Keywords: Euglenida, Kinetoplastida, Diplonemida, microbial eukaryotes, systematics, phylogeny
Abstract
Euglenozoa is a species-rich group of protists, which have extremely diverse lifestyles and a range of features that distinguish them from other eukaryotes. They are composed of free-living and parasitic kinetoplastids, mostly free-living diplonemids, heterotrophic and photosynthetic euglenids, as well as deep-sea symbiontids. Although they form a well-supported monophyletic group, these morphologically rather distinct groups are almost never treated together in a comparative manner, as attempted here. We present an updated taxonomy, complemented by photos of representative species, with notes on diversity, distribution and biology of euglenozoans. For kinetoplastids, we propose a significantly modified taxonomy that reflects the latest findings. Finally, we summarize what is known about viruses infecting euglenozoans, as well as their relationships with ecto- and endosymbiotic bacteria.
1. Introduction
It is generally accepted that Euglenozoa belong to the most unusual eukaryotes [1–3]. This is based on a substantial body of evidence showing that in a number of cellular processes and structures, these almost invariably mono- or bi-flagellated protists departed from what can be considered the ‘eukaryotic consensus'. However, this consensus was defined by the studies of just a handful of model organisms, most of which are multicellular [4]. Hence, since the majority of the extant eukaryotic diversity is hidden in protists [5], we prefer to use a ‘protist-centric’ view, which postulates that these unicellular forms actually are the eukaryotic standard, while the other lineages represent departures from the norm.
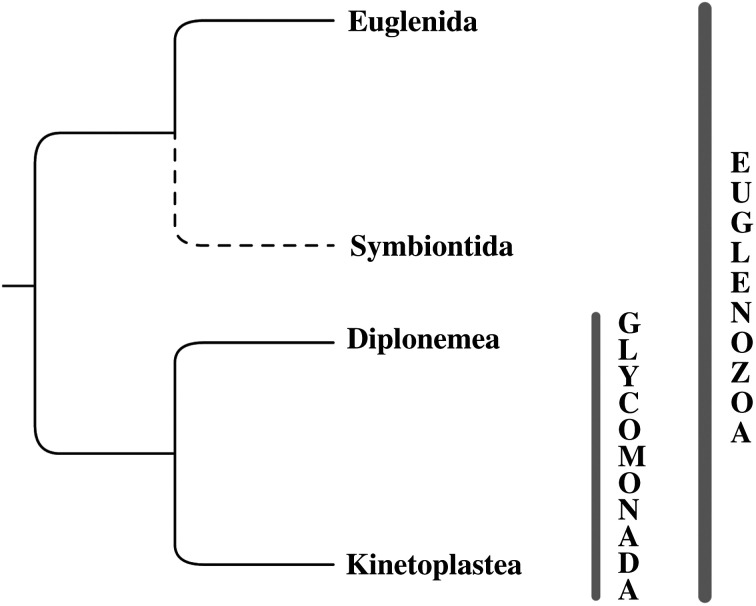
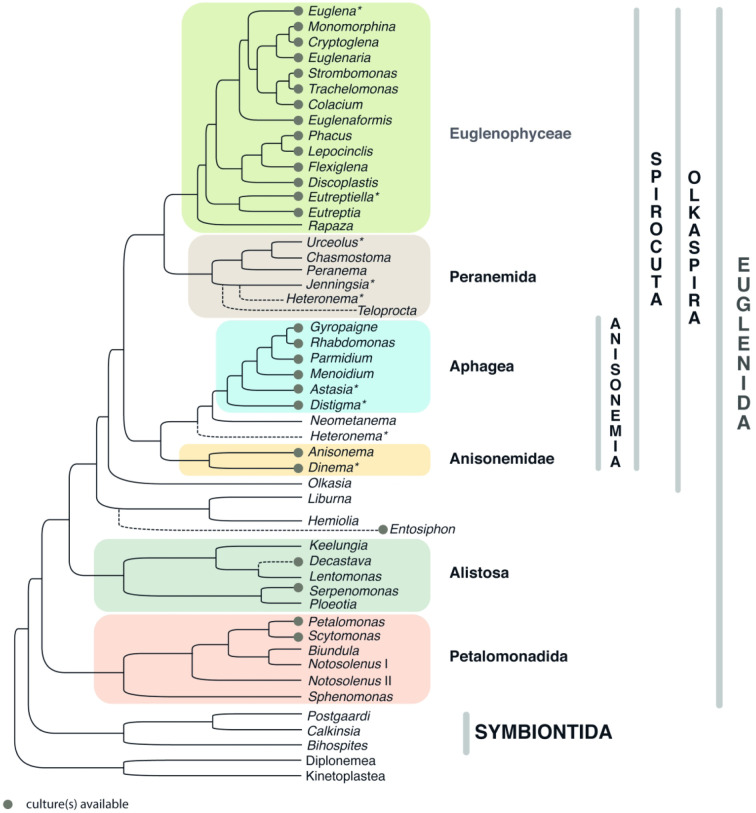
The phylum Euglenozoa splits into three well-defined lineages—euglenids, kinetoplastids and diplonemids—with different life strategies and distinct morphologies, yet still unified by a number of common features [6]. Although the euglenids are sometimes further subdivided into Euglenida and Symbiontida [3], both groups are usually treated together due to their morphological similarity, and we still cannot compare their genomic features in the absence of such data from the latter taxon [7]. A recent multigene phylogenetic reconstruction pointed to the potentially sister relationship between Symbiontida and Glycomonada (Kinetoplastea + Diplonemea) [8], suggesting that Symbiontida may become a separate group when more data become available (tree A).
Tree A.
Euglenozoa. A consensus tree based on multiple phylogenetic reconstructions showing relationships among major clades. The unstable position of Symbiontida is marked with a dotted line and further described in the section on euglenid taxonomy.
Apart from summarizing taxonomic works, the euglenozoans are almost never treated together in the literature. The kinetoplastid flagellates are by far the best-studied representatives (almost exclusively from a parasitology-centric perspective), with most attention given to the causative agents of serious diseases, such as sleeping sickness, Chagas disease and leishmaniases [9,10]. The diplonemids, as detailed below, were considered a marginal group with no ecological relevance. That has changed recently [11,12], but still very few molecular data other than 18S rRNA are available for this almost exclusively marine group. Finally, the photosynthetic and heterotrophic euglenids are ecologically significant, primarily in freshwater ecosystems, and have potential in biotechnologies [2,13].
The striking differences in lifestyles and cellular (ultra)structure obscure the significant similarities in basic molecular processes. Firstly, all these groups distinguish themselves from other eukaryotes by transcribing nuclear genes in a polycistronic manner [14]. In neither case are the co-transcribed genes functionally related, which distinguishes them from the prokaryotic operons. The usually very long polycistronic mRNA is subsequently processed into monomeric transcripts, which are subject to another process that is found in eukaryotes rather infrequently—trans-splicing. At the 5′ end of each monocistronic mRNA, short spliced leader (SL) RNA, already equipped with a methylated cap, becomes attached. The corresponding SL RNA gene is invariably multicopy, and highly conserved, yet with minor species-specific differences [15].
The similarities do not stop there. In their single or dual flagella, all euglenozoans evolved an extra-axonemal structure termed the paraflagellar rod, which supports their flagella [16]. The paraflagellar rod has a characteristic lattice-like structure, which is composed of dozens of proteins, phylogenetically restricted to euglenozoans. It is reduced only in the endosymbiont-containing trypanosomatids and the amastigotes of Leishmania [17]. Studied so far only in kinetoplastids, the paraflagellar rod not only increases propulsion of the cell [18], but also participates in morphogenetic and metabolic roles, as well as in environmental sensing [19]. While all these synapomorphies were probably present in the euglenozoan common ancestor, euglenids, diplonemids and kinetoplastids have acquired significant differences over the course of evolution. This is particularly striking in the case of cis-splicing, since spliceosomal introns are almost absent in the latter group [20], while they are abundantly present in euglenids and diplonemids, many being seemingly non-canonical [11,21]. Another clear difference rests in the size of both nuclear and mitochondrial genomes. The dearth of high-quality data for nuclear genomes of euglenids and their absence in the case of diplonemids are due to the large size and repetitive character of the latter. The transcriptomes from both groups contain an extremely high number of protein-coding genes, probably reflecting their metabolic versatility [6,13]. The situation is quite different in kinetoplastids, the parasitic lifestyle of which led to gene reduction and streamlining [6]. Moreover, due to their small and compact genomes, they belong to the most sequenced eukaryotes [22].
Unexpected differences among the main euglenozoan lineages recently became apparent for their mitochondrial genomes and transcriptomes. Kinetoplastids harbour in their mitochondrial DNA in the form of relaxed (rarely supercoiled) circular molecules, either catenated or free, of two types—maxicircles and minicircles, with the former carrying all protein-coding genes, while the latter encode guide RNA genes required for the editing of the maxicircle transcripts [23]. The size of maxicircles is rather uniform, while the minicircles come in different variants [24]. In diplonemids, the single type of non-catenated circles uniquely encodes fragments of protein-coding genes, the transcripts of which have to be massively trans-spliced and edited in order to become translatable [25]. However, in both groups, the mitochondrial DNA is inflated, and its transcripts are extensively edited [26]. This contrasts with euglenids that lack any form of editing in their mitochondrion, which also contains a small genome composed of heavily fragmented linear molecules [27]. Probably, the most important difference among these groups is the presence of a secondary green plastid solely in euglenids, which have acquired it after their divergence from other euglenozoans [2,28].
Until recently, our knowledge of different groups within euglenozoans was much influenced by the availability of full-size nuclear genome sequences. While hundreds of high-quality genomes are available for trypanosomatids [22], only one such genome is available for bodonids [29] and euglenids [13], respectively, and none for diplonemids. However, this is bound to change soon, mostly due to the ever-decreasing costs and improving sequencing technologies. Recent comparative analyses of molecular features among kinetoplastids, euglenids and diplonemids were based on transcriptomes available for all of them [30].
Future studies of euglenozoans will be heavily influenced by the accessibility of their representatives to (efficient) genetic manipulations. The amenability of trypanosomatids to a range of genetic tools turned them into arguably the functionally best-studied protists [31], while most other groups significantly lag behind. However, this unfavourable situation has changed recently, as first reports of genetic modifications of bodonids, diplonemids and euglenids have been published [32–36]. Anticipated improvement of the methodologies of forward and reverse genetics, which would allow medium- or high-throughput functional analyses in these taxonomic groups, almost guarantee major discoveries.
Euglenozoa is a very peculiar group, encompassing organisms strikingly dissimilar in their ecology, ranging from autotrophy to obligate parasitism. This inevitably influenced their classification in the era of the two-kingdoms-of-life paradigm. Kinetoplastids and diplonemids were historically considered predominantly as protozoa, and thus the International Code of Zoological Nomenclature (ICZN) was used for their nomenclature, while euglenids have been classified by different authors as either protozoa or algae. This ambiguity is reflected in their nomenclature, which has been governed in parallel by the ICZN as well as the International Code of Botanical Nomenclature (ICBN) and the International Code of Nomenclature for algae, fungi and plants (ICN), which replaced the latter in 2011. Apart from formal differences, such as the rules on citing authorship of names and emendations, this led to significant issues that include certain taxa having different names depending on the selected system (zoological or botanical). This concerns names of family-group taxa, which have different suffixes depending on the system and, more importantly, names of genera, which may be valid according to one code, but regarded as junior homonyms, and therefore replaced with different names. In addition, the ICZN jurisdiction does not extend above the family-group level, whereas ICN does not have such a restriction. Here, the nomenclature of kinetoplastids and diplonemids follows the ICZN, while for some euglenid groups, the ICN is used by default with the valid names according to the ICZN indicated.
2. Kinetoplastea
2.1. Biology
2.1.1. Free-living kinetoplastids
The common ancestor of Kinetoplastea apparently was a free-living benthic bacterivorous organism using the anterior flagellum for motion and transporting food particles to the cytostome, while the posterior one ensured gliding on the substrate (plate A, 1,2,4–10,12–14). This lifestyle is still preserved by a large proportion of kinetoplastids [37]. They inhabit permanent and temporary water bodies with various levels of salinity (freshwater to hypersaline) and some species were shown to tolerate the transition from marine water to freshwater, and vice versa [38–41]. Kinetoplastids are very numerous in benthic communities, where they constitute 5–20% of the total biomass of all heterotrophic flagellates, second only to euglenids, suggesting their important role in controlling bacterial growth [42]. They are abundant in seawater ice and some species can be cultured even from the pelagic zone, demonstrating their presence there at least at the cystic stage [43,44]. Recent studies using molecular methods demonstrated that in many water bodies, most of the kinetoplastid biomass is created by neobodonids, of which Neobodo, Rhynchomonas and Dimastigella are the most frequent ones (plate A, 7,9,14) [45–47]. An extensive analysis of free-living kinetoplastids in hundreds of globally collected oceanic samples revealed their abundance being 0.14%, with highest abundance in the mesopelagic zone. Their community structure and richness are significantly influenced by oxygen concentration, salinity and temperature [48].
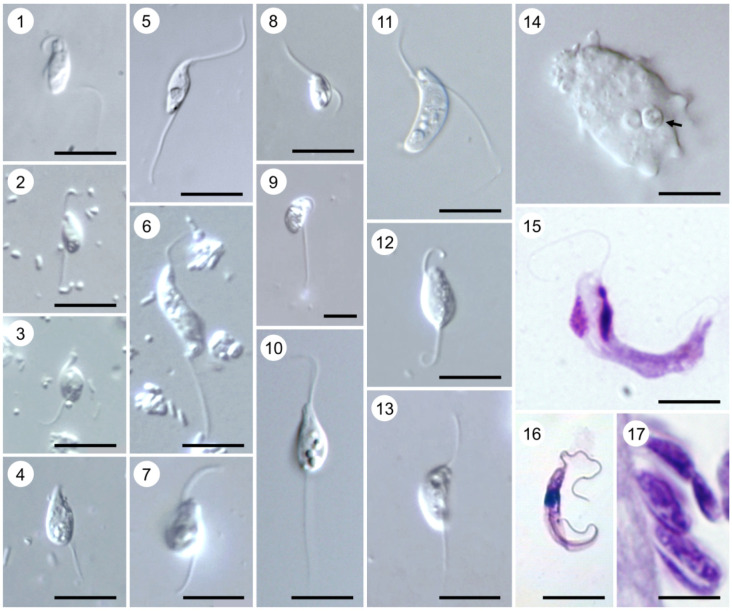
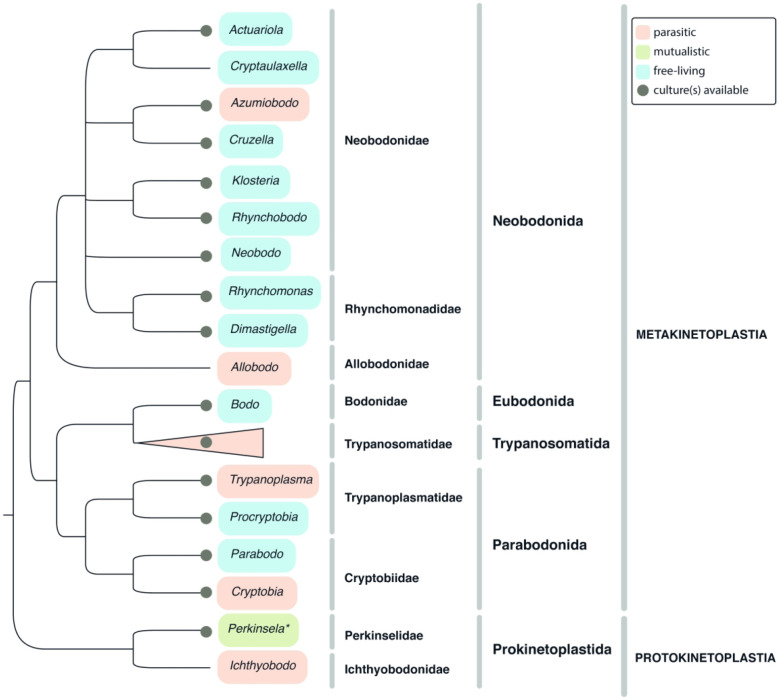
Plate A.
Bodonids. Light micrographs of cultured (1) Actuariola framvarensis (provided by Thorsten Stoeck); (2) Neobodo curvifilus (provided by Kristina Prokina and Denis Tikhonenkov); (3) Rhynchomonas nasuta (provided by Kristina Prokina and Denis Tikhonenkov); (4) Rhynchobodo sp. (provided by Kristina Prokina and Denis Tikhonenkov); (5) Azumiobodo hoyamushi (provided by Shinichi Kitamura and Euichi Hirose); (6) Dimastigella mimosa (provided by Kristina Prokina and Denis Tikhonenkov); (7) Bordnamonas tropicana (provided by Kristina Prokina and Denis Tikhonenkov); (8) Klosteria bodomorphis (provided by Kristina Prokina and Denis Tikhonenkov); (9) Bodo saltans (provided by Kristina Prokina and Denis Tikhonenkov); (10) Cruzella marina; (11) Allobodo chlorophagus (provided by Alastair Simpson and Yana Eglit); (12) Procryptobia sorokini (provided by Kristina Prokina and Denis Tikhonenkov); (13) Parabodo caudatus (provided by Kristina Prokina and Denis Tikhonenkov); (14) Perkinsela sp. (arrow indicates its position inside Paramoeba pemaquidensis) (provided by Ivan Fiala); (15) Giemsa-stained Trypanoplasma borreli; (16) Cryptobia vaginalis (provided by Marina N. Malysheva); (17) Toluidine-stained semi-thin section of fish gill with attached Ichthyobodo necator (provided by Iva Dyková). Scale bar, 10 µm (1–8; 10–17); 5 µm (9).
Moreover, many kinetoplastids live in the soil and readily settle on various organic substrates, such as faeces, composts, etc. [49,50]. Most of the free-living flagellates graze on bacteria with the help of cytostomal lips or, as in Rhynchomonadidae, a flagellum-attached motile proboscis. In addition to bacterial cells, their digestive vacuoles can contain microalgae or detritus particles. Parabodo caudatus (plate A, 13), being a relatively large (up to 20 µm long) species, exerts both bacterivory and predation, while Rhynchobodo spp. (plate A, 4) are obligatory predators devouring other flagellates [37].
Some kinetoplastids are known to form cysts, which help them to survive adverse conditions, for example, pass through the digestive system of an animal and settle in its faeces after their discharge [37]. Moreover, some of these flagellates become very tolerant to harsh environments even at the active (non-encysted) stage. This resulted in a series of records of free-living kinetoplastids (Parabodo caudatus, Dimastigella trypaniformis and Procryptobia tremulans) from stool and urine samples or urine-impregnated animal cage bedding sometimes misinterpreted as evidence of their parasitic nature [51–54]. Interestingly, such tolerance in parabodonids apparently preadapted them to parasitism, which originated in this group at least twice [55].
2.1.2. Parasitic, mutualistic and commensal non-trypanosomatid kinetoplastids
Multiple transitions to various forms of symbiosis can be observed in all orders of Kinetoplastea except Eubodonida (tree B). The earliest branch within this group, Prokinetoplastida, does not contain any described free-living forms. The ectoparasitic Ichthyobodo, affecting both freshwater and marine fish, is generally similar to free-living kinetoplastids (plate A, 17). In contrast with other symbiotic forms, it anchors on the host epithelium with its rostrum forming an attachment disc and a cytostome process, which is inserted directly into the cytoplasm of the host cell and feeds by myzocytosis [56]. Accumulation of parasites on the epithelium of gills and fins leads to tissue necrosis, which often entails the death of fish, especially fingerlings. Dissemination of Ichthyobodo spp. occurs using a free-swimming stage lacking the rostrum [57].
Another prokinetoplastid genus, Perkinsela, represents one of the most simplified symbiotic eukaryotes, permanently living in the cytoplasm of amoebae, such as Paramoeba (plate A, 14) [58]. The nature of their relationships is mutualistic as judged by reciprocal metabolic dependence of the two partners, evident from the study of their genomes [59]. Since both Ichthyobodo and Paramoeba live on fish gills, it was proposed that an ancestral Ichthyobodo-like flagellate had been engulfed, but not digested, by an amoeba, and eventually evolved into the endosymbiont Perkinsela [58]. Perkinsela is tightly associated with cosmopolitan Paramoeba, lacks any traces of flagellum (plate A, 14), and has an extremely reduced metabolism, as well as the largest known mitochondrial DNA [59].
The currently unclassified flagellate Desmomonas prorhynchi, a parasite of the turbellarian Prorhynchus, shares two features with Ichthyobodo: polykinetoplast DNA and attachment to the host cell by an appendage at the anterior end. However, this structure performs an exclusively mechanical function, while feeding is supposed to occur via osmotrophy [60]. Another flagellate with an uncertain taxonomic position, Cephalothamnium cyclopum, is the only described colonial kinetoplastid, which attaches to freshwater copepods [61]. Like its free-living relatives, this flagellate feeds by intercepting bacterial cells with its anterior flagellum and directing them to cytostomal opening. Given that C. cyclopum uses its host only as a substrate and the only inconvenience from its presence may consist of decreased hydrodynamic characteristics of the crustacean, this kinetoplastid is considered as ectocommensal.
Azumiobodo hoyamushi is a neobodonid parasite of ascidians, of which the most important is the sea pineapple Halocynthia roretzi, a cultivated edible species popular in Korea and Japan (plate A, 5). By invading the tunic, this flagellate is responsible for the so-called soft tunic syndrome, associated with high mortality rates [62]. Being seasonal, it survives the period of high temperatures in resistant cysts attached to the substrate [63,64]. Another parasitic neobodonid is the recently described Allobodo chlorophagus, invading the utricles and the main filaments of the green siphonal alga Codium fragile and feeding on its chloroplasts and starch granules (plate A, 11) [65].
Parasitic parabodonids are represented by the genera Trypanoplasma and Cryptobia, which often used to be combined into one genus due to morphological similarity. Trypanoplasma spp. are extracellular parasites of fish bloodstream transmitted by haematophagous leeches during blood-feeding (plate A, 15) [66]. However, at least two species, T. salmositica and T. bullocki, can exit to the body surface where they reside in the mucus and can be transmitted to other fish by direct contact [57]. After ingestion with the blood, parasites multiply in the leech crop without significant changes in morphology and migrate to the proboscis sheath, wherefrom they are transmitted to the bloodstream of another fish [67]. The severity of infection—acute to chronic—apparently depends more on the level of mutual adaptation as well as the individual variation of host immunity between host and parasite than on parasitaemia [68–70]. There is only a single record of a trypanoplasma in a non-fish host, namely in a salamander [71].
The genus Cryptobia can be subdivided into three distinct ecological groups: (i) ectoparasites of fish, (ii) endoparasites of invertebrates and (iii) endoparasites of fish. Members of the first group, represented by Cryptobia carassii, live on fish gills and are regarded as commensals feeding on dead epithelium and microorganisms [57]. However, gill infections by C. branchialis are associated with high mortality in adult cultured carps, goldfish and catfish as well as in juvenile grass carp [72]. Lamellasoma bacillaria, described as a monoflagellated kinetoplastid living on the fish gills, may also belong to this group of cryptobiae [73].
In invertebrates, Cryptobia infections are quite diverse in terms of localization within the host and taxonomic groups parasitized. Some of them (Cryptobia helicis, C. innominata and C. carinariae) were found attached to the epithelium of spermatheca of floating sea snails and various pulmonates [74–76]. The vagina of haematophagous leeches often serves as a habitat of C. vaginalis (plate A, 16) [77], while C. udonellae was described from the excretory system of an ectoparasitic marine worm [78]. Other species were described from the intestine of a chaetognath (C. sagittae) and a freshwater planarian (C. dendrocoeli), the latter of which was also detected in the eggs, pointing to a potential transovarial transmission [79,80]. It is presumed that Cryptobia spp. from the reproductive system are transmitted via sexual contacts [75,81], while the ectoparasites of aquatic animals should have free-swimming swarmers, although this has not been confirmed [73]. The cryptobiae found in the intestinal contents of frogs and lizards appear to be accidentally ingested parasites of invertebrates, as judged by the morphology of the flagellates and uniqueness of such records [82,83].
In the third group of cryptobiae, encompassing piscine intestinal flagellates, six out of seven described species are known as specific parasites of marine fish. These species do not display any pathogenic effect and therefore are usually considered as commensals [57]. The only known freshwater representative of this group, C. iubilans, infects various cichlid fishes and causes gastroenteritis often associated with invasion of other organs, leading to high mortality [84–86]. Сryptobiae belonging to this group can be transmitted directly by ingestion from water and by feeding on infected corpses [57].
Jarrelia atramenti, a flagellate described from the blowhole mucus of a pygmy sperm whale, appears to be a harmless commensal feeding on detritus and/or bacteria [87]. Its resemblance to parasitic parabodonids, proposed to be evidence of their relatedness, may be in fact a parallelism caused by similar living conditions. Indeed, flexible body and flagellar attachment evolved independently in Cryptobia, Trypanoplasma and Dimastigella.
2.1.3. Trypanosomatids
The family Trypanosomatidae contains exclusively obligate parasites and represents the most diverse kinetoplastid group in terms of the number of species described and/or revealed using molecular typing [88–90]. Among parasitic protists, it has the widest host range: animals (predominantly insects and vertebrates), flowering plants and even ciliates [91]. Based on the type of life cycle, trypanosomatids are usually subdivided into two non-taxonomic groups. Monoxenous species develop in a single host, whereas dixenous switch between two, of which one serves as a vector. Molecular phylogenies suggest that the most recent common ancestor of trypanosomatids was a monoxenous parasite of insects [92,93], with the dixenous lifestyle emerging independently at least three times in distantly related lineages of these flagellates [55].
2.1.3.1. Monoxenous trypanosomatids
Most trypanosomatid genera are monoxenous and the overwhelming majority of their species parasitize two large groups of insects: Diptera and Heteroptera (i.e. flies and true bugs, respectively) [91,94]. Among other insects, used by them as hosts are Hymenoptera (bees, bumblebees, wasps and sawflies), Siphonaptera (fleas), Blattodea (cockroaches), Lepidoptera (moths) and Trichoptera (caddis flies). The single records of monoxenous trypanosomatids from a louse (Anoplura), a planthopper (Homoptera), a scorpion fly (Mecoptera) and a domestic cricket (Orthoptera) may refer to accidental non-specific infections [91]. The adaptation to insects, which are omnipresent, extremely diverse and abundant animals, probably predetermined the transition of these flagellates to other hosts. Trypanosomatids invaded Acari (ticks and mites) and freshwater ciliates living side-by-side with insects, vertebrates and plants. The two latter host groups are associated with dixenous trypanosomatids, although monoxenous species have also been occassionally reported from them [95,96]. The presence of trypanosomatids in nematodes and molluscs [97] may indicate a more complex evolutionary pathway of these flagellates, but first it requires confirmation with modern methods.
The ancestral and still most common lifestyle of monoxenous trypanosomatids includes stages that inhabit insect gut, usually being attached to its wall, and some either active (i.e. flagellate) or inactive (endomastigote or cyst-like amastigote) cells are discharged with faeces. Other insects become infected by feeding on contaminated substrates or directly on fresh faeces (coprophagy) [98]. In addition, the parasites can be transmitted between insects via cannibalism and predation, although the latter way is probably responsible only for the transmission of non-specific transient infections [99]. Some monoxenous trypanosomatids can migrate within insects to other locations in order to facilitate transmission [89]. Thus, parasitism in Malpighian tubules of female firebugs ensures timing the mass production of infective cyst-like amastigotes of Blastocrithidia papi to oviposition [100]. Haemocoel invasion allows the inheritance of Herpetomonas swainei between developmental phases of the host saw fly [101], while in the case of Leptomonas pyrrhocoris, this increases the efficiency of transmission by cannibalism [102]. The role of intracellular stages, which are very rare in life cycles of monoxenous trypanosomatids, is uncertain [103]. However, the potential to live intracellularly probably preconditioned transition of these flagellates to dixeny (see below) and parasitism in ciliates. The latter has been repeatedly described from various ciliate species where it was always associated with the macronucleus and, at least in some cases, effective transmission between host cells was observed pointing to specific relationships [104–107].
Although most monoxenous trypanosomatids are considered non-pathogenic or even commensals [98], this view is influenced by the fact that their effects on the hosts are poorly known and have been investigated in only a few practically important or model insect species. It was shown that trypanosomatid infections lead to elevated mortality rates in triatomine bugs, honeybees, sawflies, eye gnats, fruit flies, firebugs and water striders [101,108–112]. Other adverse consequences of trypanosomatid infections on insects include delayed development, decrease in body weight, disturbed digestion and excretion, lower endurance, impaired foraging efficiency and lower fecundity [113–118]. The above effects have a significant impact on host fitness, and thus trypanosomatids play an important role in controlling the population sizes of their hosts.
2.1.3.2. Phytomonas
Some trypanosomatids acquired the ability to live in plants, on which their bug hosts feed and, thus, became dixenous. These flagellates belong to the genus Phytomonas and parasitize phloem, fruits, latex or seeds of various plants [119,120]. The bug hosts serve as vectors and, since the contaminative route of transmission to plants is not very effective, the parasites migrate from the intestine through haemocoel to salivary glands [96]. Here, the infective endomastigotes are formed, which are inoculated into plant juices with the bug's saliva during feeding [103,121,122]. Interestingly, in some species, no development occurs in the host gut, which is then used only for the transit of flagellates [103,123]. At least one phytomonad species, P. nordicus, became secondarily monoxenous, since it inhabits a predatory pentatomid bug [124]. The pathogenicity of Phytomonas for insects remains unknown, while their effect on plants ranges from asymptomatic infections to serious diseases of cultural plants [120]. Phytomonas francai living in lactiferous ducts of manioc is associated with root dystrophy; P. leptovasorum causes phloem necrosis and subsequent lethal wilt of coffee trees; P. staheli obstructing phloem of oil and coconut palms accounts for acute wilt in these plants; and an unnamed phytomonad is responsible for the withering of red ginger [96]. These diseases have a high impact on agriculture in developing countries and result in serious economic losses [119].
2.1.3.3. Leishmania and related dixenous genera
The genera Leishmania, Porcisia and Endotrypanum represent a monophyletic group, whose parasitism in blood-sucking sandflies (Phlebotominae) allowed them to become dixenous parasites of mammals [125]. Secondarily, some Leishmania spp. changed either the vertebrate host or the vector: the subgenus Sauroleishmania switched from mammals to lizards and snakes, while the subgenus Mundinia started using biting midges (Ceratopogonidae) instead of sandflies [126–128]. Leishmania is most species-rich genus and many of its members are human parasites, which drew most attention to this group, while the information about Porcisia and Endotrypanum is scarce. The development of leishmaniae in vectors is confined to the intestine, although there are some differences between subgenera in the localization of the proliferative procyclic promastigotes (midgut, pylorus and/or hindgut) [127]. However, they eventually migrate to the anterior midgut, where they destroy the chitin lining of the stomodaeal valve and secrete a gel plug obstructing the alimentary canal, thus disturbing the normal sucking process [129]. An infected sandfly regurgitates the plug with metacyclic flagellates into the vertebrate bloodstream and due to the inability to swallow the blood makes more attempts increasing chances of spreading the parasites [130]. In the vertebrate, the metacyclic promastigotes are quickly taken up by phagocytic cells and proliferate in their phagolysosomes as amastigotes [131]. Depending on the behaviour of infected macrophages, leishmaniasis manifests itself as either cutaneous (skin ulcers), mucocutaneous (sores in the mucosa of nose, mouth or throat) or visceral, which affects internal organs such as the liver, spleen and bone marrow and is usually fatal without treatment [127].
About 20 species of Leishmania, belonging to the subgenera Leishmania, Viannia and Mundinia parasitize humans. They are responsible for up to one million new cases of leishmaniasis annually, of which up to 90 000 correspond to the visceral form [132]. The visceral form of the disease can be spread even outside the endemic areas either venereally or congenitally [133–136]. In addition to humans, Leishmania was reported to infect about 70 species of mammals (rodents, carnivores, xenartrans, hyraxes, marsupials, chyropterans, ungulates lagomorphs and primates), with most cases being asymptomatic. The only notable exceptions are canine visceral leishmaniasis, with severe symptoms in over 50% of cases [137], and rare cases of atypical cutaneous leishmaniasis in cows and horses [138,139]. Porcisia living in porcupines and Endotrypanum parasitizing sloths and squirrels do not appear to produce any symptoms [125]. However, Leishmania colombiensis (now assigned to Endotrypanum) is known to cause both cutaneous and visceral leishmaniasis-like diseases in humans [140,141].
2.1.3.4. Trypanosoma
Trypanosoma is a very speciose genus enclosing approximately 500 species or over 60% of all described species of the family Trypanosomatidae (plates B and C, 18–40). While the frog-infecting type species (T. sanguinis = T. rotatorium) described by Gruby already in 1843 may be of marginal importance, ever since trypanosomes became the best-known protists. Life cycles of these flagellates vary considerably as they parasitize all classes of vertebrates (from agnathans to mammals) and are transmitted by a wide range of vectors including blood-sucking insects (flies, bugs, fleas and lice), ticks, leeches and even vampire bats [9,91]. In vertebrates, they occur most frequently as trypomastigotes, rarely as epimastigotes or amastigotes, while in invertebrates, they predominantly exhibit most trypomastigote or epimastigote morphology, or infrequently occur as promastigotes and amastigotes [92].
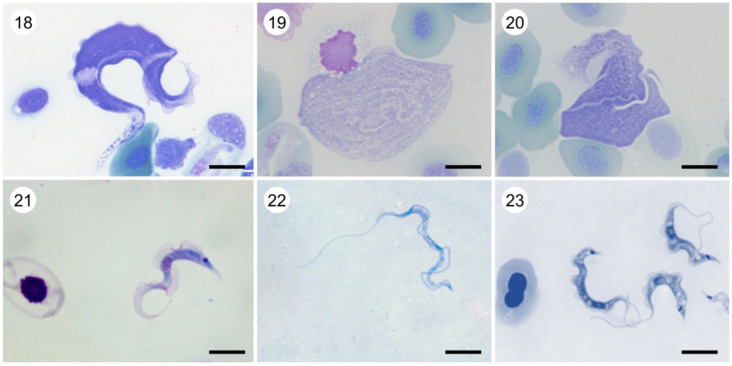
Plate B.
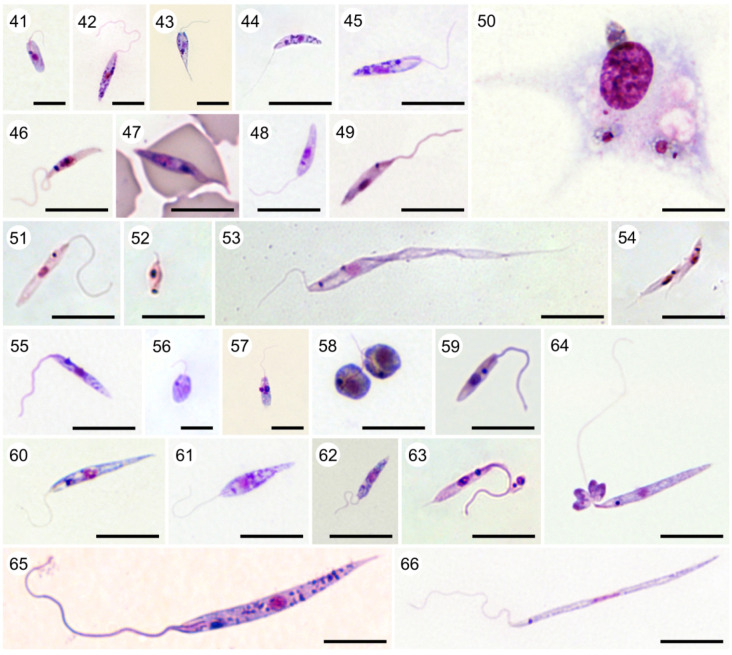
Trypanosoma (aquatic clade). Light micrographs of Giemsa-stained (18) T. (Trypanosoma) rotatorium ex Pelophylax kl. esculentus (provided by Klára Poloprudská); (19) T. (Trypanosoma) loricatum ex Pelophylax kl. esculentus (provided by Klára Poloprudská); (20) T. (Trypanosoma) ranarum ex Pelophylax kl. esculentus (provided by Klára Poloprudská); (21) T. (Haematomonas) clandestinus ex Caiman yacare (experimental infection) (provided by Erney Camargo and Marta Teixeira); (22) T. (Haematomonas) cf. cobitis ex Cobitis ‘taenia'; (23) T. (Haematomonas) sp. ex cichlid (provided by Iva Dyková). Scale bar, 10 µm (18–23).
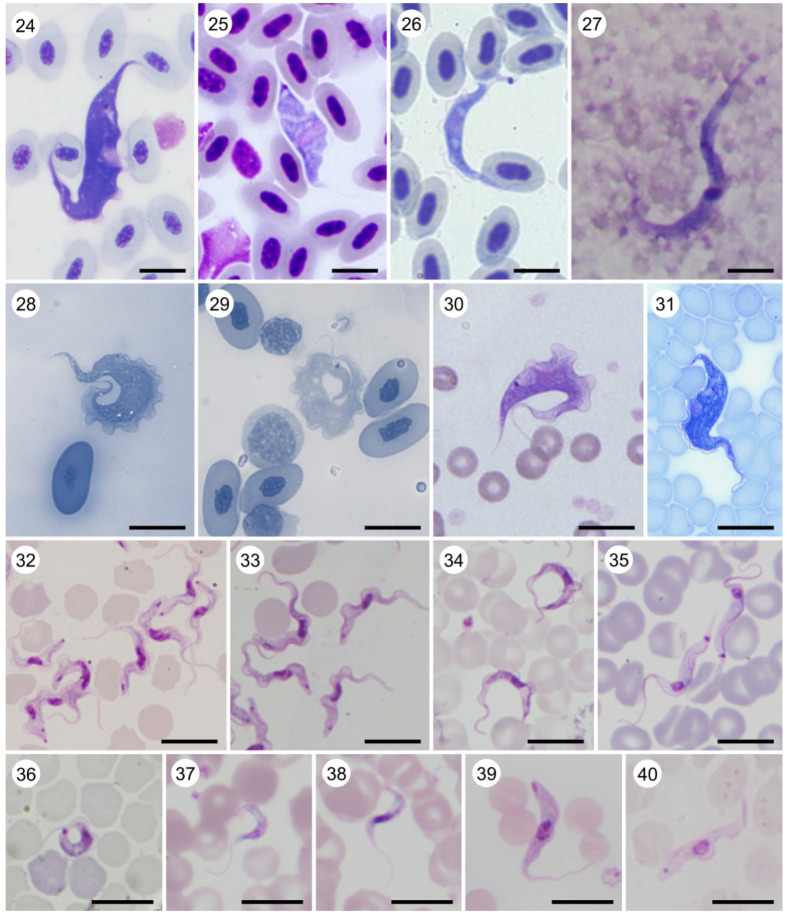
Plate C.
