Abstract
Background & objectives:
Early case detection is essential to interrupt transmission and to prevent further spread of tuberculosis (TB) in high endemic settings. Nucleic acid amplification tests (NAATs) with visual read-outs are ideal as point-of-care tests. Truenat™ MTB is an indigenous chip-based NAAT for detection of Mycobacterium tuberculosis, which involves extraction of DNA and real-time polymerase chain reaction (PCR) using portable, automated, battery-operated instruments. The current multicentric study was aimed to evaluate Truenat for detection of MTB in sputum samples obtained from patients with presumptive pulmonary TB with reference to culture as gold standard and Xpert as a comparator.
Methods:
The study was conducted at four sites, namely ICMR-National Institute for Research in Tuberculosis, Chennai; All India Institute of Medical Sciences, New Delhi; ICMR-National JALMA Institute for Leprosy and Other Mycobacterial Diseases, Agra; and National Institute of TB and Respiratory Diseases, New Delhi. Patients suspected to have TB were screened for eligibility. Two sputum samples were collected from each patient. Tests included smear, Xpert and Truenat directly from the sputum sample and culture by Lowenstein-Jensen (L-J) medium and MGIT960 from decontaminated pellets. Sample used for Truenat assay was coded. Resolution of Truenat false positives was done using an in-house PCR with TRC4 primers.
Results:
The study enrolled 2419 presumptive TB patients after screening 2465 patients, and 3541 sputum samples were collected from the enrolled patients. Results of 2623 samples were available for analysis. Truenat showed a positivity rate of 48.5 per cent as compared to 37.0 per cent by Xpert. The sensitivities of Truenat and Xpert were was 88.3 and 79.7 per cent, respectively in comparison with culture.
Interpretation & conclusions:
Truenat MTB identified more positives among culture-confirmed samples than Xpert and had higher sensitivity. In addition, other advantageous operational features of Truenat MTB were identified which would be useful in field settings.
Keywords: Battery-operated tuberculosis detection kit, molecular detection, MTB, POC test, rapid detection of MTB, Truenat, tuberculosis
Tuberculosis (TB) control in disease endemic high-burden countries poses a challenge and requires rapid, cost-effective, reliable and user-friendly diagnostics1. Although TB is treatable, the disease burden in India remains worrisome and the mortality rate continues to be high2. The National TB Control Programme under the Government of India has been pushing various agencies to focus their efforts in search of reliable and speedier diagnostic tools. At present, the primary diagnostic tool used in most peripheral settings is sputum smear. However, due to its low and variable sensitivity, almost 50 per cent of cases are missed3. Thus, limitation in diagnosis becomes the source for new infections allowing false-negative cases to spread the disease4. Culture methods including the commercially available fully automated liquid mycobacterial growth indicator tube (MGIT) are complex, slow, require specialized laboratories5 and are still not widely available in high-endemic countries. In addition, the high contamination rate observed in liquid culture poses another challenge6.
Affordable, high-sensitivity nucleic acid amplification (NAA)-based technologies using closed systems could be the answer to improve access to diagnosis in high-burden settings. In addition, technologies requiring minimal infrastructure, especially those that are portable, automated with visual readout of results, could bring testing as near to the patient as possible. Commercial NAA tests (NAATs) such as Xpert MTB/RIF assay (Xpert, Cepheid, USA)7,8,9,10,11 and line probe assay (LPA) (GenoType MTBDRplus, Hain Lifescience, Germany)12,13,14 have been widely studied, and the WHO endorsed these tests for detection of Mycobacterium tuberculosis complex (MTBC) and rifampicin resistance (RR)15,16. Xpert simultaneously detects both MTBC and RR within two hours8 while LPA detects MTBC, RR and resistance to isoniazid (INH) in 48 h14. Both these technologies have been introduced and are being scaled up within the National Tuberculosis Elimination Programme (NTEP) of India17. With the successful implementation of these technologies, many new technologies were developed along similar lines as fast followers that require stringent evaluation for use in India.
