Abstract
We propose a new paradigm for dense functional imaging of brain activity to surmount the limitations of present methodologies. We term this approach integrated neurophotonics; it combines recent advances in microchip-based integrated photonic and electronic circuitry with those from optogenetics. This approach has the potential to enable lens-less functional imaging from within the brain itself - to achieve dense, large-scale stimulation and recording of brain activity with cellular resolution at arbitrary depths. We perform a computational study of several prototype 3D architectures for implantable probe-array modules that are designed to provide fast and dense single-cell resolution, e.g., within a 1-mm3 volume of mouse cortex comprising ~100,000 neurons. We describe progress toward realizing integrated neurophotonic imaging modules, which can be produced en masse with current semiconductor foundry protocols for chip manufacturing. Implantation of multiple modules can cover extended brain regions.
Massively parallel interrogation of brain activity
“Within the central nervous system, the events in each unit are not so important. We are more concerned with the interactions of large numbers, and our problem is to find the way in which such interactions can take place.”
-Edward D. Adrian (1926)
These final lines from Lord Adrian’s Nobel lecture (Adrian, 1926) illustrate the extraordinary prescience of this researcher who first discovered neuronal spiking. He anticipated that understanding brain computation is not likely to be achieved only by studies of individual neurons but, instead, by observing coordinated interactions of neurons and their collective activity patterns.
Realizing instrumentation to monitor population activity within the brain with single-neuron resolution is a profoundly difficult challenge; Figure 1 provides a sense of the scale involved. The slow rate of technological development in neuroscience is elucidated in Figure 2; it charts the evolution of our ability to simultaneously resolve and track the activity of a multiplicity of neurons in vivo, over the six decades since the invention of whole-cell recording (Stevenson and Kording, 2011). Today, the state-of-the-art permits simultaneous, full bandwidth recording in vivo in awake rodents from multi-shank neural probe modules, each with up to 1,024 channels (Rios et al., 2016; Shobe et al., 2015). With implantation of multiple probes of these types, many thousands of neurons are now being simultaneously recorded (Steinmetz et al., 2019). Although it is unequivocal that these advances open exciting research frontiers, the number of observable neurons has continued to remain comparable to the electrode count. This is consistent with the empirical observation that multi-site extracellular electrodes yield, on average, just one or two units per site, even with optimal spike-sorting algorithms (Marblestone et al., 2013). At this rate of development, another 90 years must elapse before the activity of an entire mouse brain, containing roughly 75 million neurons, will become observable (Figure 2). Clearly, we must significantly accelerate this rate of development.
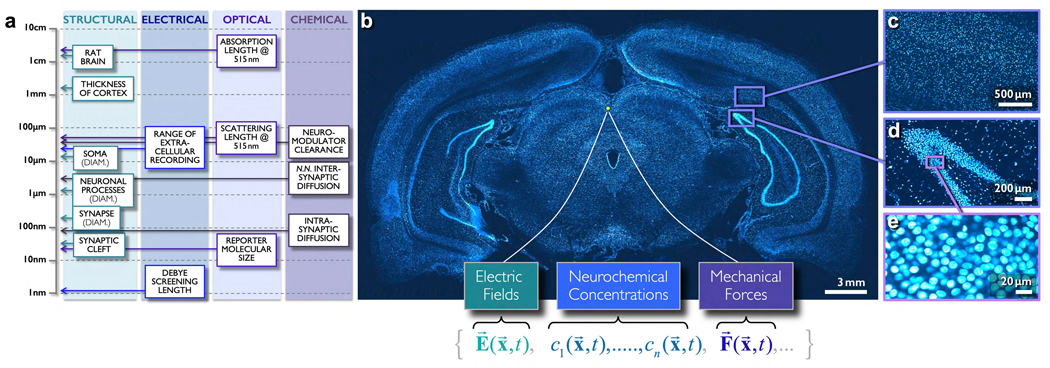
Figure 1. Brain complexity, “brain fields”, and structural length scales vis-à-vis cell-body location, density and heterogeneity in the rodent brain.
Strong light scattering and absorption in brain tissue make it extremely difficult to achieve dense, volumetric functional imaging with cellular resolution. a) Biophysical scales for electrical, neurochemical and optical domain recordings, and relative sizes of brain structures. b) A ~2μm thick section of an adult rat brain slice, stained with a fluorescent nuclear stain, wet-mounted, and imaged by large-scale serial two-photon microscopy (L. Moreaux, 2010). Beneath this image we enumerate three “brain fields”, that is, neural activity domains: the electrical, neurochemical, and mechanical. c,d,e) Cellular nuclear density at multiple scales, from the macroscopic down to the level of individual cells.
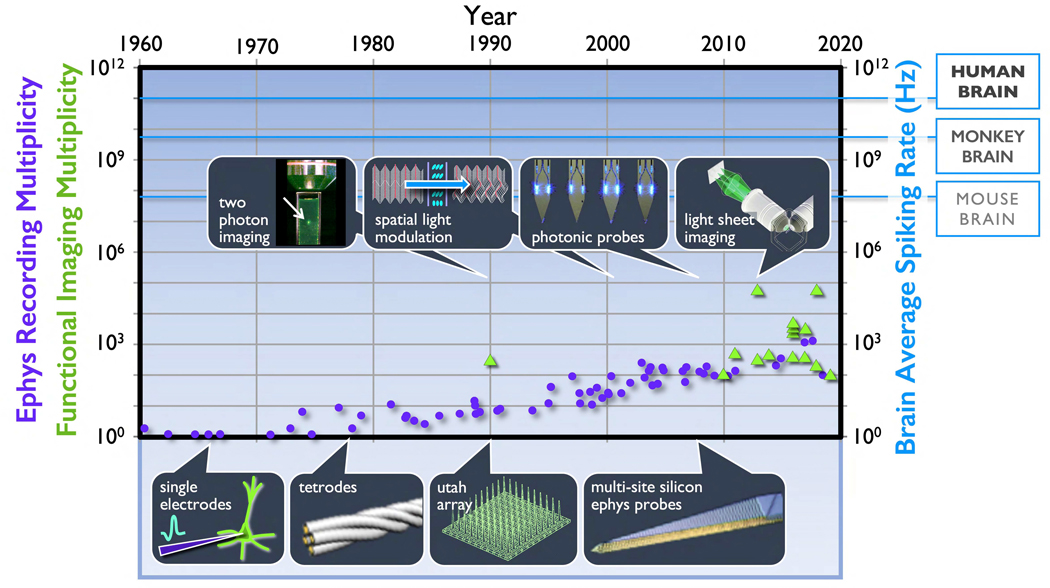
Figure 2. Evolution of recording multiplicity for electrophysiology and functional imaging vis-à-vis overall brain activity (spiking rates).
Left ordinate: (violet dots) The evolution of multiplicity for individual electrodes, implantable multi-site probes, and multi-probe modules since the invention of whole cell recording (Stevenson and Kording 2011; Steinmetz, et al. 2018). (green triangles) Also shown are recording multiplicities for multi-photon functional imaging (from Lecoq et al., 2019) and light sheet microscopy (Ahrens et al., 2013, Chen et al., 2018). For a current review of recording capabilities with multi-photon techniques, see (Lecoq et al., 2019). Right ordinate: To compare the evolution of the technology against large-scale volumetric activity, we show the average spiking rate over entire brains is estimated as the product of the number of neurons and the brain average firing rate per neuron (~2Hz).
This was a central aim of our proposal (Alivisatos et al., 2012) that eventually culminated in the launching of the U.S. BRAIN Initiative (Bargmann and Newsome, 2014). Our initial vision, which still remains true, is that advances in nanotechnology, molecular reporters, and large-scale integration of semiconductor devices now make it feasible to precipitously upscale the rate of progress toward massively multiplexed interrogation of brain circuits (Alivisatos et al., 2012).
Here, we focus in more depth on these prospects. Our aim in this Perspective is not solely to identify ways to increase the total number of neurons that can be recorded from simultaneously. Instead, we explore the possibility of achieving dense recording from within a targeted tissue volume, to ultimately achieve complete interrogation of local brain circuit activity. We use the word interrogation to denote recording and direct causal manipulation of a brain circuit’s individual neurons by the application of patterned, deterministic stimulation with single-neuron resolution. To achieve this, we are pursuing a new approach, which we term integrated neurophotonics, that offers significant potential for accelerating progress toward Lord Adrian’s vision. This technological path offers the prospect of dense functional imaging of neuronal activity in highly scattering neural tissue - providing cellular-scale resolution at arbitrary depths in the brain. Our approach is based on implanting an entire lens-less imaging system within the brain itself, by distributing dense arrays of microscale photonic emitter and detector pixels (hereafter, E- and D-pixels) positioned on a 3D spatial lattice (Roukes, 2011; Roukes et al., 2016). These pixel arrays are integrated onto narrow silicon shanks (needles), which leverage recent advances in silicon-nanoprobe-based fabrication (Rios et al., 2016; Shobe et al., 2015; Steinmetz et al., 2018). Used with functional molecular reporters (Andreoni et al., 2019; Chen et al., 2013; Lin and Schnitzer, 2016) and optogenetic actuators (Boyden, 2011; Miesenböck, 2011), this novel instrumentation offers the prospect of approaching the interrogation of all neuronal activity from within a 1-mm3 volume (~100,000 neurons in mouse cortex). The approach leverages recent breakthroughs in molecular reporters that can enable multimodal and multi-physical sensing (Figure 1), advances in optogenetic actuators that enable optical control of neural activity, and the genetically encoded delivery of reporters and actuators that provide specificity of cell type. Further, the methodology is potentially scalable - multiple modules can be tiled to densely cover extended regions deep within the brain. We anticipate this will ultimately permit interrogation, that is, simultaneous recording and patterned stimulation of millions of neurons, at arbitrary positions and depths in the brain, to unveil dynamics of neural networks - with single-cell resolution and specificity of cell type. Like their contemporary counterparts for highly multiplexed electrophysiology (Ephys), ultranarrow photonic neural probes perturb brain tissue minimally given their small cross-sections and passivated surfaces. They impose negligible tissue displacement upon implantation, while dissipating low power during operation - comparable to today’s active, multi-site Ephys probes that also employ complementary metal-oxide-semiconductor (CMOS) technology. And, importantly, they offer near-term prospects for wide deployment to the neuroscience research community, as they are mass producible by well-validated semiconductor foundry (microchip- production factory) methods.
Conventional electrophysiology
Background: intracellular v. extracellular Ephys
Intracellular or “whole-cell” recording was pioneered by Hamill et al. in 1981 (Hamill et al., 1981), and this technique has remained the gold standard in Ephys. Its singular advantages are its high signal-to-noise ratio (SNR), the ability to directly target specific cells, and the important ability to deduce connection strengths between neurons directly, in what are often termed “multi-patch” (i.e., multi-electrode) experiments. However, there are significant technical challenges associated with whole cell recording, particularly in vivo (Hulse et al., 2016). These arise from the delicate manipulations involved in patching, that prevent upscaling the technique to enable simultaneous recording from more than roughly a dozen neurons, the practical limit achieved by today’s expert practitioners (Jiang et al., 2015; Jiang et al., 2013; Perin et al., 2011). Recent efforts have focused on the automation of patching protocols (Kodandaramaiah et al., 2012), but these have not yet yielded a substantial increase in the multiplicity of simultaneous whole-cell recording.
Extracellular electrophysiological recording, as first pioneered by Lord Adrian, is more amenable to upscaling. Microwire electrodes in twisted pairs (McNaughton et al., 1983) (“stereotrodes”) and quadruples (“tetrodes”) (Recce and O’Keefe, 1989), enable single-unit recordings from several adjacent neurons (Gray et al., 1995) when combined with spike-sorting protocols such as first developed by Gerstein and Clark in 1964 (Gerstein and Clark, 1964). Over the past several decades, arrays of tetrodes have been successfully employed to observe patterns of activity over distributed regions of the brain (Wilson and McNaughton, 1993). However, scaling upward to the regime of thousands of channels has proven challenging.
Electrical recording: The present state-of-the-art
In 1970, well prior to the development of tetrodes, Wise et al. first pioneered use of silicon-based microfabrication techniques to upscale extracellular recording multiplicity (Wise et al., 1970). Here, by multiplicity we mean the number of active neurons that can be simultaneously resolved and recorded. Massively-multiplexed silicon neural probes have since been widely validated (Wise et al., 2008), commercialized, and upscaled to permit multiplexing of hundreds of recording sites within an individual probe (Berenyi et al., 2014). However, despite the technology’s potential and the advances made, the multiplicity of extracellular recording has increased only by a factor of two roughly every seven years over the past sixty years (Figure 2). As mentioned, silicon neural probes have recently been upscaled to contain over one thousand simultaneously active, full bandwidth recording electrodes within a single functional multi-probe module. Here, nanofabrication methods permitting nanowire interconnects along the shanks enable the attainment of narrow shank cross sections (Rios et al., 2016; Shobe et al., 2015; Steinmetz et al., 2018) that are comparable to the diameter of tetrodes (typically, ~35μm) that permit chronic implantation. The expectation here is that (cross sectional) size matters.
To evaluate the practical and ultimate limits of the recording multiplicity attainable with multi-site electrophysiological recording, the physical origin of extracellular currents and potentials must be considered. Extracellular currents arise from the activity of ion channels and pumps occurring both at soma and at neural processes. The slow spatiotemporal evolution of the local field potential (LFP) arises from ion exchange between soma, processes, synapses, and their extracellular environment - that is, throughout the vast regions over which neural processes extend. By contrast, rapid local waveforms arising from spiking are dominated by much faster somatic ion currents. (Berenyi et al., 2014; Buzsaki, 2004; Buzsaki et al., 2012) Accordingly, despite the nomenclature, so-called “local field potentials” tend to be spatially diffuse in character, while fast transients associated with action potentials are much more spatially localized. Spike sorting protocols with multi-site electrodes take advantage of the latter; they permit separation and extraction of the fast temporal activity of individual “units” (arising from single-neuronal spiking) from the LFP signals (that arise from complex, ensemble-average, multi-neuronal activity.) Recently, the effectiveness of spike sorting protocols in various experimental contexts have been assessed qualitatively (Marblestone et al., 2013). This meta-analysis of their empirical limits is sobering. It’s conclusion is that - while future improvements might, in principal, yield up to 10 neurons per electrode for multi-site probes - experiments today typically yield a recording multiplicity that is smaller, i.e., much closer to 1:1. In other words, on average, one electrode is required for every neuron recorded. An important caveat here is that the activity of the brain region probed matters; regions with sparse activity will yield smaller multiplicities that can be obtained from regions with less-sparse activity.
This low multiplicity obtained from most multi-site Ephys recordings has important practical implications, both for the potential level of scale-up (multiplexing) that is attainable, and for the likely density of neuronal coverage that is practicable. Scaling up the number of recording sites with 3D multi-shank architectures was first demonstrated in the 1990’s (Campbell et al., 1991; Hoogerwerf and Wise, 1994). To minimize perturbation of brain activity, the consensus is that implantable probes should altogether volumetrically displace only a fraction of a percent of neural tissue (Marblestone et al., 2013). However, this asserted limit is just an estimate; controlled, direct, and systematic studies have not yet been carried out.
