![]()
Keywords: cell division, SsgB, mutagenesis, ectopic septa, crystal structure
Abstract
In most bacteria, cell division begins with the polymerization of the GTPase FtsZ at mid-cell, which recruits the division machinery to initiate cell constriction. In the filamentous bacterium Streptomyces, cell division is positively controlled by SsgB, which recruits FtsZ to the future septum sites and promotes Z-ring formation. Here, we show that various amino acid (aa) substitutions in the highly conserved SsgB protein result in ectopically placed septa that sever spores diagonally or along the long axis, perpendicular to the division plane. Fluorescence microscopy revealed that between 3.3% and 9.8% of the spores of strains expressing SsgB E120 variants were severed ectopically. Biochemical analysis of SsgB variant E120G revealed that its interaction with FtsZ had been maintained. The crystal structure of Streptomyces coelicolor SsgB was resolved and the key residues were mapped on the structure. Notably, residue substitutions (V115G, G118V, E120G) that are associated with septum misplacement localize in the α2–α3 loop region that links the final helix and the rest of the protein. Structural analyses and molecular simulation revealed that these residues are essential for maintaining the proper angle of helix α3. Our data suggest that besides altering FtsZ, aa substitutions in the FtsZ-recruiting protein SsgB also lead to diagonally or longitudinally divided cells in Streptomyces.
1. Introduction
During bacterial cell division, the tubulin homologue FtsZ is the first component recruited to the division site, where it polymerizes into a ring-like structure known as the Z-ring, which contracts during cytokinesis [1]. FtsZ is a self-activating GTPase that undergoes guanosine-5-triphosphate (GTP)-dependent assembly into protofilaments by the head-to-tail association of individual monomers [2–4]. Super-resolution microscopy revealed that the Z-ring is discontinuous and consists of FtsZ assemblies [5,6]. Recent studies suggested that FtsZ filaments undergo treadmilling circumferentially around the division plane and drive the movement of the peptidoglycan (PG) -synthesizing machinery [7–9]. New cell wall components are inserted progressively at the septum, ultimately resulting in cell constriction. The treadmilling is associated with the conformational change and the kinetic polarity of the FtsZ filament and controls the rate of PG synthesis and cell division [8,10]. Septum-site localization in unicellular bacteria depends on FtsA and ZipA, which anchor FtsZ polymers to the cell membrane, with ZapA stabilizing the FtsZ filaments and promoting lateral interactions [11–13]. In Escherichia coli, control of Z-ring timing and localization is governed by the Min system and by nucleoid occlusion, which negatively regulate FtsZ polymerization [14–17].
Owing to its central role in cell division, FtsZ is essential in nearly all bacteria. Two exceptions are the parasite Mycoplasma [18], which has a reduced genome size and no cell walls, and Streptomyces [19,20]. Streptomycetes are filamentous Gram-positive bacteria in the phylum of Actinobacteria that have a complex mycelial lifestyle [21]. These bacteria produce over 60% of all known antibiotics and many other bioactive natural products [22,23]. Streptomycetes are model organisms for the study of multicellularity and bacterial morphogenesis [24,25]. Exponential growth of the multi-nucleoid vegetative hyphae is achieved by apical growth and branching. At this stage of the life cycle, cell division does not affect physical separation of the cells, but instead long syncytial cells are formed that are separated by cross-walls [26]. When the developmental programme is switched on, streptomycetes produce aerial hyphae that ultimately differentiate into chains of unigenomic spores. Recent studies highlight that local membrane synthesis and branching may be an important divisome-independent mechanism for cell proliferation in Streptomyces [27,28].
Streptomycetes lack the canonical cell-division regulation systems such as Min and Noc [29]. Instead, a positive control system has evolved, whereby FtsZ is actively recruited to the septum sites by SsgB, in concert with its paralogue SsgA [30]. The onset of sporulation and the correct localization of SsgB to the future septum sites are regulated by a transmembrane protein SepG [31]. In addition, two dynamin-like proteins and SepF are involved in stabilization of the Z-ring during Streptomyces sporulation [32], where the assembly of Z-ring is influenced by a small protein called CrgA [33]. The SsgA-like proteins (SALPs) are a family of regulatory proteins that is unique in sporulating actinobacteria [34,35]. SsgB is the archetypical SALP, with a conserved function in the development of actinomycetes [36]. While the SsgB protein sequence varies strongly between less related Actinobacteria, the protein is extremely well conserved within a genus, with a maximum of one amino acid (aa) variation, a feature that has been applied for the phylogenetic analysis of closely related Actinobacteria [37]. Actinobacteria that form single spores, such as Micromonospora or Thermobifida, only have one SALP, namely the FtsZ-recruiting protein SsgB, while up to 14 SALPs can be found in those genera that form chains of spores, such as Streptomyces [38]. The crystal structure of a SsgB orthologue from Thermobifida fusca was resolved, which revealed a ‘bell-shape’ trimer that is assembled mainly through α-helix interactions.
In this work, we report on the importance of individual residues in the localization of the septum during sporulation-specific division, by creating a library of SsgB mutants and studying their effect on cell division and morphogenesis. Single aa changes in SsgB had major effects on cell division, spore-wall synthesis and DNA condensation and/or segregation. Remarkably, specific mutations led to the formation of additional septa with 10° to 90° rotation of the division plane. Altered division planes have so far only been observed in mutants of FtsZ itself [39–41], and in worm-associated bacteria Candidatus Thiosymbion oneisti and Thiosymbion hypermnestrae. In the latter two bacteria, cell growth and longitudinal division are polarized by their symbiotic nematode hosts [42]. X-ray crystallography revealed that SsgB from Streptomyces coelicolor resembles its distant orthologue from T. fusca (PDB ID: 3CM1). Key mutational residues were mapped onto the structure of SsgB. Further structural analyses and molecular simulation were conducted that highlight the important role of α2–α3 loop region to the function of SsgB. Our data support the predominant role of SsgB in the accurate positioning of the division site and the placement of the Z-ring.
2. Results
2.1. Mutational analysis of ssgB
SsgB shows unusual conservation, with near complete conservation within a genus and high divergence even between related actinobacterial genera (electronic supplementary material, figure S1). To investigate this further, we analysed the effect of amino acid substitutions in ssgB on cell division and morphogenesis, using S. coelicolor as the model system. For this, we first created a random mutant library using error-prone PCR, similar to the approach used previously for the mutational analysis of SsgA [43]. Mutant genes, as determined by sequencing, preceded by (and transcribed from) the original ssgB promoter region, were cloned into the low-copy number vector pHJL401 and introduced into the ssgB null mutant, followed by scrutiny of sporulation and cell division. To ascertain that the observed phenotypes were not owing to differences in SsgB expression, the mutant was also complemented with pHJL401 expressing wild-type SsgB from its original promoter, which gave a full wild-type sporulation phenotype. To exclude plasmid loss, plasmids from transformants expressing SsgB variants were isolated and sequenced before further analysis. Additionally, Western analysis was performed using anti-SsgB antibodies (electronic supplementary material, figure S2). Samples were equalized for protein load and normalized based on the levels of elongation factor EF-Tu1 [44]. This revealed an average expression level of 77 ± 10% of the wild-type level.
Spores of S. coelicolor are grey-pigmented owing to the production of the WhiE spore pigment [45], while colonies developing non-sporogenic aerial hyphae are white; intermediate phenotypes (reduced sporulation results in a light-grey pigmentation) also occur. This feature was used to subcategorize all transformants into three groups: white, light grey and grey. The mean grey level of growing patches was analysed based on the scanner images. By this approach, the degree of sporulation could be readily monitored (electronic supplementary material, figure S3 and table S1). 232 clones were isolated from the transformants and sequenced. Of these, 84 had no or silent mutations, 39 had multiple mutations and 65 had insertions or deletions. Of the 42 remaining clones, 35 unique single substitutions were identified and these were analysed further. Out of 35 SsgB variants, six failed to sporulate and the others showed significant sporulation defects or reduced sporulation (electronic supplementary material, figure S4 and table S1).
To obtain more detailed insights into the morphological changes correlating to the substitution mutants, the transformants expressing SsgB variants were subjected to transmission electron microscopy (TEM) (figure 1). Wild-type spores were homogeneous in size, with a thick electron-dense spore wall and condensed DNA in the centre of the spores. Conversely, spores from transformants expressing SsgB variants generally showed high variation in spore-wall thickness, spore size and shape, and frequently also aberrant DNA segregation and/or condensation (figure 1 and electronic supplementary material, table S2). Much to our surprise, in some cases up to 90° rotation of the septal plane was seen, dividing the spores parallel to the growth direction of the hyphae. This suggests that mutation of single SsgB residues may affect the coordination of cell division in aerial hyphae of Streptomyces, as detailed below.
Figure 1.
