Abstract
BACKGROUND
The genetic heterogeneity of Leishmania parasites is a major factor responsible for the wide variety of Leishmania-associated manifestations. Consequently, understanding the genetic make-up of Leishmania species using suitable molecular markers is an important component of realising local and regional scale disease risk. The cytochrome b (cytb) is frequently used to type New World Leishmania species. However, its potential to discriminate Leishmania species and variants requires further evaluation.
OBJECTIVES
To explore the capacity of cytb gene to identify New World Leishmania species and variants and to develop an approach able to type local Leishmania species and variants.
METHODS
We retrieved 360 partial and complete Leishmania cytb gene sequences publicly available in GenBank database to study all single nucleotide polymorphisms (SNPs) across the cytb gene that differentiate New World Leishmania species. This information was used to develop an approach based upon the polymorphisms found in a DNA segment of 948bp. We also compared the typing results found with this technique with the polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) profiling obtained using HSP70 gene as target. One hundred Panamanian isolates were used to both typed Leishmania species and assess local genetic variability.
FINDINGS
We found complete agreement between our cytb approach and the PCR-RFLP profiling method based on HSP70 for Leishmania species identification. Ninety-two isolates were identified as L. panamensis, although other Viannia species were found circulating at a lower frequency. Three L. panamensis haplotypes were identified in Panamanian provinces. We also provide an initial report of L. guyanensis haplotypes circulating in Panama.
MAIN CONCLUSIONS
Cytb gene sequence encompasses key main SNPs that aid to identify Leishmania species. The cytb approach developed with this information was able to identify and assess genetic variability of local Leishmania species found in this study.
Key words: leishmaniasis, cytochrome b gene, haplotypes, Leishmania panamensis, Leishmania guyanensis, Panama
Leishmaniasis, a neglected tropical disease transmitted by female sandflies and caused by kinetoplastic protozoa parasites of the genus Leishmania, is endemic in 98 countries worldwide. Due to the high number of cases and its geographical expansion, the World Health Organization (WHO) considers leishmaniasis as an emerging/reemerging vector borne parasitic disease. 1 Regarding its impact on public health, leishmaniasis has an estimated annual incidence of 2.0 million cases and an approximated prevalence of 12,000,000 cases. 2 In addition, this disease is responsible for 20,000 to 40,000 deaths occurring in rural and suburban populations all year round. 1 , 2
Visceral and tegumentary leishmaniasis are the main clinical forms of the disease which show different clinical expressions depending upon the species of Leishmania responsible for the disease, the genetic background and immunological status of the host, and factors in the sand fly vector saliva. 3 , 4 According to data reported by WHO in 2017, the cutaneous leishmaniasis is widely spread across the American continent and it is 15 times more frequent than the visceral form of leishmaniasis. 1 After its first description in Panama more than one hundred years ago in 1910, leishmaniasis has now become the most prevalent vector-borne parasitic disease in the country with annual incidence as high as 3,000 new cases and afflicting people of all ages, but mainly those younger than 14 years old. 5 , 6 , 7 Related to its neglected status, the transmission of cutaneous leishmaniasis in Panama is concentrated in sylvatic and rural areas affecting primarily socially and economically marginalised people who primarily present with cutaneous leishmaniasis (CL) and less than 3% with the mucocutaneous form (MCL). 1
Leishmania panamensis is principal etiological agent of leishmaniasis in Panama as it has been found infecting anthropophilic vectors, mammalian reservoirs, and causing CL. 8 , 9 , 10 However, other species of Viannia subgenera, such as Leishmania braziliensis, Leishmania guyanensis and Leishmania naiffi also circulate at a minor frequency in regions endemics for CL. 11 Furthermore, an enzootic cycle of Leishmania mexicana and few human cases caused by this Leishmania species have been reported in Panama. In general, the evidence gathered so far, indicates that Leishmania Viannia species are more prevalent in Panamanian endemic areas. 8 , 10 , 12 , 13
The genetic make-up of Leishmania defines biological traits responsible for parasite survival in mammals and vector hosts and its capacity to evoke a particular clinical form. The broad spectrum of Leishmania-associated symptoms described in an endemic area are determined by both parasite and hosts’ genetic composition. On the other hand, clonal diversity, and genetic heterogeneity responsible for variability in parasite virulence are quite common in Leishmania. 14 , 15 Thus, it is important to gather information on the genetic background of Leishmania species circulating in specific endemic areas to strengthen local epidemiological surveillance programs and provide proper disease treatment.
L. panamensis has been associated to diverse cutaneous forms of the disease, a behavior that might be the result of local interplay between L. panamensis variants and human populations with different genetic backgrounds. 16 , 17 , 18 , 19 Little is known about the population structure of L. panamensis in Panama. Local studies focus mainly on discriminating Leishmania species on vectors, hosts, and humans rather than inferring their genetic diversity. 10 , 20 , 21 Therefore, it is important to conduct further studies in our country using molecular markers that can differentiate Leishmania at the subspecies level. Since haplotypes might arise as an intrinsic attribute of population genetic variation, it is feasible to use these targets as molecular markers in studies aiming to assess the genetic structure of a particular etiological agent. 22 As a result, it is worth to explore the potential of molecular targets for uncovering different haplotypes that could be circulating in different Leishmania transmission cycles. In this regard, some molecular approaches based on cytb gene have already been used in some Latin American countries to discriminate species and haplotypes of Leishmania. 23 , 24 , 25 However, most of these studies were based on partial cytb sequences and no description of the polymorphic sites used to discriminate Leishmania species and variants were reported. Thus, it might be necessary a further evaluation of the cytb gene-based methods or develop new ones assessing the extent of polymorphic sites and their capacity to type species and variants of Leishmania. In this sense, we developed a molecular approach based upon the polymerase chain reaction (PCR)-amplification and sequencing of a 948 bp segment of cytb gene that showed enough single nucleotide polymorphisms (SNPs) to discriminate New World Leishmania species. Also, to shed light on the local genetic variability of Leishmania, we evaluated one-hundred isolates from eastern and western side of Panama.