The Truenat™ MTB (Molbio Diagnostics, Goa) is a chip-based NAAT developed for detection of M. tuberculosis from sputum samples. The test involves sputum processing using Trueprep-MAG™ (a nanoparticle-based protocol run on a battery-operated device) and real-time polymerase chain reaction (PCR) performed on the Truelab-UNO™ analyzer (handheld battery-operated thermal cycler). The Truenat™ MTB has been evaluated in India in comparison with acid-fast bacillus (AFB) smear, culture and an in-house PCR as a comparator18. The test was reported to have a sensitivity and specificity of 91.1 [confidence interval (CI): 89.1-94.7%] and 100 per cent (CI: 90.0-100.0%), respectively, as compared to the in-house nested PCR with 90.58 per cent (CI: 85.5-94.3%) sensitivity and 91.4 per cent (CI: 76.9-98.2%) specificity18.
The present multicentre study was aimed to evaluate Truenat™ MTB assay for rapid detection of MTB from sputum samples of presumptive TB patients in comparison with culture as a gold standard and Xpert as a comparator and to resolve discrepancies using a comprehensive reference standard.
Material & Methods
The blinded, cross-sectional, multicentre study was conducted at four sites in India [ICMR-National Institute for Research in Tuberculosis (NIRT), Chennai; All India Institute of Medical Sciences (AIIMS), New Delhi; National Institute for Tuberculosis and Respiratory Diseases (NITRD), New Delhi; and National JALMA Institute for Leprosy and Other Mycobacterial Diseases (NJILOMD), Agra]. The study protocol was approved by the individual Institutional Ethics Committees. All four sites were certified by the NTEP to perform culture by solid, liquid and molecular methods and participated in intra-laboratory quality assurance for all methods.
Study design: Sputum samples from presumptive TB patients were collected and subjected to smear, Xpert and Truenat from raw sample followed by culture on solid Lowenstein-Jensen (L-J) medium and MGIT. Performance of Truenat in comparison with culture was analyzed and discrepancy resolution of culture-negative - Truenat-positive samples was done using an established in-house PCR. In the current study, in-house PCR using TRC4 primers was used instead of the widely tested IS6110 primers considering its discriminating limitations with regard to south Indian MTB strain19. Following discrepancy resolution and identification of true positives defined by demonstration of MTB in any test other than culture (smear or Xpert or TRC4 PCR), further analysis to evaluate the ability of Truenat to identify true positives was done using a comprehensive reference standard including all TB diagnostic tests used in the study.
Sample size: Based on an earlier study18, the sample size was calculated to achieve critical performance targets among groups with (i) sensitivity of 99 per cent (CI of 2%) among smear-positive, culture-positive patients requiring 150 patients; (ii) sensitivity of 75 per cent among smear-negative, culture-positive patients and CI of 7.5 per cent requiring 180 patients; and (iii) specificity of 98 per cent among symptomatic non-TB patients and CI of two per cent requiring 250 patients. Hence, the minimum required group size was 700 after adjusting for either 20 per cent refusal or dropout rate. All values were calculated based on the estimation procedure. In order to enrol 700 patients, an estimated 2000 presumptive TB patients were needed to be screened and recruited across the four sites.
Inclusion and exclusion criteria: Adults (≥18 yr) with clinical suspicion of pulmonary TB and persistent productive cough for ≥two weeks were included. Patients who had received one or more doses of anti-TB medication in the 60 days before screening were excluded7.
Sample population: During the study period between December 2014 and April 2017, consecutive adult presumptive pulmonary TB patients were screened at the four centres for eligibility and written informed consent was taken from each participant before sample collection. Less than two per cent of patients refused consent. Two sputum samples of 4 ml each were collected from each patient - spot specimen on day 1 and morning specimen on day 2. Samples were transported to the laboratories and processed on the same day except on holidays when the samples were stored at 4-10°C in the laboratories. Whenever the patient failed to provide a second sample or if the second sample was found to be less than 4 ml, all tests were done with the first sample. A detailed study workflow is presented in Figure 1.
Fig. 1.
