Abstract
Optimized and stringent chemical methods to profile nascent RNA expression are still in demand. Herein, we expand the toolkit for metabolic labeling of RNA through application of inverse electron demand Diels-Alder (IEDDA) chemistry. Structural examination of metabolic enzymes guided the design and synthesis of vinyl-modified nucleosides, which we systematically tested for their ability to be installed through cellular machinery. Further, we tested these nucleosides against a panel of tetrazines to identify those which are able to react with a terminal alkene, but are stable enough for selective conjugation. The selected pairings then facilitated RNA functionalization with biotin and fluorophores. We found that this chemistry is not only amenable to preserving RNA integrity but also endows the ability to both tag and image RNA in cells. These key findings represent a significant advancement in methods to profile the nascent transcriptome using chemical approaches.
Introduction
RNA plays critical roles in the regulation of nearly all biological processes: from controlling chromatin state, to directing transcription, to regulation of cell proliferation.1, 2 Profiling nascent RNA expression can allow researchers to identify potentially functional RNAs, and this is aided by the availability of metabolic chemical reporters.3 As such, several chemical functionalities have been adapted for RNA nucleoside analogs. 4-thiouridine is a conventionally used probe, which allows for enrichment of nascent RNA through disulfide exchange chemistry.4 More recently, alkynyl-5–7 and azido-containing8, 9 nucleosides have been introduced into cells, wherein nascent RNA transcripts can be tracked through click chemistry.4 Both the Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC) and strain-promoted azide-alkyne cycloaddition (SPAAC) can be utilized for enrichment as well as imaging.
Despite the utility of these analogs, their chemistries present several drawbacks. 4-thiouridine enrichment is often inefficient due to the transient nature of disulfide bonds, which further impedes RNA imaging in cells.10 Likewise, CuAAC reactions are known to dramatically compromise the integrity of RNA, where spurious fragmentation of transcripts has altered the results of downstream applications such as RNA sequencing.11, 12 Methods that are damaging to cells or RNA integrity can confound interpretation of biologically meaningful results.13–15 Lastly, discovering novel analogs for labeling is further hindered by the selectivity of metabolic enzymes, which often have limited functional group tolerance.16 These challenges demonstrate the need to further expand the scope of different bioorthogonal chemistries applicable for RNA metabolic labeling (Figure 1A).
Figure 1. Schematic of metabolic labeling of RNA.
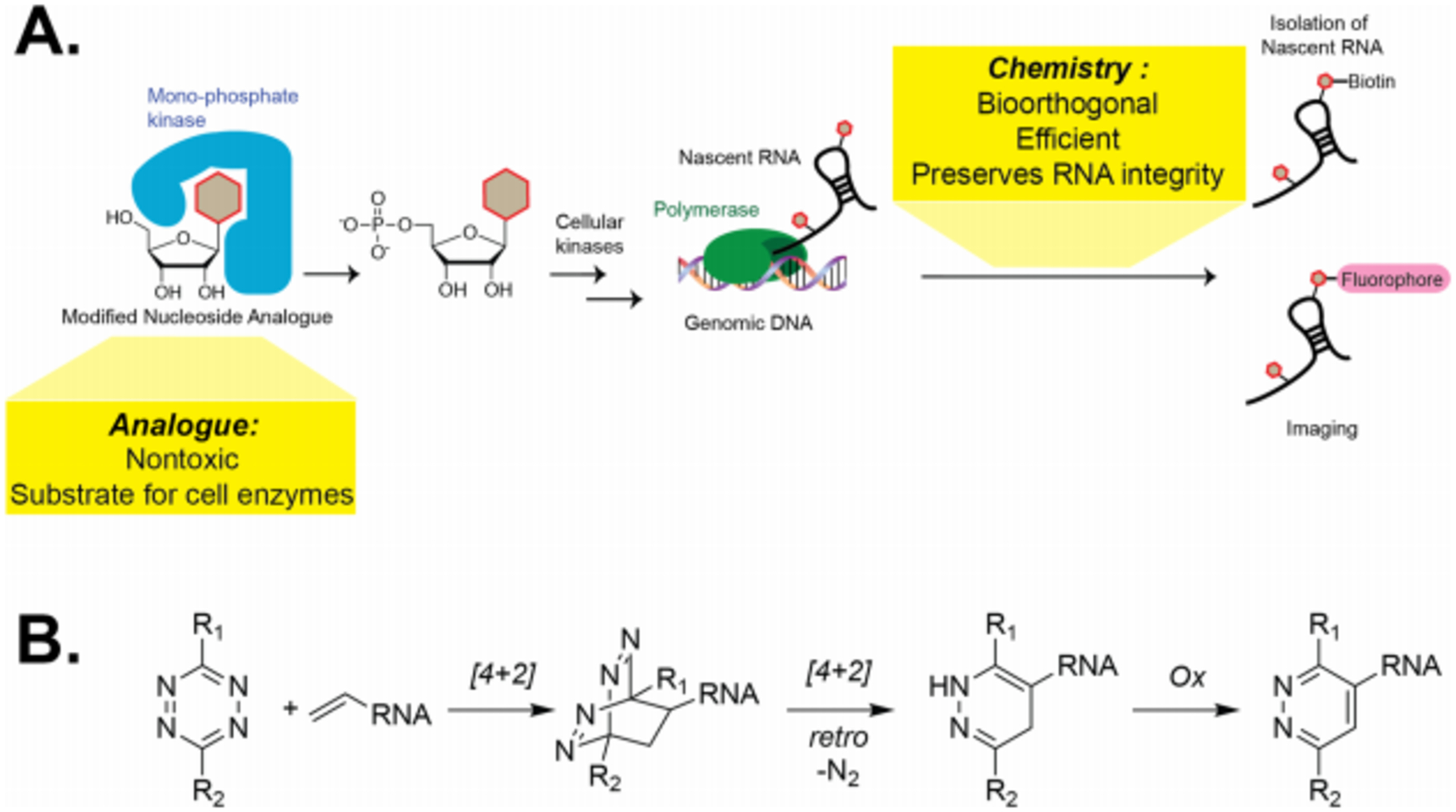
A. Representative steps that are needed to be optimized in metabolic labeling of RNA. B. Schematic of the inverse electron-demand Diels-Alder reaction with vinyl-modified RNA.
The inverse electron-demand Diels-Alder (IEDDA) reaction presents an attractive alternative to the currently adopted chemistries.17, 18 It has been widely used for labeling biomolecules due to its fast kinetics, high yields and exclusion of metal catalysts (Figure 1B). Traditionally, installation or genetic encoding of a strained alkene such as norbornene, trans-cyclooctene and cyclopropene have facilitated this rapid labeling.19–21 However, this presents a significant challenge to reporter strategies that rely solely on cellular machinery for installation, wherein the enzymes in these metabolic pathways may not accommodate large or even moderately-sized functional groups. We have observed that the size and placement of biorthogonal handles on nucleosides often dictates their utility as metabolic reporters. Furthermore, there is the possibility for cross-reactivity with nucleophiles in the cellular milieu.22 Thus, applying this chemistry requires smaller and more stable alkenes for nucleoside functionalization. These challenges present a unique opportunity to explore IEDDA reaction pairs that could be used to form cycloadducts on acceptable timescales, but at the same time are stable enough to provide selectivity.
While the application of vinyl-deoxynucleosides and vinyl-nucleotides has been explored, there has been no investigation of vinyl-nucleosides for RNA metabolic labeling in living cells.23, 24 Furthermore, there has been no analysis of pairing vinyl-nucleosides with optimized tetrazines that balance the required reactivity with stability for labeling.
Herein, we report a holistic analysis of vinyl-nucleoside reactivity with a larger subset of tetrazines. We examine which nucleoside analogs can be incorporated into nascent RNA following addition to cellular media. We then synthesized and characterized optimal tetrazine/vinyl-nucleoside pairs based on kinetics and molecular orbital analyses. Optimized pairs enable tagging and imaging of nascent RNA without compromising RNA integrity. The methods described herein are also orthogonal to conventional CuAAC reactions used for RNA analysis. Overall, this analysis greatly expands our understanding of IEDDA reactions with nucleoside analogs and underscores the utility of biorthogonal methods that are amenable to RNA labeling.
Results
Metabolic Incorporation of vinyl-nucleoside analogs.
Chemical reporter strategies that rely on host cell machinery naturally also depend on the modified analog being a substrate. In many cases, such as for sugars and amino acids, modifications can be placed or sized more liberally; that is, the enzymes in these metabolic pathways accept many configurations.25 However, the design of successful nucleoside analog reporters is more challenging, requiring specific analysis of enzyme structures.
