysregulate PAD activity, drive hypercitrullination, and generate a prominent citrullinome in neutrophils that mimics citrullination patterns found in the RA joint.16, 72 In the case of citrullination of extracellular substrates, the presence of PAD4 agonistic antibodies is the only known mechanism identified, thus far, that may explain PAD4 activity in the hostile extracellular oxidizing environment.18, 100 As these antibodies are found in only one fifth of patients with RA, other unknown mechanisms are likely to sustain extracellular citrullination in the RA joint.
MS analysis of complex samples is a powerful tool for the study of diseases linked to dysregulated PAD activation. In the case of RA, MS analysis offers a unique opportunity to identify and characterize potential extraarticular sites of disease initiation, to validate pathways thought to dysregulate PAD activation (e.g. NETs and membranolytic damage), to discover novel mechanisms that sustain both intra- and extracellular citrullination, and potentially to identify the causal triggers of RA.
ACKNOWLEDGMENTS:
Funding for this project was provided by the Jerome L. Greene Foundation, Rheumatology Research Foundation, and the National Institutes of Health (NIH) grants number R01 AR069569, R01 AR050026–12A1, R01HL111362. The content of this paper is solely the responsibility of the author and does not represent the official views of the NIH.
Footnotes
CONFLICT OF INTEREST
ED and FA are authors on licensed patent no. 8,975,033, entitled “Human autoantibodies specific for PAD3 which are cross-reactive with PAD4 and their use in the diagnosis and treatment of rheumatoid arthritis and related diseases” and on provisional patient no. 62/481,158 entitled “Anti-PAD2 antibody for treating and evaluating rheumatoid arthritis”. FA serves as consultant for Bristol-Myers Squibb, has received a grant from Medimmune, and personal fees from Celgene, outside of this submitted work. ED has received a grant from Pfizer, Celgene, and Medimmune and personal fees from Celgene, outside of this submitted work. The remaining authors declare no competing interests.
REFERENCES
- 1.Coons AH, Creech HJ, Jones RN, Berliner E. The demonstration of pneumococcal antigen in tissues by the use of fluorescent antibody. Journal of Immunology.1942;45:159–170. [Google Scholar]
- 2.Holman HR, Kunkel HG. Affinity between the lupus erythematosus serum factor and cell nuclei and nucleoprotein. Science.1957;126:162–163. [DOI] [PubMed] [Google Scholar]
- 3.Beck JS. Variations in the morphological patterns of “autoimmune” nuclear fluorescence. Lancet.1961;1:1203–1205. [DOI] [PubMed] [Google Scholar]
- 4.Pollak VE, Mandema E, Kark RM. Antinuclear factors in the serum of relatives of patients with systemic lupus erythematosus. Lancet.1960;2:1061–1063. [DOI] [PubMed] [Google Scholar]
- 5.Hazenberg BP, Gruys E, van Rijswijk MH. In memoriam Enno Mandema, MD (1921–2010). Amyloid-Journal of Protein Folding Disorders.2011;18:249–250. [Google Scholar]
- 6.Mandema E, Pollak VE, Kark RM, Rezaian J. Quantitative observations on antinuclear factors in systemic lupus erythematosus. J Lab Clin Med.1961;58:337–352. [PubMed] [Google Scholar]
- 7.Nienhuis RL, Mandema E. A New Serum Factor in Patients with Rheumatoid Arthritis; The Antiperinuclear Factor. Ann Rheum Dis.1964;23:302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young BJ, Mallya RK, Leslie RD, Clark CJ, Hamblin TJ. Anti-keratin antibodies in rheumatoid arthritis. Br Med J.1979;2:97–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simon M, Girbal E, Sebbag M, et al. The cytokeratin filament-aggregating protein fillagrin is the target of the co-called “antikeratin antibodies,” autoantibodies specific for rheumatoid arthritis. J Clin Invest.1993;92:1387–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sebbag M, Simon M, Vincent C, et al. The antiperinuclear factor and the so-called antikeratin antibodies are the same rheumatoid arthritis-specific autoantibodies. J Clin Invest.1995;95:2672–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schellekens GA, de Jong BA, van den Hoogen FH, van de Putte LB, van Venrooij WJ. Citrulline is an essential constituent of antigenic determinants recognized by rheumatoid arthritis-specific autoantibodies. J Clin Invest.1998;101:273–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ge C, Holmdahl R. The structure, specificity and function of anti-citrullinated protein antibodies. Nat Rev Rheumatol.2019;15:503–508. [DOI] [PubMed] [Google Scholar]