Najafi and Wise first explored the possibilities for massively upscaling the recording multiplicity of silicon-based multi-site neural probes for Ephys (Najafi et al., 1990). They consider the mechanical robustness of silicon probes and conclude that, to readily permit penetration of the pia, shanks 5–10mm long (permitting access to deep brain structures in small mammals) should maintain a cross-sectional area of ~200μm2, e.g. 20μm width × (at least) 10μm thickness. Maintaining <1% volumetric exclusion of brain tissue for a 3D neural probe array constructed from such shanks would then impose a minimum inter-shank pitch of ~140μm, assuming a square grid architecture. With this configuration, and using the fact that state-of-the art, low-noise single-unit recording requires an electrode geometric surface area of ~180μm2 (~15μm diameter for a circular electrode) (Ludwig et al., 2011), the maximum practical number of electrodes per unit volume of brain tissue can be estimated. Assuming each shank comprises a linear array of electrodes with a pitch of 1.5 times the electrode diameter (i.e., 22.5μm center- to-center spacing), a maximum fill-factor of approximately 2,100 electrodes/mm3 is attainable. One cubic millimeter of mouse cortex contains approximately 100,000 neurons and, being (overly) optimistic by assuming that a maximum of two units are extracted per recording site, we conclude that a volume coverage of only ~4% of the neuronal population is the upper limit of attainable coverage. Again, this must be qualified that that coverage obtained will be dependent on the relative level of activity within the brain region probed.
These considerations can be revised somewhat in light of the new generation of ultra-flexible neural probes emerging from multiple laboratories (Jeong et al., 2015; Liu et al., 2015; Rousche et al., 2001). Thinner probes - which, for implantation, require removable stiffeners - could, in principle, permit closer probe spacing and, thereby, increase coverage of the neural population. Current efforts pursuing flexible probe technology either focus on devices based on polymeric materials or ultra-thinned silicon. The latter make use of fabrication protocols permitting conventional semiconductors - usually considered brittle, inelastic materials - to be rendered extremely pliable by making them extremely thin. Thinning the silicon substrate has the two very significant benefits: it yields much thinner devices than their polymeric counterparts, and it enables use of state-of-the-art silicon CMOS circuitry (Navaraj et al., 2018; Shahrjerdi and Bedell, 2013). A recent analysis for these new flexible probe designs, in the same vein as that of Najafi and Wise, arrives at somewhat more optimistic conclusions (Kleinfeld et al., 2019).
Ultra-thinned CMOS silicon neural probes can thus be made almost an order of magnitude thinner than those estimated by Najafi et al. (Najafi et al., 1990) - and this could reduce tissue displacement by a similar factor. Following the logic above, it could permit reducing shank spacing down to a ~50μm pitch. However, there are significant challenges to achieving reproducible implantation of such flexible 3D arrays. The requisite stiffeners, used to facilitate probe implantation, will likely displace a comparable amount of tissue as the somewhat thicker silicon probes that the flexible devices are intended to replace. Nonetheless, it is interesting to note that a 50μm shank separation would be commensurate with the empirically deduced range of multi-site electrical recording (Buzsaki, 2004). The limitation on Ephys range arises from the combined biophysical effects of electrostatic screening in the ionic cerebrospinal fluid, the presence of other distributed sources of electrophysiological activity (which can raise the practical noise floor for detection) and the need to maintain sufficient SNR in extracellular recordings to permit efficacious spike-sorting. Following the logic outlined above to ensure that volume exclusion of neural tissue kept below 1%, a ten-fold increase in electrode density over the estimate above would permit volumetric coverage of up to 40% of adjacent neurons with an ultrathin probe array. But we emphasize that this is an optimistic estimate; it is predicated on implanting an immense number of flexible probes (with their requisite stiffeners) without damaging adjacent tissue - a challenging task that may be difficult to achieve in practice.
Within the last few years, introduction of modern microelectronics technology, particularly through the development of the (rigid) Neuropixel probes, has resulted in significant upscaling of multiplexed electrophysiology (Jun et al., 2017; Steinmetz et al., 2019). Recent efforts by the Neuralink team (Musk, 2019) are also upscaling flexible probes that are packaged with application-specific integrated circuits (ASICs) for neural recording. The approaches used to connect the probes to the electronics today limit the number of achievable channels, but this will continually improve as packaging technologies evolve. This scaling, however, has largely been devoted to studying multiple or otherwise spatially disparate brain regions, rather than to realize dense brain circuit coverage within a specific region. For this latter and very important challenge, practical and fundamental limits constrain the multiplicity of multi-site silicon probes for Ephys. These limitations result from the inherent locality of electrical recording, which makes only a few neurons accessible to any specific electrode within an array. For this reason, if dense recording (full volumetric coverage of active neurons) appears to be very challenging to achieve by conventional extracellular Ephys. Accordingly, it is natural to ask if alternative technologies exist that might provide dense coverage of brain circuits with far fewer implanted elements. The key here is to vastly increase the recording multiplicity obtainable from each recording element within an implanted multiplexed array, thereby minimizing volumetric displacement, while massively upscaling recording density and volumetric coverage. We will focus on this overarching challenge throughout of the remainder of this article.
Beyond conventional Ephys: Free-space functional imaging
Functional imaging is an alternative approach to Ephys that enables both recording and localization of neurons in anatomical space. It also provides the very important added capability of specificity of cell type. However, as brain functions are not optical processes, neuronal activity must be transduced from its intrinsic domain (Figure 1b) into the optical domain; this is achieved by activity reporters. These reporters can be specific macromolecules or nanoparticles that comprise two moieties: one, the sensor, reacts to a targeted physical domain of local activity - be it electrical potential, molecular recognition, or mechanical forces; the other, the chromophore, provides optical functionality - it fluoresces at a particular wavelength when excited by an incoming photon within a specific band of wavelengths. In response to local neural activity from one of the aforementioned multiphysical measurement domains (Figure 1), the sensor in turn, modulates the optical susceptibility of the chromophore. These bipartite reporters are then continuously interrogated optically to determine their instantaneous state of activity. Hence, local neuronal activity is directly reflected by changes in the chromophore’s optical susceptibility.
Functional imaging at depth
Currently, functional imaging of neuronal activity in the rodent cortex is widely achieved using free-space multi-photon laser-scanning microscopy (Denk et al., 1990; Lecoq et al., 2019), with brain tissue that is labeled by molecular reporters (Grienberger and Konnerth, 2012). This combination readily provides cellular resolution of neural activity. Among such reporters are exogenous synthetic molecules, providing no cellular specificity; or genetically encoded proteins that, as described below, provide cellular specificity through restriction of their expression to specific cell types.
Free-space, two- and three- photon, laser-scanning microscopy (Denk et al., 1990; Tolias) together with fluorescent calcium reporters (Chen et al., 2013) enable functional imaging with sub-cellular resolution. However, at increasing depths within the brain, scattering and absorption ultimately preclude delivery of ballistic (i.e., unscattered) light with sufficient intensity and focus to achieve multi-photon excitation of specific reporter-labeled neurons. The ultimate depth of delivery is limited by the optical attenuation length, , where Lsc and Lab are the wavelength-dependent scattering and absorption lengths, respectively (Wang et al., 2018). Further compounding this is the challenge of extracting the information- bearing visible-wavelength fluorescent photons emitted by reporters. They are even more strongly scattered; La at green wavelengths is (Figure 6a). This results from Mie scattering (Bohren and Huffman, 2004), which is predominant in this regime; it greatly diminishes the photon yield available to free-space optics placed outside the brain. Accordingly, even state-of-the-art three-photon functional imaging provides cellular resolution solely at depths less than ~1.7mm (Ouzounov et al., 2017). Despite significant effort and investment, the growing consensus among experts is that it’s unlikely this range can be extended much further. All current methods for free-space imaging are therefore applicable solely at rather shallow (< 2mm) tissue depths (e.g., cortex), or to transparent organisms (e.g., zebrafish larvae).
Figure 6. Mesoscopic light scattering and photon transport within the brain.
a) Absorption, scattering, and attenuation in brain tissue versus wavelength. (After N.G. Horton, et al., Nature Photonics 7, 205 (2013). b) Forward Mie scattering in brain tissue is overwhelmingly forward directed. Polar diagram of the scattering of blue light (λ=480nm, unpolarized) from a sphere with radius r ~15μm, index difference </di>from the environment. This closely approximates scattering from a soma in extracellular media. Each concentric circle represents a ten-fold increase in intensity. The forward peak at 0° (cyan trace) is generally more than 5 orders higher in intensity than scattered light. Adapted from (Laven, 2020). c) Schematic depicting illumination impinging upon a neuron after propagating a distance, z, in scattering tissue. For simplicity, the E-pixel is idealized as a point emitter (d=0) d) Heatmap showing beam intensity versus distance from microscopic emitter. For this analysis the emitted beam is assumed to start with zero width. e) Lateral beam profile for five distances from the emitter shown as dashed lines in panel c. The microscale beam remains highly collimated even 200μm from the emitter. f) Comparison between ballistic photons (blue trace) collected by the “neuron” (15μm-diamater disc, representing a somatic cross-section) as depicted panel c, with those arriving after scattering (orange trace). The horizontal dashed line exemplifies that, because of strong forward scattering in the mesoscopic regime, a given total photon flux (green trace) for, e.g., a million photons, can be collected a significant distance,, further from the source than is the case considering only the ballistic contribution.
Microendoscopy and microfiber-based imaging
The complications outlined above have motivated the development of microendoscopy. This method employs an optical fiber implanted in targeted brain regions, sometimes with a miniature lens or prism, to achieve, e.g., calcium functional imaging at the fiber’s distal end via one-photon (1p) or two-photon (2p) fluorescence excitation. (Liberti et al., 2017; Zong et al., 2017) Although such direct implantation resolves the issue of light delivery and recovery from deep within the brain, the approach has several limitations: i) imaging is achieved only within the optical plane near the endoscope tip, ii) tissue along the path of implanted cannula/fiber (typically 0.3–2mm dia.) is destroyed and, hence, iii) current implementations of this approach do not permit functional imaging along extended vertical regions (e.g., multiple cortical layers) simultaneously. Accordingly, the approach is generally feasible only for acute measurements around the fiber/lens tip, using direct CMOS imaging (Inscopix) or confocal laser microendoscopy (Mauna Kea). Finally, iv) it seems unlikely that this method can be scaled up to achieve the dense volumetric coverage of neural activity that we consider here.
Another strategy, closely related to the integrated neurophotonics paradigm described herein, involves use of implantable tapered, optical fibers patterned to enable multi-point illumination. These are coupled to an external laser source to enable passive, multi-point brain illumination at depth (Pisanello et al., 2017; Pisanello et al., 2014). By adjusting the incident light angle at the input fiber facet, various optical modes within the fiber can be addressed. A number of these modes are preferentially coupled to patterned optical windows along the length of the tapered fiber, so modal selection enables, in turn, site-selective light delivery. Similarly-patterned tapered optical fibers can also can permit passive local collection of light from a small number of sites along the fiber, when coupled to an external photodetector (Pisano et al., 2019). Here again, microscale structuring of the fiber permits modal selectivity of the collected light. This approach enables depth profiling by fluorimetry for structures in close proximity to the fiber. This approach has recently been combined with a multi-electrode array (Sileo et al., 2018), and more recently a wireless system (Emara et al., 2018).
These advancements provide considerable advantages over conventional optical fibers, including the smaller, tapered form factor that is more amenable to tissue insertion, as well as finer-scale light delivery, i.e., finely patterned multi-point sources of light. A drawback, particularly for photodetection, is the fact that the brain interrogation area achievable by this approach is restricted to cells in close proximity to the fiber. Also, the passive light collection used in this approach, i.e., guiding photons from the light source within the brain via an optical fiber to an external photomultiplier, differs from the active, highly multiplexed in situ light collection employed for photonic neural probes in our work, described below. The results of Pisannelo, et al. represent an early validation of the new paradigm of implantable imaging systems with microscale dimensions. However, as tapered/window-bearing fiber fabrication is carried out one-by-one, this technology is not directly compatible with foundry-based mass production. Thus, it appears difficult to adapt this approach to permit the massive upscaling of multiplicity required to enable dense, volumetric-scale interrogation of brain activity over extended brain regions that we envisage here.
Status quo: Large-scale, volumetric functional imaging
One prominent recent example of large-scale volumetric functional imaging in vivo is the multi-institution IARPA MICrONS project. In this effort, functional calcium imaging of all excitatory neurons expressing GCaMP6 within a ~1 mm3 volume spanning the mouse primary visual cortex and higher visual areas was obtained using a wide field “mesoscope” (Sofroniew et al., 2016; Walker et al., 2019). For each mouse studied, multiple scans tiling the visual areas and cortical layers were obtained; these comprised many imaging planes acquired at a spatial resolution of 0.4μm/pixel and a temporal resolution of 6.3 Hz. Over 5000 neurons were imaged simultaneously, thereby enabling functional characterization of approximately 70,000 cells within each mouse. Once functionally imaged, the mice were sectioned and imaged by electron microscopy (EM) with nanometer-scale resolution at the Allen Institute of Brain Science. For one mouse, sectioning and imaging the complete ~1mm3 volume spanning these regions by EM was performed. The EM data were subsequently provided to Princeton University where the separate sectional images were aligned, segmented (to identify every soma, axon, dendrite, and synapse of the ~100,000 cells within this tissue sample), and reconstructed in 3D. This combination of dense functional imaging and EM-based anatomical reconstructions within the same tissue volume is an important first step towards understanding relationships between the structure and function of neural circuits. Such efforts will ultimately permit deciphering circuit-level mechanisms that connect brain computations with behavior.
Molecular reporters and optogenetic actuators
To date, the most widely employed approach for functional imaging involves intracellular Ca2+ sensing (Charpak et al., 2001; Yuste and Katz, 1991). The temporal evolution of intracellular calcium concentration provides a robust proxy for direct electrophysiological measurements (Charpak et al., 2001; Ding et al., 2017; Moreaux and Laurent, 2007), but the approach has important limitations (Moreaux and Laurent, 2007). These molecular reporters operate by sensing the calcium influx to the cell following an action potential; the resulting change in concentration modulates binding of the calcium to the reporter’s Ca-sensing moiety and, thereby, this induces a change to the optical susceptibility of its chemically attached chromophore. The resulting stereotypical fluorescent transient that results is interrogated optically to provide a “report” on calcium influx after the neuron fires (Moreaux and Laurent, 2007, 2008). This has become widely adopted (Grienberger and Konnerth, 2012; Yuste, 2010) owing to the development of the excellent new class of fast optogenetically-based calcium reporters exemplified by GCaMP6 (Chen et al., 2013).
Optical stimulation of neural activity requires optogenetic actuators (Miesenböck, 2011). The most successful and widely deployed of these are derived from the Opsin family (Shichida and Matsuyama, 2009). Opsins are light-sensitive ion channels and pumps that transport specific ions across membranes in response to optical stimuli (Zhang et al., 2007)(Boyden, 2011). Embedded within the cell membrane, these actuators can induce or block action potentials when irradiated with light within a specific wavelength band.