It is reported that mono-phosphate nucleoside kinases utilized in nucleoside salvage pathways are the most selective for their substrates. These enzymes have the highest level of substrate specificity due to very specific active site structures and substrate binding contacts.26 Therefore, to identify positions that could accommodate vinyl functionality, we examined the crystal structures of uridine-cytidine kinase (PDB: 1BX4)27 and adenosine kinase (PDB: 1UEJ)28. Uridine-cytidine kinase revealed an open space at position C5 (Figure 2A). Previous work has demonstrated that 5-ethynyluridine is well tolerated and incorporated into RNA, thus we synthesized 5-vinyluridine (5-VU) and 5-vinylcytidine (5-VC) as possible substrates.24, 29 Adenosine kinase has cavernous pockets about C2, N6 and N7 of the canonical substrate. Alkyl modifications of adenosine at all three of these positions have been previously demonstrated.5, 7 However, we did not wish to perturb the Watson-Crick face of the nucleoside, which is responsible for base pairing, thus we synthesized only 2-vinyladenosine (2-VA) and 7-deazavinyladenosine (7-dVA).30 There have been no successful reports of incorporation of C8-functionalized purine nucleosides, thus we synthesized 8-vinyladenosine (8-VA) and 8-vinylguanosine (8-VG) to test the limits of modification. This brought the final total to six vinyl-modified nucleosides, as shown in Figure 2B.
Figure 2. Initial analysis of vinyl nucleoside incorporation into RNA.
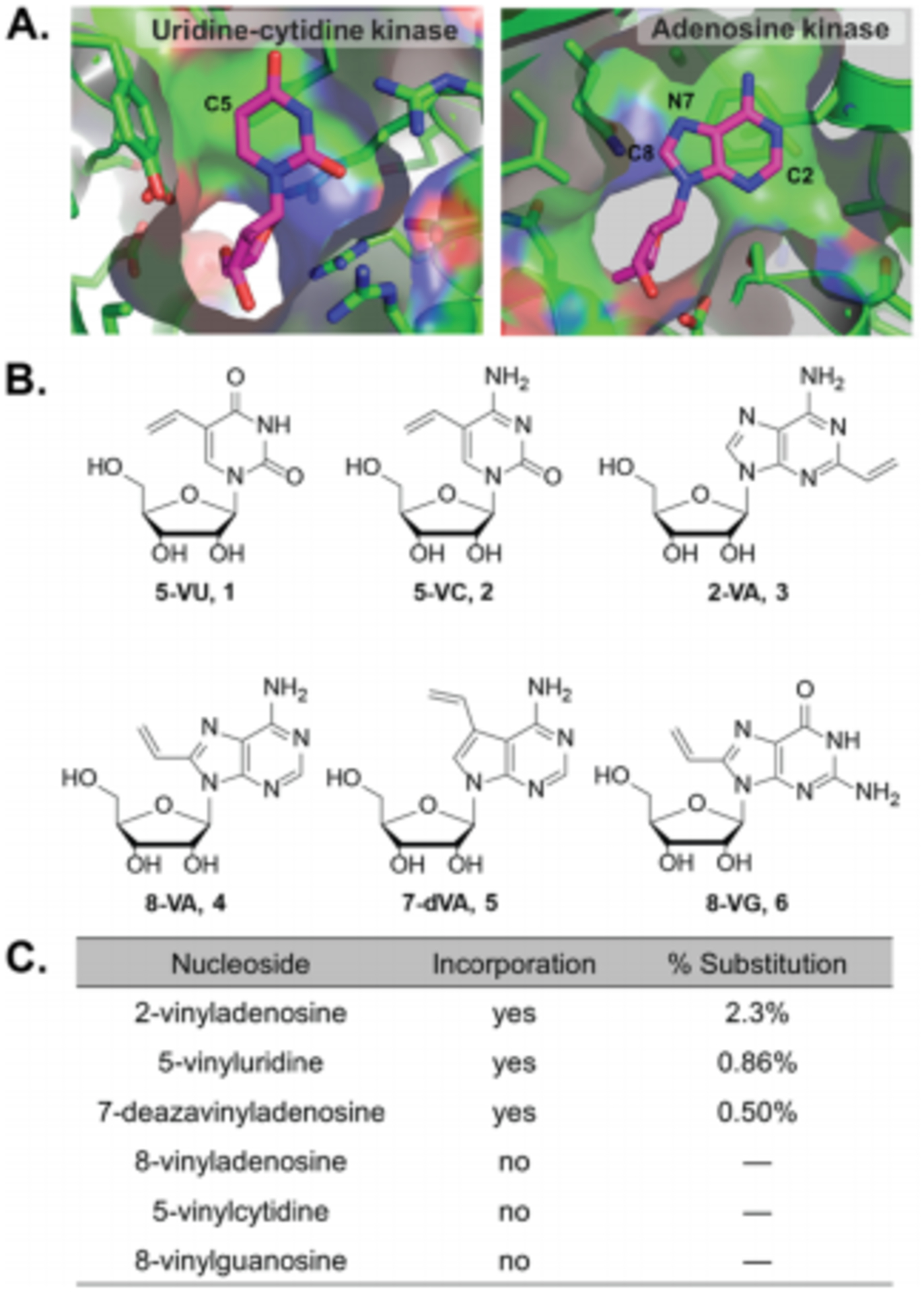
A. Crystal structures of uridine-cytidine kinase and adenosine kinase bound to cytidine and adenosine, respectively. PDB: 1UEI, 1BX4 B. Structures of vinyl-nucleosides C. Incorporation levels of vinyl-nucleosides. HEK293T cells were treated for 12 h at 1 mM final concentration, after which RNA was isolated. Percent incorporation was determined through total RNA digestion and LC-MS/MS analysis of mono-nucleoside mixtures.
To understand the extent of nucleoside substitution, we performed LC-MS/MS to obtain quantitative analysis of percent incorporation for each analog (Table S1). RNA was isolated from cells treated with each analog, followed by digestion to mono-nucleosides. LC-MS/MS analysis showed that 2-VA had the highest incorporation at 2.3% (Figure 2C). 5-VU showed 0.86% incorporation, while 7-dVA showed 0.50% incorporation. These percent incorporation are similar to other metabolic probes, such as 5-ethynyuridine.6 The markedly higher incorporation of 2-VA is likely consistent with other reports that adenosine analogs with small modifications at C2 are highly incorporated into messenger RNA through polyadenylation.8
We were unable to detect incorporation of 8-VA, 8-VG, and 5-VC. Aside from the structure of the kinases, the solution-structure of modified nucleosides could be a reason for lack of incorporation. Two of the analogs (8-VA and 8-VG) may adopt a primarily syn conformation about their N9-C1 glycosidic bond. Analysis of other C8 modified nucleosides has demonstrated that modifications at this position can dramatically alter syn-anti ratios of purine nucleosides.31, 32 Therefore, these analogs may not be suitable substrates for initial kinase steps. A greater surprise was the lack of 5-VC incorporation. LC-MS analysis of the 5-VC incubated with Trizol reagent revealed 5-VC was completely degraded, and the Michael adduct with thiocyanate is observed (Figure S1). Therefore, we believe 5-VC is susceptible to other Michael donors in the cellular environment.
Our initial analysis provides an extended framework for understanding the design and successful incorporation of vinyl-modified nucleosides in cells. Furthermore, ascertaining the limitations of modification placement and size will afford greater insights into the potential design of completely orthogonal, nucleoside-enzyme systems.
Kinetic Analysis of vinyl-nucleosides and tetrazines.
Having established the structures of vinyl nucleosides tolerated by the cellular machinery, we turned our attention to identifying and characterizing paired tetrazines for IEDDA reactions. Acceptable chemical structures and functionalities of nucleoside analogs are dictated by host-cell enzymes in the metabolic pathways. As such, modifying them to achieve faster IEDDA rates through HOMO-LUMO gap narrowing can be challenging because terminal alkenes, the least reactive dienophiles, were installed. Therefore, we reasoned that modifying the structure of tetrazines with electron withdrawing groups would lower the LUMO and thereby narrow the HOMO-LUMO energy gap. After characterization of IEDDA kinetics, we can identify nucleoside-tetrazine pairs.
We worked toward the synthesis of six tetrazines (Figure 3A), each of them having unique structures that could systematically lower the LUMO+1 orbital energy. Tz-1, Tz-3, Tz-5, and DP-Tz have been previously synthesized for use in IEDDA reactions with dienophile-bearing amino acids in proteins.33–36 Tz-2 and Tz-4 were pursued due to the examination of the LUMO+1 orbital energies calculated with B3LYP, 6–31+G* (Figure S2). From these calculations, Tz-2 showed a similar but slightly lower LUMO+1 orbital energy than Tz-1. Structurally, Tz-1 and Tz-2 differ in placement of the nitrogen of the pyridine ring, which is ortho- or meta- to the tetrazine, respectively. Overall, we surmised this broad range of LUMO+1 orbital energies and structures would allow us to identify tetrazine-nucleoside pairs for IEDDA reactions.
Figure 3. Screening tetrazines for IEDDA reactions with vinyl nucleosides.