MATERIALS AND METHODS
Design of the PCR approach
In silico design of the PCR approach - We developed a PCR approach able to discriminate new world Leishmania species based on the amplification by PCR and Sanger-sequencing of a 948 bp cyt b gene sequence. To develop this PCR-approach, we retrieved 360 partial and complete Leishmania cytochrome b (cytb) gene sequences publicly available in GenBank database including: L. braziliensis (110), L. peruviania (29), L. guyanensis (80), L. panamensis (6), L. lainsoni (22), L. naiffi (5), L. shawi (3), L. mexicana (39), L. amazonensis (17), L. infantum/L. donovani (39), L. tarentolae (1), L. major(1), L. aethiopica (1), L. tropica (3), L. aristidesi (1), L. garnhami (1) and L. pifanoi (1). Twenty-eight out of the 360 cytb sequences corresponding to the pre-edited version of cytb gene were used to design specific primers (Table I). The rest of the sequences allowed us to analyse polymorphic sites [Supplementary data (555.9KB, pdf) (Table I)]. All sequences were downloaded to the bioinformatic software UGENE v.34 version and were aligned using the MAFFT algorithm included on the software. 26 Sequence alignment was used to find consensus regions for all Leishmania species which were then employed to design five primers sets using the algorithm of primer3 incorporated in UGENE. Important characteristic of primers sets, like formation of secondary structures and hairpins was evaluated in silico by OligoAnalyzer tool (Integrated DNA technology) and their specificity was assessed using primer-Blast tool (NCBI).
TABLE I. Complete cytochrome b (cytb) sequences retrieved from GenBank to design Leishmania specific primers.
| Length | GenBank accession number | Leishmania specie |
| 1398pb | M10126.1 | Leishmania tarentolae |
| 1078pb | BK010875.1 | Leishmania panamensis |
| 1078pb | AB095968.1 | Leishmania panamensis |
| 1078pb | MK570510.1 | Leishmania panamensis |
| 1078pb | AB434682.1 | Leishmania braziliensis |
| 1078pb | AB434681.1* | Leishmania braziliensis |
| 1078pb | AB095967.1* | Leishmania braziliensis |
| 1078pb | AB095966.1 | Leishmania braziliensis |
| 1078pb | AB095969.1 | Leishmania guyanensis |
| 1078pb | BK010883.1 | Leishmania shawi |
| 1078pb | AB434680.1 | Leishmania shawi |
| 1079pb | AB095963.1* | Leishmania mexicana |
| 1078pb | AB095964.1 | Leishmania amazonensis |
| 1079pb | AB434679.1 | Leishmania pifanoi |
| 1079pb | AB095965.1 | Leishmania garnhami |
| 1078pb | AB434678.1 | Leishmania aristidesi |
| 1079PB | HQ908261.1* | Leishmania infantum |
| 1079pb | HQ908267.1* | Leishmania donovani |
| 1079pb | HQ908262.1* | Leishmania donovani |
| 1079pb | AB095957.1 | Leishmania donovani |
| 1079pb | AB095959.1* | Leishmania infantum |
| 1079pb | AB095958.1 | Leishmania infantum |
| 1079pb | AB434677.1 | Leishmania archibaldi |
| 1080pb | AB095961.1 | Leishmania major |
| 1080pb | AB095962.1 | Leishmania aethiopica |
| 1080pb | AB095960.1 | Leishmania tropica |
| 1080pb | HQ9082270.1 | Leishmania tropica |
| 1080pb | HQ9082270.1 | Leishmania tropica |
| 1080pb | HQ908257.1 | Leishmania tropica |
*: reference Leishmania sequences used for the phylogenetic analysis.
The best set of primers based on the characteristics evaluated were cytb-VF 5’-AAGCGGAGAGAAAAGAAAAGG-3’ and cytb-VR 5’-AATGAATAAGTAAATCATAATAAC-3’ which align to regions 1-21 and 925-948 of cytb gene, respectively, and amplify a PCR-product of 948 bp. This set of primers was then employed to standardise a PCR-sequencing approach aiming to identify Leishmania species and cytb haplotypes.
Average of genetic distances between groups of cytb sequences - The average of genetic distances between nine groups of Leishmania species depicted on Table II were inferred by Mega X software using cytb gene reference sequences depicted on Table I. 27 The rate variation among sites was modeled with a gamma distribution (shape parameter = 4) and all positions including coding and noncoding ones were included in the analysis. All ambiguous positions were removed for each sequence pair.
TABLE II. Estimates of evolutionary divergency over sequence pairs between groups of Leishmania cytochrome b (cytb) sequences.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| 1. L. tarentolae | |||||||||
| 2. L. panamensis | 0.10 | ||||||||
| 3. L. braziliensis | 0.10 | 0.01 | |||||||
| 4. L. guyanensis | 0.10 | 0.01 | 0.01 | ||||||
| 5. L. shawi | 0.10 | 0.01 | 0.01 | 0.01 | |||||
| 6. L. mexicana complex | 0.11 | 0.12 | 0.12 | 0.12 | 0.11 | ||||
| 7. L. donovani complex | 0.12 | 0.12 | 0.11 | 0.11 | 0.11 | 0.10 | |||
| 8. L. major | 0.13 | 0.13 | 0.12 | 0.13 | 0.12 | 0.11 | 0.08 | ||
| 9. L. tropica complex | 0.13 | 0.13 | 0.12 | 0.13 | 0.12 | 0.10 | 0.08 | 0.07 |
Ethical statement - This research was considered by the Comité de Bioética de la Investigación del Instituto Conmemorativo Gorgas de Estudios de la Salud (CBI-ICGES), and deemed exempt (Note No. 1084/CBI/ICGES/16).
Evaluation of the PCR approach
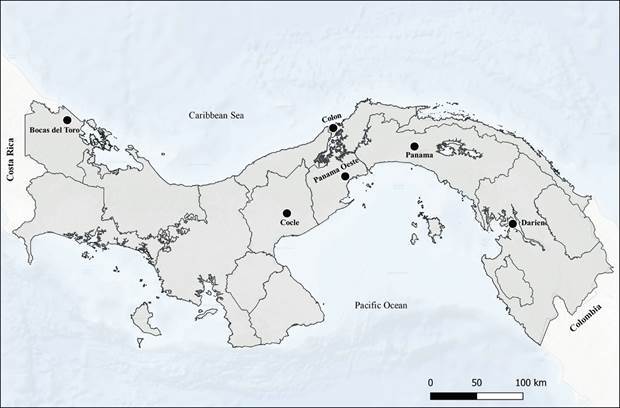
Isolates - One hundred Leishmania isolates from both eastern border and western borders of Panama Canal were used in this study (Fig. 1). Isolates were collected during the last five years (2015-2020) and stored in liquid nitrogen at the Departamento de Investigación en Parasitología, Instituto Conmemorativo Gorgas de Estudios de la salud (ICGES) situated in Panama City. Parasites were isolated from patients living in Provinces located east (Panama and Darien Provinces) and west (Colon, Panama Oeste, Cocle and Bocas del Toro) of the Panama Canal (Fig. 1). These locations are recognised endemic areas of leishmaniasis in the country with a long-term average of CL incidence rates between 5-25 new cases per 10,000. 28
Fig. 1: geographical positions of the Panamanian Provinces where Leishmania spp. isolates were obtained.