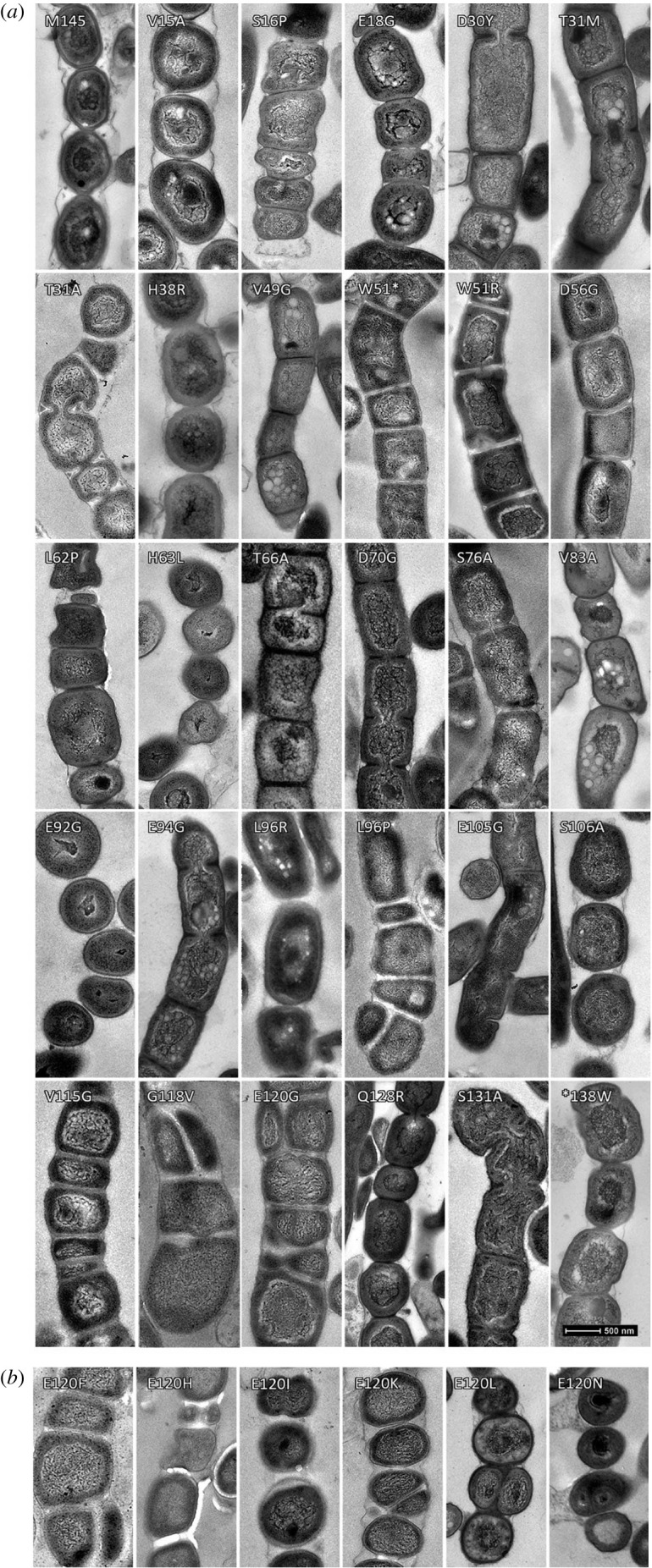
Transmission electron micrographs of sporogenic hyphae from the wild-type and single sporulating SsgB mutants. (a) D30Y, V49G, T66A, D70G, E94G and S131A result in thinner cell walls. L96P, V115G, G118V and E120G give rise to septum rotation, which also affect DNA condensation. Disturbed DNA segregation was observed in L88R (see electronic supplementary material, figure S4). Mutants expressing SsgB variants V15A, S16P, E18G, T31A, T31M, H38R, W51R, L62P, H63L, S76A, V83A, E92G, L96R, E105G, S106A and Q128R showed aberrant spore sizes (see also electronic supplementary material, table S2). (b) Six additional amino acid substitutions of E120 that lead to longitudinal division. (E120G is shown in figure 1a). Bars: 500 nm for TEM micrographs.
2.2. Rotation of the division plane owing to single amino acid substitutions in SsgB
Based on the outcome of the random mutant library, 22 residues were selected for site-saturated mutagenesis, namely W51, L88, A95, L96, L97 and the C-terminal residues 115–131 that are centred around E120 that correlated to the surprising longitudinal division (electronic supplementary material, table S3). Each of these residues was changed into on average 14 different aa residues using DNA synthesis. Mutants for the hydrophobic residues W51, L88, A95, L97, V115, P116 and P117 frequently had non-sporulating phenotypes (electronic supplementary material, table S3). Aberrant spore sizes were seen in most of the mutants, with some also showing irregular cell wall thickening (i.e. D70G; figure 1) and abnormal DNA segregation (L88R) (electronic supplementary material, figure S5 and table S2). Importantly, seven mutants wherein E120 was replaced by F, G, H, I, K, L or N produced septa with significant rotation of the division plane—in addition to canonical septa. The angles of these ectopically positioned septa ranged from diagonal to longitudinal (i.e. 90° rotation, with septa parallel to the hyphal wall), of which 22.7% were positioned diagonally (336 rotated septa out of 1479, 22.7%). Out of 336 rotated septa, 7.3% were longitudinally positioned. See figure 1b; electronic supplementary material, figure S6. In addition to mutants expressing SsgB E120 variants, longitudinal division was also seen in mutants expressing SsgB variants V115G, G118V, L96I, L96P and L96S, whereby the latter three produced relatively few ectopic septa, with an average of 1.7% rotated septation. To ascertain that longitudinal division does not occur in the wild-type strain under the chosen conditions, over 1011 spores of the wild-type strain were checked by scanning electron microscopy (SEM) and TEM, and not a single rotated septum was observed.
In order to see if the longitudinal septation also resulted in physical separation of the severed spores, we made impression prints of the strain expressing SsgB variant E120G, which had been grown for 7 days on soya flour mannitol (SFM) agar plates. These spores were then fixed with 1.5% glutaraldehyde in PBS, followed by dehydration using a graded series of acetone (70–100%). This experiment clearly demonstrated that the strain expressing SsgB-E120G produces spores that are longitudinally sectioned in two and that this process is completed by spore fission (figure 2b, panels (iii–iv)). Panel (ii) shows the size variability of SsgB-E120G mutant spores, and panels (iii) and (iv) present evidence of longitudinal indentations and a later stage of longitudinal division.
Figure 2.
Impression prints of spores from cells expressing wild-type SsgB or SsgB E120 mutants. (a) Longitudinal cell division (pointed by arrowhead) that are shown by fluorescent microscope from different E120 mutants. Left, bright-field images; middle, Syto9/PI stained images; right, overlays of the two images. All the spores were obtained after 7 days of growth from wild-type cells or from transformants of its ssgB null mutant expressing SsgB E120 mutants. Bars, 1 µm. (b) SEM imaging of wild-type SsgB and its E120G mutant. Panel (ii) shows the variability in spore size, whereas panels (iii) and (iv) show the start of longitudinal indentations and a later stage of longitudinal division, respectively.
Viability of the spores of mutants in which the residue E120 had been replaced by other aa was compared to those of wild-type spores. For this, impression prints were stained with Syto9 for viable spores and propidium iodide (PI) for dead spore, and imaged via fluorescence microscopy (FM). While wild-type SsgB spores were nearly all viable, those obtained from E120 substitution mutants varied a lot, with 5–70% dead spores, depending on the mutant (electronic supplementary material, figure S7). Like in the SEM experiments, longitudinal septation could also be seen from the outside using light microscopy, indicative of unique cell fission parallel to the hyphal wall (figure 2a, panels (ii–vi)). Some empty spore compartments were seen in the live/dead staining in the longitudinally septated spores (panels (ii) and (iv)), which can be explained by the unequal DNA septation during cell division in ssgB mutant. This resembles the minicompartments that were seen in parA deletion mutant, where the prespores are anucleate [46]. The rate of the physical separation of the severed spores was counted for E120 substitution mutant, which revealed a range of 3.3–9.8% split spores as compared to the normal separation spores (electronic supplementary material, table S4).
2.3. Localization and dynamics of SsgB variants
To test whether longitudinal division in the aerial hyphae correlated to the localization of SsgB, chimaeric SsgB-eGFP, SsgB-E120G-eGFP and SsgB-G118V-eGFP fusions were created as described [30]. While wild-type SsgB showed the typical pattern of foci on either side of the hyphal wall (electronic supplementary material, figure S8, panel a), the G118V variants localized more centrally and also longitudinally (electronic supplementary material, figure S8, panels b). This resulted in both canonical septal rings (perpendicular to the hyphal wall) and with a certain frequency also septa that were tilted by 90o (marked by arrowheads in electronic supplementary material, figure S8, panel b).
