Abstract
In recent years, different acquired resistance mechanisms, including transposons, bacteriophages, plasmids, and integrons have been identified as involved in the spread of resistance genes in bacteria. The role of integrons as mobile genetic elements playing a central role in antibiotic resistance has been well studied and documented. Integrons are the ancient structures that mediate the evolution of bacteria by acquiring, storing, disposing, and resorting to the reading frameworks in gene cassettes. The term integron describes a large family of genetic elements, all of which are able to capture gene cassettes. Integrons were classified into three important classes based on integrase intI gene sequence. Integrons can carry and spread the antibiotic resistance genes among bacteria and are among the most significant routes of distribution of resistance genes via horizontal transfer. All integrons have three essential core features. The first feature is intI, the second one is an integron-associated recombination site, attI, and an integron-associated promoter, Pc, is the last feature. Among them, the class 1 integron is a major player in the dissemination of antibiotic resistance genes across pathogens and commensals. Various classes of integrons possessing a wide variety of gene cassettes are distributed in bacteria throughout the world. This review thus focuses on the distribution of integrons among important bacteria.
Key Words: Antibiotic resistance, Gene cassettes, Integrases, Integrons, Mobile elements
Introduction
Integrons are the ancient structures that mediate the evolution of bacteria by acquiring, storing, disposing, and resorting to the reading frameworks in mobile elements called cassettes. They are present in approximately 17% of the bacterial chromosomes (1). These structures are found in different environments such as forest, desert soils, river sediments, antarctic soils, hot springs, biofilms, plant surfaces, marine sediments, and deep-sea sediments. Nowadays, the term integron describes a large family of genetic elements, all of which are able to capture gene cassettes. Although about one-third of integrons have been found in bacterial genome that do not carry gene cassettes (empty integrons) (2). Although the first antibiotic-resistant bacteria were reported in the mid-1950s, until the 1970s it was not clear that resistance phenotypes were associated with transmissible plasmids or elements. In the late 1980s, integrons were identified. Integrons play an important role in the distribution of antibiotic resistance, especially in Gram-negative pathogens. In resistance integrons, an action plan is associated with genetic moving elements such as transposons or plasmids, so interspecies and intraspecies transmission are increased. It has now been well established that integrons act as the main reason for multiple resistance in Gram negative more than in Gram-positive bacteria (3).
Classification of integrons
Integrons can be discriminated based on the relative homology of intI, although the cutoff point is not clear (2). Initially, it was suggested that integrons can be divided into two categories: 1) mobile integrons: have a small number of cassettes, usually encode antibiotic resistance, have different attC sites and their mobility is dependent on transposons or plasmids, 2) super integrons: have many cassettes, homogenous attC sites and located on the chromosome (4). Also, phylogenetic studies showed that a branch of integrons is associated with organismal phylogeny. These integrons are divided into three groups based on the phylogeny of the integrase genes: (i) a group of isolated Proteobacteria from freshwater and soil, including clinical integrons in classes I and III. (ii) a group found in Gamma-Proteobacteria in marine environments that includes class II, SXT integrative conjugative elements integrons and pRSV1 plasmids in Vibrio; and (iii) the integrons whose integrase genes are inverted. These inverse integrons have been found in Spirochaetes, Planctomycetes, Cyanobacteria, and Chlorobi spp. (4). Up to now, more than 9 classes of integrons have been identified based on 16 amino acids conserved in Gram-negative bacteria (5), but only 4 main classes are associated with clinical isolates. As stated above, the amino acid sequencing of the integrase gene is used as a marker for the classification of integrons into different classes, and members having the same integrase have the same class but can carry different gene cassettes. Of all the different classes of integrons, the class I integrons and then class II are the most common classes among clinical isolates (6). Classes I to III are also integrons with multiple resistance. The class IV integrons are considered a distinct type of integron called super integron, which is found on the small chromosome Vibrio cholera (7).
Integrons structure
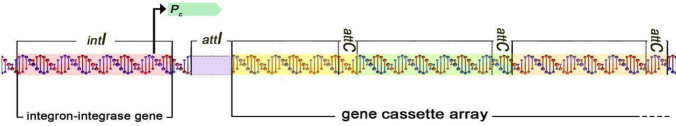
Structurally, all integrons consist of three main components, including 5′ and 3’ conserved segment and a central variable region between the 5’ and 3’ zone, in which integrons are responsible for the capture and expression of exogenous genes, which are part of the gene cassette (1). Essential components of the 5’ and 3’ zone in all integrons include 1) integrase gene (intI) encoding a specific recombinant site and a part of the family of tyrosine recombinase (8), 2) the attI receptor site identified by integrase and adjacent to the intI gene, so that attI is in the upstream of intI, except for the Triponama spp. (9). Also, integrase protein can catalyse a recombinant between the input gene cassette and the attI. 3) the promoter sequence consists of Pc and Pint which is placed inside intI or between intI and attI and causes expression of the existing genes in the integrated gene cassette in integrons and integrase (9). The 3’ conserved segment of integrons has different structures that differ in integron classes. The gene cassettes are located between the 3’ and 5’ zone, where integrons receive new genes through these cassettes. The integron system has two important benefits as a genetic innovation: the new genetic material is integrated into the bacteria genome at attI, therefore, they do not cause abnormalities in the existing genes; second, the new integrated genes are expressed by the integrons promoter (Pc) (Figure 1) (10).