Genes that encode for these molecular reporters and optogenetic actuators are introduced into neurons, either by gene delivery methods using viral vectors or through genetic engineering to create transgenic animal lines (Luo et al., 2018). This process enables neurons to express exogenous GCaMP and Opsins. Selective expression of optogenetic actuators and molecular reporters restricted to specific cell types is achieved through the use of promoters that provide specificity of cell type. This offers enhanced selectivity and enables controlled or sparse expression of optical reporters within brain tissue.
Advanced calcium reporters
One- and two-photon imaging of neurons expressing genetically encoded fluorescent reporters of calcium concentration has become widely adopted in neuroscience. This is because calcium signals are robust, with intracellular calcium rising dramatically in concentration in many neuron types as a byproduct of firing action potentials, and because bright, high dynamic range, fast, genetically encoded reporters can be created by fusing fluorescent proteins to well-known calcium-binding protein motifs. The widely-employed GCaMP family of calcium reporters (Chen et al., 2013; Dana et al., 2019; Tian et al., 2009), for example, are based upon green fluorescent protein (GFP). Modern versions of these reporters, such as GCaMP6 and GCaMP7, can reliably report both well-separated action potentials and enable estimation of the frequencies of fast series of action potentials. However, expressing a calcium reporter throughout a neuron results in fluorescent light not only being generated from cell bodies, which many investigators want to focus on, but also from neuronal processes, i.e., from any axons and dendrites that are also illuminated. As many axons and dendrites pass within an optical diffraction limit of a cell body, this can lead to neuropil contamination during dense brain circuitry imaging. In this situation, optical signals from nearby axons and dendrites contribute artifactual spikes to a cell body of interest, and thus lead to artifactual activity correlations between neurons. Much effort has been invested in focusing or patterning light to improve the collection of calcium signals from specific cells. However, complementary recent efforts have pursued what one might call molecular focusing - that is, fusing calcium reporters to protein motifs that will localize them preferentially within the cell body. Two recent efforts on such somatic localization fuse GCaMP calcium reporters to different proteins, including a coiled-coil peptide set that restricts GCaMP to the cell body (Figure 4a, lower panel) (Shemesh et al., 2020), and a protein that tethers GCaMP to ribosomes, which also restricts GCaMP to the soma (Chen et al., 2020). In both cases, neuropil contamination is significantly suppressed due to reduced axonal and dendritic GCaMP, while somatic GCaMP brightness remains high and its kinetics remain fast. This improvement serves to suppress incorrectly attributed spikes and artifactual correlations between neurons. These benefits are observed in the mouse brain as well as in other species, and they enhance functional imaging with both one- and two-photon instrumentation (including microendoscopy). Thus, by lessening the reliance on optics for selecting information to be obtained from specific cells, these “molecular focusing” strategies help to clean up signals in a way that is complementary to optical focusing methods. This can facilitate use of simpler, more scalable optical systems than currently employed for imaging of neural dynamics in vivo. This “molecular focusing” approach has also been applied to fluorescent reporters of transmembrane potential (Fig. 4), as discussed in the following section.
Figure 4. estricted sub-cellular localization of genetically engineered optical reporters of neural activity.
(a) Optical calcium reporters (GCaMP family). Representative time-averaged projection images of GCaMP6f (top panel) and its respective fusion protein variant (bottom) expressed in mouse dorsal striatum. Images were acquired with a 1P epi-fluorescent microscope. The fusion protein variant was identified in a screen designed to identify GCaMP fusion proteins with enhanced localization within 50 μm of the cell body with no effect on toxicity and GCaMP kinetics. Fusion of GCaMP6f to a de novo designed coiled-coil peptide to realize SomaGCaMP6f2 provides better SNR and fewer artifact spikes from neuropil than its non-fusion counterparts (bottom panel). Coiled-coil motifs, comprised of amino acid repeats that can assemble into complexes by “coiling” around one another via cognate sequence-structure pairing, were hypothesized by the authors to potentially slow diffusion of the GCaMP fusion proteins out of the cell body (Shemesh et al., 2020). (b) Representative confocal images of neurons in cortex layer 2/3 (left), hippocampus (middle), and striatum (right) expressing Archon1 (top) and SomArchon (bottom). Scale bar, 50 microns (Piatkevich et al., 2019). (c) Optical voltage indicator (ASAP reporters). Expression of ASAP2s (left panel) and ASAP2s fused to a cytosolic segment of the potassium voltage-gated channel Kv2.1 (right panel) in Cux2+ neurons in mouse cortex. ASAP voltage reporters are based on a circularly permuted GFP variant inserted within the voltage sensitive domain of a voltage-sensing phosphatase (Villette et al., 2019). (d) Optogenetic dopamine reporters (dLight1 sensors). (left panel) Simulated protein structure of the Dopamine D1 receptor (DRD1)-based dLight1 sensor, color-coded to denote key modules and components: inert DRD1 (purple), circularly permutated GFP (green), transmembrane regions (red, yellow) and linkers (white, black). (right panel) dLight1 plasma membrane localization in HEK cells (Patriarchi et al., 2018).
Voltage reporters
As with optical calcium reporters, genetic approaches have also been used to design optical voltage reporters, often referred to as genetically encoded fluorescent voltage indicators (GEVIs). GEVIs are capable of reporting subthreshold voltage dynamics, which are not resolvable using extracellular electrodes placed adjacent to individual neurons (Herreras, 2016). Further, as with all of the genetically encoded optical reporters described here, genetic restriction - both in terms of cell type and subcellular location - can greatly facilitate data extraction and analysis (i.e. optical de-mixing and back-end computational analysis, as described below). To this end, in 2018 Daigle et al. reported the first somatically targeted GEVI, which was achieved by fusing ASAP2s to a cytosolic segment of a potassium voltage-gated channel (Kv2.1); this provided subcellular localization (Figure 4c) (Daigle et al., 2018). This approach was subsequently employed with ASAP3 and Archon, to create ASAP3-Kv (Villette et al., 2019) and SomArchon (Piatkevich et al., 2019) (Fig. 4b), respectively.
Additional parameters essential for optical de-mixing of GEVI signals include the reporter’s fluorescence amplitude in response to single action potentials, and its molar brightness. The largest relative fluorescence responses to individual action potentials are currently provided by the red-excitable SomArchon (ΔF/F = 20–50% per AP) (Piatkevich et al., 2019) and the blue-excitable ASAP3-Kv (ΔF/F = 10–30% per AP) (Villette et al., 2019). Due to a difference in molar brightness between the two (0.076 mM−1 cm−1 for SomArchon versus 15 mM−1 cm−1 for ASAP3-Kv), illumination of ASAP3-Kv at ~25 mW/mm2 at the focal plane achieves the same SNR as illumination of SomArchon at 400 mW/mm2 (Villette et al., 2019). ASAP3-Kv and SomArchon thus provide two GEVI options at different wavelengths, with ASAP3-Kv requiring less power delivery to tissue.
One key drawback of GEVIs is the relatively fast sampling rate required. Actually, this drawback originates from the relative slowness of present-day instrumentation; fast-responding GEVIs such as Ace-NeonGreen or ASAP3 require sampling rates of > 500 Hz dynamics to optimally track their fast temporal response (Gong et al., 2015; Villette et al., 2019). Used in conjunction with 2p excitation, the instrumentation’s sampling rates restrict the number of points that can be sampled over a spatially-limited plane given the rather slow, serial nature of 2p microscopy (Villette et al., 2019; Wu et al., 2020). By contrast, genetically encoded calcium reporters respond to calcium transients triggered by action potentials that last for >100ms. For these slower responding reporters, frame rates of 15 to 30 Hz are sufficient to track reporter dynamics.
Integrated neurophotonic probes offer the possibility to record GEVIs “at speed,” given their intrinsically fast temporal dynamics, which (as described below) are sufficiently fast to follow even the ns-scale temporal decay of the chromophores (Choi et al., 2019). In addition, since light delivery by neurophotonic probes is much more strategically delivered within the illuminated tissue volume, background epifluorescence will be reduced compared to free-space 1p methodologies.
Neurochemical reporters
The development of genetically encoded, intensiometric and ratiometric fluorescence-based neurochemical reporters make it possible to perform direct, long-term, and chemically specific functional imaging of neurotransmitters and neuromodulation dynamics. A range of targets are now accessible, including dopamine, norepinephrine, serotonin, melatonin and opioid peptides (Leopold et al., 2019; Oe et al., 2020; Patriarchi et al., 2018; Sun et al., 2018). Generally speaking, there are two main types, i.e. design approaches, used to develop neurochemical reporters: G protein-coupled receptor (GPCR)-based reporters and periplasmic binding protein (PBP)-based reporters (Andreoni et al., 2019; Leopold et al., 2019; Ravotto et al., 2020). A recent example of the former, the GPCR-based dLight1 chemical reporter family (Patriarchi et al., 2018) couples conformational changes of inert human dopamine receptors to changes in the fluorescence intensity of circularly permuted GFP (cpGFP) (Fig. 4d, left panel). This provides a direct read-out of dopamine kinetics with broadly tunable affinity and dynamic range, relatively rapid kinetics (10 ms on and 100 ms off), and fast temporal resolution that matches the performance of electrochemical methods for detecting monoamines - while also providing subcellular resolution and molecular specificity. Additionally, the presence of the dopamine receptor transmembrane domain provides cell membrane targeting (Figure 4d, right panel).
In addition to GPCRs, bacterial PBPs have also been adapted for use as scaffolds for engineering small molecule reporters. Their use exploits the conformational change that occurs following binding of small molecules by its ligand binding domain (Leopold et al., 2019), sometimes referred to as a “Venus Flytrap” domain (by analogy), which shares similarity with binding domains of many eukaryotic chemical receptors (including GPCRs) (Acher and Bertrand, 2005; Felder et al., 1999; O’Hara et al., 1993). As with GPCR-based reporters, when coupled to a chromophore, this conformational change can be translated into a change in the chromophore’s optical susceptibility and, thus, its fluorescence (Leopold et al., 2019). PBP-based neurochemical probes, which include probes for glutamate (iGluSnFR), GABA (iGABASnFR), acetylcholine (iAchSnFR) and serotonin (iSeroSnFR), are brighter, have larger dynamic range, lower affinity, and faster kinetics compared to GPCR sensors. These attributes may mitigate problems such as buffering of native chemistry within the cytosol and interference with endogenous receptors (Marvin et al., 2018; Marvin et al., 2019).
The optical cross-sections of these chemical reporters are now similar to those of GCaMP reporters; hence, future advancements in subcellular targeting (for example, dendritic targeting of dopamine sensors) offers further potential for new insights. As the toolbox of genetically encoded optical reporters continues to grow, combining them with other reporters for simultaneous measurement of different functional read-outs becomes a way to dissect multiphysical information processing within brain circuits occurring in diverse physical domains (Figure 1). With the development of red-shifted dLight1 variants and the new classes of calcium and voltage reporters, use of a multicolor approach offers the possibility of investigating, in real-time, the simultaneous correlated activity of neurotransmitters, neuromodulators and spiking.
Chromophore excitation
Currently, serial scanning methods based on two-photon microscopy are widely employed to excite the chromophores within optical reporters. This involves simultaneous absorption of two photons to induce nonlinear excitation of the reporter. Subsequently, decay of the excited chromophore back to its ground state results the emission of a fluorescent photon in the visible spectrum. Near-infrared excitation wavelengths are typically used for biological microscopy given their longer attenuation lengths (Figure 6a). However, as the 2p optical cross-section is very small, extremely high photon density is required to induce 2p absorption. Accordingly, to achieve requisite intensities, 2p excitation requires use of a single, tightly spatially focused beam of pulsed light that is also temporally focused into femtosecond-scale pulses. To achieve volumetric sampling under these conditions, a serial point-scanning methodology becomes necessary. The typical two-photon interrogation voxel, generally of order ~0.5 × 0.5 × 2μm3, is thus scanned in 3D, one point at a time, to spatially map the activity-dependent fluorescence of reporters within an ensemble of neurons one-by-one. Today’s 2p-microscopes employing state- of-the-art acousto-optic deflectors (AODs) enable provide down to ~1μs point-access time and optical spike detection in multiple neighboring cells (~20) (Villette et al., 2019). This approach currently permits routine mapping of ~400 neurons in a 3D volume of 200 × 200 × 100μm3 with the requisite signal-to-noise-ratio (SNR) to track spiking activity via the resulting modulation of somatic calcium signals (Cotton et al., 2013; Grewe et al., 2010; Katona et al., 2012).
The aforementioned approach has two important limitations that greatly complicate attempts to scale it up to enable functional imaging of large neuronal ensembles over extended brain regions: (i) serial optical interrogation, and (ii) SNR degradation with depth. We discuss each in turn below.
Multiplexing limits of free-space optical interrogation
Serial point-scanning optical techniques can provide sub-cellular resolution, but they have the significant disadvantage that the total number of scanned voxels is limited by scanner speed. This is exacerbated by the photometric requirement to dwell at each voxel long enough to collect enough photons to attain requisite SNR. Simultaneous use of multiple excitation beams has enabled multiplexing by in-plane parallelization of two-photon microscopy with regular wide-field detection. In this implementation, each beam is encoded with specific binary amplitude modulation to identify the location where fluorescence is generated (Ducros et al., 2013). Depth multiplexing using four pulsed laser beams, has also been developed; this has been successfully applied to mapping cortical activity in four optical planes at four different depths (Beaulieu et al., 2020; Cheng et al., 2011). While these approaches enable significant multiplexing, in practice only a relatively small number of beams can be implemented. The maximum benefit obtained is ultimately determined by the number of beams multiplexed, the laser repetition rate, the reporter fluorescence decay time, and the total optical power that can be absorbed by brain tissue without undue perturbation to neuronal activity (or the tissue itself).
Signal-to-noise ratio limits to the depth of imaging
Scattering and absorption limit the ability to deliver ballistic (i.e., unscattered) light with sufficient intensity to achieve two-photon excitation deep within the brain. Ultimately, absorption limits the depth of delivery; in the near infrared (NIR) the maximum attenuation length is La ~500μm (Figure 6a). To overcome this significant limitation, several approaches have been explored. In one, the instantaneous laser power is increased to enable deeper two- photon excitation, while the pulse repetition rate is reduced to minimize the average power delivered to the tissue. This approach enables recording neuronal activity in populations of L5 neuronal soma up to ~800μm deep (Mittmann et al., 2011). However, collecting the visible- wavelength fluorescence photons from brain tissue becomes especially problematic at increasing depths. For these photons, scattering becomes the predominant limitation - as described below.