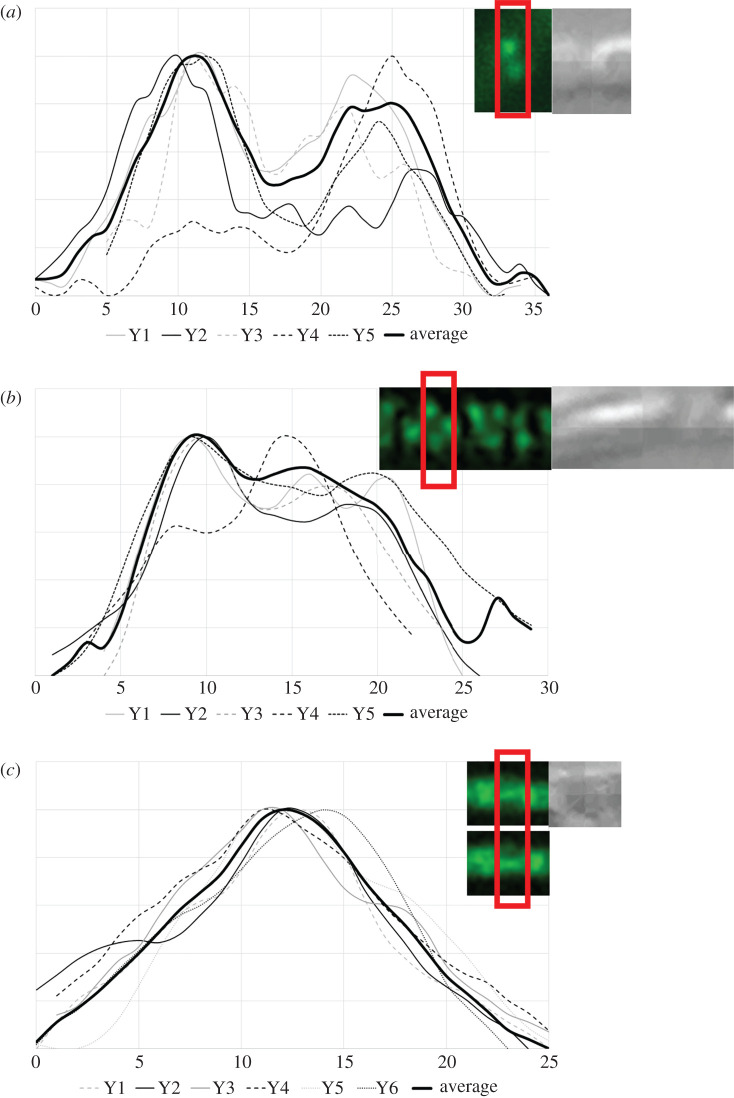
Fluorescence intensity distribution of SsgB-E120G-eGFP and SsgB-G118V-eGFP was compared with SsgB-eGFP. For comparison the same width of the hyphae was selected and five different septa were measured (Y1–5, figure 3). The plotted graph of wild-type SsgB indicates its localization on either side of the hyphae wall. Variants of SsgB-E120G and SsgB-G118V showed aberrant localization, which occasionally resulted in longitudinal septation; this correlated to localization of SsgB in the middle of the hyphae and perpendicular to the canonical septal plane. To gain insights into the dynamic association/dissociation of SsgB with the divisome, monomeric exchange was examined via fluorescence recovery after photobleaching (FRAP) for the E120G and G118V mutant. The recovery time after photobleaching was determined both on pre-septation foci as well as on septa. No difference in dynamics was seen between wild-type SsgB and its G118V variant. Both showed a recovery time of around 60 s (electronic supplementary material, figure S9), while the E120 variant differed slightly from the wild-type SsgB, with a recovery time of around 42 s, which is similar to previously reported data [30].
Figure 3.
Intensity plots of SsgB foci on the septa. (a) The wild-type SsgB-localization during early cell division shows two foci on either side of the hyphal wall; (b) Mutants expressing SsgB (E120G) showed aberrant localization, whereby SsgB was located all over the hyphal wall; (c) Occasional longitudinal septation was seen in SsgB (G118V), whereby eGFP fusions of SsgB mutant proteins localized parallel to the hyphal wall and in the middle of the hyphae. The red box indicates the width of the box that was used for making the profiles. Y1–Y5, single intensity plots of selected foci; X-axis, the distance between two foci on either side of hyphal wall; Y-axis, the fluorescent intensity.
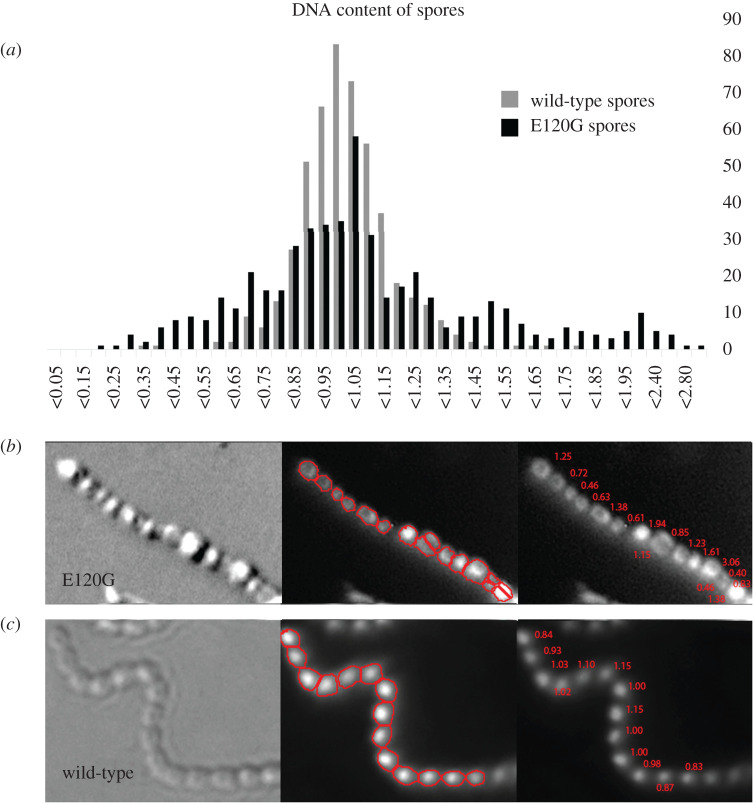
The relative DNA content per spore of over 500 spores of wild-type strain and SsgB-E120G mutant was studied. While wild-type spores showed a normal DNA distribution, SsgB-E120G mutant showed major variation in the relative DNA content per spore (figure 4). As an illustration, one spore chain containing longitudinal divisions is shown with the respective DNA content in each spore, which revealed 0.4–3.0 chromosomes for each spore in SsgB-E120G mutant (figure 4b). Conversely, 0.83–1.15 chromosomes were observed in the wild-type strains for each spore (figure 4c).
Figure 4.
DNA content analysis of SsgB variants. (a) The DNA content distribution in both wild-type (light grey) and E120G spores (black) is shown. The median DNA content of the spores was set to 1 to normalize the data. A normal distribution was observed for wild-type strain, whereas the strain expressing SsgB (E120G) had much more variation in DNA content. Y-axis, the frequency at which each DNA content was observed; X-axis, the DNA content in each spore. (b) To illustrate, one spore chain containing longitudinal divisions is shown with the respective DNA content in each spore (between 0.4 and 3.0 chromosomes). (c) In comparison, wild-type spores showed relatively little variation (between 0.83 and 1.15 in the spore chain).
To establish whether SsgB-E120G had retained the ability to interact with FtsZ, wild-type SsgB, SsgB variants E120G and E120A, as well as a C-terminally truncated version of SsgB (SsgBΔC, spanning 1–114 aa) were expressed and purified and then tested using a pelleting assay. FtsZ polymers formed in the presence of GTP. When wild-type SsgB or its E120A, E120G and SsgBΔC variants were added to the reaction mixture, both FtsZ and all SsgB variants were enriched in the pellet fractions, whereby the amount of FtsZ that pelleted was the same in all fractions, with or without SsgB (electronic supplementary material, figure S10A). The pelleting assay provides strong evidence that all of the SsgB variants can interact with FtsZ. We then tested the polymerization of FtsZ by the same SsgB variants using negative staining. Bundles were formed when wild-type SsgB or SsgB-E120G was mixed with FtsZ (electronic supplementary material, figure S10B). However, in the presence of SsgB E120A and SsgBΔC, less extensive bundling was observed. In addition, bacterial two-hybrid studies (BACTH) revealed that the interaction of SsgB with itself was not hampered by the E120G substitution, while the interaction with SsgA was reduced (electronic supplementary material, figure S11). BACTH of wild-type SsgB or SsgA with SsgB variants V115G, G118V and L96P were also measured. While SsgB V115G and G118V showed similar interaction with wild-type SsgB and its E120G variant, the interaction of SsgB L96P with either SsgB or SsgA was largely reduced.
