Keywords: γ-tubulin, microtubules, γ-TuRC
Abstract
Cryo-electron microscopy recently resolved the structure of the vertebrate γ-tubulin ring complex (γ-TuRC) purified from Xenopus laevis egg extract and human cells to near-atomic resolution. These studies clarified the arrangement and stoichiometry of γ-TuRC components and revealed that one molecule of actin and the small protein MZT1 are embedded into the complex. Based on this structural census of γ-TuRC core components, we developed a recombinant expression system for the reconstitution and purification of human γ-TuRC from insect cells. The recombinant γ-TuRC recapitulates the structure of purified native γ-TuRC and has similar functional properties in terms of microtubule nucleation and minus end capping. This recombinant system is a central step towards deciphering the activation mechanisms of the γ-TuRC and the function of individual γ-TuRC core components.
1. Introduction
Microtubules are essential cytoskeletal components with multiple important functions in chromosome segregation, cellular transport, cell organization and cell motility. Microtubules are polymers of the heterodimer αβ-tubulin that dynamically assemble via lateral and longitudinal subunit interactions into a cylindric structure composed of 13 αβ-tubulin protofilaments [1]. Formation of the initial αβ-tubulin oligomers is the rate-limiting step in the de novo assembly of microtubules [2]. In cells, complexes containing the tubulin family member γ-tubulin facilitate this step in a controlled manner and thus determine the sites of microtubule formation [3,4].
In higher eukaryotes, nucleation of microtubules is mediated by the γ-tubulin ring complex (γ-TuRC) [5]. High-resolution structural information on the γ-TuRC was limited to a few isolated components [6,7], until recent advances in cryo-electron microscopy (cryo-EM) facilitated resolving the structure of purified vertebrate γ-TuRCs to near–atomic resolution [8–10]. These analyses showed that the γ-TuRC contains γ-tubulin, five paralogous gamma-ring complex proteins (GCPs, GCP2 to 6), mitotic spindle organizing protein 1 (MZT1), MZT2 and actin in a defined stoichiometry [8–11]. The γ-TuRC comprises 14 asymmetrically arranged spokes (pairs of GCP and γ-tubulin; figure 1a) with the order (GCP2-3)4-GCP4-GCP5-GCP4-GCP6-(GCP2-3)1. The GCP proteins interact with each other via their N-terminal gamma-tubulin ring protein 1 (GRIP1) domains, while their C-terminal GRIP2 domains bind one molecule of γ-tubulin each. A prominent feature of the γ-TuRC is a structural scaffold in the lumen of the complex, formed by two copies of MZT1, actin and the N-termini of GCP6 and one copy of GCP3. This ‘luminal bridge' follows the inner surface of the spiral on the level of the GRIP1 domains from spokes 2 to 11 [12]. Further proteins that have been established to cooperate and co-purify with the γ-TuRC in a defined stoichiometry, such as NEDD1 and NME7 [13,14], could not be detected in the cryo-EM structures, probably because these proteins are flexibly associated [8].
Figure 1.
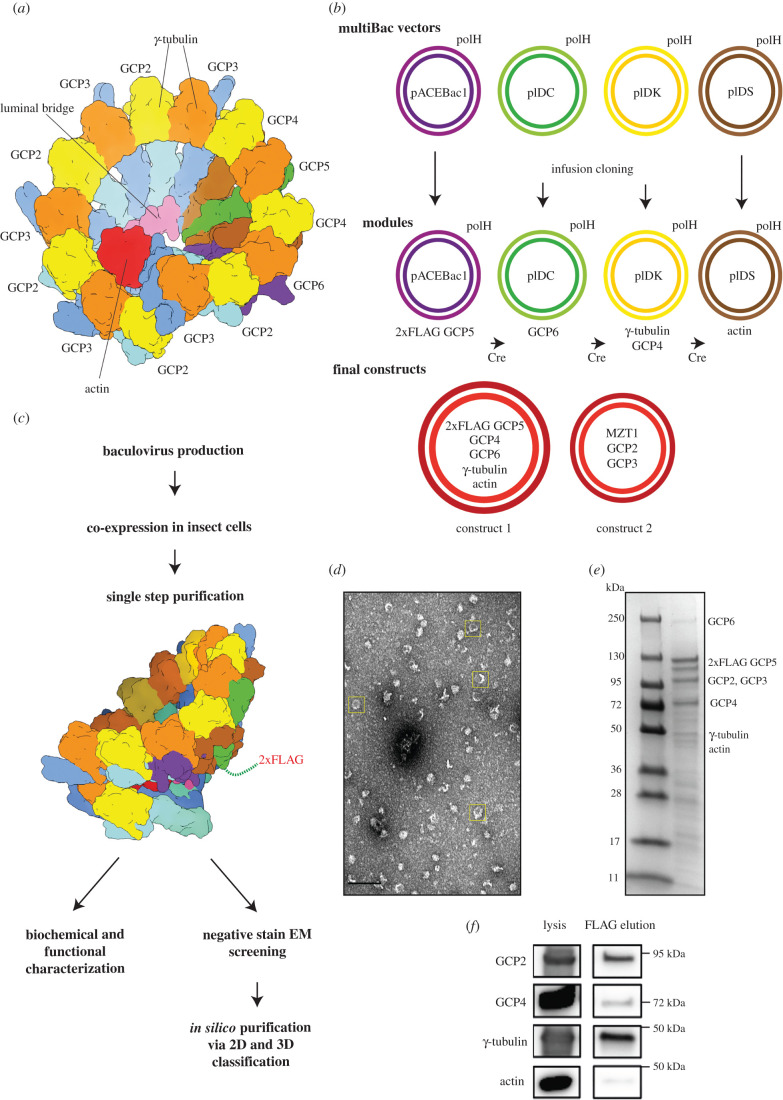
A modularized expression system enables purification of 2xFLAG-tagged recombinant γ-TuRC. (a) Schematic representation of the 14-spoke vertebrate γ-TuRC. Colours: GCP2 (light blue), GCP3 (dark blue), γ-tubulin (orange/yellow), GCP4 (brown), GCP5 (green), GCP6 (purple), actin (red) and luminal bridge (pink). (b) Cloning strategy for the recombinant γ-TuRC using the MultiBac vectors pACEBac1, PIDC, pIDK and pIDS with polyhedrin (polH) expression cassette. Human genes of the γ-TuRC were inserted via Infusion cloning. The ‘modules', plasmids with one or two genes of interest, were combined via subsequent Cre-recombination. Construct 1 for γ-TuRC expression consists of 2xFLAG-GCP5, GCP6, GCP4, TUBG1 and ACTB. This construct was used for bacmid and virus production and for protein expression. It was complemented with construct 2 coding for MZT1, GCP2 and GCP3. (c) Recombinant γ-TuRC was isolated via FLAG affinity purification and FLAG peptide elution in a single-step protocol and used for subsequent characterization using negative stain EM as well as biochemical approaches. Scheme of the γ-TuRC indicating the approximate position of the 2xFLAG tag at the N-terminus of GCP5. Colours as in (a). (d) Section of a negative stain EM micrograph of human recombinant γ-TuRC after FLAG affinity purification. Yellow boxes indicate exemplary γ-TuRC particles. Scale bar: 100 nm. (e) Section of Coomassie Blue-stained SDS-PAGE gel of γ-TuRC elution after FLAG affinity purification. (f) Cell lysate and FLAG affinity-purified recombinant γ-TuRC (FLAG elution) were probed using immunoblotting against the indicated antibodies. See electronic supplementary material, figure S2 for uncropped images.