Extending this approach to achieve even deeper functional imaging becomes increasingly challenging; among issues are the generation of out-of-focus fluorescence (even with moderate spatial confinement along the beam), and the onset of nonlinear photodamage in neural tissue. A recent alternative approach involves using longer excitation wavelengths in the near infrared (NIR) around 1.6mm. This becomes possible if three-photon absorption processes are harnessed, and remarkable progress in this area has been achieved (Horton et al., 2013). Here, the principal complication is the significantly smaller 3p cross-sections for existing reporters; this imposes limits on the utility of this methodology, as much higher illumination intensities are required. An alternative approach employs adaptive optical corrections to rectify wavefront aberrations arising from spatially inhomogeneous optical scattering and absorption in brain tissue (Yaqoob et al., 2008) (Girkin et al., 2009). This can effectively restore optical resolution in the 2p modality and can significantly improve deep-imaging capability. The approach is contingent upon measuring and employing the precise aberration matrix for a large volume of heterogeneous tissue. This is a challenging prospect; it requires complex multi-point measurements and subsequent computations. As brain topology is dynamical, the correction matrix remains effective only for the interval over which tissue is, in effect, stationary. Future work will elucidate the realm of applicability of this approach.
Structured Illunimination
To separate fluorescent signal sources, laser-scanning microscopy localizes the illumination in space and time. We term this precisely structured illumination. Other fluorescence microscopy modalities structure their collection fields, sorting emitted photons by the place of their origin. For example, in wide-field microscopy signals are localized by focusing emitted light to form a spatial image. When neither the illumination fields nor collection fields can be structured with sufficient spatial and temporal resolution, fluorescent signals become mixed. In this case, computational approaches can be employed to separate or infer the underlying signals. Several recording and imaging modalities (including some fluorescence imaging modalities) use computational approaches to infer the image or optical signals from insufficiently structured illumination and collection.
In diffuse optical tomography (DOT), multiple spatially separated light emitters illuminate the tissue (usually one at a time), and the resulting scattered light is subsequently collected by optical detectors at many different spatial locations. In this modality, spatial resolution is increased by increasing the number of sources and detectors. This technique has yielded resolution of ~ 4mm for imaging blood oxygenation level dependent (BOLD) effects through the skull (Dehghani et al., 2008; Wheelock et al., 2019). Time-dependent diffusive optical tomography (TD-DOT) approaches, in which the emitters are pulsed and the time-of-flight (ToF) of the photons is determined at each of the detectors, improves the imaging capabilities by allowing better separation of scattering effects from those resulting from absorption or fluorescence (Azizi et al., 2009; Painchaud et al., 1999; Puszka et al., 2013).
Light-sheet fluorescence microscopy
(LSFM), also known as selective-plane illumination microscopy, is one example of a compromise in which some degree of structured illumination is maintained while achieving higher frame rates than are possible in a point-based laser-scanning system, and use of focused imaging to separate collected light. LSFM is a rapid, wide-field, volumetric imaging technique that enables volumetric imaging with optical sectioning (Chen et al., 2014; Hillman et al., 2019). In LSFM, a thin sheet of excitation light is generated, either by cylindrically focusing a beam or by digitally scanning a Gaussian or Bessel beam (Keller et al., 2008; Mertz, 2011; Power and Huisken, 2017). The sheet is scanned linearly across the sample as fluorescence images are sequentially collected perpendicular to the illumination plane. Stacking these sheet images then forms the desired volumetric image (Huisken et al., 2004; Keller and Ahrens, 2015). A complication of conventional LSFM is its requirement for two orthogonal objective lenses that must be specifically positioned spatially. This has largely constrained application of the technique to quasi-transparent organisms (e.g., larval zebrafish, C. elegans, Drosophila embryos) (Ahrens et al., 2013; Chen et al., 2018), chemically cleared mammalian brains (Keller and Ahrens, 2015), and brain slices (Haslehurst et al., 2018). Recently, swept confocally-aligned planar excitation (SCAPE) microscopy, an LSFM method requiring only a single objective, has been developed to circumvent these constraints. With SCAPE, in vivo calcium neural imaging has been demonstrated in mice (Bouchard et al., 2015; Voleti et al., 2019).
Acousto-optical techniques
Another approach to improving imaging depth is scattering tissue is to employ ultrasound- modulated optical tomography (UOT), also known and acousto-optic imaging (Resnik et al., 2012; Wang and Zhao, 1997). This has been used successfully to imaging absorption (Wang et al., 1995) and scattering (Kothapalli et al., 2007) at depth in tissue. Light is passed through an ultrasound beam. When it does so, it undergoes a frequency shift by multiples of the ultrasound frequency; by detecting the frequency-shifted light, resolution can be determined by the properties of the ultrasound (Wang, 2001). This tagging can be done at the source of emission. This approach is very similar to photoacoustic tomography (PAT) (Xu and Wang, 2006). In this case, acousto-optical effects are used to produce ultrasound upon optical illumination and this ultrasound is used for imaging. While ultrasound is much more penetrative than light, these techniques are still limited in depth by absorption of ultrasound energy, which increases with carrier frequencies. Higher frequencies are required to improve wavelength-determined resolutions. While imaging is possible at depths up to several centimeters, cellular resolution is not attainable at depths beyond ~3mm (Liu and Li, 2020).
Implantable microscopes
Several realizations of head-mounted microscopes for 1p and 2p calcium imaging in mice have proven the feasibility of fluorescence microscopy in compact form factors (Corder et al., 2019; de Groot et al., 2020; Ghosh et al., 2011; Jacob et al., 2018; Shuman et al., 2020; Skocek et al., 2018; Zong et al., 2017). To achieve imaging at depth, these instruments require implantation of the requisite GRIN lenses for these microscopes (typically 0.3 – 2 mm dia.), which results in rather significant displacement of brain tissue. Extending these devices to support LSFM requires generating light sheets parallel to the surface of the brain at arbitrary depths while ensuring tissue damage is minimal after implantation of the requisite elements. In (Ye et al., 2016), a light sheet was generated by a microchip using a nanophotonic grating coupler, a rather thick (>100 μm) glass spacer element, and a metallic slit lens. In another demonstration, a sizable millimeter-scale prism coupled to a GRIN lens for light sheet delivery was implanted alongside a second imaging GRIN lens (Engelbrecht et al., 2010). These examples remain limited by the significant tissue displacement of their implantable elements and their capability to generate only a single, static, light sheet.
Emission-related limitations
As mentioned, scattering in neural tissue drastically suppresses the yield of fluorescence photons that can be collected outside the brain via free-space optics. Fluorescent photons originating deep from within the brain are multiply scattered as they pass through tissue to its periphery. To efficiently capture them, free-space collection optics with large angular acceptance, i.e. large field of view and low magnification have been employed (Oheim et al., 2001). These can be rather costly and physically immense. To date, they have provided rather modest benefits.
The integrated neurophotonics paradigm
To surmount the limitations of free-space and endoscopic functional imaging described above, we have conceived of a new paradigm that we term integrated neurophotonics. It can provide the basis to enable fast and dense volumetric mapping of brain activity. It leverages recent advances in integrated silicon nanophotonics, nanoelectronics, and optogenetics, to enable massively multiplexed functional imaging arbitrarily deep within the brain. Employed together with optogenetic actuators and molecular reporters, these photonic neural probe arrays - realized by integrating all elements of a lens-less imaging system onto ultranarrow implantable silicon shanks - can enable dense interrogation of brain activity with minimal tissue displacement (Figures 5 & 7). As such, it contains elements of many of the approaches described above including leverage implantable CMOS electronics (as in scaled Ephys approaches), structured illumination (as in LFSM or scanning microscopy), and computation approaches that attempt to exact as much information as possible from scattered photon (as in diffuse optical tomography). Unlike DOT, however, this brings the emitters and detectors closer to the fluorescent sources, producing finer structure in local illumination and collection fields to resolve local signals even in a turbid medium such as brain tissue.
Figure 5. The integrated neurophotonics paradigm via photonic neural probe arrays.
Left) Schematic representation of a 625-shank photonic probe array module. a) Architecture 1, described in the text and Fig. 9a, is designed to record from 1mm3 of mouse cortex. b) We decompose the brain region bounded by four adjacent shanks into unit volumes delineated by the repeat distance of E- and D- pixels along the shank. c) For Architecture 1, each unit volume is surrounded by a small ensemble of E- and D- pixels that illuminate soma and collect fluorescent photons in their proximity. Right) Time-domain interrogation of reporters. d) After an action potential, the optical susceptibility of calcium reporters within a labeled neuronal cell changes. This is read out by a blue-wavelength excitation-pulse that produces a green-wavelength fluorescence transient. e) Photonic probes operate in the mesoscopic regime where proximal emitters and detectors are separated by only a few scattering lengths. This circumvents issues with functional imaging in highly scattering brain tissue. f) The emission peak for a typical GCaMP-family calcium reporter is separated by only ~20nm from its absorption peak, making continuous measurements essentially impossible; the excitation light is overwhelmingly more intense than the neuron’s fluorescence. For this reason, we operate in the time domain. g) Implementation of time-gating to reject excitation light to enable detection of the much weaker neuronal fluorescence.
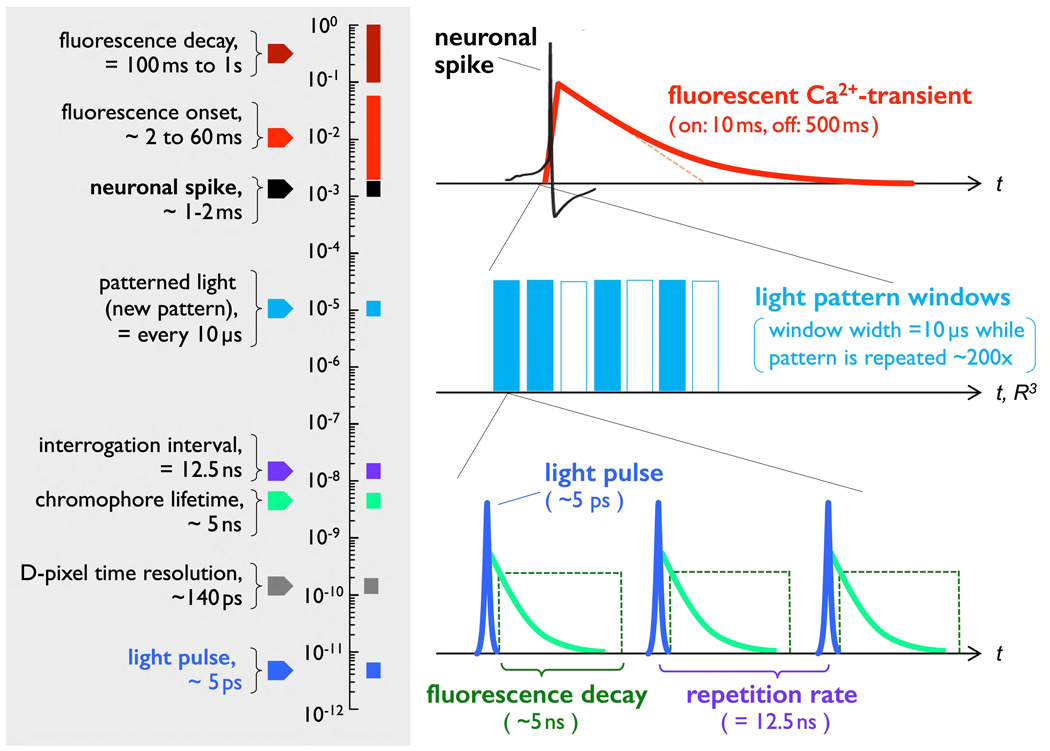
Figure 7. Temporal scales and the time-domain acquisition protocol.
The system’s fastest time scale is the duration of individual E-pixel emission pulses (~5ps) and the temporal resolution of the SPAD D-pixels (~140ps). As mentioned, together they enable resolving the fast temporal decay of reporter chromophore fluorescence following an excitation pulse (~5ns). Data acquisition sequences - a geometric pattern of pulsed, multiple-E-pixel light emission (5ps), followed by a D-pixel acquisition window (~10ns gate) - are repeated every 12.5ns (80 MHz repetition rate). Given the 5ns typical fluorescence lifetime of the reporter chromophore (green trace), this interval allows for a sufficient recovery period before the next interrogation. A specific light pattern is repeated as a train of ~800 data acquisition sequences are acquired and averaged; the stationary light pattern used during one ~10μs data acquisition window is then changed for the subsequent 10μs window. Thus, on the (relatively) slow time scale of a single action potential (~1–2ms), several hundred light patterns can be imposed, each of which is repeatedly signal-averaged roughly one thousand times to suppress photon statistics to enable acquisition of high SNR data and minimize energy deposited within the tissue.
Photonic-probe-based interrogation is unique in its ability to circumvent the scattering limitations of brain tissue. This is achieved by distributing the imaging components - specifically, geometrically-ordered arrays (architectures) of microscale photon emitters (E-pixels) and detectors (D-pixels) - within the brain, separated only by distances of order a few times LA (Figure 5e). The depth limitation for photonic probe functional imaging then solely arises from the readily-engineerable probe length and the actual depth of their implantation. This new paradigm requires the integration of a lens-less imaging system onto narrow implantable shanks. It also requires the creation of new back-end hardware - to control the instrumentation and stream the vast amounts of data it will produce. And, it requires new and efficient computational algorithms that transform the immense cache of raw data the system produces into a succession of time-sequenced, cell-specific functional recordings of neuronal activity. Together, these elements form a complete and ultrafast lens-less functional imaging system with an implantable front-end having microscale dimensions. Probe modules comprise an ordered array of shanks that can be readily implanted at arbitrary depths anywhere within the brain - either as an individual module, or as coherent module arrays to permit dense and extended volumetric coverage.
A fundamental concept: Source localization in highly scattering brain tissue
A pervasive mindset is that achieving cellular level resolution of sources in diffuse media, even at modest distances, is impossible because (as the thought goes) light scattering in neural tissue quickly and completely randomizes the direction of emerging photons. These challenges, for example, lead to the limitations of conventional DOT. For integrated neurophotonic systems, this problem is surmounted by positioning microscale emitters and detectors within the brain tissue separated by distances of order a few times LA. This is schematically depicted in Figure 5e. We term this the mesoscopic regime for light scattering, and it governs the performance of photonic neural probe array architectures. Several concepts clarify the underlying physics and provide intuitive understanding of this regime. First, structural imaging with high spatial resolution is not required to achieve functional imaging. Instead, it is sufficient to simply obtain sufficient information for source separation and localization, that is, to be able to deduce the cellular origin of photons collected by the D-pixels across an ensemble of diverse illumination patterns. Our goal, in contrast to that of conventional imaging, is to track the functional activity of individual soma. To facilitate this process, it is critical to employ the latest generation of optogenetic reporters that are somatically localized (Piatkevich et al., 2019; Shemesh et al., 2020; Villette et al., 2019). They serve to preclude dilution or complete obfuscation of somatic fluorescence signals by contaminating background light that would otherwise be generated by the neuropil. We seek to faithfully track the fluorescent photons emitted from these somatically localized reporters; they convey information about each labeled cell’s instantaneous state and activity. In this paradigm, we thus solely need to uniquely separate one neuron’s information from that of adjacent soma. This criterion significantly relaxes the requisite spatial resolution, making resolution at the cellular scale (~15μm) sufficient. Second, whereas imaging at high resolution unequivocally requires minimal scattering of a scene’s photons to avoid distortion or degradation, source separation and localization requires only a moderately faithful extrapolation back to the particular soma from which the photons originate.