Figure1.
Integron structure intI, a gene for the integron integrase; attI, recombination site; Pc, an integron promoter. Gene cassettes, sequentially inserted into an array via recombination between attI and the cassette associated recombination sites (attC)
Gene cassette
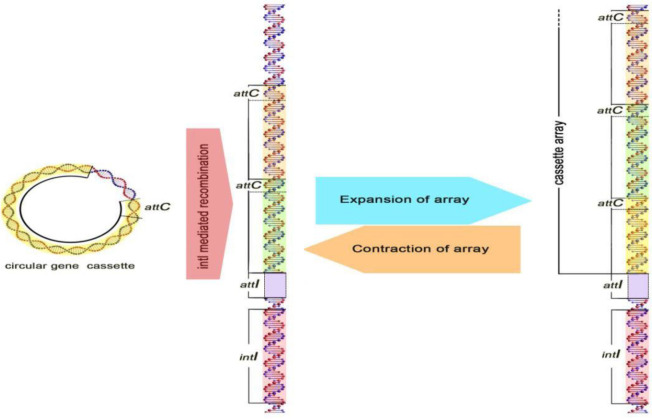
The number of cassettes reported differ from zero to 100 in different studies, this difference in the number of cassettes is seen in Vibrio cholerae and some other species of Vibrio. A great variety is created in integron classes through the acquisition of various gene cassettes (11). Many studies have been done on the origin of the gene cassettes, but so far with no result. However, the size and frequency of gene cassettes suggest that some organisms, using single or pair genes, can create a gene cassette, the mechanism of which method has not yet been identified. These cassettes are mobile elements that code multiple genes, including antibiotic resistance genes. These antibiotic classes include all known β-lactams, all aminoglycosides, chloramphenicol, trimethoprim, streptothricin, erythromycin, quinolones, rifampin, lincomycin, fosfomycin, and antiseptics of the quaternary ammonium-compound family (1, 3). According to previous studies, gene cassettes are randomly combined in the region between the 3’ and 5’ conserved segment of integrons (12). The integration of gene cassettes is done with the integrase gene between two recombination sites (attI, attC) which process is reversible and cassettes can be released in the form of free DNA from integrons (Figure 2) (11). In general, gene cassettes are compact and dense DNA that have a simple structure, consisting of two components of the recombination site and the open reading frame (ORF) (13). Gene cassettes lack the promoter sequences in their structure, thus expressing the gene cassettes associated with an external promoter (Pc) in the structure of integrons. Because integrons have promoter sequences, they can express the genes in the gene cassettes, so integrons act as both the expression vector of the gene and as a natural cloning system (12). There are areas at the end and beginning of a gene cassette that consist of a protected sequence including GTTRRRY (representing recombination activity). Although attC has distinct sizes between 57-141 bp, they have a set of common characteristics. These elements have a core site (CS) with the sequence GTTRRRY and an inverse core site (ICS) with the sequence RYYYAAC. The attC sequence was formerly known as 59bp element (14). All 59bp elements have a symmetrical central axis in their structure, on both sides of this axis of symmetry, there are ICS and CS sites, which has led to the creation of hairpins in the 59bp element. The attC sequences are categorized into different groups, based on the size of the first and largest group of 12 members and dependent on aacA(Iia), aadA, aadB, CatB3, and orfD genes. The factors present in these genes are only different in some base pairs (15).
Figure 2.
Acquisition of gene cassettes. Integrons acquire new gene cassettes by recombination between attC of a circular cassette and the attI site of integrons
Mobility
The mobility of integrons has been considered to be a major concern in clinical pathogens of spreading antibiotic resistance, and this mobility is related to mobile DNA elements (transposon and plasmid) (16). However, integrons do not have the ability to move, but integrons (mostly Class I) are known as mobile genetic elements that are most commonly found on transferable plasmids. Therefore, these moving plasmids carry gene cassettes that can transfer to other integrons or even to the bacterial genome. The integron system allows microorganisms to combine gene cassettes and convert them into functional proteins by expressing the genes correctly (2). Mobile genetic elements containing plasmids, transposons, secretion sequences, and genetic islands can act as extensive reservoirs of information for integrons, which are shared among bacteria. With the motion of the gene cassettes, the integrons play an important role in the distribution and spread of resistance genes. In addition to clinical aspects, there are many reports of the presence of integrons in environmental microorganisms, which indicate their high diversity in various functions. This explains exactly that integrons are old genetic elements within the genome and play an important role in evolution and adaptation (13).