γ-tubulin complexes have been proposed to promote de novo formation of microtubules by acting as structural templates [15]. The arrangement of the first eight γ-tubulin molecules of the γ-TuRC spiral indeed almost recapitulates the arrangement of αβ-tubulin heterodimers in microtubules. However, from spoke 9 onwards, a clear deviation of γ-tubulin arrangement from microtubule symmetry could be observed, possibly ‘intentionally' limiting the ability of the γ-TuRC to function as a microtubule nucleation template [8–10]. Regulation of γ-TuRC activity thus might be realized by a ‘simple' conformational change of the γ-TuRC diameter. However, how the γ-TuRC is activated in detail remains unclear.
While reconstitution of the simpler heterotetrameric γ-tubulin small complex (γ-TuSC) from lower eukaryotes was achieved [15–18], our structural and basic functional understanding of the vertebrate γ-TuRC to date exclusively relies on purified native complexes [8–10]. The lack of a recombinant reconstitution system for the γ-TuRC particularly hampers experiments addressing the mechanistic role of γ-TuRC structural determinants in complex assembly, activation and microtubule nucleation. Based on the structural census of γ-TuRC core components obtained from recent cryo-EM studies, we have developed a reconstitution system for the human γ-TuRC that is based on the co-expression of all γ-TuRC subunits in insect cells and a gentle one-step purification strategy. We show that the recombinant γ-TuRC has the same structural organization as the native purified γ-TuRC and is active in microtubule nucleation and microtubule minus end capping. This system is a major step towards closing the gap between structural and functional/mechanistic understanding of γ-TuRC-mediated microtubule nucleation.
2. Material and methods
2.1. Molecular cloning
GCP2, GCP3 and GCP4 cDNA were a gift of Andreas Merdes (Toulouse), GCP5 cDNA was from the DKFZ, Heidelberg collection, GCP6 from Ingrid Hoffmann (DKFZ, Heidelberg) [19], human γ-tubulin (TUBG1) from Tim Stearns (Stanford), MZT1 from Jens Lüders (Barcelona) and the beta-actin (ACTB) gene was obtained from AddGene. Cloning was based on the MultiBac (Geneva Biotech) expression system [20], with adapted vectors, where all plasmids contain the polH expression cassette as described in Eustermann et al. [21]. Each of the human genes, TUBG1, GCP2, GCP3, GCP4, 2xFLAG-GCP5, GCP6, ACTB and MZT1 were cloned via InFusion cloning (Takara) using Q5 DNA polymerase (NEB) with standardized primer design for a direct insertion following the polH promoter (figure 1b; electronic supplementary material, table S1 and S2). Two gene cassettes were combined via infusion cloning using the primers listed in (electronic supplementary material, table S2). GCP5 was tagged with 2xFLAG-TEV side in two subsequent PCR reactions. The generated modules were combined via subsequent Cre-recombination using Cre-recombinase (NEB) following the MultiBac manual (Geneva Biotech, version 5.1). Constructs for Cre-recombination were pACEBac1 (2xFLAG-GCP5 or MZT1), pIDC (GCP6 or GCP2, GCP3) pIDK (TUBG1, GCP4) and pIDS (ACTB). Genes of the different modules were verified via PCR amplification and sequencing.
2.2. Protein expression and purification
Bacmid production was performed as described in the MultiBac manual. Bacmids were used for virus production in Sf21 insect cells with cellfectin II reagent (Thermo Fisher Scientific). Viruses were amplified in 30 ml (1 × 106 cells ml−1) and diluted 1 : 100 in 100–400 ml (1 × 106 cells ml−1) expression culture in Sf21 or Hi5 cells using Sf-900 III medium (Thermo Fisher Scientific). Insect cells were infected with constructs 1 and 2 (figure 1b). In some experiments, human γ-tubulin with a C-terminal Myc-His6 tag was co-expressed using a third virus [7,22] (figure 3a). Infected cells were kept at 27°C for 60 h, harvested via centrifugation (800g for 5 min), flash frozen in liquid N2 and stored at −80°C until protein purification.
Figure 3.