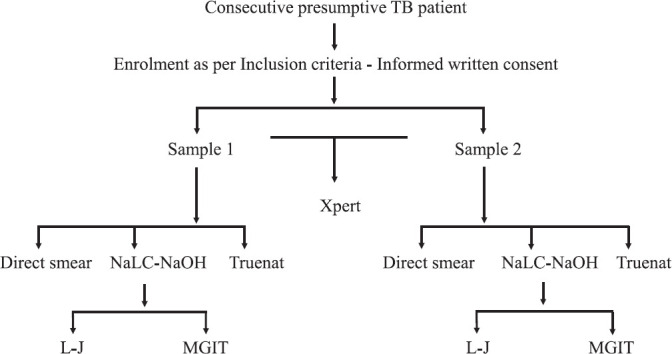
Workflow of the study samples. NaLC-NaOH, N-acetyl-L-cysteine-sodium hydroxide; L-J, Lowenstein-Jensen medium; MGIT, mycobacterial growth indicator tube; ICT, immunochromatography test. All positive cultures confirmed by ICT.
Methods: Sputum samples were homogenized using sterile glass beads. Five hundred microlitres of sputum from each sample from the same patient was pooled and subjected to Xpert testing9. Another 0.5 ml aliquot of the sample was used for Truenat testing. The aliquot for Truenat testing was coded and the technician performing Truenat was blinded to the results from other tests. Briefly, to 0.5 ml of homogenized sputum two drops of liquefaction buffer provided with the Trueprep®Auto Sample Pre-treatment Pack (TSPP) was added, shaken gently and allowed to stand for five minutes with intermittent shaking. The liquefied mixture was transferred to 2.5 ml of lysis buffer from TSPP, shaken vigorously and allowed to stand for 3-5 minutes. The whole content was transferred to the sample chamber of the cartridge provided in the Trueprep®Auto Sputum Sample Prep Kit using a Pasteur pipette. The cartridge was placed in the Trueprep®Auto Sample Prep Device. The device automatically extracted DNA in 25 min which was collected in the DNA elution chamber covered with aluminium foil. The DNA was transferred to a screw-capped cryovial. Six microlitres of the DNA was placed on the Truenat™ MTB micro-PCR chip placed on the chip tray of the Truelab™ Uno (V1.5) analyzer. The test was completed in 35 min and the displayed results were recorded. The remaining DNA was stored at −20°C. Samples that yielded invalid or error results were repeated once again to obtain interpretable results and those which gave similar results on repeat testing were considered invalid/error and were excluded from the analysis.
The remaining sputum sample was subjected to smear by fluorescence microscopy and subsequent decontamination using the standard N-acetyl-L-cysteine-sodium hydroxide (NaLC-NaOH) method20. The resultant pellet was used for inoculating MGIT960 tube (Becton Dickinson, USA) and L-J slopes (in-house preparation) Figure 1 as per the standard protocols. Positive cultures were tested by the standard identification tests including AFB smear, contamination check by inoculation on brain-heart infusion (BHI) agar and confirmation of the presence of MTBC using the MPT64 antigen-based immunochromatographic lateral flow assay21. Cultures that showed growth on BHI agar (HiMedia, Mumbai) were considered as contaminated. Cultures that showed the presence of MTBC along with contamination were decontaminated, and a repeat culture was done in MGIT system22. Once flagged positive by MGIT instrument, all three identification tests were repeated on the culture and final results were based on the second culture. Results from all the tests were compared after excluding invalids/errors/contaminations. Evaluation of the Truenat MTB test was performed in comparison with culture as the gold standard and Xpert as a comparator for detection of M. tuberculosis.
Performance characteristics such as sensitivity, specificity and kappa were calculated using the Statistical Package for the Social Sciences (SPSS) version 25.0 (IBM, Corp., Armonk, NY, USA). P=0.05 was considered statistically significant for all tests.
For resolution of false positives, DNA elution was subjected to an in-house PCR using TRC4 primers. The primers used were TRC4 primers 1 (5'-GACAACGACGTGCGCCTACT-3') and 2 (5'-GACCGAATTAGCGTAGCTCC-3'). The 18-mer primers amplify 173 bp long single target of the repetitive element of TRC4. The fragment is located in the open reading frame of RV0697 in the MTB genome23. The two-step PCR conditions for DNA amplification were an initial denaturation step at 95°C for one minute, followed by 45 cycles of 95°C for 10 seconds and 65°C for 34 seconds. The final product assessed by two per cent gel electrophoresis was a 173 bp fragment24 as shown in Figure 2.
Fig. 2.