Figure 6 shows that while ballistic photons are indeed scattered over short propagation distances, this scattering is overwhelmingly dominated by small-angle scattering. Hence, even after multiple scattering events, fluorescence photons propagating within the mesoscopic regime largely remain forward directed, and it is thus feasible to trace their origin back to a distinct soma.
Computational lens-less functional source separation and localization
The raw data acquired from lens-less photonic-probe-array imagers requires back-end computation to arrive at the desired information. For this paradigm, the overarching question is whether it is possible to computationally de-mix the ensembles of simultaneously recorded somatic fluorescence signals. The answer is obviously affirmative if the individual soma are each separately and sequentially illuminated - this is precisely how random-access, point-scanning, multi-photon functional imaging is achieved. Similar issues have recently been encountered, and solved, for Bessel-beam-based functional imaging (Lu et al., 2020; Wu et al., 2020), where multiple soma may be simultaneously illuminated along the beam’s path and contribute to the fluorescence signal. Here, the key is for our experimental instrumentation to provide sufficient spatial selectivity. By this we mean that the individual pixels’ illumination and collection fields (Figure 3c) are geometrically reduced in scale to provide tight, microscopic spatial resolution. By sufficiently increasing what we term the spatial diversity of the pixels, we would circumvent the need for de-mixing, in the same manner as for other microscopy imaging modalities with microscopic (diffraction-limited) point spread functions. However, increasing our system’s spatial selectivity to enable this ultimate level of resolution would require an immense number of E- and D- pixels with very high spatial diversity that strongly restricts of their individual angular ranges of emission and collection. Instead, we seek a more practical solution that maximally simplifies system architecture and complexity (and, ultimately, cost) - while attaining the more relaxed imaging goal of simply achieving signal separation. We have carried out detailed computations of linear de-mixing for a variety of E- and D- pixel architectures that are summarized in Figure 8 and discussed below. These initial efforts show that relatively simple configurations can indeed achieve this goal (Yatsenko et al., 2020).
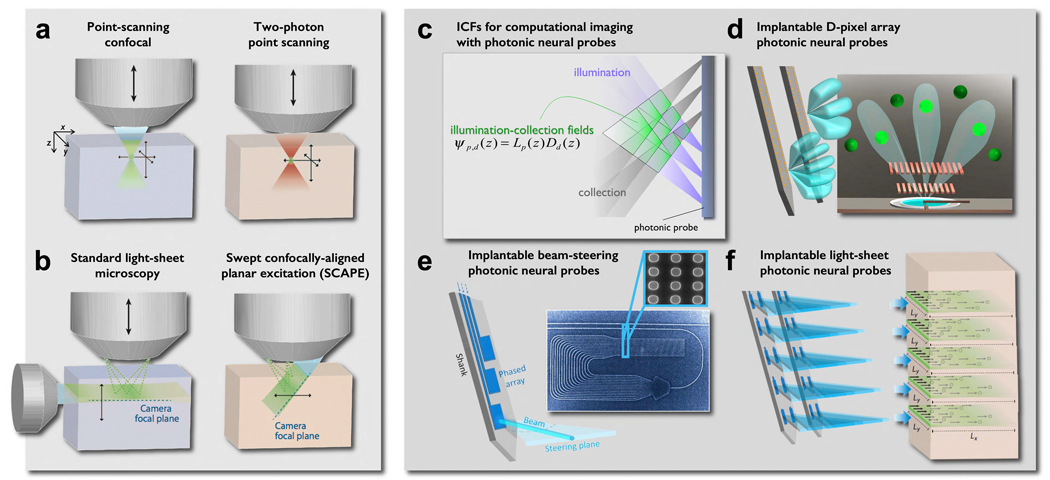
Figure 3. Functional imaging methodologies: free-space versus implantable, lens-less photonic neural probes.
– Left panel, Free-space Microscopy: a) Epifluorescence and confocal; multi-photon. b) Light- sheet microscopy in transparent tissues (denoted by bluish blocks), and oblique confocal scanning (SCAPE) in opaque tissues (brownish blocks). Panels a and b are adapted from (Hillman et al., 2019). - Right panel: Paradigm and components of photonic neural probes: (c) Concept of fluorescence interrogation voxels - illumination collection fields (ICF), overlay of illumination angular-fields produced by micro-sized emitter pixels (E-pixels) with detector angular-fields of micro-sized photodetectors. ICFs are analogous to the point-spread function, or optical-transfer function in optical imaging. (d) An angle-selective single-photon avalanche diode (AS-SPAD) detector pixel (D-pixel) arrays, where each D-pixel is equipped with off-axis Talbot gratings to yield an angle-restricted detection field. The diversity in spatial frequency, phase and direction in the Talbot gratings of each pixel allows maximally randomized spatial sampling of the tissue volume, allowing computational reconstruction. (e) Implantable beam- steering photonic probe. Using coherent light in the blue spectrum and an optical phased array, an implantable photonic probe enables micro-sized collimated beams to be scanned within the brain tissue by optical spectral addressing. Combining spectral (beam scan-angle, k) and spatial addressing (pixel number, i.e. phase-array element) enables scanning at different addressable depths. (f) Implantable light-sheet photonic probes imaging within opaque tissue. Photonic probes deliver blue light sheets enabling 2D-interrogation of fluorescently labeled neurons within selective and individually addressable planes. As photonic probes can be implanted at arbitrary depths, they provide access to regions that are impossible to image with free-space methodologies.
Figure 8. Computational approach to de-mix detector photon counts to obtain multiplexed time records of individual neuron activity.
a) Schematic depicting a top view of one 24-shank photonic probe array module, “design 1”, implanted in a 0.416mm3 volume of mouse cortex that is labeled with increasing density. b) Schematic of a probe-array architecture (designs 2, 3, and 4) providing dense coverage. c) Family of results at various labeling densities for three photonic probe architectures. d) Summary of the evolution of separability with increasing labeling density for the three module architectures. Here, we assume SNR>1 as the criterion for separability.
It is important to draw the distinction between inverse imaging (e.g., as achieved with DOT), and source separation and localization - which are critical to functional imaging. The goal of inverse imaging is to obtain an image as a function of spatial coordinates. If the image happens to be of a population of cells labeled with a fluorescent calcium indicator, the image can be segmented to measure the calcium signals for its distinct spatial location: in this case the signals are already separated based on their spatial origin. In contrast, signal separation refers to techniques that extract individual fluorescent signals from mixtures without ever producing an image. If sufficiently many independent recordings are made, each with differently mixed signals, special algorithms known as blind source separation (BSS) methods can isolate the individual voices from the cacophony (Comon and Jutten, 2010). This problem is related to the famous cocktail party problem that our brain solves, quite successfully, when we listen to a friend tell a story in a noisy bar. Blind source separation is a difficult problem to solve generally: it relies on the innate properties of the signals making the mixture: sparsity and synchrony, temporal structure and amplitude distribution. Finally, source localization solves the problem of assigning a spatial location to a signal extracted by source separation. In our evaluations through simulation, we focused on the necessary conditions for effective source separability.
The formalism
We have developed a mathematical formalism and numerical code to explore a variety of possible system architectures and quantitatively evaluate their performance for lens-less functional imaging. The ultimate metric for performance is signal separability, and we must understand how this evolves as fluorescent labeling density is increased. We define it as the percentage of labeled neurons that can be de-mixed, that is, isolated and spatially localized, to provide separate activity time records from large ensembles of neurons interacting in realistic measurement scenarios.
For our initial effort we set an ambitious target goal of recording, in real time, the activity of all neurons within a 1-mm3 volume of mouse cortex, comprising ~100,000 neurons. Although even partial coverage will be transformational, we have identified several specific probe-array architectures that permit such dense functional imaging (Figure 8b). Briefly, our computational approach involves, first, direct numerical simulation of photon propagation and scattering from spiking, fluorescently labeled neurons within a model of cortical tissue that captures the essential physics. With a prescribed 3D system architecture input (which can be varied), the simulations then permit evaluation of the photon counts received at each D-pixel within the array. These arrive from the labeled and active neurons within the target volume, which fluoresce in response to a programmed variety of illumination patterns generated by the E-pixel arrays. With this simulated data in hand, the second step is to assess the numeric properties of optical mixing to establish the feasibility of linearly de-mixing the acquired data to extract the functional activity of each individual neuron in the ensemble. The de-mixed output constitutes the desired collection of activity time records from each neuron within the targeted volume of brain tissue.
In Ephys, the spiking activity of individual neurons, termed single units, can be effectively isolated despite being mixed in a cacophony of many other signals picked up in multichannel electrical recordings. This de-mixing of spikes, or spike sorting, is possible thanks to each neuron’s precise and stereotypical spatiotemporal depolarization-repolarization pattern in the course of its action potential. These events are fast, on the order of 1–2 ms, and are separated in time by a refractory period; this further aids the ability of algorithms to recognize and isolate spikes.
dBy contrast, neuronal spiking activity measured optically with fluorescent calcium reporters is temporally blurred due to the relatively slow kinetics of both the intracellular calcium response and the Ca-reporter binding. This blurring can erase both the separation between spikes in time and confound any distinguishing characteristics of individual neuronal responses. Further complicating this situation is the shot-noise limited nature of low-light imaging, which results in much lower SNRs than for Ephys recording. These challenges preclude direct application of standard Ephys spike sorting algorithms for calcium imaging. Similar challenges exist for imaging with voltage reporters. Instead of spike sorting, for Ca functional imaging we employ modalities aimed at isolating individual cells optically, by focusing the illumination fields (e.g. laser-scanning microscopy), focusing the collection fields (wide-field microscopy), or focusing both (e.g. confocal and light sheet microscopy). Even for relatively good optical isolation, additional linear de-mixing may be required to isolate cellular signals that are conflated with those of the neuropil. To achieve this, a de-mixing matrix inferred directly from the data, by methods such as blind-source separation, is employed (Mukamel et al., 2009; Pnevmatikakis et al., 2016; Zhou et al., 2018). The aforementioned new classes of somatically restricted reporters help to greatly simplify these issues. For integrated neurophotonics we employ all of the advances described above, to enable lens-less functional imaging.
The role and importance of system architecture
Architectures and subcomponents for photonic neural probes can be configured to provide varying degrees of spatial selectivity. The simplest instantiations, however, can be too coarse to directly isolate cell bodies optically. Yet the probe’s illumination and collection fields (Figure 5e) can produce strong spatial gradients, thereby yielding distinct combinations of illumination intensities and collection probabilities from the arrays of E-pixels and D-pixels, respectively. Together they generate high-dimensional and highly mixed optical responses. The total number of linearly independent signals (the rank of the mixing matrix) from a photonic neural probe is equal to the product of the number of D-pixels and the number of linearly independent illumination patterns employed. With hundreds of emitter and detector pixels implanted within a cubic millimeter volume of neural tissue, the number of such linearly independent measurements can be immense - reaching hundreds of thousands, and thereby exceeding the total number of cell bodies in the same volume.
The question then becomes: Can the photon counts acquired by a D-pixel array, using optimal cycles of patterned illumination, be de-mixed computationally to isolate every neuron’s optical signal? To resolve this, we simulate signal acquisition from a population of neurons within the (approximately) one cubic millimeter tissue volume embedding a photonic-probe-array module. Further, we investigate how signal separability is affected by both module architecture and the attributes of their constituent D- and E- pixels. To be specific, we systematically alter the number, shapes, and spatial diversity of the pixels. The latter determines the geometric arrangements of the illumination and collection fields of the individual detectors and emitters and determines the spatial selectivity they provide as an ensemble (Figure 3c). For these different architectures and elements, the total fraction of neurons that can be effectively separated varies. To enhance separability, we also design specific algorithms to optimize the illumination cycles, i.e., sequences of emitter “on” times and the spatial patterns of light employed during a single acquisition window. Full details of this work are presented elsewhere (Yatsenko et al., 2020); here we provide an overview of the computational procedure, the prototype architectures explored, and the salient points we’ve gleaned. Model parameters are summarized in Panel A.
Architecture 1: Partial separability
The first design we consider comprises 24 shanks, forming a square array with a pitch of 283 μm X 283 (Figure 8a). Each of these shanks supports 72 E-pixels, arranged as nine separate rings, each comprising eight E-pixels. Along the length of the shank (z-axis) and between these rings of E-pixels are opposite-facing pairs of D-pixels. In total, the design features 1296 E-pixels and 288 D-pixels. The E-pixels measure 10 μm X 10 μm in size, whereas the D-pixels measure 10 μm X 50 The rings of E-pixels and the pairs of D-pixels are arranged uniformly along the 400 μm length of the shanks. The E-pixels have Lambertian emission profiles, thereby illuminating with a cosine intensity distribution in all directions over the hemisphere normal to the shank surface. Similarly, in Architecture 1 the D-pixels are receptive to all incident angles, resulting in a Lambertian collection profile. The total convex hull volume spanned by the shanks is 0.416 mm3.
We then estimate the ability of the probe-array architecture to separate the acquired optical signals by first carrying out a Monte-Carlo simulation of E-pixel-array light emission, propagation, fluorescence, and D-pixel-array detection. We start with the idealized case in which all fluorescence signals emanate from a number of discrete sources with a specified cell density. For example, these may be thought of as sparsely labeled cell bodies with the fluorescent reporters constrained within the soma. Although we model the cells as small spheres, all considerations below apply equally well to sources (neurons) of considerable spatial complexity as long as the optical signal they produce exhibits temporal coherence, i.e., appears as a single source, distinct from other sources, rather than a mixture of several sources with distinct spatial distributions.
The de-mixing process
The relationship between the instantaneous fluorescence states of the ensemble of labeled neurons and the photon counts received by the D-pixel array is represented by what we term the mixing matrix. To permit interpretation of the data acquired from an experiment using a photonic probe module, we seek its inverse, the de-mixing matrix. It estimates the configuration and time records of fluorescent signals from the sources, given the photon counts acquired by the D-pixel array. In a real-life recording, the mixing matrix is generally unavailable. Accordingly, well-established numerical methods for blind source separation are used to estimate the de-mixing matrix directly from the data (Comon and Jutten, 2010). In our simulations, however, since we impose both the architecture and the source properties directly, we have complete knowledge of ground truth. In this situation, we can obtain the de-mixing matrix using a regularized pseudo-inverse of the mixing matrix. This allows us to evaluate the numerical properties of the mixing process, and use it to subsequently iterate and optimize the effectiveness of various probe-array architectures and the characteristics (spatial diversity) of the E- and D- pixels they comprise, and devise specific illumination patterns and evaluate the results generated.