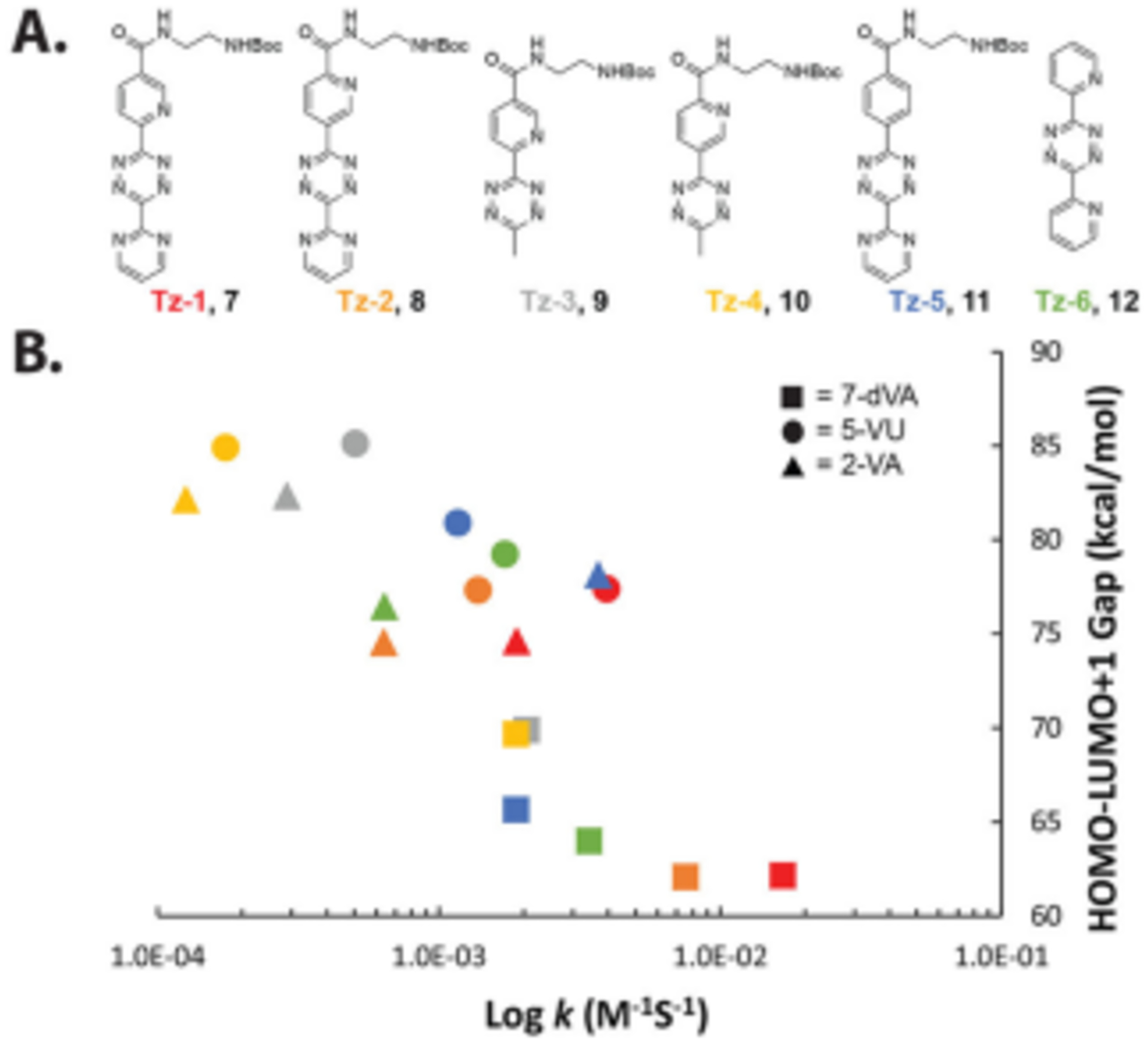
A. Structures of tetrazines synthesized for this study. B. Comparison between HOMO-LUMO+1 energy gap and IEDDA rate constants. Second- order rate constants were determined from reactions containing 1 mM tetrazine and 10, 12.5, 15, 17.5 and 20 mM vinyl nucleoside or nucleobase.
Kinetic analysis was performed by measuring the change in absorbance of tetrazine over time. The reactions exhibited a color change of red-to-yellow, which was used to determine second-order rate constants through the initial rates method (Figure S3).37 The cycloadduct and the oxidized tetrazine products were also observed by LC-MS0 (Figure S4). As expected, addition of a polar solvent (water) resulted in faster reaction kinetics. Rates of these reactions were found to be between 10−4 – 10−2 M−1s−1. Tz-1 and 7-dVA showed the fastest rate with 1.67 ± 0.68 × 10−2 M−1s−1.
We compared the HOMO-LUMO+1 gap and rate constants directly (Figure 3B). Overall, narrower HOMO-LUMO+1 gaps resulted in faster IEDDA reaction kinetics. Notable outliers were observed with Tz-1 and Tz-2 and their reaction kinetics. The rate of reactions with each vinyl nucleoside showed that Tz-1 were 2 to 10 factors faster than that of Tz-2 despite having similar LUMO+1 energies. (Figure S2).
In an attempt to reconcile these differences, we modeled the transition states between Tz-1 and Tz-2 with 5-VUb and 7-dVA (Figure S5) using an exhaustive conformational search. The computed barrier heights closely agree with the observed trends in rate constants, predicting, for example, that for 5-VUb, Tz-1 is faster than Tz-2 by a factor 10.1, compared to a measured factor of 10 (Figure S5). The HOMO-LUMO+1 gap, on the other hand, predicts the reaction would be faster with Tz-2. The qualitative discrepancy between the HOMO-LUMO+1 gap results and the barrier height results indicates that the frontier orbital picture is incomplete, presumably because it ignores effects of long-range noncovalent interactions. Our calculations suggest that the tetrazine adopts a slightly bent boat conformation, which may be helping to assist in the formation of a transition state to encourage the cycloaddition reaction. Although we observed this structure with many of the tetrazines, we did not use this criterion to distinguish the differences in kinetics at this time. Future work on this topic in our lab will be more focused on distortion interaction models38 and how they could explain key differences.
Overall, we have provided a comprehensive analysis of reaction rates between vinyl nucleosides and tetrazine pairs, which nicely underscores the spectrum of reactivity amongst these reagents. The rates of reaction were measured to be comparable to SPAAC reactions (~10−2 M−1s−1). This result is especially exciting as it suggests that IEDDA reactions could be a sufficient replacement for CuAAC chemistry, and IEDDA could be orthogonal to other biorthogonal chemistry approaches for studying RNA.
Establishing reaction conditions to stabilize tetrazines during IEDDA with modified RNA.
Our kinetic analysis suggested that reactions between Tz-1 and the vinyl-nucleosides would be the most efficient for IEDDA labeling of metabolic RNA. However, we questioned its stability in contexts where it would be used for cell imaging and RNA enrichment. A major challenge to utilizing more reactive tetrazines for IEDDA chemistry is their susceptibility to be hydrolyzed, restricting their utility in biological applications.39 We therefore tested whether each tetrazine would be amenable to conditions used for RNA labeling. Addition of water to the tetrazines resulted in a color shift of red-to-yellow over time, indicating disruption of the extended pi-conjugation. Hydrolysis of all tetrazines was monitored by LC-MS (Figure S4). While there were changes noted for all tetrazines, we found that Tz-1 was the most susceptible to hydrolysis (Figure S6). In light of this finding, we surmised it would not be amenable to RNA tagging reactions, despite its fast IEDDA kinetics. Further attempted syntheses of a biotinylated version failed under multiple conditions due to degradation by nucleophilic addition (data not shown).
We next sought to identify reaction conditions that could reduce hydrolysis of the tetrazines, and thus improve the efficiency. We hypothesized that the rate determining step of the hydrolysis pathway is expulsion of nitrogen after addition of water. With the addition of water being reversible, we thought that the equilibrium of the addition could be shifted through lowering the pH (Figure 4A). Hydrolysis was tested with 10% acetic acid added to reactions of Tz-1 in a DMSO:H2O (1:1) mixture (Figure 4B). A striking reduction of hydrolysis was observed in reactions with the addition of acetic acid (pH 5.0).
Figure 4. Establishing reaction conditions to stabilize tetrazines during IEDDA.