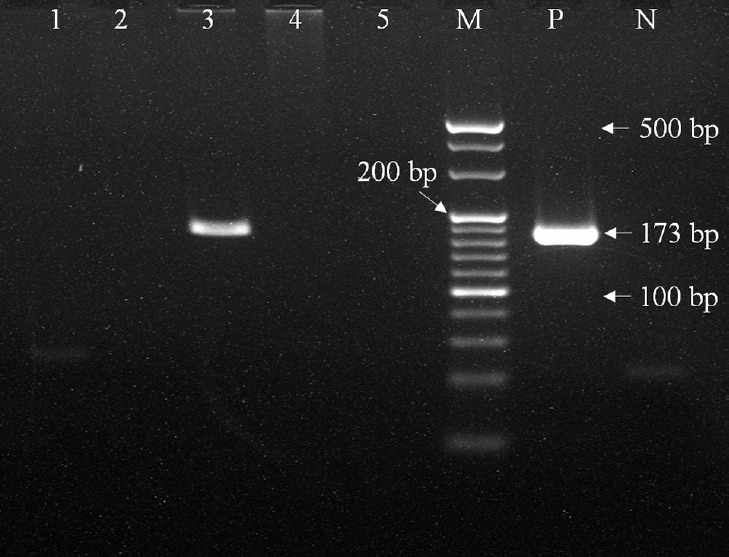
Polymerase chain reaction amplification of 173 bp region in MTB genomic DNA using TRC4 primers. Lanes 1, 2, 4, 5: samples negative for 173 bp product; Lane 3: sample positive for 173 bp product; Lane M: 20 bp molecular weight marker; Lane P: positive control; Lane N: negative control.
Results
The study enrolled 2419 adult presumptive TB patients after screening 2465 patients, and 3541 sputum samples were collected (Table I). Among 3541 samples, valid results for all tests were available for comparison from 2623 samples after elimination of errors, invalids and contaminations and these were considered for analysis. The results for all samples tested and among the analyzable set are presented in Table II.
Table I.
Site-wise distribution of patients enrolled and samples collected
| Site | Patients | Samples |
|---|---|---|
| All India Institute of Medical Sciences, New Delhi | 747 | 747 |
| National JALMA Institute for Leprosy and Other Mycobacterial Diseases, Agra | 550 | 550 |
| ICMR-National Institute for Research in Tuberculosis, Chennai | 572 | 1144 |
| National Institute for Tuberculosis and Respiratory Diseases, New Delhi | 550 | 1100 |
| Total | 2419 | 3541 |
Four sites collectively enrolled 2419 patients including smear positives and smear negatives
Table II.
Distribution of all test results in 2623 samples
| Test | Overall samples (n=3541), n (%) | Samples taken for comparative analysis (n=2623), n (%) | ||||
|---|---|---|---|---|---|---|
| Positive | Negative | Invalid/error/contamination | Positive | Negative | Invalid/error/contamination | |
| Smear | 999 (28.2) | 2542 (71.8) | 0 (0.0) | 777 (29.6) | 1846 (70.4) | 0 (0.0) |
| Culture | 1113 (31.4) | 2317 (65.4) | 111 (3.1) | 941 (35.9) | 1682 (64.1) | 99 (2.8) |
| Xpert@ | 1055 (29.8) | 1904 (53.8) | 32 (0.9) | 972 (37.1) | 1651 (62.9) | 32 (0.9) |
| Truenat | 1590 (44.9) | 1707 (48.2) | 244 (6.9) | 1272 (48.5) | 1351 (51.5) | 237 (6.7) |
@550 samples could not be processed due to non-availability of Xpert cartridges at the time of study and were excluded from the analysis. Truenat yielded maximum positivity of 44.9% among all samples tested and 48.5% among those considered for comparative analysis while the same for Xpert was 29.8 and 37.1%, respectively
Among the target groups, the sensitivity of Truenat for identifying MTBC was 92.9 per cent among smear- and culture-positive samples and 75 per cent among smear-negative culture-positive samples in comparison with 86 and 60.6 per cent, respectively, by Xpert. The differences in sensitivities between the two tests in these target groups were significant (Table III). Among the 80 smear-positive culture-negative samples, Truenat detected MTBC in 65 (81.2%) while Xpert was positive in 62 (77.5%), however, the difference was not significant. Truenat and Xpert were also positive in 376 (23.4%) and 160 (9.98%) samples, respectively, among 1602 smear- and culture-negative samples (Table III). Direct comparison with Xpert showed that among 972 samples that were positive by Xpert, Truenat was positive for 859 (88.3%), and among 1651 samples that were Xpert negative, 1238 (75%) were negative by Truenat.