Class I integron
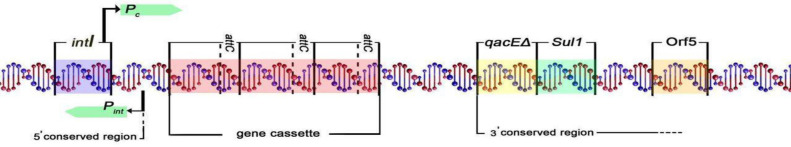
In 1998, class I integron was found for the first time in Corynebacterium glutamicum, which is a Gram-positive microorganism (7), first discovered by Hall and Stoke in 1989 (17), and then it was observed in Corynebacterium, Streptococcus, Enterococcus, Staphylococcus, Aerococcus and Brevibacterium. Class I integron has the highest frequency among the integrons (11, 18). IntI is able to identify three types of recombinant sites (attI, attC, and secondary sites). Therefore, these types of integrons are able to receive gene cassettes through the recombination of dedicated sites. Also, class I integron has a direct relationship with Tn402 and the Tn3 family of transposons (1, 18). In this class of integrons, the gene cassettes can be further expressed through the promoters Pc andP2 (second promoter that is usually inactive). Pc plays an important role in integron function because it ensures the correct expression of the gene cassettes. Class I integron has been studied in different microorganisms. The prevalence of class I integron is about 22 to 55%, among the Gram-negative bacteria isolated from the clinic, which include Acinetobacter, Aeromonas, Alcaligenes, Burkholderia, Campylobacter, Citrobacter, Enterobacter, Escherichia, Klebsiella, Mycobacterium, Providencia, Pseudomonas, Salmonella, Serratia, Shigella, Stenotrophomonas, and Vibrio (19), and it is also found in about 22 to 59% of Campylobacter jejuni, Providencia stuartii, Serratia marcescens, Stenotrophomonas maltophilia (7). Class I integron acts as a common factor in the distribution and spread of antimicrobial resistance. This class carries over 40 resistance genes related to aminoglycosides, beta-lactams, chloramphenicol, macrolides, sulfonamides, disinfectants, and dexophthane (20). The 3′-conserved segment in class I integron consists of the following components: 1) qacEΔ1 gene encoding resistance to quaternary ammonium salts and dexophthane, 2) sul1 gene encoding resistance to sulfonamides (14), 3) orf5 has no known function but is similar to puromycin acetyltransferase in Streptomyces albonier, this suggests that it leads to resistance to puromycin through the mechanism of acetyltransferase (Figure 3) (21).
Figure 3.
Structure of class I integron. The following cassettes are a part of the 3′conserved region and not mobile: sul1 gene encoding resistant to sulfonamides, qacEΔ encoding resistant to quaternary ammonium compounds, and orf5 unknown function
Class II integron
Class II integrons indicate a high prevalence in clinical isolates in Gram-negative bacteria such as Acinetobacter, Shigella, Salmonella, Pseudomonas (7). Class II integrons, similar to class I, are also associated with the Tn7 family of transposons (Tn7 and its derivatives such as Tn1825, Tn1826, and Tn4132) which carry the recombinant site attI2 and Pc. The 3′-conserved segment includes 5 tns (tnsA, tnsB, tnsC, tnsD, and tnsD) genes that play a role in the transposon movement. Class II integrons contain gene cassettes including dfrA1 (dihydrofolate reductase), sat1 (streptothricin-acetyl transferase), and aadA1 (aminoglycoside adenyltransferase) which are resistant to trimethoprim, streptomycin, and streptomycin/spectinomycin, respectively. The ereA gene (erythromycin esterase) has also been found in the class II integron (7, 16). The integrase gene in class II integrons is about 46% similar to the integrase gene in class I integrons. One of the most important differences between intI1 and intI2 is that the integrase gene in the class II integrons (intI2) is stopped early by the end codons (TAA), resulting in the 178 amino acid protein synthesis being deactivated. Therefore, class II integrons are weaker in moving gene cassettes than class I integrons. However, mutations in end codons lead to reactivity of these amino acids, which results in the activation of the integrase gene (3).
Class III integron
In 1993, class III integrons were first identified in Japan by Arakawa and colleagues in S. marcescens. These types of integrons rarely present in clinical specimens, they have been found in a small number of bacteria such as Acinetobacter spp., Alcaligenes, Citrobacter freundii, Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, Pseudomonas putida, Salmonella spp. and S. marcescens (7). The 3′-conserved segment in these integrons is similar to that of class I integrons: it contains four genes, qacEΔ1, sul1, orf5, orf6, and their only difference is the lack of transcription genes. In these integrons, there are several gene cassettes including blaIMP-1 and aacA4 that encode metallo-lactamase enzymes and the aminoglycoside resistance gene (tobramycin), respectively. The aacA4 cassette was originally found in class I integrons but the blaIMP-1 cassette was first recognized in class III integrons, however due to the extensive recombination that occurred between class I and III integrons, this gene is also found in class I integrons and has been widely reported worldwide (22).
Class IV integron
In fact, class IV integrons are super integrons that were first detected in Vibriocholera. Mazel in 1998, for the first time, called them super integron (19). These integrons are seen in microorganisms including Vibrionaceae, Shewanella, Xanthomonas, Pseudomonas, and other proteobacteria. To date, class IV integrons have been found to carry gene cassettes imparting resistance to the antibiotic’s chloramphenicol and fosfomycin (23).