- 13.Derksen V, Huizinga TWJ, van der Woude D. The role of autoantibodies in the pathophysiology of rheumatoid arthritis. Semin Immunopathol.2017;39:437–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Beers JJ, Schwarte CM, Stammen-Vogelzangs J, et al. The rheumatoid arthritis synovial fluid citrullinome reveals novel citrullinated epitopes in apolipoprotein E, myeloid nuclear differentiation antigen, and beta-actin. Arthritis Rheum.2013;65:69–80. [DOI] [PubMed] [Google Scholar]
- 15.Wang F, Chen FF, Gao WB, et al. Identification of citrullinated peptides in the synovial fluid of patients with rheumatoid arthritis using LC-MALDI-TOF/TOF. Clin Rheumatol.2016;35:2185–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romero V, Fert-Bober J, Nigrovic PA, et al. Immune-mediated pore-forming pathways induce cellular hypercitrullination and generate citrullinated autoantigens in rheumatoid arthritis. Sci Transl Med.2013;5:209ra150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tutturen AE, Fleckenstein B, de Souza GA. Assessing the citrullinome in rheumatoid arthritis synovial fluid with and without enrichment of citrullinated peptides. J Proteome Res.2014;13:2867–2873. [DOI] [PubMed] [Google Scholar]
- 18.Darrah E, Andrade F. Rheumatoid arthritis and citrullination. Curr Opin Rheumatol.2018;30:72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raijmakers R, Zendman AJ, Egberts WV, et al. Methylation of arginine residues interferes with citrullination by peptidylarginine deiminases in vitro. J Mol Biol.2007;367:1118–1129. [DOI] [PubMed] [Google Scholar]
- 20.Kearney PL, Bhatia M, Jones NG, et al. Kinetic characterization of protein arginine deiminase 4: a transcriptional corepressor implicated in the onset and progression of rheumatoid arthritis. Biochemistry.2005;44:10570–10582. [DOI] [PubMed] [Google Scholar]
- 21.Fujisaki M, Sugawara K. Properties of peptidylarginine deiminase from the epidermis of newborn rats. J Biochem.1981;89:257–263. [DOI] [PubMed] [Google Scholar]
- 22.Kinloch A, Lundberg K, Wait R, et al. Synovial fluid is a site of citrullination of autoantigens in inflammatory arthritis. Arthritis Rheum.2008;58:2287–2295. [DOI] [PubMed] [Google Scholar]
- 23.Foulquier C, Sebbag M, Clavel C, et al. Peptidyl arginine deiminase type 2 (PAD-2) and PAD-4 but not PAD-1, PAD-3, and PAD-6 are expressed in rheumatoid arthritis synovium in close association with tissue inflammation. Arthritis Rheum.2007;56:3541–3553. [DOI] [PubMed] [Google Scholar]
- 24.Chang X, Yamada R, Suzuki A, et al. Localization of peptidylarginine deiminase 4 (PADI4) and citrullinated protein in synovial tissue of rheumatoid arthritis. Rheumatology (Oxford).2005;44:40–50. [DOI] [PubMed] [Google Scholar]
- 25.Senshu T, Sato T, Inoue T, Akiyama K, Asaga H. Detection of citrulline residues in deiminated proteins on polyvinylidene difluoride membrane. Anal Biochem.1992;203:94–100. [DOI] [PubMed] [Google Scholar]
- 26.Bicker KL, Subramanian V, Chumanevich AA, Hofseth LJ, Thompson PR. Seeing citrulline: development of a phenylglyoxal-based probe to visualize protein citrullination. J Am Chem Soc.2012;134:17015–17018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neeli I, Radic M. Current Challenges and Limitations in Antibody-Based Detection of Citrullinated Histones. Frontiers in Immunology.2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholas AP, Whitaker JN. Preparation of a monoclonal antibody to citrullinated epitopes: its characterization and some applications to immunohistochemistry in human brain. Glia.2002;37:328–336. [PubMed] [Google Scholar]
- 29.Shi J, Willemze A, Janssen GM, et al. Recognition of citrullinated and carbamylated proteins by human antibodies: specificity, cross-reactivity and the ‘AMC-Senshu’ method. Ann Rheum Dis.2013;72:148–150. [DOI] [PubMed] [Google Scholar]
- 30.Shi J, Knevel R, Suwannalai P, et al. Autoantibodies recognizing carbamylated proteins are present in sera of patients with rheumatoid arthritis and predict joint damage. Proc Natl Acad Sci U S A.2011;108:17372–17377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altelaar AF, Munoz J, Heck AJ. Next-generation proteomics: towards an integrative view of proteome dynamics. Nat Rev Genet.2013;14:35–48. [DOI] [PubMed] [Google Scholar]