2.4. Crystal structure of Streptomyces coelicolor SsgB
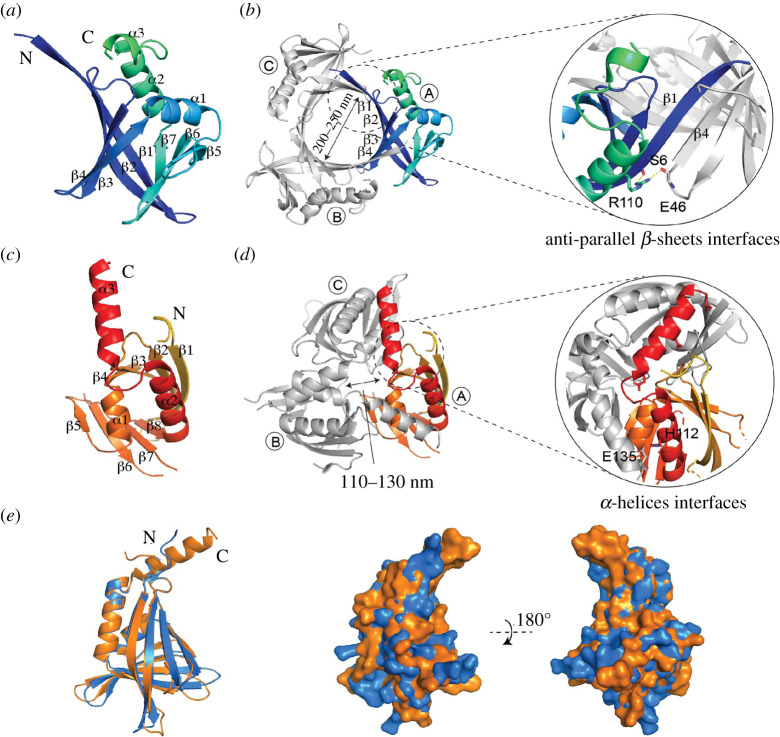
In order to gain more insights into the structure–function relationship for key SsgB residues, the structure of S. coelicolor SsgB (ScSsgB) was resolved via X-ray crystallography. For this, tetragonal crystals of ScSsgB were obtained, and based on this, a homo-trimer was resolved at 2.1 Å (PDB ID Code 6SLC) with eight molecules per asymmetric unit (electronic supplementary material, table S6). The 13 aa residues at the C-terminus were highly mobile and could therefore not be modelled, owing to lack of electron density. Each subunit was arranged as an α + β fold, with seven β-strands packed into a barrel structure, covered by three α-helices (figure 5a,b), which strongly resemble those of T. fusca SsgB (Tf SsgB) structure [36] (PDB ID: 3CM1, figure 5c,d). The root-mean-square deviation (r.m.s.d.) is 1.9 Å with 92% of all residues aligned in ScSsgB and 87% in Tf SsgB (aa sequence identity between these two proteins is 46%). A superimposition of ScSsgB and Tf SsgB subunits is shown in figure 5e. ScSsgB trimer adopts a ‘whirly’ shape and is assembled through an antiparallel β-sheet interaction between β1 from one subunit and β4 from the neighbouring subunit (figure 5b; electronic supplementary material, figure S12). By contrast, the Tf SsgB trimer is assembled mainly through α-helices (figure 5d). The ScSsgB trimer forms a 12-stranded beta-barrel with an inner diameter of about 20–25 Å (figure 5b).
Figure 5.
Crystal structure of the ScSsgB trimer. (a) Ribbon diagrams showing the monomer structure of SsgB from S. coelicolor. (b) The overall structure of ScSsgB reveals a trimer. Structure statistics are listed in electronic supplementary material, table S6. The interface between adjacent monomers is formed by two antiparallel β-sheets. (c) The monomer structure of SsgB from T. fusca (PDB code 3CM1). (d) The interface between adjacent monomers of Tf SsgB is formed by α-helices. (e) Overlap of ScSsgB (blue) and Tf SsgB (orange) subunits. Left, side view of the electrostatic surface alignment of ScSsgB and Tf SsgB structure. Right, the same electrostatic figure but rotated by 180°.
Crystallographic data obtained for SsgB from T. fusca [36] and for S. coelicolor (this work) suggested that SsgB forms trimers. To ascertain this, size-exclusion chromatography (SEC) of wild-type ScSsgB and ScSsgBΔC was conducted. SEC results revealed that purified ScSsgB and ScSsgBΔC mainly existed as a monomer in solution (electronic supplementary material, figure S13), which is consistent with our previous multimerization studies of T. fusca SsgB [36], indicating that a possible equilibrium exists between the monomeric and trimeric states.
2.5. Analysis of single aa substitutions and mapping of key residues
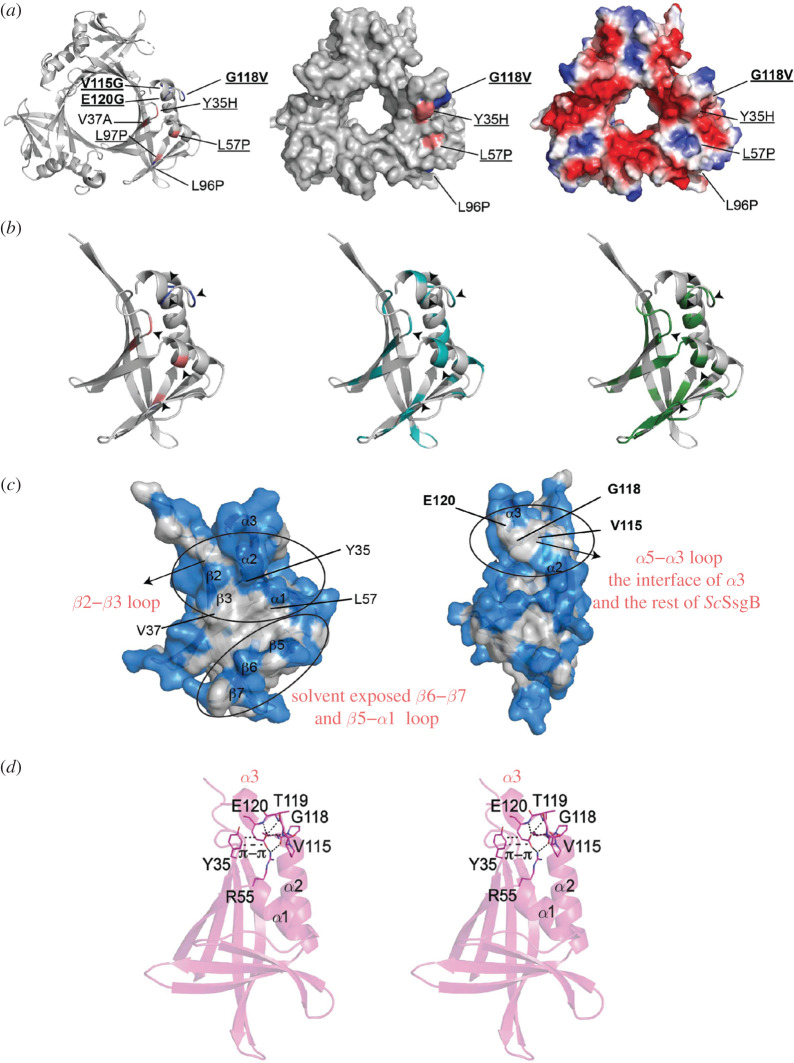
Key residues that correlated to the occurrence of longitudinal cell division were mapped onto the ScSsgB trimer structure (figure 6a; electronic supplementary material, figure S14A). Mutations that correlate to residues that are evolutionarily conserved in all SALPs are underlined (figure 6a; electronic supplementary material, figure S14B). Residues V115, G118 and E120 cluster together, and are centered on the lid of the beta-barrel consisting of α1, α2–α3 and β1–β2 loop, which is close to the interface between α3 and the rest of the subunit (figure 6a,b). Consistent with their strategic localization in the α2–α3 loop (figure 6c), aa substitutions V115G, G118V or E120G resulted in the formation of aberrant tilted septa in addition to the canonical septa perpendicular to the hyphal wall, with some septa showing full 90° rotation of the division plane. Residue E120 plays a key role in maintaining the proper angle between α3 and the rest of the protein. Three hydrogen bonds are formed between the E120 side chain and the main chains of E120, T119 and G118 in the α2–α3 loop region (figure 6d). Besides, E120 and V115 provide two additional salt bridges to R55. Interestingly, a π–π interaction and a hydrogen bond were observed for E120-Y35 and H121-Y35. All these interactions stabilize the angle of α3, supporting the importance of the proper angle between α3 and the rest of the protein to the function of ScSsgB.
Figure 6.
Key mutations and their interactions in SsgB trimer structure. (a) Left, aa substitutions L96P, V115G, G118V and E120G show tilted division; aa substitutions of Y35H, V37A, L57P and L97P (deep salmon) result in non-sporulating phenotype; middle, right, top view of all the functional aa substitutions mapped on the surface structures. Conserved residues are underlined. (b) Left, key residues as shown by (a); middle, residues corresponding to all substitutions in this study (cyan); right, conserved residues (green). (c) Key residues (Y35H, V37A, L57P and L97P) are located in the hydrophobic patch, while V115, G118 and E120 are clustered on the α2–α3 loop region that mediates the interactions of α3 and the globular domain of SsgB. White patches in the surface structure indicate hydrophobic residues. (d) Stereo-view of E120 in the monomer structure and its interactions with the surrounding residues.