Table III.
Performance of molecular tests among the target groups
| Target group | Total | TN positive (%) | Xpert positive (%) | P |
|---|---|---|---|---|
| Sm+ Cult+ | 697 | 648 (92.9) | 602 (86) | <0.001 |
| Sm− Cult+ | 244 | 183 (75) | 148 (60.6) | <0.001 |
| Sm+ Cult− | 80 | 65 (81.2) | 62 (77.5) | 0.702 |
| Sm− Cult− | 1602 | 376 (23.4) | 160 (9.98) | <0.001 |
TN: Truenat; Sm+ Cult+, smear-positive/culture-positive samples; Sm− Cult+, smear-negative/culture-positive samples; Sm+ Cult−, smear-positive/culture-negative samples; Sm−Cult−, smear-negative/culture-negative samples. MTBC, Mycobacterium tuberculosis complex
Concordance analysis of Truenat and Xpert with culture yielded an overall sensitivity of 88.3 (95% CI: 86.1-90%) and 79.7 per cent (95% CI: 77-82.2%) for Truenat and Xpert, respectively (Table IV). The overall specificities of Truenat and Xpert were 73.8 per cent (95% CI: 71.6-75.9%) and 86.8 per cent (95% CI: 85.1-88.4%). Analysis of smear-positive and smear-negative samples independently yielded similar patterns of higher sensitivity for Truenat and higher specificity for Xpert in both types of samples Tables V and VI.
Table IV.
Performances of Truenat and Xpert in comparison with culture
| Truenat | Culture | Total | Xpert | Culture | Total | ||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||||
| Positive | 831 | 441 | 1272 | Positive | 750 | 222 | 972 |
| Negative | 110 | 1241 | 1351 | Negative | 191 | 1460 | 1651 |
| Total | 941 | 1682 | 2623 | Total | 941 | 1682 | 2623 |
| Sensitivity: 88.3% (86.1-90.3%) | Sensitivity: 79.7% (77.0-82.2%) | ||||||
| Specificity: 73.8% (71.6-75.9%) | Specificity: 86.8% (85.1-88.4%) | ||||||
Among all samples tested, Truenat demonstrated higher sensitivity than Xpert and difference was significant (P<0.001). Xpert showed significantly higher specificity (P<0.001) than Truenat
Table V.
Performances of Truenat and Xpert in comparison with culture among smear-positive samples
| Truenat | Culture | Total | Xpert | Culture | Total | ||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||||
| Positive | 648 | 65 | 713 | Positive | 602 | 62 | 664 |
| Negative | 49 | 15 | 64 | Negative | 95 | 18 | 113 |
| Total | 697 | 80 | 777 | Total | 697 | 80 | 777 |
| Sensitivity: 93% (90.8-94.8%) | Sensitivity: 86.4% (83.6-88.8%) | ||||||
| Specificity: 18.8% (10.9-29%) | Specificity: 22.5% (13.9-33.2%) | ||||||
Sensitivities of Truenat and Xpert for detection of MTB among smear-positive samples in comparison with culture were 93 and 86.4%, respectively. The difference in sensitivity was significant (P<0.001) while specificities were 18.8 and 22.5%, and the difference was not significant
Table VI.