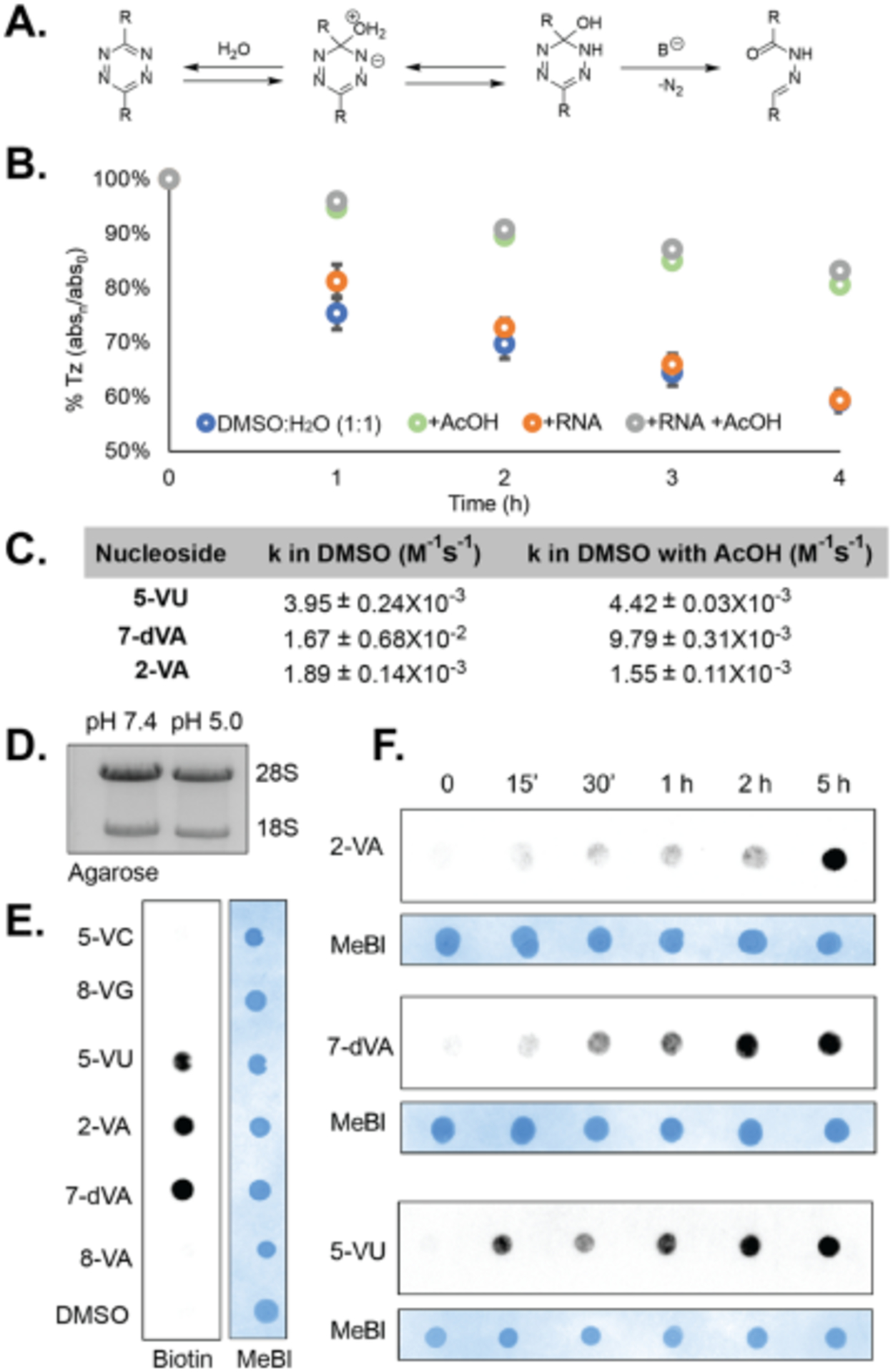
A. Proposed hydrolysis reaction of tetrazines B. Analysis of tetrazine stability in different buffer conditions. C. Comparison of rate constants in DMSO and DMSO with acetic acid. D. Agarose gel demonstrating maintained RNA integrity post IEDDA labeling at lower pH. E. Streptavidin dot blot detection of analogs 2-VA, 7-dVA and 5-VU demonstrates biotinylation through IEDDA. HEK293T cells were treated with each analog for 5 h at 1 mM concentrations. F. Time course analysis of 2-VA, 7-dVA and 5-VU incorporation at times 0 min, 15 min, 30 min, 1 h, 2 h and 5 h treated. At 1 mM concentrations. All IEDDA biotinylation reactions were performed with 50 μM Tz-4 biotin incubated for 2 h at 37 °C.
Kinetic analysis was then performed on reactions of Tz-1 and vinyl-nucleosides and vinyl-nucleobases in the presence of 10% acetic acid (Figure 4C; Figure S2, S3). Interestingly, increased rates are observed in 5-VU and 5-VC under acidic conditions, which may be due to protonation of the nucleobase, thereby lowering the HOMO of the dienophile. These results suggest that lowered pH conditions could provide greater tetrazine stability and in turn, efficiency, while at the same time enabling robust reactivity for labeling.
We then compared RNA integrity and IEDDA biotinylation at pH 7.4 and 5.0. We observed that lowering the pH did not cause spurious RNA degradation, and may indeed be protective to RNA degradation through preventing cleavage of the phosphodiester bonds via the 2’ hydroxyl (Figure 4D). We also note that at pH 5.0, the rate of depurination is ~10−8 M−1s−1,40 which is far slower than the rate of IEDDA, making this a viable method for preserving RNA stability and conjugation.
After comparing reaction kinetics and tetrazine hydrolysis, we decided to pursue synthesis of Tz-4 biotin (Supplementary Information) due to its stability and ease of synthesis. Further, kinetic analyses in Figure 3 demonstrated that this tetrazine exhibited reasonable rate constants with the all nucleosides. We then utilized our modified reaction conditions to perform dot-blot analysis of vinyl nucleoside incorporation (Figure 4E). Nucleosides were incubated in cells for 5 h, after which total RNA was extracted and reacted with Tz-4 biotin to enable detection of nascent transcripts through streptavidin-HRP. Notably, reactions with vinyl-modified RNA were completed in 2 h, in contrast to previous reports of IEDDA reactions taking >24 h for completion.23, 24 Dot blot analysis confirmed that 7-dVA, 2-VA, and 5-VU were incorporated into nascent RNA, with the greatest signal resulting from 7-dVA (Figure S7).
We also analyzed the time-dependence of incorporation through performing a time course of analog treatment followed by dot blot analysis (Figure 4F; Figure S7). Notably, biotinylation signal was visible after only 15 min, and the greatest incorporation was noted at 5 h. While 2-VA was the most highly incorporated according to the LC-MS/MS analysis, it is not the most reactive. We found that 5-VU, which had second highest incorporation, also demonstrated efficient biotinylation and was more easily detected at lower time points.
The addition of acid to IEDDA markedly reduced tetrazine hydrolysis, which serves to improve conjugation. To the best of our knowledge, IEDDA reactions with tetrazines and olefins have typically been performed at neutral pH. As such, the possibility of augmenting IEDDA reactions through lowering the pH offers a unique improvement to contexts where the dienophile is a terminal alkene and the tetrazine is highly reactive. It is also exciting to note that reactions between modified RNA and tetrazines can be performed under conditions that preserve RNA integrity.
Utilizing vinyl nucleosides for in-cell analyses.
We have observed that many of our vinyl nucleosides can be incorporated into RNA. To further test their utility in tracking RNA synthesis, we aimed to establish: (1) vinyl analogs are non-toxic and compare them to other more widely-used analogs for RNA labeling, (2) vinyl analogs can be imaged through IEDDA, and (3) that the IEDDA conditions developed herein are indeed orthogonal to other biorthogonal methods of labeling.
Metabolic labeling of RNA requires that the modified analogs be substrates for a wide variety of enzymes and pathways. As such, there is always the possibility that analogs could exert stress upon cells and cause cytotoxicity or inhibited growth. This is especially concerning when incubating analogs in cells for longer timescales. Furthermore, this limits their use in living animals where it can take several days for analogs to be incorporated at rates suitable for downstream analysis.
Other analogs such as 4-thiouridine41 have been shown to induce significant amounts of toxicity in cells; however, analysis of other conventional analogs has not been extensively explored. MTT assays demonstrated that all incorporated vinyl analogs had no significant effect on cell proliferation after incubations as long as 12 h (Figure S8). We compared this to 5-ethynyluridine (5-EU)6, a well-established, commercially available nucleoside used in metabolic labeling of RNA. Interestingly, we found that it had modest effects on cell proliferation at 12 h, but after 48 h, cell proliferation had decreased by nearly 50% (Figure 5A). Even more striking, a direct comparison with 5-VU at these same conditions showed no significant changes in cell proliferation. Cell viability assays indicated that growth inhibition, and not direct cytotoxicity was the primary effect of extended 5-EU incubations, as the membrane remained intact (Figure S9). This striking difference has similarly been observed with some alkynyl analogs of DNA.42 We also observed changes to cell morphology, including cell detachment and aggregation (Figure 5B), which is consistent with problems in cellular respiration and toxicity.
Figure 5. Effects of vinyl-nucleosides on cell viability and use for RNA imaging.