2.6. Molecular dynamic simulation of SsgB-E120G
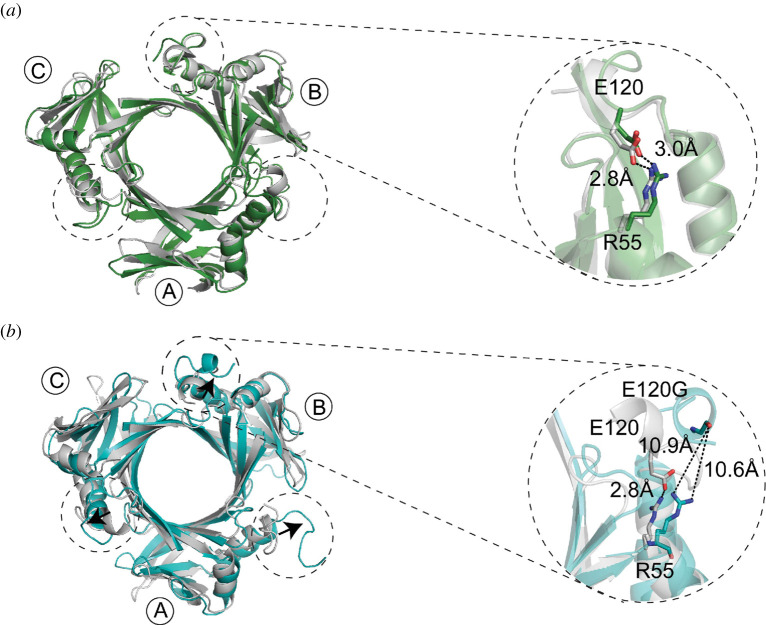
Our work demonstrated that residue E120 plays a key role in maintaining the proper angle of α3 relative to the rest of the protein. Mutation of this residue would disrupt the interaction, and changing the angle of α3 may drive rotation of the septum plane, and thus explain the observed longitudinal cell division. The heading and the high B-factor of the existing residues in the α3-helix (electronic supplementary material, figure S15) suggest that it can extend flexibly to the centre of the trimer. Despite many attempts under different crystallization conditions, we failed to obtain crystals for SsgB-E120G or SsgB E120A. In silico molecular dynamics revealed that while the α3 helix of the wild-type SsgB protein is not affected significantly by the simulations, with a distance of 2.7 and 3.0 Å, respectively, between E120 and R55 (figure 7a), the α3 helix of SsgB-E120G flips some 90° outwards of the tight trimer, with the distance increasing to 10.6 and 10.9 Å, respectively (figure 7b). This provides supportive evidence that the angle of the α3 helix plays a key role in the proper functionality of SsgB.
Figure 7.
Molecular dynamic simulation of SsgB wild-type and E120G mutant. (a) MD result of SsgB wild-type structure. The α3 helix stays at the same orientation and the distance between R55 and E120 keeps at 2.8 Å and 3.0 Å, before (green) and after MD (grey). (b) The arrows indicate the changed angle of SsgB-E120G mutant (cyan) compared to the wild-type SsgB (grey) after MD.
3. Discussion
SALPs play a central role in controlling the steps of sporulation-specific cell division in Streptomyces. Inspired by the extremely high conservation of the SsgB protein in Streptomyces species, with natural variants only found in aa 128 (Q, R or T), we studied the effect of point mutations in the protein on cell division and morphogenesis of the model strain S. coelicolor. As expected, many mutants showed morphological defects relating to cell division and sporulation, including varying spore sizes, aberrant DNA segregation and condensation, and cell wall thickening. However, SsgB substitutions L96P, V115G, G118V and various aa substitutions in residue E120 caused rotated to longitudinal cell division in addition to the canonical perpendicular septa. As a result, spores were formed that were diagonally or longitudinally sectioned, visualized both inside spores (via TEM and FM) and outside (via SEM and FM). These results highlight the dominant role of SsgB in the control of cell division and Z-ring formation in Streptomyces, which is also consistent with the observation of the extremely high level of conservation of SsgB within Streptomyces species.
It is important to note that diagonal or longitudinal septa always coincided with canonically oriented septa as seen by TEM and FM. Furthermore, the longitudinal septa always connected two canonical septa: we never observed these additional Z-rings formed between septa and the hyphal wall. And finally, the longitudinal septa could be formed with different spacing relative to the hyphal wall, allowing asymmetric cleavage of spores. This suggests that during normal septation, a second septum is formed that grows sideways, under different angles. Evidence of longitudinal septation was also frequently seen as diagonally or longitudinally severed spores. This shows that these ectopic cell division events were completed via cytokinesis. However, the mechanism of longitudinal cell division is unclear and needs further analysis; we cannot exclude that rotated septa formed in prespore compartments could be essentially spherical or cylindrical. DNA content and viability studies of SsgB E120 substitution mutants revealed abnormal chromosome distribution (0.4–3.0) and relatively reduced viability (5–70% dead spores) compared to the wild-type strain. Besides affecting septum positioning, mutations of SsgB also resulted in defects in spore viability, spore shape, wall thickness and DNA condensation and/or segregation. Analysis of substitutions of residue L96 revealed that exactly two thirds of all spores had disturbed DNA condensation. Aberrant DNA segregation was also seen frequently in SsgB L88R mutants. These data indicate that SsgB may play key roles beyond septum positioning. BACTH studies revealed that the interactions of SsgB variants V115G, G118V and E120G with wild-type SsgB have been retained, while the interactions of these variants with SsgA were largely reduced. Notably, for SsgB variant L96P, its interactions with SsgB and SsgA were all blocked. SALPs such as SsgA–G play important and highly specific roles in the control of the sporulation process [34]. Previous studies showed that mutants lacking ssgC are disturbed in DNA segregation/condensation, those lacking ssgD are defective in spore-wall synthesis, those lacking either ssgE or ssgF have defects in autolytic spore separation and ssgG is required for correct septum localization [34]. It is very possible that altered interaction between SsgB and one or more other SALPs, or perhaps other components of the divisome, may contribute to the ectopic septum placement. This requires further verification.
SsgB-E120G, E120A and SsgBΔC had retained the ability to interact with FtsZ in vitro. However, instead of promoting the filamentation of FtsZ, SsgB and its variants may assist in the bundling of FtsZ filaments as observed by negative staining. In vivo localization studies showed that the foci of SsgB-E120G-eGFP and G118V-eGFP simultaneously formed both canonical and longitudinal septa. FRAP studies on SsgB-E120G-eGFP and G118V-eGFP further confirmed the in vivo studies by showing that these variants had the same dynamics as wild-type SsgB; this was true for foci that were localized canonically (adjacent to the cell wall) and for those that were centrally located.
Notably, protein structural analyses revealed that residues that are associated with septum misplacement are located in the α2–α3 loop region that link the α3 helix and the rest of the protein. Several interactions, including hydrogen bonds (E120–E120, E120–T119 and E120–G118, H121–Y35), salt bridges (E120–R55 and V115–R55) and π–π interaction (E120–Y35) likely stabilize and maintain the proper angle of the α3 helix with respect to the rest of the protein. Mutating E120 likely disrupts this critical interaction, and thus results in major changes in the orientation of α3, by up to 90°. Mutations of the interacting partners all showed functional defects, as seen from the blocked cell division (Y35H), septum rotation (V115G, G118V), DNA segregation (V115G, G118V) and heterogeneity in spore sizes (V115G, G118A, H121I). Moreover, some longitudinal septa were seen in L96P mutants, which again can be explained by changes in the orientation of helix α3, as the mutation will disrupt the interaction between the neighbouring α2 helix and β 7-strand. Molecular dynamic (MD) simulation further confirmed that the E120 substitutions are correlated to the motion of α3 helix of SsgB. These data support the significance of the orientation of helix α3 to the function of SsgB. However, whether changing the position of helix α3 is the direct factor that causes the observed diagonal and longitudinal cell division events remains to be established.
Naturally occurring longitudinal fission had been reported in the insect endosymbiont Spiroplasma poulsonii and two marine nematode-attached Gammaproteobacteria, Candidatus Thiosymbion oneisti and Thiosymbion hypermnestrae [42,47]. In Spiroplasma, cells are divided in a Y-shape mode that links to Z-ring reorientation, while in the Gammaproteobacteria, the machineries for growth and division are not reoriented; instead, they mesh to a point at which they are considered as ‘squeezed’ E. coli cells. Mutants in FtsZ that alter the division plane have also been reported in E. coli, B. subtilis and Streptomyces spp. expressing mutant FtsZ species [39–41]. In temperature-sensitive ftsz26 (Ts) mutants of E. coli, FtsZ forms spirals that further contract and form spirally invaginating septa [40]. The abnormal septal invaginations were also observed in a similar ftsZ (ts1) mutant in B. subtilis [41]. Specific aa substitutions F37I and F37R of FtsZ led to spiraling septation in Streptomyces spp. [39]. These results suggest that the geometry of division could occur in the perpendicular plane as seen in Staphylococcus aureus [48]. Our work shows that rather than aa substitutions in FtsZ itself, changes in FtsZ-recruiting proteins such as SsgB in Streptomyces also lead to diagonally or longitudinally divided spores. Further studies to localize FtsZ in SsgB-G118V, E120G and E120A mutants are underway to understand the mechanism behind longitudinal cell division in Streptomyces. It will also be important to study the interactions between SsgB-G118V, E120G and E120A proteins and other cell division components and to determine whether mutations in the α2–α3 loop affect interactions with other proteins.
4. Conclusion
Our work shows that single aa substitutions in SsgB affect cell wall synthesis, septum orientation and DNA condensation and segregation during sporulation of S. coelicolor, including unique longitudinal division events that have not been seen previously and lead to horizontally severed spores. The crystal structure of S. coelicolor SsgB was resolved and key mutational residues were mapped onto the structure. Notably, residues that are associated with septum misplacement are located in α2–α3 loop region that links the α3 helix and the rest of the protein. Further protein structural analyses and MD simulation showed that residues V115, G118 and E120 are essential to maintain the proper angle of the α3 helix, whereby substitution of aa V115G, G118V or E120G ultimately leads to cell fission along the long axis of the spores. Our work provides important new structural and biochemical insights into the function of the key cell division control protein SsgB during sporulation-specific cell division in Streptomyces.