Performances of Truenat and Xpert in comparison with culture among smear-negative samples
| Truenat | Culture | Total | Xpert | Culture | Total | ||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||||
| Positive | 183 | 376 | 559 | Positive | 148 | 160 | 308 |
| Negative | 61 | 1226 | 1287 | Negative | 96 | 1442 | 1538 |
| Total | 244 | 1602 | 1846 | Total | 244 | 1602 | 1846 |
| Sensitivity: 75% (69.1-80.3%) | Sensitivity: 60.7% (54.2-66.8%) | ||||||
| Specificity: 76.5% (74.4-78.6%) | Specificity: 90.0% (88.4-91.4%) | ||||||
Sensitivity of Truenat and Xpert in detecting MTB among smear-negative samples in comparison with culture was 75 and 60.7% while specificities were 76.5 and 90.0%, respectively. The differences in sensitivity and specificity between Truenat and Xpert were significant (P<0.001)
Among all 441 samples that were culture-negative Truenat positive, 311 samples showed evidence of TB by other tests (smear +ve, 18; Xpert +ve, 160; smear and Xpert +ve, 62 and PCR (TRC4 primer) +ve, 71) and were considered true MTB positives. The ability of Truenat to identify true positives in comparison with a comprehensive reference standard (CRS) including smear, culture, Xpert and TRC4 PCR showed an increase in sensitivity and specificity to 91.2 and 90.5 per cent, respectively (Table VII).
Table VII.
Performances of Truenat in comparison with comprehensive reference standard
| Truenat | CRS | Total | |
|---|---|---|---|
| Positive | Negative | ||
| Positive | 1142 | 130 | 1272 |
| Negative | 110 | 1241 | 1351 |
| Total | 1252 | 1371 | 2623 |
| Sensitivity: 91.2% (89.5-92.7%) | |||
| Specificity: 90.5% (88.8-92.0%) | |||
CRS: comprehensive reference standard included smear, culture, Xpert and TRC4 PCR
Discussion
Truenat was evaluated earlier in India by Hinduja Hospitals, Mumbai. Sputum samples from 226 presumptive TB patients were tested by smear, culture, Truenat and an in-house PCR. A CRS including smear, culture, clinical findings and response to treatment was used to evaluate Truenat with the in-house PCR as a comparator. Truenat demonstrated a sensitivity of 91.1 per cent (CI: 86.1-94.7%) and a specificity of 100 per cent (CI: 90.0-100%) in comparison with the CRS18. The current evaluation on a larger sample size showed similar findings with Truenat yielding a sensitivity of 91.2 per cent and a specificity of 90.5 per cent in comparison with a CRS including a battery of diagnostic tests for TB. Direct comparison of Truenat with the comparator Xpert in the current study yielded a specificity of 75 per cent similar to the earlier study18 where, in comparison with the in-house PCR, Truenat yielded a specificity of 72 per cent.
Truenat demonstrated the highest overall positivity of 44.9 per cent among all the tests performed. The overall invalid rate of Truenat was 6.9 per cent in comparison with 0.9 per cent with Xpert. This finding was similar to the early error rates reported for Xpert25 following implementation.
In the present study, the higher sensitivity of Truenat in comparison with culture among the target groups demonstrated the assay's capability to detect higher number of true positives. In comparison with culture, the assay achieved a sensitivity of 92.9 per cent as against the expected 99 per cent among smear-positive culture-positive samples, while among smear-negative culture-positive samples, the expected sensitivity of 75 per cent was achieved. These values were significantly higher than those of Xpert. However, Xpert showed a higher specificity of 90 per cent among the target group smear-negative culture-negative than Truenat (76.5%) while the overall specificities of Xpert and Truenat were 87 and 74 per cent, respectively. The ideal way to eliminate false positivity by a TB diagnostic test is to demonstrate tubercle bacilli or its component in the same sample by other standard tests or by looking for clinical evidence for TB in the patient including response to treatment. Following discrepancy resolution, performance of Truenat was compared to a comprehensive reference standard including all available TB diagnostic tests such as smear, culture, Xpert and in-house TRC4 PCR as done in an earlier work by Nikam et al18. The sensitivity and specificity of Truenat increased to 91.2 and 90.5 per cent, respectively. Truenat failed to identify 11.6 per cent samples that were positive by culture, but it was less than those missed by Xpert (20.2%).
One key limitation of the study was the use of an in-house PCR for discrepancy resolution instead of sequencing. This was unavoidable as the only sample available for any further investigation was the DNA from the Truenat assay in the case of Truenat-positive/culture-negative samples.
The present multicentre study helped identify the operational advantages of Truenat. The assay required minimal training as technicians with minimal or nil exposure to molecular tests could perform the assay following a short training session. Truenat instruments were battery operated and temperature stable thereby demonstrating the robustness of the instruments while Xpert required continuous power supply and air-conditioning26. No instrument failures were reported from any of the sites during the study period. Functional remote monitoring of the devices was another notable feature. Another study27 has shown Truenat to be the most cost-effective test in a point-of-care setting.