- 32.Larsen MR, Trelle MB, Thingholm TE, Jensen ON. Analysis of posttranslational modifications of proteins by tandem mass spectrometry. Biotechniques.2006;40:790–798. [DOI] [PubMed] [Google Scholar]
- 33.Hensen SMM, Pruijn GJM. Methods for the Detection of Peptidylarginine Deiminase (PAD) Activity and Protein Citrullination. Molecular & Cellular Proteomics.2014;13:388–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Creese AJ, Grant MM, Chapple LLC, Cooper HJ. On-line liquid chromatography neutral loss-triggered electron transfer dissociation mass spectrometry for the targeted analysis of citrullinated peptides. Anal Methods-Uk.2011;3:259–266. [DOI] [PubMed] [Google Scholar]
- 35.Verheul MK, van Veelen PA, van Delft MAM, et al. Pitfalls in the detection of citrullination and carbamylation. Autoimmun Rev.2018;17:136–141. [DOI] [PubMed] [Google Scholar]
- 36.Hagiwara T, Hidaka Y, Yamada M. Deimination of histone H2A and H4 at arginine 3 in HL-60 granulocytes. Biochemistry.2005;44:5827–5834. [DOI] [PubMed] [Google Scholar]
- 37.Hao G, Wang DC, Gu J, et al. Neutral Loss of Isocyanic Acid in Peptide CID Spectra: A Novel Diagnostic Marker for Mass Spectrometric Identification of Protein Citrullination. J Am Soc Mass Spectr.2009;20:723–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen YF, Tolic N, Xie F, et al. Effectiveness of CID, HCD, and ETD with FT MS/MS for Degradomic-Peptidomic Analysis: Comparison of Peptide Identification Methods. Journal of Proteome Research.2011;10:3929–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steckel A, Uray K, Turiak L, et al. Mapping the tandem mass spectrometric characteristics of citrulline-containing peptides. Rapid Commun Mass Sp.2018;32:844–850. [DOI] [PubMed] [Google Scholar]
- 40.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proceedings of the National Academy of Sciences of the United States of America.2004;101:9528–9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin Z, Fu Z, Yang J, et al. Identification and characterization of citrulline-modified brain proteins by combining HCD and CID fragmentation. Proteomics.2013;13:2682–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee CY, Wang D, Wilhelm M, et al. Mining the Human Tissue Proteome for Protein Citrullination. Mol Cell Proteomics.2018;17:1378–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kovacs JM, Mant CT, Hodges RS. Determination of intrinsic hydrophilicity/hydrophobicity of amino acid side chains in peptides in the absence of nearest-neighbor or comformational effects. Biopolymers.2006;84:283–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orgovan G, Noszal B. The complete microspeciation of arginine and citrulline. J Pharmaceut Biomed.2011;54:965–971. [DOI] [PubMed] [Google Scholar]
- 45.Hermansson M, Artemenko K, Ossipova E, et al. MS analysis of rheumatoid arthritic synovial tissue identifies specific citrullination sites on fibrinogen. Proteomics Clin Appl.2010;4:511–518. [DOI] [PubMed] [Google Scholar]
- 46.Krasny L, Bland P, Kogata N, et al. SWATH mass spectrometry as a tool for quantitative profiling of the matrisome. Journal of Proteomics.2018;189:11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu JX, Song XM, Pascovici D, et al. SWATH Mass Spectrometry Performance Using Extended Peptide MS/MS Assay Libraries. Molecular & Cellular Proteomics.2016;15:2501–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fert-Bober J, Venkatraman V, Hunter CL, et al. Mapping Citrullinated Sites in Multiple Organs of Mice Using Hypercitrullinated Library. Journal of Proteome Research.2019;18:2270–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stensland M, Holm A, Kiehne A, Fleckenstein B. Targeted analysis of protein citrullination using chemical modification and tandem mass spectrometry. Rapid Commun Mass Sp.2009;23:2754–2762. [DOI] [PubMed] [Google Scholar]
- 50.Choi M, Song JS, Kim HJ, Cha S, Lee EY. Matrix-assisted laser desorption ionization-time of flight mass spectrometry identification of peptide citrullination site using Br signature. Analytical Biochemistry.2013;437:62–67. [DOI] [PubMed] [Google Scholar]
- 51.Thompson DA, Ng R, Dawson PE. Arginine selective reagents for ligation to peptides and proteins. J Pept Sci.2016;22:311–319. [DOI] [PubMed] [Google Scholar]
- 52.Sanborn BM, Hein GE. The interaction of trypsin with neutral substrates and modifiers. Biochemistry.1968;7:3616–3624. [DOI] [PubMed] [Google Scholar]
- 53.Bennike TB, Ellingsen T, Glerup H, et al. Proteome Analysis of Rheumatoid Arthritis Gut Mucosa. J Proteome Res.2017;16:346–354. [DOI] [PubMed] [Google Scholar]