5. Material and methods
5.1. Strains and culturing conditions
All strains described in the paper are listed in electronic supplementary material, table S7. S. coelicolor M145 was obtained from the John Innes centre strain collection. Its ssgB null mutant was described previously [49]. Transformants harbouring pHJL401 plasmids were grown on SFM agar plates containing apramycin (for the ssgB deletion) and thiostrepton (to maintain the plasmid) to final concentrations of 50 µg ml−1 and 25 µg ml−1 at 30°C. For growth in liquid medium, the recombinants were grown in a 1 : 1 mix of TSBS and YEME at 30°C. Escherichia coli JM109 was used for amplification of plasmids. Escherichia coli Rosetta™ 2 (DE3) pLysS strain was used for overexpression of all the proteins used in this paper. Escherichia coli BTH101 was used for BACTH assay. Protoplasts used for transformation were prepared based on previously described protocols [50,51] (see electronic supplementary material, method).
5.2. Random mutagenic PCR
The SsgB promoter region and coding sequence were amplified separately from the S. coelicolor chromosome using oligonucleotide pairs pSsgB_fw + pSsgB_rv and SsgB_fw + SsgB_rv, respectively (electronic supplementary material, table S8). An NdeI restriction site was introduced overlapping the translational start codon to enable ligation of these fragments after mutagenic PCR. Because pUC19 and pHJL401 already contain an NdeI site, these were removed prior to construct creation using the DNA polymerase I Klenow fragment method. The * behind the plasmid indicates that an NdeI site was removed. The ssgB gene (EcoRI-HindIII) was cloned into a variant of pUC19*, thereby creating pJPM1. Construct pJPM2 was based on the low-copy number shuttle vector pHJL401* and contained the PCR-amplified ssgB promoter fragment (cloned as EcoRI-HindIII fragment). Mutations in S. coelicolor ssgB were introduced by random mutagenic PCR using pJPM1 as template, as described [43]. The desired mutation frequency of 1–3 mutations was checked both on gel and by sequencing. The 500 bp long fragments were purified and amplified by a standard PCR. The mixture of mutagenized ssgB genes produced by error-prone PCR was then ligated as NdeI-HindIII fragments under the control of the natural ssgB promoter in pJPM2. The DNA was subsequently transformed into protoplasts of the S. coelicolor ssgB null mutant, thereby generating a collection of Streptomyces colonies each expressing a variant of SsgB from the natural ssgB promoter. For the eGFP fusions, we amplified the ssgB-eGFP fusion from the adapted cosmid STL2 by using primers pSsgB_fw and GFP_rv. The centre part of ssgB was then replaced by SacI, XhoI digestion (two naturally occurring restriction sites within ssgB) with the substitution version of ssgB. Mutations were verified by DNA sequencing.
5.3. Scanner-based imaging
Plates were incubated at 30°C on a flat-bed CCD scanner and imaged every 30 min for 7 days. Automated scanning was performed using Quickscan (www.burrotech.com) activated by the Windows task scheduler. Hereafter the image stack was analysed for grey values using imageJ/FIJI. This was achieved by drawing an equal sized circle in the middle of the grown colonies and measuring the grey level intensity of the stack via the Measure stack plugin of ImageJ.
5.4. Automated spore measurements
For each image in the folder, the scale is set to correspond to the microscope's settings, hereafter the image size is increased to optimize for averaging of pixels values at later stages of the macro. After increasing the image size by 5% in 20 consecutive operations, the edge detection filter of ImageJ is applied. Everything above the default threshold is defined as a proper edge. The holes are filled and the spores and hyphae that are detected in this manner are defined as ‘in focus’. The original file is thresholded with default settings and combined with an AND operation with the sharp spores. Particles that fall within the range of spores are analysed, with minimum size of 0.65 μm2, and a roundness value between 0.75 and 1.
5.5. Microscopy
5.5.1. Live/dead staining and DNA quantification
For live/dead staining, Streptomyces strains were grown on SFM agar plates and after 7 days coverslips were pressed onto the colony and mounted in PBS containing 10 µM syto 9 and 10 µg ml−1 PI. Fluorescence and light microscopy were performed as described previously [52].
For DNA quantification, Streptomyces colonies were grown against coverslips (at 45° angle) on SFM agar, and after 7 days the agar samples were fixed with 2% paraformaldehyde for 5 min and washed with 70% ethanol. Subsequently spores were stained with 1 µM Syto green (ThermoFischer) and imaged with a zeiss Axioplan 2, with 470/40 excitation and 525/50 emission. For localization studies, coverslips were immediately imaged without pretreatment, using the same microscope. For DNA quantification the total intensity of each separate spore in a spore chain was measured and to circumvent staining variation the median value of each spore chain was set to 1 to normalize the data. To have enough data for normalization, we used spore chains that consisted of a minimum of 10 spores. All images were background-corrected, setting the signal outside the hyphae to 0 to obtain a sufficiently dark background. Images were processed using Adobe Photoshop CS4 and FIJI.
5.5.2. Fluorescence recovery after photobleaching
FRAP was performed with a Zeiss Imager LSM 510, using 488 nm excitation and 505–550 nm detection as described [30].
5.5.3. Electron microscopy
Cryo-scanning electron microscopy (cryo-SEM) was performed as described [53]. For SEM imaging of individual spores, impression prints of 7-day old confluent plates were obtained and fixed with 1.5% glutaraldehyde in PBS. After 15 min fixation, samples were dehydrated using a graded series of acetone (70%, 80%, 90%, 96%, 100%, 15 min each) and subsequently critical point dried. Before examination, 10 nm Platinum/Paladium was sputter coated on the sample to prevent charging during imaging. All images were obtained with a Jeol 7600 at 5 kV and a working distance of 8 mm.
For TEM, small cubes of colonies were fixed with 1.5% glutaraldehyde in PBS for 1 h, postfixed with osmium tetroxide (1%) for 1 h and dehydrated with a graded ethanol series (70%, 80%, 90%, 100% 15 min each). Ultrathin sections of 70 nm were cut and examined using a Jeol 1010. Purified FtsZ (15 µM) was combined with an equimolar amount of wild-type SsgB, or one of its mutants (SsgB-E120G, SsgB E120A and SsgBΔC) in a reaction buffer of 20 mM HEPES, pH 7.5, 150 mM KCl, 2.5 mM MgCl2. Polymerization was initiated by the addition of 2 mM GTP to the assembly reaction. 50 µl reaction mixture was incubated for 2 min at 37°C. 15 µl aliquot was placed on a carbon-coated copper grid and negatively stained with 2% uranyl acetate for 10 min, then washed and dried. Images were collected using transmission electron microscope (Jeol 1010) operated at 70 kV with 670 pA cm−2 density and recorded on a camera.
5.6. Co-pelleting assay
For pelleting experiments, purified FtsZ, with or without equimolar ratios of wild-type SsgB, or its mutants (SsgB-E120G, SsgB E120A and SsgBΔC) were mixed at a final concentration of 15 µM. The samples were pre-spun at 50 k r.p.m. in a Beckman 70 Ti rotor for 30 min. Then supernatants were transferred to new tubes and 2 mM MgCl2 and GTP were added. After 10 min of incubation at 30°C, the tubes were centrifuged at 65 k r.p.m. at 20°C for 30 min. The supernatants were removed for analysis and pellets were washed with the same buffer aside from the proteins. Pellets were dissolved in SDS gel loading buffer. All the samples were analysed on a 4–16% Pre-cast SDS-PAGE gel (BIO-RAD).
5.7. Protein expression and purification
For expression of His6-tagged fusions of SsgB and FtsZ in E. coli, the entire coding region of ssgB or its mutants was cloned from the genomic DNA of S. coelicolor as NdeI-HindIII fragments and ligated into pET28a (Novagen). Sequences of S. coelicolor ftsZ were codon optimized prior to cloning into pET28a. SsgB was overexpressed in E. coli Rosetta™ 2 (DE3) pLysS strain as a 157 aa-long fusion protein containing the 137 aa native polypeptide and an N-terminal tag, GSSHHHHHHSSG. SsgB point mutants E120G and E120A, or its deletion mutant (ΔSsgB, 1–114 aa; lacking the C-terminal 11 aa), were expressed in the same way. FtsZ was produced as C-terminal His-tag fusion. His6-tagged FtsZ proteins were purified using routine methods as described [54].
5.8. Oligomerization studies of SsgB
The polymerization state of SsgB was analysed by size-exclusion chromatography on an ÄKTA-pure system using column Superdex™ 75 Increase 10/300 GL. The elution solvent was 20 mM HEPES, 150 mM KCl, 2.5 mM MgCl2, 5% Glycerol, 1 mM DTT, at pH 7.5, and the flow rate was 0.1 ml min−1. Absorbance at 280 nm was monitored. Conalbumin, γ-globulin, ovalbumin, myoglobin and vitamin B12 were used as molecular weight standards.