Parasite culture and DNA extraction - Leishmania stocks were cultured using Schneider’s insect medium (Sigma Aldrich, Inc., St. Louis, USA) supplemented with 20% heat-inactivated fetal bovine serum (Gibco, Grand Island, USA) at 26ºC. Total genomic DNA was extracted from promastigotes using a commercial kit (Wizard Genomic DNA Purification Kit, Promega, Madison, WI) according to the manufacturer’s instructions.
PCR conditions - The optimal conditions for amplifying the selected cytb gene fragment consisted of the addition of one unit of Platinum high fidelity taq polymerase, 0.2 μM of dNTs, 2.5 mM of MgSO4, 0.3 μM of primers forward and reverse, 2.5 μL of 10X high fidelity buffer, 50 ng of genomic DNA and water up to a final volume of 25 μL. Target DNA was amplified after a primary denaturation of 95ºC for 5 min following 35 cycles consisting of 96ºC for 10 s, 50ºC for 30 s and 68ºC for 60 s and a final extension of 68ºC for 10 min.
Cytb gene sequencing and phylogenetic analysis - The cytb amplification products were run in 1.2% agarose gels prepared with 0.5X TBE buffer and stained with GelRed® (Biotium). PCR products were cleaned up by PureLinkTM PCR purification kit (Invitrogen) following manufacturer’s instructions. DNA sequencing of both strands was carried out using PCR-primers abovementioned and BigDye Terminator 3.1 cycle sequencing kit (Applied Biosystems). Primers and deoxynucleotide triphosphates were removed using Big dye XterminatorTM (Applied Biosystems) and sequencing reactions were run on ABI 3500XL sequencer. Sequencing chromatograms were edited using the assembling-to-reference tool of UGENE toolkit using a trimming quality value of 25. After edition, a segment of 792 bp of cytb sequence were used to perform the phylogenetic analysis. Sequences were then multiple aligned using the MAFFT algorithm also included in UGENE with a maximum number of iterative refinements of 3 and a gap penalty of 1.53. The HKY (G + I) model was found as the best DNA evolution model by the software JModelTest 2. 29 A phylogenetic tree reconstruction of Leishmania was implemented applying Bayesian inference (BI) with the Mr. Bayes v.3.2 software. 30 Ten Markov chains were proceeded for ten million of generations, and trees were sampled for every 7,000 generations setting the program to run the substitution model with the option invgamma and permit different rates of transition and transversion (K80 or HKY85 model). Twenty-five percent of the sampled trees were discarded, and the remaining were used to build up a consensus tree and to calculate posterior probabilities of clades. The result of Bayesian analyses was visualised using Figtree v1.4.2. The cytb gene sequence of Leishmania siamensis (JX195634.1) was used as outgroup. Cytb gene reference sequences used to perform this analysis are showed in Table I and Supplementary data (555.9KB, pdf) (Table I) with the GenBank accession number marked by an asterisk. Transformation of the leaves and a schematic representation of the root were applied for visualisation purposes.
HSP70-RFLP analysis - Eighty isolates of Leishmania spp. were used to typed Leishmania species by PCR-RFLP approach based on HSP70 gene. PCR was performed with oligonucleotides F25 and R1310, that amplify a 1286 bp product from the repeated gene heat shock protein 70 (hsp70) as previously described. 31 Amplification reactions were performed in a final volume of 50 μL containing 25 μ L of Go Taq Green Master Mix 2X (Promega), 0.6 μmol/L of each primer and 5 μL of DNA. The resulted amplicons were digested with Hae III and subsequently with BccI or RsaI endonucleases. Obtained restriction patterns were analysed by electrophoresis and compared with reference strain patterns.
Haplotype analyses - The sequences recognised as haplotypes herein, were identified from aligned cytb sequences of L. panamensis displayed in UGENE Alignment Editor. Sequences from identified haplotypes were then evaluated in PopART software v.1.7 to construct a median joining network among haplotypes. 32
RESULTS
Evaluation of partial and complete cytb sequences - Fifteen percent (15%) of the evaluated 948 bp cytb sequences consist of 145 main polymorphic sites, including 42 singletons and 103 parsimony-informative sites that can discriminate members of the L. Viannia subgenus, species of L. mexicana complex and distinguish the three complexes of New World Leishmania [Supplementary data (555.9KB, pdf) (Table II)]. Furthermore, this cytb sequence encompassed 35 polymorphic specific sites for species of subgenus Viannia, 15 SNPs that differentiate L. mexicana from L. mexicana amazonensis, 50 SNPs that aid to discriminate members of L. mexicana complex from subgenus Viannia complexes, and 24 singletons that are found in L. infantum as well as L. donovani.
Table II shows estimates of evolutionary divergency over sequence pairs between groups consisting of the pre-edited version of cytb sequences from nine groups of Leishmania species. A low rate of divergency was found in Leishmania species of the subgenus Viannia as the total genetic distance between groups of them was 0.01. However, we found enough SNPs across the 948 bp cytb sequences to discriminate species of this subgenus except for L. peruviania and L. braziliensis which had great similarity in nucleotide sequence. As we analysed partial cytb sequences of L. peruviana starting from position 74 onwards, we do not rule out the presence of any SNPs that permit to discriminate these species somewhere upward this position. Conversely, we found high genetic distances ranging from 0.11 to 0.13 between cytb sequences from Viannia subgenus and grouped cytb sequences belonging to L. mexicana complex, L. donovani complex, L. tropica complex, L. major and L. tarentolae. This finding demonstrates that the cytb gene segment selected for our PCR-approach is useful to distinguish species belonging to the different Leishmania complexes.
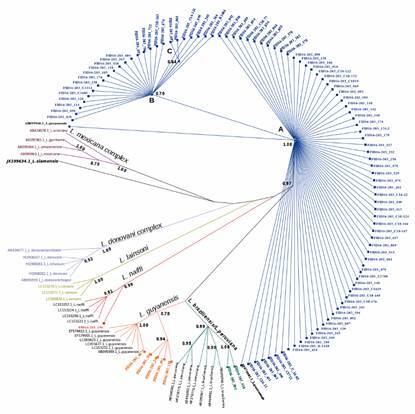
Phylogenetic inference - The phylogenetic inference using the cyt b sequences clearly separated and grouped in clusters Leishmania species from L. mexicana complex, L. donovani complex and subgenus Viannia (Fig. 2). All local isolates analysed in this study were found to belong to subgenus Viannia. The partial cytb gene sequences obtained from these isolates were deposited in GenBank under accession numbers MW117326 to MW117425. Ninety-two percent of all field isolates clustered with L. panamensis reference sequences, branching into two major groups that include three L. panamensis haplotypes. The remaining 8% of the field isolates were identified as L. guyanensis, L. braziliensis/L. peruviana and L. naiffi, according to the phylogenetic analysis (Fig. 2). As for L. guyanensis local isolates, one of them clustered with L. guyanensis reference group and the rest of the isolates clustered into a sister subclade laying close to this group on the tree. The presence of two L. guyanensis different subclades after the phylogenetic analysis indicates the circulation of at least two different haplotypes of this Leishmania specie in Panama. On the other hand, local L. braziliensis and L. naiffi isolates clustered with their respective reference groups conformed by cytb gene sequences retrieved from GenBank.