Trypanosoma (terrestrial clade). Light micrographs of Giemsa-stained (24) T. (Trypanomorpha) avium ex Lanius collurio; (25) T. (Ornithotrypanum) everetti (provided by Gediminas Valkiūnas); (26) T. (Avitrypanum) culicavium (experimental infection) (provided by Milena Svobodová); (27) T. (Megatrypanum) theileri ex cattle (provided by Andrei Mihalca); (28) T. (Squamatrypanum) cascaveli ex Crotalus durissus (provided by Erney Camargo and Marta Teixeira); (29) T. (Crocotrypanum) terena ex Caiman yacare (provided by Erney Camargo and Marta Teixeira); (30) T. (Australotrypanum) copemani ex Bettongia penicillata (provided by Sarah Keatley and Andrew Thompson) (31) Trypanosoma livingstonei ex African bat (provided by Erney Camargo and Marta Teixeira); (32) T. (Trypanozoon) brucei brucei ex mouse (experimental infection); (33) T. (Trypanozoon) brucei equiperdum ex mouse (experimental infection); (34) T. (Trypanozoon) brucei evansi ex mouse (experimental infection); (35) T. (Herpetosoma) lewisi ex Rattus sp.; (36) T. (Aneza) vespertilionis ex Pipistrelus pipistrelus; (37) T. (Schizotrypanum) cruzi (C-shape; experimental infection); (38) T. (Schizotrypanum) cruzii (S-shape; experimental infection); (39) T. (Duttonella) vivax; (40) T. (Nannomonas) congolense. Scale bar, 10 µm (24–40).
From the practical point of view, the most important mammalian trypanosomes were traditionally subdivided into two sections or intrageneric taxons that follow distinct developmental programmes [142]: Salivaria (derived from saliva), with the best-known member being the Trypanosoma brucei complex causing human African sleeping sickness and nagana in livestock and other animals (plate C, 32–34), terminate development in the salivary glands mouthparts of the vector and are transmitted to a vertebrate host by bite. Stercoraria (stercus = dung) exemplified by Trypanosoma cruzi complex species causing Chagas disease (plate C, 37–38), terminate development in the rear part of the digestive tract of the vector with the transmission to the vertebrate host being contaminative by excrements. With the advent of molecular phylogenetics, it became obvious that neither mammalian trypanosomes in general nor any of the two proposed sections represent monophyletic groups, and therefore they do not deserve a taxonomic status [89,143,144]. Nevertheless, the words salivarian/stercorarian still can refer to the type of development within the vector. Non-mammalian trypanosomes generally follow the stercorarian developmental programme, but in leeches, parasites migrate to the proboscis sheath to be transmitted during blood-sucking [57].
Trypanosoma brucei evansi and T. b. equiperdum are two notable exceptions (plate C, 33–34), as they lost the capacity to survive in the gut of an insect vector [145]. The former subspecies therefore switched to mechanical transmission, which allowed it to use non-specific vectors, while the latter adapted to the direct (venereal) transmission and thus became a monoxenous parasite [146]. In other species, direct transmission can also occur, but is facultative [147–150]. The most (in)famous species are Trypanosoma brucei and T. cruzi, which cause serious human diseases—sleeping sickness and Chagas disease, respectively [151]. The first one is transmitted by tsetse flies in Africa and invades various tissues, but primarily the blood and adipose tissue [152], as free-swimming trypomastigotes and eventually infects cerebrospinal fluid with fatal consequences [153]. Being a serious public health threat in the past, this disease is now on the way to elimination [154]. Trypanosoma cruzi is transmitted by triatomine bugs among a wide range of mammalian hosts, in which it develops in various organs and tissues as intracellular amastigotes [155]. In most cases, the disease does not manifest clinical signs at the beginning, but during the prolonged chronic phase, it significantly undermines health in the human population of South and Central America leading to increased mortality rates [156]. Trypanosoma rangeli has the same geographical distribution and vectors as T. cruzi and is also able to infect humans but appears to be non-pathogenic [157]. Some tsetse-transmitted African trypanosomes, such as T. vivax, T. congolense and T. brucei brucei, cause serious diseases in livestock, collectively named African animal trypanosomiasis. These diseases are associated with high mortality rates and lead to significant damage in animal husbandry, although some local breeds and wild animals acquired tolerance to them [158]. For the overwhelming majority of trypanosome species, their effects on the host are not known and they are often considered as non- or subpathogenic and can cause observable disease only under stress conditions. This is exemplified by piscine trypanosomes, which seem to be well tolerated in wild fish populations [159]. However, in farmed fish or wild juvenile individuals, infections are associated with high mortality rates due to anaemia, anorexia and tissue damage [160–163].
2.2. Taxonomy
This section contains nomenclatural changes and according to the ICZN requirements for publications in online-only journals, this work has been registered in Zoobank: urn:lsid:zoobank.org:pub:81EA01C5-8989-4BBD-9C64-04D81132307D.
Class Kinetoplastea Honigberg, 1963 emend. Vickerman, 1976 (tree B).
Tree B.
Kinetoplastea. A tree summarizing multiple phylogenetic reconstructions, mostly 18S rRNA gene-based. Highlighting denotes lifestyles (see graphical legend). Asterisk denotes that the protist can be cultivated only within its host. Host Trypanosomatidae clade is collapsed (shown with a triangle) and is presented in detail on a separate figure.
Possess kinetoplast, represented by one or several large masses of mitochondrial DNA termed kinetoplast DNA (kDNA). Four kinetoplast types are distinguished: eukinetoplast—dense network of interlocked DNA circles, prokinetoplast—single compact mass not organized into a network, polykinetoplast—several clusters scattered over the mitochondrial lumen, and pankinetoplast—a diffuse mass occupying a large portion of a mitochondrial lumen [164]. Ancestrally, kinetoplastids bear two heterodynamic flagella, of which one or both were lost in some lineages; mitochondrial RNA undergoes editing represented by deletions and insertions of uridine residues. Some features considered for a long time to be defining (e.g. polycistronic transcription of nuclear genes, trans-splicing via spliced leader RNA, compartmentalized glycolysis, base J, etc.) were recently shown to be present also in other euglenozoan lineages [6].
Note: Until relatively recently, all kinetoplastids have been classified into two large groups—bodonids (free-living, ectocommensals, ecto- or endoparasitic biflagellate species) and trypanosomatids (exclusively endoparasitic uniflagellate species). However, the 18S rRNA gene-based molecular phylogenetic analysis showed paraphyly of bodonids, which were subsequently separated into four orders [165]. Thus, now the term ‘bodonids’ for the designation of non-trypanosomatid kinetoplastids is deprecated. As judged by available environmental sequences, the diversity of kinetoplastids is much broader than that described to date, and there are some undiscovered lineages potentially representing new high-level taxa (up to the subclass) [166].
-
• Subclass Prokinetoplastia Vickerman, 2004. The phylogenetic group enclosing the genera Ichthyobodo and Perkinsela as judged by 18S rRNA gene-based trees [165].
-
○ Order Prokinetoplastida Vickerman, 2004. With the same definition as the subclass.
-
▪ Family Ichthyobodonidae Isaksen et al., 2007. Ectoparasitic on freshwater and marine fish; polykinetoplastic; biflagellate; the flagellar pocket extends to the lateral cell surface as a longitudinal groove; the modified anterior end (rostrum), present in trophozoites, is used for attachment [167]. Single genus.
-
▪ Genus Ichthyobodo Pinto, 1928. With the same definition as the family.Type species: Costia necatrix Henneguy, 1883 (= Ichthyobodo necator) (plate A, 17).
-
▪
-
▪ Family Perkinselidae Kostygov, fam. nov.Diagnosis: The phylogenetic group comprising Perkinsela Dyková, Fiala and Peckova, 2008 (type genus) and related endosymbiotic forms as judged from 18S rRNA gene-based trees [168,169].
-
▪ Genus Perkinsela Dyková, Fiala and Pecková, 2008. Permanently endosymbiotic in the cytoplasm of various amoebae (Paramoeba, Neoparamoeba, Janickina, etc.), parasitophorous vacuole not formed; oval aflagellate cells; massive prokinetoplast; usually binucleate; microtubular corset reduced and present only in a thin layer on both sides of the kinetoplast; no oral apparatus, no flagellum [58,169].Type species: Perkinsiella amoebae Hollande, 1980 (= Perkinsela amoebae). Monotypic (plate A, 14).Note: Axenic cultivation is impossible, but can be grown in the host amoebae.
-
▪
-
▪
-
○
-
• Subclass Metakinetoplastia Vickerman, 2004. The phylogenetic group enclosing Neobodonida, Parabodonida, Eubodonida and Trypanosomatida as judged by 18S rRNA gene-based trees [165].
-
○ Order Eubodonida Vickerman 2004. Free-living; biflagellate, with non-tubular mastigonemes on the anterior flagellum; prokinetoplastic; phagotrophic, with anterolateral cytostome bordered by lappets and no conspicuous preoral ridge, cytopharynx traversing body [165]. Single family.
-
▪ Family Bodonidae Bütschli, 1883. With the same definition as the order. Single genus.
-
▪
-
○ Order Neobodonida Vickerman, 2004. Free-living, or parasitic; solitary; biflagellate, usually without mastigonemes, both flagella free or the posterior one attached to the cell body; pro- or polykinetoplastic; apical cytostome on preflagellar rostrum; phagotrophic [3,165].
-
▪ Family Allobodonidae Goodwin et al., 2018. The phylogenetic group enclosing Allobodo and related forms on 18S rRNA gene-based trees [65].
-
▪ Genus Allobodo Goodwin et al., 2018. Parasitic in seaweeds; both flagella free; with apical rostrum; phagotrophic, short tubular cytopharynx not supported by a microtubular rod; pankinetoplastic [65].Type species: Allobodo chlorophagus Goodwin, Lee, Kugrens and Simpson, 2018. Monotypic (plate A, 11).
-
▪
-
▪ Family Neobodonidae Cavalier-Smith, 2016. Free-living or parasitic in animals; biflagellate, flagella free or attached; rostrum rigid; pro-, poly- or pankinetoplastic; phagotrophic, bacterivorous or eukaryovorous [170].Note: There is no evidence that the family is monophyletic.
-
▪ Genus Actuariola Stoeck, Schwarz, Boenigk, Schweikert, von der Heyden and Behnke, 2005. Free-living; solitary, phagotrophic; both flagella free and without mastigonemes; prokinetoplastic; cytopharynx supported by a non-prismatic microtubular rod [171].Type species: Actuariola framvarensis Stoeck, Schwarz, Boenigk, Schweikert, von der Heyden and Behnke, 2005. Monotypic (plate A, 1).
-
▪ Genus Azumiobodo Hirose, Nozawa, Kumagai and Kitamura, 2012. Parasitic in ascidians; anterior flagellum attached to the rostrum in basal part, posterior flagellum usually attached to the cell body; polykinetoplastic; cytostome at the apex of the long rostrum; curved cytopharynx, presence of supporting rod not assessed; unique globular bodies with electron-dense bands of various shapes [62].Type species: Azumiobodo hoyamushi Hirose, Nozawa, Kumagai and Kitamura, 2012. Monotypic (plate A, 5).
-
▪ Genus Cruzella Faria, Cunha and Pinto, 1922, emend Kostygov.Diagnosis: Free-living, solitary, two mastigoneme-free flagella originating under beak-shaped rostrum; phagotrophic, cytostome on rostrum tip, well-developed tubular cytopharynx without supporting microtubular rod; polykinetoplastic; intensive metaboly [172–174].Type species: Cruzella marina Faria, Cunha and Pinto, 1922. Monotypic (plate A, 10).
-
▪ Genus Cryptaulaxella Kostygov, nom. nov.Diagnosis: Free-living, solitary; both flagella free; prominent spiral groove on the surface; the presence of extrusomes questionable; ultrastructure not studied; type of kinetoplast uncertain [49,175].Type species: Spiromonas akopos Skuja, 1939 (= Cryptaulaxella akopos comb. nov.).Justification: This newly proposed name refers to the genus previously known as: (i) Spiromonas Skuja, 1939—homonym of Spiromonas Perty, 1852 [176] (Dinoflagellata); (ii) Cryptaulax Skuja, 1948—homonym of Cryptaulax Tate, 1869 [177] (Gastropoda) and Cryptaulax Cameron, 1906 [178] (Insecta); and (iii) Cryptaulaxoides Novarino, 1996—homonym of Cryptaulaxoides Uchida 1940 (Insecta).Etymology: The new name and the two previous ones share the Greek roots κρυπτός (hidden) and αὖλαξ (furrow), referring to the distinctive feature of the genus, and the feminine gender.
-
▪ Genus Klosteria Mylnikov and Nikolaev, 2003. Free-living, solitary; both flagella free, arise from a subapical flagellar pocket and bear short acronemes, anterior one with mastigonemes; rostrum not prominent; phagotrophic, cytopharynx tubular, without supporting microtubular rod; cytostome lips absent; pankinetoplastic; trichocysts near ventral side of the flagellar pocket [181].Type species: Klosteria bodomorphis Mylnikov and Nikolaev, 2003. Monotypic (plate A, 8).
-
▪ Genus Neobodo Vickerman, 2004. Free-living; solitary, phagotrophic; biflagellate with free posterior flagellum; prokinetoplastic; cytopharynx supported by a prismatic microtubular rod [165].Type species: Bodo designis Skuja, 1948 (= Neobodo designis) (plate A, 2).
-
▪ Genus Rhynchobodo Vørs, 1992. Free-living, solitary; flagella exit subapically and bear acronemes; phagotrophic; well-developed rostrum with apical cytostome, tubular cytopharynx and multiple extrusomes; conspicuous spiral groove on the body surface; polykinetoplastic [182] (plate A, 4).Type species: Cryptaulax taeniata Skuja, 1956 (= Rhynchobodo taeniata).
-
▪
-
▪ Family Rhynchomonadinae Cavalier-Smith, 2016. Solitary, free-living; biflagellate, the anterior flagellum adheres to the flexible proboscis and they move together, the posterior flagellum is used for gliding and attached to the body at least in its proximal part; cytopharynx not supported by a microtubular rod [170].
-
▪ Genus Dimastigella Sandon, 1928. Free-living, in soil or freshwater; anterior flagellum significantly longer than the proboscis, posterior flagellum attached to the cell body across the whole length of the latter; cytostome on or under rostrum; phagotrophic; polykinetoplastic [184,185] (plate A, 6).Type species: Dimastigella trypaniformis Sandon, 1928.
-
▪ Genus Rhynchomonas Klebs, 1892. Free-living, short anterior flagellum is attached to a long rostrum representing a motile proboscis, posterior flagellum attached to the cell body in the proximal part, both flagella with mastigonemes; phagotrophic; prokinetoplastic [186].Type species: Heteromita nasuta Stokes, 1888 (= Rhynchomonas nasuta) (plate A, 3).
-
▪
-
▪
-
○ Order Parabodonida Vickerman, 2004. Clade enclosing the genera Cryptobia, Parabodo, Procryptobia and Trypanoplasma. Free-living, commensal or parasitic; biflagellate, without mastigonemes, posterior flagellum attached or free; pro-, poly- or pankinetoplastic; phagotrophic or osmotrophic; anterolateral cytostome with or without developed cytopharynx [3,165]. Previously, parasitic representatives of this group were considered a single lineage and often lumped into one genus (Cryptobia), but molecular phylogenetic analyses showed their polyphyly [187].
-
▪ Family Cryptobiidae Poche, 1911 emend. Kostygov. Clade uniting the genera Cryptobia and Parabodo based on 18S rRNA gene phylogenies [68,188].
-
▪ Genus Cryptobia Leidy, 1846. Parasites/commensals of fish (on gills or in the gut) or various invertebrates (in the lumen of reproductive, digestive or excretory organs) [73,78,189]; recurrent flagellum attached to cell body without the formation of undulating membrane, its posterior part is used for attachment to host epithelium; conspicuous ventral furrow; phagotrophic with well-developed but miniaturized cytopharynx in most species; subpellicular microtubules extend to whole-cell length; pro- or pankinetoplastic [81,164] (plate A, 16).Type species: Cryptobia helicis Leidy, 1846.Note: The genus may be paraphyletic with respect to Parabodo as judged by sequence data on two species from invertebrates and fish, although the relationships are poorly resolved [190].
-
▪ Genus Parabodo Skuja, 1939 emend. Vickerman, 2004. Free-living, solitary; posterior flagellum free; the cytostome is placed at the anterior end of the cell making the latter bifurcate, well-developed cytopharynx; subpellicular microtubules extend to whole-cell length; prokinetoplastic [191,192] (plate A, 13).Type species: Parabodo nitrophilus Skuja, 1939.
-
▪
-
▪ Family Trypanoplasmatidae Hartmann and Chagas, 1910 emend. Kostygov. The clade uniting the genera Procryptobia and Trypanoplasma based on 18S rRNA gene phylogenies [68,188].
-
▪ Genus Procryptobia Vickerman, 1978. Solitary, free-living, prokinetoplastic; recurrent flagellum attached to the cell surface, ventral groove absent; short anterolateral rostrum; cell bears subpellicular microtubules only in the anterior portion and easily changes shape; phagotrophic [52,193] (plate A, 12).Type species: Procryptobia vorax Vickerman, 1978.
-
▪ Genus Trypanoplasma Laveran and Mesnil, 1901. Leech-transmitted obligate hemoparasites of fish; posterior flagellum attached to the cell body forming a conspicuous undulating membrane bordering a ventral furrow; osmotrophic, cytopharynx reduced; subpellicular microtubules extend to whole-cell length; megakinetoplast [66,164].Type species: Trypanoplasma borreli Laveran and Mesnil, 1901 (plate A, 15).
-
▪
-
▪
-
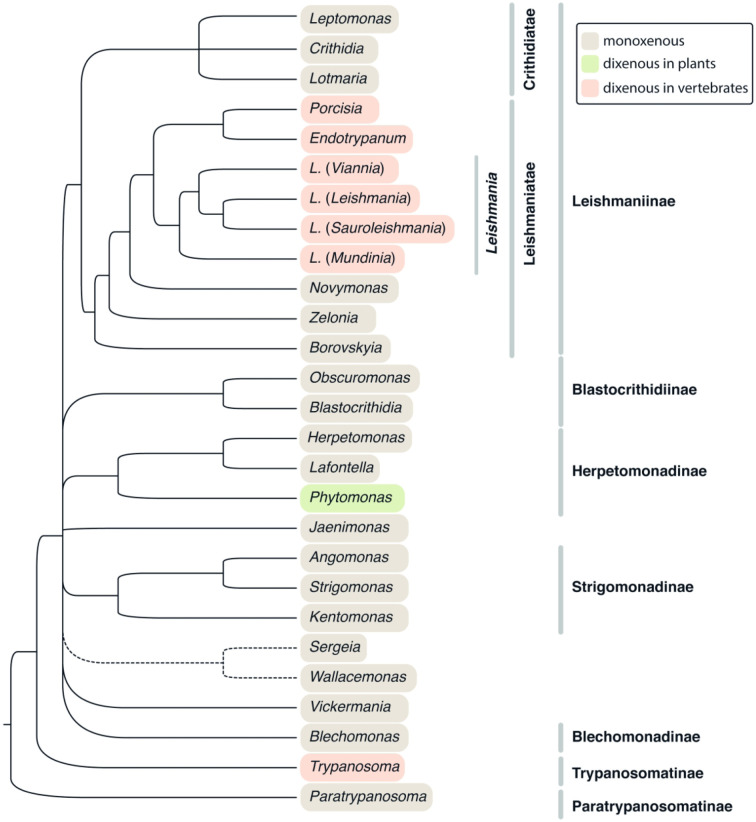
○ Order Trypanosomatida Kent, 1880. Monoxenous or dixenous obligatory endoparasites of arthropods, leeches, vertebrates, plants and ciliates; single flagellum, emerging from flagellar pocket apically or laterally, is mastigoneme-free and oriented anteriorly; eukinetoplastic with the kDNA network attached to the basal body of the flagellum [194]; phagotrophic or osmotrophic; cytostome–cytopharyngeal complex fully developed only in a few representatives, while the majority has no cytopharynx, and cytostome is present as a shallow pit or completely absent [195,196]. For a long time, the classification was based on the presence of the following morphotypes in the cell cycle: promastigote (elongated with apical flagellum and prenuclear kinetoplast), choanomastigote (shortened, with apical flagellum and prenuclear kinetoplast), opisthomastigote (elongated, with apical flagellum and postnuclear kinetoplast), opisthomorph (shortened, with apical flagellum and postnuclear kinetoplast), epimastigote (with lateral flagellum attached to the cell body and prenuclear kinetoplast), trypomastigote (with lateral flagellum attached to the cell body and postnuclear kinetoplast), amastigote/endomastigote (flagellum not emerging from the pocket, the first variant predominantly used when flagellum is very short) and cyst-like amastigote (compact cells with dense cytoplasm, completely lacking flagellum). Single family. The taxonomic changes introduced here follow the guidelines specified for this group previously [197].
-
▪ Family Trypanosomatidae Doflein, 1901. With the same definition as the order (tree C).Note: Historically, all monoxenous genera were often termed ‘lower trypanosomatids', but after the switch to the phylogeny-oriented paradigm, this concept has been abandoned [92]. As an alternative, a colocation ‘insect trypanosomatids’, which has no evolutionary connotations, is often used.
-
▪ Subfamily Trypanosomatinae Doflein, 1901. A distinct clade on phylogenetic trees is based on 18S rRNA, gGAPDH and multiple protein-coding genes enclosing the genus Trypanosoma [10]. Single genus.
-
▪
-
▪
-
○
Tree C.
Trypanosomatidae. A tree summarizing multiple phylogenetic reconstructions, mostly 18S rRNA gene-based. Dashed line denotes an unstable clade, which is disrupted when certain taxa are included into the analysis. For the genus Leishmania, relationships between its four subgenera are also shown. Highlighting denotes types of the life cycles (see graphical legend).
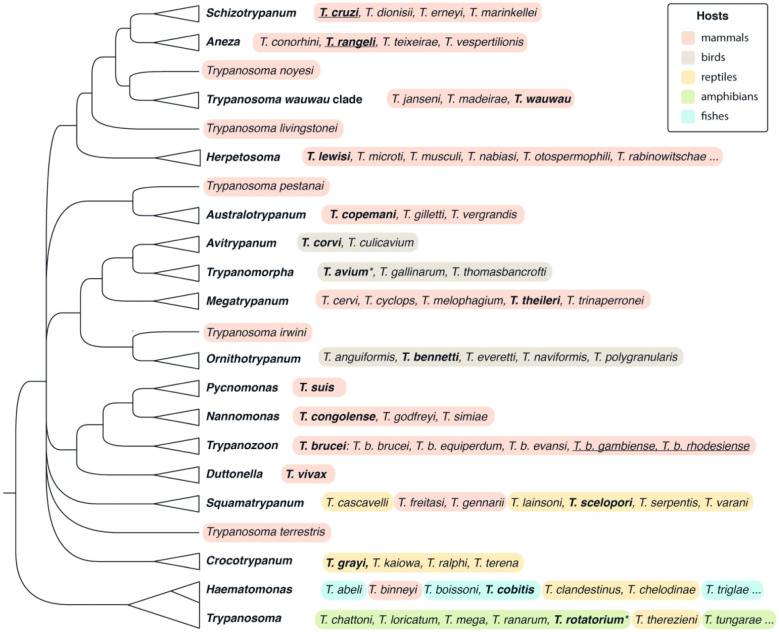
Genus Trypanosoma Gruby, 1843. Dixenous parasites of all classes of vertebrates (in blood and tissues) transmitted by blood-sucking arthropods or leeches (digestive tract and salivary glands); trypomastigotes and amastigotes (in vertebrates) or epimastigotes and trypomastigotes (in invertebrates) (plates B and C, 18–40) (tree D).
Tree D.
Trypanosoma. A tree summarizing multiple phylogenetic reconstructions, mostly 18S rRNA gene-based. All species or a selection of the most important ones (marked with three dots at the end), for which affiliation with a given subgenus and/or clade was confirmed using molecular phylogenetic methods, are listed. Highlighting denotes orders of vertebrate hosts (see graphical legend). Human parasites are underlined. Type species are shown in bold and the names accepted as senior subjective synonyms are marked with an asterisk.
Type species: Trypanosoma sanguinis Gruby, 1843 (junior subjective synonym of T. rotatorium (Mayer, 1843)).
Note: Historically, mammalian trypanosomes were subdivided into the sections Stercoraria (Herpetosoma, Megatrypanum and Schizotrypanum) and Salivaria (Duttonella, Nannomonas, Pycnomonas and Trypanozoon). Being long-established and of practical importance for the community of medical and veterinary doctors, this mammalian-centred classification is incorrect from the phylogenetic point of view (see Biology of Kinetoplastea), as it not only does not correspond with the known diversity of trypanosomes but also cannot accommodate species transmitted by other modes. Moreover, mammalian trypanosomes are not monophyletic and the names stercoraria and salivaria, which were proposed to reflect the differences between the modes of transmission, are non-taxonomical and have only historical value.
In parallel, the genus Trypanosoma was divided into several subgenera based on rather subtle morphological differences. Interestingly, this historical taxonomical classification in general corresponds well with the current phylogenetic analysis. However, despite the indisputable usefulness of this classification, in recent decades, the usage of subgenera was largely omitted (probably because of anticipated—but not materialized—conflicts between the morphology- and phylogeny-based systems). As a consequence, newly emerging clades in the expanding phylogenetic trees were named after the best-known representatives, causing confusion. Here, we revive the original subgeneric concept, and by erecting several new subgenera achieve mutual harmonization of the old morphological and the modern phylogenetic approaches. We believe that this taxonomical system reflects best the true diversity of these important parasites.
Species for which only morphological but no sequence information is available are marked with an asterisk (*). We have mapped the hosts (classes of vertebrates) onto the phylogenetic trees, highlighting associations among clades and their vertebrate host.
-
•
‘the aquatic clade’ (monophyletic) (plate B, 18–23).
Note: Molecular phylogenies confirmed the monophyly of the genus Trypanosoma [198] and its subdivision into the aquatic and terrestrial clades [199–201]. In most reconstructions, the aquatic clade was further split into the ‘fish/turtle’ and ‘amphibian’ subgroups. While the former was invariably monophyletic, depending on the taxonomic set used, the latter appeared either as monophyletic or unresolved until an analysis including some key frog species demonstrated its clear paraphyly [202–204]. The ‘fish/turtle’ subgroup, which also includes trypanosomes from a platypus (T. binneyi Mackerras, 1959) and a crocodile (T. clandestinus Teixeira and Camargo, 2015) is here designated as the subgenus Haematomonas. The remaining aquatic trypanosomes, mostly parasitizing frogs (with T. therezieni Brygoo, 1963 infecting chameleons being a single known exception), fall into the paraphyletic subgenus Trypanosoma.
-
▪
Subgenus Haematomonas Mitrophanow, 1883 emend. Votýpka and Kostygov. Diagnosis: Leech-transmitted parasites of aquatic vertebrates. Morphologically variable medium to large conspicuously elongated trypomastigotes with a notably bent body, undulating membrane including free flagellum, and with the kinetoplast situated close to the posterior end of the body. Defined by 18S rRNA-based phylogenetic analyses.
Type species: Haematomonas cobitis Mitrophanow, 1883 (= Trypanosoma cobitis).
Note: Mitrophanow described two monoflagellates from the European freshwater fish and placed them into the new genus Haematomonas as H. cobitis (from a weatherfish, Misgurnus fossilis; formerly genus Cobitis) and H. carassii (from a crucian carp, Carassius carassius) which were both later reclassified into the genus Trypanosoma [205]. Since then, more than 190 trypanosome species have been described from both marine and freshwater jawless, cartilaginous and bony fish worldwide [206]. Phylogenetic analyses [202,204,207,208] revealed three monophyletic clades within this subgenus: (i) freshwater fish trypanosomes (including T. clandestinus Teixeira and Camargo, 2016 from crocodiles); (ii) marine fish trypanosomes (including T. rajae Laveran and Mesnil, 1902 from rays); (iii) turtle trypanosomes (including T. binneyi from a platypus). The following species are included in published phylogenetic trees: T. abeli, T. binneyi, T. boissoni, T. cobitis, T. chelodinae, T. clandestinus, T. epinepheli, T. fulvidraco, T. granulosum, T. mocambicum, T. murmanensis, T. nudigobii, T. ophiocephali, T. pleuronectidium, T. pseudobagri, T. rajae, T. sinipercae and T. triglae.
-
▪
Subgenus Trypanosoma Gruby, 1843 emend. Votýpka and Kostygov.
Diagnosis: Morphologically variable medium to large trypomastigotes characterized by a wide range of forms with remarkable morphological plasticity; besides classical fusiform trypomastigotes, there are rounded, oval, claviform, fan-shaped, leaf-like, or irregular cells with or without a free flagellum, and longitudinal or spiral striations. Defined by 18S rRNA-based phylogenetic analyses.
Type species: Trypanosoma sanguinis Gruby, 1843 (junior subjective synonym of T. rotatorium [209]).
Note: Mayer in 1843 found in the blood of a frog (Rana esculenta) captured in Germany two organisms that he named Amoeba rotatoria and Paramaecium loricatum, which were later recognized as first-ever described trypanosomes [209]. Gruby published later the same year a description of a haemoflagellate from the blood of a frog in France and named it Trypanosoma sanguinis (from Greek trypanon, an auger; soma, body) [210]. In 1901, Doflein created the family Trypanosomidae, in which the genus Trypanosoma was subdivided into three subgenera including the nominotypical one—Trypanosoma with T. sanguinis Gruby 1843 as a type [205]. In 1926, International Commission on Zoological Nomenclature accepted T. rotatorium Mayer 1843 as the senior synonym of T. sanguinis Gruby 1843 [211].
The following described species are included in published phylogenetic trees: T. chattoni, T. fallisi, T. herthameyeri, T. loricatum, T. mega, T. neveulemairei, T. percae, T. ranarum, T. rotatorium, T. therezieni and T. tungarae.
-
•
‘the terrestrial clade’ (monophyletic) (plate C, 24–40).
Note: The internal classification of trypanosomes from terrestrial hosts is rather confusing. Mammalian trypanosomes, subdivided into the sections stercoraria and salivaria, were mostly singled out, followed by the avian trypanosome branch(es). Additionally, new clades appeared gradually in phylogenetic studies, the inclusion of which is a significant problem. Moreover, these above-mentioned groups are not monophyletic, making the internal system of the terrestrial clade unstable. We have attempted to rectify the situation by building a system that accommodates taxonomic units, for which sequence information is available.-
○ avian subgenera (paraphyletic)Based on comparative morphology and developmental cycles, avian trypanosomes were hypothesized to be closely related to the subgenus Megatrypanum that infects ruminants [212,213]. This assumption is now supported by phylogenetic and phylogenomic studies that have also shown paraphyly of avian trypanosomes, represented by at least three distinct lineages (T. avium, T. corvi and T. bennetti clades) [144,214,215]. These groups are distinguishable by the morphology of bloodstream stages and the kinetoplast thickness [216,217].
-
▪ Subgenus Trypanomorpha Woodcock, 1906 emend. VotýpkaDiagnosis: Medium to large size trypomastigotes (40–100 μm) with longitudinal striations (myonemes), central oval nucleus, prominent undulating membrane and small kinetoplast to which the free flagellum is anterior. Cells in culture have kinetoplast exceptionally thick (greater than 500 nm). Defined by 18S rRNA-based phylogenetic analyses; cosmopolitan distribution.Type species: Trypanosoma noctuae Schaudinn, 1904 (junior subjective synonym of T. avium Danilewsky, 1885, see note).Note: For the type species described from a European little owl Athene noctua [218], neither culture nor sequence data are available. Both exist for T. thomasbancrofti Šlapeta, 2016 and also for less clearly defined T. avium Danilewsky, 1885 and T. gallinarum Bruce et al., 1911 [214–217,219]. The vaguely defined species Trypanosoma avium was described in 1885 by Danilewsky from birds, but the type material was not preserved [220]. In 1903, Laveran proposed to restrict this species name to parasites of owls [221]; however, the name was often used to designate any bird trypanosome.
-
▪ Subgenus Avitrypanum Votýpka, subgen. nov.Diagnosis: Medium to large trypomastigotes (about 40–80 μm) in the bloodstream of bird hosts with longitudinal striations (myonemes), nucleus positioned centrally and posterior kinetoplast; the thickness of kinetoplast in cultured cells is less than 500 nm. Defined by 18S rRNA-based phylogenies; cosmopolitan distribution.Type species: Trypanosoma corvi Stephens and Christophers, 1908, here designated.Etymology: The generic name refers to the fact that trypanosomes come from bird hosts, the order Aves (the Latin name for a bird is avis).Note: Baker [222] emended T. corvi and restricted the use of the name to large trypanosomes from non-American corvids and also from other bird families. The species was re-described [223] and phylogenetically characterized [214] and forms the T. corvi clade along with T. culicavium Votýpka et al., 2012. Although morphologically indistinguishable from the subgenus Trypanomorpha (T. avium clade), Aviotrypanum (T. corvi clade) is not directly related to it [144,216,217], justifying separate treatment.
-
▪ Subgenus Ornithotrypanum Votýpka, subgen. nov.Diagnosis: Small to medium-size non-striated avian trypanosomes (less than 40 µm in length) with kinetoplast situated close to the posterior end of the body. Kinetoplast thickness of cells in culture below 500 nm. Defined by 18S rRNA-based phylogenetic analyses; cosmopolitan distribution.Type species: Trypanosoma bennetti Kirkpatrick, Terway-Thompson and Iyengar, 1986, here designated.Etymology: The generic name refers to the fact that trypanosomes come from bird hosts, the order Aves (the ancient Greek name for a bird is órnῑs; ὄρνῑς).Note: Out of a number of morphologically described species parasitizing wild birds, for five of them (T. anguiformis Valkiūnas et al., 2011, T. bennetti, T. everetti Molyneux, 1973, T. naviformis Sehgal et al., 2015 and T. polygranularis Valkiunas et al., 2011) sequence data are available [144,224,225]. Their divergence justifies the establishment of a new subgenus—Ornithotrypanum.Phylogenomic approach revealed that the T. bennetti (= Ornithotrypanum) and T. avium (= Trypanomorpha) clades are nested within the mammalian clade and are paraphyletic with respect to the ruminants-infecting Trypanosoma theileri Laveran, 1902 [144], thus breaking the monophyly of mammalian trypanosomes. Interestingly, the host generalism of avian trypanosomes contrasts with the host specificity observed for some mammalian flagellates.
-
▪
-
○ ‘mammalian subgenera (stercoraria)’ (polyphyletic)
-
▪ Subgenus Herpetosoma Doflein, 1901.Diagnosis: Medium-size trypomastigotes (20–40 µm) with long pointed posterior extremity, large rod-like subterminal kinetoplast but well away from the posterior end and long free flagellum. Parasitizing a wide range of rodents and lagomorphs, as amastigote and/or epimastigotes.Type species: Herpetosoma lewisi Kent, 1880 (= Trypanosoma lewisi).Note: This globally distributed species found in more than 100 rodent species (predominantly Rattus spp.) and rarely also in humans [226], is transmitted by the ingestion of fleas Xenopsylla cheopis and Nosopsyllus fasciatus or in their faeces. Considered largely non-pathogenic for rodents, it can cause serious disease in unnatural hosts. After its introduction in synanthropic rats to Christmas Island, it drove the endemic rat Rattus macleari to extinction, being the only known cases of a trypanosomatid responsible for the extinction of its host species.Molecular phylogenies revealed the polyphyly of Herpetosoma [143,227–229], excluding T. rangeli Tejera, 1920 into a newly established subgenus Tejeraia (now Aneza), keeping Herpetosoma (also named T. lewisi-like clade) monophyletic. 18S rRNA sequences are available for the following species: T. niviventerae (rat, Niviventer confusianus), T. musculi (mouse, Mus musculus/domesticus), T. grosi (field mouse, Apodemus spp.), T. microti (vole, Microtus spp.), T. evotomys (bank vole, Clethrionomys glareolus), T. rabinowitschae (hamster, Cricetus cricetus), T. blanchardi (dormouse, Eliomys quercinus), T. kuseli (squirrel, Pteromys volans), T. ostospermophili (squirrel, Urocitellus spp.) and T. nabiasi (rabbit, Oryctolagus cuniculis), while molecular data are not available for approximately 30 more species, including *T. acomys, *T. acouchii, *T. ellobii and *T. lemmi.The questions whether three phylogenetically related species with Megatrypanum-like morphology and basal phylogenetic position, T. talpae (European mole, Talpa europaea), T. sapaensis (white-toothed shrew, Crocidura dracula) and T. anourosoricus (mole shrew, Anourosorex yamashinai) should be included into the subgenus Herpetosoma remains unresolved [230].
-
▪ Subgenus Megatrypanum Hoare, 1964Diagnosis: Large trypomastigotes (40–100 µm) with long pointed posterior extremity; medium nonterminal kinetoplast located near the nucleus and far from the posterior end of the body; long free flagellum; reproduction as epimastigotes in the mammalian host.Type species: Trypanosoma theileri Laveran, 1902; a cosmopolitan non-pathogenic bovine trypanosome transmitted by tabanids.Note: Other members are ovine T. melophagium Flu, 1908 using a sheep ked (Melophagus ovinus) as a vector, while caprine *T. theodori Hoare, 1931 is transmitted by a goat ked (Lipoptena capreoli). Two species of cervid trypanosomes, European and North American T. cervi Kingston and Morton, 1975 and newly described Pan-American T. trinaperronei Teixeira, Camargo and García, 2020, are transmitted by deer keds (Lipoptena cervi and L. mazamae) [231]. Tabanids probably transmit *T. stefanskii Kingston et al., 1992 from a roe deer (Capreolus capreolus) [232] and *T. ingens Bruce et al., 1909 from African antelopes [233]. Vectors remain unknown for the closely related simian T. cyclops Weinman, 1972 and *T. lucknowi Weinman, White and Antipa, 1984 from Macaca monkeys. Morphological [234,235] and phylogenetic studies [199,236] justify inclusion of these two species into the subgenus Megatrypanum.
- Note: Probably least settled within the former stercoraria is the taxonomy within the so-called T. cruzi superclade. Its members have been hypothesized to primarily be parasites of bats (Chiroptera), from which they have expanded into other mammals. Two species, T. cruzi Chagas, 1909 and T. rangeli, both restricted to the New World, are capable of infecting humans. The majority of known invertebrate vectors of these trypanosomes belong to true bugs (Heteroptera). The T. cruzi superclade incorporates two subgenera: Schizotrypanum and Aneza, as well as several (un)named clades and species complexes [237–239].