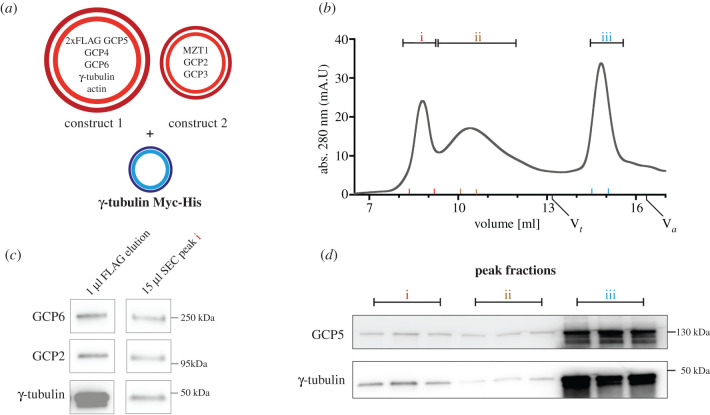
Size exclusion chromatography of recombinant human γ-TuRC. (a) For SEC experiments, construct 1 (2xFLAG-GCP5, GCP6, GCP4, TUBG1 and ACTB) and construct 2 (MZT1, GCP2 and GCP3) were co-expressed with C-terminal Myc-His tagged γ-tubulin and purified via FLAG purification. (b) Chromatogram of a SEC run with ‘Superose 6 Increase (10/300) GL' column. Peak fractions (i (γ-TuRC peak), ii, iii) were analysed via immunoblotting (c,d). Red, brown and blue markers on x-axes (b) indicate borders of analysed fractions (i, ii, iii) in (d). Size markers thyroglobulin 669 kDa (Vt) and aldolase 158 kDa (Va) are indicated. (c) FLAG elution left and ‘γ-TuRC peak' fraction (i, b) were probed by immunoblotting against the indicated antibodies. (d) Immunoblot analysis of ethanol precipitated peak fractions (i, ii, iii) from SEC experiment shown in (b). Equal amount of fraction volumes was loaded. See electronic supplementary material, figure S2 for uncropped images.
For protein purification, cells were resuspended in cold lysis buffer (50 mM HEPES, 150 mM KCl, 5 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.1 mM GTP, pH 7.4 + 0.02% (v/v) Brij-35, and 250 units Benzonase (Sigma Aldrich), 1 complete EDTA-free protease inhibitor tablet (Roche) per 15 ml lysis buffer) and kept on ice. Resuspended cells were sonicated (3 × 1 min with 0.6 amplitude, Hielscher UP50H) and centrifuged at 20 000g for 60 min at 4°C. Anti-FLAG M2 Affinity Gel (Sigma Aldrich) was equilibrated in lysis buffer and incubated with the centrifuged and filtered lysate (Whatman sterile filters 0.45 µm pore size) for 90 min rotating at 4°C. Afterwards beads were washed once with lysis buffer and twice with wash buffer (50 mM HEPES, 150 mM KCl, 5 mM MgCl2, 1 mM EGTA, 0.5 mM DTT, 0.1 mM GTP, pH 7.4). Elution was done with one bead volume of elution buffer (wash buffer plus 0.2 mg ml−1 3xFLAG peptide; Gentaur) and 20 min incubation at 4°C. Elution was repeated once, and samples were used for subsequent experiments or flash frozen in liquid N2 and stored at −80°C. For samples which were purified for size exclusion chromatography (SEC), all buffers contained 5% (w/v) glycerol. Native γ-TuRC purified from X. laevis egg extract was performed as described previously [8]. SDS gels were run together with Page Ruler Plus (Thermo Fisher Scientific) on precast gradient gels (4–20% BIO-RAD) and stained with Coomassie Brilliant Blue G250 (Sigma Aldrich) or used for immunoblot analysis of the γ-TuRC components as described previously [8] with the antibodies listed in electronic supplementary material, table S3 and figures S2 and S3.
2.3. Fluorescence-based tubulin polymerization assay
MT nucleation was performed using the fluorescence-based Tubulin Polymerization assay kit (Cytoskeleton, Denver Com cat. no. BK011P) following the manufacturer's instructions. Briefly, 5 µl or 10 µl of the samples (buffer control (elution buffer/SEC buffer), human recombinant γ-TuRC, human recombinant γ-TuRC after size exclusion chromatography (SEC), X. laevis γ-TuRC or 3 µM paclitaxel (final concentration)) was pipetted into a 96-well microtiter plate, prewarmed and mixed with 45 µl of tubulin buffer (2 mg ml−1 porcine tubulin in 80 mM PIPES pH 6.9, 2 mM MgCl2, 0.5 mM EGTA, 1 mM GTP, 15% (w/v) glycerol). The concentration of human recombinant γ-TuRC after SEC and native X. laevis γ-TuRC were normalized to γ-tubulin signal in immunoblot analysis and diluted in SEC buffer prior to nucleation measurements accordingly. The polymerization reaction was triggered by transferring the plate to the 37°C chamber of the plate reader. Fluorescence was measured at 37°C using the plate reader for 60 min at 1 min intervals (CLARIOstar, BMG Labtech, excitation, F: 360-10, emission, F: 450-10). Data were processed using PRISM software (GraphPad v. 8).
2.4. In vitro microtubule nucleation assay for electron microscopy analysis
Thirty micromolar of porcine brain tubulin containing 4% Cy3-labelled tubulin in 1x BRB80 buffer (80 mM PIPES/KOH pH 6.8, 1 mM MgCl2, 1 mM EGTA and 12.5% (w/v) glycerol) was centrifuged (5 min, 352 860g, 4°C) with S100-AT3 rotor (Thermo Fisher Scientific). Supernatant was mixed with γ-TuRC premix (1 : 20 dilution of γ-TuRC, 1 mM GTP in 1x BRB80, 12.5% (w/v) glycerol) in ratio 1 : 1. Samples were incubated for 15–30 min on ice and transferred to a 37°C water bath for 3 min for MT polymerization. After MT polymerization samples were immediately cross-linked with 1% glutaraldehyde and used for negative stain EM.