Fig. 2: phylogenetic tree inferred from Bayesian analysis of cytochrome b (cytb) gene using Mr. Bayes v. 3.2. The numbers at the node represent Bayesian posterior probabilities; Leishmania siamensis JX195634.1 was used as the outgroup. Different labels colours in L. panamensis (blue), L. guyanensis (orange), L. braziliensis (green) and L. naiffi (red) phylogenetic groups represent isolates of these species found herein. The letters A, B and C on the tree depict L. panamensis haplotypes.
PCR-RFLP analysis of HSP70 gene - We found 100% of concordance between cytb approach described here and the PCR-RFLP based on HSP70 when used to identify eighty Leishmania isolates from different regions of Panama (Table III). All isolates considered by cytb approach as L. braziliensis / L. peruviana were confirmed by the HSP70 based approach as L. braziliensis isolates.
TABLE III. Leishmania identification by polymerase chain reaction-cytochrome b (PCR-cytb) gene sequencing and PCR-restriction fragment length polymorphism (PCR-RFLP) based on HSP70 gene.
| Leishmania Viannia species | Cyt b approach (n = 80) | Hsp70 approach (n = 80) |
| L. panamensis | 72 | 72 |
| L. braziliensis | 5* | 5 |
| L. guyanensis | 2 | 2 |
| L. naiffi | 1 | 1 |
*: isolates identified as L. braziliensis after confirmation by PCR-RFLP approach based on HSP70 gene.
Leishmania haplotypes - We found three cytb haplotypes of L. panamensis circulating in the studied areas that were assigned as haplotype A, haplotype B, and haplotype C as depicted on Fig. 3. Only two non-synonymous SNPs at positions 357 and 669 of the L. panamensis cytb gene complete sequence distinguished these haplotypes. One of them is a transition C↔T at position 357, where haplotype B and C showed the nucleotide cytosine and the haplotype A thymine instead. Additionally, at position 669 of cytb sequence we found an A↔G transition. In this nucleotide position, haplotypes A and B showed the base guanine and the haplotype C adenine instead. As a result, two SNPs distinguish haplotype A from C and only one SNP differentiate haplotype A from B and haplotype B from C.
Fig. 3: haplotype network inferred by a median-joining method using cytochrome b (cytb) sequences from Leishmania panamensis. Circles represent different haplotypes. Circle size is proportional to haplotype frequency; colours indicate geographical origins. Hatch marks correspond to nucleotide substitutions.
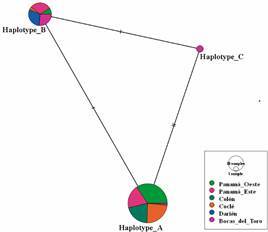
The number of L. panamensis haplotypes found by province is shown in Table IV. All L. panamensis isolates (n = 92) were used to identify haplotypes from provinces located to the west and east side of the Panama Canal. Three haplotypes of L. panamensis were found distributed across the studied provinces (Fig. 3). The most frequent haplotype (A) was found in 67 out of 92 L. panamensis cytb sequences and occurred in almost all evaluated provinces, except in Darien where only haplotype B was identified. Conversely, haplotypes B and C seem to circulate at a lower frequency since only 25 and 2 cytb sequences from both haplotypes, respectively, were detected in this study. Haplotype B was found in all provinces except for Colon, and haplotype C was only observed in Bocas del Toro Province. However, because a low number of isolates from Bocas del Toro (9) and Darien (7) were evaluated, we do not rule out the circulation of other haplotypes at frequencies different from the ones described in this study.
TABLE IV. Total number of Leishmania panamensis haplotypes found in the study by localities.
| Localities | Haplotype A | Haplotype B | Haplotype C | Haplotypes found by province |
| Panama Oeste | 23 | 2 | 0 | 25 |
| Colón | 14 | 0 | 0 | 14 |
| Bocas del Toro | 1 | 6 | 2 | 9 |
| Coclé | 16 | 1 | 0 | 17 |
| Panamá Este | 13 | 7 | 0 | 20 |
| Darién | 0 | 7 | 0 | 7 |
| Total number of haplotypes | 67 | 23 | 2 | 92 |
Two L. guyanensis haplotypes were found circulating in Cocle and Panama Oeste province in this study. Remarkably, these haplotypes showed greater diversity than L. panamensis haplotypes as we found six polymorphic sites at position 119 (T↔C), 241 (G↔T), 313 (A↔T), 619 (G↔A), 686 (A↔G) and 860 (C↔T). Only one SNP at position 686 was nonsynonymous causing the amino acid replacement isoleucine to valine in cytb protein. Since only five isolates of L. guyanensis were detected in this study, there is a high probability that other L. guyanensis haplotypes might be circulating in these endemic areas.
DISCUSSION
Cytb is one of the set of cytochromes responsible for the electron transport process initially used for the phylogenetic analysis of Leishmania genus. 33 As molecular marker for Leishmania spp., cytb is a good typing option providing its good resolution for all tested species across the globe. It is also sensitive enough for use on clinical and environmental samples, and it is readily analysed. 34 However, to increase its reliability, it is important to validate its performance using a more extended set of sequences from strains belonging to current Leishmania complexes and from different geographical regions, including parasites isolated from reservoirs and vectors.