- Trypanosoma wauwau Lima et al., 2015 from Pteronotus bats in South America constitutes a potentially novel subgenus, now termed the T. wauwau clade. Other members of this clade are unnamed species from the Neotropical bats, T. janseni Lopes et al., 2018 from inner organs of a Brazilian opossum Didelphis aurita, and T. madeirae Battos et al., 2019 from a Neotropical vampire Desmodus rotundus. The other two candidate subclades are represented by Trypanosoma noyesi Botero and Cooper, 2016 found in Australian marsupials (woylie, wallabies, kangaroos and possums) and the genetically highly diverse (a complex of species) infecting various African bats with the only described member, Trypanosoma livingstonei Teixeira and Camargo, 2013.
-
▪ Subgenus Schizotrypanum Chagas, 1909Diagnosis: Relatively small trypomastigotes (15–25 μm), typically C- or S-shaped in blood smears with short pointed posterior extremity, a large subterminal kinetoplast and long free flagellum. In mammals, reproduction takes place in form of the intracellular amastigotes.Type species: Trypanosoma cruzi Chagas, 1909; a causative agent of Chagas disease in humans, transmitted by triatomine bugs (e.g. Triatoma, Rhodnius) [240].Note: Trypanosoma cruzi (also known as T. cruzi cruzi or T. cruzi sensu stricto) has a very high molecular and phenotypic heterogeneity, reflected by the existence of seven genetically distinct lineages (or discrete typing units, DTUs) termed TcI–TcVI and Tcbat [241]. An impartial comparison of this conspicuous genetic diversity, which corresponds very well with life cycles, clinical manifestations and host specificity, with the situation of the T. brucei complex, reveals a striking and untenable discrepancy between these two key species complexes. While in the T. brucei complex, five DTUs have the status of five different (sub)species (see below), the T. cruzi complex has so far not been split into subspecies and sticks to the DTU code. Therefore, we propose that the same system, based on subspecies, should be applied for both species complexes. We urge our colleagues working with T. cruzi to implement such a system.The current T. cruzi complex is accompanied by three (sub)species from bats with typical Schizotrypanum morphology: T. marinkellei Baker et al., 1978 (restricted to South America), T. dionisii Bettencourt and França, 1905 (occurring in the Old and New World) and T. erneyi Lima and Teixeira, 2012 (found in Africa).
-
▪ Subgenus Aneza Özdikmen, 2009 (= Tejeraia Añez, 1982 [preoccupied]).Diagnosis: Small to medium-size trypomastigotes (25–35 µm) with long pointed posterior extremity, medium subterminal kinetoplast and long free flagellum, are all similar to the subgenus Herpetosoma. At least, some species (e.g. T. rangeli) produce metacyclic stages in the salivary glands of its triatomine bug vectors, and therefore are not strictly speaking stercorarians (although transmission via faeces also occurs).Type species: Trypanosoma rangeli Tejera, 1920.Note: T. rangeli is restricted to South America and has a wide mammalian host range including humans, for which it is non-pathogenic; the only known vectors are triatomine bugs of the genus Rhodnius. While another member of the subgenus, T. conorhini Donovan, 1909, is found worldwide in rats and Indonesian primates and is transmitted by triatomine bug T. vespertilionis Edmond and Etienne Sergent, 1905 infecting bats is widely distributed in Africa and Europe, where it is transmitted by Cimex spp. Sequence data are also available for T. teixeirae Barbosa et al., 2016 found in the blood of Australian flying foxes.
-
▪
-
○ ‘mammalian subgenera (salivaria)’ (monophyletic)
-
▪ Subgenus Duttonella Chalmers, 1908Diagnosis: Small to medium-size trypomastigotes (21–26 µm) with large and usually terminal kinetoplast, small to rounded posterior extremity and long free flagellum. Development in the insect vector is confined to the proboscis and the adjacent cibarial pump.Type species: Trypanosoma vivax Ziemann, 1905; a causative agent of souma in cattle and other ungulates in Africa and South America (following its import from western Africa).Note: Although only one described species has been formally assigned to this subgenus, phylogenetic analyses revealed a complex of species related to T. vivax [242]. Previously described *T. uniforme Bruce et al., 1911 and *T. vivax ellipsiprymni Keymer, 1969, termed ‘T. vivax-like’ may be a part of this complex.
-
▪ Subgenus Nannomonas Hoare, 1964Diagnosis: Small trypomastigotes (12–20 μm) with medium-sized subterminal marginal kinetoplast, blunt posterior extremity and free flagellum either absent or very short. Vector development takes place in the midgut and proboscis.Type species: Trypanosoma congolense Broden, 1904; a causative agent of nagana in cattle and other ungulates.Note: T. congolense is further split into three types/‘subspecies’ (Savannah, Forest and Kilifi) that are, arguably, sufficiently different to warrant species status due to different virulency for domestic animals. Other species infect ungulates and monkeys (T. simiae Bruce et al., 1909 which is represented by two ‘good’ species) or were detected in tsetse flies like T. godfreyi McNamara, Mohammed and Gibson, 1994 and several unnamed species [243,244].
-
▪ Subgenus Pycnomonas Hoare, 1964Diagnosis: Small trypomastigotes (8–20 µm) with very short pointed posterior extremity, small subterminal kinetoplast and short free flagellum. Vector development takes place in the midgut and salivary glands of tsetse flies (Glossina spp.).Type species: Trypanosoma suis Ochmann, 1905; a causative agent of chronic porcine trypanosomiasis, infects Suidae in sub-Saharan Africa.
-
▪ Subgenus Trypanozoon Lühe, 1906Diagnosis: Pleomorphic trypomastigotes represented by long slender (mean length 30 μm) with long free flagellum and short stumpy forms (mean length 18 µm) with no free flagellum; both have a small subterminal kinetoplast.Type species: Trypanosoma brucei Plimmer and Bradford, 1899.Note: According to the life cycle, transmission mode, vectors, vertebrate hosts and clinical manifestations, five (sub)species are recognized and widely accepted: Trypanosoma brucei brucei (in ungulates, the causative agent of nagana in cattle; transmitted by tsetse that restrict its distribution to sub-Saharan Africa), T. brucei rhodesiense (causative agent of acute sleeping sickness in humans; game animals and livestock are primary reservoir; vectored by tsetse, sub-Saharan Africa), T. brucei gambiense (chronic sleeping sickness in humans; some domestic animals are reservoir; vectored by tsetse, sub-Saharan Africa), T. brucei evansi (causative agent of trypanosomiasis in camels, horses, cattle, buffalo, dogs and pigs, called surra in Africa and Asia and murrina in South America; transmitted mechanically by blood-sucking insects and vampires) and T. brucei equiperdum (causes dourine in horses in Asia, Africa, South America and Europe; transmitted sexually). The latter two subspecies are closely related and are unique in being so-called petite mutants of T. brucei [145].
-
▪
-
○ ‘other terrestrial subgenera’ (paraphyletic)
-
▪ Subgenus Australotrypanum Votýpka and Kostygov, subgen. nov.Diagnosis: Morphologically highly variable trypomastigotes in the blood of marsupials and bats in Australia. Defined by 18S rRNA-based phylogenetic analyses.Type species: Trypanosoma copemani Austen, Jefferies, Friend, Ryan, Adams and Reid, 2009, here designated.Etymology: The generic name refers to the origin from Australian mammals.Note: A distinct monophyletic clade composed of T. copemani, T. gilletti McInnes et al., 2011, and T. vegrandis Thompson et al., 2013 that infect marsupials and bats (in case of T. copemani) [245–248]. They seem to be transmitted by ticks [249] and have been implicated in the decreased survival of koalas (Phascolarctos cinereus) [248]. T. copemani exhibits polymorphic ‘slender’ and ‘broad’ trypomastigote stages in the bloodstream [245] and intracellular amastigotes [250]. Sphaeromastigotes, amastigotes and promastigotes were present in vitro. On the other hand, T. vegrandis is believed to be the smallest species formally described from mammals, with trypomastigotes below 10 μm of length [247]. Trypanosoma gilletti, described from koalas based on 18S rRNA sequences only, is 50 µm long [246].
-
▪ Subgenus Crocotrypanum Votýpka and Kostygov, subgen. nov.Diagnosis: Large striated trypomastigotes (up to 100 µm) occurring in very small numbers in peripheral blood of crocodiles and caimans in the Neotropic and Afrotropic. The conspicuous undulating membrane forms a well-marked frill along the edge of the cell and continues to free flagellum. In tsetse flies (Glossina) and horse flies (Tabanidae), epimastigotes and promastigotes develop in the midgut and hindgut; transmission occurs via contaminative way. Defined by 18S rRNA-based phylogenetic analyses.Type species: Trypanosoma grayi Novy, 1906, here designated.Etymology: The generic name refers to the fact that trypanosomes come from hosts of the order Crocodilia.Note: T. grayi transmitted by tsetse [251–253] clusters together with three recently described species, T. terena Teixeira and Camargo, 2013, T. ralphi Teixeira and Camargo, 2013 and T. kaiowa Teixeira and Camargo, 2019 transmitted by insect vectors [254,255], into a strongly supported monophyletic clade [255]. Based on morphology, *T. cecili Lainson, 1977 could also belong to this subgenus. All described crocodilian trypanosomes form the monophyletic crocodilian clade (subgenus Crocotrypanum) of the terrestrial lineage and are transmitted by insect vectors. Trypanosoma clandestinus Teixeira and Camargo, 2016, transmitted among caimans by leeches, is not related to this group and is nested within the aquatic lineage (subgenus Haematomonas) [255].
-
▪ Subgenus Squamatrypanum Votýpka and Kostygov, subgen. nov.Diagnosis: Morphologically variable medium to large-sized trypomastigotes with multi-folded undulating membrane including free flagellum and kinetoplast located near the nucleus. Defined by 18S rRNA-based phylogenetic analyses.Type species: Trypanosoma scelopori Ayala, 1970, here designated.Etymology: The unusual combination of hosts (see below) was used for the subgenus name combining the Latin name of reptiles (Squamata) and mammals (Mammalia).Note: This clade brings together trypanosomes from diverse hosts, namely lizards, snakes, rodents and marsupials. Based on their morphology, T. lainsoni Naiff and Barrett, 2013 from rodents and T. freitasi Rêgo, Magalhães and Siqueira, 1957 from marsupials used to belong to the subgenus Megatrypanum; however, this taxonomic classification does not reflect their phylogenetic position. While T. varani Wenyon, 1908 described from a Nile monitor lizard (Varanus niloticus) in Sudan [256] and later found in a Ghanaian ball python (Python reginus) [257] represents the only Afrotropical species, other three species within this subgenus were described in American reptiles: T. serpentis Viola et al., 2009 from Brazilian snake Pseudoboa nigra [258], T. scelopori Ayala, 1970 from North American western fence lizard (Sceloporus occidentalis) [259] and T. cascavelli Pessôa and Da Biasi, 1971 from a South American rattlesnake (Crotalus durissus) [260]. The latter species also survives in the blood of Neotropical marsupials [261]. T. freitasi and T. gennarii Marcili, 2017, were described from Didelphis and Monodelphis opossums, respectively [262,263], yet their host spectrum is even broader [261]. Finally, T. lainsoni, originally described from Amazonian rodents [264], can infect South American marsupials and bats [261].
-
▪
-
○ ‘incertae sedis species'Note: Three species—Trypanosoma irwini McInnes et al., 2009, T. pestanai Bettencourt and Franca, 1905 and T. terrestris Marcili, 2013—do not fall into any of the above-listed subgenera and constitute separate branches.Trypanosoma irwini from Australian koala (Phascolarctos cinereus), with middle-sized (approx. 40 µm) trypomastigotes with prominent kinetoplast, undulating membrane, pointed posterior end and long free flagellum [265], is closely related to the avian trypanosomes of the subgenus Ornithotrypanum [265,266].Trypanosoma pestanai from Eurasian badgers was, based on morphology (middle-sized trypomastigotes approx. 35 µm long, subterminal kinetoplast) [267,268], associated with the subgenus Megatrypanum, yet phylogenetic analyses indicate its affiliation rather with the subgenus Australotrypanum [266,269].Finally, T. terrestris infecting South American lowland tapir (Perissodactyla) is not closely related to any subgenera [270].
-
▪ Subfamily Leishmaniinae Maslov and Lukeš, 2012. Group identified by 18S rRNA and GAPDH gene-based phylogenies. Includes the monoxenous genera Borovskyia, Crithidia, Leptomonas, Lotmaria, Novymonas and Zelonia, as well as the dixenous genera Endotrypanum, Porcisia and Leishmania [271].
-
○ Infrafamily Crithidiatae Kostygov and Yurchenko, 2017. The clade comprises genera Crithidia, Leptomonas and Lotmaria, which currently cannot be reliably separated from each other [271].
-
▪ Genus Leptomonas Kent, 1880. Parasites of the gut of invertebrates; promastigotes as the only motile form [94] (plate D, 42).Type species: Leptomonas butschlii Kent, 1880.
-
▪ Genus Crithidia Léger, 1902. Parasites of the gut of insects (dipterans, heteropterans, hymenopterans); choanomastigotes as the only motile form [94] (plate D, 41).Type species: Crithidia fasciculata Léger, 1902.
-
▪ Genus Lotmaria Evans and Schwarz, 2014. The clade comprising Lotmaria passim and related species, based on the concatenation of 18S rRNA and gGAPDH genes; promastigotes in the gut of bees [272] (plate D, 43).Type species: Lotmaria passim Schwarz, 2014. Monotypic (see note).
-
▪
-
○
-
○
-
○ Infrafamily Leishmaniatae Maslov and Lukeš, 2012. Comprises the dixenous genera of the subfamily along with Novymonas, Zelonia and Borovskyia [271].
-
▪ Genus Leishmania Ross, 1903. Dixenous parasites of mammals and reptiles infecting cells of the mononuclear phagocyte system, where they multiply as amastigotes. Widely distributed in tropical and subtropical regions. In mammals, depending on the preferred type(s) of phagocytes, they cause different clinical forms of the disease: cutaneous, mucocutaneous and visceral. Transmitted predominantly by phlebotomine sandflies (Diptera: Psychodidae: Phlebotominae), in whose gut they develop as promastigotes [127] (plate D, 49,50).Type species: Piroplasma donovani Laveran and Mesnil, 1903 (= Leishmania donovani).
-
• Subgenus Leishmania Ross, 1903. Cause cutaneous and visceral forms of leishmaniasis in mammals. Distributed in Africa, Eurasia and Americas. Transmitted by sandflies, in which they develop in the midgut [127].Type species: same as for the genus.
-
• Subgenus Viannia Lainson and Shaw, 1987. Cause cutaneous and mucocutaneous forms of leishmaniasis in mammals; restricted to South America; transmitted by sandflies, in which they develop in the midgut and hindgut [127].Type species: Leishmania braziliensis Vianna, 1911.
-
• Subgenus Sauroleishmania Ranque, 1973. Live in the blood cells of lizards and snakes, predominantly in the mononuclear phagocyte system, but reported also from erythrocytes and thrombocytes [276]. Transmitted by sandflies, in which they develop in the hindgut [127].Type species: Leishmania tarentolae Wenyon, 1920.
-
• Subgenus Mundinia Shaw, Camargo and Teixeira, 2016. Cause cutaneous and visceral leishmaniases in mammals [125,277]; recorded on all continents except Antarctica; transmitted by biting midges [126,278].Type species: Leishmania enriettii Muniz and Medina, 1948.
-
▪ Genus Porcisia Shaw, Camargo and Teixeira, 2016. Leishmania-like dixenous flagellates parasitizing skin and visceral organs of porcupines as intracellular amastigotes; transmitted by phlebotomine sand flies as promastigotes [125,279,280] (plate D, 46).Type species: Leishmania hertigi Herrer, 1971 (= Porcisia hertigi).
-
▪ Genus Endotrypanum Mesnil and Brimont, 1908. May represent a mixture of two distinct dixenous taxa, one of which is defined morphologically (intraerythrocytic trypomastigotes and/or epimastigotes in sloths) and another phylogenetically (Leishmania-like flagellates related to former L. herreri and parasitizing skin and visceral organs of various mammals as amastigotes [125,271,281]. The latter is transmitted by phlebotomine sand flies as promastigotes [125] (plate D, 48).Type species: Endotrypanum schaudinni Mesnil and Brimont, 1908.
-
▪ Genus Novymonas Kostygov and Yurchenko, 2020. Monoxenous, insect host unknown; promastigotes and choanomastigotes; the only known species bears multiple vacuole-enclosed β-proteobacterial cells in the cytoplasm [275].Type species: Novymonas esmeraldas Votýpka, Kostygov, Maslov and Lukeš, 2020. Monotypic (plate D, 44).
-
▪ Genus Zelonia Shaw, Camargo and Teixeira, 2017. Monoxenous; promastigotes parasitizing true bugs and dipterans; represent a distinct lineage that cannot be associated with any other described genus [125].Type species: Leptomonas costaricensis Yurchenko, Lukeš, Jirků, Zeledon and Maslov, 2006 (= Zelonia costaricensis) (plate D, 45).
-
▪ Genus Borovskyia Kostygov and Yurchenko, 2017. Monoxenous; parasites of true bugs; only promastigotes are known; represents the earliest branch within Leishmaniatae [271].Type species: Leptomonas barvae Maslov and Lukeš, 2010 (= Borovskyia barvae). Monotypic.
-
•
-
▪ Subfamily Herpetomonadinae Alexeieff, 1911, stat. nov., emend. Kostygov and Yurchenko (= Phytomonadinae Yurchenko, Kostygov, Votýpka and Lukeš, 2015; unavailable name).Diagnosis: Clade of monoxenous parasites of insects and dixenous parasites of insects and plants defined by phylogenetic analyses based on 18S rRNA and gGAPDH gene sequences; promastigotes or choanomastigotes are dominant morphotypes; may also form opisthomastigotes, opisthomorphs and endomastigotes. Arginase absent.Type genus: Herpetomonas Kent, 1880.Note: Herpetomonas was designated as a type genus of the subfamily Phytomonadinae [282], making the latter unavailable according to article 11.7.1.1 of ICZN. At the same time, the name Herpetomonadidae, a synonym of Trypanosomatidae Doflein, 1901 at the family level, is available as a name of the subfamily (with the ending -inae), being the only suitable one for a clade containing its type genus.
-
▪ Genus Herpetomonas Kent, 1880. Monoxenous; parasites of dipterans, true bugs, fleas, cockroaches and ciliates; polymorphic: predominant promastigotes varying in size and shape as well as non-mandatory opisthomastigotes, opisthomorphs and endomastigotes [106,282,283] (plate D, 51,52).Type species: Bodo muscarum Leidy, 1856 (= Herpetomonas muscarum).
-
▪ Genus Lafontella Kostygov and Yurchenko, 2015. Monoxenous; parasitic in the gut of flies; promastigotes, opisthomastigotes and long endomastigotes with elongated coiled flagellum [282,284].Type species: Herpetomonas mariadeanei Yoshida, Freymuller and Wallace, 1978 (= Lafontella mariadeanei). Monotypic.
-
▪ Genus Phytomonas Donovan, 1909 emend. Kostygov.Diagnosis: long (often twisted) promastigotes and endomastigotes; most species alternate between plants and phytophagous true bugs, some switched to predatory bugs and became secondary monoxenous; obligate development in salivary glands [103,119,122,123] (plate D, 53,54).Type species: Leptomonas davidi Lafont, 1909 (= Phytomonas davidi).
-
▪
-
▪ Subfamily Strigomonadinae Votýpka, Yurchenko, Kostygov and Lukeš, 2014. Monoxenous, with several apomorphic traits: single β-proteobacterial endosymbiont not enclosed in a vacuole, extensively branched mitochondrion disrupting subpellicular corset of microtubules, rudimentary paraflagellar rod [274].
-
▪ Genus Angomonas Souza and Corte-Real, 1991. Monoxenous parasites in the gut of blowflies; choanomastigotes and opisthomorphs; kinetoplast nearly rectangular, with kinetoplast minicircles greater than 4 kb [285].Type species: Crithidia deanei Carvalho, 1973 (= Angomonas deanei) (plate D, 56).
-
▪ Genus Strigomonas Lwoff and Lwoff 1931. Monoxenous parasites of the gut of dipterans and true bugs; polymorphic: epimastigotes and trypomastigotes or choanomastigotes and opisthomorphs; kinetoplast lens-shaped, usually with only one side convex and another flat or concave; minicircles less than 3 kb [285].Type species: Strigomonas oncopelti Lwoff and Lwoff, 1931 (plate D, 58).
-
▪ Genus Kentomonas Votýpka, Yurchenko, Kostygov and Lukeš, 2014. Monoxenous parasites of the gut of flies; mitochondrial branches press on the plasmatic membrane forming ridges on the cell surface; kinetoplast nearly rectangular [274].Type species: Kentomonas sorsogonicus Votýpka and Lukeš, 2014. Monotypic (plate D, 57).
-
▪
-
▪ Subfamily Blastocrithidiinae Votýpka, Yurchenko and Lukeš, 2021. A well-supported monophyletic group (as judged by the analyses based on 18S rRNA gene) of monoxenous trypanosomatids inhabiting the gut of true bugs (Heteroptera) [286].
- ▪
- Type species: Crithidia gerridis Patton, 1908 (= Blastocrithidia gerridis).
-
▪ Genus Obscuromonas Votýpka and Lukeš, 2021. In 18S rRNA-based phylogenies, a sister group to Blastocrithidia; monoxenous parasites in different organs of heteropterans; some members produce cyst-like straphangers [286] (plate D, 64).Type species: Obscuromonas modryi Votýpka and Lukeš, 2021.
-
▪ Subfamily Blechomonadinae Votýpka and Suková, 2013. A clade comprising the genus Blechomonas according to phylogeny inferred using 18S rRNA and gGAPDH genes [288].
-
▪ Subfamily Paratrypanosomatinae Votýpka and Lukeš, 2013. The earliest-branching lineage within the family as judged by the phylogenies inferred using 18S rRNA and multiple protein-coding genes [289]. Single genus.
-
• Genera not assigned to subfamilies
-
▪ Genus Jaenimonas Votýpka and Hamilton, 2020. Distinct monoxenous lineage in 18S rRNA and gGAPDH gene-based phylogenies; parasite of the gut of fruit flies; monotypic [112].Type species: Jaenimonas drosophilae Votýpka and Hamilton, 2020 (plate D, 60).
-
▪ Genus Vickermania Kostygov and Yurchenko, 2020. Monoxenous parasites of the gut of flies; promastigotes with two anteriorly oriented flagella of unequal length, typically attached to each other and separated during cell division; uniflagellate cells appear only shortly after division; flagellar tips have rounded or elongated apex and lateral extensions; large and loosely arranged kDNA [291].Type species: Herpetomonas muscarum ingenoplastis Rogers and Wallace, 1971 (= Vickermania ingenoplastis) (plate D, 65).
-
▪ Genus Sergeia Svobodová et al., 2007. Distinct monoxenous lineage in 18S rRNA and gGAPDH gene-based trees; parasite of the gut of biting midges; promastigotes as the only motile stage [292].Type species: Sergeia podlipaevi Svobodová et al., 2007. Monotypic (plate D, 59).
-
▪ Genus Wallacemonas Kostygov and Yurchenko, 2014. Distinct monoxenous lineage in 18S rRNA and gGAPDH gene-based trees; parasites of dipterans and true bugs; promastigotes as well as non-mandatory opisthomorphs and endomastigotes [293,294].Type species: Leptomonas collosoma Wallace, Clark, Dyer and Collins, 1960 (= Wallacemonas collosoma) (plate D, 61).
-
▪
-
• Trypanosomatidae incertae sedis (none is available in culture)
-
▪ Genus Cercoplasma Roubaud, 1911. Monoxenous; in the gut of flies; epimastigotes and trypomastigotes; flagellum without free part, accompanied by a filamentous cell processus [295].Type species: Cercoplasma caulleryi Roubaud, 1911. Monotypic.
-
▪ Genus Malacozoomonas Nicoli, Penaud and Timon-David, 1971. Monoxenous; in the gut and hepatopancreas of molluscs; promastigotes and amastigotes [296].Type species: Herpetomonas patellae Porter, 1914 (= Malacozoomonas patellae).
-
▪ Genus Nematodomonas Nicoli, Penaud and Timon-David, 1971. Monoxenous; in the gut of nematodes; promastigotes only [297].Type species: Nematodomonas goodeyi Nicoli, 1971. Monotypic.
-
▪ Genus Rhynchoidomonas Patton, 1910 (= Cystotrypanosoma Roubaud, 1911). Monoxenous; in the gut and Malpighian tubules of flies; trypomastigotes without free flagellum and conspicuous undulating membrane; cyst-like amastigotes observed in some species [94,298];Type species: Rhynchomonas luciliae Patton, 1910 (= Rhynchoidomonas luciliae).
-
▪
-
• Protists erroneously assigned to Kinetoplastea (none is available in culture)
-
▪ Genus Trypanophis Keysselitz, 1904 emend. Kostygov.Diagnosis: Parasites of the gastrovascular cavity of siphonophores; biflagellate, with striated rootlet, short free anterior flagellum, long posterior flagellum attached to the cell body and situated in shallow longitudinal groove; subpellicular microtubules present only under groove; membranous sacs under plasmalemma; micropores; elongated mitochondrion parallel to the flagellar groove, with tubular cristae, possesses anterior dilation (‘kinetoplast’) with multiple osmiophilic bodies; no traces of oral apparatus [299].Type species: Trypanosoma grobbeni Poche, 1903 (= Trypanophis grobbeni).Note: Assignment of this genus to kinetoplastids is not justified, since its ‘kinetoplast’, as judged by Feulgen staining, does not contain DNA. The presence of cortical membranous sacs, micropores and a striated rootlet [299] strongly suggest that this flagellate is a member of Alveolata Cavalier-Smith, 1991.
-
▪
-
• Kinetoplastea incertae sedis
-
▪ Genus Bordnamonas Larsen and Patterson, 1990. Free-living, solitary, with pliable body; biflagellate, anterior flagellum forms arc extending in front of the cell, while posterior flagellum is trailing; phagotrophic, cytostome is anterior to flagella [300]; fine structure and type of kinetoplast unknown.Type species: Bordnamonas tropicana Larsen and Patterson, 1990. Monotypic (plate A, 7).
-
▪ Genus Cephalothamnium Stein, 1878. Ectocommensal on freshwater copepods; biflagellate, both flagella with mastigonemes, the posterior flagellum attached to cell body; forms sedentary colonies with up to 30 cells attached to secreted stalk by the distal end of the recurrent flagellum; large prokinetoplast; subpellicular microtubules only in the anterior part; phagotrophic, with apical cytostome and funnel-shaped cytopharynx [61].Type species: Cephalothamnium cyclopum Stein, 1878. Monotypic.
-
▪ Genus Desmomonas Williams, 1999. Parasitic in turbellarian parenchyma, either free or attached to host cell masses via anterior processus with desmosomes; biflagellate, both flagella unattached to the cell body, oriented posteriorly and lacking prominent paraflagellar rod; corset of subpellicular microtubules has breaches allowing body shape changes; no cytostome detected (osmotrophic); compact mitochondrion with polykinetoplast distant from the flagellar base; microneme-like osmiophilic bodies [60].Type species: Desmomonas prorhynchi Williams, 1999. Monotypic.
-
▪ Genus Jarrelia Poynton, Whitaker and Heinrich, 2001. Parasite of blowhole mucus of pygmy sperm whale; biflagellate, posterior flagellum forms undulating membrane and can attach to host material by its tip; polykinetoplastic; osmotrophic [87].Type species: Jarrellia atramenti Poynton, Whitaker and Heinrich, 2001. Monotypic.Note: previously assigned to Parabodonida based on superficial resemblance with Trypanoplasma [3]; however, no reliable evidence supporting such an assignment is available.
-
▪ Genus Lamellasoma Davis, 1947. Parasite of fish gills; uniflagellate, single flagellum oriented posteriorly and attached to cell body; type of kinetoplast uncertain; epibiotic bacteria on the surface [81].Type species: Lamellasoma bacillaria Davis, 1947. Monotypic.Note: May represent an unusual species of piscine Cryptobia with a very short or completely reduced anterior flagellum [81].
-
▪
Plate D.
Trypanosomatids other than Trypanosoma. Light micrographs of Giemsa-stained (41) Crithidia thermophila (culture) (42) Leptomonas seymouri (culture); (43) Lotmaria passim (culture); (44) Novymonas esmeraldas (culture); (45) Zelonia costaricensis (culture); (46) Porcisia hertigi (culture) (provided by Jovana Sádlová); (47) Endotrypanum sp. ex sloth; (48) Endotrypanum monterogeii (culture); (49) Leishmania major, metacyclic promastigote (culture) (provided by JS); (50) L. major, amastigotes in a macrophage (provided by Tereza Leštinová); (51) Herpetomonas nabiculae, promastigote (culture) (provided by Marina N. Malysheva and Alexander O. Frolov); (52) H. nabiculae, opistomastigote (provided by Marina N. Malysheva and Alaxander O. Frolov); (53) Phytomonas lipae, promastigote ex Coreus marginatus (provided by Marina N. Malysheva and Alaxander O. Frolov); (54) P. lipae, endomastigotes ex C. marginatus (provided by Marina N. Malysheva and Alaxander O. Frolov); (55) Lafontella sp. (culture); (56) Angomonas deanei (culture) (provided by Anna I. Ganyukova); (57) Kentomonas sorsogonicus (culture); (58) Strigomonas oncopelti (culture); (59) Sergeia podlipaevi (culture); (60) Jaenimonas drosophilae (culture); (61) Wallacemonas collosoma (culture); (62) Paratrypanosoma confusum (culture); (63) Blastocrithidia frustrata (culture); (64) Obscuromonas oborniki (culture); (65) Vickermania ingenoplastis (culture); (66) Blechomonas englundi (culture). Scale bar, 5 µm (41–43, 56, 57); 10 µm (44–55, 58–66).
3. Diplonemea
3.1. Biology
The body of knowledge on the biology of diplonemids comprises a large number of environmental 18S rRNA sequences, few cultured and sequenced species, as well as several formally described species lacking sequence data and not available in culture. Until recently, diplonemids have been perceived as a small and unimportant group of euglenozoans. However, deep-sea sampling and extensive metabarcoding surveys in the past two decades uncovered extraordinary diversity of marine planktonic diplonemids [12,302,303]. Their discovery began with the recovery of environmental 18S rRNA sequences from deep-sea planktonic and hydrothermal samples, which together formed a novel well-supported clade, sister to Diplonemidae [304,305]. Subsequently, a diplonemid-focused study of several oceanic regions uncovered considerable diversity within the new clade by amplifying 18S rRNA, designated as deep-sea pelagic diplonemids (DSPD) I clade [302]. The same study identified another small lineage called DSPD II.
A breakthrough came with a V9 region metabarcoding survey by the Tara Oceans expedition, which uncovered remarkable abundance and diversity of DSPD in the tropical and subtropical sunlit ocean, expanding the number of potential diplonemid species to over 12 300 [303]. Further analysis of combined datasets from photic and mesopelagic zones identified as many as approximately 45 000 diplonemid species, thus qualifying them among the most species-rich planktonic eukaryotes in the ocean [12]. In the most recent study, extended with smaller datasets from the Arctic, Adriatic Sea and anoxic Cariaco Basin, the number of species increased to approximately 67 000, designating diplonemids as the most diverse and fifth most abundant eukaryotic clade [48]. However, fluorescence in situ hybridization studies or those based on the V4 region of 18S rRNA reported significantly lower abundance [306–308].
Analysis of extended Tara Oceans datasets identified that 97% of diplonemid diversity is confined to the DSPD I clade, or eupelagonemids, whereas classic diplonemids (or Diplonemidae), hemistasiids and DSPD II accounted for 1% each [12,48]. The distribution of eupelagonemids showed clear depth stratification: although their sequences were recovered from the surface water down to the abyssopelagic zone [309], they are most diverse and abundant in the mesopelagic zone. However, multiple lineages of eupelagonemids show cosmopolitan distribution without a clear biogeographic pattern and a rather weak relation to abiotic factors [310]. Eupelagonemids generally show preference for dark and moderately oxygenated environments, but were occasionally detected under anoxic conditions [48]. It is still not known what drives the high diversity of eupelagonemids given the relative homogeneity of the physico-chemical conditions in the deep ocean, especially in dysphotic and aphotic zones. It has been suggested that different species might use different nutrient resources [302]. Indeed, the co-occurrence analyses showed very few obvious patterns of interaction with other components of marine plankton, among which are positive correlations with parasitic dinoflagellates and stramenopiles as well as with bacteria and bacterivorous stramenopiles, indicating possible bacteriovory and parasitic lifestyle of some eupelagonemids [12] (plate E, 67–76).
Plate E.
Diplonemids. Light micrographs of cultured (67) Diplonema papillatum; (68) Rhynchopus euleeides; (69) Sulcionema specki; (70) Artemidia motanka; (71) Eupelagonemid sp. (provided by Noriko Okamoto and Patrick Keeling); (72) Diplonema aggregatum; (73) Lacrimia lanifica; (74) Flectonema neradi; (75) Namystinia karyoxenos; (76) Hemistasia phaeocysticola. Scale bar, 10 µm (67–75).
Unlike the deep-sea planktonic clades, which inhabit nutrient-poor, dark and cold ocean zones [12,302], classic diplonemids seem to prefer a variety of nutrient-rich environments, such as benthos, coastal surface waters, artificial water bodies and aquaria [311]. Classical diplonemids have been widely considered as benthic organisms [11] apparently due to the sampling bias. Their sequences have been indeed recovered from various benthic environments, including cold anoxic seeps [312], hydrothermal vents [304] and the sea floor [302], in addition to several species that were observed in tropical shallow-water [300,313] and deep-sea sediments [311]. However, diplonemids are also a common component of the plankton in photic layer of the temperate to tropical zones [12]. In addition, their representatives are known from coastal planktonic communities, including Rhynchopus coscinodiscivorus [314], Diplonema breviciliata [315], D. papillatum [316], D. nigricans [317], Lacrimia lanifica, Rhynchopus serpens and Sulcionema specki [311]. Classical diplonemids were also frequently isolated from aquaria, such as D. japonicum, D. aggregatum, D. ambulator ATCC 50223, Rhynchopus humris, Flectonema neradi and Rhynchopus ATCC 50230 [311] (plate E, 67–70,72–76).
Diplonemids have been occasionally reported from freshwater ecosystems, such as the case of Rhynchopus amitus [318] and Diplonema ambulator from a freshwater aquarium [319]. Subsequently, metabarcoding approach identified a low number of diplonemids in geographically isolated deep lakes, such as Baikal [320], and lakes in Japan [321], Switzerland and the Czech Republic [322]. Further systematic metabarcoding screening of lakes using diplonemid-specific primers might uncover so far overlooked diversity of freshwater diplonemids.
The fourth diplonemid clade, Hemistasiidae, is represented by several hundred species, so far found in the photic zone [12]. Hemistasia-like flagellates have been frequently detected in coastal waters of North and Baltic seas, Mediterranean, the Sea of Japan and around Australia and Antarctica [313,315,323–325], pointing to their cosmopolitan distribution from cold to tropical regions. All hemistasiids were described from planktonic samples, except for Artemidia motanka isolated from an aquarium [311] and a hemistasiid associated with shallow-water sediments [313].
Classic diplonemids and hemistasiids are exclusively heterotrophic organisms, mostly known as eukaryovores, and displaying a wide array of lifestyles, such as ectocommensalism, predation, scavenging and opportunistic endoparasitism. Several species were found to parasitize lobsters, clams [180,326] and plants [319], while others were referred to as epibionts of crabs [327], lobsters [180], plants [300,315,316] and algal biofilms (Diplonema sp. 4 ATCC 50232). Another common trophic mode for both groups is predation and/or scavenging on planktonic algae and small invertebrates, including diatoms, dinoflagellates, green algae, prymnesiophytes and copepods [314,315,318,323]. Bacteriovory was described from only two species and seems rather uncommon [300,328].
3.2. Taxonomy
Class Diplonemea and order Diplonemida Cavalier-Smith, 1993 (tree E).
Tree E.
Diplonemids. A tree summarizing phylogenetic reconstructions based on 18S rRNA gene. Circles denote genera with cultured representatives.
Naked colourless biflagellates with apical papilla; subapical flagellar pocket; plasma membrane subtended by dense microtubular corset; peripheral mitochondria with giant lamellar cristae and multiple interspersed DNA aggregates; equally thick flagella; tubular extrusomes in several species; likely phagotrophic feeding.
-
• Family Diplonemidae Cavalier-Smith, 1993; also known as classic diplonemids with metaboly always present.
-
▪ Genus Diplonema Griesmann, 1914. Based on 18S rRNA gene, the genus is paraphyletic [180]; morphological and 18S rRNA discrepancies (D. papillatum is 87% different from D. ambulator) justify possible division of this genus into two genera. Elongated body with constricted anterior end; equal to subequal flagella; gliding movement and ambulation of flagella in trophic stage; tubular Euglenozoa-type extrusomes and heterodynamic flagella with paraflagellar rods (PFR) in swimming stage (if present) (applies to Diplonema ambulator, D. japonicum and D. aggregatum).
- Type species: Diplonema (Isonema) papillatum Porter, 1973 (short flagella of equal length lacking PFR; nearly apical flagellar pocket; big papilla) (plate E, 67).
-
▪ Genus Rhynchopus Skuja, 1948. Flagellar stubs with disorganized axoneme microtubules, concealed inside flagellar pocket and gliding motion in trophic stage; flagella are gradually built in actively gliding cells; long heterodynamic flagella with PFR in swimming stage, anterior flagellum forming a lasso and the posterior one stretched along the body (plate E, 68).
- Type species: Rhynchopus amitus Skuja, 1948.
-
▪ Genus Lacrimia Tashyreva, Prokopchuk, Horák and Lukeš, 2018. Permanently long subequal flagella with PFR; teardrop-shaped body; big posterior digestion vacuole; rotation movement and oscillating swimming pattern.
- Type species: Lacrimia lanifica Tashyreva, Prokopchuk, Horák and Lukeš, 2018 (plate E, 73).
-
▪ Genus Flectonema Tashyreva, Prokopchuk, Horák and Lukeš, 2018. Short flagella of equal length; elongated, slender, crooked body reminiscent to D. ambulator type; distinguished by the presence of PFR in trophic stage; gliding and rotation motion, swimming absent, dispersal swimming stage not described.
- Type species: Flectonema neradi Tashyreva, Prokopchuk, Horák and Lukeš, 2018 (plate E, 74).
-
▪ Genus Sulcionema Tashyreva, Prokopchuk, Horák and Lukeš, 2018. Short flagella of equal length, containing PFR; long flat body with conspicuous cytoplasmic inclusions, pleomorphic; writhing motion, but swimming and gliding absent; highly metabolic.