- 54.Vossenaar ER, Despres N, Lapointe E, et al. Rheumatoid arthritis specific anti-Sa antibodies target citrullinated vimentin. Arthritis Res Ther.2004;6:R142–R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Beers JJ, Willemze A, Stammen-Vogelzangs J, et al. Anti-citrullinated fibronectin antibodies in rheumatoid arthritis are associated with human leukocyte antigen-DRB1 shared epitope alleles. Arthritis Res Ther.2012;14:R35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Darrah E, Rosen A, Giles JT, Andrade F. Peptidylarginine deiminase 2, 3 and 4 have distinct specificities against cellular substrates: novel insights into autoantigen selection in rheumatoid arthritis. Ann Rheum Dis.2012;71:92–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Harlow L, Rosas IO, Gochuico BR, et al. Identification of citrullinated hsp90 isoforms as novel autoantigens in rheumatoid arthritis-associated interstitial lung disease. Arthritis Rheum.2013;65:869–879. [DOI] [PubMed] [Google Scholar]
- 58.Dwivedi N, Upadhyay J, Neeli I, et al. Felty’s syndrome autoantibodies bind to deiminated histones and neutrophil extracellular chromatin traps. Arthritis Rheum.2012;64:982–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Konig MF, Giles JT, Nigrovic PA, Andrade F. Antibodies to native and citrullinated RA33 (hnRNP A2/B1) challenge citrullination as the inciting principle underlying loss of tolerance in rheumatoid arthritis. Ann Rheum Dis.2016;75:2022–2028. [DOI] [PubMed] [Google Scholar]
- 60.Kinloch A, Tatzer V, Wait R, et al. Identification of citrullinated alpha-enolase as a candidate autoantigen in rheumatoid arthritis. Arthritis Res Ther.2005;7:R1421–R1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Matsuo K, Xiang Y, Nakamura H, et al. Identification of novel citrullinated autoantigens of synovium in rheumatoid arthritis using a proteomic approach. Arthritis Res Ther.2006;8:R175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suzuki A, Yamada R, Ohtake-Yamanaka M, et al. Anti-citrullinated collagen type I antibody is a target of autoimmunity in rheumatoid arthritis. Biochem Biophys Res Commun.2005;333:418–426. [DOI] [PubMed] [Google Scholar]
- 63.Burkhardt H, Sehnert B, Bockermann R, et al. Humoral immune response to citrullinated collagen type II determinants in early rheumatoid arthritis. Eur J Immunol.2005;35:1643–1652. [DOI] [PubMed] [Google Scholar]
- 64.Okazaki Y, Suzuki A, Sawada T, et al. Identification of citrullinated eukaryotic translation initiation factor 4G1 as novel autoantigen in rheumatoid arthritis. Biochem Biophys Res Commun.2006;341:94–100. [DOI] [PubMed] [Google Scholar]
- 65.Goeb V, Thomas-L’Otellier M, Daveau R, et al. Candidate autoantigens identified by mass spectrometry in early rheumatoid arthritis are chaperones and citrullinated glycolytic enzymes. Arthritis Res Ther.2009;11:R38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vossenaar ER, Nijenhuis S, Helsen MM, et al. Citrullination of synovial proteins in murine models of rheumatoid arthritis. Arthritis Rheum.2003;48:2489–2500. [DOI] [PubMed] [Google Scholar]
- 67.Gully N, Bright R, Marino V, et al. Porphyromonas gingivalis Peptidylarginine Deiminase, a Key Contributor in the Pathogenesis of Experimental Periodontal Disease and Experimental Arthritis. Plos One.2014;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cantaert T, Teitsma C, Tak PP, Baeten D. Presence and role of anti-citrullinated protein antibodies in experimental arthritis models. Arthritis Rheum.2013;65:939–948. [DOI] [PubMed] [Google Scholar]
- 69.Konig MF, Darrah E, Andrade F. Insights into the significance of peptidylarginine deiminase 4 and antibodies against citrullinated antigens in the absence of “true ACPAs” in an experimental model of arthritis: comment on the article by Shelef et al et al. Arthritis Rheumatol.2014;66:2642–2644. [DOI] [PubMed] [Google Scholar]
- 70.Shelef MA, Sokolove J, Robinson WH, Huttenlocher A. Insights into the significance of peptidylarginine deiminase 4 and antibodies against citrullinated antigens in the absence of “true ACPAs” in an experimental model of arthritis: comment on the article by Shelef et al Reply. Arthritis & Rheumatology.2014;66:2644–2645. [DOI] [PubMed] [Google Scholar]
- 71.Vossenaar ER, van Boekel MAM, van Venrooij WJ, et al. Absence of citrulline-specific autoantibodies in animal models of autoimmunity. Arthritis and Rheumatism.2004;50:2370–2372. [DOI] [PubMed] [Google Scholar]