The availability of Leishmania genomic sequences increases as more studies are carried out using specific molecular markers or even complete genomes. This the case of cytb gene, which has been utilised successfully to discriminated new world and old world Leishmania species and to assess their populational structure, a fact that has increased the number of cytb sequences publicly available in databases. 25 , 33 , 35 , 36 Herein, we describe 145 main SNPs found across the 948 bp segment selected of the cytb gene after evaluating 360 complete and partial sequences from New World Leishmania species belonging to different complexes. Altogether, these SNPs are responsible for the genetic divergence seen between L. Viannia species and species from L. mexicana and L. donovani complexes, particularly considering that no size differences were observed between complete cytb sequences of these species. The first report of cytb gene variation in Leishmania evaluated 15 strains, and found 245 polymorphic sites and an average sequence identity between species from Leishmania complexes of 90%. 35 In an additional study by these authors, 30 additional strains of Leishmania were evaluated highlighting the usefulness of cytb for phylogenetic analysis. In that study, Lugo and collaborators were able to group Leishmania cytb sequences in five main complexes using completes cytb sequences. 33
Our results suggest that the 948bp segment contains 60% of the polymorphic sites described across the cytb gene in the first report, exhibiting enough sequence divergence to discriminate species from New World Leishmania complexes. Moreover, its usefulness is supported by the availability of a larger set of Leishmania sequences and the presence of an adequate amount of SNPs as source of phylogenetical signal necessary to discriminate Leishmania species as reported elsewhereusing completes cytb gene sequences. 33 As depicted in Fig. 2, singletons and parsimony informative sites across the 948 bp cytb segment allowed a clear separation of L. Viannia species from species of both L. donovani and L. mexicana complexes. Nonetheless, our phylogenetic approach based upon the 948 bp cytb sequence did not separate L. donovani and L. infantum nor L. braziliensis and L. peruviana. The same results were described using a phylogenetical analysis based upon the complete cytb sequence. 34 The species status of L. peruviana within the L. braziliensis complex is a current matter of discussion. However, a comparative genomic analysis of both species found interspecific SNP/Indel differences and chromosome copy number variations that support that they are closely related but distinct species. 37 As approaches based on HSP70 gene or Multilocus typing are able to differentiate both species, any of these tools could be used in conjunction with cytb to confirm the circulation of these species and identify others. 31 , 38 Alternatively, methods based on PCR and sequencing of genes HSP70, miniexon and ITS1can differentiate L. donovani complexes, making it easy to use in combination with the cytb gene as complementary tool in places where tegumentary and visceral leishmaniasis overlapped. 39 Nonetheless, additional set of markers with a better discriminatory power should be applied to clarify any nuclear-mitochondrial disagreement, a fact that already has been reported in Leishmania typing. 25 Also, our PCR approach was designed to perform molecular typing of Leishmania isolates using a known quantity of genomic DNA, a fact that might affect the sensitivity of this methodology if used with clinical samples because of the low-copy number of cytb gene.
We found a high grade of agreement between the cytb approach developed in this study and the one based on PCR/RFLP of HSP70 gene when typing Panamanian isolates using both techniques. Indeed, the PCR-RFLP analysis of HSP70 gene aid to confirmed five isolates typed as L. braziliensis/L. peruviana by cytb approach as L. braziliensis. This fact points out that our cytb approach has enough discriminatory power to discriminate L. Viannia species that it is the case of the ones causing CL in Panama. However, some cases of disagreement have been reported when used HSP70 based molecular tools and approaches based on mitochondrial gene to identify New World Leishmania species. 25 In spite of this divergence, we believe that the use of the approach presented herein in combination with HSP70 based molecular tools might improve Leishmania species assignment as showed in different Leishmania typing studies. 25 , 40
L. panamensis was the predominate species isolated throughout all geographic regions studied in Panama (Fig. 2). Previous studies carried out in Panama support the wide distribution and prevalence of human cases of CL caused by L. panamensis. 9 , 10 Besides, this species has been found naturally infecting all anthropophilic sand flies, 41 and it is also responsible for the high infection rates found in the two-toed sloth; a major sylvatic reservoir widely distributed in Leishmania endemic areas of Panama. 8 , 42 The rest of the isolates from this study belongs to L. guyanensis (5), L. braziliensis (2) and L. naiffi (1). These species have been recently reported circulating at low frequencies in the country. 11 This fact has important epidemiological implications for the local surveillance and control programs as well as for the clinical management of patients. In this line, it would be necessary initiatives aiming to monitor and determine geographical distribution, dynamic of infections, frequency of circulation of new reported species and to establish their transmission cycles. From clinical point of view, disease outcome and treatment prognosis are influenced by Leishmania species. 43 In this context, L. braziliensis and L. guyanensis have been associated with localised cutaneous leishmaniasis, disseminated cutaneous leishmaniasis and mucosal lesions. 44 In the case of L. naiffi, no association between this species and mucosal leishmaniasis has been observed. Moreover, cutaneous lesions occasioned by L. naiffi are single, smalls and located on hands, arms or legs. 45 On the other hand, L. braziliensis isolates with reduced drug uptake have been recovered from atypical lesions, and some mutant variants of L. guyanensis strains are resistant to pentavalent antimonials, the first-line treatment option for CL in Panama. 46 , 47 Interestingly, a poor response to antimonial or pentamidine therapy has already been described in patients infected with L. naiffi. 48 Thus, the sympatric circulation of different Leishmania species from subgenus Viannia in Panama should be further studied because it might have significance regarding human disease pathogenesis and control.
Three L. panamensis haplotypes were found circulating in the study areas (Fig. 3). A previous study conducted in Colombia using a partial sequence of cytb as a barcode, analysed 201 L. panamensis cytb sequences. A great variability was found supported by the existence of 80 SNPs across the cytb sequence. 23 Herein, we only identified three SNPs along the L. panamensis cytb sequence responsible for the haplotype assignments. This might reflect a low genetic diversity of L. panamensis due to the expansion and fitness of few variants in both anthropophilic vectors and human beings. In this line, a pilot study conducted in Morocco using ITS1 as marker suggested that the overrepresentation of one haplotype over the rest in an endemic focus might be result of this fitness. 49
It seems that there are differences in abundance and frequency of L. panamensis haplotypes in the studied sites. Haplotype A predominated in the studied sites as it was found in almost all evaluated Provinces except for Darien. Conversely, haplotype B showed a high frequency in Bocas del Toro and Darien Provinces, and only two cytb sequences belonging to Haplotype C were found in Bocas del Toro. Apparently, there is an association between genetic diversity and geographical origins of the L. panamensis isolates. In this sense, haplotype A might be involved in most of the current leishmaniasis transmission cycles in Panama along with local anthropophilic species of Lutzomyia and Choleopus hoffmani. While haplotype B and C might be circulating only in sylvatic transmission cycles, where human settlements are surrounded by tropical rain forest at the human-forest interface. To confirm this finding, further studies are necessary considering all components in the transmission cycles in which L. panamensis haplotypes participate. It is also possible that the low number of isolates evaluated from some Provinces have led to a sample bias that keep us from suggesting the overall predominance of haplotype A or even ruling out the circulation of other haplotypes.