5.9. Crystallization and structure determination
SsgB trimer crystals were grown at 18°C by using the sitting-drop vapour-diffusion method. In a typical experiment, 1 µl of the protein stock (6.2 mg ml−1 protein) was mixed with 1 µl of a reservoir buffer consisting of 0.2 M sodium chloride, 0.1 M sodium/potassium phosphate (pH 6.2), 50% PEG200. Crystals were moved to the same condition supplemented with 20% glycerol and flash-frozen in liquid nitrogen. Data were collected at beam line ID30B [55] at ESRF (Grenoble, France, 2017). Images were collected with a 0.15° oscillation angle and an exposure time of 0.037 s per frame at 100 K. Crystals form diffracted to 2.1 Å for SsgB trimer. The data were processed with XDS [56] and scaled using AIMLESS [57] from CCP4 package [58]. Phases of SsgB trimer were calculated by molecular replacement with SsgBTfus PDB entry 1C3M [36] as a model using MOLREP [59]. The structures were finalized by manual building in COOT [60] and refined with REFMAC [61]. Residues were in the most favoured regions of the Ramachandran plot [62] as determined by PROCHECK [63]. Crystallographic data are summarized in electronic supplementary material, table S6. SsgB trimer structure was deposited in the PDB with code 6SLD.
5.10. Molecular simulation
All atom molecular dynamics (AAMD) simulation of SsgB wild-type and SsgB-E120G were prepared and run using Gromacs 2016 package [64]. The simulated protein, wild-type or mutant, was centred in a cubic box surrounded by water molecules and counter ions Na+ were added to keep the total charge of the simulation box to be zero. The bonded and non-bonded parameters were obtained from AMBER99SB force field [65]. The SPCE [66] water model was used. The particle-mesh Ewald method was used to treat the long-range electrostatic interactions [67]. A cutoff of 12 Å was used for non-bonded interactions. Temperature was maintained at 300 K using v-rescale thermostat [68]; pressure was maintained at 1 bar Parrinello–Rahman barostat [69,70]. The simulation box was cubic of length around 9.2 nm, with periodic boundary conditions applied to all dimensions. All simulated systems were stabilized though energy minimization, short NVT (10 ps) and short NPT (5 ns) MD simulations to relax to favourable conformations. NPT simulations at 300 K and 1.0 bar were performed upon the stabilized structures and 50 ns trajectories were collected for our analysis.
Acknowledgements
We thank Helga van der Heul for assistance in bacteria two-hybrid analysis. We gratefully acknowledge the European Synchrotron Radiation Facility, Grenoble, France.
Contributor Information
Jan Pieter Abrahams, Email: jan-pieter.abrahams@psi.ch.
Gilles P. van Wezel, Email: g.wezel@biology.leidenuniv.nl.
Data accessibility
The atomic coordinates of SsgB from Streptomyces coelicolor have been deposited in the Protein Data Bank (PDB) with accession code 6SLC.
Authors' contributions
G.P.v.W. and J.W. conceived the project. J.W. conducted the mutational studies and imaging analysis. X.X. performed the protein overexpression, purification and crystallography experiments. X.X. and N.P. collected the crystal data and solved the SsgB structure, in close collaboration with J.P.A. and M.L. X.X. performed the biochemical studies and P.V. the oligomerization studies. X.L. performed molecular dynamic simulations. G.P.v.W., J.P.A. and N.P. supervised the project. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Funding
This work was supported by a grant from the Chinese Scholarship Council (CSC).
References
- 1.Bi EF, Lutkenhaus J. 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354, 161-164. ( 10.1038/354161a0) [DOI] [PubMed] [Google Scholar]
- 2.Mukherjee A, Lutkenhaus J. 1994. Guanine nucleotide-dependent assembly of FtsZ into filaments. J. Bacteriol. 176, 2754-2758. ( 10.1128/JB.176.9.2754-2758.1994) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erickson HP, Taylor DW, Taylor KA, Bramhill D. 1996. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc. Natl Acad. Sci. USA 93, 519-523. ( 10.1073/pnas.93.1.519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Boer P, Crossley R, Rothfield L. 1992. The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359, 254-256. ( 10.1038/359254a0) [DOI] [PubMed] [Google Scholar]
- 5.Lyu Z, Coltharp C, Yang X, Xiao J. 2016. Influence of FtsZ GTPase activity and concentration on nanoscale Z-ring structure in vivo revealed by three-dimensional Superresolution imaging. Biopolymers 105, 725-734. ( 10.1002/bip.22895) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holden SJ, Pengo T, Meibom KL, Fernandez Fernandez C, Collier J, Manley S. 2014. High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc. Natl Acad. Sci. USA 111, 4566-4571. ( 10.1073/pnas.1313368111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang X, Lyu Z, Miguel A, McQuillen R, Huang KC, Xiao J. 2017. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355, 744-747. ( 10.1126/science.aak9995) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bisson-Filho AW, et al. 2017. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739-743. ( 10.1126/science.aak9973) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strauss MP, Liew AT, Turnbull L, Whitchurch CB, Monahan LG, Harry EJ. 2012. 3D-SIM super resolution microscopy reveals a bead-like arrangement for FtsZ and the division machinery: implications for triggering cytokinesis. PLoS Biol. 10, e1001389. ( 10.1371/journal.pbio.1001389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramirez-Diaz DA, Garcia-Soriano DA, Raso A, Mucksch J, Feingold M, Rivas G, Schwille P. 2018. Treadmilling analysis reveals new insights into dynamic FtsZ ring architecture. PLoS Biol. 16, e2004845. ( 10.1371/journal.pbio.2004845) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pichoff S, Lutkenhaus J. 2005. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol. Microbiol. 55, 1722-1734. ( 10.1111/j.1365-2958.2005.04522.x) [DOI] [PubMed] [Google Scholar]
- 12.Hale CA, deBoer PAJ. 1997. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88, 175-185. ( 10.1016/S0092-8674(00)81838-3) [DOI] [PubMed] [Google Scholar]
- 13.Pichoff S, Lutkenhaus J. 2002. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21, 685-693. ( 10.1093/emboj/21.4.685) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adams DW, Errington J. 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7, 642-653. ( 10.1038/nrmicro2198) [DOI] [PubMed] [Google Scholar]
- 15.Margolin W. 2005. FtsZ and the division of prokaryotic cells and organelles. Nat. Rev. Mol. Cell Biol. 6, 862-871. ( 10.1038/nrm1745) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lutkenhaus J. 2007. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 76, 539-562. ( 10.1146/annurev.biochem.75.103004.142652) [DOI] [PubMed] [Google Scholar]
- 17.Shapiro L, McAdams HH, Losick R. 2009. Why and how bacteria localize proteins. Science 326, 1225-1228. ( 10.1126/science.1175685) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lluch-Senar M, Querol E, Pinol J. 2010. Cell division in a minimal bacterium in the absence of ftsZ. Mol. Microbiol. 78, 278-289. ( 10.1111/j.1365-2958.2010.07306.x) [DOI] [PubMed] [Google Scholar]
- 19.McCormick JR. 2009. Cell division is dispensable but not irrelevant in Streptomyces. Curr. Opin. Biotechnol. 12, 689-698. ( 10.1016/j.mib.2009.10.004) [DOI] [PubMed] [Google Scholar]
- 20.McCormick JR, Su EP, Driks A, Losick R. 1994. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol. Microbiol. 14, 243-254. ( 10.1111/j.1365-2958.1994.tb01285.x) [DOI] [PubMed] [Google Scholar]
- 21.Barka EA, et al. 2016. Taxonomy, physiology, and natural products of the Actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1-43. ( 10.1128/MMBR.00019-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hopwood DA. 2007. Streptomyces in nature and medicine: the antibiotic makers. New York, NY: Oxford University Press. [Google Scholar]
- 23.van der Heul HU, Bilyk BL, McDowall KJ, Seipke RF, van Wezel GP. 2018. Regulation of antibiotic production in Actinobacteria: new perspectives from the post-genomic era. Nat. Prod. Rep. 35, 575-604. ( 10.1039/C8NP00012C) [DOI] [PubMed] [Google Scholar]
- 24.Claessen D, Rozen DE, Kuipers OP, Sogaard-Andersen L, van Wezel GP. 2014. Bacterial solutions to multicellularity: a tale of biofilms, filaments and fruiting bodies. Nat. Rev. Microbiol. 12, 115-124. ( 10.1038/nrmicro3178) [DOI] [PubMed] [Google Scholar]
- 25.Flärdh K, Buttner MJ. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7, 36-49. ( 10.1038/nrmicro1968) [DOI] [PubMed] [Google Scholar]
- 26.Wildermuth H, Hopwood D. 1970. Septation during sporulation in Streptomyces coelicolor. J. Gen. Microbiol. 60, 51-59. ( 10.1099/00221287-60-1-51) [DOI] [PubMed] [Google Scholar]
- 27.Celler K, Koning RI, Willemse J, Koster AJ, van Wezel GP. 2016. Cross-membranes orchestrate compartmentalization and morphogenesis in Streptomyces. Nat. Commun. 7, 11836. ( 10.1038/ncomms11836) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yagüe P, et al. 2016. Subcompartmentalization by cross-membranes during early growth of Streptomyces hyphae. Nat. Commun. 7, 12467. ( 10.1038/ncomms12467) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jakimowicz D, van Wezel GP. 2012. Cell division and DNA segregation in Streptomyces: how to build a septum in the middle of nowhere? Mol. Microbiol. 85, 393-404. ( 10.1111/j.1365-2958.2012.08107.x) [DOI] [PubMed] [Google Scholar]