- 72.Konig MF, Abusleme L, Reinholdt J, et al. Aggregatibacter actinomycetemcomitans-induced hypercitrullination links periodontal infection to autoimmunity in rheumatoid arthritis. Sci Transl Med.2016;8:369ra176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spengler J, Lugonja B, Jimmy YA, et al. Release of Active Peptidyl Arginine Deiminases by Neutrophils Can Explain Production of Extracellular Citrullinated Autoantigens in Rheumatoid Arthritis Synovial Fluid. Arthritis Rheumatol.2015;67:3135–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Malinin TI, Pekin TJ Jr, Zvaifler NJ. Cytology of synovial fluid in rheumatoid arthritis. Am J Clin Pathol.1967;47:203–208. [DOI] [PubMed] [Google Scholar]
- 75.Smolen JS, Aletaha D, McInnes IB. Rheumatoid arthritis. Lancet.2016;388:2023–2038. [DOI] [PubMed] [Google Scholar]
- 76.Khandpur R, Carmona-Rivera C, Vivekanandan-Giri A, et al. NETs Are a Source of Citrullinated Autoantigens and Stimulate Inflammatory Responses in Rheumatoid Arthritis. Sci Transl Med.2013;5:178ra140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mihalache CC, Yousefi S, Conus S, et al. Inflammation-associated autophagy-related programmed necrotic death of human neutrophils characterized by organelle fusion events. J Immunol.2011;186:6532–6542. [DOI] [PubMed] [Google Scholar]
- 78.Makrygiannakis D, af KE, Lundberg IE, et al. Citrullination is an inflammation-dependent process. Ann Rheum Dis.2006;65:1219–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chapuy-Regaud S, Sebbag M, Baeten D, et al. Fibrin deimination in synovial tissue is not specific for rheumatoid arthritis but commonly occurs during synovitides. J Immunol.2005;174:5057–5064. [DOI] [PubMed] [Google Scholar]
- 80.Pieterse E, Rother N, Yanginlar C, et al. Cleaved N-terminal histone tails distinguish between NADPH oxidase (NOX)-dependent and NOX-independent pathways of neutrophil extracellular trap formation. Ann Rheum Dis.2018;77:1790–1798. [DOI] [PubMed] [Google Scholar]
- 81.Li P, Li M, Lindberg MR, et al. PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J Exp Med.2010;207:1853–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thalin C, Daleskog M, Goransson SP, et al. Validation of an enzyme-linked immunosorbent assay for the quantification of citrullinated histone H3 as a marker for neutrophil extracellular traps in human plasma. Immunol Res.2017;65:706–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kenny EF, Herzig A, Kruger R, et al. Diverse stimuli engage different neutrophil extracellular trap pathways. Elife.2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Konig MF, Andrade F. A critical reappraisal of neutrophil extracellular traps (NETs) and NETosis mimics based on differential requirements for protein citrullination. Front Immunol.2016;7:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carmona-Rivera C, Carlucci PM, Moore E, et al. Synovial fibroblast-neutrophil interactions promote pathogenic adaptive immunity in rheumatoid arthritis. Sci Immunol.2017;2:eaag3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kleinnijenhuis AJ, Hedegaard C, Lundvig D, et al. Identification of multiple post-translational modifications in the porcine brain specific p25alpha. J Neurochem.2008;106:925–933. [DOI] [PubMed] [Google Scholar]
- 87.Ordonez A, Martinez-Martinez I, Corrales FJ, et al. Effect of citrullination on the function and conformation of antithrombin. FEBS J.2009;276:6763–6772. [DOI] [PubMed] [Google Scholar]
- 88.Zhao X, Okeke NL, Sharpe O, et al. Circulating immune complexes contain citrullinated fibrinogen in rheumatoid arthritis. Arthritis Res Ther.2008;10:R94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Keefe D, Shi L, Feske S, et al. Perforin Triggers a Plasma Membrane-Repair Response that Facilitates CTL Induction of Apoptosis. Immunity.2005;23:249–262. [DOI] [PubMed] [Google Scholar]
- 90.Luzio JP, Daw RA, Hallett MB, Richardson PJ, Campbell AK. The rapid increase in intracellular free calcium ion concentration induced by complement and its role in cell damage [proceedings]. Biochem Soc Trans.1979;7:1066–1068. [DOI] [PubMed] [Google Scholar]
- 91.Morgan BP, Luzio JP, Campbell AK. Intracellular Ca2+ and cell injury: a paradoxical role of Ca2+ in complement membrane attack. Cell Calcium.1986;7:399–411. [DOI] [PubMed] [Google Scholar]