To carry out such optimizations, we examine the signal-to-noise ratios (SNRs) of simulated calcium spikes extracted from the signals de-mixed by a regularized pseudoinverse. Regularization allows estimating the de-mixed signals, the photon shot (quantal) noise in the de-mixed signals even when the mixing matrix is ill-conditioned for inversion. The computation also yields channel cross-contamination levels that arise from the regularization procedure so that source with excessive contamination can be excluded. When this contamination exceeds 1% of the de-mixed signal power for any of the fluorescent sources, we deem such sources inseparable from the signals of the surrounding population. We find the fraction of such sources is negligible for most of the architectures we have explored. For the remaining unbiased signals, we compute the SNR of a single calcium spike, or the de-mixed spike SNR. Showing that an effective demixing matrix exists is a necessary but insufficient condition for demonstrating that numerical demixing is possible in the absence of a ground-truth mixing matrix. Here, we employ de-mixing effectiveness as a principal design criterion for evaluating possible photonic probe-array architectures.
Time-domain data acquisition
Compared to the time scale of neuronal dynamics, our lens-less imaging system is extremely fast. Figure 7 shows the temporal hierarchy that applies to our paradigm.
We describe a typical temporal data acquisition sequence. For Architecture 1 we employ a cycle of 277 illumination patterns, in which specific collections of the 1296 E-pixels switch on for just ~5ps, i.e., just a fraction of the 10μs light pattern windows (Figure 7). After each light pattern pulse, D-pixel counts received are integrated for ~10ns. The same pattern is repeated for ~1000 times during the 10 μs light pattern window, to permit significant averaging of the photons counts received during each 10 ns data acquisition window. This serves to suppress shot noise and thereby yield good SNR. The volumetric and temporal average light intensity irradiating the tissue is set to correspond to a level commonly employed for multi-photon imaging. The light patterns we employ are designed to minimize overlap between simultaneously active illumination fields for uniform coverage and better signal separability. For Architecture 1, this illumination cycle yields a mixing matrix with 79,776 rows (277 illumination patterns X 288 D-pixels), yielding the effective number of measurements, or effective channels, in each recorded sample. For linear de-mixing, the number of channels also sets the upper limit for the number of separable sources.
Operating with pulsed illumination is important for circumventing what might otherwise result in an accumulation of diffusively propagating photons that create a contaminating light background. Such considerations are especially important for architectures where many multi-shank modules are tiled to cover extended brain regions. To circumvent a diffusive (multiply scattered) light background emanating from remote modules that could otherwise potentially contaminate a given module, only a single module’s emitters will be activated at any given time. This is feasible. while still preserving rapid data acquisition, because the base acquisition time is orders of magnitude shorter than the typical interval between action potentials (~1ms.) By temporally interleaving data acquisition from multiple modules sequentially, the background generated from an individual module’s excitation light will decay before an adjacent module is queried. In this manner, essentially all the background fluorescence generated by a specific activated module will decay sufficiently to avoid light contamination in subsequent measurements within adjacent modules. For the wavelengths employed, the lifetime of the cloud of multiply scattered photons is ~7.5ns, during which they diffuse ~2cm. With a repetition rate of 80 MHz (12.5ns intervals), the diffusive light contamination from one module to those adjacent to it will decay by a factor of 108, and thus contribute a negligible background.
Computational modeling: Interpretation and insights
To gain a deeper understanding of how the properties of the probe design affect the separability of signals, we decomposed the de-mixed spike SNRs into two factors: the normalized spike amplitude and the de-mixing factor. In Figure 8c, each point on the graphs represents a single fluorescent signal source (e.g., a neuronal soma), represented by its normalized spike amplitude (on the x-axis) and its de-mixing factor (y-axis). Normalized spike amplitude is calculated as the SNR of a single calcium spike due to one action potential, while all the other cells (sources) are kept at a fixed, average level of fluorescence. In this case, noise arises solely from the finite photon statistics. The de-mixing factor on the y-axis is the cosine of the angle between the given cell’s signal vector and the hyperplane formed by the activity of all other cells in the multi-dimensional signal space of all recorded channels. If a given neuron has a high normalized spike amplitude, it indicates the cell is well positioned with respect to a small ensemble of nearby E- and D- pixels (i.e., their spatial separations are within several optical attenuation lengths). A high de-mixing factor indicates that the cell’s activity produces a distinct pattern across the recorded channels, and that it can effectively be de-mixed from other signals. The de-mixed SNR is the product of the normalized spike amplitude and the de-mixing factor. In Figure 8c, we provide an isoline indicating de-mixed SNR levels of 1.0 (thick diagonal red line), and two thinner, parallel red lines demarcating SNR levels of 0.3 and 3.0.
When the fluorescent source density is low, e.g., 5000 sources/mm3 (~3000 sources within the embedding volume in Architecture 1), about 18.5% of cells yield calcium spike SNRs, after demixing, in excess of 1.0 (Figure 8c). As the density of sources increases with higher percentage somatic labeling, both factors of de-mixed spike signal quality deteriorated. First, the normalized spike amplitude decreases because of the more intense background fluorescence from the other cells. Second, the de-mixing factor also decreases since the distinctiveness of signal distributions across all the recorded channels is reduced with densely spaced sources. In other words, with increasing labeling density, the rising intensity of background light “washes out” the desired fluorescence signals. As a result, when labeling density approaches 100% (i.e., 100,000 neurons per mm3, the typical cell density in mouse cortex), only than 1.2% of all the neurons provide a SNR > 1.0 for Architecture 1, our initial and most simplistic system configuration (Figures 8c, d).
Architectures 2,3, and 4: Enhanced separability
For Architecture 1, the fast drop in signal separability with labeling density arises from both the fast decrease in illumination intensity from the Lambertian sources, and the lack of spatial selectivity in the emission and detection fields (given their broad and uniform intensity profiles.) These shortcomings can be addressed by enhancing spatial selectivity by making three modifications to increase what we term the spatial diversity of the pixels: narrowing the emission beams from the E-pixels, enabling steering of the emitted beams, and spatially modulating the angular detection selectivity of the D-pixels. Increasing the spatial diversity of both E- and D- pixels results in sharper focusing of the resulting illumination-collection fields.
The second design we present, Architecture 2, comprises 34 E-pixels and 33 D-pixels spaced uniformly along the full 1-mm active length of its nineteen shanks. In this case the shanks are arranged 200 μm apart, along rows spaced with 200 μm pitch, to form a triangular lattice (Figure 8b). The volume of tissue spanned by one module becomes 0.332 mm3. We assume the E-pixels measure 10 μm × 10 μm in size, and the D-pixels measure 10 μm × 20 μm Each pixel’s field is oriented 112.5° clockwise from pixel immediately above it on the shank, to form a helical pattern (Figure 8b). In this case the beams from the E-pixels are narrowed to 60° cones. Further, the beams from the E-pixels are steerable in the shank’s plane, in the direction along its length, by up to 60°. We again follow the same procedure, here with a pixel count totaling 850 E-pixels and 825 D-pixels, to design an optimal illumination cycle conveying a total optical power, again conveying 40 mW into the tissue. We separately integrate over signals acquired by imposing 127 distinct illumination patterns, which yields 104,775 recorded channels (127 × 825). The detection fields of the D-pixels were narrowed with a sensitivity profile modeled as the eighth power of the cosine of the angle between photons’ incident direction and the unit vector normal to the D-pixel surface. This restriction of the D-pixel spatial sensitivity profile reduces detection efficiency by 80% as compared to the case without beam narrowing - but it nonetheless results in significant improvement in de-mixing. This second architecture produces higher de-mixed spike SNRs that persist up to much higher source labeling densities (Figures 8c, d) — with 99.8% of cells producing single-spike SNR > 1.0 at cell labeling density of 5000 mm−3. Yet with higher labeling densities, the fractions of separable cells drop again so that, at 50,000 mm−3 labeling density, only ~20% (or about 10,000 cells) express demixed spike SNR>1.0 and, by 100,000 mm−3, this fraction drops to 5%.
In Architecture 3, further narrowing the E-pixels’ light emission profiles from 60° to 15° produces another sharp jump in performance approaching 95.5% demixing rate with spike SNR > 1.0 at 20,000 mm−3 labeling density, 73.1% at 50,000 mm−3, only dropping to 19.3% at 100,000 mm−3.
Thus, the probe design provides multiple control knobs for tuning the signal separability, including the numbers and shapes of the illumination fields and the number and shapes of the collection fields. To see what it would take to record a complete population, we transformed Architecture 3 into Architecture 4 by reducing the shank spacings to 150 120 μm (from the original 200 μm in Architecture 3), without changing other geometric properties. This reduced the recorded volume to 56.25%, but the spike SNR of spikes increased by a much greater degree, nearly 5-fold in Architecture 4, on average. Thus, in Architecture 4, 49.0% of cells yielded single-spike SNR > 1.0 after demixing at the full labeling density of 100,000 mm−3 (Figure 8d).
We find these results to be only slightly diminished when the scattering length, Lsc, is reduced by a factor of two, i.e. from 100 down to 50 μm - whereas decreasing Lsc precipitously (by a factor of 1,000) reduces separability to zero, as expected.
We note again that here we measure the SNR of spikes once the optimal demixing matrix is obtained through a source separation method. We do not yet tackle the problem of finding the demixing matrix. The mathematical formalism in these simulations allows us to select optimal designs that, at least in theory, can record from dense populations. These simulations demonstrate that the geometric arrangements and shapes of the illumination and detection fields are much more effective at controlling the SNR of demixed signals than the illumination intensity. The analysis highlights the key advantages of integrated neurophotonics over related methods such as DOT by bringing the E-pixels and D-pixels close to the imaged tissue and shaping the fields of illumination and light collection for increased spatial selectivity. Much work still remains in finding the optimal combinations of these arrangements and tradeoffs against other factors such as tissue displacement.
Photonic neural probes: The technological building blocks
The elements required to achieve the integrated neurophotonics paradigm are now being realized. Over the past 15 years, silicon integrated photonics has rapidly matured as a technology. Advances in this field now position it to engender new classes of mass producible and widely deployable nanophotonic technologies for neuroscience. Silicon integrated photonics leverages the worldwide CMOS manufacturing infrastructure to enable fabrication of photonic devices and circuits on large-diameter silicon wafers (200mm or 300mm), by following well-established protocols for very large-scale integration (VLSI) of electronic devices. These technological developments are commercially driven by the continually increasing bandwidth demands for data communication, and also by the increasing number of commercial and R&D scale semiconductor foundries now invested in and producing silicon photonic systems.
Although significant investment has been made worldwide to advance integrated photonics for telecommunications, the wavelengths employed by the telecom industry are in the near infrared (typically >1μm). This is unfortunate for neuroscience; it renders this extensive global technological manufacturing base largely incompatible with the requirements for addressing optogenetic actuators and molecular reporters operating at visible wavelengths. However, there are other CMOS-compatible materials beyond silicon that are transparent at visible and near- infrared wavelengths; these are now being carefully engineered to be compatible with the processes and protocols used in CMOS foundries. These provide the basis for visible wavelength integrated photonic technology, which is advancing steadily, but is still in its infancy. We and others are currently pursuing this avenue to enable mass-producible visible wavelength integrated photonic and neurophotonic systems (Fowler, 2019; Jans et al., 2018; Sacher et al., 2019c). Realizing visible-wavelength photonic devices and circuits is challenging compared with infrared photonics. Given the smaller visible wavelengths, they necessitate smaller feature sizes and much tighter control over fabrication variations and surface roughness. Nonetheless, these challenges are now being surmounted through careful design and a succession of engineering refinements. The field of visible-wavelength integrated neurophotonics is beginning to leverage robust industrial-grade silicon integrated photonic technologies to produce miniature implantable imaging systems, en masse. Only through use of robust and stable foundry processing can photonic components be fabricated in the requisite volume, precision, and sophistication required to meet the needs of integrated neurophotonic systems.
Passive E-pixel arrays
Microscale light sources typically operate only with modest efficiency; hence they can dissipate significant power when operated. Neural activation thresholds (Goldin and Mindlin, 2017; Moser et al., 1993; Owen et al., 2019; Thompson et al., 1985) and blood flow (Rungta et al., 2017) can be significantly altered by only small temperature changes. Accordingly, for our systems we produce and control light off-probe for optogenetic actuation and chromophore excitation, to preclude risk of local tissue heating. The output optical power of a single E-pixel used in our proposed architectures (20μW, 200mW/mm2) is within the range of commercial optogenetic hardware (Senova et al., 2017), and would be expected to correspond to only a modest local elevation of temperature based on thermal imaging experiments of optical stimulation of brain tissue (Senova et al., 2017). In addition, on average, per illumination frame (2ms) only 75 E-pixels are on simultaneously, and the same pattern of ON pixels is not repeated consecutively. Each illumination pattern has a duty-cycle, roughly once every ~100 frames which leads to an average power of 7.4mW in mm3 of brain tissue at most (Yatsenko et al., 2020). We have already experimentally validated that our power levels (~10–100μW per emitter pixel) are non-damaging, both in vitro (with brain slices) and in vivo (in mouse) (Segev et al., 2017). The power range of 10–100 |aW per E-pixel corresponds to, or is below, the typical power levels used in photostimulation of opsins. For the simulations, we have kept to the low end of this range.
Our present approach for realizing implantable photonic probes E-pixel arrays thus focuses on passive, on-probe nanophotonic components. Our specific needs in this regard do not detract from the excellent advances in neurotechnology recently made with on-probe light sources. Here we mention the significant achievement of combining LEDs and Ephys electrodes for concurrent optogenetic stimulation and electrophysiological recording (Mohanty et al., 2020; Wu et al., 2015). For our present application, however, many hundreds of microscale light sources are required within small tissue volumes - and this probably precludes use of active emitters, unless heavily duty-cycled operation can be employed to keep average power sufficiently low. As mentioned, today a minority of efforts in photonics VLSI focus on visible wavelength components. For this effort, we have developed a complete fabrication process for photonic components using thin-film silicon nitride (SiN). This material is transparent at visible wavelengths and is compatible with CMOS processes.
Optogenetic simulation with microscale beams: First experiments
We first demonstrated the functionality of the first photonic probe prototypes by achieving single cell optogenetic stimulation (Segev et al., 2017). A single E-pixel photonic probe was implanted within a mouse with cortical neurons co-expressing both ChR2 and GCaMP6s. The photonic neural probe provided local, cellular-scale illumination to induce optogenetic stimulation, while neuronal activity was recorded simultaneously via free-space, two-photon calcium functional imaging. The photonic probe was inserted into cortical layer 2/3 of an awake, head-fixed mouse. The probe-generated illumination beam was directed upwards from the probe surface into the brain tissue, and a local population of neurons was imaged approximately 130 μm above the probe tip. The microscale beam width at the imaging plane was only ~20 μm FWHM, i.e., roughly of cellular dimensions.