2.5. Size exclusion chromatography
SEC was performed either with ‘Superose 6 (10/300) GL' or ‘Superose 6 Increase (10/300) GL' (GE Healthcare) equilibrated with SEC buffer (50 mM HEPES, 150 mM KCl, 5 mM MgCl2, 1 mM EGTA, 0.5 mM DTT, 0.1 mM GTP, 5% glycerol (v/w), pH 7.4). Only for runs with the ‘Superose 6 (10/300) GL' column, FLAG elutions were concentrated (Amicon, 30 K) before injection into the column. Runs with size markers thyroglobulin (669 kDa and aldolase 158 kDa) were performed separately. Peak fractions were aliquoted and flash frozen in liquid N2 for storage at −80°C. Samples of peak fractions were also used for ethanol precipitation (90% EtOH, −20°C, overnight) for subsequent immunoblot analysis.
2.6. Negative stain electron microscopy grid preparation and data collection
Five microliters of sample was applied on glow-discharged copper-palladium 400 EM mesh grids covered with an approximately 10 nm-thick continuous carbon layer. After 30 s incubation at 23°C, grids were blotted with a Whatman filter paper 50 (cat no. 1450-070) and washed on 3 drops of water. Sample on grids was stained with 3% uranyl acetate in water. Grids with nucleated microtubules were imaged at a Jeol JE-1400 (Jeol Ltd., Tokyo, Japan) operating at 80 kV equipped with a 4 k × 4 k digital camera (F416, TVIPS, Gauting, Germany). Micrographs were adjusted in brightness and contrast using ImageJ. Negative stain EM data for two- and three-dimensional class averaging of native X. laevis γ-TuRC [8] and recombinant human γ-TuRC were acquired on a Talos L120C TEM equipped with 4 k × 4 K Ceta CMOS camera (Thermo Fisher Scientific). To compensate for the preferred orientation of γ-TuRC particles on the continuous carbon grids, data were partially acquired at a stage tilting angle of 20 degrees. Data were acquired using EPU (Thermo Fischer Scientific) at a nominal defocus of −3 µm and an object pixel size of 0.4125 nm.
2.7. Negative stain electron microscopy data processing
In total, we acquired approximately 1600 micrographs of the recombinant human γ-TuRC (957 micrographs untilted, 640 micrographs at 20° stage tilt), approximately 1100 micrographs of the native X. laevis γ-TuRC (500 micrographs not tilted, 600 micrographs at 20° stage tilt) and 500 micrographs of recombinant human γ-TuRC after SEC without tilt. Image processing for all datasets was performed in Relion 3.1 Beta [23]. The contrast transfer function (CTF) of micrographs was estimated using gCTF [24]. For all datasets, approximately 1000 particles were selected manually and extracted at a pixel size of 0.825 nm with a box size of 64 pixels. Extracted particles were subjected to two-dimensional classification with 50 classes, a translational search range of 20 pixels at 2 pixels increment and a mask diameter of 450 Å. Two-dimensional classes unambiguously representing γ-TuRCs were selected and used as references for automated particle picking on all micrographs. The overall number of picked particles was 374 389 for the recombinant human γ-TuRC, 192 845 for the native X. laevis γ-TuRC and 38 139 for the recombinant human γ-TuRC after SEC. Extracted particles from each dataset were subjected to several rounds of to two-dimensional classification with 100–200 classes, a translational search range of 20 pixels at 2 pixels increment and 450 Å mask size. For three-dimensional classification particles included in true-positive classes were merged, yielding 12 402 particles of the recombinant human γ-TuRC and 12 271 particles of the native X. laevis γ-TuRC. Next, particles were aligned in three-dimensional using three-dimensional classification with 1 class and a translation search range of 20 pixels at 2 pixels increment. As a reference, we used a strongly low-pass filtered cryo-EM density of the γ-TuRC [8], from which we removed the GRIP2 domains and γ-tubulins of spokes 5 and 6. The aligned particles were subjected to the second round of three-dimensional classification into 6 classes without image alignment, focused on the spokes that were artificially removed from the reference in the first classification run. This allowed us to separate correctly aligned true-positive particles (with density for spokes 5 and 6) from false positive or incorrectly aligned particles (without density for spokes 5 and 6). The retained particles (6253 particles for the recombinant human γ-TuRC; 6827 particles for the X. laevis γ-TuRC) were submitted to another round of three-dimensional classification into 3 classes with image alignment and the best classes were selected for further structural analysis (2064 particles of the recombinant human γ-TuRC; 2490 particles of the native X. laevis γ-TuRC).
3. Results
3.1. A recombinant system for the expression of gamma-ring complex proteins, γ-tubulin, actin and mitotic spindle organizing protein 1 in insect cells
At least eight different proteins assemble in defined stoichiometry to form the γ-TuRC [25]. In order to achieve expression of all subunits, we decided to subclone human γ-TuRC component genes into a modified MultiBac expression system [20]. Instead of using the multiple cloning site, we followed an InFusion cloning strategy with vectors carrying the polyhedrin (polH) expression cassette (figure 1b) [21]. Either two gene cassettes were combined into one vector, or single gene vectors were used for subsequent Cre-recombination. This strategy allows for easy modification and shuffling of the expressed genes (figure 1b). As GCPs are large proteins, genes of interest were arranged on two final plasmids to avoid construction of one oversized plasmid. The two plasmids were used for virus production: construct 1 contained 2xFLAG-GCP5, GCP4, GCP6, TUBG1 (coding for γ-tubulin) and ACTB (coding for β-actin), which were suggested to form an initial stable core complex during the assembly of γ-TuRC [8,26]; construct 2 contained MZT1, GCP2 and GCP3, the ‘minimal set' of additional subunits required to reconstitute the 14-spoke γ-TuRC including the luminal bridge as observed in cryo-EM studies [8–10,12].