The transmission cycle of L. guyanensis has been mainly associated with the presence of human beings in forest regions. 47 In this sylvatic environment, animals such as sloths, anteater, marsupials and rodents are responsible for maintaining parasite populations. 50 Two L. guyanensis haplotypes were found circulating in communities located through the central mountain range of Panama specifically in Cocle and the mountainous regions of Panama Oeste. These regions are considered wet/rain environments with a high proportion of forest coverage and where CL shows a high incidence. 28 In these regions, as other endemic regions of the country, a part of the original forest environment has been preserved, with an abundance of anthropophilic sand flies and the presence of several Leishmania reservoirs, with a high rate of human infection. 41 Therefore, it is feasible that under this eco-epidemiological scenario a zoonotic cycle has been established in which different lineages of L. guyanensis are participating. In this context, our results suggest the circulation of two variants of L. guyanensis that somehow might be associated with transmission cycles of CL in Coclé and Panama Oeste Provinces. This Leishmania species displays great genetic plasticity due to a high level of sexual recombination in vectors, complemented with a clonal expansion in natural hosts which are the main responsible for its diffusion in endemic regions. 51 Thus, sylvatic cycles currently established in Cocle and Panama Oeste combined with a high abundance and diversity of parasite vectors and reservoirs meet all necessary characteristics for the emergency and dissemination of different L. guyanensis genotypes. However, entomological, and zoological data are required to support this hypothesis.
Using ribosomal fingerprinting, Rotureau et al. observed two different non sympatric lineages of L. guyanensis in French Guiana associated with regions characterised by dense primary rain forest spotted with gold-digging sites where anthropogenic changes were occurring. 52 In Panama, opencast copper mining activities in Cocle and Colon Provinces has led to major man-made environmental changes including boosting human settlements and human exposure to already established zoonotic cycles of Leishmania parasites. Therefore, human infections with different L. guyanensis genotypes already circulating in these areas are plausible and might become a public health concern in these regions of Panama.
The genetic background of Leishmania species is thought to be one of the main factors influencing leishmaniasis clinical outcomes. Therefore, it is important to associate existent clinical effects with genetic variants of Leishmania spp. circulating in different epidemiological scenarios using appropriate molecular markers. There is limited data regarding the genetic variation of Leishmania spp. based on the sequencing of cytb gene. A study carried out in southern Iran found that typical and atypical cutaneous lesions were caused by different Leishmania major variants. 53 Approaches based on the sequencing of cytb gene have been used mainly to identify New World Leishmania species without making an association with clinical outcomes. 23 , 25 , 54 , 55 As the goal of our study was to assess the potential of cytb gene sequence to discriminate Leishmania species and variants, we did not look for an association between the haplotypes found herein and the clinical manifestations seen in Panamanian patients. However, we do not rule out that this association might exist as one approach based on HSP70 found a strong correlation between genetic variant of L. braziliensis and different clinical manifestations. 56
Altogether, our results highlight the presence of different L. Viannia species and genetic variants within L. panamensis and L. guyanensis circulating in the country. The plethora of clinical characteristics seen in the country might be the result of infections with these species or their variants. To evaluate possible associations between genetic variants and clinical expression of the disease, it is necessary to use suitable molecular markers. Sequence variations of the cytb gene have already been related with different CL clinical outcomes. 43 Consequently, an approach like the one presented in this article based on cytb gene that shows enough SNPs to identify New World Leishmania species and their variants represent a good option for Leishmania species identification in Latin American countries afflicted by leishmaniasis.
ACKNOWLEDGMENTS
To the technical staff of the Sanger sequence facility located in the Instituto Conmemorativo Gorgas de Estudios de la Salud for its support in performing the capillary electrophoresis related to this study.
Footnotes
Financial support: The National Secretary of Sciences and technology of Panama (SENACYT) grant No. FID16-203 as a part of the Research for Development initiative (I + D) in Panama.
REFERENCES
- 1.PAHO - Pan American Health Organization . Leishmaniasis: epidemiological report in the Americas. Washington: DC; 2019. [Google Scholar]
- 2.Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J. Leishmaniasis worldwide and global estimates of its incidence. PLoS One. 2012;7(5):e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Moura TR, Oliveira F, Novais FO, Miranda JC, Clarêncio J, Follador I. Enhanced Leishmania braziliensis infection following pre-exposure to sandfly saliva. PLoS Negl Trop Dis. 2007;1(2):e84. doi: 10.1371/journal.pntd.0000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vanloubbeeck Y, Jones DE. The immunology of Leishmania infection and the implications for vaccine development. Ann NY Acad Sci. 2004;1026:267–272. doi: 10.1196/annals.1307.041. [DOI] [PubMed] [Google Scholar]
- 5.Darling ST. Autocthonous oriental sore in Panama. Trans R Soc Trop Med Hyg. 1910;4(2):60–63. [Google Scholar]
- 6.Vázquez A, Paz H, Alvar J, Perez D, Hernandez C. Informe final. Estudios sobre la epidemiología de la leishmaniasis en la parte Occidental de la República de Panamá. Panama. 1998 [Google Scholar]
- 7.PAHO - Pan American Health Organization . Leishmaniasis: epidemiological report in the Americas. Washington: DC; 2017. [Google Scholar]