- 92.Law RH, Lukoyanova N, Voskoboinik I, et al. The structural basis for membrane binding and pore formation by lymphocyte perforin. Nature.2010;468:447–451. [DOI] [PubMed] [Google Scholar]
- 93.Masson-Bessiere C, Sebbag M, Girbal-Neuhauser E, et al. The major synovial targets of the rheumatoid arthritis-specific antifilaggrin autoantibodies are deiminated forms of the alpha- and beta-chains of fibrin. J Immunol.2001;166:4177–4184. [DOI] [PubMed] [Google Scholar]
- 94.Zhou Y, Chen B, Mittereder N, et al. Spontaneous Secretion of the Citrullination Enzyme PAD2 and Cell Surface Exposure of PAD4 by Neutrophils. Front Immunol.2017;8:1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Damgaard D, Bjorn ME, Steffensen MA, Pruijn GJ, Nielsen CH. Reduced glutathione as a physiological co-activator in the activation of peptidylarginine deiminase. Arthritis Res Ther.2016;18:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Damgaard D, Senolt L, Nielsen MF, Pruijn GJ, Nielsen CH. Demonstration of extracellular peptidylarginine deiminase (PAD) activity in synovial fluid of patients with rheumatoid arthritis using a novel assay for citrullination of fibrinogen. Arthritis Res Ther.2014;16:498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ottaviano FG, Handy DE, Loscalzo J. Redox regulation in the extracellular environment. Circ J.2008;72:1–16. [DOI] [PubMed] [Google Scholar]
- 98.Harris ML, Darrah E, Lam GK, et al. Association of autoimmunity to peptidyl arginine deiminase type 4 with genotype and disease severity in rheumatoid arthritis. Arthritis Rheum.2008;58:1958–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Darrah E, Giles JT, Davis RL, et al. Autoantibodies to Peptidylarginine Deiminase 2 Are Associated With Less Severe Disease in Rheumatoid Arthritis. Front Immunol.2018;9:2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Darrah E, Giles JT, Ols ML, et al. Erosive Rheumatoid Arthritis Is Associated with Antibodies That Activate PAD4 by Increasing Calcium Sensitivity. Sci Transl Med.2013;5:186ra165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Giles JT, Darrah E, Danoff S, et al. Association of cross-reactive antibodies targeting peptidyl-arginine deiminase 3 and 4 with rheumatoid arthritis-associated interstitial lung disease. PLoS One.2014;9:e98794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Navarro-Millan I, Darrah E, Westfall AO, et al. Association of anti-peptidyl arginine deiminase antibodies with radiographic severity of rheumatoid arthritis in African Americans. Arthritis Res Ther.2016;18:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shi J, Darrah E, Sims GP, et al. Affinity maturation shapes the function of agonistic antibodies to peptidylarginine deiminase type 4 in rheumatoid arthritis. Ann Rheum Dis.2018;77:141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Darrah E, Kim A, Zhang X, et al. Proteolysis by Granzyme B Enhances Presentation of Autoantigenic Peptidylarginine Deiminase 4 Epitopes in Rheumatoid Arthritis. J Proteome Res.2017;16:355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Trapani JA, Smyth MJ. Functional significance of the perforin/granzyme cell death pathway. Nat Rev Immunol.2002;2:735–747. [DOI] [PubMed] [Google Scholar]
- 106.Romero V, Andrade F. Non-apoptotic functions of granzymes. Tissue Antigens.2008;71:409–416. [DOI] [PubMed] [Google Scholar]
- 107.Rantapaa Dahlqvist S, Andrade F. Individuals at risk of seropositive rheumatoid arthritis: the evolving story. J Intern Med.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Holers VM, Demoruelle MK, Kuhn KA, et al. Rheumatoid arthritis and the mucosal origins hypothesis: protection turns to destruction. Nat Rev Rheumatol.2018;14:542–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Potempa J, Mydel P, Koziel J. The case for periodontitis in the pathogenesis of rheumatoid arthritis. Nat Rev Rheumatol.2017;13:606–620. [DOI] [PubMed] [Google Scholar]