For protein expression, insect cells were infected with equal amounts of viruses and harvested after 60 h. One of the GCP subunits, GCP5, was tagged with 2xFLAG at its N-terminus for gentle single-step affinity purification of the complex from a relatively small cell culture volume (see methods) (figure 1c). Negative stain EM showed regular ring-shaped structures on the micrographs (figure 1d, boxed particles), indicating complex formation. This is supported by the detection of other γ-TuRC components co-purifying with 2xFLAG-GCP5 (figure 1e). As observed in other studies before [8,10], the signal intensity of the γ-tubulin band in Coomassie Blue-stained SDS gels does not reflect its high stoichiometry in the complex. The identity of several γ-TuRC components was further confirmed by immunoblotting (figure 1f). In conclusion, we have established an insect cell-based recombinant expression system for reconstitution of the human γ-TuRC, which allows complex purification in a one-step protocol.
3.2. Recombinant and native γ-tubulin ring complex are structurally indistinguishable by negative staining electron microscopy
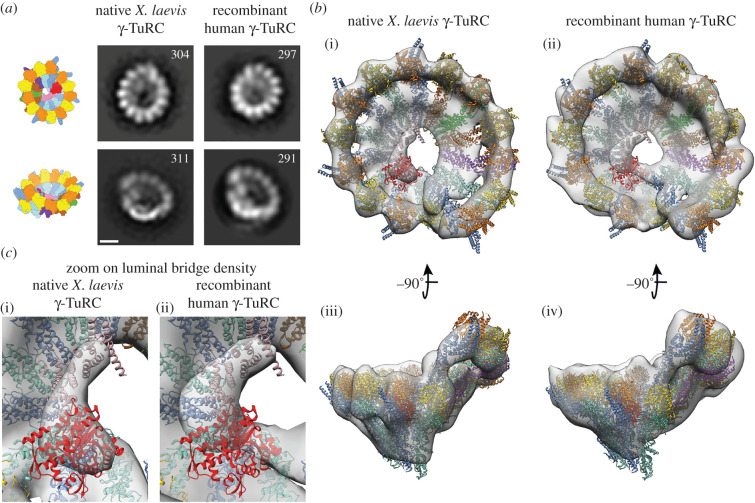
To verify the integrity and correct assembly of the reconstituted γ-TuRC, we performed negative stain EM. The structures of recombinant human and native X. laevis γ-TuRCs were compared by two-dimensional (figure 2a) and three-dimensional class averaging (figure 2b). The EM densities recapitulate the structure of the γ-TuRC as previously determined using cryo-EM, including the 14-spoke overall organization of the complex and the luminal bridge on the inner surface of the γ-TuRC (figure 2b). Visibility of the ordered ‘luminal bridge' structure (figure 2c) indicates that MZT1 and actin were correctly integrated into the complex. Importantly, the native and recombinant γ-TuRCs were structurally indistinguishable in the resolution range accessible with negative stain EM, as judged by the fit of atomic models into the two densities (figure 2b).
Figure 2.
Negative stain EM analysis of recombinant human and native X. laevis γ-TuRCs. (a) Comparison of representative two-dimensional classes from the native X. laevis (left) and recombinant human γ-TuRC (right) in two different views, as indicated in the cartoons. Number of particles in each class is given. Scale bar: 10 nm. (b) Three-dimensional EM densities of (i,iii) the native X. laevis (2490 particles) and (ii,iv) recombinant human γ-TuRC (2064 particles) in two different views. The atomic model of the human γ-TuRC (PDB-6V5 V) [9] was docked as a rigid body and superposed to the EM densities. Colouring as in figure 1a. (c) Zooms focused on the luminal bridge density of the native X. laevis (i) and recombinant human γ-TuRC (ii). The same colouring as in figure 1a.
3.3. Size exclusion chromatography enables separation of recombinant γ-tubulin ring complex from sub-complexes
To further optimize γ-TuRC reconstitution, we repeated the recombinant expression of γ-TuRC components with additional co-expression of C-terminal tagged (Myc-His6) human γ-tubulin [7] (figure 3a) that appeared substoichiometric in figure 1. In addition, we included size exclusion chromatography (SEC) as an analysis step to judge the ratio of fully assembled γ-TuRC relative to sub-complexes (figure 3b; electronic supplementary material, figure S1). The size exclusion chromatogram of the recombinant γ-TuRC (figure 3b) showed an early high molecular weight peak (figure 3b,i), which contained γ-TuRC components, as judged by immunoblotting (figure 3c,d). Adjacent lower molecular weight peak fractions (figure 3d,ii) also contained γ-TuRC components but due to their running behaviour probably correspond to γ-TuRC sub-complexes (electronic supplementary material, figure S1a,b). The presence of intact γ-TuRC in early SEC elution fractions was further confirmed by negative stain EM from another γ-TuRC preparation that after concentration contained fewer medium to low molecular weight γ-tubulin sub-complexes (electronic supplementary material, figure S1c–e). SEC analysis thus confirms the integrity of the recombinant γ-TuRC and additionally suggests the presence of stable γ-TuRC assembly intermediates that will be central for dissecting the assembly pathway of this megadalton complex.