- 8.Herrer A, Christensen HA. Epidemiological patterns of cutaneous leishmaniasis in Panama III. Endemic persistence of the disease. Am J Trop Med Hyg. 1976;25(1):54–58. doi: 10.4269/ajtmh.1976.25.54. [DOI] [PubMed] [Google Scholar]
- 9.Christensen HA, De Vásquez AM, Petersen JL. Short report epidemiologic studies on cutaneous leishmaniasis in eastern Panama. Am J Trop Med Hyg. 1999;60(1):54–57. doi: 10.4269/ajtmh.1999.60.54. [DOI] [PubMed] [Google Scholar]
- 10.Miranda A, Carrasco R, Paz H, Pascale JM, Samudio F, Saldaña A. Molecular epidemiology of American tegumentary leishmaniasis in Panama. Am J Trop Med Hyg. 2009;81(4):565–571. doi: 10.4269/ajtmh.2009.08-0265. [DOI] [PubMed] [Google Scholar]
- 11.Sosa N, Pascale JM, Jiménez AI, Norwood JA, Kreishman-Detrick M, Weina PJ. Topical paromomycin for New World cutaneous leishmaniasis. PLoS Negl Trop Dis. 2019;13(5):e0007253. doi: 10.1371/journal.pntd.0007253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrer A, Christensen HA, Beumer RJ. Reservoir hosts of cutaneous leishmaniasis among Panamanian forest mammals. Am J Trop Med Hyg. 1973;22(5):585–591. doi: 10.4269/ajtmh.1973.22.585. [DOI] [PubMed] [Google Scholar]
- 13.Vásquez AM, Saenz RE, Petersen JL, Johnson CM. Leishmania mexicana complex human infections in the Republic of Panama. Am J Trop Med Hyg. 1990;43(6):619–622. doi: 10.4269/ajtmh.1990.43.619. [DOI] [PubMed] [Google Scholar]
- 14.Woolhouse MEJ, Taylor LH, Haydon DT. Population biology of multihost pathogens. Science. 2001;292(5519):1109–1112. doi: 10.1126/science.1059026. [DOI] [PubMed] [Google Scholar]
- 15.Garin YJF, Sulahian A, Pratlong F, Pascale M, Gangneus JP, Prina E. Virulence of Leishmania infantum is expressed as a clonal and dominant phenotype in experimental infections. Infect Immun. 2001;69(12):7365–7373. doi: 10.1128/IAI.69.12.7365-7373.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hashiguchi Y, Gomez EL, Kato H, Martini LR, Velez LN, Uezato H. Diffuse and disseminated cutaneous leishmaniasis clinical cases experienced in Ecuador and a brief review. Trop Med Health. 2016;44(1):1–9. doi: 10.1186/s41182-016-0002-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osorio LE, Castillo CM, Ochoa MT. Mucosal leishmaniasis due to Leishmania (Viannia) panamensis in Colombia clinical characteristics. Am J Trop Med Hyg. 1998;59(1):49–52. doi: 10.4269/ajtmh.1998.59.49. [DOI] [PubMed] [Google Scholar]
- 18.Grimaldi G, Tesh RB, McMahon-Pratt D. A review of the geographic distribution and epidemiology of leishmaniasis in the New World. Am J Trop Med Hyg. 1989;41(6):687–725. doi: 10.4269/ajtmh.1989.41.687. [DOI] [PubMed] [Google Scholar]
- 19.Freites CO, Gundacker ND, Pascale JM, Saldaña A, Diaz-Suarez R, Jimenez G. First case of diffuse leishmaniasis associated with Leishmania panamensis. Open Forum Infect Dis. 2018;5(11):1–3. doi: 10.1093/ofid/ofy281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miranda A, Saldaña A, González K, Paz H, Santamaria G, Samudio F. Evaluation of PCR for cutaneous leishmaniasis diagnosis and species identification using filter paper samples in Panama, Central America. Trans R Soc Trop Med Hyg. 2012;106(9):544–548. doi: 10.1016/j.trstmh.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Miranda A, Samudio F, González K, Saldaña A, Brandão A, Calzada JE. Calmodulin polymerase chain reaction-restriction fragment length polymorphism for Leishmania identification and typing. Am J Trop Med Hyg. 2016;95(2):383–387. doi: 10.4269/ajtmh.15-0709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark AG. The role of haplotypes in candidate gene studies. Genet Epidemiol. 2004;27(4):321–333. doi: 10.1002/gepi.20025. [DOI] [PubMed] [Google Scholar]
- 23.Ramírez JD, Hernández C, León CM, Ayala MS, Flórez C, González C. Taxonomy, diversity, temporal and geographical distribution of cutaneous leishmaniasis in Colombia a retrospective study. Sci Rep. 2016;6:28266–28266. doi: 10.1038/srep28266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mondolfi AEP, Garmendia AC, Pérez YM, Hernández-Pereira CE, Medina C, Vargas F. Autochthonous cutaneous leishmaniasis in urban domestic animals (Felis catus/Canis lupus familiaris) from central-western Venezuela. Acta Trop. 2019;191:252–260. doi: 10.1016/j.actatropica.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 25.Kato H, Gómez EA, Seki C, Furomoto H, Martini-Robles L, Muzzio J. PCR-RFLP analyses of Leishmania species causing cutaneous and mucocutaneous leishmaniasis revealed distribution of genetically complex strains with hybrid and mito-nuclear discordance in Ecuador. PLoS Negl Trop Dis. 2019;13(5):1–18. doi: 10.1371/journal.pntd.0007403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okonechnikov K, Golosova O, Fursov M. the UGENE team Unipro UGENE a unified bioinformatics toolkit. Bioinformatics. 2012;28(8):1166–1167. doi: 10.1093/bioinformatics/bts091. [DOI] [PubMed] [Google Scholar]
- 27.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamada K, Valderrama A, Gottdenker N, Cerezo L, Minakawa N, Saldaña A. Macroecological patterns of American cutaneous leishmaniasis transmission across the health areas of Panamá (1980-2012) Parasit Epidemiol Control. 2016;1(2):42–55. doi: 10.1016/j.parepi.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Posada D jModelTest: phylogenetic model averaging Mol Biol. Evol. 2008;25(7):1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- 30.Ronquist F, Teslenko M. Van Der Mark P.Ayres DL.Darling A.Höhna S Mrbayes 3 2: efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Montalvo AM, Fraga J, Maes I, Dujardin J-C Van der Auwera G Three new sensitive and specific heat-shock protein 70 PCRs for global Leishmania species identification. Eur J Clin Microbiol Infect Dis. 2012;31(7):1453–1461. doi: 10.1007/s10096-011-1463-z. [DOI] [PubMed] [Google Scholar]
- 32.Leigh JW, Bryant D. POPART full-feature software for haplotype network construction. Methods Ecol Evol. 2015;6(9):1110–1116. [Google Scholar]
- 33.Asato Y, Oshiro M, Myint CK, Yamamoto Y, Hirotomo K, Marco JD. Phylogenic analysis of the genus Leishmania by cytochrome b gene sequencing. Exp Parasitol. 2009;121(4):352–361. doi: 10.1016/j.exppara.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 34.Van der Auwera G.Dujardin J-C Species typing in dermal leishmaniasis. Clin Microbiol Rev. 2015;28(2):265–294. doi: 10.1128/CMR.00104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luyo-Acero GE, Uezato H, Oshiro M, Takey K, Kariya K, Katakura K. Sequence variation of the cytochrome b gene of various human infecting members of the genus Leishmania and their phylogeny. Parasitology. 2004;128:483–491. doi: 10.1017/s0031182004004792. [DOI] [PubMed] [Google Scholar]