- Type species: Sulcionema specki Tashyreva, Prokopchuk, Horák and Lukeš, 2018 (plate E, 69).
-
▪
-
• Family Hemistasiidae Cavalier-Smith, 2016. Fast swimming, long flagella with prominent PFR; tubular Euglenozoa-type extrusomes; peripheral lacunae; highly asymmetrical apex with flexible pointy rostrum; metaboly always present; anterior groove; subapically inserted flagella inside a deep invagination; large posterior digestion or lipid vacuole is common; cylindrical to pyriform body; establishment of three genera is justified by substantial differences in 18S rRNA gene [325].
-
▪ Genus Hemistasia Griesmann, 1914. Invariable presence of tubular extrusomes; distinguished from related genera by acute rostrum and smaller body.
- Type species: Oxyrrhis phaeocysticola Scherffel, 1900 (= Hemistasia phaeocysticola) (plate E, 76).
-
▪ Genus Namystinia Prokopchuk, Tashyreva and Lukeš, 2019. Broader rostrum; tubular extrusomes only in starved cells; morphologically indistinguishable from Artemidia.
- Type species: Namystinia karyoxenos Prokopchuk, Tashyreva and Lukeš, 2019 (plate E, 75).
-
▪ Genus Artemidia Prokopchuk, Tashyreva and Lukeš, 2019. Broader rostrum; invariable presence of tubular extrusomes.
- Type species: Artemidia motanka Prokopchuk, Tashyreva and Lukeš, 2019 (plate E, 70).
-
▪
-
• Family Eupelagonemidae Okamoto and Keeling, 2019. Formerly known as ‘deep sea pelagic diplonemids 1’ (DSPD I), possibly non-metabolic.
-
▪ Genus Eupelagonema Okamoto and Keeling, 2019. Elongated elliptical body, round on one end and constricted on the other (plate E, 71).
- Type species: Eupelagonema oceanica Okamoto and Keeling, 2019.
- Note: DSPD II (deep sea pelagic diplonemids II)—small planktonic clade, well-supported phylogenetically, known exclusively from sequences of the V9 region of 18S rRNA [12], without cultured or formally described representatives; morphology and ultrastructure not known.
-
▪
4. Euglenida and Symbiontida
4.1. Biology
Since most of the 18S rRNA phylogenies placed symbiontids either within euglenids, or as a sister clade to them (e.g. [170,329,330]), they are often regarded as derived euglenids (e.g. [329,331]). For those reasons, here we discuss the biology of euglenids and symbiontids together. However, a recent phylogenomic reconstruction [8] placed the latter group as a sister to diplonemid-kinetoplastid clade (Glycomonada), suggesting that they should be treated as a separate group within Euglenozoa.
Euglenida (plate F, 77–96) and Symbiontida (plate F, 97) inhabit aquatic environments, but they dominate in different ecological niches. Phagotrophic euglenids (plate F, 88–96) are widespread in shallow marine, brackish and freshwater sediments, and are presumably important predators in these ecosystems [42,332,333]. Recently, they have also been reported from deep-sea samples [334]. Osmotrophic (plate F, 82) and phototrophic euglenids (euglenophytes; plate F, 77–81, 83–87) mainly inhabit the water column of freshwater environments. In the temperate zone, euglenophytes are abundant in small eutrophic reservoirs where the water warms up quickly. They might form blooms, including toxic blooms caused by Euglena sanguinea [335,336]. In the tropical climate, euglenophyte blooms are also commonly reported, especially from aquaculture ponds [337,338]. Several lineages of typically freshwater genera—Discoplastis (plate F, 83), Phacus (plate F, 79), Lepocinclis (plate F, 78), Euglena (plate F, 85) and Euglenaria, have been detected in the coastal environments in low abundances [339]. Moreover, some species (Euglena rustica and E. obtusa) have been reported to migrate vertically in marine sand in coordination with tidal and diurnal cycles. They are usually highly abundant and form green patches in marine sand during low tides [340]. The three earliest-branching lineages of photosynthetic euglenids, Rapaza (plate F, 87), Eutreptia and Eutreptiella (plate F, 86), belong to marine plankton. Although the known diversity of marine species of euglenophytes is low, blooms of Eutreptiales have been reported from eutrophic coastal waters, where they can make up to approximately 46% of the total biomass of the phytoplankton population [341,342]. Symbiontids, inhabiting both shallow and deep anoxic marine sediments, host sulfur-oxidizing or sulfide-oxidizing epsilonproteobacterial epibionts, which detoxify their immediate surrounding environment [41]. They might be a dominant group in certain environments, such as in the protist community associated with bacterial mats in oxygen-depleted sediment in Monterey Bay [343].
Plate F.
Euglenids. Light micrographs of cultured (77) Cryptoglena sp. (provided by Bożena Zakryś); (78) Lepocinclis autumnalis (provided by Bozena Zakrys); (79) Phacus acuminatus (provided by Bozena Zakrys); (80) Trachelomonas armata (provided by Bozena Zakrys); (81) Monomorphina sp. (provided by Bozena Zakrys); (82) Menoidium sp. (provided by Bozena Zakrys); (83) Discoplastis spathirhyncha (provided by Bozena Zakrys); (84) Euglenaformis proxima (provided by Bozena Zakrys); (85) Euglena gracilis (provided by Bozena Zakrys); (86) Eutreptiella pomquetensis (provided by Bozena Zakrys); (87) Rapaza viridis (provided by Naoji Yubuki); (88) Jenningsia sp. (provided by Gordon Lax); (89) Peranema sp. (provided by Gordon Lax); (90) Anisonema sp. (provided by Gordon Lax); (91) Heteronema vittatum (provided by Gordon Lax); (92) Dinema sp. (provided by Gordon Lax); (93) Olkasia polycarbonata (provided by Gordon Lax); (94) Notosolenus ostium (provided by Gordon Lax); (95) Sphenomonas teres (provided by Gordon Lax); (96) Lentomonas corrugata (provided by Gordon Lax); (97) Calkinsia aureus (provided by Naoji Yubuki). Scale bar, 10 µm (77–97).
It has been demonstrated that both phagotrophic (e.g. Distigma) and photosynthetic euglenids (e.g. Euglena gracilis; plate F, 85) are remarkably tolerant to various kinds of pollution with heavy metals such as cadmium, chromium or lead, as well as capable to remove these ions from the environment, making these protists potentially suitable for use in bioremediation of heavy metal-rich industrial wastewater [344,345]. The genetic background of heavy metal resistance in euglenids has been examined in detail only in the genus Peranema. Interestingly, although the investigated Peranema sp. strain exhibited the capability for efficient removal of cadmium from wastewater samples, the study revealed that it possesses genes responsible for resistance to a variety of other heavy metals, but not cadmium [346]. Euglenophytes have also been found in waters polluted with diesel oil [347], phenol [348], herbicides and insecticides [349,350], and can survive in highly radioactive water [351]. Some Euglenophytes are also extremophiles (e.g. Euglena mutabilis), as they tolerate very high salinity [352] in extremely acidic environments [353] or in hot mud pools [354].
Euglenids and symbiontids are predominantly free-living, exhibiting a remarkably wide range of nutrition modes, including phagotrophy, osmotrophy and photoautotrophy. Phagotrophic euglenids consume bacteria or microbial eukaryotes, and their prey size correlates with the euglenids' cell size and flexibility. Some phototrophs are capable of pinocytosis, or even phagotrophy of algae in the case of the deep-branching phototroph Rapaza [355] (plate F, 87). Symbiontids are marine heterotrophs, presumably phagotrophs, as suggested by their ultrastructure. Additionally, the bacteria on their surface probably exchange metabolites with the hosts' mitochondria-related organelles, and it is also possible that they provide a food source for the symbiontids [7].
In contrast with the dominant free-living euglenids, there is an assemblage of eight heterotrophic genera incertae sedis (Michajlowastasia, Parastasiella, Dinemula, Paradinemula, Mononema, Ovicola, Naupliicola and Embryocola) that exhibit obligate parasitic lifestyles. Although their host range is rather narrow, encompassing exclusively freshwater, free-living copepods (specifically the eggs, larvae and digestive tracts of adults), their geographical range spans across the eutrophic freshwater bodies of all continents and nearly all climate zones, covering the range of their host group. Unfortunately, the phylogeny of these eight genera remains unresolved, as virtually all studies of the parasitic euglenids, however extensive, were carried out in the pre-sequencing era [356]. Occasionally, other photosynthetic (Euglenamorpha) and heterotrophic (Heteronema) euglenids have been identified in the gastrointestinal tracts of a very wide range of vertebrate and invertebrate animal hosts; however, it remains disputed whether they are symbionts or parasites [357,358]. Several species of euglenophytes (mainly genus Colacium) have been recognized as parasites or, more likely, epibionts of zooplankton [359–363], while others (e.g. Euglena mutabilis, Trachelomonas hispida) have also been identified in the traps of carnivorous plants, such as Genlisea [364] or Utricularia [365]. Whether euglenophytes are prey, accidental inhabitants, or a part of the specialized community of the carnivorous plants' traps, remains unresolved.
As most aquatic microbial eukaryotes, euglenids and symbiontids are considered cosmopolitan. Our understanding of their distribution is hampered by the limited number of environmental sequencing projects focused on those groups. Despite clear microscopical evidence of phagotrophic euglenids in sediments and phototrophic euglenids in ponds, they are suspiciously rare in environmental sequencing datasets [166,366–368]. It was suggested that euglenids' 18S rRNA is divergent and often longer than the typical eukaryotic one [369,370], and the universal primers are not working efficiently for euglenids. Environmental sequences might be, however, obtained with specific primers designed for a certain group [371]. The same as for diplonemids, the V9 region of 18S rRNA seems to be a more suitable metabarcoding marker, and phototrophic euglenids have been surveyed in the environmental sequences from the TARA Oceans dataset and OSD dataset [339]. Although euglenophytes are overall quite rare in the marine plankton, their distribution is quite broad in the global ocean, with preference for the coastal upwelling zones, where the nutrient availability is higher than in other parts of the ocean [372]. Symbiontids have been investigated in several environmental 18S rRNA-based surveys in different geographical regions, suggesting their cosmopolitan distribution [166,373–376].
4.2. Taxonomy
Class Euglenida Bütschli, 1884 emend. Simpson, 1997 (tree F).
Tree F.
Euglenids and symbiontids. A tree summarizing multiple phylogenetic reconstructions, primarily based on 18S rRNA gene sequences, genes encoded by plastid genomes (Euglenophyceae), and a set of nuclear protein-coding genes retrieved from transcriptomes. Polyphyletic genera are marked with an asterisk (*). Lineages with highly unstable position are marked with a dotted line. Possible paraphyly of clades has been further described in the section on Euglenids taxonomy.
Euglenids’ synapomorphy is a pellicle build of proteinaceous strips beneath the plasma membrane. The strips can be fused together in certain genera; otherwise, the organisms are capable of characteristic ‘euglenoid motion’ also known as ‘metaboly’. Another characteristic feature, shared with Kinetoplastida, is the presence of flagella inserted at the base of the flagellar pocket, and the flagella are conspicuously thickened due to the presence of paraxonemal (paraflagellar) rods. Additionally, the main storage polymer of most euglenids is paramylon, a distinctive β-1,3-glucan.
Note: Historically, species belonging to class Euglenida were described according to the rules of the ICZN or the ICN due to the presence of photosynthetic organisms within this ancestrally non-photosynthetic group. This has created some confusion, as several taxa bear two different, equally valid names. Moreover, many clades are unstable and assigning a rank and a name to them would be unproductive, since these names are likely to become obsolete very soon. The taxonomy proposed here is a consensus between a strictly organized taxonomy for stable clades and informal nomenclature. Additionally, Euglenophyceae and their subordinate taxa are treated as algae and classified according to ICN (with alternative names consistent with ICZN provided in notes), while all other subordinate taxa of Euglenida are classified according to ICZN (with alternative names consistent with ICN provided in notes).
-
• Clade Olkaspira Lax and Simpson, 2020. This robustly supported monophyletic clade includes organisms with pellicle composed of S-shaped proteinaceous strips with overhangs, and chisel-shaped feeding apparatus (if the apparatus is present; see below) [8]. Flexible cells belong to the subordinate clade Spirocuta; rigid cells belong to the subordinate genus Olkasia:
-
▪ Genus Olkasia Lax, Lee, Eglit and Simpson, 2019. Rigid, flattened, biflagellate cells with 10 pellicle strips; consists of only one species (O. polycarbonata), formerly classified as Ploeotia [377].
- Type species: Olkasia polycarbonata Lax, Lee, Eglit and Simpson, 2019 (plate F, 93). No culture available; several sequences of SSU rDNA and transcriptomes obtained from single-cell isolates.
-
▪
-
○
Clade Spirocuta Cavalier-Smith, 2016. The monophyletic group encompassing all flexible euglenids including phototrophs (Euglenophyceae), primary osmotrophs (Aphagea) and various phagotrophs [378]. The synapomorphy of this group is the capability for ‘metaboly’ also known as ‘euglenoid motion’.
Note: The other name used for this assemblage is Helicales [170,330,379,380].
-
▪
Clade Euglenophyceae Schoenichen, 1925 emend. Marin and Melkonian, 2003. The monophyletic group [170,381–383] comprising the basal monotypic genus Rapaza [355]. A predominantly photosynthetic group with plastids derived from secondary endosymbiosis with green alga, some species secondarily osmotrophic, most species with photosensory eyespot.
- Note: The other name used for this group is Euglenea Bütschli, 1884 emend. Busse and Preisfeld, 2002; however, as pointed out by Cavalier-Smith in the work cited above, this name is shared with a beetle genus.
-
○ Order Euglenales Leedale, 1967 emend. Marin and Melkonian, 2003. Cells with one emergent flagellum and one vestigial within the cell; feeding by phototrophy or secondarily by osmotrophy, mostly freshwater [3]; 18S rRNA gene has C in the first position of the Helix 7/8 spacer [381]; introns highly abundant in the plastid genome (always more than 28 and usually more than 51); intron maturase mat1/ycf13 always present in the plastid genome, with mat2, mat5 or both of them usually present [384].
-
• Family Euglenaceae Dujardin, 1841 emend. Kim et al., 2010. Solitary or colonial, mostly free-living, but some inhabit the digestive tracts of animals; usually possess one emergent flagellum and one non-emergent; some may possess mineralized external shells (lorica); size, number and presence of pyrenoids in chloroplasts varies with the species [3]; ribosomal operon may be present in the plastid genome in one copy or more, but never as two identical inverted repeats [384].Note: valid name under ICZN is Euglenidae Dujardin, 1841.
-
▪ Genus Colacium Ehrenberg, 1834. Solitary or colonial cells with envelopes that also form stalks for surface attachment [385]; often sessile (epizoic; attached to copepods) [386]; ribosomal operon present in the plastid genome in one full and one incomplete copy with the same orientation [387].Type species: Colacium vesiculosum Ehrenberg, 1834. Type species and other species available in the culture collections; multiple 18S rRNA sequences and a full plastid genome sequence of the type species are available.
-
▪ Genus Cryptoglena Ehrenberg, 1831. Rigid, solitary, laterally compressed cells with a longitudinal furrow and one or two chloroplasts (plate F, 77); 18S rRNA gene has AT base pair in the third position of the Helix 40 [381].Type species: Cryptoglena pigra Ehrenberg, 1832. Type species and other species available in the culture collections; multiple 18S rRNA sequences and a full plastid genome sequence of C. skujae (non-type species) are available.
-
▪ Genus Euglena Ehrenberg, 1830. Solitary cells with very visible metaboly; chloroplasts with pyrenoids (plate F, 85); at least two species (E. longa, E. quartana) secondarily non-photosynthetic, feeding by osmotrophy; 18S rRNA gene has T (rarely A or C) in the seventh position of the Helix 47/33 spacer [381]; ribosomal operon present in the plastid genome in one copy or in consecutive, tandemly repeated copies [384]; mitochondrial genome consists of seven linear chromosomes and encodes seven proteins which constitute components of complexes I, III and IV of the respiratory chain [27].Type species: Cercaria viridis O.F. Müller, 1786 (= Euglena viridis).Note: the genus Euglena is polyphyletic—on the phylogenetic trees Euglena archaeplastidiata and Euglena velata do not group with the main clade of Euglena [382,383]. Type species and other species available in culture collections, multiple sequences of 18S rDNA available; nuclear genome, plastid genome and mitochondrial genome of E. gracilis (non-type species), as well as multiple plastid genomes of other species, are available.
-
▪ Genus Euglenaformis Bennett and Triemer, 2014. Cryptic genus, morphologically undistinguishable from Euglena, but phylogenetically basal to all Euglenaceae; ribosomal operon present in the plastid genome in one copy [388]. Euglenaformis is a monospecific genus.Type species: Euglena proxima Dangeard, 1902 (= Euglenaformis proxima) (plate F, 84). Available in culture collections; 18S rRNA and a full plastid genome sequence of the type species are available.
-
▪ Genus Euglenaria Karnkowska, Linton and Kwiatowski, 2010. Solitary, metabolic cells with parietal, lobed chloroplasts with single pyrenoids and bilateral paramylon caps; distinguished from the genus Euglena by its distant phylogenetic position, sister to Monomorphina [389]; ribosomal operon present in the plastid genome in one copy [390].Type species: Euglena caudata Hübner, 1886 (= Euglenaria caudata). Type species and other species available in culture collections; 18S rDNA of multiple species and a full plastid genome sequence of Ea. anabaena (non-type species) are available.
-
▪ Genus Monomorphina Mereschkovsky, 1877. Rigid or slightly metabolic cells with pellicle-formed tail, 2–4 large paramylon plates and one or few large, spherical chloroplasts [381,391] (plate F, 81); 18S rRNA gene has T in the third position of the terminal loop of the Helix 27 [381]; ribosomal operon present in the plastid genome in one copy [390].Type species: Euglena pyrum Ehrenberg, 1832 (= Monomorphina pyrum). Type species and other species available in culture collections; 18S rRNA of multiple species and two full plastid genome sequences (M. aenigmatica and M. parapyrum; non-type species) are available.
-
▪ Genus Strombomonas Deflandre, 1930. Cells of variable shape and size with discoid or flat chloroplasts with pyrenoids and smooth lorica without collar [392]; ribosomal operon present in the plastid genome in one full and one incomplete copy with opposite orientation [387].Type species: Trachelomonas hispida var. verrucosa E. Daday, 1905 (= Strombomonas verrucosa). Type species and other species available in culture collections; 18S rRNA of multiple species and a full plastid genome sequence of S. acuminata (non-type species) are available.
-
▪ Genus Trachelomonas Ehrenberg, 1834. Cells of variable shape and size, with ornamented lorica with collar (plate F, 80); some species osmotrophic [381]; ribosomal operon present in the plastid genome in one copy [390].Type species: Microglena volvocina Ehrenberg, 1831 (= Trachelomonas volvocina). Type species and other species available in culture collections; 18S rRNA of multiple species and a full plastid genome sequence of the type species are available.
-
▪
-
• Family Phacaceae Kim, Triemer and Shin 2010. Solitary, free-living, with large paramylon grains and numerous small, discoid chloroplasts without pyrenoids.Note: valid name under ICZN is Phacidae Kim, Triemer and Shin 2010.
-
▪ Genus Discoplastis Triemer, 2006. Cells capable of metaboly, with a sharp, colourless tail [393,394]; two ribosomal operon-containing inverted repeats present in the plastid genome [384].Type species: Euglena spathirhyncha Skuja, 1948 (= Discoplastis spathirhyncha) (plate F, 83). Type species and other species available in culture collections; 18S rRNA of multiple species and a full plastid genome sequence of the type species are available.
-
▪ Genus Flexiglena Zakryś and Łukomska, 2020. Highly metabolic cells with numerous small paramylon grains and a distinct large, single grain located near the stigma [394].Type species: Euglena variabilis Klebs, 1883 (= Flexiglena variabilis). Currently not available in culture collections, but deposition of the type species in a culture collection is in progress; 18S rRNA of multiple species available.
-
▪ Genus Lepocinclis Perty, 1849. Rigid, unflattened cells with ring-shaped paramylon grains (plate F, 78); some (L. cyclidiopsis) secondarily non-photosynthetic, feeding by osmotrophy; 18S rRNA gene has GC base pair in the sixth position from the end of the Helix 12 and T in the second position of the Helix 23/27 spacer [381]; two ribosomal operon-containing inverted repeats present in the plastid genome [384].Type species: Lepocinclis globulus Perty, 1849. Type species and other species available in culture collections; 18S rRNA and full plastid genome sequences of multiple non-type species are available.
-
▪ Genus Phacus Dujardin, 1841. Rigid, laterally or triangularly compressed cells with ring-shaped paramylon grains (plate F, 79); some (P. ocellatus) secondarily non-photosynthetic, feeding by osmotrophy [381]; ribosomal operon present in one copy in the plastid genome [384].Type species: Euglena longicauda Ehrenberg, 1830 (= Phacus longicauda). Type species and other species available in culture collections; 18S rRNA and full plastid genome sequences of multiple non-type species are available.
-
▪
-
•
-
○ Order Eutreptiales Leedale, 1967 emend. Marin and Melkonian, 2003. Solitary, free-living cells with two or four flagella of equal or unequal length, capable of metaboly [3]. Predominantly marine. Introns are present in their plastid genomes, but not abundant (usually fewer than 28 and never more than 51); intron maturase mat1/ycf13 always present in the plastid genome, but mat2 and mat5 absent [384].
-
• Family Eutreptiaceae Hollande, 1942. With the same definition as the order.Note: valid name under ICZN is Eutreptiidae Hollande, 1942.
-
▪ Genus Eutreptia Perty, 1852. Two emergent flagella of almost equal length [395]; ribosomal operon present in one copy in the plastid genome [384].Type species: Eutreptia viridis Perty, 1852. Type species and other species available in culture collections; 18S rRNA and a full plastid genome sequence of the type species are available.
-
▪ Genus Eutreptiella da Cunha, 1914. Two emergent flagella of notably unequal length [395] or four flagella composed of longer and shorter pairs [396] (plate F, 86); may possess epi- or endobiotic bacteria [397]; mostly psychrotolerant or psychrophilic [342,396]; 18S rRNA gene possesses a CA insertion after the second position in the loop of Helix 18 [381]; ribosomal operon present in two copies with opposite orientation in the plastid genome, but one copy may be split [170,381]. Type species: Eutreptiella marina da Cunha, 1914. Type species and other species available in culture collections; 18S rRNA sequences of multiple non-type species, two full plastid genome sequences (Etl. gymnastica and Etl. pomquetensis; non-type species) and a transcriptomic dataset of Etl. gymnastica (non-type species) are available.
-
▪
-
•
-
○ Order Rapazida Cavalier-Smith, 2016. Solitary, free-living cells with two flagella of unequal length, feeding by phagotrophy on microalgae such as Tetraselmis; marine, capable of metaboly [355].
-
▪ Family Rapazidae Cavalier-Smith, 2016. With the same definition as the order.
-
▪ Genus Rapaza Yamaguchi, 2012. With the same definition as the family.Type species: Rapaza viridis Yamaguchi, Yubuki and Leander, 2012 (plate F, 87); 18S rRNA sequence of the type species available.
-
▪
-
▪
-
○ Euglenophyceae incertae sedis—genera with unresolved position due to lack of molecular data, and therefore questionable status, are:
-
▪ Genus Ascoglena Stein, 1878. Small, solitary cells with lorica, often sessile (attached to filamentous algae) [392].Type species: Ascoglena vaginicola Stein, 1878.
-
▪ Genus Euglenamorpha Wenrich, 1924. Elongated, metabolic cells of highly variable size with 3–6 flagella of equal length; inhabit the intestinal tracts of Rana spp. tadpoles [357].Type species: Euglenamorpha hegneri Wenrich, 1924.
-
▪ Genus Euglenopsis Klebs, 1892. Sessile cells with four long flagella and transverse cell division, forming colonies of branched filaments attached to the surface [398].Type species: Euglenopsis vorax Klebs, 1892.
-
▪ Genus Glenoclosterium Carter, 1869. Spindle-shaped cells with visible eyespot and at least four longitudinally elongated chloroplasts, but without a notable emergent flagellum [399].Type species: Glenoclosterium varians Carter, 1869.
-
▪ Genus Hegneria Brumpt and Lavier, 1924. Elongated, colourless cells with six flagella of equal length; inhabits the intestinal tract of tadpoles [400].Type species: Hegneria leptodactyli Brumpt and Lavier, 1924.
-
▪ Genus Klebsina Silva, 1961. Sessile, loricate cells, inhabiting marine habitats; originally described as Klebsiella [392], renamed by Silva due to conflicting name with a bacterial genus [401].Type species: Klebsiella alligata Pascher, 1931 (= Klebsina alligata).
-
▪ Genus Euglenocapsa Steinecke, 1932. Small, oval-shaped, faintly coloured cells with multiple disc-shaped, pyrenoid-lacking chloroplasts adjacent to the cell wall and a single emergent flagellum of length up to three times the length of the cell [402].Type species: Euglenocapsa ochracea Steinecke, 1932.Note: No representative of the incertae sedis genera is available in culture collections.
-
▪
-
▪ Clade Anisonemia Cavalier-Smith, 2016. The monophyletic group comprising predominantly flexible (metabolic), mostly biflagellate heterotrophs (phago- and osmotrophs), capable of skidding motility or gliding using posterior flagellum [378].
-
○ Order Anisonemida Cavalier-Smith, 2016. Feeding by phagotrophy, capable of gliding motility using posterior flagellum; paraphyletic with respect to Aphagea [8,378].
-
• Family Anisonemidae Kent, 1880. With the same definition as the order.
-
▪ Genus Anisonema Dujardin, 1841. Weakly metabolic cells with two unequal flagella (posterior one is longer), occurs in brackish waters (plate F, 90); during mitosis, basal body duplication and replication occurs in the late stages [403]; many species/morphotypes distinguished [377].Type species: Anisonema acinus Dujardin, 1841. Not available in culture collections; 18S rRNA sequence of the type species and single-cell transcriptomes of multiple species are available.
-
▪ Genus Dinema Perty, 1852. Usually strongly metabolic cells (with few weakly metabolic or rigid species, e.g. Dinema inaequale) with a thick pellicle and two unequal flagella [300] (plate F, 92); paraphyletic with respect to Anisonema [378].Type species: Dinema griseolum Perty, 1852. 18S rRNA and single-cell transcriptomic data of several species available.Note: valid name under ICN is Dinematomonas Silva, 1960, since the name Dinema Perty, 1852 is a synonym of Dinema Lindley, 1826 (Plantae: Magnoliophyta).
-
▪
-
○ Clade Aphagea Cavalier-Smith, 1993 emend. Busse and Preisfeld, 2002. Feeding by osmotrophy, without ingestion apparatus; monophyletic.
-
▪ Genus Astasia Dujardin, 1830. Cells without ingestion apparatus or stigma, with one emergent flagellum and visible metaboly; paraphyletic [170,404,405].Type species: Astasia limpida Dujardin, 1841. Multiple non-type species available in culture collections; 18S rRNA sequences of multiple non-type species are available.
-
▪ Genus Distigma Ehrenberg, 1831. Cells without ingestion apparatus, with two emergent flagella and intense metaboly; some (D. proteus) possess endobiotic bacteria; despite the name, no stigma present [170,379,404,405]; paraphyletic.Type species: Distigma proteus Ehrenberg, 1831. Type species and other species available in culture collections; 18S rRNA sequences of multiple species, including the type species, are available.
-
▪ Genus Gyropaigne Skuja, 1939. Rigid cells with prominent keels, fused pellicle and one emergent flagellum with hairs [404]; microtubule scroll present [404].Type species: Gyropaigne kosmos Skuja, 1939. G. lefevrei (non-type species) available in culture collection; 18S rRNA sequence of the same species is available.
-
▪ Genus Menoidium Perty, 1852. Rigid, flattened and elongated cells with fused pellicle and one emergent flagellum without hairs [407]; microtubule scroll present [404] (plate F, 82).Type species: Menoidium pellucidum Perty, 1852. Multiple non-type species available in culture collection; 18S rRNA sequences of multiple species, including the type species, are available.
-
▪ Genus Parmidium Christen, 1962. Rigid cells with deep indentations, fused pellicle and one emergent flagellum without hairs [407]; microtubule scroll present [404].Type species: Parmidium circulare Christen, 1962. Type species and other species available in culture collection; 18S rRNA sequences of multiple species, including the type species, are available.
-
▪ Genus Rhabdomonas Fresenius, 1858. Rigid cells with fused pellicle and one emergent flagellum; microtubule scroll present; a rather disputable, paraphyletic genus with no distinct synapomorphy, encompassing multiple species sharing different traits of other genera [404].Type species: Rhabdomonas incurva Fresenius, 1858. Type species and other species available in culture collections; 18S rRNA sequences of multiple species, including the type species, are available.
-
▪
-
○ Order Peranemida Cavalier-Smith, 1993. Uniflagellate or biflagellate cells, capable of gliding motility using anterior (or single) flagellumNote: this clade is resolved on some trees as polyphyletic, encompassing four clades (themselves monophyletic), scattered across the tree of Spirocuta [378]. Only a recent multigene phylogeny resolves them as a monophyletic sister clade to Euglenophyceae, but with very weak support [8]. Regardless of the phylogenetic uncertainties, these organisms have been informally referred to as ‘peranemids’ due to their common morphological traits.
-
▪ Genus Peranema Dujardin, 1841. Biflagellate, highly metabolic cells with longer and thicker anterior flagellum and protruding feeding apparatus, capable of cutting into other cells and sucking in its contents (plate F, 89); can and will attempt to feed on anything, including bacteria, yeast, microalgae, ink, raw starch and other euglenids [408]; very rapid response to light by rhodopsin-mediated phototaxis [409]; in recent phylogeny resolved as monophyletic [378].Type species: Peranema globulosum Dujardin, 1841. Non-type species (P. trichophorum) available in an educational resources repository (Carolina Biological Supply Co.), but not in culture collections; 18S rRNA sequence and single-cell transcriptomes of P. trichophorum (non-type species) are available.Note: valid name under ICN is Pseudoperanema Christen, 1962, since Peranema Dujardin, 1841 is synonym of Peranema D. Don 1825 (Plants: Polypodiopsida)
-
▪ Genus Chasmostoma Massart, 1920. Uniflagellate, metabolic cells with a pronounced flagellar cavity [378,410].Type species: Chasmostoma nieuportense Massart, 1920. Not available in culture collections; a single-cell transcriptome of the type species is available.
-
▪ Genus Jenningsia Schaeffer, 1918. Uniflagellate, metabolic cells [410] (plate F, 88); polyphyletic, currently comprising two separate monophyletic clades: one (Jenningsia fusiforme) branching at the base of Euglenophyceae, and another branching at the base of all non-peranemid Spirocuta [378]; in a recent multigene phylogeny, still resolved as polyphyletic, but both clades are placed within the monophyletic Peranemida [8].Type species: Jenningsia diatomophaga Schaeffer, 1918. Not available in culture collections; 18S rRNA sequences and single-cell transcriptomes of multiple non-type species strains are available.
-
▪ Genus Teloprocta Cavalier-Smith, 2016. Cylindrical or spindle-shaped cells with two long flagella (ventral and dorsal) and 28 pellicle strips; consists of species formerly classified as Heteronema, e.g. Teloprocta scaphurum [411].Type species: Heteronema scaphurum Skuja, 1934 (= Teloprocta scaphurum). Not available in culture collections; 18S rRNA sequence of the type species available.
-
▪ Genus Urceolus Mereschkovsky, 1877. Highly metabolic, sack-shaped cells with one emergent flagellum and a flattened anterior collar [300,378]; polyphyletic genus, with two strains (ABLN1 and WBF1) branching separately, with weak support, from the otherwise monophyletic sister clade to Teloprocta [378]; resolved as monophyletic in a recent multigene phylogeny; however, the two divergent strains mentioned above were not included in the analysis [8].Type species: Urceolus alenizinii Mereschkovsky, 1879. Not available in culture collections; 18S rRNA sequences and single-cell transcriptomes of multiple non-type species strains are available.
-
▪
-
▪ Genera unassigned to any subordinate taxa within Spirocuta:
-
▪ Genus Neometanema Lee and Simpson, 2014. Biflagellate, flattened cells with two equally long flagella, visible feeding apparatus, weak metaboly and 22 helical pellicle strips; distinguished from Heteronema/Anisonema by skidding motility involving the use of both flagella; a taxonomic replacement for Metanema [331,378].Type species: Neometanema parovale Lee and Simpson, 2014. Not available in culture collections; single-cell transcriptomes of multiple species, including the type species, are available.
-
▪ Genus Heteronema Dujardin, 1841. Biflagellate cells, capable of metaboly and gliding movement using the longer and thicker anterior flagellum, freshwater (plate F, 91).Type species: Heteronema marina Dujardin, 1841. Not available in culture collections; 18S rRNA sequences and single-cell transcriptomes of multiple non-type species strains are available.Note: Heteronema is a highly disputable genus, comprising species with varied morphological features [331,411]; recent phylogeny resolves this taxon as polyphyletic, with Heteronema globuliferum branching within Peranemida, and Heteronema vittatum (monophyletic) branching within Anisonemida, neither of which is the type species [378].
-
▪
-
•
-
○
-
• Clade Alistosa Lax et al., 2020. Oval-shaped, biflagellate cells, usually with 10–12 pellicle strips with keels; using the longer posterior flagellum for gliding motility; monophyletic [8,377].
-
▪ Genus Ploeotia Dujardin, 1841. Rigid, biflagellate cells (posterior one trailing against the substrate) with non-protrusible ingestion apparatus [300,412].Type species: Ploeotia vitrea Dujardin, 1841. Not available in culture; 18S rRNA sequences of multiple strains and two single-cell transcriptomes, both including the type species, are available.
-
▪ Genus Serpenomonas Triemer, 1986. Small, slightly flattened cells with two flagella (posterior one is longer) and a non-retractive feeding apparatus, inhabiting salt marshes [413].Type species: Serpenomonas costata Triemer, 1986. Type species available in culture; 18S rRNA sequences of multiple strains of the type species are available.
-
▪ Genus Keelungia Chan, 2013. Biflagellate, very small cells [414] with 10 flat pellicle strips [377].Type species: Keelungia pulex Chan and Moestrup, 2013. Not available in culture; 18S rRNA sequences of multiple species, including the type species, are available.
-
▪ Genus Lentomonas Farmer and Triemer, 1994. Rigid biflagellate cells with thicker and longer posterior flagellum, and straight, longitudinal pellicle strips [415], out of which seven dorsal strips are prominent, while three ventral strips are flat [377] (plate F, 96).Type species: Entosiphon applanatum Preisig, 1979 (= Lentomonas applanatum). Not available in culture; 18S rRNA sequences of multiple non-type species are available.
-
▪ Genus Decastava Cavalier-Smith, 2016. Long anterior flagellum and short posterior flagellum; 10 longitudinal pellicle strips [411].Type species: Decastava edaphica Cavalier-Smith and Vickerman, 2016. Type species available in culture; 18S rRNA sequences of multiple species, including type species, are available.
-
○ Order Petalomonadida Cavalier-Smith, 1993. Uniflagellate or biflagellate cells, using the longer, anterior flagellum (or the single flagellum) for gliding motility; recent phylogeny resolves Petalomonadida as monophyletic with strong support [378].
-
▪ Genus Petalomonas Stein, 1859. Rigid, flattened cells with one emergent gliding flagellum, mostly freshwater [300].Type species: Cyclidium abcissum Dujardin 1841 (= Petalomonas abcissa). P. cantuscygni (non-type species) available in culture collection; 18S rRNA sequences and single-cell transcriptomes of multiple non-type species are available.
-
▪ Genus Scytomonas Stein, 1878. Possesses five pellicle strips, a single flagellum and centriole, feeds when sessile [411].Type species: Scytomonas pusilla Stein, 1878. Sc. saepesendens (non-type species) available in culture collection; 18S rRNA sequence of the same species is available.
-
▪ Genus Notosolenus Stokes, 1884. Rigid, flattened cells with long anterior and short posterior flagellum [300] (plate F, 94).Type species: Solenotus apocamptus Stokes, 1884 (= Notosolenus apocamptus). Not available in culture collections; 18S rRNA sequences of multiple non-type species strains and a single-cell transcriptome of N. urceolatus (non-type species) are available.
-
▪ Genus Biundula Cavalier-Smith, 2016. Possesses a single emergent flagellum, consists of four species formerly classified as Petalomonas, e.g. Biundula sphagnophila, Biundula sulcata, distinguished by pellicle structure (2–8 smooth undulations on dorsal and ventral surface, continuous pellicle without sutures between strips) [411].Type species: Petalomonas sphagnophila Christen, 1962 (= Biundula sphagnophila). Not available in culture collections; 18S rRNA sequence of the type species available.
-
▪ Genus Sphenomonas Stein, 1878. Small, rigid, biflagellate cells with a large hyaline inclusion [378] (plate F, 95); no phagotrophy observed, probably osmotrophic [416].Type species: Sphenomonas quadrangularis Stein, 1878. Not available in culture collections; a single-cell transcriptome of the type species is available.
-
▪
-
▪
-
• Genera unassigned to any subordinate taxa within Euglenida:
-
▪ Genus Hemiolia Lax, Lee, Eglit and Simpson, 2019. Oblong, moderately flattened cells with very long (over three times cell length) posterior flagellum, hardly notable pellicle strips and feeding apparatus not visible in light microscopy; consists of only one species (H. trepidum), formerly classified as Anisonema [377].Type species: Anisonema trepidum J. Larsen, 1987 (= Hemiolia trepidum). Not available in culture; 18S rRNA sequences of multiple strains of the type species are available.
-
▪ Genus Liburna Lax, Lee, Eglit and Simpson, 2019. Rigid, oblong cells with very long (about three times cell length), hooked posterior flagellum, hardly noticeable pellicle strips and feeding apparatus not visible in light microscopy; consists of only one species (L. glaciale), formerly classified as Anisonema [377].Type species: Anisonema glaciale Larsen and Patterson, 1990 (= Liburna glaciale). Not available in culture; 18S rRNA sequences of multiple strains of the type species and two single-cell transcriptomic datasets are available.
-
▪ Genus Entosiphon Stein, 1878. Cells with protrusible ingestion apparatus and 12 pellicle strips [377].Type species: Anisonema sulcatum Dujardin, 1841 (= Entosiphon sulcatum). Two species, including the type species, available in culture; 18S rRNA sequences of multiple species, including the type species, are avaiable.Note: the position of this genus is neither strongly supported nor stable, as depending on methods and datasets used for phylogeny, it may either branch off together with Hemiolia and Liburna, or form a separate branch in various positions among ‘rigid’ euglenids (i.e. Euglenida excluding Olkaspira). Therefore, despite the abundance of molecular data, Entosiphon cannot be classified as a member of any major group within Euglenida, and should be regarded as orphan genus among phagotrophic euglenids [8,377,378].