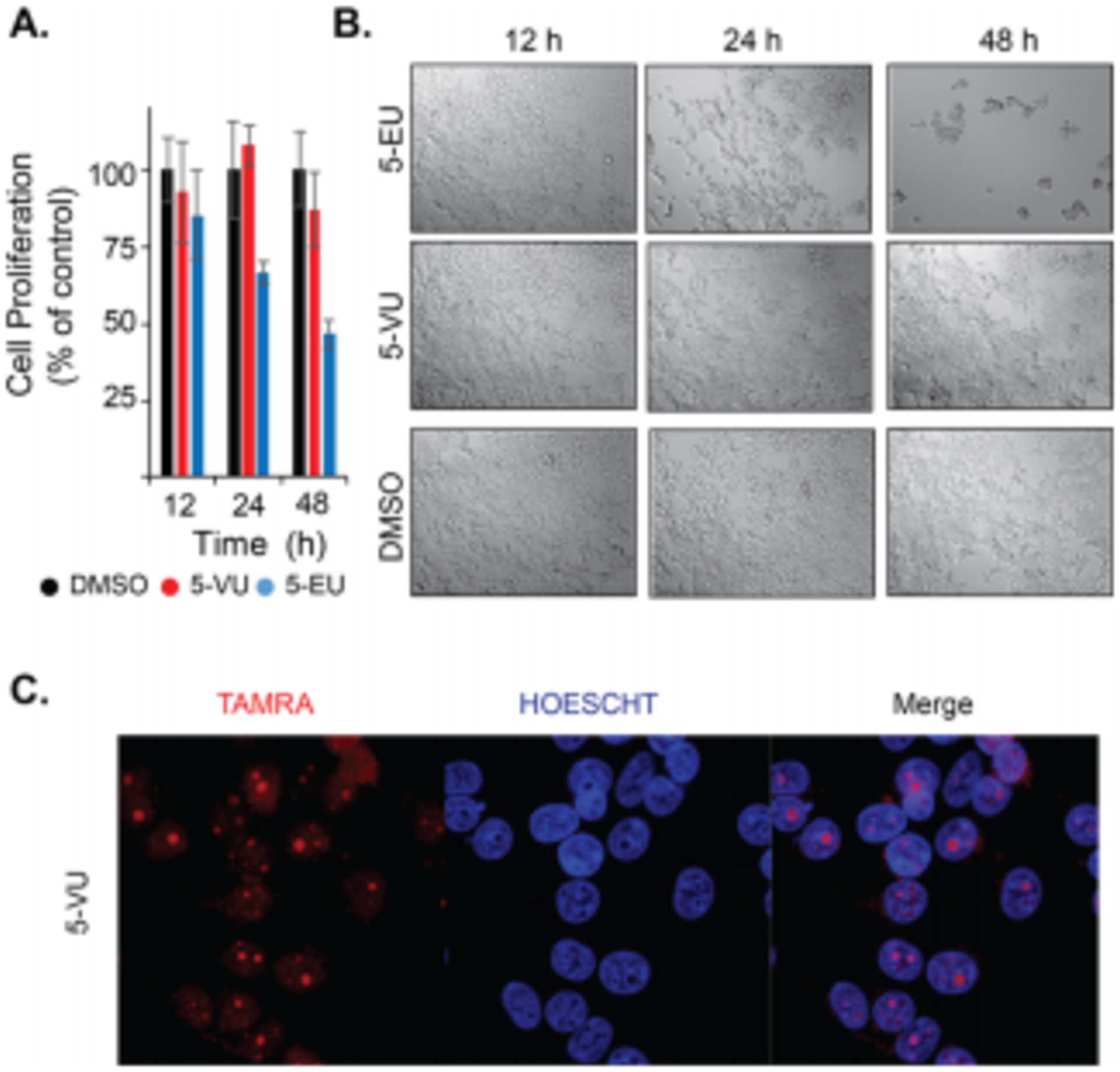
A. MTT assays demonstrate that 5-VU does not have an effect on cell growth, whereas 5-EU produces significant growth inhibition relative to the DMSO-treated control at 1 mM for 12, 24 and 48 h. B. DIC imaging of cells after various incubation lengths confirms the effects on cell growth. C. IEDDA-based imaging demonstrates that 5-VU can be used for visualizing nascent transcription. HEK293T cells were treated with 1 mM 5-VU for 5 h, following by fixation, permeabilization and incubation with Tz-3 TAMRA.
To further understand the effects of 5-EU and 5-VU, we performed RNA sequencing after cells were treated with each analog for 16 hours (Methods). Analysis of differential expression between either analog and DMSO-treated controls revealed that 5-EU labeling resulted in significant differential expression of nearly 200 genes (greater than 2-fold, up or down), while only 18 genes were differentially expressed with 5-VU (Figure S10, A & B). Interestingly, we noticed that differentially-expressed genes preferentially clustered to specific chromosomes (Figure S10, C). It has been recently reported that treatment with other toxic nucleoside analogs can alter RNA processing and expression in the nucleolus (site of ribosomal RNA synthesis).41 We therefore hypothesized that the patterns of gene alteration we observed may be due to certain chromosomes having preferential association with nucleolar domains. To test this, we compared chromosomes known to have preferential association with the nucleolus43 and the chromosomes with the highest altered gene expression. As shown in Figure S10, C, chromosomes that harbor ribosomal DNA and have higher contact with the nucleolus also have greater changes in gene expression. As such, 5-EU may be altering gene expression through a mechanism similar to the more toxic analog 4-thiouridine. These findings, coupled with the toxicity analyses, demonstrate that 5-VU is less toxic and is significantly less perturbative to RNA expression in comparison to 5-EU. Additionally, these results further demonstrate that vinyl analogs are a safer option for in vivo applications, due to the potential for secondary effects of the analog used. This is an incredibly important result for researchers focused on performing RNA labeling experiments in animals, which require longer incubation times.
We next investigated using vinyl-analogs for imaging of nascent transcription. We synthesized a fluorophore-conjugated tetrazine (Tz-3 TAMRA; Supplementary Information). As shown in Figure 5C, robust RNA labeling is observed in the nucleolus for 5-VU, which is the location of rRNA biosynthesis. This pattern has been observed with other analogs that can be used for RNA imaging.6 We saw similar imaging results for 2-VA and 7-dVA, and treatment with actinomycin D, an RNA polymerase I/II inhibitor, significantly diminished nucleolar staining for all three analogs (Figure S11).
Finally, we wanted to test if our IEDDA reactions would be mutually orthogonal to a CuAAC reaction with alkyne-modified RNA. That is, we wished to ascertain that the tetrazine was selective only for the alkene and the azide only for the alkyne. After incubation of 5-VU or 5-EU in cells and isolation of the vinyl- or alkynl-RNA, reactions with biotin-N3 and Tz-4 biotin were carried out. Dot blot analysis showed no cross talk between functional groups (Figure S12). To our knowledge, this is the first example of mutually orthogonal reactions performed on RNA transcripts, offering the ability to perform multiple bioorthogonal reactions in the same experiment for the study of RNA. This approach may be useful on shorter time scale experiments performed in vitro.
Conclusions
Elucidating the utility and limitations of biorthogonal reactions is critical to expanding their use for studying all classes of biomolecules. We have established that vinyl nucleosides can be utilized to metabolically label nascent cellular RNA. Our data demonstrates the successful metabolic incorporation of 2-VA, 7-dVA, and 5-VU, which were intelligently designed through enzyme structural examination. We have also performed an exhaustive evaluation of IEDDA kinetics between all of our synthesized analogs and accompanying tetrazines. Our study highlights the importance of performing these context-dependent analyses to identify optimized reagents and conditions. Using these comparisons, we have identified fast-acting tetrazines that could provide great utility for IEDDA reactions with vinyl analogs of any biomolecule.
Through our studies, we are also the first to report the use of acidic reaction conditions to supplant tetrazine hydrolysis, without severely compromising reaction kinetics or altering RNA integrity. Using our modified protocols, we have demonstrated that vinyl-nucleosides and tetrazines can be used to detect and image nascent transcripts. Vinyl nucleosides do not inhibit cellular proliferation like the most common nucleosides used for in vivo work, such as 4-thiouridine and 5-EU. In addition, vinyl-nucleosides can be used in tandem with alkyne functionalized nucleosides and for multicomponent experiments to study RNA.
We envision these vinyl nucleosides will be used extensively both in vitro and in vivo, as they are easily handled, non-toxic, are able to be functionalized through selective chemistry, and can preserve RNA integrity. Future work in our lab will be to use these exciting new tools to evaluate differences in RNA expression and biology within living systems, such as whole animals.
Materials and Methods
Synthetic Procedures.
All synthetic procedures and characterization of compounds is reported in the supplementary information.
Vinyl-nucleoside TRIzol Reaction Analysis by LC-MS
To analyze the stability of vinyl-nucleosides in Trizol, 5-VU, 5-VC, 7-dVA, and 2-VA were placed in TRIzol and analyzed by LC-MS. 20 μL of 2 mM stock solution of Vinyl-nucleosides in DMSO were added to 80 μL of Trizol (Life Technologies) in a HPLC vials. The mixtures were placed on a shaker at r.t. for 5 h and then analyzed by LC-MS using a C18 column and eluted with a gradient of H2O/MeCN 5–95% with 0.1% formic acid. The mass spectrums were searched for the exact mass of each vinyl-nucleoside and the subsequent thiocyanate addition product.
HOMO-LUMO Energy Calculation
Vinyl-nucleoside HOMO energy and tetrazine LUMO+1 energies were calculated using density functional theory (DFT). Calculations were performed with Spartan 14, using the B3LYP level of theory and the basis set 6–31+G*.