Antibiotic resistance
In recent years, excessive consumption of antibiotics has led to increased antibiotic resistance. Genetic mutations and acquisition of resistance genes are involved in the development of resistance. One of the most important causes of the development and spread of antibiotic resistance of genes is the acquisition of resistance genes that occur through the horizontal transduction of the gene or genetic moving elements such as plasmids and transposons (24). These integrons share a collection of genetic cassettes most of which encode antibiotic resistance. In general, about 130 resistant genetic cassettes have been identified with varying patterns of codon and attC sites (3). Resistance integrals have several common features: they usually have motion, their cassette arrangements are short, and typically only carry antibiotic resistance genes. However, these common features in them are not inherent characteristics of the ancestors of integrons but they are created by strong selective pressure during the use of antibiotics by humans.
Integrons in various bacteria
Escherichia coli
E. coli is known as the head of the large family of Enterobacteriaceae. Based on diversity in pathogenesis and clinical symptoms, the strains of E. coli are divided into two intestinal and extra-intestinal. E. coli is one of the most important causes of gastrointestinal and urinary tract infections in humans that has been resistant to a wide range of antibiotics over the past years (25). Acquiring mobile elements, including plasmids, transposons, and integrons among Gram-negative bacteria, it plays an important role in the development of antibiotic resistance (2). Class I integron was reported in 1973 (26). Common cassettes in this class of integron result in resistance to aminoglycosides (aadA1, aadA2, aadB, aadA5), trimethoprim (dfrA1, dfrA5, dfrA7, dfrA12, dfrA17, dfrB2, dfrA1-gcuC, dfrA17-aadA5, dfrA1-aadA1, dfrA12-gcuF-aadA2), erythromycin (ere2) and broad-spectrum beta-lactams (ESBL) (blaOXA-101-aac(6’)–Ib) (27). Also, class II integron has been reported less frequently. This integron contains gene cassettes dfrA1-sat1-aadA1, dfrA1-sat2-aadA1 and estX-sat2-aadA1 (28). Also, recently, a new class II integron has been identified (29), which includes two gene cassettes. One of them is the dfrA14 gene. There are also reports available on the availability of Class III integrons that are commonly seen with class I and III integrons in this bacterium (30). (Table 1) in our study in 2015, we collected E. coli from clinical isolates of patients in the north of Iran, which showed 22% of E. coli isolates carried class I integron (Table 2) (31).
Table 1.
Common gene cassette arrays in types of integrons in diverse bacterial species and their role in antimicrobial resistance
| References | Antibiotics associated with gene cassettes | gene cassettes | Integrons | Bacteria |
|---|---|---|---|---|
| (28, 30) | Aminoglycosides, Trimethoprim, Extended Spectrum Beta-Lactamase, Erythromycin. | aadA1, aadA2, aadA5 aadB, dfrA1, dfrA5, dfrA7 dfrA12, dfr14, dfrA17, dfrB2, dfrA1-gcuC, dfrA1-aadA1, dfr17-aadA5, dfr12-gcuF-aadA2, dfrA1-sat1-aadA1, dfrA1-sat2-aadA1, estX-sat2-aadA1, blaOXA-101-aac (6')–Ib, ere2. |
I, II, III | Escherichia coli |
| (27, 32, 33) | Extended Spectrum Beta-Lactamase, Aminoglycosides, Trimethoprim. | blaCARB-2, aadA1, aadA2, aadB, dfrA1, dfrA7, dfrA1-gcuF, dfrA1- aadA1, dfr17-aadA5, dfr12-gcuF-aadA2, sat1. | I, II | Acinetobacterbaumannii |
| (27, 36) | Aminoglycosides, Trimethoprim, Extended Spectrum Beta-Lactamase. | aadA, aadA1a, aadA2, aadA5, aadB, dfrA1, dfrA7, dfrA12, dfrA17, dfrA1-gcuF, dfrA1-aadA1a, dfr17-aadA5, dfr12-gcuF-aadA2, blaCARB-2. | I, II | Salmonella spp. |
| (4, 27) | Extended Spectrum Beta-Lactamase, Trimethoprim, Aminoglycosides, | blaCARB-2 blaGES-1, aadA, aadA1, aadB, dfrA1, dfrA7, dfrA1-gcuF, dfrA1-aadA1a, dfr17-aadA5, dfr12-gcuF-aadA2. | I, II, III | Klebsiella spp. |
| (16) | Aminoglycosides, Trimethoprim | aadA2, aadB, dfr17-aadA5, dfr12-gcuF-aadA2. | I | Pseudomonas aeruginosa |
| (46) | Aminoglycosides, Trimethoprim, Chloramphenicol | aadA1, aadA2, dfr17-aadA5, dfr12-gcuF-aadA2, aacA4-cmlA1 | I | Staphylococcus aureus |
| (43) | Aminoglycosides, Trimethoprim | aadA1a, dfr12-gcuF-aadA2, dfrA1-sat1-aadA1. | I | Enterococcus faecalis |
| (44) | Aminoglycosides, Trimethoprim | aadA1a, aadA2, dfrA7, dfrA1-aadA1a, dfr17-aadA5, dfr12-gcuF-aadA | I | Enterobacter spp. |
Table 2.