-
▪
-
• Euglenida incertae sedis—15 genera with unresolved position due to lack of molecular data, and therefore questionable status, are:
-
▪ Genus Atraktomonas Christen, 1962. Possesses a single emergent flagellum, closely related to Petalomonas [331,417].Type species: Atraktomonas laevis Christen, 1962.
-
▪ Genus Calycimonas Christen, 1959. Non-metabolic cells, possesses a single emergent flagellum [417,418].Type species: Calycimonas physaloides Christen, 1959.
-
▪ Genus Dolium Larsen and Patterson, 1990. Rigid, sessile cells with one emergent flagellum [300]; generic name shared with an animal genus—Dolium Lamarck, 1801 (Mollusca: Gastropoda).Type species: Dolium sedentarium Larsen and Patterson, 1990.
-
▪ Genus Dylakosoma Skuja, 1964. Distinguished from Petalomonas due to the presence of epibiotic bacteria [419].Type species: Dylakosoma pelophilum Skuja, 1964.
-
▪ Genus Peranemopsis Lackey, 1940. Uniflagellate, metabolic, wedge-shaped cells, with only one rod in the feeding apparatus and no eyespot [183].Type species: Peranemopsis striata Lackey, 1940.
-
▪ Genus Tropidoscyphus Stein, 1878. Slightly plastic cells with eight strips and two unequal flagella, but both are described as anterior [420].Type species: Tropidoscyphus octocostatus Stein, 1878.
-
▪ Genus Michajlowastasia Krell and Shabalin, 2008. Organisms with two-stage life cycle: a free-living reproductive phase, and a parasitic feeding phase, taking place in the intestines or other body cavities of copepods and ending with formation of cyst-like structures. In free-living stage, cells are indistinguishable from genus Astasia; in parasitic stage, cells lose the emergent flagellum, become larger in size and enriched with paramylon grains [356].Type species: Astasia cyclopis Michajłow, 1956 (= Michajlowastasia cyclopis).Note: this genus had been originally described by Michajłow under the name Parastasia in order to distinguish the assemblage of parasitic forms from the exclusively free-living Astasia spp. That name, however, was recognized as invalid due to its homonymity with an earlier described beetle genus Parastasia Westwood, 1841 (Coleoptera: Scarabaeidae), and subsequently renamed by Krell & Shabalin [356,421].
-
▪ Genus Parastasiella Michajłow, 1965. Organisms with two-stage life cycle, similar to Michajlowastasia, but the parasitic stage involves copepod eggs and larvae (nauplii) as hosts instead of adults. Cells are among the smallest of all known euglenids, reaching a maximum length of 5 μm, and form heterogenous paramylon grains of different size [356].Type species: Astasiella velox Michajłow, 1965 (= Parastasiella velox).
-
▪ Genus Dinemula Michajłow, 1965. Organisms with two-stage life cycle (see Parastasiella). Spindle-shape cells with two unequal flagella; the anterior flagellum is the longer one and is formed earlier (during parasitic stage), while the posterior one is formed during the free-living stage [356].Type species: Dinemula celer Michajłow, 1965.
-
▪ Genus Paradinemula Monchenko, 1967. Organisms with two-stage life cycle (see Parastasiella). Morphologically similar to Dinemula, but more oval-shaped, with longer anterior flagellum, a stiff, laterally protruding flagellum turned to the back, and a large translucent nucleus [356].Type species: Paradinemula polonica Monchenko, 1967.
-
▪ Genus Mononema Michajłow, 1967. Organisms with two-stage life cycle (see Parastasiella). Similar to Paradinemula, but with a single emergent flagellum, protruding from a swelling in the anterior part of the cell and directed towards the back of the cell [356].Type species: Mononema reptans Michajłow, 1967.
-
▪ Genus Ovicola Michajłow, 1965. Organisms with two-stage life cycle, similar to Parastasiella, but reproduction occurs in parasitic stage within copepod eggs, with the free-living stage's role limited only to invasion of new hosts. Egg-shaped, uniflagellate cells with a thick, arched flagellum which makes rowing movements only with its distal part. In free-living stage, each cell contains only one large paramylon grain [356].Type species: Ovicola abyssinicus Michajłow, 1965.
-
▪ Genus Naupliicola Michajłow, 1965. Organisms with two-stage life cycle, similar to Ovicola, but reproduction occurs in body cavities of copepod nauplii instead of eggs. Morphologically similar to Ovicola, but with multiple paramylon grains in free-living stage [356].Type species: Naupliicola necans Michajłow, 1965.
-
▪ Genus Embryocola Michajłow, 1969. Organisms with two-stage life cycle, similar to Naupliicola. Morphologically similar to Naupliicola, but develops specifically inside the eyes of copepod nauplii in the parasitic stage of its life cycle [356].Type species: Embryocola ocelli Michajłow, 1969.
-
▪ Genus Copromonas Dobell, 1908. Rigid, pyriform, colourless cells with one long emergent flagellum and clearly visible cytopharynx, feeding by phagotrophy; isolated from intestines of frogs (Rana temporaria) and toads (Bufo vulgaris); described as resembling Petalomonas and Scytomonas, but also observed to conjugate, which makes its affiliation to Euglenida disputable [422].Type species: Copromonas subtilis Dobell, 1908.Note: no representative of the incertae sedis genera is available in culture collections.
-
▪
Class Symbiontida Yubuki, Edgcomb, Bernhard and Leander, 2009.
This group has two shared synapomorphies: a thick mantle of rod-shaped epibiotic bacteria covering almost the entire cell, and a layer of mitochondria-derived organelles with reduced or absent cristae located beneath the cell membrane. Despite superficially similar morphology (with the exception of the pellicle strips), their relationship with euglenids has not been fully resolved. Most commonly found in hypoxic zones of marine habitats.
-
▪
Genus Bihospites Breglia, Yubuki, Hoppenrath and Leander, 2010. Cells with a rudimentary pellicle, robust feeding rod and two morphotypes of epibiotic bacteria (large, rod-shaped ones arranged in longitudinal bands and small, spherical ones with extrusive apparatuses) [329,423].
Type species: Bihospites bacati Breglia, Yubuki, Hoppenrath and Leander, 2016. Not available in culture; 18S rRNA sequence of the type species available.
-
▪
Genus Calkinsia Lackey, 1960. Cells with reduced feeding rod and without pellicle, but with elaborate extracellular matrix, orange in colour; only rod-shaped epibiotic bacteria present [423,424].
Type species: Calkinsia aureus Lackey, 1960 (plate F, 97). Not available in culture; 18S rRNA sequence of the type species available.
-
▪
Genus Postgaardi Fenschel, Bernard, Esteban, Findlay, Hansen and Iversen, 1995. Cells with complex feeding apparatus with an oval-shaped gutter, covered by the anterior lip overlapping a reinforced ridge, but with less developed extracellular matrix [423,425]; phylogenetic position unresolved due to lack of molecular data.
Type species: Postgaardi mariagerensis Fenschel, Bernard, Esteban, Findlay, Hansen and Iversen, 1995. Not available in culture; no sequence data available.
5. Viruses in Euglenozoa
Viruses are the most abundant and widespread life form on our planet. During several billion years of coevolution, viruses have developed specific mechanisms allowing them to infect virtually any cellular organism [426]. It was estimated that viruses lyse about 20% of oceanic protists daily and, thus, play a major role in regulating the Earth's biogeochemical cycle [427]. Euglenozoa are no exception to this rule, although viral diversity was thoroughly investigated only in kinetoplastids [428]. The reason for such discrepancy is obvious—this is by far the best-studied group, which includes several parasites of medical or economic importance [10]. There is no doubt that representatives of Euglenida, Diplonemea and Symbiontida can be infected by viruses, but this has not been verified experimentally.
Kinetoplastids possess DNA and RNA viruses. The only documented case of a DNA virus is the one infecting free-living Bodo saltans [429]. This Bodo saltans virus belongs to the family Mimiviridae and its genome of about 1.39 Mb is among the largest described genomes of giant viruses. The functional role this virus may play in the biology of bodonids remains to be elucidated, but the plethora of acquired adaptation traits (such as the mechanism to facilitate membrane fusion, interference competition, contracted translation machinery and inflated genome with numerous genome rearrangements) makes this virus an interesting model for future studies. Also of note is that the abundance of such nucleo-cytoplasmic large DNA viruses was estimated at 104–105 genomes ml−1 in the photic zone and 102–103 genomes ml−1 of water in the oxygen minimum zone of the World Ocean [430].
The situation with RNA viruses is more complex. Here, we will only discuss viruses with known genetic structure and will not cover older reports of the mere presence of virus-like particles (reviewed by Grybchuk et al. [428]). The best-studied cases are of Leishmania RNA viruses (Leishmaniavirus spp., LRVs of the family Totiviridae). Discovered in the late 1980s in representatives of the Leishmania subgenus Viannia [431], the very first Leishmaniavirus LRV1 was sequenced in the early 1990s [432], and its biological role was uncovered about 20 years later [433]. Its presence is linked to the increased metastatic potential, parasite burden, immune response in mouse models of leishmaniasis and frequent treatment failures [434,435]. Notably, Old World leishmanias L. (Leishmania) possess a phylogenetically related Leishmaniavirus, LRV2, which is widespread in isolates of L. major [436,437], but its role in the disease progression is unknown. The phylogenies of viruses and their respective hosts are mainly congruent, suggesting long-term coevolution [438]. In addition to Leishmania, representatives of Leishmaniavirus LRV3 and LRV4 have been documented in another group of trypanosomatids, Blechomonas spp. [439]. These viruses have probably been acquired from Leishmania during co-infections.
The most successful group of viruses infecting trypanosomatids are bunyaviruses (LBVs, proposed family Leishbunyaviridae). They infect multiple Crithidia and Leptomonas spp. [440,441], Leishmania (Mundinia) martiniquensis [442], and at least one isolate of Phytomonas sp. [441]. The wide distribution of these viruses can be explained by their encapsulated structure, which promotes easy dispersion in co-infections.
Narnaviruses (family Narnaviridae) were detected in Leptomonas seymouri and Phytomonas serpens [95,441,443]. The viral load in L. seymouri is extremely high, indicating that this virus may enhance Leishmania virulence in the case of Leishmania donovani–Leptomonas seymouri co-infections [444]. Other viruses are less widespread and, in many instances, appear to be restricted to a particular trypanosomatid host, as can be exemplified by Tombus-like viruses and Ostravirus in Leptomonas pyrrhocoris [441].
6. Endo- and ectosymbioses in Euglenozoa
6.1. Kinetoplastid endosymbionts
Reports on endosymbionts in kinetoplastids are rare, but a few cases have been described in detail. Endosymbiosis between a bacterium and a trypanosomatid host occurred independently at least twice in the evolutionary history of trypanosomatids, in which neither hosts nor bacteria are closely related. The members of subfamily Strigomonadinae engaged in endosymbiotic relationship with Ca. Kinetoplastibacterium spp., representatives of Alcaligenaceae family (Burkholderiales; β-proteobacteria). Three genera of Strigomonadinae—Angomonas, Strigomonas and Kentomonas—are considered to share an endosymbiont-bearing ancestor, in which the reductive evolution of the endosymbiont genome occurred prior to the radiation of the host genera [274,445]. Each member of Strigomonadinae carries a different Kinetoplastibacterium species, which co-evolved together with its host [285]. Another trypanosomatid, Novymonas esmeraldas (most closely related to Leishmania), established endosymbiosis with Ca. Pandoraea novymonadis, representing another family (Burkholderiaceae) of Burkholderiales [275].
The presence of endosymbionts in all these cases likely compensates for the inability of their hosts to synthesize certain metabolites, such as haem, nucleotides, and several amino acids and vitamins, which are provided by the bacteria [275,285,445]. Both P. novymonadis and Kinetoplastibacterium spp. feature genomes, which are strongly reduced compared to related free-living β-proteobacteria, nevertheless preserving genes necessary for nutritional provisioning of their hosts [195]. Both endosymbiotic associations are permanent with bacteria being transmitted vertically; however, the association between Kinetoplastibacterium and Strigomonadinae is considered more ancient than that of N. esmeraldas and P. novymonadis [89,195]. The latter partnership is characterized by the lack of stringent control over the number of bacteria, less extensive genome reduction, higher GC content, and the presence of TCA and amino acid synthesis pathways [89].
Outside of these two systems, the presence of endosymbiotic bacteria was reported from a free-living freshwater kinetoplastid Bodo curvifilus [446], while the recently studied endosymbionts of Bodo saltans have been assigned to Paracaedibacter, with possible role in defensive endosymbiosis [447]. Finally, the trypanosomatid Phytomonas borealis isolated from the midgut of spiked shieldbugs also harbours endosymbionts [448], although their taxonomic identity and function remain unknown.
6.2. Diplonemid endosymbionts
Diplonemids are known for establishing symbiosis with members of Holosporaceae and Rickettsiaceae families, which are exclusively parasitic/endosymbiotic lineages of α-proteobacteria [449]. At present, there are only two reports on endosymbionts in diplonemids: Holosporaceae bacteria inside two Diplonema species [450], and a hemistasiid Namystinia karyoxenos with a Rickettsiaceae endosymbiont [325]. While D. aggregatum and N. karyoxenos contain a single endosymbiont, D. japonicum harbours two species of bacteria from closely related genera, the genomes of which have been sequenced, assembled and analysed [451]. They are severely reduced with similar gene content retained, and lack all energy metabolism pathways, including glycolysis, pentose-P pathway, the TCA cycle and oxidative phosphorylation. Although complete synthesis pathways for amino acids or vitamins are absent, the nutritional role of the endosymbionts cannot be ruled out due to the large number of proteins without known functions. A large portion of their highly reduced genomes is dedicated to secreted proteins that are possibly involved in manipulation of the host metabolism. Similar to Holosporaceae and Rickettsiaceae in other protist hosts, the role of the diplonemid endosymbionts is not clear. However, it was hypothesized that due to the presence of various secretion/toxin systems, the endosymbionts might take part in Defence against bacterial pathogens [451].
6.3. Symbiontid symbionts
As suggested by their very name, the distinctive trait of the Symbiontida is their capability of forming permanent, probably obligatory symbiotic relationships [7,41]. Thus far, three distinctive kinds of epibiotic bacteria have been described to thrive on the surface of symbiontid cells: rod-shaped, sulfide-oxidizing ε-proteobacteria associated with symbiontid genera Calkinsia and Bihospites [41], extrusive apparatus-bearing cocci with strong resemblance to hypotrich ciliate-associated Verrucomicrobia, endemic to the genus Bihospites [329], and magnetotactic Deltaproteobacteria, associated with multiple unclassified environmental strains of symbiontids [452].
Although the role of magnetosome-bearing, but non-motile δ-proteobacterial symbionts is clearly to provide their hosts with magnetotaxis [452], the functions of other microorganisms in their relationships with symbiontids seem to be more complex. It has been suggested that the ε-proteobacterial symbionts detoxify the local surroundings to limit the inhibitory effect of sulfide on the cellular respiration of the symbiontids [41], while the Verrucomicrobia-like bacteria provide their hosts with a Defence mechanism against predators [329]. Additionally, the epibiotic bacteria can be used by their hosts as an auxiliary food source [329,452]. In exchange, the eukaryotic hosts' role is to provide their epibionts with various metabolites, such as hydrogen, as all symbiontids possess hydrogenosomes [7], and to serve as efficient means of transport along the oxycline in the deep-sea environment [41]. It remains uncertain, however, whether the metabolic coupling between the symbiontids and ε-proteobacteria is limited to the outward hydrogen flux, and, perhaps more importantly, if any metabolic exchange occurs between the Verrucomicrobia-like bacteria and their symbiontid hosts.
6.4. Euglenid symbionts
Curiously, the observable affinity of the symbiontids towards prokaryotic partners is, to some extent, shared by their postulated close relatives—the euglenids. In their case, the tight relationships with bacteria are not as widespread, as a majority of the described euglenid genera have never been observed to harbour any symbionts, but the diversity of these relationships may be substantially greater [397]. Unfortunately, our knowledge of the euglenid–bacteria associations remains superficial due to the fact that they have mostly been reported in the pre-genomics and transcriptomics era [419,453,454]. Nonetheless, it is undeniable that these relationships are widespread, as they involve both phagotrophic (genera Petalomonas and Dylakosoma; [419,455]) and photosynthetic euglenids (genera Euglena, Phacus, Lepocinclis and others; [454]), and of rather diverse nature, as euglenids have been observe to harbour bacteria within their cells [389,454], on their surface [419,456], or even both [397]. What is more, a single heterotrophic euglenid species (Anisonema platysomum) has been observed to harbour magnetosomes. It remains unclear whether this typically prokaryotic trait has been acquired by Anisonema from magnetotactic bacteria (similar to those associated with symbiontids), or evolved independently [457].
As indicated by the so far most elaborate studies of euglenid–bacteria associations, involving a phagotrophic euglenid Petalomonas sphagnophila and a strain of photosynthetic Eutreptiella sp., it is evident that euglenids are capable of harbouring multiple distinct bacterial symbionts simultaneously [397,455]. Petalomonas sphagnophila from Canadian peatlands has been observed to carry six different bacteria within its cells, namely two strains of Rickettsiales, one representative of Firmicutes, one γ-proteobacterium, one δ-proteobacterium and one enigmatic, pigmented prokaryote with unidentified affiliation [455]. Moreover, Eutreptiella from the Long Island Sound possesses epibionts classified as Roseovarius, Oceanicaulis (Alphaproteobacteria) and Marinobacter (Gammaproteobacteria), as well as an endobiotic representative of Rickettsiales, though phylogenetically distant from those associated with P. sphagnophila [397]. Unfortunately, except for the hypothesis that the epibiotic bacteria of Eutreptiella supply their host with vitamin B12, well supported by cultivation experiments and transcriptomic data, very little is known about the nature and purpose of the relationships between bacteria and the two aforementioned euglenid hosts [397,455]. In fact, it is uncertain whether these associations are symbiotic at all, especially considering that Rickettsiales are common intracellular parasites of a vast variety of eukaryotes [458].
Note added in proof
After the acceptance of this paper for publication, a taxonomic description of two new kinetoplastid flagellates, marine Papus ankaliazontas and freshwater Apiculatamorpha spiralis, has been published [459]. These new taxa represent free-living representatives of the order Prokinetoplastida. The available 18S rRNA gene-based phylogeny does not allow estimating reliably the relationships of these flagellates with the previously characterized genera Perkinsela and Ichthyobodo due to low statistical supports [459]. Moreover, the inferred position of these new taxa contradicts with the well-supported topology of the recently published phylogenomic tree, which, however, does not include Ichthyobodo [30]. Thus, it is currently premature to classify the two new genera and they are considered as Prokinetoplastida incertae sedis. Below is a short characterization of these forms.
P. ankaliazontas: free-living, solitary, eukaryvorous; two unattached flagella, the anterior one with thin undulate mastigonemes; flagellar pocket connected to oblique groove; pronounced rostrum with apical cytostome, tubular cytopharynx supported by prismatic microtubular rod; trichocysts at the anterior end.
A. spiralis: free-living, solitary, eukaryvorous; two unattached flagella the anterior one with fine fiber layer; cell surface with spherical and lamellate scales; pronounced rostrum with apical cytostome, tubular cytopharynx supported by prismatic microtubular rod; trichocysts at the anterior end.
Acknowledgements
Some photos (as indicated in the plates) were kindly provided by the following colleagues: Euichi Hirose (University of Ryukyus, Okinawa, Japan), Thorsten Stoeck (University of Kaiserslautern, Kaiserslautern, Germany), Alastair Simpson and Yana Eglit (Dalhousie University, Halifax, Canada), Shinichi Kitamura (Ehime University, Matsuyama, Japan), Kristina Prokina and Denis Tikhonenkov (Papanin Insitute for Biology of Inland Waters, Borok, Russia), Ivan Fiala (Institute of Parasitology, České Budejovice, Czech Republic), Iva Dyková (Masaryk University, Brno, Czech Republic), Gordon Lax, Noriko Okamoto and Patrick Keeling (University of British Columbia, Vancouver, Canada), Gediminas Valkiūnas (Nature Research Centre, Vilnius, Lithuania), Andrei Mihalca (University of Agricultural Sciences and Veterinary Medicine, Cluj-Napoca, Romania), Erney Plessmann Camargo & Marta Teixeira (University of Sao Paolo, Sao Paolo, Brazil), Sarah Keatley and Andrew Thompson (Murdoch University, Perth, Australia), Bożena Zakryś (University of Warsaw, Warsaw, Poland), Naoji Yubuki (Paris-Sud University, Orsay, France), Anna I. Ganyukova, Marina N. Malysheva and Alexander O. Frolov (Zoological Institute, St Petersburg, Russia) and Milena Svobodová, Jovana Sádlová, Tereza Leštinová and Klára Poloprudská (Charles University, Prague, Czech Republic). We are also grateful to Andreu Saura (University of Ostrava, Ostrava, Czech Republic) for the preparation of smears of some trypanosomatid species as well as Alexander O. Frolov, Petr Kment (National Museum, Prague, Czechia) and Alastair Simpson (Dalhousie University, Halifax, Canada) for the discussion of some issues, which arose during the work on the manuscript.
Data accessibility
This article has no additional data.
Authors' contributions
J.L. wrote the abstract and Introduction, conceived the idea for this manuscript and supervised the work. A.Y.K. prepared plate D and trees B–D, and wrote the section on Kinetoplastea except the section on taxonomy of Trypanosoma, which was written by J.V. J.V. also prepared plates B and C. V.Y. wrote the section on Viruses in Euglenozoa. D.T. and J.L. wrote the section on Diplonemea and subsections on diplonemid and kinetoplastid endosymbionts, and prepared plates A and E and tree E. A.K. and K.M. wrote the section on Euglenida and Symbiontida, and subsections on euglenid symbionts and symbiontid symbionts, and prepared plate F and trees A and F. All authors reviewed, corrected and accepted the manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by the Czech Science Foundation grants 20-07186S, 20-22689S and 18-15962S (to V.Y. and J.L.), ERC CZ grant LL1601 (to J.L.), ZIN RAS State Assignment АААА-А19-119031200042-9 (to AYK), the ERD funds of the Czech Ministry of Education (16_019/0000759) (to J.L., D.T., A.Y.K., J.V. and V.Y.), the Czech Bioimaging grant LM2018129 (to D.T.), National Science Centre, Poland Sonata grant 2016/21/D/NZ8/01288 (to A.K.), Preludium grant 2018/31/N/NZ8/01840 (to K.M.), EMBO Installation Grant (to A.K.) and L'Oreal Poland—UNESCO Scholarship for Women in Science (to A.K.).
References
- 1.Lukeš J, Leander BS, Keeling PJ. 2009. Cascades of convergent evolution: the corresponding evolutionary histories of euglenozoans and dinoflagellates. Proc. Natl Acad. Sci. USA 106, 9963-9970. ( 10.1073/pnas.0901004106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leander BS, Lax G, Karnkowska A, Simpson AGB. 2017. Euglenida. In Handbook of the protists (ed. Archibald JM), pp. 1-42. Cham, Switzerland: Springer International Publishing. [Google Scholar]
- 3.Adl SM, et al. 2019. Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 66, 4-119. ( 10.1111/jeu.12691) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldstein B, King N. 2016. The future of cell biology: emerging model organisms. Trends Cell Biol. 26, 818-824. ( 10.1016/j.tcb.2016.08.005.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pawlowski J, et al. 2012. CBOL Protist Working Group: barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms. PLoS Biol. 10, e1001419. ( 10.1371/journal.pbio.1001419) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butenko A, et al. 2020. Evolution of metabolic capabilities and molecular features of diplonemids, kinetoplastids, and euglenids. BMC Biol. 18, 23. ( 10.1186/s12915-020-0754-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yubuki N, Leander BS. 2018. Diversity and evolutionary history of the Symbiontida (Euglenozoa). Front. Ecol. Evol. 6, 100. ( 10.3389/fevo.2018.00100) [DOI] [Google Scholar]
- 8.Lax G, et al. 2021. Multigene phylogenetics of euglenids based on single-cell transcriptomics of diverse phagotrophs. Mol. Phylogenet. Evol. 159, 107088. ( 10.1016/j.ympev.2021.107088) [DOI] [PubMed] [Google Scholar]
- 9.Gibson W. 2017. Kinetoplastea. In Handbook of the protists (eds Archibald JM, Simpson AGB, Slamovits CH), pp. 1089-1138. Cham, Switzerland: Springer International Publishing. [Google Scholar]
- 10.Maslov DA, Opperdoes FR, Kostygov AY, Hashimi H, Lukeš J, Yurchenko V. 2019. Recent advances in trypanosomatid research: genome organization, expression, metabolism, taxonomy and evolution. Parasitology 146, 1-27. ( 10.1017/S0031182018000951) [DOI] [PubMed] [Google Scholar]
- 11.Gawryluk RMR, del Campo J, Okamoto N, Strassert JFH, Lukeš J, Richards TA, Worden AZ, Santoro AE, Keeling PJ. 2016. Morphological identification and single-cell genomics of marine diplonemids. Curr. Biol. 26, 3053-3059. ( 10.1016/j.cub.2016.09.013) [DOI] [PubMed] [Google Scholar]
- 12.Flegontova O, Flegontov P, Malviya S, Audic S, Wincker P, de Vargas C, Bowler C, Lukeš J, Horák A. 2016. Extreme diversity of diplonemid eukaryotes in the ocean. Curr. Biol. 26, 3060-3065. ( 10.1016/j.cub.2016.09.031) [DOI] [PubMed] [Google Scholar]
- 13.Ebenezer TE, et al. 2019. Transcriptome, proteome and draft genome of Euglena gracilis. BMC Biol. 17, 11. ( 10.1186/s12915-019-0626-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clayton CE. 2016. Gene expression in Kinetoplastids. Curr. Opin. Microbiol. 32, 46-51. ( 10.1016/j.mib.2016.04.018) [DOI] [PubMed] [Google Scholar]
- 15.Campbell DA, Thomas S, Sturm NR. 2003. Transcription in kinetoplastid protozoa: why be normal? Microb. Infect. 5, 1231-1240. ( 10.1016/j.micinf.2003.09.005) [DOI] [PubMed] [Google Scholar]
- 16.Portman N, Gull K. 2010. The paraflagellar rod of kinetoplastid parasites: from structure to components and function. Int. J. Parasitol. 40, 135-148. ( 10.1016/j.ijpara.2009.10.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sunter J, Gull K. 2017. Shape, form, function and Leishmania pathogenicity: from textbook descriptions to biological understanding. Open Biol. 7, 170165. ( 10.1098/rsob.170165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheeler RJ. 2017. Use of chiral cell shape to ensure highly directional swimming in trypanosomes. PLoS Comput. Biol. 13, e1005353. ( 10.1371/journal.pcbi.1005353) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horáková E, Changmai P, Vancová M, Sobotka R, van den Abbeele J, Vanhollebeke B, Lukeš J. 2017. The Trypanosoma brucei TbHrg protein is a heme transporter involved in the regulation of stage-specific morphological transitions. J. Biol. Chem. 292, 6998-7010. ( 10.1074/jbc.M116.762997) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang XH, Haritan A, Uliel S, Michaeli S. 2003. trans and cis splicing in trypanosomatids: mechanism, factors, and regulation. Eukaryot. Cell 2, 830-840. ( 10.1128/EC.2.5.830-840.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Milanowski R, Gumińska N, Karnkowska A, Ishikawa T, Zakryś B. 2016. Intermediate introns in nuclear genes of euglenids—are they a distinct type? BMC Evol. Biol. 16, 49. ( 10.1186/s12862-016-0620-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.del Campo J, Sieracki ME, Molestina R, Keeling P, Massana R, Ruiz-Trillo I. 2014. The others: our biased perspective of eukaryotic genomes. Trends Ecol. Evol. 29, 252-259. ( 10.1016/j.tree.2014.03.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen RE, Englund PT. 2012. Network news: the replication of kinetoplast DNA. Annu. Rev. Microbiol. 66, 473-491. ( 10.1146/annurev-micro-092611-150057) [DOI] [PubMed] [Google Scholar]
- 24.Li SJ, Zhang X, Lukeš J, Li BQ, Wang JF, Qu LH, Hide G, Lai DH, Lun ZR. 2020. Novel organization of mitochondrial minicircles and guide RNAs in the zoonotic pathogen Trypanosoma lewisi. Nucleic Acids Res. 48, 9747-9761. ( 10.1093/nar/gkaa700) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burger G, Valach M. 2018. Perfection of eccentricity: mitochondrial genomes of diplonemids. IUBMB Life 70, 1197-1206. ( 10.1002/iub.1927) [DOI] [PubMed] [Google Scholar]
- 26.Lukeš J, Wheeler R, Jirsová D, David V, Archibald JM. 2018. Massive mitochondrial DNA content in diplonemid and kinetoplastid protists. IUBMB Life 70, 1267-1274. ( 10.1002/iub.1894) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobáková E, Flegontov P, Skalický T, Lukeš J. 2015. Unexpectedly streamlined mitochondrial genome of the euglenozoan Euglena gracilis. Genome Biol. Evol. 7, 3358-3367. ( 10.1093/gbe/evv229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novák Vanclová AMG, et al. 2020. Metabolic quirks and the colourful history of the Euglena gracilis secondary plastid. New Phytol. 225, 1578-1592. ( 10.1111/nph.16237) [DOI] [PubMed] [Google Scholar]
- 29.Jackson AP, et al. 2016. Kinetoplastid phylogenomics reveals the evolutionary innovations associated with the origins of parasitism. Curr. Biol. 26, 161-172. ( 10.1016/j.cub.2015.11.055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butenko A, Hammond M, Field MC, Ginger ML, Yurchenko V, Lukeš J. 2021. Reductionist pathways for parasitism in euglenozoans? Expanded datasets provide new insights. Trends Parasitol. 37, 100-116. ( 10.1016/j.pt.2020.10.001) [DOI] [PubMed] [Google Scholar]
- 31.Matthews KR. 2015. 25 years of African trypanosome research: from description to molecular dissection and new drug discovery. Mol. Biochem. Parasitol. 200, 30-40. ( 10.1016/j.molbiopara.2015.01.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaur B, Valach M, Peña-Diaz P, Moreira S, Keeling PJ, Burger G, Lukeš J, Faktorová D. 2018. Transformation of Diplonema papillatum, the type species of the highly diverse and abundant marine microeukaryotes Diplonemida (Euglenozoa). Environ. Microbiol. 20, 1030-1040. ( 10.1111/1462-2920.14041) [DOI] [PubMed] [Google Scholar]
- 33.Nomura T, Inoue K, Uehara-Yamaguchi Y, Yamada K, Iwata O, Suzuki K, Mochida K. 2019. Highly efficient transgene-free targeted mutagenesis and single-stranded oligodeoxynucleotide-mediated precise knock-in in the industrial microalga Euglena gracilis using Cas9 ribonucleoproteins. Plant Biotechnol. J. 17, 2032-2034. ( 10.1111/pbi.13174) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faktorová D, et al. 2020. Genetic tool development in marine protists: emerging model organisms for experimental cell biology. Nat. Methods 17, 481-494. ( 10.1038/s41592-020-0796-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faktorová D, Kaur B, Valach M, Graf L, Benz C, Burger G, Lukeš J. 2020. Targeted integration by homologous recombination enables in situ tagging and replacement of genes in the marine microeukaryote Diplonema papillatum. Environ. Microbiol. 22, 3660-3670. ( 10.1111/1462-2920.15130) [DOI] [PubMed] [Google Scholar]
- 36.Gomaa F, Garcia PA, Delaney J, Girguis PR, Buie CR, Edgcomb VP. 2017. Toward establishing model organisms for marine protists: successful transfection protocols for Parabodo caudatus (Kinetoplastida: Excavata). Environ. Microbiol. 19, 3487-3499. ( 10.1111/1462-2920.13830) [DOI] [PubMed] [Google Scholar]
- 37.Vickerman K. 1991. Organization of the bodonid flagellates. In The biology of free-living heterotrophic flagellates. The systematics association special volume (eds Patterson DJ, Larsen J), pp. 159-176. Oxford, UK: Clarendon Press. [Google Scholar]
- 38.Patterson DJ, Simpson AGB. 1996. Heterotrophic flagellates from coastal marine and hypersaline sediments in Western Australia. Eur. J. Protistol.y 32, 423-448. ( 10.1016/S0932-4739(96)80003-4) [DOI] [Google Scholar]
- 39.Arndt H, Dietrich D, Auer B, Cleven E-J, Gräfenhan T, Weitere M, Mylnikov AP. 2000. Functional diversity of heterotrophic flagellates in aquatic ecosystems. In The flagellates (eds Leadbeater BSC, Green JC), pp. 240-268. London, UK: Taylor & Francis Ltd. [Google Scholar]
- 40.Ekelund F. 2002. Tolerance of soil flagellates to increased NaCl levels. J. Eukaryot. Microbiol. 49, 324-328. ( 10.1111/j.1550-7408.2002.tb00378.x) [DOI] [PubMed] [Google Scholar]
- 41.Edgcomb VP, Breglia SA, Yubuki N, Beaudoin D, Patterson DJ, Leander BS, Bernhard JM. 2011. Identity of epibiotic bacteria on symbiontid euglenozoans in O2-depleted marine sediments: evidence for symbiont and host co-evolution. ISME J. 5, 231-243. ( 10.1038/ismej.2010.121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boenigk J, Arndt H. 2002. Bacterivory by heterotrophic flagellates: community structure and feeding strategies. Antonie van Leeuwenhoek, Int. J. Gen. Mol. Microbiol. 81, 465-480. ( 10.1023/A:1020509305868) [DOI] [PubMed] [Google Scholar]
- 43.Patterson DJ, Nygaard K, Steinberg G, Turley CM. 1993. Heterotrophic flagellates and other protists associated with oceanic detritus throughout the water column in the mid North Atlantic. J. Mar. Biol. Assoc. UK 73, 67-95. ( 10.1017/S0025315400032653) [DOI] [Google Scholar]
- 44.Vørs N, Buck KR, Chavez FP, Eikrem W, Hansen LE, Østergaard JB, Thomsen HA. 1995. Nanoplankton of the equatorial Pacific with emphasis on the heterotrophic protists. Deep-Sea Res. Part II Top. Stud. Oceanogr. 42, 585-602. ( 10.1016/0967-0645(95)00018-L) [DOI] [Google Scholar]
- 45.Flegontova O, Flegontov P, Malviya S, Poulain J, de Vargas C, Bowler C, Lukeš J, Horák A. 2018. Neobodonids are dominant kinetoplastids in the global ocean. Environ. Microbiol. 20, 878-889. ( 10.1111/1462-2920.14034) [DOI] [PubMed] [Google Scholar]
- 46.Salani FS, Arndt H, Hausmann K, Nitsche F, Scheckenbach F. 2012. Analysis of the community structure of abyssal kinetoplastids revealed similar communities at larger spatial scales. ISME J. 6, 713-723. ( 10.1038/ismej.2011.138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mukherjee I, Hodoki Y, Nakano S-I. 2015. Kinetoplastid flagellates overlooked by universal primers dominate in the oxygenated hypolimnion of Lake Biwa, Japan. FEMS Microbiol. Ecol. 91, fiv083. ( 10.1093/femsec/fiv083) [DOI] [PubMed] [Google Scholar]
- 48.Flegontova O, Flegontov P, Londoño PAC, Walczowski W, Šantić D, Edgcomb VP, Lukeš J, Horák A. 2020. Environmental determinants of the distribution of planktonic diplonemids and kinetoplastids in the oceans. Environ. Microbiol. 22, 4014-4031. ( 10.1111/1462-2920.15190) [DOI] [PubMed] [Google Scholar]
- 49.Ekelund F, Patterson DJ. 1997. Some heterotrophic flagellates from a cultivated garden soil in Australia. Arch. Protistenk. 148, 461-478. ( 10.1016/S0003-9365(97)80022-X) [DOI] [Google Scholar]
- 50.von der Heyden S, Cavalier-Smith T. 2005. Culturing and environmental DNA sequencing uncover hidden kinetoplastid biodiversity and a major clade within ancestrally freshwater Neobodo designis. Int. J. Syst. Evol. Microbiol. 55, 2605-2621. ( 10.1099/ijs.0.63606-0) [DOI] [PubMed] [Google Scholar]
- 51.Hassall AH. 1859. On the development and signification of Vibrio lineola, Bodo urinarius, and on certain fungoid and other organic productions generated in alkaline and albuminous urine. Lancet 74, 503-506. ( 10.1016/S0140-6736(02)74345-6) [DOI] [Google Scholar]
- 52.Vickerman K. 1978. The free-living trypanoplasms: descriptions of three species of the genus Procryptobia n. g., and redescription of Dimastigella trypaniformis Sandon, with notes on their relevance to the microscopical diagnosis of disease in man and animals. Trans. Am. Microsc. Soc. 97, 485-502. ( 10.2307/3226165) [DOI] [PubMed] [Google Scholar]
- 53.Vandersea MW, Birkenheuer AJ, Litaker RW, Vaden SL, Renschler JS, Gookin JL. 2015. Identification of Parabodo caudatus (class Kinetoplastea) in urine voided from a dog with hematuria. J. Vet. Diag. Invest. 27, 117-120. ( 10.1177/1040638714562827) [DOI] [PubMed] [Google Scholar]
- 54.Kaczmarek A, Śledź A, Cielecka D, Sałamatin R. 2019. Diagnostic traps: Parabodo cf. caudatus. Ann. Parasitol. 65, s133. [Google Scholar]
- 55.Lukeš J, Skalický T, Týč J, Votýpka J, Yurchenko V. 2014. Evolution of parasitism in kinetoplastid flagellates. Mol. Biochem. Parasitol. 195, 115-122. ( 10.1016/j.molbiopara.2014.05.007) [DOI] [PubMed] [Google Scholar]
- 56.Isaksen TE, Karlsbakk E, Watanabe K, Nylund A. 2011. Ichthyobodo salmonis sp. n. (Ichthyobodonidae, Kinetoplastida), an euryhaline ectoparasite infecting Atlantic salmon (Salmo salar L.). Parasitology 138, 1164-1175. ( 10.1017/S0031182011000916) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woo PTK. 1994. Flagellate parasites of fish. In Parasitic protozoa, vol. 8 (ed. Kreier JP), pp. 1-80. London, UK: Academic Press. [Google Scholar]
- 58.Dyková I, Fiala I, Lom J, Lukeš J. 2003. Perkinsiella amoebae-like endosymbionts of Neoparamoeba spp., relatives of the kinetoplastid Ichthyobodo. Eur. J. Protistol. 39, 37-52. ( 10.1078/0932-4739-00901) [DOI] [Google Scholar]
- 59.Tanifuji G, et al. 2017. Genome sequencing reveals metabolic and cellular interdependence in an amoeba-kinetoplastid symbiosis. Sci. Rep. 7, 11688. ( 10.1038/s41598-017-11866-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams JB. 1999. Description of a new flagellate protist Desmomonas prorhynchi gen. et sp. n. associated with problematical cell masses, parasitic in the turbellarian Prorhynchus sp. (Lecithoepitheliata). Fol. Parasitol. 46, 248-256. [Google Scholar]
- 61.Hitchen ET. 1974. The fine structure of the colonial kinetoplastid flagellate Cephalothamnium cyclopum Stein. J. Protozool. 21, 221-231. ( 10.1111/j.1550-7408.1974.tb03645.x) [DOI] [PubMed] [Google Scholar]
- 62.Hirose E, Nozawa A, Kumagai A, Kitamura SI. 2012. Azumiobodo hoyamushi gen. nov. et sp. nov. (Euglenozoa, Kinetoplastea, Neobodonida): a pathogenic kinetoplastid causing the soft tunic syndrome in ascidian aquaculture. Dis. Aquat. Organ. 97, 227-235. ( 10.3354/dao02422) [DOI] [PubMed] [Google Scholar]
- 63.Nam KW, Shin YK, Park KI. 2015. Seasonal variation in Azumiobodo hoyamushi infection among benthic organisms in the southern coast of Korea. Parasites Vectors 8, 569-575. ( 10.1186/s13071-015-1179-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nawata A, Hirose E, Kitamura SI, Kumagai A. 2015. Encystment and excystment of kinetoplastid Azumiobodo hoyamushi, causal agent of soft tunic syndrome in ascidian aquaculture. Dis. Aquat. Organ. 115, 253-262. ( 10.3354/dao02897) [DOI] [PubMed] [Google Scholar]
- 65.Goodwin JD, Lee TF, Kugrens P, Simpson AGB. 2018. Allobodo chlorophagus n. gen. n. sp., a kinetoplastid that infiltrates and feeds on the invasive alga Codium fragile. Protist 169, 911-925. ( 10.1016/j.protis.2018.07.001) [DOI] [PubMed] [Google Scholar]
- 66.Lom J. 1979. Biology of the trypanosomes and trypanoplasms of fish. In Biology of the kinetoplastida, vol. 2 (eds Lumsden WHR, Evans DA), pp. 269-337. London, UK: Academic Press London. [Google Scholar]
- 67.Kruse P, Steinhagen D, Körting W. 1989. Development of Trypanoplasma borreli (Mastigophora: Kinetoplastida) in the leech vector Piscicola geometra and its infectivity for the common carp, Cyprinus carpio. J. Parasitol. 75, 527-530. ( 10.2307/3282901) [DOI] [PubMed] [Google Scholar]
- 68.Losev A, Grybchuk-Ieremenko A, Kostygov AY, Lukeš J, Yurchenko V. 2015. Host specificity, pathogenicity, and mixed infections of trypanoplasms from freshwater fishes. Parasitol. Res. 114, 1071-1078. ( 10.1007/s00436-014-4277-y) [DOI] [PubMed] [Google Scholar]
- 69.Steinhagen D, Kruse P, Körting W. 1990. Some haematological observations on carp, Cyprinus carpio L., experimentally infected with Trypanoplasma borreli Laveran & Mesnil, 1901 (Protozoa: Kinetoplastida). J. Fish Dis. 13, 157-162. ( 10.1111/j.1365-2761.1990.tb00768.x) [DOI] [Google Scholar]
- 70.Saeij JPJ, Stet RJM, de Vries BJ, van Muiswinkel WB, Wiegertjes GF. 2003. Molecular and functional characterization of carp TNF: a link between TNF polymorphism and trypanotolerance? Dev. Comp. Immunol. 27, 29-41. ( 10.1016/S0145-305X(02)00064-2) [DOI] [PubMed] [Google Scholar]
- 71.Rankin JS. 1937. An ecological study of parasites of some North Carolina salamanders. Ecol. Monogr. 7, 169-269. ( 10.2307/1943289) [DOI] [Google Scholar]
- 72.Woo PTK. 1987. Cryptobia and cryptobiosis in fishes. Adv. Parasitol. 26, 199-237. ( 10.1016/S0065-308X(08)60297-3) [DOI] [PubMed] [Google Scholar]
- 73.Vickerman K. 1976. The diversity of the kinetoplastid flagellates. In Biology of the kinetoplastida, vol. 1 (eds Lumsden WHR, Evans DA), pp. 1-34. London, UK: Academic Press. [Google Scholar]
- 74.Bradbury PC. 1994. Parasitic protozoa of molluscs and crustacea. In Parasitic protozoa, Vol. 8 (ed. Kreier JP), pp. 139-264. Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 75.Kozloff EN. 2004. Redescription of Cryptobia helicis Leidy, 1846 (Kinetoplasta: Bodonea: Cryptobiidae), disposition of flagellates mistakenly assigned to this species, and description of a new species from a North American pulmonate snail. Acta Protozool. 43, 123-132. [Google Scholar]
- 76.Lukeš J, Jirků M, Avliyakulov N, Benada O. 1998. Pankinetoplast DNA structure in a primitive bodonid flagellate, Cryptobia helicis. EMBO J. 17, 838-846. ( 10.1093/emboj/17.3.838) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hesse E. 1910. Trypanoplasma vaginalis n. sp., parasite du vagin de la sangsue . C. R. Hebd. Séances Acad. Sci. Paris 151, 504-505. [Google Scholar]
- 78.Frolov AO, Kornakova EE. 2001. [Cryptobia udonellae sp. n. (Kinetoplastidea: Cryptobiida)—parasites of the excretory system of Udonella murmanica (Udonellida)] (In Russian). Parazitologiia 35, 454-459. [PubMed] [Google Scholar]
- 79.Fantham HB, Porter A. 1910. On a new trypanoplasm, T. dendrocoeli sp. n. from Dendrocoelum lacteum . Proc. Zool. Soc. Lond. 3, 670-671. [Google Scholar]
- 80.Hovasse R. 1924. Trypanoplasma sagittae nov. sp. Comptes Rendus des Séances et Mémoires de la Société de Biologie, Paris 91, 1254–1255.