- 110.Scher JU, Abramson SB. The microbiome and rheumatoid arthritis. Nat Rev Rheumatol.2011;7:569–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sokolove J, Bromberg R, Deane KD, et al. Autoantibody epitope spreading in the pre-clinical phase predicts progression to rheumatoid arthritis. PLoS One.2012;7:e35296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gomez-Banuelos E, Mukherjee A, Darrah E, Andrade F. Rheumatoid Arthritis-Associated Mechanisms of Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans. J Clin Med.2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Schwenzer A, Quirke AM, Marzeda AM, et al. Association of Distinct Fine Specificities of Anti-Citrullinated Peptide Antibodies With Elevated Immune Responses to Prevotella intermedia in a Subgroup of Patients With Rheumatoid Arthritis and Periodontitis. Arthritis & Rheumatology.2017;69:2303–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol.2012;10:717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Goulas T, Mizgalska D, Garcia-Ferrer I, et al. Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase. Sci Rep.2015;5:11969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Montgomery AB, Kopec J, Shrestha L, et al. Crystal structure of Porphyromonas gingivalis peptidylarginine deiminase: implications for autoimmunity in rheumatoid arthritis. Ann Rheum Dis.2016;75:1255–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Tanner AC, Haffer C, Bratthall GT, Visconti RA, Socransky SS. A study of the bacteria associated with advancing periodontitis in man. J Clin Periodontol.1979;6:278–307. [DOI] [PubMed] [Google Scholar]
- 118.Slots J, Reynolds HS, Genco RJ. Actinobacillus actinomycetemcomitans in human periodontal disease: a cross-sectional microbiological investigation. Infect Immun.1980;29:1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zambon JJ, Christersson LA, Slots J. Actinobacillus actinomycetemcomitans in human periodontal disease. Prevalence in patient groups and distribution of biotypes and serotypes within families. J Periodontol.1983;54:707–711. [DOI] [PubMed] [Google Scholar]
- 120.Zambon JJ, Slots J, Genco RJ. Serology of oral Actinobacillus actinomycetemcomitans and serotype distribution in human periodontal disease. Infect Immun.1983;41:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Slots J, Bragd L, Wikstrom M, Dahlen G. The Occurrence of Actinobacillus-Actinomycetemcomitans, Bacteroides-Gingivalis and Bacteroides-Intermedius in Destructive Periodontal-Disease in Adults. Journal of Clinical Periodontology.1986;13:570–577. [DOI] [PubMed] [Google Scholar]
- 122.Johansson A. Aggregatibacter actinomycetemcomitans leukotoxin: a powerful tool with capacity to cause imbalance in the host inflammatory response. Toxins (Basel).2011;3:242–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Claesson R, Hoglund-Aberg C, Haubek D, Johansson A. Age-related prevalence and characteristics of Aggregatibacter actinomycetemcomitans in periodontitis patients living in Sweden. J Oral Microbiol.2017;9:1334504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zambon JJ. Actinobacillus actinomycetemcomitans in human periodontal disease. J Clin Periodontol.1985;12:1–20. [DOI] [PubMed] [Google Scholar]
- 125.Chen C, Wang T, Chen W. Occurrence of Aggregatibacter actinomycetemcomitans serotypes in subgingival plaque from United States subjects. Mol Oral Microbiol.2010;25:207–214. [DOI] [PubMed] [Google Scholar]
- 126.Pahumunto N, Ruangsri P, Wongsuwanlert M, et al. Aggregatibacter actinomycetemcomitans serotypes and DGGE subtypes in Thai adults with chronic periodontitis. Arch Oral Biol.2015;60:1789–1796. [DOI] [PubMed] [Google Scholar]
- 127.Kim TS, Frank P, Eickholz P, Eick S, Kim CK. Serotypes of Aggregatibacter actinomycetemcomitans in patients with different ethnic backgrounds. J Periodontol.2009;80:2020–2027. [DOI] [PubMed] [Google Scholar]
- 128.Minguez M, Pousa X, Herrera D, et al. Characterization and serotype distribution of Aggregatibacter actinomycetemcomitans isolated from a population of periodontitis patients in Spain. Arch Oral Biol.2014;59:1359–1367. [DOI] [PubMed] [Google Scholar]
- 129.Yang HW, Huang YF, Chan Y, Chou MY. Relationship of Actinobacillus actinomycetemcomitans serotypes to periodontal condition: prevalence and proportions in subgingival plaque. Eur J Oral Sci.2005;113:28–33. [DOI] [PubMed] [Google Scholar]
- 130.Arenas Rodrigues VA, de Avila ED, Nakano V, Avila-Campos MJ. Qualitative, quantitative and genotypic evaluation of Aggregatibacter actinomycetemcomitans and Fusobacterium nucleatum isolated from individuals with different periodontal clinical conditions. Anaerobe.2018;52:50–58. [DOI] [PubMed] [Google Scholar]