In conclusion, the study demonstrated that the precision of detecting MTB was higher with Truenat as compared with Xpert. The study also identified other parameters of the assay that may prove more advantageous over Xpert. Demonstration of these features through feasibility studies at peripheral field settings under routine NTEP conditions will be required for inclusion of the assay in the diagnostic algorithm of the national programme.
Acknowledgment:
Authors acknowledge the Department of Biotechnology, India, for the funding the study and being a constant support for smooth conduct and completion of the study; Central TB Division for constant support and guidance in developing the protocol and approval to enrol patients attending NTEP clinics; State TB Officers and District TB officers of Tamil Nadu, Uttar Pradesh and New Delhi for providing support and coordinating with the patients; Scientific Advisory and Ethics Committees of the institutes in guiding the study to completion; and staff of all the reference laboratories for their cooperation, participation and technical assistance in the conduct of the study.
Footnotes
Financial support & sponsorship: The study was financially supported by the Department of Biotechnology, Government of India (BT/PR11915/MED/01/Diagnostics/2010, dt.: 27.06.2014).
Conflicts of Interest: None.
References
- 1.Parsons LM, Somoskövi A, Gutierrez C, Lee E, Paramasivan CN, Abimiku A, et al. Laboratory diagnosis of tuberculosis in resource-poor countries: challenges and opportunities. Clin Microbiol Rev. 2011;24:314–50. doi: 10.1128/CMR.00059-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organization. Global Tuberculosis Report 2019. Geneva: WHO; 2019. [Google Scholar]
- 3.Reid MJ, Shah NS. Approaches to tuberculosis screening and diagnosis in people with HIV in resource-limited settings. Lancet Infect Dis. 2009;9:173–84. doi: 10.1016/S1473-3099(09)70043-X. [DOI] [PubMed] [Google Scholar]
- 4.Sreeramareddy CT, Qin ZZ, Satyanarayana S, Subbaraman R, Pai M. Delays in diagnosis and treatment of pulmonary tuberculosis in India: a systematic review. Int J Tuberc Lung Dis. 2014;18:255–66. doi: 10.5588/ijtld.13.0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. Use of liquid TB culture and drug susceptibility testing (DST) in low and medium income settings. Summary report of the Expert Group Meeting on the use of liquid culture media. Geneva: WHO; 2007. [Google Scholar]
- 6.World Health Organization. Implementing tuberculosis diagnostics: A policy framework. Geneva: WHO; 2015. [Google Scholar]
- 7.Catharina CB, Pamela N, Doris H, Nicol MP, Shubhada S, Fiorella K, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363:1005–15. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott LE, McCarthy K, Gous N, Nduna M, Van Rie A, Sanne I, et al. Comparison of Xpert MTB/RIF with other nucleic acid technologies for diagnosing pulmonary tuberculosis in a high HIV prevalence setting: a prospective study. PLoS Med. 2011;8:e1001061. doi: 10.1371/journal.pmed.1001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steingart KR, Schiller I, Horne DJ, Pai M, Boehme CC, Dendukuri N. Xpert® MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database Syst Rev. 2014;21:CD009593. doi: 10.1002/14651858.CD009593.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reddy R, Alvarez-Uria G. Molecular epidemiology of rifampicin resistance in Mycobacterium tuberculosis using the GeneXpert MTB/RIF assay from a rural setting in India. J Pathog. 2017;2017:6738095. doi: 10.1155/2017/6738095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramachandran R, Muniyandi M. Rapid molecular diagnostics for multi-drug resistant tuberculosis in India. Expert Rev Anti Infect Ther. 2018;16:197–204. doi: 10.1080/14787210.2018.1438262. [DOI] [PubMed] [Google Scholar]