- 81.Vickerman K. 1977. DNA throughout the single mitochondrion of a kinetoplastid flagellate: observations on the ultrastructure of Cryptobia vaginalis (Hesse, 1910). J. Protozool. 24, 221-233. ( 10.1111/j.1550-7408.1977.tb00970.x) [DOI] [Google Scholar]
- 82.Walker EL. 1910. Trypanoplasma ranæ n. sp. and its life-cycle in cultures. J. Med. Res. 23, 391-406. [PMC free article] [PubMed] [Google Scholar]
- 83.Bovee EC, Telford SR. 1962. Protozoan inquilines from Florida reptiles. III. Rigidomastix scincorum n. sp.; Cercobodo stilosomorum n. sp.; and Cryptobia geccorum n. sp. Q. J. Florida Acad. Sci. 25, 180-191. [Google Scholar]
- 84.Nohýnková E. 1984. A new pathogenic Cryptobia from freshwater fishes: light and electron microscopic study. Protistologica 20, 181-195. [Google Scholar]
- 85.Dyková I, Lom J. 1985. Histopathological changes due to infections with Cryptobia iubilans Nohýnková 1984, in two cichlid fishes. J. Appl. Ichthyol. 1, 34-38. ( 10.1111/j.1439-0426.1985.tb00409.x) [DOI] [Google Scholar]
- 86.Yanong RPE, Curtis E, Russo R, Francis-Floyd R, Klinger RE, Berzins I, Kelley K, Poynton SL. 2004. Cryptobia iubilans infection in juvenile discus. J. Am. Vet. Med. Assoc. 224, 1644-1650. ( 10.2460/javma.2004.224.1644) [DOI] [PubMed] [Google Scholar]
- 87.Poynton SL, Whitaker B, Heinrich A. 2001. A novel trypanoplasm-like flagellate Jarrellia atramenti n. g., n. sp. (Kinetoplastida: Bodonidae) and ciliates from the blowhole of a stranded pygmy sperm whale Kogia breviceps (Physeteridae): morphology, life cycle and potential pathogenic. Dis. Aquat. Organ. 44, 191-201. ( 10.3354/dao044191) [DOI] [PubMed] [Google Scholar]
- 88.d'Avila-Levy CM, et al. 2015. Exploring the environmental diversity of kinetoplastid flagellates in the high-throughput DNA sequencing era. Mem. Inst. Oswaldo Cruz 110, 956-965. ( 10.1590/0074-02760150253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lukeš J, Butenko A, Hashimi H, Maslov DA, Votýpka J, Yurchenko V. 2018. Trypanosomatids are much more than just trypanosomes: clues from the expanded family tree. Trends Parasitol. 34, 466-480. ( 10.1016/j.pt.2018.03.002) [DOI] [PubMed] [Google Scholar]
- 90.Podlipaev S. 2001. The more insect trypanosomatids under study-the more diverse Trypanosomatidae appears. Int. J. Parasitol. 31, 648-652. ( 10.1016/S0020-7519(01)00139-4) [DOI] [PubMed] [Google Scholar]
- 91.Podlipaev SA. 1990. Catalogue of world fauna of Trypanosomatidae (Protozoa) (in Russian). Leningrad, Russia: Zoologicheskii Institut AN SSSR. [Google Scholar]
- 92.Maslov DA, Votýpka J, Yurchenko V, Lukeš J. 2013. Diversity and phylogeny of insect trypanosomatids: all that is hidden shall be revealed. Trends Parasitol. 29, 43-52. ( 10.1016/j.pt.2012.11.001) [DOI] [PubMed] [Google Scholar]
- 93.Kaufer A, Ellis J, Stark D, Barratt J. 2017. The evolution of trypanosomatid taxonomy. Parasit. Vectors 10, 287-303. ( 10.1186/s13071-017-2204-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wallace FG. 1966. The trypanosomatid parasites of insects and arachnids. Exp. Parasitol. 18, 124-193. ( 10.1016/0014-4894(66)90015-4) [DOI] [PubMed] [Google Scholar]
- 95.Kraeva N, et al. 2015. Leptomonas seymouri: adaptations to the dixenous life cycle analyzed by genome sequencing, transcriptome profiling and co-infection with Leishmania donovani. PLoS Pathog. 11, e1005127. ( 10.1371/journal.ppat.1005127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Camargo EP. 1999. Phytomonas and other trypanosomatid parasites of plants and fruit. Adv. Parasitol. 42, 29-112. ( 10.1016/s0065-308x(08)60148-7) [DOI] [PubMed] [Google Scholar]
- 97.Nicoli RM, Penaud A. 1971. Sur la definition du genre Leptomonas Saville Kent, 1880 (Trypanosomatida). Bull. Soc. Zool. France 96, 15-17. [Google Scholar]
- 98.Wallace FG. 1976. Biology of the Kinetoplastida of arthropods. In Biology of the Kinetoplastida, vol. 2 (eds Lumsden WHR, Evans DA), pp. 213-240. London, UK: Academic Press. [Google Scholar]
- 99.Králová J, Grybchuk-Ieremenko A, Votýpka J, Novotný V, Kment P, Lukeš J, Yurchenko V, Kostygov AY. 2019. Insect trypanosomatids in Papua New Guinea: high endemism and diversity. Int. J. Parasitol. 49, 1075-1086. ( 10.1016/j.ijpara.2019.09.004) [DOI] [PubMed] [Google Scholar]
- 100.Frolov AO, Malysheva MN, Ganyukova AI, Yurchenko V, Kostygov AY. 2018. Obligate development of Blastocrithidia papi (Trypanosomatidae) in the Malpighian tubules of Pyrrhocoris apterus (Hemiptera) and coordination of host-parasite life cycles. PLoS ONE 13, e0204467. ( 10.1371/journal.pone.0204467) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Smirnoff WA, Lipa JJ. 1970. Herpetomonas swainei sp. n., a new flagellate parasite of Neodiprion swainei (Hymenoptera: Tenthredinidae). J. Invertebr. Pathol. 16, 187-195. ( 10.1016/0022-2011(70)90059-5) [DOI] [Google Scholar]
- 102.Frolov AO, Skarlato SO. 1987. [Light and electron microscopy studies of Leptomonas pyrrhocoris (Trypanosomatidae)] (In Russian). Parazitologiya 21, 3-9. [Google Scholar]
- 103.Frolov AO, Malysheva MN, Ganyukova AI, Spodareva VV, Yurchenko V, Kostygov AY. 2019. Development of Phytomonas lipae sp. n. (Kinetoplastea: Trypanosomatidae) in the true bug Coreus marginatus (Heteroptera: Coreidae) and insights into the evolution of life cycles in the genus Phytomonas. PLoS ONE 14, e0214484. ( 10.1371/journal.pone.0214484) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wille JJ, Weidner EJ, Steffens WL. 1981. Intranuclear parasitism of the ciliate Euplotes by a trypanosomatid flagellate. J. Protozool. 28, 223-227. ( 10.1111/j.1550-7408.1981.tb02837.x) [DOI] [Google Scholar]
- 105.Görtz HD, Dieckmann J. 1987. Leptomonas ciliatorum n. sp. (Kinetoplastida, Trypanosomatidae) in the macronucleus of a hypotrichous ciliate. J. Protozool. 34, 259-263. ( 10.1111/j.1550-7408.1987.tb03171.x) [DOI] [Google Scholar]
- 106.Fokin SI, Schrallhammer M, Chiellini C, Verni F, Petroni G. 2014. Free-living ciliates as potential reservoirs for eukaryotic parasites: occurrence of a trypanosomatid in the macronucleus of Euplotes encysticus. Parasit. Vectors 7, 203. ( 10.1186/1756-3305-7-203) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gillies C, Hanson ED. 1963. A new species of Leptomonas parasitizing the macronucleus of Paramecium trichium. J. Protozool. 10, 467-473. ( 10.1111/j.1550-7408.1963.tb01707.x) [DOI] [PubMed] [Google Scholar]
- 108.Schaub GA. 1994. Pathogenicity of trypanosomatids on insects. Parasitol. Today 10, 463-468. ( 10.1016/0169-4758(94)90155-4) [DOI] [PubMed] [Google Scholar]
- 109.Gómez-Moracho T, et al. 2020. Experimental evidence of harmful effects of Crithidia mellificae and Lotmaria passim on honey bees. Int. J. Parasitol. 50, 1117-1124. ( 10.1016/j.ijpara.2020.06.009) [DOI] [PubMed] [Google Scholar]
- 110.Bailey CH, Brooks WM. 1972. Effects of Herpetomonas muscarum on development and longevity of the eye gnat, Hippelates pusio (Diptera: Chloropidae). J. Invertebr. Pathol. 20, 31-36. ( 10.1016/0022-2011(72)90077-8) [DOI] [PubMed] [Google Scholar]
- 111.Arnqvist G, Mäki M. 1990. Infection rates and pathogenicity of trypanosomatid gut parasites in the water strider Gerris odontogaster (Zett.) (Heteroptera: Gerridae). Oecologia 84, 194-198. ( 10.1007/BF00318271) [DOI] [PubMed] [Google Scholar]
- 112.Hamilton PT, Votýpka J, Dostálová A, Yurchenko V, Bird NH, Lukeš J, Lemaitre B, Perlman SJ. 2015. Infection dynamics and immune response in a newly described Drosophila-trypanosomatid association. mBio 6, e01356-15. ( 10.1128/mBio.01356-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shykoff JA, Schmid-Hempel P. 1991. Incidence and effects of four parasites in natural populations of bumble bees in Switzerland. Apidologie 22, 117-125. ( 10.1051/apido:19910204) [DOI] [Google Scholar]
- 114.Brown MJF, Schmid-Hempel R, Schmid-Hempel P. 2003. Strong context-dependent virulence in a host-parasite system: reconciling genetic evidence with theory. J. Animal Ecol. 72, 994-1002. ( 10.1046/j.1365-2656.2003.00770.x) [DOI] [Google Scholar]
- 115.Gegear RJ, Otterstatter MC, Thomson JD. 2006. Bumble-bee foragers infected by a gut parasite have an impaired ability to utilize floral information. Proc. R. Soc. B 273, 1073-1078. ( 10.1098/rspb.2005.3423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Otterstatter MC, Gegear RJ, Colla SR, Thomson JD. 2005. Effects of parasitic mites and protozoa on the flower constancy and foraging rate of bumble bees. Behav. Ecol. Sociobiol. 58, 383-389. ( 10.1007/s00265-005-0945-3) [DOI] [Google Scholar]
- 117.Schaub GA. 2009. Interactions of trypanosomatids and triatomines. Adv. Insect Physiol. 37, 177-242. ( 10.1016/S0065-2806(09)37004-6) [DOI] [Google Scholar]
- 118.Klingenberg CP, Barrington Leigh RH, Keddie BA, Spence JR. 1997. Influence of gut parasites on growth performance in the water strider Gerris buenoi (Hemiptera: Gerridae). Ecography 20, 29-36. ( 10.1111/j.1600-0587.1997.tb00344.x) [DOI] [Google Scholar]
- 119.Jaskowska E, Butler C, Preston G, Kelly S. 2015. Phytomonas: trypanosomatids adapted to plant environments. PLoS Pathog. 11, e1004484. ( 10.1371/journal.ppat.1004484) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schwelm A, et al. 2018. Not in your usual Top 10: protists that infect plants and algae. Mol. Plant Pathol. 19, 1029-1044. ( 10.1111/mpp.12580) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jankevicius JV, Jankevicius SI, Campaner M, Conchon I, Maeda LA, Teixeira MMG, Freymüller E, Camargo EP. 1989. Life cycle and culturing of Phytomonas serpens (Gibbs), a trypanosomatid parasite of tomatoes. J. Protozool. 36, 265-271. ( 10.1111/j.1550-7408.1989.tb05361.x) [DOI] [Google Scholar]
- 122.Freymüller E, Milder R, Jankevicius JV, Jankevicius SI, Camargo EP. 1990. Ultrastructural studies on the trypanosomatid Phytomonas serpens in the salivary glands of a phytophagous hemipteran. J. Protozool. 37, 225-229. ( 10.1111/j.1550-7408.1990.tb01132.x) [DOI] [Google Scholar]
- 123.Seward EA, Votýpka J, Kment P, Lukeš J, Kelly S. 2017. Description of Phytomonas oxycareni n. sp. from the salivary glands of Oxycarenus lavaterae. Protist 168, 71-79. ( 10.1016/j.protis.2016.11.002) [DOI] [PubMed] [Google Scholar]
- 124.Frolov AO, Malysheva MN, Yurchenko V, Kostygov AY. 2016. Back to monoxeny: Phytomonas nordicus descended from dixenous plant parasites. Eur. J. Protistol. 52, 1-10. ( 10.1016/j.ejop.2015.08.002) [DOI] [PubMed] [Google Scholar]
- 125.Espinosa OA, Serrano MG, Camargo EP, Teixeira MMG, Shaw JJ. 2018. An appraisal of the taxonomy and nomenclature of trypanosomatids presently classified as Leishmania and Endotrypanum. Parasitology 145, 430-442. ( 10.1017/S0031182016002092) [DOI] [PubMed] [Google Scholar]
- 126.Dougall AM, Alexander B, Holt DC, Harris T, Sultan AH, Bates PA, Rose K, Walton SF. 2011. Evidence incriminating midges (Diptera: Ceratopogonidae) as potential vectors of Leishmania in Australia. Int. J. Parasitol. 41, 571-579. ( 10.1016/j.ijpara.2010.12.008) [DOI] [PubMed] [Google Scholar]
- 127.Akhoundi M, Kuhls K, Cannet A, Votýpka J, Marty P, Delaunay P, Sereno D. 2016. A historical overview of the classification, evolution, and dispersion of Leishmania parasites and sandflies. PLoS Negl. Trop. Dis. 10, e0004349. ( 10.1371/journal.pntd.0004349) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Butenko A, et al. 2019. Comparative genomics of Leishmania (Mundinia). BMC Genomics 20, 726. ( 10.1186/s12864-019-6126-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dostálová A, Volf P. 2012. Leishmania development in sand flies: parasite-vector interactions overview. Parasit. Vectors 5, 276-288. ( 10.1186/1756-3305-5-276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bates PA. 2007. Transmission of Leishmania metacyclic promastigotes by phlebotomine sand flies. Int. J. Parasitol. 37, 1097-1106. ( 10.1016/j.ijpara.2007.04.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Antoine JC, Prina E, Courret N, Lang T. 2004. Leishmania spp.: on the interactions they establish with antigen-presenting cells of their mammalian hosts. Adv. Parasitol. 58, 1-68. ( 10.1016/S0065-308X(04)58001-6) [DOI] [PubMed] [Google Scholar]
- 132.WHO. 2020. Leishmaniasis. See https://www.who.int/news-room/fact-sheets/detail/leishmaniasis (accessed on 2 March 2020).
- 133.Symmers WS. 1960. Leishmaniasis acquired by contagion: a case of marital infection in Britain. Lancet 275, 127-132. ( 10.1016/S0140-6736(60)90052-0) [DOI] [PubMed] [Google Scholar]
- 134.Pagliano P, Carannante N, Rossi M, Gramiccia M, Gradoni L, Faella FS, Gaeta GB. 2005. Visceral leishmaniasis in pregnancy: a case series and a systematic review of the literature. J. Antimicrob. Chemother. 55, 229-233. ( 10.1093/jac/dkh538) [DOI] [PubMed] [Google Scholar]
- 135.Boehme CC, Hain U, Novosel A, Eichenlaub S, Fleischmann E, Löscher T. 2006. Congenital visceral leishmaniasis. Emerg. Infect. Dis. 12, 359. ( 10.3201/eid1202.050449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zinchuk A, Nadraga A. 2010. Congenital visceral leishmaniasis in Ukraine: case report. Ann. Trop. Paediatr. 30, 161-164. ( 10.1179/146532810X12703902516400) [DOI] [PubMed] [Google Scholar]
- 137.Ribeiro RR, Michalick MSM, da Silva ME, dos Santos CCP, Frézard FJG, da Silva SM. 2018. Canine leishmaniasis: an overview of the current status and strategies for control. BioMed Res. Int. 2018, 3296893. ( 10.1155/2018/3296893) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lobsiger L, Müller N, Schweizer T, Frey CF, Wiederkehr D, Zumkehr B, Gottstein B. 2010. An autochthonous case of cutaneous bovine leishmaniasis in Switzerland. Vet. Parasitol. 169, 408-414. ( 10.1016/j.vetpar.2010.01.022) [DOI] [PubMed] [Google Scholar]
- 139.Müller N, et al. 2009. Occurrence of Leishmania sp. in cutaneous lesions of horses in Central Europe . Vet. Parasitol. 166, 346-351. ( 10.1016/j.vetpar.2009.09.001) [DOI] [PubMed] [Google Scholar]
- 140.Kreutzer RD, et al. 1991. Characterization of Leishmania colombiensis sp. n. (Kinetoplastida: Trypanosomatidae), a new parasite infecting humans, animals, and phlebotomine sand flies in Colombia and Panama. Am. J. Trop. Med. Hyg. 44, 662-675. ( 10.4269/ajtmh.1991.44.662) [DOI] [PubMed] [Google Scholar]
- 141.Rodriguez-Bonfante C, Bonfante-Garrido R, Grimaldi G, Momen H, Cupolillo E. 2003. Genotypically distinct Leishmania colombiensis isolates from Venezuela cause both cutaneous and visceral leishmaniasis in humans. Infect. Genet. Evol. 3, 119-124. ( 10.1016/S1567-1348(03)00012-1) [DOI] [PubMed] [Google Scholar]
- 142.Hoare CA. 1972. The trypanosomes of mammals. A zoological monograph. Oxford, UK: Blackwell Scientific Publications. [Google Scholar]
- 143.Stevens JR, Teixeira MM, Bingle LE, Gibson WC. 1999. The taxonomic position and evolutionary relationships of Trypanosoma rangeli. Int. J. Parasitol. 29, 749-757. ( 10.1016/s0020-7519(99)00016-8) [DOI] [PubMed] [Google Scholar]
- 144.Galen SC, Borner J, Perkins SL, Weckstein JD. 2020. Phylogenomics from transcriptomic ‘bycatch’ clarify the origins and diversity of avian trypanosomes in North America. PLoS ONE 15, e0240062. ( 10.1371/journal.pone.0240062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lai DH, Hashimi H, Lun ZR, Ayala FJ, Lukeš J. 2008. Adaptations of Trypanosoma brucei to gradual loss of kinetoplast DNA: Trypanosoma equiperdum and Trypanosoma evansi are petite mutants of T. brucei. Proc. Natl Acad. Sci. USA 105, 1999-2004. ( 10.1073/pnas.0711799105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Brun R, Hecker H, Lun ZR. 1998. Trypanosoma evansi and T. equiperdum: distribution, biology, treatment and phylogenetic relationship (a review). Vet. Parasitol. 79, 95-107. ( 10.1016/s0304-4017(98)00146-0) [DOI] [PubMed] [Google Scholar]
- 147.Deane MP, Lenzi HL, Jansen A. 1984. Trypanosoma cruzi: vertebrate and invertebrate cycles in the same mammal host, the opossum Didelphis marsupialis. Mem. Inst. Oswaldo Cruz. 79, 513-515. ( 10.1590/S0074-02761984000400021) [DOI] [PubMed] [Google Scholar]
- 148.Rocha G, Martins A, Gama G, Brandão F, Atouguia J. 2004. Possible cases of sexual and congenital transmission of sleeping sickness. Lancet 363, 247. ( 10.1016/S0140-6736(03)15345-7) [DOI] [PubMed] [Google Scholar]
- 149.Howard EJ, Xiong X, Carlier Y, Sosa-Estani S, Buekens P. 2014. Frequency of the congenital transmission of Trypanosoma cruzi: a systematic review and meta-analysis. BJOG 121, 22-33. ( 10.1111/1471-0528.12396.Frequency) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gomes C, Almeida AB, Rosa AC, Araujo PF, Teixeira ARL. 2019. American trypanosomiasis and Chagas disease: sexual transmission. Int. J. Infect. Dis. 81, 81-84. ( 10.1016/j.ijid.2019.01.021) [DOI] [PubMed] [Google Scholar]
- 151.Stuart K, Brun R, Croft S, Fairlamb A, Gürtler RE, McKerrow J, Reed S, Tarleton R. 2008. Kinetoplastids: related protozoan pathogens, different diseases. J. Clin. Invest. 118, 1301-1310. ( 10.1172/JCI33945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Trindade S, et al. 2016. Trypanosoma brucei parasites occupy and functionally adapt to the adipose tissue in mice. Cell Host Microbe 19, 837-848. ( 10.1016/j.chom.2016.05.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kennedy PGE. 2013. Clinical features, diagnosis, and treatment of human African trypanosomiasis (sleeping sickness). Lancet Neurol. 12, 186-194. ( 10.1016/S1474-4422(12)70296-X) [DOI] [PubMed] [Google Scholar]
- 154.Barrett MP. 2018. The elimination of human African trypanosomiasis is in sight: report from the third WHO stakeholders meeting on elimination of gambiense human African trypanosomiasis. PLoS Negl. Trop. Dis. 12, e0006925. ( 10.1371/journal.pntd.0006925) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pérez-Molina JA, Molina I. 2018. Chagas disease. Lancet 391, 82-94. ( 10.1016/S0140-6736(17)31612-4) [DOI] [PubMed] [Google Scholar]
- 156.Kirchhoff LV. 2011. Epidemiology of American trypanosomiasis (Chagas disease). Adv. Parasitol. 75, 1-18. ( 10.1016/B978-0-12-385863-4.00001-0) [DOI] [PubMed] [Google Scholar]
- 157.Guhl F, Vallejo GA. 2003. Trypanosoma (Herpetosoma) rangeli Tejera, 1920—an updated review. Mem. Inst. Oswaldo Cruz 98, 435-442. ( 10.1590/S0074-02762003000400001) [DOI] [PubMed] [Google Scholar]
- 158.Yaro M, Munyard KA, Stear MJ, Groth DM. 2016. Combatting African Animal Trypanosomiasis (AAT) in livestock: the potential role of trypanotolerance. Vet. Parasitol. 225, 43-52. ( 10.1016/j.vetpar.2016.05.003) [DOI] [PubMed] [Google Scholar]
- 159.Becker CD. 1977. Flagellate parasites of fish. In Parasitic protozoa Vol. 8. Taxonomy, kinetoplastids, and flagellates of fish (ed. Kreier JP), pp. 357-416. New York, NY: Academic Press. [Google Scholar]
- 160.Dyková I, Lom J. 1979. Histopathological changes in Trypanosoma danilewskyi Laveran & Mesnil, 1904 and Trypanoplasma borelli Laveran & Mesnil, 1902 infections of goldfish, Carassius aurata (L.). J. Fish Dis. 2, 381-390. ( 10.1111/j.1365-2761.1979.tb00390.x) [DOI] [Google Scholar]
- 161.Khan RA. 1985. Pathogenesis of Trypanosoma murmanensis in marine fish of the northwestern Atlantic following experimental transmission. Can. J. Zool. 63, 2141-2144. ( 10.1139/z85-315) [DOI] [Google Scholar]
- 162.Islam A, Woo P. 1991. Anemia and its mechanism in goldfish Carassius auratus infected with Trypanosoma danilewskyi. Dis. Aquat. Organ. 11, 37-43. ( 10.3354/dao011037) [DOI] [Google Scholar]
- 163.Ahmed MS, Shafiq K, Ali H, Ollevier F. 2011. Pathogenic effects associated with Trypanosoma danilewskyi strain FCc 1 infection in juvenile common carp, Cyprinus carpio L. J. Anim. Plant Sci. 21, 800-806. [Google Scholar]
- 164.Lukeš J, Guilbride DL, Votýpka J, Zíková A, Benne R, Englund PT. 2002. Kinetoplast DNA network: evolution of an improbable structure. Eukaryot. Cell 1, 495-502. ( 10.1128/EC.1.4.495-502.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Moreira D, López-García P, Vickerman K. 2004. An updated view of kinetoplastid phylogeny using environmental sequences and a closer outgroup: proposal for a new classification of the class Kinetoplastea. Int. J. Syst. Evol. Microbiol. 54, 1861-1875. ( 10.1099/ijs.0.63081-0) [DOI] [PubMed] [Google Scholar]
- 166.Kolisko M, et al. 2020. EukRef-excavates: seven curated SSU ribosomal RNA gene databases. Database 2020, baaa080. ( 10.1093/database/baaa080) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Isaksen TE, Karlsbakk E, Nylund A. 2007. Ichthyobodo hippoglossi n. sp. (Kinetoplastea: Prokinetoplastida: Ichthyobodonidae fam. nov.), an ectoparasitic flagellate infecting farmed Atlantic halibut Hippoglossus hippoglossus . Dis. Aquat. Organ. 73, 207-217. ( 10.3354/dao073207) [DOI] [PubMed] [Google Scholar]
- 168.Dyková I, Fiala I, Pecková H. 2008. Neoparamoeba spp. and their eukaryotic endosymbionts similar to Perkinsela amoebae (Hollande, 1980): coevolution demonstrated by SSU rRNA gene phylogenies . Eur. J. Protistol. 44, 269-277. ( 10.1016/j.ejop.2008.01.004) [DOI] [PubMed] [Google Scholar]
- 169.Sibbald SJ, Cenci U, Colp M, Eglit Y, O'Kelly CJ, Archibald JM. 2017. Diversity and evolution of Paramoeba spp. and their kinetoplastid endosymbionts. J. Eukaryot. Microbiol. 64, 598-607. ( 10.1111/jeu.12394) [DOI] [PubMed] [Google Scholar]
- 170.Cavalier-Smith T. 2016. Higher classification and phylogeny of Euglenozoa. Eur. J. Protistol. 56, 250-276. ( 10.1016/j.ejop.2016.09.003) [DOI] [PubMed] [Google Scholar]
- 171.Stoeck T, Schwarz MVJ, Boenigk J, Schweikert M, von der Heyden S, Behnke A. 2005. Cellular identity of an 18S rRNA gene sequence clade within the class Kinetoplastea: the novel genus Actuariola gen. nov. (Neobodonida) with description of the type species Actuariola framvarensis sp. nov . Int. J. Syst. Evol. Microbiol. 55, 2623-2635. ( 10.1099/ijs.0.63769-0) [DOI] [PubMed] [Google Scholar]
- 172.Zíková A, Vancová M, Jirků M, Lukeš J. 2003. Cruzella marina (Bodonina, Kinetoplastida): non-catenated structure of poly-kinetoplast DNA. Exp. Parasitol. 104, 159-161. ( 10.1016/j.exppara.2003.08.002) [DOI] [PubMed] [Google Scholar]
- 173.Frolov AO, Malysheva MN. 2002. [Ultrastructure of the flagellate Cruzella marina (Kinetoplastidea)] (In Russian). Tsitologiia 44, 477-484. [PubMed] [Google Scholar]
- 174.Faria J, da Cunha AM, Pinto C. 1922. Estudos sobre Protozoarios do mar. Mem. Inst. Oswaldo Cruz 15, 186-208. ( 10.1590/S0074-02761922000200013) [DOI] [Google Scholar]
- 175.Novarino G. 1996. Notes on flagellate nomenclature. I. Cryptaulaxoides nom. n., a zoological substitute for Cryptaulax Skuja, 1948 (Protista incertae sedis) non Cryptaulax Tate, 1869 (Mollusca, Gastropoda) non Cryptaulax Cameron, 1906 (Insecta, Hymenoptera), with remarks on botanical nomenclature . Acta Protozool. 35, 235-238. [Google Scholar]
- 176.Perty M. 1852. Zur Kenntniss Kleinster Lebensformen: Nach Bau, Funktionen, Systematik, mit Specialverzeichniss der in der Schweiz beobachteten. Bern, Switzerland: Verlag von Jent & Reinert. [Google Scholar]
- 177.Tate R. 1869. Contributions to Jurassic palaeontology. I. Cryptaulax, a new genus of Cerithiadae. Ann. Mag. Nat. Hist 4, 417-419. ( 10.1080/00222936908696088) [DOI] [Google Scholar]
- 178.Cameron P. 1906. Descriptions of new species of parasitic Hymenoptera chiefly in the collection of the South African Museum, Cape Town. Ann. South Afr. Mus. 5, 17-186. [Google Scholar]
- 179.Bernard C, Simpson AGB, Patterson DJ. 2000. Some free-living flagellates (protista) from anoxic habitats. Ophelia 52, 113-142. ( 10.1080/00785236.1999.10409422) [DOI] [Google Scholar]
- 180.von der Heyden S, Chao EE, Vickerman K, Cavalier-Smith T. 2004. Ribosomal RNA phylogeny of bodonid and diplonemid flagellates and the evolution of Euglenozoa. J. Euk. Microbiol. 51, 402-416. ( 10.1111/j.1550-7408.2004.tb00387.x) [DOI] [PubMed] [Google Scholar]
- 181.Nikolaev SI, Mylnikov AP, Fahrni J, Petrov N, Pawlowski J. 2003. The taxonomic position of Klosteria bodomorphis gen. and sp. nov. (Kinetoplastida) based on ultrastructure and SSU rRNA gene sequence analysis . Protistology 3, 126-135. [Google Scholar]
- 182.Vørs N. 1992. Heterotrophic amoebae, flagellates and heliozoa from the Tvärminne area, Gulf of Finland, in 1988-1990. Ophelia 36, 1-109. ( 10.1080/00785326.1992.10429930) [DOI] [Google Scholar]
- 183.Lackey JB. 1940. Some new flagellates from the Woods Hole area. Am. Midland Natural. 23, 463-471. ( 10.2307/2420679) [DOI] [Google Scholar]
- 184.Breunig A, König H, Brugerolle G, Vickerman K, Hertel H. 1993. Isolation and ultrastructural features of a new strain of Dimastigella trypaniformis Sandon 1928 (Bodonina, Kinetoplastida) and comparison with a previously isolated strain. Eur. J. Protistol. 29, 416-424. ( 10.1016/S0932-4739(11)80404-9) [DOI] [PubMed] [Google Scholar]
- 185.Frolov AO, Mylnikov AP, Malysheva MN. 1997. [Description and electron microscopical study of the free-living cryptobiid flagellate Dimastigella mimosa sp. n. (Kinetoplastida, Cryptobiidae)] (In Russian). Tsitologiia 39, 447-448. [Google Scholar]
- 186.Swale EMF. 1973. A study of the colourless flagellate Rhynchomonas nasuta (Stokes) Klebs. Biol. J. Linn. Soc. 5, 255-264. ( 10.1111/j.1095-8312.1973.tb00705.x) [DOI] [Google Scholar]
- 187.Doležel D, Jirků M, Maslov DA, Lukeš J. 2000. Phylogeny of the bodonid flagellates (Kinetoplastida) based on small-subunit rRNA gene sequences. Int. J. Syst. Evol. Microbiol. 50, 1943-1951. ( 10.1099/00207713-50-5-1943) [DOI] [PubMed] [Google Scholar]
- 188.Todal JA, Karlsbakk E, Isaksen TE, Plarre H, Urawa S, Mouton A, Hoel E, Koren CWR, Nylund A. 2004. Ichthyobodo necator (Kinetoplastida)—a complex of sibling species. Dis. Aquat. Organ. 58, 9-16. ( 10.3354/dao058009) [DOI] [PubMed] [Google Scholar]
- 189.Lom J, Dyková I. 1992. Protozoan parasites of fishes. Amsterdam, The Netherlands: Elsevier Science Publishers New York. [Google Scholar]
- 190.Freeman MA, Kristmundsson A. 2018. A closer look at Cryptobia dahli: a parabodonid flagellate from the stomach of the Atlantic lumpfish. Bull. Eur. Assoc. Fish Pathol. 38, 195-201. [Google Scholar]
- 191.Brooker BE. 1971. Fine structure of Bodo saltans and Bodo caudatus (Zoomastigophora: Protozoa) and their affinities with the Trypanosomatidae. Bull. Br. Mus. Nat. Hist. 22, 89-102. [Google Scholar]
- 192.Mylnikov AP. 1986. [Ultrathin structure of the flagellar apparatus in the bacteriotrophic flagellate Parabodo nitrophilus Skuja, 1948 (Kinetoplastea, Excavata)] (In Russian). Tsitologiia 28, 1056-1060. [Google Scholar]
- 193.Frolov AO, Karpov SA, Mylnikov AP. 2001. The ultrastructure of Procryptobia sorokini (Zhukov) comb. nov. and rootlet homology in kinetoplastids. Protistology 2, 85-95. [Google Scholar]
- 194.Schneider A, Ochsenreiter T. 2018. Failure is not an option—mitochondrial genome segregation in trypanosomes. J. Cell Sci. 131, jcs221820. ( 10.1242/jcs.221820) [DOI] [PubMed] [Google Scholar]
- 195.Harmer J, Yurchenko V, Nenarokova A, Lukeš J, Ginger ML. 2018. Farming, slaving and enslavement: histories of endosymbioses during kinetoplastid evolution. Parasitology 145, 1311-1323. ( 10.1017/S0031182018000781) [DOI] [PubMed] [Google Scholar]
- 196.Frolov AO, Karpov SA. 1995. Comparative morphology of kinetoplastids. Tsitologiia 37, 1072-1096. [PubMed] [Google Scholar]
- 197.Votýpka J, d'Avila-Levy CM, Grellier P, Maslov DA, Lukeš J, Yurchenko V. 2015. New approaches to systematics of Trypanosomatidae: criteria for taxonomic (re)description. Trends Parasitol. 31, 460-469. ( 10.1016/j.pt.2015.06.015) [DOI] [PubMed] [Google Scholar]
- 198.Lukeš J, Jirků M, Doležel D, Kral'ová I, Hollar L, Maslov DA. 1997. Analysis of ribosomal RNA genes suggests that trypanosomes are monophyletic. J. Mol. Evol. 44, 521-527. ( 10.1007/PL00006176) [DOI] [PubMed] [Google Scholar]
- 199.Stevens JR, Noyes HA, Schofield CJ, Gibson W. 2001. The molecular evolution of Trypanosomatidae. Adv. Parasit. 48, 1-56. ( 10.1016/s0065-308x(01)48003-1) [DOI] [PubMed] [Google Scholar]
- 200.Hamilton PB, Gibson WC, Stevens JR. 2007. Patterns of co-evolution between trypanosomes and their hosts deduced from ribosomal RNA and protein-coding gene phylogenies. Mol. Phyl. Evol. 44, 15-25. ( 10.1016/j.ympev.2007.03.023) [DOI] [PubMed] [Google Scholar]
- 201.Fermino BR, et al. 2015. Field and experimental evidence of a new caiman trypanosome species closely phylogenetically related to fish trypanosomes and transmitted by leeches. Int. J. Parasitol. Parasit. Wildl. 4, 368-378. ( 10.1016/j.ijppaw.2015.10.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Dvořáková N, Čepička I, Qablan MA, Gibson W, Blažek R, Široký P. 2015. Phylogeny and morphological variability of trypanosomes from African pelomedusid turtles with redescription of Trypanosoma mocambicum Pienaar, 1962. Protist 166, 599-608. ( 10.1016/j.protis.2015.10.002) [DOI] [PubMed] [Google Scholar]
- 203.Bernal XE, Pinto CM. 2016. Sexual differences in prevalence of a new species of trypanosome infecting túngara frogs. Int. J. Parasitol. Parasit. Wildl. 5, 40-47. ( 10.1016/j.ijppaw.2016.01.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Spodareva VV, Grybchuk-Ieremenko A, Losev A, Votýpka J, Lukeš J, Yurchenko V, Kostygov AY. 2018. Diversity and evolution of anuran trypanosomes: insights from the study of European species. Parasit. Vectors 11, 447. ( 10.1186/s13071-018-3023-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Doflein F. 1901. Die Protozoen als Parasiten und Krankheitserreger nach Biologischen Gesichtspunkten Dargestellt. Jena, Germany: Fischer [Google Scholar]
- 206.Woo PTK. 2006. Diplomonadida (Phylum Parabasalia) and Kinetoplastea (Phylum Euglenozoa). In Fish diseases and disorders, Vol. 1: protozoan and metazoan infections (ed. Woo PTK), pp. 46-114. Wallingford, UK: CABI. [Google Scholar]
- 207.Gibson WC, Lom J, Pecková H, Ferris VR, Hamilton PB. 2005. Phylogenetic analysis of freshwater fish trypanosomes from Europe using SSU rRNA gene sequences and random amplification of polymorphic DNA. Parasitology 130, 405-412. ( 10.1017/S0031182004006778) [DOI] [PubMed] [Google Scholar]
- 208.Lemos M, Fermino BR, Simas-Rodrigues C, Hoffmann L, Silva R, Camargo EP, Teixeira MMG, Souto-Padrón T. 2015. Phylogenetic and morphological characterization of trypanosomes from Brazilian armoured catfishes and leeches reveal high species diversity, mixed infections and a new fish trypanosome species. Parasit. Vectors 8, 573-589. ( 10.1186/s13071-015-1193-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Mayer AFJK. 1843. Spicilegium observationum anatomicarum de organo electrico in raiis anelectricis et de haematozois. Bonnae Caroli Georgii.