3.4. The recombinant γ-tubulin ring complex has microtubule nucleation activity and caps the minus ends of microtubules
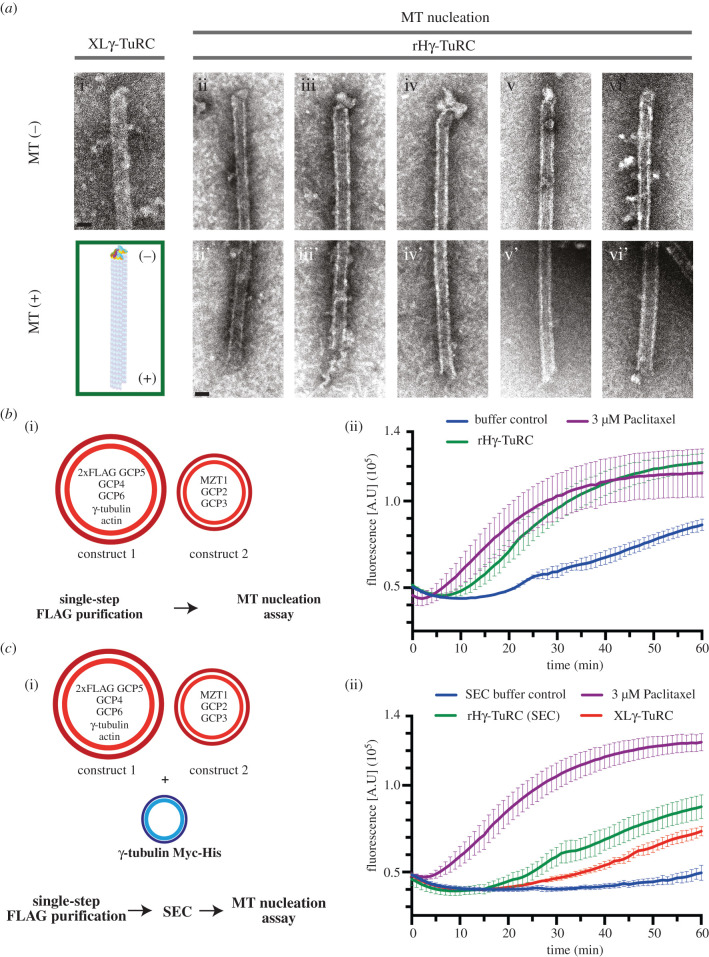
Having confirmed the structural integrity of the recombinant γ-TuRC, we next aimed at characterizing its functional properties. The γ-TuRC initiates assembly of microtubules by forming a structural template for the recruitment of the initial αβ-tubulin subunits and then remains stably associated with the minus end of the growing microtubule [10,27–29]. Consistently, when analysing microtubules nucleated by the recombinant γ-TuRC via negative stain EM, we frequently observed a cap-like structure representing the γ-TuRC on one of the microtubule ends (figure 4a). The opposite end of these microtubules always had a frayed or open appearance as typically observed for the microtubule plus end [30,31], suggesting that the γ-TuRC is localized on the microtubule minus end (figure 4a). Furthermore, to determine microtubule nucleation activity of the recombinant γ-TuRC, we used a fluorescence-based microtubule nucleation assay (figure 4b,c). Upon the addition of the recombinant γ-TuRC after FLAG purification, microtubule assembly was clearly increased compared to elution buffer without γ-TuRC, which served as a negative control (figure 4b). Next, we analysed the nucleation activity of the recombinant γ-TuRC after FLAG elution and SEC (figure 4c) and—as an additional control—compared it to the nucleation activity of affinity-purified X. laevis γ-TuRC [8], normalized to the γ-tubulin content of the two preparations. We observed that the microtubule nucleation activity of the recombinant γ-TuRC was comparable to the affinity-purified X. laevis γ-TuRC. Collectively, these data confirm that the recombinantly expressed and purified γ-TuRC has microtubule nucleation activity.
Figure 4.
The recombinant human γ-TuRC has microtubule nucleation and minus end capping activity. (a) Representative negative stain EM images from microtubules nucleated by native X. laevis γ-TuRC; (i; XLγ-TuRC) or the recombinant human γ-TuRC (ii-vi, rHγ-TuRC). For microtubules nucleated by the recombinant human γ-TuRC, the capped microtubule minus ends (ii–vi; MT-) and the flared or sheet microtubule plus ends (ii'–vi'; +MT) are shown. A schematic representation of a γ-TuRC capped microtubule is shown in the green box. Scale bars, 25 nm. (b,c) Tubulin polymerization assay where the increase in fluorescence intensity over time represents αβ-tubulin polymerization into microtubules. Shown are error bars for the standard deviation of the mean of three (b) and four (c) technical replicates. (b) The recombinant human γ-TuRC (construct 1 and construct 2 in figure 1, rHγ-TuRC) was analysed together with elution buffer without recombinant γ-TuRC as negative control, and 3 µM Paclitaxel as positive control. (c) Recombinant human γ-TuRC (construct 1, construct 2 and C-terminal Myc-His6 tagged γ-tubulin) was purified by FLAG affinity purification and subsequent SEC (figure 3b i). The γ-tubulin content of ‘γ-TuRC peak’ fraction (rHγ-TuRC SEC) and native X. laevis γ-TuRC (XLγ-TuRC) was determined by immunoblotting. Samples were diluted in SEC buffer to equal γ-tubulin concentrations and were then used for the microtubule nucleation assay with SEC buffer as a negative control and 3 µM Paclitaxel as positive control.
4. Discussion
Here, we report a strategy for recombinant expression, reconstitution and purification of the human γ-TuRC from small cell culture volumes of insect cells. This system enables fast and efficient screening of structural and functional properties of genetically modified γ-TuRC variants in follow-up experiments. The success of this strategy, which is based on FLAG-GCP5 affinity purification, indicates that the N-terminus of GCP5, which has not been completely traced in the recent cryo-EM reconstructions, is accessible for antibody binding and likely not as deeply embedded in the γ-TuRC as the GCP6 N-terminus [12]. This FLAG-tagged γ-TuRC variant thus can likely be immobilized in a defined orientation for single-molecule TIRF microtubule nucleation assays [10,29], which require the γ-tubulin face of the γ-TuRC to be accessible (figure 1c). Such an approach will allow comparing microtubule nucleation activity of γ-TuRC species with differing subunit composition or mutations.
SEC as an additional purification step furthermore indicated that our purification strategy yields not only fully assembled recombinant γ-TuRC, but also sub-complexes that may represent intermediates in γ-TuRC assembly (figure 3; electronic supplementary material, figure S1). Moreover, our experiments show that expression of TUBG1, GCP2 to GCP6, ACTB and MZT1 with or without the excess of tagged TUBG1 was sufficient for reconstitution of the γ-TuRC. Thus, the γ-TuRC targeting factor NEDD1 and the nucleation regulator NME7 seem to be not required for complex assembly, consistent with previous data [13,14]. Similarly, since the human γ-TuRC was reconstituted in the absence of MZT2, which is not encoded in insect cells [32], we can conclude that MZT2 is not essential for structural integrity and microtubule nucleation activity of the γ-TuRC. This is consistent with the recently proposed function for MZT2 as an adaptor of the γ-TuRC for MTOC binding [33]. By contrast, the two molecules of MZT1 integrated into the luminal bridge scaffold clearly fulfil a structural role [12] and are probably required for the formation of a stable complex. Such a function is consistent with the observation that MZT1 depletion affects the integrity of the γ-TuRC [17].