Rate Studies
The reactions between vinyl-nucleosides and tetrazines were monitored by the change in the tetrazine absorbance at 530 nm. Reactions were initiated in a 96-well plate by mixing 100 μL of 2 mM tetrazine solution in DMSO with 100 μL of vinyl-nucleoside in the proposed solvent (DMSO, 1:1 H2O:DMSO, and 1:5 AcOH:DMSO). The concentration of vinyl-nucleoside ranged from 10–20 mM, while tetrazine concentrations were held at 1 mM. All rate studies were performed in triplicate under pseudo-first order conditions. Absorbance measurements were recorded every min over a time period of 3 h time interval using a BioTek Epoch plate reader equipped with Gen5 software. Pseudo-first order rate constants (koba) were calculated by plotting natural log of [tetrazine] versus time (in s). Second-order rate constants were determined by plotting kobs vs nucleoside concentration.
Reaction Analysis by LC-MS
To analyze the product formation from the tetrazine-nucleoside reactions were monitored by LC-MS. 50 μL of tetrazine (1 mM) in DMSO was added to a 96 well plate. 50 μL of nucleoside/nucleobase (1 mM) in DMSO were added to the subsequent well and reacted at r.t. for 72 h. 10 μL of each reaction was diluted with 90 μL and stored in a 0 °C freezer. The reactions were then analyzed by LC-MS using a cyano-column and eluted with a gradient of H2O/MeOH 5–95%.
Structure Generation
Relaxed structures for all vinylnucleotides and tetrazines were obtained through a combination of conformational search and density functional theory (DFT) optimization. First, conformers were generated using the confab44 tool in OpenBabel45. To avoid the incorrect planarization of the tetrazine core produced by all force fields available in OpenBabel (including, for example, MMFF9446 and MMFF94s47), we then performed the subsequent optimization with DFT, using the TPSS48 density functional including D3 dispersion corrections49 and the def2-SVP basis set.50 All DFT calculations were performed using Turbomole.51 The resolution of the identity (RI) approximation was used for the Coulomb integrals with a tight integration grid (m5) and convergence criteria (keywords scfconv = 8, denconv = 10−8). Finally, the lowest energy structure obtained from the previous step was further optimized including COSMO52 (ϵ = 80.1) for solvation. Final structures for all stationary points along the reaction path were computed with identical parameters. Vibrational frequencies were computed to confirm that all structures correspond to local minima.
All stationary points (initial complexes and transition states) exhibit many low-frequency large-amplitude vibrational modes. For example, across all initial complexes computed here, the lowest frequency vibrational mode for each initial complex ranged from 6–25 cm−1 and 12–15 vibrational modes had frequencies below 100 cm−1. We follow the approach of Liu et al53 and tame the low-frequency modes by treating all modes below 100 cm−1 as 100 cm−1. Relative rates were estimated using the Arrhenius equation assuming a constant prefactor such that where R is the ideal gas constant and T = 298K.
Reaction Generation
The reaction profiles were determined for the six cycloaddition reactions involving tetrazines Tz-1 or Tz-2 and nucleotides 5-VUb, 7-dVA, and 2-VAb. For each tetrazine-vinylnucleotide pair, 8–16 distinct initial complexes can be formed depending on the relative orientation of each molecule. Transition states (TSs) were computed for each such relative orientation. In total, more than 60 TSs were determined (6 reactions × 8–16 TSs per reaction). In all cases, an asymmetric transition state was found in which the bond involving the primary vinyl carbon forms first, as seen in Figure S5M–R. Initial guesses for TSs were found by first optimizing the geometry while constraining the distance between the primary vinyl carbon and the tetrazine carbon (d1 in Figure S5S) to be 196 pm. All structures were confirmed to be proper TSs by computation of vibrational frequencies.
Tetrazine Hydrolysis Analysis
Analysis of tetrazine hydrolysis was performed by NMR. 10 mg of Tz-1 was added to an NMR tube and dissolved in DMSO-D6 (0.5 mL). An initial 1H NMR was performed to establish a baseline. H2O (0.5 mL) was then added to the NMR tube and vortexed. The NMR tube was then heated to 37 °C for 24 h. 1H NMR was then performed on the sample and hydrolysis products were determined.
Tetrazine Stability with AcOH
Evaluation of the stability of Tetrazine and RNA was performed by monitoring the change of the tetrazine absorbance at 530 nm. Reactions were performed within in a 96-well plate by mixing 300 μL of 2 mM tetrazine solution in DMSO with 300 μL of the proposed solvent (H2O, 20 μg of RNA in H2O, 20 μg of RNA in 20% AcOH in H2O, and in 20% AcOH in H2O). The concentration of tetrazines were held at 1 mM, AcOH concentrations were held at 10%, and RNA were held at 10 μg. All stability studies were performed in triplicate at 23 °C and absorbance measurements were recorded every hover a time period of 4 h time interval using a BioTek Epoch plate reader equipped with Gen5 software.
Vinyl-nucleoside labeling of cellular RNA
HEK293T cells (ATCC) were cultured in DMEM (Corning) supplemented with 10% FBS, 1% penicillin and streptomycin and grown at 37 °C, 5% CO2. Analogs were added to complete culture medium from 200 mM stocks with a final concentration of <1% DMSO.
RNA isolation and biotinylation via IEDDA
After labeling, total cellular RNA was harvested using Trizol Reagent (Invitrogen) following the manufacturer’s instructions or RIPA lysis buffer (150 mM NaCl, 5 mM EDTA pH 8.0, 50 mM Tris, pH 8.0, 1.0 % NP-40, 0.5 % sodium deoxycholate, 0.1% SDS) spiked with DNase Turbo and RNaseOut (Thermo Fisher). Briefly, ~20 million cells were scraped and suspended in 2 mL of lysis buffer, and incubated on ice for 15 min. Lysates were mixed 1:1 with phenol/chloroform pH 5.2 (Fisher) vortexed briefly and spun down at 4 °C to separate the layers. The aqueous layer was then applied to a Zymo RNA clean and concentrator IIC column (Zymo) and total RNA was isolated according to the manufacturer’s instructions, with RNA eluted in 50 μL of nuclease free water. IEDDA reactions were prepared using 10 μg of total RNA in solutions of 6% DMSO:AcOH (pH 5.0) water, and 1 mM Tz-4 biotin to a final volume of 50 μL. Reactions were incubated with at 37 °C for 2 h at 400 RPM. Biotinylation reactions were purified using Zymo RNA clean and concentrator-5 spin column according to the manufacturer instructions, with RNA eluted in 10 μL of nuclease free water.
HRP-streptavidin Dot blotting
Equal amounts of column-purified RNA were spotted onto 2x SSC equilibrated Hybond-N+ membrane (GE Healthcare), allowing spots to dry in between. The membrane was crosslinked using the auto crosslink function at 254 nm on a UV Stratalinker (Stratagene). The membrane was then blocked in a 10% SDS blocking solution followed by incubation with 1:10,000 dilution of high sensitivity streptavidin-HRP (ThermoFisher Scientific) in blocking solution. The membrane was washed x2 in a 1:10 solution of blocking solution and x2 in Tris-saline buffer. It was then incubated in SuperSignal West Pico Chemiluminescent Substrate (ThermoFisher Scientific) according to manufacturer’s instructions and imaged on a ChemiDoc MP imaging system (Bio-Rad) under the chemiluminescence channel. Following imaging, the blot was stained in a 0.04% methylene blue, 0.3 M sodium acetate solution to visualize all RNA as a loading control. The blot was imaged again using the colorimetric channel.
RNA fluorescence imaging via IEDDA
Coverslips in 6-well tissue culture plates were treated with 1x poly-D lysine solution for 3 h at 37 °C. HEK293T cells were seeded at densities of 2.5×105 and grown to ~50% confluency on glass cover slips. Cells were treated with vinyl nucleosides to final concentrations of 1 mM and incubated for 5 h. After labeling, cells were washed twice with DPBS and fixed for 10 min at room temperature with 3.7% paraformaldehyde. Cells were quenched with 50 mM glycine, DPBS for 5 min, and then washed twice more. Cells were permeabilized in 0.5% Triton-X DPBS for 15 min, and then washed x2 with DPBS. Cells were then treated with solutions of 5 μM Tz-3 TAMRA in DPBS (6% DMSO:AcOH (pH 5.0)), and incubated for 3 h at 37 °C in the dark. Cells were washed x2 with 0.25% Triton-X DPSB for 5 min, and then x5 with DPBS for 5 min each on an orbital shaker. Cells were stained with a solution of 1:3000 Hoechst 333242 for 10 min (Thermo Fisher). Coverslips were briefly washed and then mounted using VectaShield mounting medium (Vector Labs). Slides were imaged via fluorescence confocal microscopy using a 63x oil immersion objective on a Leica 700 Carl Zeiss microscope.
RNA fluorescence imaging with Actinomycin D
Cells were grown and treated with analog as previously described, with transcriptional inhibitor actinomycin D (Sigma) being co-administered to a final concentration of 20 μM. Following incubation, cells were washed x3 with DPBS and lightly fixed with 0.5% paraformaldehyde in 0.5% Triton-X DPBS. Cells were then fixed, stained, mounted and imaged as previously described.