Prevalence of different types of integron and their gene cassettes in our studies between 2011-2017
| Bacteria | Class I integron | Class II integron | Class III integron | Frequency of gene cassettes | References |
|---|---|---|---|---|---|
| Escherichia coli | 22% | --- | --- | --- | (31) |
| Acinetobacterbaumannii | 25.7% | 88.6% | 28.6% | aadA1(5.7), blaOXA30(40%), aadB(94.3%), dfrA1(77.1%), | (33, 34) |
| Salmonella infantis | 36% | --- | --- | --- | (37) |
| Klebsiellapneumoniae | 36.6% | --- | --- | BlaVIM-1(30%) | (39, 40) |
| Pseudomonas aeruginosa | 37–39.5% | --- | --- | --- | (41, 42) |
Acinetobacterbaumannii
A. baumannii is recognized as an important cause of nosocomial infections that has antibiotics resistant genes including efflux pumps, class B β-lactamase (Metallo-beta-lactamase (MBL)), class C chromosomal β-lactamase (Amp C), class D β-lactamase (OXA-type carbapenemase), integrons, and associated insertion sequence (IS) elements (32). Class I integron in this bacterium has several gene cassettes (aadA2, aadB, dfrA7, blaCARB-2, dfrA1-gauC, dfrA17-aadA5, dfrA1-aadA1, dfrA12-gcuF-aadA2) that caused resistance to aminoglycosides, ESBLs, and trimethoprim (27). Although class II integrons including dfrA1, aadA1, sat1, and aadB which are resistanct to trimethoprim, streptomycin, tobramycin, and kanamycin, respectively (Table 1)(33). In other studies, in 2017, we collected A. baumannii isolates from BAL samples of patients admitted to the ICU at Ayatollah Rouhani Hospital in Babol, Iran. The distribution analysis of intI genes showed that 25.7%, 88.6%, and 28.6% of isolates carried the intI1, intI2, and intI3genes, respectively. Also, the prevalence of aadB, dfrA1, blaOXA30, aadA1 and blaPSE1 gene cassettes were 94.3%, 77.15, 40%,5.7%, and 0%, respectively (33, 34).
Salmonella spp.
Salmonella spp. are intestinal pathogens that are usually transmitted by contaminated food, especially animals such as meat, poultry, eggs, and milk. Multidrug-resistant (MDR) salmonella has been a major public health issue since 1990. Salmonella spp. is associated with different classes of integrons, usually containing one to three gene cassettes (35). Class I integrons play a major role in antibiotic resistance in Salmonella spp. This class has several types of gene cassettes resistant to aminoglycosides (aadA, aadA1, aadA2, aadA5, aadB), beta-lactams (blaCARB-2) and trimethoprim (dfrA1, dfrA7, dfrA12, dfrA17, dfrA1-gcuC, dfrA1-aadA1, dfrA17-aadA5, dfrA12-gcuF- aadA2) (27). Although the percentage of class II integrons is relatively lower in this bacterium, it appears in different serotypes of salmonella spp. with dfrA1-sat1-aadA1 and estX-sat2-aadA1 cassettes. Class III integrons are not yet found in this bacterium (Table 1)(36). In our study in 2015, the prevalence of Class I integrons in Salmonella infantis was 36% (Table 2) (37).
Klebsiella spp.
Klebsiella spp. causes diseases such as pneumonia, meningitis, and blood and urinary tract infections. This bacterium is resistant to a wide range of antibiotics (aminoglycosides, cephalosporins, and ESBLs). Class I integrons in Klebsiella spp. has different gene cassettes, including aadA1a, aadA2, dfrA7, aadB, dfrA1-gcuC, dfrA1-aadA1, dfrA17-aadA5, dfrA12-gcuF-aadA2, aadA, and blaCARB-2. However, multiple resistance to these bacteria has been determined due to dfrA12-orfF-aadA2, and dfrA1-orfC(27). Antibiotic resistance in Klebsiella spp. Is known more to ESBLs such as TEM, SHV, CTX-M. Class I integrons in Klebsiella spp. also has genes associated with MBLs like Verona integrin (VIM) and IMP-type carbapenemases (IMP). These MBLs hydrolyze β-lactams including carbapenems, which are commonly enclosed in class I integrons. The class II integron also has dfrA1, sat1 / sat2, and aadA1 cassettes, and class III integron includes blaGES-1 (Table 1) (38). Also, we studied K. pneumonia isolates in 2011 and 2015 . Our results showed that 36.6% of isolates carried the intI1 gene and the prevalence of blaVIM-1 gene cassette was 30% (Table 2) (39, 40).
Pseudomonas aeruginosa
P. aeruginosa is one of the most important factors in hospital infections, especially in burn patients and patients with cystic fibrosis. Due to increased MDR bacteria, treatment for this bacterium is difficult to find. Class I integron is an important factor for the development of antibiotic resistance and the emergence of MDR strains (41). This class of integrons includes some gene cassettes such a aadB and aadA2 (resistance to aminoglycosides), dfrA17-aadA5 and dfrA12-gcuF-aadA2 (resistance to trimethoprim), blaCARB-2 (resistance to carbenicillin) (27). Class II integrons in P. aeruginosa include dfrA1-sat1-aadA1 (Table 1)(16). Our studies in 2012 and 2013 on P. aeruginosa isolates showed 37–40% of isolates had the intI1 gene (Table 2) (41, 42).