- 131.Hoglund AC, Haubek D, Kwamin F, Johansson A, Claesson R. Leukotoxic activity of Aggregatibacter actinomycetemcomitans and periodontal attachment loss. PLoS One.2014;9:e104095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Klareskog L, Stolt P, Lundberg K, et al. A new model for an etiology of rheumatoid arthritis: smoking may trigger HLA-DR (shared epitope)-restricted immune reactions to autoantigens modified by citrullination. Arthritis Rheum.2006;54:38–46. [DOI] [PubMed] [Google Scholar]
- 133.Makrygiannakis D, Hermansson M, Ulfgren AK, et al. Smoking increases peptidylarginine deiminase 2 enzyme expression in human lungs and increases citrullination in BAL cells. Ann Rheum Dis.2008;67:1488–1492. [DOI] [PubMed] [Google Scholar]
- 134.Damgaard D, Friberg Bruun Nielsen M, Quisgaard Gaunsbaek M, et al. Smoking is associated with increased levels of extracellular peptidylarginine deiminase 2 (PAD2) in the lungs. Clin Exp Rheumatol.2015;33:405–408. [PubMed] [Google Scholar]
- 135.Reynisdottir G, Karimi R, Joshua V, et al. Structural changes and antibody enrichment in the lungs are early features of anti-citrullinated protein antibody-positive rheumatoid arthritis. Arthritis Rheumatol.2014;66:31–39. [DOI] [PubMed] [Google Scholar]
- 136.Ytterberg AJ, Joshua V, Reynisdottir G, et al. Shared immunological targets in the lungs and joints of patients with rheumatoid arthritis: identification and validation. Ann Rheum Dis.2015;74:1772–1777. [DOI] [PubMed] [Google Scholar]
- 137.Samara KD, Trachalaki A, Tsitoura E, et al. Upregulation of citrullination pathway: From Autoimmune to Idiopathic Lung Fibrosis. Respir Res.2017;18:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bongartz T, Cantaert T, Atkins SR, et al. Citrullination in extra-articular manifestations of rheumatoid arthritis. Rheumatology (Oxford).2007;46:70–75. [DOI] [PubMed] [Google Scholar]
- 139.Lugli EB, Correia RE, Fischer R, et al. Expression of citrulline and homocitrulline residues in the lungs of non-smokers and smokers: implications for autoimmunity in rheumatoid arthritis. Arthritis Res Ther.2015;17:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Willis VC, Demoruelle MK, Derber LA, et al. Sputum autoantibodies in patients with established rheumatoid arthritis and subjects at risk of future clinically apparent disease. Arthritis Rheum.2013;65:2545–2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Demoruelle MK, Harrall KK, Ho L, et al. Anti-Citrullinated Protein Antibodies Are Associated With Neutrophil Extracellular Traps in the Sputum in Relatives of Rheumatoid Arthritis Patients. Arthritis Rheumatol.2017;69:1165–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Demoruelle MK, Bowers E, Lahey LJ, et al. Antibody Responses to Citrullinated and Noncitrullinated Antigens in the Sputum of Subjects With Rheumatoid Arthritis and Subjects at Risk for Development of Rheumatoid Arthritis. Arthritis Rheumatol.2018;70:516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kolarz G, Scherak O, Popp W, et al. Bronchoalveolar lavage in rheumatoid arthritis. Br J Rheumatol.1993;32:556–561. [DOI] [PubMed] [Google Scholar]
- 144.Garcia JG, Parhami N, Killam D, Garcia PL, Keogh BA. Bronchoalveolar lavage fluid evaluation in rheumatoid arthritis. Am Rev Respir Dis.1986;133:450–454. [DOI] [PubMed] [Google Scholar]
- 145.Pridgeon C, Lennon GP, Pazmany L, et al. Natural killer cells in the synovial fluid of rheumatoid arthritis patients exhibit a CD56bright,CD94bright,CD158negative phenotype. Rheumatology (Oxford).2003;42:870–878. [DOI] [PubMed] [Google Scholar]
- 146.Goto M, Zvaifler NJ. Characterization of the natural killer-like lymphocytes in rheumatoid synovial fluid. J Immunol.1985;134:1483–1486. [PubMed] [Google Scholar]
- 147.Young LH, Joag SV, Lin PY, et al. Expression of cytolytic mediators by synovial fluid lymphocytes in rheumatoid arthritis. Am J Pathol.1992;140:1261–1268. [PMC free article] [PubMed] [Google Scholar]
- 148.Dupuy dAA, Monier S, Jorgensen C, et al. Increased percentage of CD3+, CD57+ lymphocytes in patients with rheumatoid arthritis. Correlation with duration of disease. Arthritis Rheum.1993;36:608–612. [DOI] [PubMed] [Google Scholar]
- 149.Warrington KJ, Takemura S, Goronzy JJ, Weyand CM. CD4+,CD28- T Cells in Rheumatoid Arthritis Patients Combine Features of the Innate and Adaptive Immune Systems. Arthritis Rheum.2001;44:13–20. [DOI] [PubMed] [Google Scholar]
- 150.Ren J, Feng Z, Lv Z, Chen X, Li J. Natural killer-22 cells in the synovial fluid of patients with rheumatoid arthritis are an innate source of interleukin 22 and tumor necrosis factor-alpha. J Rheumatol.2011;38:2112–2118. [DOI] [PubMed] [Google Scholar]