- 210.Gruby D. 1843. Recherches et observations sur une nouvelle espèce d'hématozoaire, Trypanosoma sanguinis. C. R. Hebd. Séances Acad. Sci. Paris 17, 1134-1136. [Google Scholar]
- 211.International Commission on Zoological Nomenclature. 1926. Opinion 95. Two generic names of Protozoa placed in the Official List of Generic Names. Smithsonian Misc. Collect. 73, 14-15. [Google Scholar]
- 212.Baker JR. 1963. Speculations on the evolution of the family Trypanosomatidae Doflein, 1901. Exp. Parasitol. 13, 219-233. ( 10.1016/0014-4894(63)90074-2) [DOI] [PubMed] [Google Scholar]
- 213.Hoare CA. 1967. Evolutionary trends in mammalian trypanosomes. Adv. Parasitol. 5, 47-91. ( 10.1016/S0065-308X(08)60375-9) [DOI] [PubMed] [Google Scholar]
- 214.Votýpka J, Lukeš J, Oborník M. 2004. Phylogenetic relationship of Trypanosoma corvi with other avian trypanosomes. Acta Protozool. 43, 225-231. [Google Scholar]
- 215.Votýpka J, Szabová J, Rádrová J, Zídková L, Svobodová M. 2012. Trypanosoma culicavium sp. nov., an avian trypanosome transmitted by Culex mosquitoes. Int. J. Syst. Evol. Microbiol. 62, 745-754. ( 10.1099/ijs.0.032110-0) [DOI] [PubMed] [Google Scholar]
- 216.Zídková L, Čepička I, Szabová J, Svobodová M. 2012. Biodiversity of avian trypanosomes. Infect. Genet. Evol. 12, 102-112. ( 10.1016/j.meegid.2011.10.022) [DOI] [PubMed] [Google Scholar]
- 217.Šlapeta J, Morin-Adeline V, Thompson P, McDonell D, Shiels M, Gilchrist K, Votýpka J, Vogelnest L. 2016. Intercontinental distribution of a new trypanosome species from Australian endemic regent honeyeater (Anthochaera phrygia). Parasitology 143, 1012-1025. ( 10.1017/S0031182016000329) [DOI] [PubMed] [Google Scholar]
- 218.Schaudinn F. 1904. Generations-und Wirtswechsel bei Trypanosoma und Spirochaete. Arbeiten aus dem Kaiserl. Gesundheitsamte 20, 566-573. [Google Scholar]
- 219.Sehgal RNM, Valkiūnas G, Iezhova TA, Smith TB. 2006. Blood parasites of chickens in Uganda and Cameroon with molecular descriptions of Leucocytozoon schoutedeni and Trypanosoma gallinarum. J. Parasitol. 92, 1336-1343. ( 10.1645/GE-927R.1) [DOI] [PubMed] [Google Scholar]
- 220.Danilewsky B. 1885. Zur parasitologie des blutes. Biol. Z. 5, 529-537. [Google Scholar]
- 221.Laveran MA. 1903. Sur un trypanosome d'une chouette. C. R. Séances Mém. Soc. Biol. Filial 55, 528-530. [Google Scholar]
- 222.Baker JR. 1976. Biology of the trypanosomes of birds. In Biology of the Kinetoplastida, vol. 1 (eds Lumsden WHR, Evans DA), pp. 131-174. London, UK: Academic Press. [Google Scholar]
- 223.Nandi NC, Bennett GF. 1994. Re-description of Trypanosoma corvi Stephens and Christophers, 1908 emend. Baker, 1976 and remarks on the trypanosomes of the avian family Corvidae. Mem. Inst. Oswaldo Cruz 89, 145-151. ( 10.1590/s0074-02761994000200005) [DOI] [Google Scholar]
- 224.Valkiūnas G, Iezhova TA, Carlson JS, Sehgal RNM. 2011. Two new Trypanosoma species from African birds, with notes on the taxonomy of avian trypanosomes. J. Parasitol. 97, 924-930. ( 10.1645/GE-2796.1) [DOI] [PubMed] [Google Scholar]
- 225.Sehgal RNM, Iezhova TA, Marzec T, Valkiūnas G. 2015. Trypanosoma naviformis sp. nov. (Kinetoplastidae: Trypanosomatidae) from widespread African songbirds, the olive sunbird (Cyanomitra olivacea) and yellow-whiskered greenbul (Andropadus latirostris). Zootaxa 4034, 342-250. ( 10.11646/zootaxa.4034.2.6) [DOI] [PubMed] [Google Scholar]
- 226.Lun ZR, et al. 2015. Resistance to normal human serum reveals Trypanosoma lewisi as an underestimated human pathogen. Mol. Biochem. Parasitol. 199, 58-61. ( 10.1016/j.molbiopara.2015.03.007) [DOI] [PubMed] [Google Scholar]
- 227.Maia da Silva F, Marcili A, Ortiz PA, Epiphanio S, Campaner M, Catão-Dias JL, Shaw JJ, Camargo EP, Teixeira MMG. 2010. Phylogenetic, morphological and behavioural analyses support host switching of Trypanosoma (Herpetosoma) lewisi from domestic rats to primates. Infect. Genet. Evol. 10, 522-529. ( 10.1016/j.meegid.2010.02.005) [DOI] [PubMed] [Google Scholar]
- 228.Ortiz PA, et al. 2018. Diagnosis and genetic analysis of the worldwide distributed Rattus-borne Trypanosoma (Herpetosoma) lewisi and its allied species in blood and fleas of rodents. Infect. Genet. Evol. 63, 380-390. ( 10.1016/j.meegid.2017.09.001) [DOI] [PubMed] [Google Scholar]
- 229.Egan SL, Taylor CL, Austen JM, Banks PB, Ahlstrom LA, Ryan UM, Irwin PJ, Oskam CL. 2020. Molecular identification of the Trypanosoma (Herpetosoma) lewisi clade in black rats (Rattus rattus) from Australia. Parasitol. Res. 119, 1691-1696. ( 10.1007/s00436-020-06653-z) [DOI] [PubMed] [Google Scholar]
- 230.Mafie E, Saito-Ito A, Kasai M, Hatta M, Rivera PT, Ma XH, Chen ER, Sato H, Takada N. 2019. Integrative taxonomic approach of trypanosomes in the blood of rodents and soricids in Asian countries, with the description of three new species. Parasitol. Res. 118, 97-109. ( 10.1007/s00436-018-6120-3) [DOI] [PubMed] [Google Scholar]
- 231.García HA, Blanco PA, Rodrigues AC, Rodrigues CMF, Takata CSA, Campaner M, Camargo EP, Teixeira MMG. 2020. Pan-American Trypanosoma (Megatrypanum) trinaperronei n. sp. in the white-tailed deer Odocoileus virginianus Zimmermann and its deer ked Lipoptena mazamae Rondani, 1878: morphological, developmental and phylogeographical characterisation. Parasit. Vectors 13, 308. ( 10.1186/s13071-020-04169-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kingston N, Bobek B, Perzanowski K, Wita I, Maki L. 1992. Description of Trypanosoma (Megatrypanum) stefanskii sp. n. from roe deer (Capreolus capreolus) in Poland. J. Helminthol. Soc. Washington 59, 89-95. [Google Scholar]
- 233.Bruce D, Hamerton AE, Bateman HR, Mackie FP. 1909. Trypanosoma ingens, n. sp. Proc. R. Soc. Lond. B 81, 323-324. ( 10.1098/rspb.1909.0030) [DOI] [Google Scholar]
- 234.Weinman D. 1972. Trypanosoma cyclops n. sp.: a pigmented trypanosome from the Malaysian primates Macaca nemestrina and M. ira. Trans. R Soc. Trop. Med. Hyg. 66, 628-636. ( 10.1016/0035-9203(72)90309-4) [DOI] [PubMed] [Google Scholar]
- 235.Weinman D, White EA, Antipa GA. 1984. Trypanosoma lucknowi, a new species of trypanosome from Macaca mulatta with observations on its fine structure. J. Protozool. 31, 429-433. ( 10.1111/j.1550-7408.1984.tb02990.x) [DOI] [PubMed] [Google Scholar]
- 236.Stevens J, Noyes H, Gibson W. 1998. The evolution of trypanosomes infecting humans and primates. Mem. Inst. Oswaldo Cruz 93, 669-676. ( 10.1590/S0074-02761998000500019) [DOI] [PubMed] [Google Scholar]
- 237.Clément L, et al. 2020. Out of Africa: the origins of the protozoan blood parasites of the Trypanosoma cruzi clade found in bats from Africa. Mol. Phylogenet. Evol. 145, 106705. ( 10.1016/j.ympev.2019.106705) [DOI] [PubMed] [Google Scholar]
- 238.Hamilton PB, Stevens JR. 2017. Classification and phylogeny of Trypanosoma cruzi. In American trypanosomiasis Chagas disease: one hundred years of research (eds Telleria J, Tibayrenc M), pp. 321-344. Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 239.Espinosa-Álvarez O, et al. 2018. Trypanosoma rangeli is phylogenetically closer to Old World trypanosomes than to Trypanosoma cruzi. Int. J. Parasitol. 48, 569-584. ( 10.1016/j.ijpara.2017.12.008) [DOI] [PubMed] [Google Scholar]
- 240.Telleria J, Tibayrenc M. 2017. American trypanosomiasis Chagas disease: one hundred years of research, 2nd edn. Amsterdam, The Netherlands: Elsevier. [Google Scholar]
- 241.Lima L, et al. 2015. Genetic diversity of Trypanosoma cruzi in bats, and multilocus phylogenetic and phylogeographical analyses supporting Tcbat as an independent DTU (discrete typing unit). Acta Trop. 151, 166-177. ( 10.1016/j.actatropica.2015.07.015) [DOI] [PubMed] [Google Scholar]
- 242.Adams ER, Hamilton PB, Rodrigues AC, Malele II, Delespaux V, Teixeira MMG, Gibson W. 2010. New Trypanosoma (Duttonella) vivax genotypes from tsetse flies in East Africa. Parasitology 137, 641-650. ( 10.1017/S0031182009991508) [DOI] [PubMed] [Google Scholar]
- 243.Votýpka J, et al. 2015. A tsetse and tabanid fly survey of African great apes habitats reveals the presence of a novel trypanosome lineage but the absence of Trypanosoma brucei. Int. J. Parasitol. 45, 741-748. ( 10.1016/j.ijpara.2015.06.005) [DOI] [PubMed] [Google Scholar]
- 244.Rodrigues CMF, et al. 2020. Expanding our knowledge on African trypanosomes of the subgenus Pycnomonas: a novel Trypanosoma suis-like in tsetse flies, livestock and wild ruminants sympatric with Trypanosoma suis in Mozambique. Infect. Genet. Evol. 78, 104143. ( 10.1016/j.meegid.2019.104143) [DOI] [PubMed] [Google Scholar]
- 245.Austen JM, Jefferies R, Friend JA, Ryan U, Adams P, Reid SA. 2009. Morphological and molecular characterization of Trypanosoma copemani n. sp. (Trypanosomatidae) isolated from Gilbert's potoroo (Potorous gilbertii) and quokka (Setonix brachyurus) . Parasitology 136, 783-792. ( 10.1017/S0031182009005927) [DOI] [PubMed] [Google Scholar]
- 246.McInnes LM, Hanger J, Simmons G, Reid SA, Ryan UM. 2011. Novel trypanosome Trypanosoma gilletti sp. (Euglenozoa: Trypanosomatidae) and the extension of the host range of Trypanosoma copemani to include the koala (Phascolarctos cinereus). Parasitology 138, 59-70. ( 10.1017/S0031182010000971) [DOI] [PubMed] [Google Scholar]
- 247.Thompson CK, Botero A, Wayne AF, Godfrey SS, Lymbery AJ, Thompson RCA. 2013. Morphological polymorphism of Trypanosoma copemani and description of the genetically diverse T. vegrandis sp. nov. from the critically endangered Australian potoroid, the brush-tailed bettong (Bettongia penicillata (Gray, 1837)). Parasit. Vectors 6, 121. ( 10.1186/1756-3305-6-121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Cooper C, Clode PL, Peacock C, Thompson RCA. 2017. Host–parasite relationships and life histories of trypanosomes in Australia. Adv. Parasitol. 97, 47-109. ( 10.1016/bs.apar.2016.06.001) [DOI] [PubMed] [Google Scholar]
- 249.Krige AS, Thompson RCA, Clode PL. 2019. ‘Hang on a Tick’—are ticks really the vectors for Australian trypanosomes? Trends Parasitol. 35, 596-606. ( 10.1016/j.pt.2019.05.008) [DOI] [PubMed] [Google Scholar]
- 250.Cooper C, Thompson RCA, Rigby P, Buckley A, Peacock C, Clode PL. 2018. The marsupial trypanosome Trypanosoma copemani is not an obligate intracellular parasite, although it adversely affects cell health. Parasit. Vectors 11, 521. ( 10.1186/s13071-018-3092-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Novy FG. 1906. The trypanosomes of tsetse flies. J. Infect. Dis. 3, 394-411. ( 10.1093/infdis/3.3.394) [DOI] [Google Scholar]
- 252.Hoare CA. 1931. Studies on Trypanosoma grayi. III. Life-cycle in the tsetse-fly and in the crocodile. Parasitology 23, 449-484. ( 10.1017/S0031182000013858) [DOI] [Google Scholar]
- 253.Hoare CA. 1929. Studies on Trypanosoma grayi. II. Experimental transmission to the crocodile. Trans. R. Soc. Trop. Med. Hyg. 23, 39-56. ( 10.1016/S0035-9203(29)90831-2) [DOI] [Google Scholar]
- 254.Fermino BR, et al. 2013. The phylogeography of trypanosomes from South American alligatorids and African crocodilids is consistent with the geological history of South American river basins and the transoceanic dispersal of Crocodylus at the Miocene. Parasit. Vectors 6, 313-327. ( 10.1186/1756-3305-6-313) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Fermino BR, et al. 2019. Shared species of crocodilian trypanosomes carried by tabanid flies in Africa and South America, including the description of a new species from caimans, Trypanosoma kaiowa n. sp. Parasit. Vectors 12, 225. ( 10.1186/s13071-019-3463-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Wenyon CM. 1908. Report of travelling pathologist and protozoologist. In Third report of the Wellcome Research Laboratories at the Gordon Memorial College, Khartoum (ed. Balfour A), pp. 121-168. London, UK: Bailliere, Tindall and Cox. [Google Scholar]
- 257.Sato H, Takano A, Kawabata H, Une Y, Watanabe H, Mukhtar MM. 2009. Trypanosoma cf. varani in an imported ball python (Python reginus) from Ghana. J. Parasitol. 95, 1029-1033. ( 10.1645/GE-1816.1) [DOI] [PubMed] [Google Scholar]
- 258.Viola LB, Attias M, Takata CSA, Campaner M, de Souza W, Camargo EP, Teixeira MMG. 2009. Phylogenetic analyses based on small subunit rRNA and glycosomal glyceraldehyde-3-phosphate dehydrogenase genes and ultrastructural characterization of two snake trypanosomes: Trypanosoma serpentis n. sp. from Pseudoboa nigra and Trypanosoma cascavelli from Crotalus durissus terrificus. J. Eukaryot. Microbiol. 56, 594-602. ( 10.1111/j.1550-7408.2009.00444.x) [DOI] [PubMed] [Google Scholar]
- 259.Ayala SC. 1970. Two new trypanosomes from California toads and lizards. J. Protozool. 17, 370-373. ( 10.1111/j.1550-7408.1970.tb04696.x) [DOI] [Google Scholar]
- 260.Pessôa SB, de Biasi P. 1972. Trypanosoma cascavelli sp. n. parasita da cascavel: Crotalus durissus terrificus (Laurenti). Atas Soc. Biol. Rio de Janeiro 15, 67-70. [Google Scholar]
- 261.Rodrigues MS, Lima L, Xavier SC das C, Herrera HM, Rocha FL, Roque ALR, Teixeira MMG, Jansen AM. 2019. Uncovering Trypanosoma spp. diversity of wild mammals by the use of DNA from blood clots. Int. Parasitol. Parasit. Wildl. 8, 171-181. ( 10.1016/j.ijppaw.2019.02.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Rêgo SFM, Magalhães AEA, Siqueira AF. 1957. Um novo tripanossomo do gambá, Trypanosoma freitasi n. sp. Rev. Brasil Malaria 9, 277-284. [Google Scholar]
- 263.Ferreira JIGS, da Costa AP, Nunes PH, Ramirez D, Fournier GFR, Saraiva D, Tonhosolo R, Marcili A. 2017. New Trypanosoma species, Trypanosoma gennarii sp. nov., from South American marsupial in Brazilian Cerrado. Acta Trop. 176, 249-255. ( 10.1016/j.actatropica.2017.08.018) [DOI] [PubMed] [Google Scholar]
- 264.Naiff RD, Barrett TV. 2013. Trypanosoma (Megatrypanum) lainsoni n. sp. from Mesomys hispidus (Rodentia: Echimyidae) in Brazil: trypomastigotes described from experimentally infected laboratory mice. Parasite 20, 51-56. ( 10.1051/parasite/2013049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.McInnes LM, Gillett A, Ryan UM, Austen J, Campbell RSF, Hanger J, Reid SA. 2009. Trypanosoma irwini n. sp. (Sarcomastigophora: Trypanosomatidae) from the koala (Phascolarctos cinereus). Parasitology 136, 875-885. ( 10.1017/S0031182009006313) [DOI] [PubMed] [Google Scholar]
- 266.Ortiz-Baez AS, et al. 2020. Meta-transcriptomic identification of Trypanosoma spp. in native wildlife species from Australia. Parasit. Vectors 13, 447. ( 10.1186/s13071-020-04325-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Peirce MA, Neal C. 1974. Trypanosoma (Megatrypanum) pestanai in British badgers (Meles meles). Int. J. Parasitol. 4, 439-440. ( 10.1016/0020-7519(74)90055-1) [DOI] [PubMed] [Google Scholar]
- 268.Ideozu EJ, Whiteoak AM, Tomlinson AJ, Robertson A, Delahay RJ, Hide G. 2015. High prevalence of trypanosomes in European badgers detected using ITS-PCR. Parasit. Vectors 8, 480-485. ( 10.1186/s13071-015-1088-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Dyachenko V, Steinmann M, Bangoura B, Selzer M, Munderloh U, Daugschies A, Barutzki D. 2017. Co-infection of Trypanosoma pestanai and Anaplasma phagocytophilum in a dog from Germany. Vet. Parasitol. Reg. Stud. Rep. 9, 110-114. ( 10.1016/j.vprsr.2017.06.001) [DOI] [PubMed] [Google Scholar]
- 270.Acosta IDCL, da Costa AP, Nunes PH, Gondim MFN, Gatti A, Rossi JLJ, Gennari SM, Marcili A. 2013. Morphological and molecular characterization and phylogenetic relationships of a new species of trypanosome in Tapirus terrestris (lowland tapir), Trypanosoma terrestris sp. nov., from Atlantic Rainforest of southeastern Brazil. Parasit. Vectors 6, 349-361. ( 10.1186/1756-3305-6-349) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Kostygov AY, Yurchenko V. 2017. Revised classification of the subfamily Leishmaniinae (Trypanosomatidae). Folia Parasitol. 64, 020. ( 10.14411/fp.2017.020) [DOI] [PubMed] [Google Scholar]
- 272.Schwarz RS, Bauchan GR, Murphy CA, Ravoet J, de Graaf DC, Evans JD. 2015. Characterization of two species of Trypanosomatidae from the honey bee Apis mellifera: Crithidia mellificae Langridge and McGhee, and Lotmaria passim n. gen., n. sp. J. Eukaryot. Microbiol. 62, 567-583. ( 10.1111/jeu.12209) [DOI] [PubMed] [Google Scholar]
- 273.Zídková L, Čepička I, Votýpka J, Svobodová M. 2010. Herpetomonas trimorpha sp. nov. (Trypanosomatidae, Kinetoplastida), a parasite of the biting midge Culicoides truncorum (Ceratopogonidae, Diptera). Int. J. Syst. Evol. Microbiol. 60, 2236-2246. ( 10.1099/ijs.0.014555-0) [DOI] [PubMed] [Google Scholar]
- 274.Votýpka J, Kostygov AY, Kraeva N, Grybchuk-Ieremenko A, Tesařová M, Grybchuk D, Lukeš J, Yurchenko V. 2014. Kentomonas gen. n., a new genus of endosymbiont-containing trypanosomatids of Strigomonadinae subfam. n. Protist 165, 825-838. ( 10.1016/j.protis.2014.09.002) [DOI] [PubMed] [Google Scholar]
- 275.Kostygov AY, Dobáková E, Grybchuk-Ieremenko A, Váhala D, Maslov DA, Votýpka J, Lukeš J, Yurchenko V. 2016. Novel trypanosomatid-bacterium association: evolution of endosymbiosis in action. mBio 7, e01985-15. ( 10.1128/mBio.01985-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Klatt S, Simpson L, Maslov DA, Konthur Z. 2019. Leishmania tarentolae: taxonomic classification and its application as a promising biotechnological expression host. PLoS Negl. Trop. Dis. 13, e0007424. ( 10.1371/journal.pntd.0007424) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Jariyapan N, et al. 2018. Leishmania (Mundinia) orientalis n. sp. (Trypanosomatidae), a parasite from Thailand responsible for localised cutaneous leishmaniasis. Parasit. Vectors 11, 351. ( 10.1186/s13071-018-2908-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Šeblová V, Sádlová J, Vojtková B, Votýpka J, Carpenter S, Bates PA, Volf P. 2015. The biting midge Culicoides sonorensis (Diptera: Ceratopogonidae) is capable of developing late stage infections of Leishmania enriettii. PLoS Negl. Trop. Dis. 9, e0004060. ( 10.1371/journal.pntd.0004060) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Herrer A. 1971. Leishmania hertigi sp. n., from the tropical porcupine, Coendou rothschildi Thomas. J. Parasitol. 57, 626-629. ( 10.2307/3277928) [DOI] [PubMed] [Google Scholar]
- 280.Lainson R, Shaw JJ. 1977. Leishmanias of neotropical porcupines: Leishmania hertigi deanei nov. subsp. Acta Amazonica 7, 51-57. ( 10.1590/1809-43921977071051) [DOI] [Google Scholar]
- 281.Cupolillo E, Medina-Acosta E, Noyes H, Momen H, Grimaldi GJ. 2000. A revised classification for Leishmania and Endotrypanum. Parasitol. Today 16, 142-144. ( 10.1016/s0169-4758(99)01609-9) [DOI] [PubMed] [Google Scholar]
- 282.Yurchenko V, Kostygov A, Havlová J, Grybchuk-Ieremenko A, Ševčíková T, Lukeš J, Ševčík J, Votýpka J. 2016. Diversity of trypanosomatids in cockroaches and the description of Herpetomonas tarakana sp. n. J. Eukaryot. Microbiol. 63, 198-209. ( 10.1111/jeu.12268) [DOI] [PubMed] [Google Scholar]
- 283.Borghesan TC, Ferreira RC, Takata CSA, Campaner M, Borda CC, Paiva F, Milder RV, Teixeira MMG, Camargo EP. 2013. Molecular phylogenetic redefinition of Herpetomonas (Kinetoplastea, Trypanosomatidae), a genus of insect parasites associated with flies. Protist 164, 129-152. ( 10.1016/j.protis.2012.06.001) [DOI] [PubMed] [Google Scholar]
- 284.Yoshida N, Freymüller E, Wallace FG. 1978. Herpetomonas mariadeanei sp. n. (Protozoa, Trypanosomatidae) from Muscina stabulans (Falléen, 1816) (Diptera, Muscidae). J. Protozool. 25, 421-425. ( 10.1111/j.1550-7408.1978.tb04161.x) [DOI] [Google Scholar]
- 285.Teixeira MMG, et al. 2011. Phylogenetic validation of the genera Angomonas and Strigomonas of trypanosomatids harboring bacterial endosymbionts with the description of new species of trypanosomatids and of proteobacterial symbionts. Protist 162, 503-524. ( 10.1016/j.protis.2011.01.001) [DOI] [PubMed] [Google Scholar]
- 286.Lukeš J, Tesařová M, Yurchenko V, Votýpka J. 2021. Characterization of a new cosmopolitan genus of trypanosomatid parasites, Obscuromonas n. gen. Eur. J. Protistol 2021, 125778. ( 10.1016/j.ejop.2021.125778) [DOI] [PubMed] [Google Scholar]
- 287.Záhonová K, Kostygov AY, Ševčíková T, Yurchenko V, Eliáš M. 2016. An unprecedented non-canonical nuclear genetic code with all three termination codons reassigned as sense codons. Curr. Biol. 26, 2364-2369. ( 10.1016/j.cub.2016.06.064) [DOI] [PubMed] [Google Scholar]
- 288.Votýpka J, Suková E, Kraeva N, Ishemgulova A, Duží I, Lukeš J, Yurchenko V. 2013. Diversity of trypanosomatids (Kinetoplastea: Trypanosomatidae) parasitizing fleas (Insecta: Siphonaptera) and description of a new genus Blechomonas gen. n. Protist 164, 763-781. ( 10.1016/j.protis.2013.08.002) [DOI] [PubMed] [Google Scholar]
- 289.Flegontov P, et al. 2013. Paratrypanosoma is a novel early-branching trypanosomatid. Curr. Biol. 23, 1787-1793. ( 10.1016/j.cub.2013.07.045) [DOI] [PubMed] [Google Scholar]
- 290.Skalický T, et al. 2017. Extensive flagellar remodeling during the complex life cycle of Paratrypanosoma, an early-branching trypanosomatid. Proc. Natl Acad. Sci. USA 114, 11 757-11 762. ( 10.1073/pnas.1712311114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Kostygov AY, et al. 2020. Vickermania gen. nov., trypanosomatids that use two joined flagella to resist midgut peristaltic flow within the fly host. BMC Biol. 18, 187. ( 10.1186/s12915-020-00916-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Svobodová M, Zídková L, Čepička I, Oborník M, Lukeš J, Votýpka J. 2007. Sergeia podlipaevi gen. nov., sp. nov. (Trypanosomatidae, Kinetoplastida), a parasite of biting midges (Ceratopogonidae, Diptera). Int. J. Syst. Evol. Microbiol. 57, 423-432. ( 10.1099/ijs.0.64557-0) [DOI] [PubMed] [Google Scholar]
- 293.Yurchenko V, Votýpka J, Tesařová M, Klepetková H, Kraeva N, Jirků M, Lukeš J. 2014. Ultrastructure and molecular phylogeny of four new species of monoxenous trypanosomatids from flies (Diptera: Brachycera) with redefinition of the genus Wallaceina. Folia Parasitol. 61, 97-112. ( 10.14411/fp.2014.023) [DOI] [PubMed] [Google Scholar]
- 294.Kostygov AY, Grybchuk-Ieremenko A, Malysheva MN, Frolov AO, Yurchenko V. 2014. Molecular revision of the genus Wallaceina. Protist 165, 594-604. ( 10.1016/j.protis.2014.07.001) [DOI] [PubMed] [Google Scholar]
- 295.Roubaud E. 1911. Cercoplasma (n. gen.) caulleryi (n. sp.); nouveau flagellé à formes trypanosomiennes de l'intestin d’Auchmeromyia luteola Fabr. (Muscide). C. R. Séances Soc. Biol. 71, 503-505. [Google Scholar]
- 296.Nicoli RM, Penaud A, Timon-David P. 1971. Rechèrches systématiques sur les trypanosomides. II. Le genre Malacozoomonas n. gen. Bull. Soc. zool. France 96, 415-419. [Google Scholar]
- 297.Nicoli RM, Penaud A, Timon-David P. 1971. Rechèrches systématiques sur les trypanosomides. I. Le genre Nematodomonas n. gen. Bull. Soc. zool. France 96, 405-415. [Google Scholar]
- 298.Page AM, Canning EU, Barker RJ, Nicholas JP. 1986. A new species of Rhynchoidomonas Patton, 1910 (Kinetoplastida: Trypanosomatina) from Operophtera brumata (Lepidoptera: Geometridae). Syst. Parasitol. 8, 101-105. ( 10.1007/BF00009866) [DOI] [Google Scholar]
- 299.Cachon J, Cachon M, Charnier M. 1972. Ultrastructure du bodonidé Trypanophis grobbeni Poche, parasite des siphonophores. Protistologica 8, 223-236. [Google Scholar]
- 300.Larsen J, Patterson DJ. 1990. Some flagellates (Protista) from tropical marine sediments. J. Nat. Hist. 24, 801-937. ( 10.1080/00222939000770571) [DOI] [Google Scholar]
- 301.Patterson DJ, Vørs N, Simpson AGB, O'Kelly C. 2000. Residual free-living and predatory heterotrophic flagellates. In An illustrated guide to the protozoa (eds Lee JJ, Leedale GF, Bradbury P), pp. 1302-1328. Lawrence, KS: Society of Protozoologists/Allen Press. [Google Scholar]
- 302.Lara E, Moreira D, Vereshchaka A, López-García P. 2009. Pan-oceanic distribution of new highly diverse clades of deep-sea diplonemids. Environ. Microbiol. 11, 47-55. ( 10.1111/j.1462-2920.2008.01737.x) [DOI] [PubMed] [Google Scholar]
- 303.de Vargas C, et al. 2015. Ocean plankton. Eukaryotic plankton diversity in the sunlit ocean. Science 348, 1261605. ( 10.1126/science.1261605) [DOI] [PubMed] [Google Scholar]
- 304.López-García P, Vereshchaka A, Moreira D. 2007. Eukaryotic diversity associated with carbonates and fluid–seawater interface in Lost City hydrothermal field. Environ. Microbiol. 9, 546-554. ( 10.1111/j.1462-2920.2006.01158.x) [DOI] [PubMed] [Google Scholar]
- 305.López-García P, Rodríguez-Valera F, Pedrós-Alió C, Moreira D. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409, 603-607. ( 10.1038/35054537) [DOI] [PubMed] [Google Scholar]
- 306.Morgan-Smith D, Clouse MA, Herndl GJ, Bochdansky AB. 2013. Diversity and distribution of microbial eukaryotes in the deep tropical and subtropical North Atlantic Ocean. Deep Sea Res. Part I 78, 58-69. ( 10.1016/j.dsr.2013.04.010) [DOI] [Google Scholar]
- 307.Massana R, et al. 2015. Marine protist diversity in European coastal waters and sediments as revealed by high-throughput sequencing. Environ. Microbiol. 17, 4035-4049. ( 10.1111/1462-2920.12955) [DOI] [PubMed] [Google Scholar]
- 308.Pernice MC, Giner CR, Logares R, Perera-Bel J, Acinas SG, Duarte CM, Gasol JM, Massana R. 2016. Large variability of bathypelagic microbial eukaryotic communities across the world's oceans. ISME J. 10, 945-958. ( 10.1038/ismej.2015.170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Eloe EA, Shulse CN, Fadrosh DW, Williamson SJ, Allen EE, Bartlett DH. 2011. Compositional differences in particle-associated and free-living microbial assemblages from an extreme deep-ocean environment. Environ. Microbiol. Rep. 3, 449-458. ( 10.1111/j.1758-2229.2010.00223.x) [DOI] [PubMed] [Google Scholar]
- 310.Okamoto N, Gawryluk RMR, del Campo J, Strassert JFH, Lukeš J, Richards TA, Worden AZ, Santoro AE, Keeling PJ. 2019. A revised taxonomy of diplonemids including the Eupelagonemidae n. fam. and a type species, Eupelagonema oceanica n. gen. & sp. J. Eukaryot. Microbiol. 66, 519-524. ( 10.1111/jeu.12679) [DOI] [PubMed] [Google Scholar]
- 311.Tashyreva D, et al. 2018. Phylogeny and morphology of new diplonemids from Japan. Protist 169, 158-179. ( 10.1016/j.protis.2018.02.001) [DOI] [PubMed] [Google Scholar]
- 312.Takishita K, Kakizoe N, Yoshida T, Maruyama T. 2010. Molecular evidence that phylogenetically diverged ciliates are active in microbial mats of deep-sea cold-seep sediment. J. Eukaryot. Microbiol. 57, 76-86. ( 10.1111/j.1550-7408.2009.00457.x) [DOI] [PubMed] [Google Scholar]
- 313.Al-Qassab S, Lee WJ, Murray S, Simpson AGB, Patterson DJ. 2002. Flagellates from stromatolites and surrounding sediments in Shark Bay, Western Australia. Acta Protozool. 41, 91-144. [Google Scholar]
- 314.Schnepf E. 1994. Light and electron microscopical observations in Rhynchopus coscinodiscivorus spec. nov., a colorless, phagotrophic euglenozoon with concealed flagella. Arch. Protistenk. 144, 63-74. ( 10.1016/S0003-9365(11)80225-3) [DOI] [Google Scholar]
- 315.Griessmann K. 1914. Über marine Flagellaten. Arch. Protistenk. 32, 1-78. [Google Scholar]
- 316.Porter D. 1973. Isonema papillatum sp. n., a new colorless marine flagellate: a light- and electron microscopic study. J. Protozool. 20, 351-356. ( 10.1111/j.1550-7408.1973.tb00895.x) [DOI] [Google Scholar]
- 317.Schuster FL, Goldstein S, Hershenov B. 1968. Ultrastructure of a flagellate, Isonema nigricans nov. gen. nov. sp., from a polluted marine habitat. Protistologica 4, 141-149. [Google Scholar]
- 318.Skuja H. 1948. Taxonomie des Phytoplanktons einiger Seen in Uppland, Schweden. Symb. Bot. Ups. 9, 1-399. [Google Scholar]
- 319.Triemer RE, Ott DW. 1990. Ultrastructure of Diplonema ambulator Larsen & Patterson (Euglenozoa) and its relationship to Isonema. Eur. J. Protistol 25, 316-320. ( 10.1016/S0932-4739(11)80123-9) [DOI] [PubMed] [Google Scholar]
- 320.Yi Z, Berney C, Hartikainen H, Mahamdallie S, Gardner M, Boenigk J, Cavalier-Smith T, Bass D. 2017. High-throughput sequencing of microbial eukaryotes in Lake Baikal reveals ecologically differentiated communities and novel evolutionary radiations. FEMS Microbiol. Ecol. 93, fix073. ( 10.1093/femsec/fix073) [DOI] [PubMed] [Google Scholar]
- 321.Mukherjee I, Hodoki Y, Okazaki Y, Fujinaga S, Ohbayashi K, Nakano SI. 2019. Widespread dominance of kinetoplastids and unexpected presence of diplonemids in deep freshwater lakes. Front. Microbiol. 10, 2375. ( 10.3389/fmicb.2019.02375) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Mukherjee I, et al. 2020. A freshwater radiation of diplonemids. Environ. Microbiol. 22, 4658-4668. ( 10.1111/1462-2920.15209) [DOI] [PubMed] [Google Scholar]
- 323.Elbrächter M, Schnepf E, Balzer I. 1996. Hemistasia phaeocysticola (Scherffel) comb. nov., redescription of a free-living, marine, phagotrophic kinetoplastid flagellate. Arch. Protistenk. 147, 125-136. ( 10.1016/s0003-9365(96)80028-5) [DOI] [Google Scholar]
- 324.Yabuki A, Tame A. 2015. Phylogeny and reclassification of Hemistasia phaeocysticola (Scherffel) Elbrächter & Schnepf, 1996. J. Eukaryot. Microbiol. 62, 426-429. ( 10.1111/jeu.12191) [DOI] [PubMed] [Google Scholar]
- 325.Prokopchuk G, Tashyreva D, Yabuki A, Horák A, Masařová P, Lukeš J. 2019. Morphological, ultrastructural, motility and evolutionary characterization of two new Hemistasiidae species. Protist 170, 259-282. ( 10.1016/j.protis.2019.04.001) [DOI] [PubMed] [Google Scholar]
- 326.Kent ML, Elston RA, Nerad TA, Sawyer TK. 1987. An Isonema-like flagellate (Protozoa: Mastigophora) infection in larval geoduck clams, Panope abrupta. J. Invertebr. Pathol. 50, 221-229. ( 10.1016/0022-2011(87)90086-3) [DOI] [PubMed] [Google Scholar]
- 327.Bodammer JE, Sawyer TK. 1981. Aufwuchs protozoa and bacteria on the gills of the rock crab, Cancer irroratus Say: a survey by light and electron microscopy. J. Protozool. 28, 35-46. ( 10.1111/j.1550-7408.1981.tb02801.x) [DOI] [Google Scholar]
- 328.Roy J, Faktorová D, Benada O, Lukeš J, Burger G. 2007. Description of Rhynchopus euleeides n. sp. (Diplonemea), a free-living marine euglenozoan. J. Eukaryot. Microbiol. 54, 137-145. ( 10.1111/j.1550-7408.2007.00244.x) [DOI] [PubMed] [Google Scholar]
- 329.Breglia SA, Yubuki N, Hoppenrath M, Leander BS. 2010. Ultrastructure and molecular phylogenetic position of a novel euglenozoan with extrusive episymbiotic bacteria: Bihospites bacati n. gen. et sp. (Symbiontida). BMC Microbiol. 10, 145. ( 10.1186/1471-2180-10-145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Lax G, Simpson AGB. 2013. Combining molecular data with classical morphology for uncultured phagotrophic euglenids (Excavata): a single-cell approach. J. Eukaryot. Microbiol. 60, 615-625. ( 10.1111/jeu.12068) [DOI] [PubMed] [Google Scholar]