Our recombinant expression system now places us in a position to address central questions regarding γ-TuRC assembly, activation and γ-TuRC-associated microtubule nucleation from a mechanistic perspective. A prime candidate for detailed mutational analysis is the long insertion domain between the GRIP1 and GRIP2 domains of GCP6, which has been identified to interact specifically with the N-termini of GCP2 and GCP5, and therefore to potentially assist in the assembly of the γ-TuRC [8]. Our system will allow us to analyse the function and structure of γ-TuRC variants assembled in the presence of mutated versions of this GCP6 insertion domain. In a similar manner, we will be able to address the structural and functional role of actin in the γ-TuRC, which has remained elusive so far. Collectively, our recombinant γ-TuRC expression system opens up many possibilities for structure-function analyses of this microtubule nucleator and its cooperation with additional factors, such as XMAP215 and the augmin complex [34,35].
Acknowledgements
The authors thank Ursula Jäkle for support during protein expression and purification. We thank A. Merdes for GCP2, GCP3 and GCP4 cDNA, GCP5 cDNA was from the DKFZ collection, GCP6 from Ingrid Hoffmann, human γ-tubulin cDNA from Tim Stearns and MZT1 from Jens Lüders and Myc-His jagged human γ-tubulin baculovirus was a gift from David Agard. We thank the electron microscopy facility of Heidelberg University for support. We acknowledge the services SDS@hd and bwHPC supported by the Ministry of Science, Research and the Arts Baden-Württemberg, as well as the German Research Foundation (INST 35/1314-1 FUGG and INST 35/1134-1 FUGG). We also acknowledge access to the infrastructure of the Cryo-EM Network at the Heidelberg University (HDcryoNET) and support by Dirk Flemming (BZH).
Data accessibility
This article has additional data in the figures and tables of the electronic supplementary material.
Author contributions
M.W. planned and performed cloning, expressions, purifications and SEC. A.N. performed negative staining EM analysis of microtubule minus ends. A.N. and E.Z. acquired negative stain EM data of purified γ-TuRC and together with B.V. and M.W. analysed the negative stain EM data. A.B. and L.R. performed MT nucleation assays. A.B. and P.L. were involved in characterizing the purified γ-TuRC. All authors discussed the data. E.S., S.P. and S.E. planned and supervised the experiments. All authors drafted the manuscript and gave final approval for publication.
Competing interests
The authors disclose no competing interests.
Funding
This work is supported by grants of the Deutsche Forschungsgemeinschaft (DFG) to E.S. (DFG Schi 295/4-4) and to S.P. (DFG PF 963/1-4).
References
- 1.Nogales E. 2000. Structural insights into microtubule function. Annu. Rev. Biochem. 69, 277-302. ( 10.1146/annurev.biochem.69.1.277) [DOI] [PubMed] [Google Scholar]
- 2.Brouhard GJ, Rice LM. 2018. Microtubule dynamics: an interplay of biochemistry and mechanics. Nat Rev. Mol. Cell Biol. 19, 451-463. ( 10.1038/s41580-018-0009-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pereira G, Schiebel E. 1997. Centrosome-microtubule nucleation. J. Cell Sci. 110(Pt 3), 295-300. [DOI] [PubMed] [Google Scholar]
- 4.Kollman JM, Merdes A, Mourey L, Agard DA. 2011. Microtubule nucleation by γ-tubulin complexes. Nat. Rev. Mol. Cell Biol. 12, 709-721. (nrm3209 [pii] 10.1038/nrm3209). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng Y, Wong ML, Alberts B, Mitchison T. 1995. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature 378, 578-583. ( 10.1038/378578a0) [DOI] [PubMed] [Google Scholar]
- 6.Guillet V, et al. 2011. Crystal structure of γ-tubulin complex protein GCP4 provides insight into microtubule nucleation. Nat. Struct. Mol. Biol. 18, 915-919. (nsmb.2083 [pii] 10.1038/nsmb.2083). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aldaz H, Rice LM, Stearns T, Agard DA. 2005. Insights into microtubule nucleation from the crystal structure of human γ-tubulin. Nature 435, 523-527. ( 10.1038/nature03586) [DOI] [PubMed] [Google Scholar]
- 8.Liu P, et al. 2020. Insights into the assembly and activation of the microtubule nucleator gamma-TuRC. Nature 578, 467-471. ( 10.1038/s41586-019-1896-6) [DOI] [PubMed] [Google Scholar]
- 9.Wieczorek M, Urnavicius L, Ti SC, Molloy KR, Chait BT, Kapoor TM. 2020. Asymmetric molecular architecture of the human gamma-tubulin ring complex. Cell 180, 165-175. e116. ( 10.1016/j.cell.2019.12.007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Consolati T, et al. 2020. Microtubule nucleation properties of single human gamma TuRCs explained by their cryo-EM structure. Dev. Cell 53, 603-+. ( 10.1016/j.devcel.2020.04.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teixidó-Travesa N, Villen J, Lacasa C, Bertran MT, Archinti M, Gygi SP, Caelles C, Roig J, Lüders J. 2010. The γTuRC revisited: a comparative analysis of interphase and mitotic human γTuRC redefines the set of core components and identifies the novel subunit GCP8. Mol. Biol. Cell 21, 3963-3972. (E10-05-0408 [pii] 10.1091/mbc.E10-05-0408). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wieczorek M, Huang T-L, Urnavicius L, Hsia K-C, Kapoor TM. 2020. MZT proteins form multi-faceted structural modules in the γ-tubulin ring complex. Cell Rep. 31, 107791. ( 10.1016/j.celrep.2020.107791) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu P, Choi YK, Qi RZ. 2014. NME7 is a functional component of the gamma-tubulin ring complex. Mol. Biol. Cell 25, 2017-2025. ( 10.1091/mbc.E13-06-0339) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luders J, Patel UK, Stearns T. 2006. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8, 137-147. ( 10.1038/ncb1349) [DOI] [PubMed] [Google Scholar]