Spatial addressing of E-pixels
To increase the number of E-pixels that can be addressed with a single wavelength, we have successfully realized a spatial addressing strategy first devised by Zorzos, et al. (Zorzos et al., 2012). Using a MEMS scanning mirror with ~10 ms switching times and relay optics, laser light is directed to different cores of a fiber bundle comprising thousands of micron-scale optical cores. The fiber bundle is edge-coupled to the probe chip using on-chip tapered edge couplers; this enables broadband optical coupling over the visible spectrum. Once on-chip, light is then routed by integrated photonic waveguides, delivered to grating-coupler-based E-pixels, and emitted into the tissue. We have previously employed wavelength division multiplexing (Bannerjee et al., 2005) to allow selection of specific E-pixels by small changes in wavelength within the Opsin absorption band (Fowler, 2019; Segev et al., 2015). To free wavelength control to tune another parameter (emission angle, described next), instead we have employed in our recent work the aforementioned MEMS-based multiplexing scheme.
Scanning microbeams (coherent beam-forming phased arrays)
With the spatial addressing of E-pixels as described above, laser wavelength is freed to provide an independent degree of freedom for control. Employing this, we have developed beam- steerable E-pixels based on optical phased arrays, wherein modulation of the input wavelength results in a modulation of the output angle of the E-pixel beam (Figure 9). Again, to employ this strategy, the wavelength range used for tuning is kept within the absorption band (typically 40–50nm wide) of the actuator and reporter chromophores we employ. When implanted in brain tissue slices, these beam-steering E-pixel probes demonstrate highly collimated, steerable beams of light emanating from grating couplers. Their successful operation in brain tissue confirms that the emitted light remains spatially coherent in brain tissue over distances of a few hundred microns, despite strong optical scattering. Our first generation of steerable E-pixel probes are comprised of 4 shanks, each populated with 4 compact, visible wavelength phased arrays in 2 configurations employing: i) a 20 nm wavelength steering range, or ii) with larger phased arrays, a 6 nm range. Future optimization can readily provide significant performance enhancements. The requisite on-probe photonic circuitry includes: a single-mode input waveguide, a star coupler to split the input light into an array of 16 waveguides, and an array of gratings on a constant sub-micron pitch to form the compound E-pixels that emit light from each of the 16 waveguides. Our use of narrow individual grating elements results in a large emission cone along the array axis, and array emission interference that is complete within ~100 μm of the emitter.
Figure 9. Photonic neural probes.
First-generation hardware developed for implantable probes includes microscale components for cellular-scale patterning and delivery of visible coherent light, and components for lens-less functional imaging with cell-type specificity. Passive emitter-pixel arrays (E-pixels) operating in the visible spectrum: (a) Scanning E-pixels, blue light: Microscale beam-scanning phased array photonic neural probe operating with blue light (480nm). SEM of phased array E-pixel design with ~6 nm free spectral range (FSR). Top-down images of phased array emission pattern at multiple addressing wavelengths. Continuous scanning is possible by sweeping the input wavelength within the FSR (spectral addressing) (Sacher et al., 2019b). b) Light-sheet array photonic neural probes. Probe-based light patterning using the composite emission of four microscale E-pixel arrays based on grating couplers, which are fed with tightly controlled optical modes to produce addressable light sheets with ~10–20 μm thickness. The photonic probe chip is coupled with fiber bundle/array through facets of single mode waveguides of single mode fibers. Spatial addressing using a MEMS mirror allows temporal addressing of multiple planes and provides the requisite intensity per sheet to induce fluorescence by the one-photon excitation process. (Sacher et al., 2019a). Detector-pixel array (D-pixels) based upon visible spectrum angle-selective single-photon avalanche diode (SPAD) detectors: First-generation, minimally invasive photonic neural probes for lens-less functional imaging (Choi et al., 2019; Choi et al., 2020, Lee et al., 2019). (c) Images of a SPAD-based D-pixel array in clockwise order: scanning electron micrograph (SEM) of a rectangular CMOS die post-processed into shanks, waterproof flat flexible cable (FFC) assembly used for in vivo insertion, and shank with an epoxy-based 10um- thick absorption filter coating. (d) Micrograph of CMOS imager probe consisting of 512 SPADs along two 4mm-long shanks. Each pixel is masked with semi-unique off-axis Talbot gratings that vary in angular direction, angular frequency, and phase (four examples shown at top), resulting in one of sixteen distinct detection fields (bottom). The 16-pixel ensemble consists of two detection field directions (x-z, x-y), two angular frequencies, and four phases to minimize overlap between neighboring pixels. Pixel pitch: 25um x 75um, PDP (median): 16.8%, DCR (median): 40 Hz, Time-gate resolution: 140ps, Max frame rate: 50 kilo-frames/s
These prototypes have been validated in vitro using adult brain slices of transgenic mice expressing a fluorescent reporter (YFP) and an opsin (ChR2). Continuous beam-steering over ±14° is achieved in mouse brain slices with the capability to deliver 100 μW of optical power. These first-generation prototypes provide large, parallel, multi-beam illumination coverage. This will enable a multiplicity of independent illumination fields in brain tissue, as depicted in Figure 3c (Fowler, 2019; Sacher et al., 2019b).
Implantable probes for light-sheet illumination at arbitrary brain depths
We have also developed implantable neural probes that generate controllable and addressable light sheets at depth for light-sheet fluorescence microscopy (LSFM) (Fowler, 2019; Sacher et al., 2019b). Figure 9b shows these photonic neural probes deliver sheets of light parallel to the surface of the brain. The light sheets are synthesized from the emission of rows of grating couplers (GCs) that are designed to provide large emission divergences along the sheet width axis and small divergences along the sheet thickness axis. An array of input edge couplers is spatially addressed by the methods described above; they enable independent control of the multiple light sheets. Our first-generation prototypes generate five sheets that can be rapidly switched (Fowler, 2019; Sacher et al., 2019a, Sacher et al., 2020). Working with our photonic foundry collaborator, Advanced Micro Foundry (AMF), we fabricated these probes using a 200 mm wafer-scale process. Using free space beam profiling, average light sheet thicknesses between 10 and 15 μm were measured for sheet propagation distances between 100 and 300 μm.
Current limitatons and developments
Critical for scale-up and mass production are robust and reproducible methods for what is termed device packaging. While envisioning the packaging requirements for a planar electronic chip is straightforward, for integrated neurophotonic systems this is more complex. Here, we require a rather significant number of high-quality photonic and electronic connections to the outside world. Accordingly, new and sophisticated packaging methods must be innovated. Our present methodologies (as well as those of most other groups) are still more akin to handcraft than to bona fide engineering technology. While focus upon this area may at first glance seem rather pedestrian, development and investment of in packaging methods for integrated photonics is essential for this technological paradigm to become robust, reproducible, and widely deployable. Ultimately, to achieve mass production, these packaging processes must be automated. It is worth noting that microchip technology for electronics also had to carefully address similar issues to attain mass production, robustness, and reproducibility.
Currently, a principal source of optical loss in our neurophotonic probes is the fiber-to-chip edge couplers, which exhibit coupling efficiencies of ~15% at the 488 nm wavelength in our current work. In addition, the aperiodic spacing of the cores in the fiber image bundle coupled to the probe chips add insertion loss due to misalignment between the fiber cores and the edge couplers. By adopting more complex, multi-layer photonic integration - wherein multiple waveguide layers are vertically integrated on-chip - these optical transmission limits will be surmounted. Following similar strategies used for Si photonics at infrared wavelengths (Fang et al., 2010; Park et al., 2013), our simulations indicate that bilayer SiON-SiN edge couplers can achieve broadband coupling efficiencies approaching ~80%. Furthermore, integrating multiple SiN waveguide layers that can cross over one another with low optical losses and crosstalk (Sacher et al., 2017) will enable higher densities of grating coupler emitters.
Active D-pixel arrays: SPADs
Concurrent with our development of integrated photonics for E-pixel arrays, we have also designed and fabricated CMOS photon-counting imaging arrays with the form factor of an implantable shank. These permit fluorescence imaging deep within brain tissue (Figure 9) (Choi et al., 2020; Choi et al., 2019; Lee et al., 2019). In one of our recent prototypes, a D-pixel array consists of 512 single photon avalanche diodes (SPAD) aligned in a linear configuration spanning the length of the shank. Their field-of-view is to the side, to maximize the ratio of observable tissue to that displaced during insertion (Rustami et al., 2020). This configuration allows the probe to monitor neurons along its length, in contrast with typical endoscopic fibers that solely image near their tips. This advance provides an order-of-magnitude larger field-of- view. Digital addressing and data readout of these pixels also makes our solution highly scalable - in this prototype, 32,768 pixels are multiplexed using only 15 parallel wires running along the shank. As mentioned, an important design goal for miniature neural probes is attainment of small cross-sectional areas to suppress immune response and avoid gliosis after implantation. Accordingly, we post-process the acquired rectangular CMOS dies into 100μm wide, 40μm thick shanks, with length of 4.1 mm. This yields an insertion cross section similar to that of a 70 um-diameter endoscopic fiber.
To further minimize tissue displacement, we eschew the relatively thick focusing lenses typically employed for conventional microscopy. Instead, we incorporate angle-sensitive single-photon avalanche diodes (SPADs). These employ the Talbot effect, by means of CMOS metallization layers situated 3–6 um away from the SPAD active layer. Together they shape the angle-dependent collection fields of the photodetectors (Wang et al., 2009). This very compact solution is ideal for implantable photonic neural probe applications (Figure 9), as compared to other lens-less imagers, e.g., far-field instantiations that require a 200 μm spacer in the light path (Adams et al., 2017). Additionally, on top of the CMOS devices we fabricate compact single-micron thickness spectral filters, which combine absorption and interference filters (Sasagawa et al., 2018). These provide additional spectral filtering between the bright emission pulses delivered by E-pixels and the weak, photon-limited fluorescence signals detected by D- pixels. To further suppress source-detector crosstalk, we also employ fast, time-gated pixel operation in sync with a pulsed laser driving the E-pixels, as is often used for fluorescence lifetime imaging (Ulku et al., 2019; Wang et al., 1991). Together, the absorption, interference and time-gating are employed to reject the blue light employed to excite the fluorescence of the optical reporters.
The 512-pixel SPAD array is fabricated with a 6.3% fill factor, and a pixel pitch of 25.3 um along the length of the shank (Choi et al., 2020; Choi et al., 2019; Lee et al., 2019). Each D-pixel provides a photon detection probability (PDP) larger than 10% in the visible fluorescent spectral band of most biomarkers (500–600 nm), with a median dark count rate of 40 counts per second (cps) at room temperature. The power dissipated on the shank is dominated by the charging of the SPAD diode capacitance at quenching events, consuming a total of 97 μW under dark conditions and 68 mW in full output saturation lighting. There is much headroom for further optimization; 2D SPAD arrays have recently been demonstrated with a pixel pitch as small as 2.2 um (Morimoto and Charbon, 2020), 61% native fill factor (Gyongy et al., 2018). Today’s state-of-the-art 3D integration techniques will enable even higher fill factors to be attained.
Each pixel is masked with two layers of parallel Talbot gratings to create an angle-dependent field of view. Different combinations of angular modulation direction (x, y), frequency (β = 12, 20), and phase (α=0°, 90°, 180°, 270°) give rise to sixteen uniquely shaped collection fields, which are overlaid upon the 512-pixel array (Wang et al., 2009). We employ the combined information from these collection fields to increase the spatial selectivity of our system, and this greatly enhances the effectiveness of computational procedure we employ to attain source separation, as described below.
The post-processed imaging shank, wire-bonded and packaged to be waterproof, links to an field-programmable gate array (FPGA) chip through a 51-wire flat flexible cable. On the FPGA, a microcontroller, state machine, and data-FIFO control the photon collection and frame readout with a maximum rate of 50 kfps (i.e., 20 μs per frame). This rate can be readily increased in future generations. The time-gate filter, implemented as a phase-locked loop (PLL) on the FPGA, can be fine-tuned with a resolution of 140 ps for optimal signal collection. The FPGA then links to a computer through a USB-3 link that displays a real-time representation of photon counts in 3D volume (Choi et al., 2019). To illustrate the speed of our D-pixel imagers, we have recently demonstrated nanosecond-scale fluorescence imaging a standard fluorophore with one of these shanks (Choi et al., 2019; Lee et al., 2019). Accordingly, tracking the fastest of today’s voltage reporters is assured.
Co-Integration of D- and E- pixel arrays
Ultimately, photonic neural probe-array architectures must provide not only dense tissue coverage, they must also be produced by a cost-effective strategy that enables co-integration of their many electronic and photonic sub-components. We foresee four potential phases of technological evolution in this regard. Initially, imaging “modules” will be straightforwardly assembled from separate E-pixel and D-pixel probe layers that are first thinned, and then subsequently stacked back-to-back individually.
As mentioned above, optimal consolidation of all the separate microscale imaging components for mass production must eschew piecemeal assembly; instead, true wafer-scale co-integration will be required. This must ultimately be achieved at the foundry by heterogeneous (multiple materials) process integration. In the simplest, second phase, the photonic and electronic devices can be separately integrated and subsequently conjoined by wafer-scale bonding. In this case, the photonics layer will include openings to provide access for the underlying D-pixels to clear, photon-collection-path vias. This will enable functionality from both the front and back of the shanks. A third, more complex approach will involve wafer-scale production of the photonics layers in post-process, that is, after the wafers containing the CMOS imagers are foundry-fabricated. This will be executed directly upon the electronics wafer containing the D- pixels arrays and their associated circuitry. The fabrication process must make use of low- temperature SiN processes to avoid compromising the performance of the CMOS imaging elements; PECVD SiN with a 300°C deposition temperature will be employed. Such processing is compatible with CMOS circuits, and the material provides sufficiently low absorption at visible wavelengths (Gorin et al., 2008; Sriram et al., 1983). More recently, even lower temperature (75°C) SiN processes show sufficiently low infrared losses that may translate to the visible (Shao et al., 2016). In an ultimate fourth phase, where production is advanced to cost- effective large-scale manufacturing, 3D integration of separately processed photonics and electronics layers will be employed. This is the most advanced solution with significant up-front costs, but it will provide the highest level of dimensional accuracy, integration density, and device complexity - and, ultimately, enable the lowest per-unit costs once initial engineering expenses are invested. Our approach to this complex undertaking involves the previous three phases; by proceeding through the first three intermediate steps we will ensure success of this final, and most complex, undertaking.
Conclusions and projections
The integrated neurophotonics paradigm: Status quo
Over the past several years, we have achieved many significant milestones that advance us towards attaining full realization of this exciting, albeit complex, paradigm of integrated neurophotonics. We have developed reliable and low-loss wafer-scale mass-production processes for visible-wavelength integrated nanophotonics by working with foundry partners. With this in place, we have made significant advances including: demonstration of coherent optical beam formation within brain tissue; realization of beam-steerable phased arrays of micro-emitters (E-pixels) and selective-plane (light-sheet) illumination with implantable E-pixel probe arrays; and creation of implantable, angle-selective single-photon microdetector (D-pixel) arrays. These constitute the fundamental building blocks for complete implantable visible- wavelength functional imaging systems with microscale dimensions. Further, we have validated the foundational physics for the integrated photonics paradigm by implementing computational methods to evaluate the fluorescent photon yield in scattering media. This has elucidated the regime of mesoscopic light scattering, in which ensembles of closely spaced microscale imaging components - and the integrated neurophotonics paradigm - operate. We have also developed a mathematical formalism and a framework for numerical simulations to compare and optimize the expected signal quality for various system architectures. The ultimate metric for performance is signal separability, and we have explored this as a function of increasing optogenetic labeling density (from sparse to dense). With these tools we have validated our computational approach for optical source separation and localization and, thereby, have been able to identify practical, realizable architectures that can enable dense volumetric activity reconstruction at depth.