- 331.Lee WJ, Simpson AGB. 2014. Ultrastructure and molecular phylogenetic position of Neometanema parovale sp. nov. (Neometanema gen. nov.), a marine phagotrophic euglenid with skidding motility. Protist 165, 452-472. ( 10.1016/j.protis.2014.05.001) [DOI] [PubMed] [Google Scholar]
- 332.Dietrich D, Arndt H. 2000. Biomass partitioning of benthic microbes in a Baltic inlet: relationships between bacteria, algae, heterotrophic flagellates and ciliates. Mar. Biol. 136, 309-322. ( 10.1007/s002270050689) [DOI] [Google Scholar]
- 333.Lee WJ, Patterson DJ. 2000. Heterotrophic flagellates (Protista) from marine sediments of Botany Bay, Australia. J. Nat. Hist. 34, 483-562. ( 10.1080/002229300299435) [DOI] [Google Scholar]
- 334.Schoenle A, Živaljić S, Prausse D, Voß J, Jakobsen K, Arndt H. 2019. New phagotrophic euglenids from deep sea and surface waters of the Atlantic Ocean (Keelungia nitschei, Petalomonas acorensis, Ploeotia costaversata). Eur. J. Protistol 69, 102-116. ( 10.1016/j.ejop.2019.02.007) [DOI] [PubMed] [Google Scholar]
- 335.Zimba PV, Rowan M, Triemer RE. 2004. Identification of euglenoid algae that produce ichthyotoxin(s). J. Fish Dis. 27, 115-117. ( 10.1046/j.1365-2761.2003.00512.x) [DOI] [PubMed] [Google Scholar]
- 336.Zimba PV, Moeller PD, Beauchesne K, Lane HE, Triemer RE. 2010. Identification of euglenophycin—a toxin found in certain euglenoids. Toxicon 55, 100-104. ( 10.1016/j.toxicon.2009.07.004) [DOI] [PubMed] [Google Scholar]
- 337.Valadez F, Rosiles-González G, Carmona J. 2010. Euglenophytes from Lake Chignahuapan, Mexico. Cryptogamie, Algologie 31, 305-319. [Google Scholar]
- 338.Rahman MS, Shahjahan M, Haque MM, Khan S. 2012. Control of euglenophyte bloom and fish production enhancement using duckweed and lime. Iran. J. Fish. Sci. 11, 602-617. [Google Scholar]
- 339.Lukešová S, Karlicki M, Tomečková Hadariová L, Szabová J, Karnkowska A, Hampl V. 2020. Analyses of environmental sequences and two regions of chloroplast genomes revealed the presence of new clades of photosynthetic euglenids in marine environments. Environ. Microbiol. Rep. 12, 78-91. ( 10.1111/1758-2229.12817) [DOI] [PubMed] [Google Scholar]
- 340.Brown PJP, Leander BS, Farmer MA. 2002. Redescription of Euglena rustica (Euglenophyceae), a rare euglenophyte from the intertidal zone. Phycologia 41, 445-452. ( 10.2216/i0031-8884-41-5-445.1) [DOI] [Google Scholar]
- 341.Lindholm T. 1995. Green water caused by Eutreptiella gymnastica (Euglenophyceae) in a stratified Baltic Sea inlet. In Harmful marine algal blooms (ed. Lassus P), pp. 181-186. Lavoisier: Intercept. [Google Scholar]
- 342.Stonik IV. 2007. Species of the genus Eutreptiella (Euglenophyceae) from Russian waters of East/Japan Sea. Ocean Sci. J. 42, 81-88. ( 10.1007/BF03020876) [DOI] [Google Scholar]
- 343.Buck KR, Barry JP, Simpson AGB. 2000. Monterey Bay cold seep biota: Euglenozoa with chemoautotrophic bacterial epibionts. Eur. J. Protistol 36, 117-126. ( 10.1016/S0932-4739(00)80029-2) [DOI] [Google Scholar]
- 344.Rocchetta I, Ruiz LB, Magaz G, Conforti VTD. 2003. Effects of hexavalent chromium in two strains of Euglena gracilis. Bull. Environ. Contam. Toxicol. 70, 1045-1051. ( 10.1007/s00128-003-0088-z) [DOI] [PubMed] [Google Scholar]
- 345.Rehman A, Shakoori FR, Shakoori AR. 2007. Heavy metal resistant Distigma proteus (Euglenophyta) isolated from industrial effluents and its possible role in bioremediation of contaminated wastewaters. World J. Microbiol. Biotechnol. 23, 753-758. ( 10.1007/s11274-006-9291-5) [DOI] [Google Scholar]
- 346.Kamika I, Momba MNB. 2013. Assessing the resistance and bioremediation ability of selected bacterial and protozoan species to heavy metals in metal-rich industrial wastewater. BMC Microbiol. 13, 28. ( 10.1186/1471-2180-13-28) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Dennington VN, George JJ, Wyborn CHE. 1975. The effects of oils on growth of freshwater phytoplankton. Environ. Pollut 8, 233-237. ( 10.1016/0013-9327(75)90105-6) [DOI] [Google Scholar]
- 348.Werner D, Pawlitz H. 1978. Differential elimination of phenol by diatoms and other unicellular algae from low concentrations. Bull. Environ. Contam. Toxicol. 20, 303-312. ( 10.1007/BF01683525) [DOI] [PubMed] [Google Scholar]
- 349.Poorman AE. 1973. Effects of pesticides on Euglena gracilis. I. Growth studies. Bull. Environ. Contam. Toxicol. 10, 25-28. ( 10.1007/BF01684750) [DOI] [Google Scholar]
- 350.Butler GL. 1977. Algae and pesticides. In Residue reviews, vol. 66 (ed. Gunther FA), pp. 19-62. New York, NY: Springer. [Google Scholar]
- 351.Lackey JB. 1968. Ecology of Euglena. In The biology of Euglena, vol. 1 (ed. Buetow DE), pp. 27-244. New York, NY: Academic Press. [Google Scholar]
- 352.Jones DT. 1944. Two protozoans from Great Salt Lake. Bull. Univ. Utah, Biol. Ser. 35, 1-10. [Google Scholar]
- 353.Lane AE, Burris JE. 1981. Effects of environmental pH on the internal pH of Chlorella pyrenoidosa, Scenedesmus quadricauda, and Euglena mutabilis. Plant Physiol. 68, 439-442. ( 10.1104/pp.68.2.439) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Sittenfeld A, et al. 2002. Characterization of a photosynthetic Euglena strain isolated from an acidic hot mud pool of a volcanic area of Costa Rica. FEMS Microbiol. Ecol. 42, 151-161. ( 10.1111/j.1574-6941.2002.tb01004.x) [DOI] [PubMed] [Google Scholar]
- 355.Yamaguchi A, Yubuki N, Leander BS. 2012. Morphostasis in a novel eukaryote illuminates the evolutionary transition from phagotrophy to phototrophy: description of Rapaza viridis n. gen. et sp. (Euglenozoa, Euglenida). BMC Evol. Biol. 12, 29. ( 10.1186/1471-2148-12-29) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Michajłow W. 1972. Euglenoidina parasitic in Copepoda: an outline monograph. Warsaw: PWN—Polish Scientific Publishers. [Google Scholar]
- 357.Wenrich DH. 1924. Studies on Euglenamorpha hegneri n. g., n. sp., a euglenoid flagellate found in tadpoles. Biol. Bull. (Woods Hole) 47, 149-175. ( 10.2307/1536494) [DOI] [Google Scholar]
- 358.Kisielewska G, Kolicka M, Zawierucha K. 2015. Prey or parasite? The first observations of live Euglenida in the intestine of Gastrotricha. Eur. J. Protistol 51, 138-141. ( 10.1016/j.ejop.2014.12.003) [DOI] [PubMed] [Google Scholar]
- 359.Hall SR. 1931. Observations on Euglena leucops, sp. nov., a parasite of Stenostomum, with special reference to nuclear division. Biol. Bull. 60, 327-344. ( 10.2307/1536878) [DOI] [Google Scholar]
- 360.Michajłow W. 1978. Dinema antarcticum sp. n., Dinema pseudoboeckellae sp. n. and other Euglenoidina-parasites of Pseudoboeckella silvestri (Calanoida) from the Antarctica. Bull. Acad. Pol. Sci., Ser. Sci. Biol. 26, 51-54. [Google Scholar]
- 361.Wita I, Sukhanova KM. 1986. Seasonal modifications in the life cycle of Parastasia fennica (Michajłow, 1966). Acta Protozool. 25, 365-374. [Google Scholar]
- 362.Al-Dhaheri RS, Willey RL. 1996. Colonization and reproduction of the epibiotic flagellate Colacium vesiculosum (Euglenophyceae) on Daphnia pulex. J. Phycol. 32, 770-774. ( 10.1111/j.0022-3646.1996.00770.x) [DOI] [Google Scholar]
- 363.Zalocar Y, Frutos SM, Casco SL, Forastier ME, Vallejos SV. 2011. Prevalence of Colacium vesiculosum (Colaciales: Euglenophyceae) on planktonic crustaceans in a subtropical shallow lake of Argentina. Rev. Biol. Trop. 59, 1295-1306. ( 10.15517/rbt.v0i0.3400) [DOI] [PubMed] [Google Scholar]
- 364.Płachno BJ, Wołowski K. 2008. Algae commensal community in Genlisea traps. Acta Soc. Bot. Pol. 77, 77-86. ( 10.5586/asbp.2008.011) [DOI] [Google Scholar]
- 365.Gordon E, Pacheco S. 2007. Prey composition in the carnivorous plants Utricularia inflata and U. gibba (Lentibulariaceae) from Paria Peninsula, Venezuela. Rev. Biol. Trop. 55, 795-803. ( 10.15517/rbt.v55i3-4.5956) [DOI] [PubMed] [Google Scholar]
- 366.Simon M, Jardillier L, Deschamps P, Moreira D, Restoux G, Bertolino P, López-García P. 2014. Complex communities of small protists and unexpected occurrence of typical marine lineages in shallow freshwater systems. Environ. Microbiol. 17, 3610-3627. ( 10.1111/1462-2920.12591) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Forster D, et al. 2016. Benthic protists: the under-charted majority. FEMS Microbiol. Ecol. 92, fiw120. ( 10.1093/femsec/fiw120) [DOI] [PubMed] [Google Scholar]
- 368.Geisen S, Vaulot D, Mahé F, Lara E, de Vargas C, Bass D. 2019. A user guide to environmental protistology: primers, metabarcoding, sequencing, and analyses. bioRxiv, 850610. ( 10.1101/850610) [DOI]
- 369.Busse I, Preisfeld A. 2002. Unusually expanded SSU ribosomal DNA of primary osmotrophic euglenids: molecular evolution and phylogenetic inference. J. Mol. Evol. 55, 757-767. ( 10.1007/s00239-002-2371-8) [DOI] [PubMed] [Google Scholar]
- 370.Karnkowska-Ishikawa A, Milanowski R, Triemer RE, Zakryś B. 2013. A redescription of morphologically similar species from the genus Euglena: E. laciniata, E. sanguinea, E. sociabilis, and E. splendens. J. Phycol. 49, 616-626. ( 10.1111/jpy.12072) [DOI] [PubMed] [Google Scholar]
- 371.Łukomska-Kowalczyk M, Karnkowska A, Krupska M, Milanowski R, Zakryś B. 2016. DNA barcoding in autotrophic euglenids: evaluation of COI and 18S rDNA. J. Phycol. 52, 951-960. ( 10.1111/jpy.12439) [DOI] [PubMed] [Google Scholar]
- 372.Hutchings L, et al. 2009. The Benguela Current: an ecosystem of four components. Prog. Oceanogr. 83, 15-32. ( 10.1016/j.pocean.2009.07.046) [DOI] [Google Scholar]
- 373.Zuendorf A, Bunge J, Behnke A, Barger KJA, Stoeck T. 2006. Diversity estimates of microeukaryotes below the chemocline of the anoxic Mariager Fjord, Denmark. FEMS Microbiol. Ecol. 58, 476-491. ( 10.1111/j.1574-6941.2006.00171.x) [DOI] [PubMed] [Google Scholar]
- 374.Orsi W, Song YC, Hallam S, Edgcomb V. 2012. Effect of oxygen minimum zone formation on communities of marine protists. ISME J. 6, 1586-1601. ( 10.1038/ismej.2012.7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Orsi W, Edgcomb V, Jeon S, Leslin C, Bunge J, Taylor GT, Varela R, Epstein S. 2011. Protistan microbial observatory in the Cariaco Basin, Caribbean. II. Habitat specialization. ISME J. 5, 1357-1373. ( 10.1038/ismej.2011.7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Wang Y, Zhang WP, Cao HL, Shek CS, Tian RM, Wong YH, Batang Z, Al-Suwailem A, Qian PY. 2014. Diversity and distribution of eukaryotic microbes in and around a brine pool adjacent to the Thuwal cold seeps in the Red Sea. Front. Microbiol. 5, 37. ( 10.3389/fmicb.2014.00037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Lax G, Lee WJ, Eglit Y, Simpson A. 2019. Ploeotids represent much of the phylogenetic diversity of euglenids. Protist 170, 233-257. ( 10.1016/j.protis.2019.03.001) [DOI] [PubMed] [Google Scholar]
- 378.Lax G, Simpson AGB. 2020. The molecular diversity of phagotrophic euglenids examined using single-cell methods. Protist 171, 125757. ( 10.1016/j.protis.2020.125757) [DOI] [PubMed] [Google Scholar]
- 379.Busse I, Preisfeld A. 2003. Systematics of primary osmotrophic euglenids: a molecular approach to the phylogeny of Distigma and Astasia (Euglenozoa). Int. J. Syst. Evol. Microbiol. 53, 617-624. ( 10.1099/ijs.0.02295-0) [DOI] [PubMed] [Google Scholar]
- 380.Paerschke S, Vollmer AH, Preisfeld A. 2017. Ultrastructural and immunocytochemical investigation of paramylon combined with new 18S rDNA-based secondary structure analysis clarifies phylogenetic affiliation of Entosiphon sulcatum (Euglenida: Euglenozoa). Organ. Divers. Evol. 17, 509-520. ( 10.1007/s13127-017-0330-x) [DOI] [Google Scholar]
- 381.Marin B, Palm A, Klingberg M, Melkonian M. 2003. Phylogeny and taxonomic revision of plastid-containing euglenophytes based on SSU rDNA sequence comparisons and synapomorphic signatures in the SSU rRNA secondary structure. Protist 154, 99-145. ( 10.1078/143446103764928521) [DOI] [PubMed] [Google Scholar]
- 382.Karnkowska A, Bennett MS, Watza D, Kim JI, Zakryś B, Triemer RE. 2015. Phylogenetic relationships and morphological character evolution of photosynthetic euglenids (Excavata) inferred from taxon-rich analyses of five genes. J. Eukaryot. Microbiol. 62, 362-373. ( 10.1111/jeu.12192) [DOI] [PubMed] [Google Scholar]
- 383.Kim JI, Linton EW, Shin W. 2015. Taxon-rich multigene phylogeny of the photosynthetic euglenoids (Euglenophyceae). Front. Ecol. Evol. 3, 98. ( 10.3389/fevo.2015.00098) [DOI] [Google Scholar]
- 384.Karnkowska A, Bennett MS, Triemer RE. 2018. Dynamic evolution of inverted repeats in Euglenophyta plastid genomes. Sci. Rep. 8, 16071. ( 10.1038/s41598-018-34457-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Rosowski JR, Willey RL. 1977. Development of mucilaginous surfaces in euglenoids. I. Stalk morphology of Colacium mucronatum. J. Phycol. 13, 16-21. ( 10.1111/j.1529-8817.1977.tb02880.x) [DOI] [Google Scholar]
- 386.Møhlenberg F, Kaas H. 1990. Colacium vesiculosum Ehrenberg (Euglenophyceae), infestation of planktonic copepods in the Western Baltic. Ophelia 31, 125-132. ( 10.1080/00785326.1990.10430856) [DOI] [Google Scholar]
- 387.Wiegert KE, Bennett MS, Triemer RE. 2013. Tracing patterns of chloroplast evolution in euglenoids: contributions from Colacium vesiculosum and Strombomonas acuminata (Euglenophyta). J. Eukaryot. Microbiol. 60, 214-221. ( 10.1111/jeu.12025) [DOI] [PubMed] [Google Scholar]
- 388.Bennett MS, Wiegert KE, Triemer RE. 2014. Characterization of Euglenaformis gen. nov. and the chloroplast genome of Euglenaformis [Euglena] proxima (Euglenophyta). Phycologia 53, 66-73. ( 10.2216/13-198.1) [DOI] [Google Scholar]
- 389.Linton EW, Karnkowska-Ishikawa A, Kim JI, Shin W, Bennett MS, Kwiatowski J, Zakryś B, Triemer RE. 2010. Reconstructing euglenoid evolutionary relationships using three genes: nuclear SSU and LSU, and chloroplast SSU rDNA sequences and the description of Euglenaria gen. nov. (Euglenophyta). Protist 161, 603-619. ( 10.1016/j.protis.2010.02.002) [DOI] [PubMed] [Google Scholar]
- 390.Bennett MS, Triemer RE. 2015. Chloroplast genome evolution in the Euglenaceae. J. Eukaryot. Microbiol. 62, 773-785. ( 10.1111/jeu.12235) [DOI] [PubMed] [Google Scholar]
- 391.Kosmala S, Milanowski R, Brzóska K, Pe¸kala M, Kwiatowski J, Zakryś B. 2007. Phylogeny and systematics of the genus Monomorphina (Euglenaceae) based on morphological and molecular data. J. Phycol. 43, 171-185. ( 10.1111/j.1529-8817.2006.00298.x) [DOI] [PubMed] [Google Scholar]
- 392.Guiry MD, Guiry GM. 2020. AlgaeBase. World-wide electronic publication, National University of Ireland, Galway. See www.algaebase.org (accessed on 12 November 2020).
- 393.Triemer RE, Linton E, Shin W, Nudelman A, Monfils A, Bennett M, Brosnan S. 2006. Phylogeny of the Euglenales based upon combined SSU and LSU rDNA sequence comparisons and description of Discoplastis gen. nov. (Euglenophyta). J. Phycol. 42, 731-740. ( 10.1111/j.1529-8817.2006.00219.x) [DOI] [Google Scholar]
- 394.Łukomska-Kowalczyk M, Chaber K, Fells A, Milanowski R, Zakryś B. In press. Description of Flexiglena gen. nov. and new members of Discoplastis and Euglenaformis (Euglenida). J. Phycol. ( 10.1111/jpy.13107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Dawson NS, Walne PL. 1991. Structural characterization of Eutreptia pertyi (Euglenophyta). I. General description. Phycologia 30, 287-302. ( 10.2216/i0031-8884-30-3-287.1) [DOI] [Google Scholar]
- 396.McLachlan JL, Seguel MR, Fritz L. 1994. Tetreutreptia pomquetensis gen. et sp. nov. (Euglenophyceae): a quadriflagellate, phototrophic marine euglenoid. J. Phycol. 30, 538-544. ( 10.1111/j.0022-3646.1994.00538.x) [DOI] [Google Scholar]
- 397.Kuo RC, Lin S. 2013. Ectobiotic and endobiotic bacteria associated with Eutreptiella sp. isolated from Long Island Sound. Protist 164, 60-74. ( 10.1016/j.protis.2012.08.004) [DOI] [PubMed] [Google Scholar]
- 398.Davis BM. 1894. Euglenopsis: a new alga-like organism. Ann. Bot. 8, 377-390. ( 10.1093/oxfordjournals.aob.a090716) [DOI] [Google Scholar]
- 399.Carter HJ. 1869. XXXIII. Notes on filigerous green Infusoria of the Island of Bombay. J. Nat. Hist. 3, 249-260. ( 10.1080/00222936908695939) [DOI] [Google Scholar]
- 400.Brumpt E, Lavier G. 1924. Un nouvel Euglénien polyflagellé parasite du têtard de Leptodactylus ocellatus au Brésil. Ann. Parasitol. Hum. Comp. 2, 248-252. ( 10.1051/parasite/1924023248) [DOI] [Google Scholar]
- 401.Rosowski JR. 2003. Photosynthetic euglenoids. In Freshwater algae of north america: ecology and classification (eds Wehr JD, Sheath RG), pp. 383-422. San Diego, CA: Academic Press. [Google Scholar]
- 402.Khondker M, Bhuiyan RA, Yeasmin J, Alam M, Sack RB, Huq A, Colwell RR. 2008. New records of phytoplankton for Bangladesh. 5. Euglena, Euglenocapsa. Bangladesh J. Plant Taxon. 15, 39-46. ( 10.3329/bjpt.v15i1.910) [DOI] [Google Scholar]
- 403.Triemer RE. 1985. Ultrastructural features of mitosis in Anisonema sp. (Euglenida). J. Protozool. 32, 683-690. ( 10.1111/j.1550-7408.1985.tb03102.x) [DOI] [Google Scholar]
- 404.Preisfeld A, Busse I, Klingberg M, Talke S, Ruppel HG. 2001. Phylogenetic position and inter-relationships of the osmotrophic euglenids based on SSU rDNA data, with emphasis on the Rhabdomonadales (Euglenozoa). Int. J. Syst. Evol. Microbiol. 51, 751-758. ( 10.1099/00207713-51-3-751) [DOI] [PubMed] [Google Scholar]
- 405.Müllner AN, Angeler DG, Samuel R, Linton EW, Triemer RE. 2001. Phylogenetic analysis of phagotrophic, phototrophic and osmotrophic euglenoids by using the nuclear 18S rDNA sequence. Int. J. Syst. Evol. Microbiol. 51, 783-791. ( 10.1099/00207713-51-3-783) [DOI] [PubMed] [Google Scholar]
- 406.Pochmann A. 1955. Helikotropis okteres n. gen. n. spec. (Peranemataceae) und die Frage der Ätiologie der Kielbildungen bei farblosen Eugleninen. Österr. Bot. Z. 102, 1-17. ( 10.1007/BF01768757) [DOI] [Google Scholar]
- 407.Cann JP. 1986. Ultrastructural observations of taxonomic importance on the euglenoid genera Gyropaigne Skuja, Parmidium Christen, and Rhabdospira Pringsheim (Euglenida: Rhabdomonadina). Arch. Protistenk. 132, 395-401. ( 10.1016/S0003-9365(86)80032-X) [DOI] [Google Scholar]
- 408.Chen YT. 1950. Investigations of the biology of Peranema trichophorum (Euglenineae). Q. J. Microsc. Sci. 91, 279-308. [PubMed] [Google Scholar]
- 409.Saranak J, Foster KW. 2005. Photoreceptor for curling behavior in Peranema trichophorum and evolution of eukaryotic rhodopsins. Eukaryot. Cell 4, 1605-1612. ( 10.1128/EC.4.10.1605) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Lee WJ, Blackmore RB, Patterson DJ. 1999. Australian records of two lesser known genera of heterotrophic euglenids - Chasmostoma Massart, 1920 and Jenningsia Schaeffer, 1918. Protistology 1, 10-16. [Google Scholar]
- 411.Cavalier-Smith T, Chao EE, Vickerman K. 2016. New phagotrophic euglenoid species (new genus Decastava; Scytomonas saepesedens; Entosiphon oblongum), Hsp90 introns, and putative euglenoid Hsp90 pre-mRNA insertional editing. Eur. J. Protistol 56, 147-170. ( 10.1016/j.ejop.2016.08.002) [DOI] [PubMed] [Google Scholar]
- 412.Farmer MA, Triemer RE. 1988. A redescription of the genus Ploeotia Duj. (Euglenophyceae). Taxon 37, 319-325. ( 10.2307/1222141) [DOI] [Google Scholar]
- 413.Triemer RE. 1986. Light and electron microscopic description of a colorless euglenoid, Serpenomonas costata n. g., n. sp. J. Protozool. 33, 412-415. ( 10.1111/j.1550-7408.1986.tb05632.x) [DOI] [Google Scholar]
- 414.Chan YF, Moestrup Ø, Chang J. 2013. On Keelungia pulex nov. gen. et nov. sp., a heterotrophic euglenoid flagellate that lacks pellicular plates (Euglenophyceae, Euglenida). Eur. J. Protistol 49, 15-31. ( 10.1016/j.ejop.2012.04.003) [DOI] [PubMed] [Google Scholar]
- 415.Farmer MA, Triemer RE. 1994. An ultrastructural study of Lentomonas applanatum (Preisig) n. g. (Euglenida). J. Eukaryot. Microbiol. 41, 112-119. ( 10.1111/j.1550-7408.1994.tb01482.x) [DOI] [Google Scholar]
- 416.Leedale GF. 1967. Euglenoid flagellates, 1st edn. Englewood Cliffs, NJ: Prentice-Hall Press Inc. [Google Scholar]
- 417.Cann JP, Pennick NC. 1986. Observations on Petalomonas cantuscygni, n. sp., a new halo-tolerant strain. Arch. Protistenk. 132, 63-71. ( 10.1016/S0003-9365(86)80008-2) [DOI] [Google Scholar]
- 418.Christen HR. 1959. New colorless Eugleninae. J. Protozool. 6, 292-303. ( 10.1111/j.1550-7408.1959.tb04371.x) [DOI] [Google Scholar]
- 419.Wołowski K. 1995. Dylakosoma pelophilum Skuja, a rare colourless euglenophyte found in Poland. Algol. Stud. 76, 75-78. ( 10.1127/algol_stud/76/1995/75) [DOI] [Google Scholar]
- 420.Kudo RR. 1966. Protozoology, 5th edn. Springfield, IL: Charles C Thomas Publisher. [Google Scholar]
- 421.Krell FT, Shabalin S. 2008. Michajlowastasia nom. nov. for the parasitic euglenoid genus Parastasia Michajłow, 1972 (Euglenozoa: Euglenoidina: Astasiidae). Syst. Parasitol. 71, 49-52. ( 10.1007/s11230-008-9143-9) [DOI] [PubMed] [Google Scholar]
- 422.Dobell CC. 1908. The structure and life-history of Copromonas subtilis, nov. gen. et nov. spec.: a contribution to our knowledge of the Flagellata. Q. J. Microsc. Sci. 52, 75-120. [Google Scholar]
- 423.Yubuki N, Simpson AGB, Leander BS. 2013. Reconstruction of the feeding apparatus in Postgaardi mariagerensis provides evidence for character evolution within the Symbiontida (Euglenozoa). Eur. J. Protistol 49, 32-39. ( 10.1016/j.ejop.2012.07.001) [DOI] [PubMed] [Google Scholar]
- 424.Yubuki N, Edgcomb VP, Bernhard JM, Leander BS. 2009. Ultrastructure and molecular phylogeny of Calkinsia aureus: cellular identity of a novel clade of deep-sea euglenozoans with epibiotic bacteria. BMC Microbiol. 9, 16. ( 10.1186/1471-2180-9-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Simpson AGB, van den Hoff J, Bernard C, Burton HR, Patterson DJ. 1997. The ultrastructure and systematic position of the euglenozoon Postgaardi mariagerensis, Fenchel et al. Arch. Protistenk. 147, 213-225. ( 10.1016/S0003-9365(97)80049-8) [DOI] [Google Scholar]
- 426.Forterre P. 2010. Defining life: the virus viewpoint. Orig. Life Evol. Biospheres 40, 151-160. ( 10.1007/s11084-010-9194-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Suttle CA. 2005. Viruses in the sea. Nature 437, 356-361. ( 10.1038/nature04160) [DOI] [PubMed] [Google Scholar]
- 428.Grybchuk D, Kostygov AY, Macedo DH, d'Avila-Levy CM, Yurchenko V. 2018. RNA viruses in trypanosomatid parasites: a historical overview. Mem. Inst. Oswaldo Cruz 113, e170487. ( 10.1590/0074-02760170487) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Deeg CM, Chow CET, Suttle CA. 2018. The kinetoplastid-infecting Bodo saltans virus (Bsv), a window into the most abundant giant viruses in the sea. eLife 7, e33014. ( 10.7554/eLife.33014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Hingamp P, et al. 2013. Exploring nucleo-cytoplasmic large DNA viruses in Tara Oceans microbial metagenomes. ISME J. 7, 1678-1695. ( 10.1038/ismej.2013.59) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Tarr PI, Aline RF, Smiley BL, Scholler J, Keithly J, Stuart K. 1988. LR1: a candidate RNA virus of Leishmania. Proc. Natl Acad. Sci. USA 85, 9572-9275. ( 10.1073/pnas.85.24.9572) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Scheffter S, Widmer G, Patterson JL. 1994. Complete sequence of Leishmania RNA virus 1–4 and identification of conserved sequences. Virology 199, 479-483. ( 10.1006/viro.1994.1149) [DOI] [PubMed] [Google Scholar]
- 433.Ives A, et al. 2011. Leishmania RNA virus controls the severity of mucocutaneous leishmaniasis. Science 331, 775-778. ( 10.1126/science.1199326) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Rossi M, et al. 2017. Type I interferons induced by endogenous or exogenous viral infections promote metastasis and relapse of leishmaniasis. Proc. Natl Acad. Sci. USA 114, 4987-4992. ( 10.1073/pnas.1621447114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Brettmann EA, et al. 2016. Tilting the balance between RNA interference and replication eradicates Leishmania RNA virus 1 and mitigates the inflammatory response. Proc. Natl Acad. Sci. USA 113, 11 998-12 005. ( 10.1073/pnas.1615085113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Kurt Ö, Mansur N, Çavuş I, Özcan O, Batir MB, Gündüz C, Sezerman U, Özbilgın A. 2019. First report and in silico analysis of Leishmania virus (LRV2) identified in an autochthonous Leishmania major isolate in Turkey. New Microbiol. 42, 64-67. [PubMed] [Google Scholar]
- 437.Kleschenko Y, Grybchuk D, Matveeva NS, Macedo DH, Ponirovsky EN, Lukashev AN, Yurchenko V. 2019. Molecular characterization of Leishmania RNA virus 2 in Leishmania major from Uzbekistan. Genes 10, 830. ( 10.3390/genes10100830) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Widmer G, Dooley S. 1995. Phylogenetic analysis of Leishmania RNA virus and Leishmania suggests ancient virus-parasite association. Nucleic Acids Res. 23, 2300-2304. ( 10.1093/nar/23.12.2300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Grybchuk D, Kostygov AY, Macedo DH, Votýpka J, Lukeš J, Yurchenko V. 2018. RNA viruses in Blechomonas (Trypanosomatidae) and evolution of Leishmaniavirus. mBio 9, e01932-18. ( 10.1128/mBio.01932-18) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Akopyants NS, Lye LF, Dobson DE, Lukeš J, Beverley SM. 2016. A novel bunyavirus-like virus of trypanosomatid protist parasites. Genome Announc. 4, e00715-16. ( 10.1128/genomeA.00715-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Grybchuk D, et al. 2018. Viral discovery and diversity in trypanosomatid protozoa with a focus on relatives of the human parasite Leishmania. Proc. Natl Acad. Sci. USA 115, E506-E515. ( 10.1073/pnas.1717806115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Grybchuk D, et al. 2020. The first non-LRV RNA virus in Leishmania. Viruses 12, 168. ( 10.3390/v12020168) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Akopyants NS, Lye LF, Dobson DE, Lukeš J, Beverley SM. 2016. A Narnavirus in the trypanosomatid protist plant pathogen Phytomonas serpens. Genome Announc. 4, e00711-16. ( 10.1128/genomeA.00711-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Sukla S, Roy S, Sundar S, Biswas S. 2017. Leptomonas seymouri narna-like virus 1 and not leishmaniaviruses detected in kala-azar samples from India. Arch. Virol. 162, 3827-3835. ( 10.1007/s00705-017-3559-y) [DOI] [PubMed] [Google Scholar]
- 445.Alves JMP, et al. 2013. Endosymbiosis in trypanosomatids: the genomic cooperation between bacterium and host in the synthesis of essential amino acids is heavily influenced by multiple horizontal gene transfers. BMC Evol. Biol. 13, 190. ( 10.1186/1471-2148-13-190) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Burzell LA. 1975. Fine structure of Bodo curvifilus Griessmann (Kinetoplastida: Bodonidae). J. Protozool. 22, 35-39. ( 10.1111/j.1550-7408.1975.tb00942.x) [DOI] [PubMed] [Google Scholar]
- 447.Midha S, Rigden D, Siozios S, Hurst G, Jackson A. In press. The Paracaedibacter-like endosymbiont of Bodo saltans (Kinetoplastida) uses multiple putative toxin-antitoxin systems to maintain its host association. ISME J. ( 10.1038/S41396-020-00879-6) [DOI] [PMC free article] [PubMed]
- 448.Ganyukova AI, Frolov AO, Malysheva MN, Spodareva VV., Yurchenko V, Kostygov AY. 2020. A novel endosymbiont-containing trypanosomatid Phytomonas borealis sp. n. from the predatory bug Picromerus bidens (Heteroptera: Pentatomidae). Folia Parasitol. 67, 004. ( 10.14411/FP.2020.004) [DOI] [PubMed] [Google Scholar]
- 449.Muñoz-Gómez SA, Hess S, Burger G, Lang BF, Susko E, Slamovits CH, Roger AJ. 2019. An updated phylogeny of the Alphaproteobacteria reveals that the parasitic Rickettsiales and Holosporales have independent origins. eLife 8, e42535. ( 10.7554/eLife.42535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Tashyreva D, Prokopchuk G, Votýpka J, Yabuki A, Horák A, Lukeš J. 2018. Life cycle, ultrastructure, and phylogeny of new diplonemids and their endosymbiotic bacteria. mBio 9, e02447-e17. ( 10.1128/mBio.02447-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.George EE, Husnik F, Tashyreva D, Prokopchuk G, Horák A, Kwong WK, Lukeš J, Keeling PJ. 2020. Highly reduced genomes of protist endosymbionts show evolutionary convergence. Curr. Biol. 30, 925-933. ( 10.1016/j.cub.2019.12.070) [DOI] [PubMed] [Google Scholar]
- 452.Monteil CL, et al. 2019. Ectosymbiotic bacteria at the origin of magnetoreception in a marine protist. Nat. Microbiol. 4, 1088-1095. ( 10.1038/s41564-019-0432-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Leedale GF. 1969. Observations on endonuclear bacteria in euglenoid flagellates. Österr. Bot. Z. 116, 279-294. ( 10.1007/BF01379628) [DOI] [Google Scholar]
- 454.Surek B, Melkonian M. 1983. Intracellular bacteria in the Euglenophyceae: prolonged axenic culture of an alga-bacterial system. In Endocytobiology, vol. 2 (eds Schenk HEA, Schwemmler W), pp. 475-486. Berlin, Germany: de Gruyter. [Google Scholar]
- 455.Kim E, Park JS, Simpson AGB, Matsunaga S, Watanabe M, Murakami A, Sommerfeld K, Onodera NT, Archibald JM. 2010. Complex array of endobionts in Petalomonas sphagnophila, a large heterotrophic euglenid protist from Sphagnum-dominated peatlands. ISME J. 4, 1108-1120. ( 10.1038/ismej.2010.40) [DOI] [PubMed] [Google Scholar]
- 456.Leander BS, Farmer MA. 2000. Epibiotic bacteria and a novel pattern of strip reduction on the pellicle of Euglena helicoideus (Bernard) Lemmermann. Eur. J. Protistol 36, 405-413. ( 10.1016/S0932-4739(00)80046-2) [DOI] [Google Scholar]
- 457.Torres de Araujo FF, Pires MA, Frankel RB, Bicudo CEM. 1986. Magnetite and magnetotaxis in algae. Biophys. J. 50, 375-378. ( 10.1016/S0006-3495(86)83471-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Klinges JG, et al. 2019. Phylogenetic, genomic, and biogeographic characterization of a novel and ubiquitous marine invertebrate-associated Rickettsiales parasite, Candidatus Aquarickettsia rohweri, gen. nov., sp. nov. ISME J. 13, 2938-2953. ( 10.1038/s41396-019-0482-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Tikhonenkov DV, et al. 2021. First finding of free-living representatives of Prokinetoplastina and their nuclear and mitochondrial genomes. Sci. Rep. 11, 2946. ( 10.1038/s41598-021-82369-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.