Enterococcus faecalis
E. faecalis is an intestinal flora and has been identified as one of the major causes of hospital infections, with a tendency to increase antibiotic resistant class I integrons; E. faecalis contains gene cassettes dfrA12-gcuF-aadA2 (resistance to trimethoprim) and aadA1a (resistance to aminoglycosides)(43). Class II integrons has the dfrA1-sat1-aadA1 cassettes (Table 1) (44).
Enterobacter spp.
Enterobacter spp. are Gram-negative bacteria that cause gastrointestinal diseases, at present, Enterobacter spp. with MDR patterns that contain class I integron are considered one of the major concerns of physicians and infection control practitioners. This class causes resistance to aminoglycosides (dfrA7, aadA1a, and aadA2) and trimethoprim (dfrA1-aadA1, dfrA17-aadA5, dfrA12-gcuF-aadA2, and dfr12-orfF-aadA2)(Table 1)(27).
Staphylococcus aureus
S. aureus is a Gram-positive microorganism which is now considered one of the most important causes of hospital infections (45). Quantitative studies have been done on integrons in Gram-positive bacteria. Class I integrons is more common in S. aureus and causes resistance to aminoglycosides (aadA1a and aadA2), trimethoprim (dfrA17-aadA5 and dfrA12-gcuF-aadA2) and chloramphenicol (aacA4-cmlA1)( Table 1) (46).
Conclusion
Integrons being capable of integrating, expressing, and disseminating gene cassettes, carrying resistance determinants, play a critical role in facilitating the MDR phenotype in these bacteria. The ability of integrons to acquire new cassettes and their ability to recombine cassette rows emphasizes the adaptation of their diversity in bacteria. Their ability to rapidly spread resistance phenotypes makes it important to consider what other integron-mediated traits, such as increased pathogenicity, virulence, or resistance to antimicrobials might impact human health in the future. If we can control integrons and cassette formation, we could use integrons as a platform for enzyme discovery and to construct novel biochemical pathways in antimicrobial resistance. So, knowledge about the prevalence of integrons and gene cassettes is helpful for the treatment and correct use of antibiotics.
Acknowledgment
The authors thank the staff of Babol University of Medical Sciences, Babol, Iran. This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Coflicts of Interest
The authors declare that there are no conflicts of interest.
References
- 1.Osagie AE, Olalekan SH. Multiple Drug resistance: a fast-growing threat. Bio Med. 2019;21:2574–1241. [Google Scholar]
- 2.Sütterlin S, Bray JE, Miaden MCJ, Tano E. Distribution of class 1 integrons in historic and contemporary collections of human pathogenic Escherichia coli. PLoS One. 2020;15:e0233315. doi: 10.1371/journal.pone.0233315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Partridge SR, Tsafnat G, Coiera E, Iredell JR. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev. 2009;33:757–784. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 4.Wu Y-W, Doak TG, Ye Y. The gain and loss of chromosomal integron systems in the treponema species. BMC Evol Biol. 2013;13:16–24. doi: 10.1186/1471-2148-13-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ranjbar R, Taghipour F, Afshar D, Farshad S. Distribution of class 1 and 2 integrons among Salmonella enterica serovars isolated from iranian patients. Open Microbiol J. 2019;13:63–66. [Google Scholar]
- 6.Koratzanis E, Souli M, Galani I, Chryssouli Z, Armaganidis A, Giamarellou H. Epidemiology and molecular characterisation of metallo-β-lactamase-producing Enterobacteriaceae in a university hospital Intensive Care Unit in Greece. Int J Antimicrob agents. 2011;38:390–397. doi: 10.1016/j.ijantimicag.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 7.Yu G, Li Y, Liu X, Zhao X, Li Y. Role of integrons in antimicrobial resistance: A review. Afr J Microbiol Res. 2013;7:1301–1310. [Google Scholar]
- 8.Hall RM. Integrons and gene cassettes: hotspots of diversity in bacterial genomes. Ann N Y Acad Sci. 2012;1267:71–78. doi: 10.1111/j.1749-6632.2012.06588.x. [DOI] [PubMed] [Google Scholar]
- 9.Escudero JA, Loot Cl, Nivina A, Mazel D. The integron: adaptation on demand. Microbiol Spectr. 2015:139–161. doi: 10.1128/microbiolspec.MDNA3-0019-2014. [DOI] [PubMed] [Google Scholar]
- 10.Gillings MR. Integrons: past, present, and future. Microbiol Mol Biol Rev. 2014;78:257–277. doi: 10.1128/MMBR.00056-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu Z, Li L, Shi L, Shirtliff ME. Class 1 integron in staphylococci. Mol biol Rep. 2011;38:5261–5279. doi: 10.1007/s11033-011-0676-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ke X, Gu B, Pan S, Tong M. Epidemiology and molecular mechanism of integron-mediated antibiotic resistance in Shigella. Arch Microbiol. 2011;193:767–774. doi: 10.1007/s00203-011-0744-3. [DOI] [PubMed] [Google Scholar]