- 30.Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. 2011. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes Dev. 25, 89-99. ( 10.1101/gad.600211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, Willemse J, Claessen D, van Wezel GP. 2016. SepG coordinates sporulation-specific cell division and nucleoid organization in Streptomyces coelicolor. Open Biol. 6, 150164. ( 10.1098/rsob.150164) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlimpert S, Wasserstrom S, Chandra G, Bibb MJ, Findlay KC, Flardh K, Buttner MJ. 2017. Two dynamin-like proteins stabilize FtsZ rings during Streptomyces sporulation. Proc. Natl Acad. Sci. USA 114, E6176-E6183. ( 10.1073/pnas.1704612114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plocinski P, et al. 2011. Characterization of CrgA, a new partner of the Mycobacterium tuberculosis peptidoglycan polymerization complexes. J. Bacteriol. 193, 3246-3256. ( 10.1128/JB.00188-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noens EE, Mersinias V, Traag BA, Smith CP, Koerten HK, van Wezel GP. 2005. SsgA-like proteins determine the fate of peptidoglycan during sporulation of Streptomyces coelicolor. Mol. Microbiol. 58, 929-944. ( 10.1111/j.1365-2958.2005.04883.x) [DOI] [PubMed] [Google Scholar]
- 35.Traag BA, van Wezel GP. 2008. The SsgA-like proteins in actinomycetes: small proteins up to a big task. Antonie van Leeuwenhoek 94, 85-97. ( 10.1007/s10482-008-9225-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Q, et al. 2009. Structural and functional characterizations of SsgB, a conserved activator of developmental cell division in morphologically complex actinomycetes. J. Biol. Chem. 284, 25 268-25 279. ( 10.1074/jbc.M109.018564) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Girard G, Traag BA, Sangal V, Mascini N, Hoskisson PA, Goodfellow M, van Wezel GP. 2013. A novel taxonomic marker that discriminates between morphologically complex actinomycetes. Open Biol. 3, 130073. ( 10.1098/rsob.130073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Dissel D, Claessen D, Van Wezel GP. 2014. Morphogenesis of Streptomyces in submerged cultures. Adv. Appl. Microbiol. 89, 1-45. ( 10.1016/B978-0-12-800259-9.00001-9) [DOI] [PubMed] [Google Scholar]
- 39.Sen BC, Wasserstrom S, Findlay K, Soderholm N, Sandblad L, von Wachenfeldt C, Flardh K. 2019. Specific amino acid substitutions in β strand S2 of FtsZ cause spiraling septation and impair assembly cooperativity in Streptomyces spp. Mol. Microbiol. 112, 184-198. ( 10.1111/mmi.14262) [DOI] [PubMed] [Google Scholar]
- 40.Addinall SG, Lutkenhaus J. 1996. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol. Microbiol. 22, 231-237. ( 10.1046/j.1365-2958.1996.00100.x) [DOI] [PubMed] [Google Scholar]
- 41.Monahan LG, Robinson A, Harry EJ. 2009. Lateral FtsZ association and the assembly of the cytokinetic Z ring in bacteria. Mol. Microbiol. 74, 1004-1017. ( 10.1111/j.1365-2958.2009.06914.x) [DOI] [PubMed] [Google Scholar]
- 42.Pende N, et al. 2018. Host-polarized cell growth in animal symbionts. Curr. Biol. 28, 1039-1051.e5. ( 10.1016/j.cub.2018.02.028) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Traag BA, Seghezzi N, Vijgenboom E, van Wezel GP. 2007. Characterization of the sporulation control protein SsgA by use of an efficient method to create and screen random mutant libraries in streptomycetes. Appl. Environ. Microbiol. 73, 2085-2092. ( 10.1128/AEM.02755-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vijgenboom E, Woudt LP, Heinstra PW, Rietveld K, van Haarlem J, van Wezel GP, Shochat S, Bosch L. 1994. Three tuf-like genes in the kirromycin producer Streptomyces ramocissimus. Microbiology 140, 983-998. ( 10.1099/00221287-140-4-983) [DOI] [PubMed] [Google Scholar]
- 45.Kelemen GH, Brian P, Flärdh K, Chamberlin L, Chater KF, Buttner MJ. 1998. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2). J. Bacteriol. 180, 2515-2521. ( 10.1128/JB.180.9.2515-2521.1998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jakimowicz D, Żydek P, Kois A, Zakrzewska-Czerwińska J, Chater KF. 2007. Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae. Mol. Microbiol. 65, 625-641. ( 10.1111/j.1365-2958.2007.05815.x) [DOI] [PubMed] [Google Scholar]
- 47.Ramond E, Maclachlan C, Clerc-Rosset S, Knott GW, Lemaitre B. 2016. Cell division by longitudinal scission in the insect endosymbiont Spiroplasma poulsonii. mBio 7, e00881-16. ( 10.1128/mBio.00881-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tzagoloff H, Novick R. 1977. Geometry of cell division in Staphylococcus aureus. J. Bacteriol. 129, 343-350. ( 10.1128/JB.129.1.343-350.1977) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keijser BJ, Noens EE, Kraal B, Koerten HK, van Wezel GP. 2003. The Streptomyces coelicolor ssgB gene is required for early stages of sporulation. FEMS Microbiol. Lett. 225, 59-67. ( 10.1016/S0378-1097(03)00481-6) [DOI] [PubMed] [Google Scholar]
- 50.Thompson CJ, Ward JM, Hopwood DA. 1982. Cloning of antibiotic resistance and nutritional genes in streptomycetes. J. Bacteriol. 151, 668-677. ( 10.1128/JB.151.2.668-677.1982) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bibb MJ, Ward JM, Hopwood DA. 1978. Transformation of plasmid DNA into Streptomyces at high frequency. Nature 274, 398-400. ( 10.1038/274398a0) [DOI] [PubMed] [Google Scholar]
- 52.Willemse J, van Wezel GP. 2009. Imaging of Streptomyces coelicolor A3(2) with reduced autofluorescence reveals a novel stage of FtsZ localization. PLoS ONE 4, e4242. ( 10.1371/journal.pone.0004242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piette A, et al. 2005. From dormant to germinating spores of Streptomyces coelicolor A3(2): new perspectives from the crp null mutant. J. Proteome Res. 4, 1699-1708. ( 10.1021/pr050155b) [DOI] [PubMed] [Google Scholar]
- 54.Mahr K, van Wezel GP, Svensson C, Krengel U, Bibb MJ, Titgemeyer F. 2000. Glucose kinase of Streptomyces coelicolor A3(2): large-scale purification and biochemical analysis. Antonie Van Leeuwenhoek 78, 253-261. ( 10.1023/A:1010234916745) [DOI] [PubMed] [Google Scholar]
- 55.McCarthy AA, et al. 2018. ID30B—a versatile beamline for macromolecular crystallography experiments at the ESRF. J. Synchrotron Radiat. 25, 1249-1260. ( 10.1107/S1600577518007166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kabsch W. 2010. XDS. Acta Crystallogr. D 66, 125-132. ( 10.1107/S0907444909047337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Evans PR, Murshudov GN. 2013. How good are my data and what is the resolution? Acta Crystallogr. D 69, 1204-1214. ( 10.1107/S0907444913000061) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bailey S. 1994. The ccp4 suite - programs for protein crystallography. Acta Crystallogr. D 50, 760-763. ( 10.1107/S0907444993011898) [DOI] [PubMed] [Google Scholar]
- 59.Vagin A, Teplyakov A. 1997. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022-1025. ( 10.1107/S0021889897006766) [DOI] [Google Scholar]
- 60.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr. D 66, 486-501. ( 10.1107/S0907444910007493) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240-255. ( 10.1107/S0907444996012255) [DOI] [PubMed] [Google Scholar]
- 62.Ramachandran GN, Ramakrishnan C, Sasisekharan V. 1963. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 7, 95. ( 10.1016/S0022-2836(63)80023-6) [DOI] [PubMed] [Google Scholar]
- 63.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. 1993. PROCHECK - a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283-291. ( 10.1107/S0021889892009944) [DOI] [Google Scholar]
- 64.Hess B, Kutzner C, van der Spoel D, Lindahl E. 2008. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 4, 435-447. ( 10.1021/ct700301q) [DOI] [PubMed] [Google Scholar]
- 65.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. 2006. Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins 65, 712-725. ( 10.1002/prot.21123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berendsen HJC, Grigera JR, Straatsma TP. 1987. The missing term in effective pair potentials. J. Phys. Chem. 91, 6269-6271. ( 10.1021/j100308a038) [DOI] [Google Scholar]
- 67.Darden T, York D, Pedersen L. 1993. Particle Mesh Ewald: an N · log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10 089-10 092. ( 10.1063/1.464397) [DOI] [Google Scholar]
- 68.Bussi G, Zykova-Timan T, Parrinello M. 2009. Isothermal-isobaric molecular dynamics using stochastic velocity rescaling. J. Chem. Phys. 130, 074101. ( 10.1063/1.3073889) [DOI] [PubMed] [Google Scholar]
- 69.Rahman A, Stillinger FH. 1971. Molecular dynamics study of liquid water. J. Chem. Phys. 55, 3336. ( 10.1063/1.1676585) [DOI] [Google Scholar]
- 70.Parrinello M, Rahman A. 1981. Polymorphic transitions in single-crystals: a new molecular-dynamics method. J. Appl. Phys. 52, 7182-7190. ( 10.1063/1.328693) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The atomic coordinates of SsgB from Streptomyces coelicolor have been deposited in the Protein Data Bank (PDB) with accession code 6SLC.