- 15.Kollman JM, Polka JK, Zelter A, Davis TN, Agard DA. 2010. Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature 466, 879-882. ( 10.1038/nature09207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leong SL, Lynch EM, Zou J, Tay YD, Borek WE, Tuijtel MW, Rappsilber J, Sawin KE. 2019. Reconstitution of microtubule nucleation in vitro reveals novel roles for Mzt1. Curr Biol. 29, 2199-2207. e2110. ( 10.1016/j.cub.2019.05.058). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin TC, Neuner A, Flemming D, Liu P, Chinen T, Jakle U, Arkowitz R, Schiebel E. 2016. MOZART1 and gamma-tubulin complex receptors are both required to turn gamma-TuSC into an active microtubule nucleation template. J. Cell Biol. 215, 823-840. ( 10.1083/jcb.201606092) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zupa E, Zheng A, Neuner A, Würtz M, Liu P, Böhler A, Schiebel E, Pfeffer S. 2020. The cryo-EM structure of a γ-tubulin small complex provides insights into the architecture and regulation of a minimal microtubule nucleation system. Nat. Commun. 11, 5705. ( 10.1038/s41467-020-19456-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bahtz R, Seidler J, Arnold M, Haselmann-Weiss U, Antony C, Lehmann WD, Hoffmann I. 2012. GCP6 is a substrate of Plk4 and required for centriole duplication. J. Cell Sci. 125, 486-496. ( 10.1242/jcs.093930) [DOI] [PubMed] [Google Scholar]
- 20.Fitzgerald DJ, Berger P, Schaffitzel C, Yamada K, Richmond TJ, Berger I. 2006. Protein complex expression by using multigene baculoviral vectors. Nat. Methods 3, 1021-1032. ( 10.1038/nmeth983) [DOI] [PubMed] [Google Scholar]
- 21.Eustermann S, Schall K, Kostrewa D, Lakomek K, Strauss M, Moldt M, Hopfner KP. 2018. Structural basis for ATP-dependent chromatin remodelling by the INO80 complex. Nature 556, 386-390. ( 10.1038/s41586-018-0029-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy SM, Urbani L, Stearns T. 1998. The mammalian γ-tubulin complex contains homologues of the yeast spindle pole body components Spc97p and Spc98p. J. Cell Biol. 141, 663-674. ( 10.1083/jcb.141.3.663) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJH, Lindahl E, Scheres SHW. 2018. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7, e42166. ( 10.7554/eLife.42166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang K. 2016. Gctf: real-time CTF determination and correction. J. Struct. Biol. 193, 1-12. ( 10.1016/j.jsb.2015.11.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zupa E, Liu P, Würtz M, Schiebel E, Pfeffer S. 2020. The strcuture of the γ-TuRC: a 25-years-old molecular puzzle. Curr. Opin. Struct. Biol. 66, 15-21. ( 10.1016/j.sbi.2020.08.008) [DOI] [PubMed] [Google Scholar]
- 26.Haren L, Farache D, Emorine L, Merdes A. 2020. A stable sub-complex between GCP4, GCP5 and GCP6 promotes the assembly of gamma-tubulin ring complexes. J. Cell Sci. 133, jcs244368. ( 10.1242/jcs.244368) [DOI] [PubMed] [Google Scholar]
- 27.Wiese C, Zheng Y. 2000. A new function for the γ-tubulin ring complex as a microtubule minus-end cap. Nat. Cell Biol. 2, 358-364. ( 10.1038/35014051) [DOI] [PubMed] [Google Scholar]
- 28.Moritz M, Braunfeld MB, Guenebaut V, Heuser J, Agard DA. 2000. Structure of the gamma-tubulin ring complex: a template for microtubule nucleation. Nat. Cell Biol. 2, 365-370. ( 10.1038/35014058) [DOI] [PubMed] [Google Scholar]
- 29.Thawani A, Rale MJ, Coudray N, Bhabha G, Stone HA, Shaevitz JW, Petry S. 2020. The transition state and regulation of gamma-TuRC-mediated microtubule nucleation revealed by single molecule microscopy. Elife 9, e54253. ( 10.7554/eLife.54253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nogales E, Wang HW. 2006. Structural intermediates in microtubule assembly and disassembly: how and why? Curr. Opin. Cell Biol. 18, 179-184. ( 10.1016/j.ceb.2006.02.009) [DOI] [PubMed] [Google Scholar]
- 31.Mandelkow EM, Mandelkow E, Milligan RA. 1991. Microtubule dynamics and microtubule caps: a time-resolved cryoelectron microscopy study. J. Cell Biol. 114, 977-991. ( 10.1083/jcb.114.5.977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tovey CA, Conduit PT. 2018. Microtubule nucleation by gamma-tubulin complexes and beyond. Essays Biochem. 62, 765-780. ( 10.1042/Ebc20180028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cota RR, Teixido-Travesa N, Ezquerra A, Eibes S, Lacasa C, Roig J, Luders J. 2017. MZT1 regulates microtubule nucleation by linking gammaTuRC assembly to adapter-mediated targeting and activation. J. Cell Sci. 130, 406-419. ( 10.1242/jcs.195321) [DOI] [PubMed] [Google Scholar]
- 34.Thawani A, Kadzik RS, Petry S. 2018. XMAP215 is a microtubule nucleation factor that functions synergistically with the gamma-tubulin ring complex. Nat. Cell Biol. 20, 575-585. ( 10.1038/s41556-018-0091-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petry S, Vale RD. 2015. Microtubule nucleation at the centrosome and beyond. Nat. Cell Biol. 17, 1089-1093. ( 10.1038/ncb3220) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has additional data in the figures and tables of the electronic supplementary material.