- 13.Tansirichaiya S, Mullany P, Roberts AP. Promoter activity of ORF-less gene cassettes isolated from the oral metagenome. Sci Rep. 2019;9:8388. doi: 10.1038/s41598-019-44640-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Recchia GD, Hall RM. Origins of the mobile gene cassettes found in integrons. Trends Microbiol. 1997;5:389–394. doi: 10.1016/S0966-842X(97)01123-2. [DOI] [PubMed] [Google Scholar]
- 15.Larouche A, Roy PH. Effect of attC structure on cassette excision by integron integrases. Mob DNA. 2011;2:3–15. doi: 10.1186/1759-8753-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghaly TM, Geoghegan JL, Tetu SG, Gillings MR. The peril and promise of integrons: beyond antibiotic resistance. Trends Microbiol. 2019;28:455–464. doi: 10.1016/j.tim.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 17.Stokes Ht, Hall RM. A novel family of potentially mobile DNA elements encoding site-specific gene-integration functions: integrons. Mol Microbiol. 1989;3:1669–1683. doi: 10.1111/j.1365-2958.1989.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 18.Labbate M, Case RJ, Stokes HW. The integron/gene cassette system: an active player in bacterial adaptation. Methods Mol Biol. 2009:103–125. doi: 10.1007/978-1-60327-853-9_6. [DOI] [PubMed] [Google Scholar]
- 19.Deng Y, Bao X, Ji L, Chen L, Liu J, Miao J, et al. Resistance integrons: class 1, 2 and 3 integrons. Ann Clin Microbiol Antimicrob. 2015;14:45–55. doi: 10.1186/s12941-015-0100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stalder T, Barraud O, Casellas M, Dagot C, Ploy M-C. Integron involvement in environmental spread of antibiotic resistance. Front Microbiol. 2012;3:119–132. doi: 10.3389/fmicb.2012.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bissonnette L, Roy PH. Characterization of In0 of Pseudomonas aeruginosa plasmid pVS1, an ancestor of integrons of multiresistance plasmids and transposons of Gram-negative bacteria. J Bacteriol. 1992;174:1248–1257. doi: 10.1128/jb.174.4.1248-1257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendes RE, Castanheira M, Toleman MA, Sader HS, Jones RN, Walsh TR. Characterization of an integron carrying blaIMP-1 and a new aminoglycoside resistance gene, aac (6′)-31, and its dissemination among genetically unrelated clinical isolates in a Brazilian hospital. Antimicrob Agents Chemother. 2007;51:2611–2614. doi: 10.1128/AAC.00838-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fluit A, Schmitz F-J. Resistance integrons and super-integrons. Clin Microbiol Infect. 2004;10:272–288. doi: 10.1111/j.1198-743X.2004.00858.x. [DOI] [PubMed] [Google Scholar]
- 24.Xu Z, Li L, Shirtliff M, Peters B, Li B, Peng Y, et al. Resistance class 1 integron in clinical methicillin-resistant Staphylococcus aureus strains in southern China, 2001–2006. Clin Microbiol Infect. 2011;17:714–718. doi: 10.1111/j.1469-0691.2010.03379.x. [DOI] [PubMed] [Google Scholar]
- 25.Ferdosi-Shahandashti E, Javanian M, Moradian-Kouchaksaraei M, Yeganeh B, Bijani A, Motevaseli E, et al. Resistance patterns of Escherichia coli causing urinary tract infection. Caspian J Intern Med. 2015;6:148–151. [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Freijo P, Fluit A, Schmitz F, Grek V, Verhoef J, Jones M. Class I integrons in Gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J Antimicrob Chemother. 1998;42:689–696. doi: 10.1093/jac/42.6.689. [DOI] [PubMed] [Google Scholar]
- 27.Domingues S, da Silva GJ, Nielsen KM. Global dissemination patterns of common gene cassette arrays in class 1 integrons. Microbiology (Reading) 2015;161:1313–1337. doi: 10.1099/mic.0.000099. [DOI] [PubMed] [Google Scholar]
- 28.Kadlec K, Schwarz S. Analysis and distribution of class 1 and class 2 integrons and associated gene cassettes among Escherichia coli isolates from swine, horses, cats and dogs collected in the BfT-GermVet monitoring study. J Antimicrob Chemother. 2008;62:469–473. doi: 10.1093/jac/dkn233. [DOI] [PubMed] [Google Scholar]