- 36.Yang BB, Chen DL, Chen JP, Liao L, Hu XS, Xu JN. Analysis of kinetoplast cytochrome b gene of 16 Leishmania isolates from different foci of China different species of Leishmania in China and their phylogenetic inference. Parasit Vectors. 2013;6(1):1–12. doi: 10.1186/1756-3305-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valdivia HO, Reis-Cunha JL, Rodrigues-Luiz GF, Baptista RP, Baldeviano GC, Gerbasi RV. Comparative genomic analysis of Leishmania (Viannia) peruviana and Leishmania (Viannia) braziliensis. BMC Genomics. 2015;16(1):715–715. doi: 10.1186/s12864-015-1928-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsukayama P, Lucas C, Bacon DJ. Typing of four genetic loci discriminates among closely related species of New World Leishmania. Int J Parasitol. 2009;39(3):355–362. doi: 10.1016/j.ijpara.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 39.Van Der Auwera G.Ravel C.Verweij JJ.Bart A.Schönian G.Felger I Evaluation of four single-locus markers for Leishmania species discrimination by sequencing. J Clin Microbiol. 2014;52(4):1098–1104. doi: 10.1128/JCM.02936-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan D, Qin H, Zhang J, Liao L, Chen Q, Chen D. Phylogenetic analysis of HSP70 and cyt b gene sequences for Chinese Leishmania isolates and ultrastructural characteristics of Chinese Leishmania sp. Parasitol Res. 2017;116(2):693–702. doi: 10.1007/s00436-016-5335-4. [DOI] [PubMed] [Google Scholar]
- 41.Christensen HA, Fairchild GB, Herrer A, Johnson CM, Young DG, Vásquez AM. The ecology of cutaneous Leishmaniasis in the Republic of Panama. J Med Entomol. 1983;20(5):463–484. doi: 10.1093/jmedent/20.5.463. [DOI] [PubMed] [Google Scholar]
- 42.González K, Calzada JE, Saldaña A, Rigg CA, Alvarado G, Rodríguez-Herrera B. Survey of wild mammal hosts of cutaneous leishmaniasis parasites in Panamá and Costa Rica. Trop Med Health. 2015;43(1):75–78. doi: 10.2149/tmh.2014-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herwaldt BL, Berman JD. Recommendations for treating leishmaniasis with sodium stibogluconate (pentostam) and review of pertinent clinical studies. Am J Trop Med Hyg. 1992;46(3):296–306. doi: 10.4269/ajtmh.1992.46.296. [DOI] [PubMed] [Google Scholar]
- 44.Goto H, Lindoso JAL. Cutaneous and mucocutaneous leishmaniasis. Infect Dis Clin North Am. 2012;26(2):293–307. doi: 10.1016/j.idc.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Naiff RD, Freitas RA, Naiff FM, Arias JR, Barrett TV, Momem H. Epidemiological and nosological aspects of Leishmania naiffi Lainson & Shaw, 1989. Mem Inst Oswaldo Cruz. 1991;86(3):317–321. doi: 10.1590/s0074-02761991000300006. [DOI] [PubMed] [Google Scholar]
- 46.Rugani JN, Gontijo CMF, Frézard F, Soares RP, do Monte-Neto RL. Antimony resistance in Leishmania (Viannia) braziliensis clinical isolates from atypical lesions associates with increased ARM56/ARM58 transcripts and reduced drug uptake. Mem Inst Oswaldo Cruz. 2019;114:e190111. doi: 10.1590/0074-02760190111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borges AF, Gomes RS, Ribeiro-Dias F. Leishmania (Viannia) guyanensis in tegumentary leishmaniasis. Pathog Dis. 2018;76(4):1–10. doi: 10.1093/femspd/fty025. [DOI] [PubMed] [Google Scholar]
- 48.Fagundes-Silva GA, Romero GAS, Cupolillo E, Yamashita EPG, Gomes-Silva A, Guerra JAO. Leishmania (Viannia) naiffi rare enough to be neglected? Mem Inst Oswaldo Cruz. 2015;110(6):797–800. doi: 10.1590/0074-02760150128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.El Hamouchi A, Ajaoud M, Arroub H, Charrel R, Lemrani M. Genetic diversity of Leishmania tropica in Morocco does the dominance of one haplotype signify its fitness in both predominantly anthropophilic Phlebotomus sergenti and human beings? Transbound Emerg Dis. 2019;66(1):373–380. doi: 10.1111/tbed.13031. [DOI] [PubMed] [Google Scholar]
- 50.Lainson R, Shaw JJ, Ready PD, Miles MA, Povoa M. Leishmaniasis in Brazil Xvi. Isolation and identification of Leishmania species from sandflies, wild mammals and man in north Para State, with particular reference to L. braziliensis guyanensis causative agent of "pian-bois". Trans R Soc Trop Med Hyg. 1981;75(4):530–536. doi: 10.1016/0035-9203(81)90192-9. [DOI] [PubMed] [Google Scholar]
- 51.Rougeron V, De Meeûs T, Bañuls A-L Reproduction in Leishmania a focus on genetic exchange. Infect Genet Evol. 2016;50:128–132. doi: 10.1016/j.meegid.2016.10.013. [DOI] [PubMed] [Google Scholar]
- 52.Rotureau B, Ravel C, Nacher M, Couppié P, Curtet I, Dedet JP. Molecular epidemiology of Leishmania (Viannia) guyanensis in French Guiana. J Clin Microbiol. 2006;44(2):468–473. doi: 10.1128/JCM.44.2.468-473.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohammadpour I, Hatam GR, Handjani F, Bozorg-Ghalati F, Pourkamal D, Motazedian MH. Leishmania cytochrome b gene sequence polymorphisms in southern Iran relationships with different cutaneous clinical manifestations. BMC Infect Dis. 2019;19(1):1–13. doi: 10.1186/s12879-018-3667-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gómez EA, Kato H, Torres-Romero EX, Vélez LN, Villegas NV, Martillo VP. Leishmaniasis caused by Leishmania (Viannia) guyanensis in north-central Pacific region of Ecuador a clinico-epidemiological feature. Acta Trop. 2018;185:204–211. doi: 10.1016/j.actatropica.2018.05.016. [DOI] [PubMed] [Google Scholar]
- 55.Marco JD, Uezato H, Mimori T, Barroso PA, Korenaga N, Nonaka S. Are cytochrome B gene sequencing and polymorphism-specific polymerase chain reaction as reliable as multilocus enzyme electrophoresis for identifying Leishmania spp from Argentina? Am J Trop Med Hyg. 2006;75(2):256–260. [PubMed] [Google Scholar]
- 56.Quaresma PF, De Brito CFA, Rugani JMN, Freire JM, Baptista R, Moreno EC. Distinct genetic profiles of Leishmania (Viannia) braziliensis associate with clinical variations in cutaneous-leishmaniasis patients from an endemic area in Brazil. Proc Int Astron Union. 2018;145(9):1161–1169. doi: 10.1017/S0031182018000276. [DOI] [PubMed] [Google Scholar]