MTT Assay
HEK293T cells were seeded at densities of 10,000 cells into a 96-well plate and grown for 1 day. Cells were then incubated for various lengths of time with compounds 2-VA, 7-dVA, 5-VU, and 5-VC at 1 mM final concentration, 0.5% DMSO or 0.5% DMSO only. Following incubation, cell viability was assessed via the cells’ ability to reduce MTT (Life Technologies) to the insoluble formazan salt. Cell media was replaced with unsupplemented RPMI minus phenol red, and cells were incubated for 3 h at 37 °C with 1 mM MTT. Formazan solid was solubilized in DMSO for 30 min at 37 °C and absorbance readings were taken at 540 nm.
Trypan Blue Assay
HEK293T cells were seeded at densities of 1.5 – 2.5×105 into 6 well plates and grown for 18 – 24 h. Cells were then incubated for 12, 24 and 48 h with compounds 5-VU and 5-EU at 1 mM final concentration. Following incubation, cells were trypsinized, removed from the plate and spun down. The supernatant was aspirated and the cells were resuspended in 1 mL of culture medium, after which 10 uL was aliquoted for Trypan staining. To each aliquot, 10 μL of 0.4% Trypan Blue solution (Gibco) was added and 10 μL of this solution was applied to a hemocytometer for assessment of cell viability. Total and dead cell counts was recorded for each of the four corners of the grid, and repeated for biological duplicates of two.
Mass spec analysis of total RNA
Cells were treated with 1 mM analog for 12 h as previously described. After RNA extraction with TRIzol (Thermo Fischer Scientific) and purification with RNA clean & concentrator-5 columns (Zymo), 1 μg total RNA samples containing each analog were digested to nucleosides using a nucleoside digestion mix (New England Biolabs). A calibration curve of standard solutions containing Adenosine, Uridine, Guanosine, Uridine, vinyl-modified nucleoside analogues 5-VU, 5-VC, 7-dVA, 2-VA and 5-iodocytidine as an internal standard were injected on a Waters Micromass Quattro Premier XE LC-MS system equipped with a triple quadrupole (QQQ) mass detector operating in multiple reaction monitoring (MRM) and positive (+ESI) electrospray ionization modes. Liquid chromatography was carried out on a Waters Acuity UPLC equipped with a C18 column (4.6 × 150 mm, 5 μm) with a gradient mobile phase consisting of water (+0.2% acetic acid) : acetonitrile. Digested RNA sample reactions were diluted 1:10 and 10 μL was directly injected. Mass spectrometry data acquisition was recorded in MRM mode and analyzed on Waters MassLynx 4.1 software. Each nucleoside was identified according to the precursor and product ions found through QuanOpt parameters. The percent substitution of each modified nucleoside was determined by dividing the spectral counts of each analog by the total spectral counts for the canonical and modified analog.
RNA sequencing of 5-EU and 5-VU treated cells.
HEK293T cells were treated with either DMSO, 5-EU or 5-VU for 16 h at 1 mM concentrations (DMSO <1% final; biological duplicates). RNA was extracted and subjected to column purification followed by DNase TURBO treatment (Thermo Fisher) according to the manufacturer’s instructions and a second column purification and elution in nuclease free water. RNA was then used to prepare cDNA libraries following the Tru-Seq Stranded Total RNA Illumina workflow, wherein RNA was first ribosome depleted, and the first strand was synthesized using Superscript IV Reverse Transcriptase (Thermo Fisher). Following cDNA amplification, library concentrations were determined through the Qubit DNA High Sensitivity Assay (Thermo Fisher) followed by DNA analysis on the 2100 Bioanalyzer (Agilent). Libraries were pooled and sequenced on a HiSeq 4000 System (Illumina), with 100 nt single reads.
Differential expression analysis
Sequencing reads were quality checked using fastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and pseudoaligned to the GRCh38 human reference transcriptome using kallisto54. Differential expression analysis at the gene level was performed in DESeq2.55
Supplementary Material
Acknowledgements
We thank members of the Spitale lab for their careful reading and critique of the manuscript
Funding
S. Nainar is supported as a Vertex Fellow and NSF BEST-IGERT. M. Kubota is supported as an Amgen Fellow. Spitale lab is supported by startup funds from the University of California, Irvine, and the NIH (1DP2GM119164 R. Spitale) and 5R21MH113062 (R. Spitale). R. Spitale is a Pew Biomedical Scholar. This material (transition state calculations) is based upon work supported by the National Science Foundation (CHE-1800431). S. Parker is an A.O.B. Postdoctoral Fellow.
Footnotes
Supporting Information
The Supporting Information is available free of charge via the internet at http://pubs.acs.org. Supplementary figures, LC-MS, NMR spectra, imaging, HOMO-LUMO diagrams, and differential RNA expression analysis are in the Supporting Information.
References
- 1.Novikova IV, Hennelly SP, Tung CS, and Sanbonmatsu KY (2013) Rise of the RNA machines: exploring the structure of long non-coding RNAs, J Mol Biol 425, 3731–3746. [DOI] [PubMed] [Google Scholar]
- 2.Morris KV, and Mattick JS (2014) The rise of regulatory RNA, Nat Rev Genet 15, 423–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prescher JA, and Bertozzi CR (2005) Chemistry in living systems, Nat Chem Biol 1, 13–21. [DOI] [PubMed] [Google Scholar]
- 4.Miller MR, Robinson KJ, Cleary MD, and Doe CQ (2009) TU-tagging: cell type-specific RNA isolation from intact complex tissues, Nat Methods 6, 439–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng Y, and Beal PA (2016) Synthesis and evaluation of an alkyne-modified ATP analog for enzymatic incorporation into RNA, Bioorg Med Chem Lett 26, 1799–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jao CY, and Salic A (2008) Exploring RNA transcription and turnover in vivo by using click chemistry, Proc Natl Acad Sci U S A 105, 15779–15784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curanovic D, Cohen M, Singh I, Slagle CE, Leslie CS, and Jaffrey SR (2013) Global profiling of stimulus-induced polyadenylation in cells using a poly(A) trap, Nat Chem Biol 9, 671–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nainar S, Beasley S, Fazio M, Kubota M, Dai N, Correa IR Jr., and Spitale RC (2016) Metabolic Incorporation of Azide Functionality into Cellular RNA, Chembiochem 17, 2149–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nainar S, Kubota M, McNitt C, Tran C, Popik VV, and Spitale RC (2017) Temporal Labeling of Nascent RNA Using Photoclick Chemistry in Live Cells, J Am Chem Soc 139, 8090–8093. [DOI] [PubMed] [Google Scholar]
- 10.Duffy EE, Rutenberg-Schoenberg M, Stark CD, Kitchen RR, Gerstein MB, and Simon MD (2015) Tracking Distinct RNA Populations Using Efficient and Reversible Covalent Chemistry, Mol Cell 59, 858–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paredes E, and Das SR (2011) Click chemistry for rapid labeling and ligation of RNA, Chembiochem 12, 125–131. [DOI] [PubMed] [Google Scholar]
- 12.Winz ML, Samanta A, Benzinger D, and Jaschke A (2012) Site-specific terminal and internal labeling of RNA by poly(A) polymerase tailing and copper-catalyzed or copper-free strain-promoted click chemistry, Nucleic Acids Res 40, 78–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng H, Zhang X, and Zhang C (2015) mRIN for direct assessment of genome-wide and gene-specific mRNA integrity from large-scale RNA-sequencing data, Nat Commun 6, 7816–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maeda T, Date A, Watanabe M, Hidaka Y, Iwatani Y, and Takano T (2016) Optimization of Recovery and Analysis of RNA in Sorted Cells in mRNA Quantification After Fluorescence-activated Cell Sorting, Ann Clin Lab Sci 46, 571–577. [PubMed] [Google Scholar]
- 15.Nilsson H, Krawczyk KM, and Johansson ME (2014) High salt buffer improves integrity of RNA after fluorescence-activated cell sorting of intracellular labeled cells, J Biotechnol 192 Pt A, 62–65. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen K, Fazio M, Kubota M, Nainar S, Feng C, Li X, Atwood SX, Bredy TW, and Spitale RC (2017) Cell-Selective Bioorthogonal Metabolic Labeling of RNA, J Am Chem Soc 139, 2148–2151. [DOI] [PubMed] [Google Scholar]
- 17.Wu H, and Devaraj NK (2016) Inverse Electron-Demand Diels-Alder Bioorthogonal Reactions, Top Curr Chem (Cham) 374–396. [DOI] [PubMed] [Google Scholar]