- 29.Márquez C, Labbate M, Ingold AJ, Chowdhury PR, Ramírez MS, Centrón D, et al. Recovery of a functional class 2 integron from an Escherichia coli strain mediating a urinary tract infection. Antimicrob Agents Chemother. 2008;52:4153–4154. doi: 10.1128/AAC.00710-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kargar M, Mohammadalipour Z, Doosti A, Lorzadeh S, Japoni-Nejad A. High prevalence of class 1 to 3 integrons among multidrug-resistant diarrheagenic Escherichia coli in Southwest of Iran. Osong Public Health Res Perspect. 2014;5:193–198. doi: 10.1016/j.phrp.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahangarkani F, Rajabnia R, Shahandashti EF, Bagheri M, Ramez M. Frequency of class 1 integron in Escherichia coli strains isolated from patients with urinary tract infections in North of Iran. Mater Sociomed. 2015;27:10–12. doi: 10.5455/msm.2014.27.10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu C-C, Tang CY, Chang K-C, Kuo H-Y, Liou M-L. A comparative study of class 1 integrons in Acinetobacter baumannii. Gene. 2014;544:75–82. doi: 10.1016/j.gene.2014.04.047. [DOI] [PubMed] [Google Scholar]
- 33.Akrami F, Shahandashti EF, Yahyapour Y, Sadeghi M, Khafri S, Pournajaf A, et al. Integron types, genecassettes and antimicrobial resistance profile of Acinetobacter baumannii isolated from BAL samples in Babol, North of Iran. Microb Pathog. 2017;109:35–38. doi: 10.1016/j.micpath.2017.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Deylam Salehi M, Ferdosi-Shahandashti E, Yahyapour Y, Khafri S, Pournajaf A, Rajabnia R. Integron-mediated antibiotic resistance in Acinetobacter baumannii isolated from intensive care unit patients, Babol, North of Iran. Bio Med Res Int. 2017;2:1–8. doi: 10.1155/2017/7157923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gal-Mor O, Boyle EC, Grassl GA. Same species, different diseases: how and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front Microbiol. 2014;5:391–400. doi: 10.3389/fmicb.2014.00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antunes P, Machado J, Peixe L. Characterization of antimicrobial resistance and class 1 and 2 integrons in Salmonella enterica isolates from different sources in Portugal. J Antimicrob Chemothe. 2006;58:297–304. doi: 10.1093/jac/dkl242. [DOI] [PubMed] [Google Scholar]
- 37.Asgharpour F, Rajabnia R, Shahandashti EF, Marashi MA, Khalilian M, Moulana Z. Investigation of class I integron in Salmonella infantis and its association with drug resistance. Jundishapur J Microbiol. 2014:7. doi: 10.5812/jjm.10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu H, Wang M, Liu Y, Wang X, Wang Y, Lu J, et al. Characterization of antimicrobial resistance in Klebsiella species isolated from chicken broilers. Int J Food Microbiol. 2016;232:95–102. doi: 10.1016/j.ijfoodmicro.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 39.Molana Z, FERDOSI SE, Gharavi S, Shafii M, Norkhomami S, Ahangarkani F, et al. Molecular investigation of class i integron in Klebsiella pneumoniae isolated from intensive care unit (shahid beheshti hospital of babol 2010) J Babol Univ Med Sci. 2011;13:7–13. [Google Scholar]
- 40.Rajabnia R, Asgharpour F, Shahandashti EF, Moulana Z. Nosocomial emerging of (VIM1) carbapenemase-producing isolates of Klebsiella pneumoniae in North of Iran. Iran J Microbiol. 2015;7:88–93. [PMC free article] [PubMed] [Google Scholar]
- 41.Shahandashti EF, Molana Z, AsgharpourF , Mojtahedi A, Rajabnia R. Molecular detection of Integron genes and pattern of antibiotic resistance in Pseudomonas aeruginosa strains isolated from intensive care unit, Shahid Beheshti Hospital, North of Iran. Int J Mol Cell Med. 2012;1:209–217. [PMC free article] [PubMed] [Google Scholar]
- 42.Moulana Z, Rajabnia R, Asgharpour F, Ferdosi Shahandashti E, Khalilian M, Norkhomami S. Class 1 integron in Pseudomonas aeruginosa isolates from different places and devices of ICU in Babol, Iran. Jundishapur J Microbiol. 2013;6:138–143. [Google Scholar]
- 43.Sabbagh P, Ebrahimzadeh-Namvar A, Ferdosi-Shahandashti E, Javanian M, Khafri S, Rajabnia M. Molecular characterization of Staphylococcus aureus strains isolated among hospital staff nasal carriers of Babol, Iran. Caspian J Intern Med. 2017;8:311–316. doi: 10.22088/cjim.8.4.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marathe N, Nagarkar S, Vaishampayan A, Rasane M, Samant S, Dohe V, et al. High prevalence of class 1 integrons in clinical isolates of methicillin-resistant Staphylococcus aureus from India. Indian J Med Microbiol. 2015;13:231–236. doi: 10.4103/0255-0857.154905. [DOI] [PubMed] [Google Scholar]
- 45.Guo Y, Song G, Sun M, Wang J, Wang Y. Prevalence and Therapies of Antibiotic-Resistance in Staphylococcus aureus. Front Cell Infect Microbiol. 2020;10:107. doi: 10.3389/fcimb.2020.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sow AG, Aïdara-Kane A, Barraud O, Gatet M, Denis F, Ploy M-C. High prevalence of trimethoprim-resistance cassettes in class 1 and 2 integrons in Senegalese Shigella spp isolates. J Infect Dev Ctries. 2010;4:207–212. doi: 10.3855/jidc.583. [DOI] [PubMed] [Google Scholar]