- 12.Hillemann D, Gerdes SR, Richter E. Evaluation of the GenoTypeMTBDRplus assay for rifampin and Isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol. 2007;45:2635–40. doi: 10.1128/JCM.00521-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singhal R, Arora J, Lal P, Bhalla M, Myneeedu VP, Behera D. Comparison of line probe assay with liquid culture for rapid detection of multi-drug resistance in Mycobacterium tuberculosis. Indian J Med Res. 2012;136:1044–7. [PMC free article] [PubMed] [Google Scholar]
- 14.Yadav RN, Singh BK, Sharma SK, Sharma R, Soneja M, Sreenivas V, et al. Comparative Evaluation of GenoTypeMTBDRplus line probe assay with solid culture method in early diagnosis of multidrug resistant tuberculosis (MDR-TB) at a tertiary care centre in India. PLoS One. 2013;8:e72036. doi: 10.1371/journal.pone.0072036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.World Health Organization. Policy Statement: Molecular line probe assays for rapid screening of patients at risk of multidrug resistant tuberculosis (MDR-TB) Geneva: WHO; 2008. [Google Scholar]
- 16.World Health Organization. Policy Statement: Automated real-time nucleic acid amplification technology for rapid and simultaneous detection of tuberculosis and rifampicin resistance: Xpert MTB/RIF system. Geneva: WHO; 2011. [PubMed] [Google Scholar]
- 17.Central TB Division, Directorate General of Health Services, Ministry of Health & Family Welfare, Government of India. Guidelines on Programmatic Management of Drug Resistant TB in India. [accessed on November 23, 2019]. Available from: http://www.tbcindia.nic.in/showfile.php?lid=3155 .
- 18.Nikam C, Jagannath M, Narayanan MM, Ramanabhiraman V, Kazi M, Shetty A, et al. Rapid diagnosis of Mycobacterium tuberculosis with TrueNAT MTB: a near-care approach. PLoS One. 2013;8:e51121. doi: 10.1371/journal.pone.0051121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das S, Paramasivan CN, Lowrie DB, Prabhakar R, Narayanan PR. IS6110 restriction fragment length polymorphism typing of clinical isolates of Mycobacterium tuberculosis from patients with pulmonary tuberculosis in Madras, south India. Tuberc Lung Dis. 1995;76:550–4. doi: 10.1016/0962-8479(95)90533-2. [DOI] [PubMed] [Google Scholar]
- 20.Global Laboratory Initiative. Mycobacteriology Laboratory Manual. [accessed on November 23, 2019]. Available from: https://www.who.int/tb/laboratory/mycobacteriology-laboratory-manual.pdf .
- 21.Gomathi NS, Devi SM, Lakshmi R, Ramachandran R, Wares DF, Kumar V, et al. Capilia test for identification of Mycobacterium tuberculosis in MGITTM positive cultures. Int J Tuberc Lung Dis. 2012;16:788–92. doi: 10.5588/ijtld.11.0356. [DOI] [PubMed] [Google Scholar]
- 22.Siddiqi S, Rusch-Gerdes S. MGITTM Procedure Manual for BACTECTM MGIT 960TM TB System: Mycobacteria Growth Indicator Tube Culture and Drug Susceptibility Demonstration Projects. Geneva: FIND; 2006. pp. 1–52. [Google Scholar]
- 23.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–44. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 24.Narayanan S, Parandaman V, Narayanan P, Venkatesan P, Girish C, Mahadevan S, et al. Evaluation of PCR using TRC4 and IS6110 primers in detection of tuberculous meningitis. J Clin Microbiol. 2001;39:2006–8. doi: 10.1128/JCM.39.5.2006-2008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foundation for Innovative New Diagnostics. Performance of Xpert MTB/RIF version G4 assay. Geneva, Switzerland: Foundation for Innovative New Diagnostics; 2011. [Google Scholar]
- 26.Creswell J, Codlin AJ, Andre E, Micek MA, Bedru A, Cater EJ, et al. Results from early programmatic implementation of Xpert MTB/RIF testing in nine countries. BMC Infect Dis. 2014;14:2. doi: 10.1186/1471-2334-14-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee DJ, Kumarasamy N, Resch SC, Sivaramakrishnan GN, Mayer KH, Tripathy S, et al. Rapid, point-of-care diagnosis of tuberculosis with novel truenat assay: cost-effectiveness analysis for India's public sector. PLoS One. 2019;14:e0218890. doi: 10.1371/journal.pone.0218890. [DOI] [PMC free article] [PubMed] [Google Scholar]