- 18.Oliveira BL, Guo Z, and Bernardes GJL (2017) Inverse electron demand Diels-Alder reactions in chemical biology, Chem Soc Rev 46, 4895–4950. [DOI] [PubMed] [Google Scholar]
- 19.Lang K, Davis L, Wallace S, Mahesh M, Cox DJ, Blackman ML, Fox JM, and Chin JW (2012) Genetic Encoding of bicyclononynes and trans-cyclooctenes for site-specific protein labeling in vitro and in live mammalian cells via rapid fluorogenic Diels-Alder reactions, J Am Chem Soc 134, 10317–10320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patterson DM, Nazarova LA, Xie B, Kamber DN, and Prescher JA (2012) Functionalized cyclopropenes as bioorthogonal chemical reporters, J Am Chem Soc 134, 18638–18643. [DOI] [PubMed] [Google Scholar]
- 21.Lang K, Davis L, Torres-Kolbus J, Chou C, Deiters A, and Chin JW (2012) Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction, Nat Chem 4, 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossin R, van den Bosch SM, Ten Hoeve W, Carvelli M, Versteegen RM, Lub J, and Robillard MS (2013) Highly reactive trans-cyclooctene tags with improved stability for Diels-Alder chemistry in living systems, Bioconjug Chem 24, 1210–1217. [DOI] [PubMed] [Google Scholar]
- 23.Busskamp H, Batroff E, Niederwieser A, Abdel-Rahman OS, Winter RF, Wittmann V, and Marx A (2014) Efficient labelling of enzymatically synthesized vinyl-modified DNA by an inverse-electron-demand Diels-Alder reaction, Chem Commun (Camb) 50, 10827–10829. [DOI] [PubMed] [Google Scholar]
- 24.George JT, and Srivatsan SG (2017) Vinyluridine as a Versatile Chemoselective Handle for the Post-transcriptional Chemical Functionalization of RNA, Bioconjugate Chem 28, 1529–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grammel M, and Hang HC (2013) Chemical reporters for biological discovery, Nat Chem Biol 9, 475–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lutz S, Liu LF, and Liu YC (2009) Engineering Kinases to Phosphorylate Nucleoside Analogs for Antiviral and Cancer Therapy, Chimia 63, 737–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mathews II, Erion MD, and Ealick SE (1998) Structure of human adenosine kinase at 1.5 angstrom resolution, Biochemistry-Us 37, 15607–15620. [DOI] [PubMed] [Google Scholar]
- 28.Suzuki NN, Koizumi K, Fukushima M, Matsuda A, and Inagaki F (2004) Structural basis for the specificity, catalysis, and regulation of human uridine-cytidine kinase, Structure 12, 751–764. [DOI] [PubMed] [Google Scholar]
- 29.Farina V, and Krishnan B (1991) Large Rate Accelerations in the Stille Reaction with Tri-2-Furylphosphine and Triphenylarsine as Palladium Ligands - Mechanistic and Synthetic Implications, J Am Chem Soc 113, 9585–9595. [Google Scholar]
- 30.Hottin A, Betz K, Diederichs K, and Marx A (2017) Structural Basis for the KlenTaq DNA Polymerase Catalysed Incorporation of Alkene-versus Alkyne-Modified Nucleotides, Chem-Eur J 23, 2109–2118. [DOI] [PubMed] [Google Scholar]
- 31.Dudycz L, Stolarski R, Pless R, and Shugar D (1979) H-1-Nmr Study of the Syn-Anti Dynamic Equilibrium in Adenine Nucleosides and Nucleotides with the Aid of Some Synthetic Model Analogs with Fixed Conformations, Z Naturforsch C 34, 359–373. [DOI] [PubMed] [Google Scholar]
- 32.Rosemeyer H, Toth G, Golankiewicz B, Kazimierczuk Z, Bourgeois W, Kretschmer U, Muth HP, and Seela F (1990) Syn Anti-Conformational Analysis of Regular and Modified Nucleosides by 1d H-1 Noe Difference Spectroscopy - a Simple Graphical-Method Based on Conformationally Rigid Molecules, J Org Chem 55, 5784–5790. [Google Scholar]
- 33.Lang K, Davis L, Torres-Kolbus J, Chou CJ, Deiters A, and Chin JW (2012) Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction, Nature Chemistry 4, 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li ZQ, Wang DY, Li L, Pan SJ, Na ZK, Tan CYJ, and Yao SQ (2014) “Minimalist” Cyclopropene-Containing Photo-Cross-Linkers Suitable for Live-Cell Imaging and Affinity-Based Protein Labeling, J Am Chem Soc 136, 9990–9998. [DOI] [PubMed] [Google Scholar]
- 35.Horner S, Uth C, Avrutina O, Frauendorf H, Wiessler M, and Kolmar H (2015) Combination of inverse electron-demand Diels-Alder reaction with highly efficient oxime ligation expands the toolbox of site-selective peptide conjugations (vol 51, pg 11130, 2015), Chem Commun 51, 11727–11727. [DOI] [PubMed] [Google Scholar]
- 36.Yang J, Karver MR, Li WL, Sahu S, and Devaraj NK (2012) Metal-Catalyzed One-Pot Synthesis of Tetrazines Directly from Aliphatic Nitriles and Hydrazine, Angew Chem Int Edit 51, 5222–5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knall AC, Hollauf M, and Slugovc C (2014) Kinetic studies of inverse electron demand Diels-Alder reactions (iEDDA) of norbornenes and 3,6-dipyridin-2-yl-1,2,4,5-tetrazine, Tetrahedron Lett 55, 4763–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bickelhaupt FM, and Houk KN (2017) Analyzing Reaction Rates with the Distortion/Interaction-Activation Strain Model, Angew Chem Int Edit 56, 10070–10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blizzard RJ, Backus DR, Brown W, Bazewicz CG, Li Y, and Mehl RA (2015) Ideal Bioorthogonal Reactions Using A Site-Specifically Encoded Tetrazine Amino Acid, J Am Chem Soc 137, 10044–10047. [DOI] [PubMed] [Google Scholar]
- 40.An R, Jia Y, Wan BH, Zhang YF, Dong P, Li J, and Liang XG (2014) Non-Enzymatic Depurination of Nucleic Acids: Factors and Mechanisms, Plos One 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.(2013) 4-thiouridine inhibits rRNA synthesis and triggers a nucleolar stress response, Nucleus-Austin 4, 422–422. [Google Scholar]
- 42.Triemer T, Messikommer A, Glasauer SMK, Alzeer J, Paulisch MH, and Luedtke NW (2018) Superresolution imaging of individual replication forks reveals unexpected prodrug resistance mechanism, Proc Natl Acad Sci U S A 115, 1366–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nemeth A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Peterfia B, Solovei I, Cremer T, Dopazo J, and Langst G (2010) Initial Genomics of the Human Nucleolus, Plos Genet 6, 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Boyle NM, Vandermeersch T, Flynn CJ, Maguire AR, and Hutchison GR (2011) Confab - Systematic generation of diverse low-energy conformers, J Cheminformatics 3, 8–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O’Boyle NM, Morley C, and Hutchison GR (2008) Pybel: a Python wrapper for the OpenBabel cheminformatics toolkit, Chem Cent J 2, 9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Halgren TA (1996) Merck molecular force field .1. Basis, form, scope, parameterization, and performance of MMFF94, J Comput Chem 17, 490–519. [Google Scholar]
- 47.Halgren TA (1999) MMFF VI. MMFF94s option for energy minimization studies, J Comput Chem 20, 720–729. [DOI] [PubMed] [Google Scholar]
- 48.Tao JM, Perdew JP, Staroverov VN, and Scuseria GE (2003) Climbing the density functional ladder: Nonempirical meta-generalized gradient approximation designed for molecules and solids, Phys Rev Lett 91, 146401–146405. [DOI] [PubMed] [Google Scholar]
- 49.Grimme S, Antony J, Ehrlich S, and Krieg H (2010) A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu, J Chem Phys 132–147. [DOI] [PubMed] [Google Scholar]
- 50.Weigend F, and Ahlrichs R (2005) Balanced basis sets of split valence, triple zeta valence and quadruple zeta valence quality for H to Rn: Design and assessment of accuracy, Phys Chem Chem Phys 7, 3297–3305. [DOI] [PubMed] [Google Scholar]
- 51.Kuhn M, and Weigend F (2013) Implementation of Two-Component Time-Dependent Density Functional Theory in TURBOMOLE, J Chem Theory Comput 9, 5341–5348. [DOI] [PubMed] [Google Scholar]
- 52.Klamt A, and Schuurmann G (1993) Cosmo - a New Approach to Dielectric Screening in Solvents with Explicit Expressions for the Screening Energy and Its Gradient, J Chem Soc Perk T 2, 799–805. [Google Scholar]
- 53.Liu F, Liang Y, and Houk KN (2014) Theoretical Elucidation of the Origins of Substituent and Strain Effects on the Rates of Diels-Alder Reactions of 1,2,4,5-Tetrazines, J Am Chem Soc 136, 11483–11493. [DOI] [PubMed] [Google Scholar]
- 54.Bray NL, Pimentel H, Melsted P, and Pachter L (2016) Near-optimal probabilistic RNA-seq quantification, Nat Biotechnol 34, 525–527. [DOI] [PubMed] [Google Scholar]
- 55.Love MI, Huber W, and Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol 15, 550–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