Together these developments validate the potential of the integrated neurophotonics paradigm for achieving fast, dense, and deep functional imaging in brain tissue. This new technological approach has promise to surmount the limitations of existing methodologies. It is being developed to enable: i) optically-based electrophysiological recording and stimulation in real time - with cellular resolution, in all regions of the brain, no matter how deep; ii) dense coverage of neuronal populations within large target volumes; iii) cell-specific interrogation (via targeted optogenetic actuators and molecular reporters) permitting complex and finely tuned activity control. Finally, these systems are based on mass-production processes routinely carried out in existing electronic- and photonic-chip foundries. The ultimate prospect of wide dissemination of complete measurement systems to the neuroscience community, given requisite support, is thus assured.
What is the requisite technological “basis” for mapping brain circuits?
A public debate emerged in conjunction with the launch of the U.S. BRAIN Initiative concerning the optimal path forward for unraveling fundamental mechanisms, or computational motifs, that underlie brain function. Identifying a specific and sufficiently complete basis set of experimental measurements to enable such elucidation is essential. Knowledge of this basis will, in turn, enable us to identify the requisite neurotechnologies to acquire such data. A prevalent view is that connectomics and large-scale measurements of brain activity - among the most widely applied of modern approaches that provide anatomical and functional maps, respectively - are both complementary and necessary. But it has also asserted that even this combined approach will likely ultimately prove insufficient. The quest is further complicated by the fact that brain circuits employ other physicochemical activity domains for control (Figure 1b) - among which are dynamical alteration of neurons and circuits through spatiotemporal neuromodulation of channels, synapses, and local biochemistry. And compounding the complexity of this measurement challenge is the immense variety of distinct cell types encompassed within brain circuits, each of which can carry out unique, complementary, and coherent functions through interactions with others.
With this perspective, it seems that the requisite technological basis for mapping brain circuitry will ultimately likely include:
acquiring knowledge of anatomical connections (connectomics);
assessing synaptic connection strengths;
monitoring localized electrophysiological activity on a massively parallel scale;
precisely stimulating neural activity at the cellular, if not synaptic, levels (i.e., circuit “interrogation”);
identifying the individual cell types that participate; and,
observing, simultaneously, the spatiotemporal dynamics within multiple activity domains – among which include the biochemical (for neuromodulation, as mentioned); and the mechanical (to assess biological forces that arise, for example, in synaptogenesis.)
We especially wish to highlight the latter point. The necessity of acquiring real-time multiphysical data, simultaneously, from a multiplicity of measurement domains underscores the need for new hardware (and associated software) for brain activity mapping that profoundly transcends present-day capabilities. Further, by multiplexing the emission and excitation wavelengths of the chromophores comprised within molecular reporters, and assembling multicolor integrated neurophotonic systems to interrogate them, activity from different domains (Figure 1) can be simultaneously monitored (Cohen et al., 2018). This will enable the much more comprehensive multiphysical mapping of brain activity we envisage. As we have emphasized, integrated neurophotonics can be readily adapted for such investigations.
Projections
The convergence of these new technologies - optogenetic actuators and reporters of brain activity with specificity of cell-type, and mass-manufacturable photonic and electronic integrated circuitry enabling microscale implantable systems - now provide an unprecedented opportunity to realize a new class of multiphysical functional imaging tools for neuroscience. We anticipate these powerful tools will transform brain activity mapping, will enable new types of closed-loop brain interfaces with unparalleled recording capabilities, and will enable a powerful paradigm for multiphysical interrogation of the brain activity with spatial and temporal resolutions that cannot be attained by existing methodologies.
Panel A: Computational Analysis.
We model photon propagation by trajectory calculations that are limited by an absorption length, Z.ab, a scattering length Lsc, and a Henyey-Greenstein anisotropy factor, g, which accounts for the strong forward Mie scattering that occurs in brain tissue. These parameters are shown in the table at the right.
Characteristics of the probe modules that are employed in these calculations - specifically, the E- and D- pixel densities and their angular profiles, and the probe array architectures - are fully described in the text.
We assume the same transport parameters for both excitation and fluorescent light. Physical parameters used in these calculations vary minimally from the peak of the absorption band (480nm) at the peak of the emission band (515nm).
Parameters we employ for our signal separability calculations are also shown at the right; the ΔF/F value employed corresponds to the performance of new soma-restricted calcium indicators. The effective absorption cross-section of a neuron at maximum fluorescence is estimated by assuming a uniform 50 μM somatic dye concentration within a spherical volume of 7 μm radius. This corresponds to 3.3 × 107 molecules per cell, a 2.3 × 10−16 cm2 cross-section for each fluorophore molecule, and a quantum yield of ϕ = 0.6. The E-pixel emission level is comparable to power commonly employed in other imaging modalities. We have validated it is below threshold for tissue damage and phototoxicity by experiments both in vitro (brain slices) and in vivo (mouse).
Full details underlying these signal separability calculations are reported in (Yatsenko et al., 2020).
| Parameter | Value |
|---|---|
| Photon scattering length, Lsc | 50 μm |
| Photon absorption length, Lab | 1.4 cm |
| Anisotropy coefficient, g | 0.88 |
| Photons/joule (480nm) | 2.4 × 1018 |
| Fluorophore optical cross-section | 5 × 10−9 cm2 |
| Reporter resting/maximum fluorescence levels | 0.05 |
| Δf/f for a single action potential | 40% |
| Calcium transient time constant | 1.5 s |
| D-pixel detector quantum efficiency | 0.65 |
| D-pixel dark noise | 300 counts/sec |
| E-pixel emission level | 20μW / emitter |
Acknowledgements
MLR, AST, and KLS gratefully acknowledge funding from the NIH (Grant Nos. NS090596, NS009717), NSF (Grant Nos. 1265055, 1403817), DARPA (Grant Nos. N66001-17-C-4002, N66001-17-C-4012), IARPA (Grant No. DP1EY023176), and the Kavli Foundation through Caltech’s Kavli Nanoscience Institute’s “Fill the gap” award program. JKSP acknowledges the Natural Sciences and Engineering Research Canada, Canadian Institutes of Health Research, and Canada Research Chair program.
We thank the following individuals for their participation in various phases of this work: Joe Redford (initial numerical simulations); Eran Segev, Derrick Chi, Trevor Fowler, Xinyu Liu, Kukjoo Kim, Warren Fon (nanofabrication); Fu-Der Chen, John Straguzzi, Thomas Lordello, and Ilan Felts Almog (photonic probe characterization); Andres Lozano, Anton Fomenko, Taufik Valiante, Homeira Moradi-Chameh, Prajay Shah, Zach Blumenfeld (in vitro and in vivo experiments).
We thank François Berger, Miyoung Chun, Andrei Faraon, Leslie Greengard, Gilles Laurent, Konrad Kording, and Liam Paninski for helpful discussions. We especially thank our reviewers for their insightful comments.
We are grateful for the participation of our foundry partners, including scientists and engineers at: CEA-Leti (Grenoble, France): Laurent Duraffourg, Bruno Paing, Salim Boutami, Jean-Marc Fedeli, Benoit Giffard Pierre Labeye, Hughes Metras, Eric Rouchouze, Denis Renaud, and Alexei Tchelnokov; and at Advanced Micro Foundry (AMF, Singapore): Patrick Lo, Xianshu Luo; and at TSMC (Taiwan).
Three patents owned by the California Institute of Technology have been pursued in connection with this work: (1) Roukes, M.L. (2011). Brain-machine interface based on photonic neural probe arrays. U.S.P.T.O. Patent No. 10,638,933, Issued May 5, 2020, Priority Date: December 8, 2011. (2) Segev, E., Moreaux, L.M., Fowler, T.M. Faraon, A., Roukes, M.L. Implantable, highly collimated light-emitters for biological applications. U.S.P.T.O. Patent No. 10,471,273, Issue date: 12 November 2019; Priority date: 16 October 2015. (3) Roukes, M.L., Cotton, R.J., Moreaux, L.C., Shepard, K.L., Siapas, A., Tolias, A. (2016). One-photon integrated neurophotonic systems. U.S.P.T.O. Patent Application 20160150963, Filing date: November 5, 2014; Publication date: June 2, 2016.
Footnotes
Competing Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acher FC, and Bertrand HO (2005). Amino acid recognition by Venus flytrap domains is encoded in an 8-residue motif. Biopolymers 80, 357–366. [DOI] [PubMed] [Google Scholar]
- Adams JK, Boominathan V, Avants BW, Vercosa DG, Ye F, Baraniuk RG, Robinson JT, and Veeraraghavan A. (2017). Single-frame 3D fluorescence microscopy with ultraminiature lensless FlatScope. Science Advances 3, e1701548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrian ED (1926). The impulses produced by sensory nerve endings: Part I. J Physiol 61, 49–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens MB, Orger MB, Robson DN, Li JM, and Keller PJ (2013). Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nature Methods 10, 413–420. [DOI] [PubMed] [Google Scholar]
- Alivisatos AP, Chun M, Church GM, Greenspan RJ, Roukes ML, and Yuste R. (2012). The brain activity map project and the challenge of functional connectomics. Neuron 74, 970–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreoni A, Davis CMO, and Tian L. (2019). Measuring Brain Chemistry Using Genetically Encoded Fluorescent Sensors. Curr Opin Biomed Eng 12, 59–67. [Google Scholar]
- Azizi L, Zarychta K, Ettori D, Tinet E, and Tualle J-M (2009). Ultimate spatial resolution with Diffuse Optical Tomography. Optics Express 17, 12132–12144. [DOI] [PubMed] [Google Scholar]
- Bannerjee A, Park Y, Clarke F, Song H, Yang S, Kramer G, Kim K, and Mukherjee B. (2005). Wavelength-division-multiplexed passive optical network (WDM-PON) technologies for broadband access: a review. J Opt Networking 4, 737–747. [Google Scholar]
- Bargmann C, and Newsome W. (2014). In BRAIN 2025, A Scientific Vision (Advisory Committee to the Director, NIH). [Google Scholar]
- Beaulieu DR, Davison IG, Kilic K, Bifano TG, and Mertz J. (2020). Simultaneous multiplane imaging with reverberation two-photon microscopy. Nature Methods 17, 283–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenyi A, Somogyvari Z, Nagy AJ, Roux L, Long JD, Fujisawa S, Stark E, Leonardo A, Harris TD, and Buzsaki G. (2014). Large-scale, high-density (up to 512 channels) recording of local circuits in behaving animals. J Neurophysiol 111, 1132–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohren CF, and Huffman DR (2004). Absorption and scattering of light by small particles., Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim. ISBN 0–471-29340–8. [Google Scholar]
- Bouchard MB, Voleti V, Mendes CS, Lacefield C, Grueber WB, Mann RS, Bruno RM, and Hillman EM (2015). Swept confocally-aligned planar excitation (SCAPE) microscopy for high speed volumetric imaging of behaving organisms. Nat Photonics 9, 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES (2011). A history of optogenetics: the development of tools for controlling brain circuits with light. F1000 Biol Rep 3, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzsaki G. (2004). Large-scale recording of neuronal ensembles. Nature Neuroscience 7, 446–451. [DOI] [PubMed] [Google Scholar]
- Buzsaki G, Anastassiou CA, and Koch C. (2012). The origin of extracellular fields and currents--EEG, ECoG, LFP and spikes. Nat Rev Neurosci 13, 407–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PK, Jones KE, Huber RJ, Horch KW, and Normann RA (1991). A silicon-based, three-dimensional neural interface: manufacturing processes for an intracortical electrode array. IEEE Trans Biomed Eng 38, 758–768. [DOI] [PubMed] [Google Scholar]
- Charpak S, Mertz J, Beaurepaire E, Moreaux L, and Delaney K. (2001). Odor-evoked calcium signals in dendrites of rat mitral cells. Proceedings of the National Academy of Sciences of the United States of America 98, 1230–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA 3rd, Liu Z, et al. (2014). Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346, 1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Mu Y, Hu Y, Kuan AT, Nikitchenko M, Randlett O, Chen AB, Gavornik JP, Sompolinsky H, Engert F, et al. (2018). Brain-wide Organization of Neuronal Activity and Convergent Sensorimotor Transformations in Larval Zebrafish. Neuron 100, 876–890 e875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Jang H, Spratt PWE, Kosar S, Taylor DE, Essner RA, Bai L, Leib DE, Kuo TW, Lin YC, et al. (2020). Soma-Targeted Imaging of Neural Circuits by Ribosome Tethering. Neuron. 3, 454–469.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Goncalves JT, Golshani P, Arisaka K, and Portera-Cailliau C. (2011). Simultaneous two-photon calcium imaging at different depths with spatiotemporal multiplexing. Nature Methods 8, 139–U158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Taal AJ, Meng WL, Pollmann EH, Stanton EH, Lee C, Moazeni S, Moreaux LC, Roukes ML, and Shepard KL (2020). Fully Integrated Time-Gated 3D Fluorescence Imager for Deep Neural Imaging. IEEE Transactions on Biomedical Circuits and Systems. 14(4), 636–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Taal AJ, Pollmann EH, Lee C, Kim K, Moreaux LC, Roukes ML, and Shepard KL (2019). A 512-Pixel, 51-kHz-Frame-Rate, Dual-Shank, Lens-less, Filter-less Single Photon Avalanche Diode CMOS Neural Imaging Probe. IEEE J Solid-State Circuits 54, 2957–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Valm AM, and Lippincott-Schwartz J. (2018). Multispectral Live-Cell Imaging. Curr Protoc Cell Biol 79, e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comon P, and Jutten C. (2010). Handbook of Blind Source Separation: Independent component analysis and applications (Academic Press; ). 10.1016/C2009-0-19334-0 [DOI] [Google Scholar]
- Corder G, Ahanonu B, Grewe BF, Wang D, Schnitzer MJ, and Scherrer G. (2019). An amygdalar neural ensemble that encodes the unpleasantness of pain. Science 363, 276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotton RJ, Froudarakis E, Storer P, Saggau P, and Tolias AS (2013). Three-dimensional mapping of microcircuit correlation structure. Frontiers in Neural Circuits 7, 151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle TL, Madisen L, Hage TA, Valley MT, Knoblich U, Larsen RS, Takeno MM, Huang L, Gu H, Larsen R, et al. (2018). A Suite of Transgenic Driver and Reporter Mouse Lines with Enhanced Brain-Cell-Type Targeting and Functionality. Cell 174, 465–480 e422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Sun Y, Mohar B, Hulse BK, Kerlin AM, Hasseman JP, Tsegaye G, Tsang A, Wong A, Patel R, et al. (2019). High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nature Methods 16, 649–657. [DOI] [PubMed] [Google Scholar]
- de Groot A, van den Boom BJ, van Genderen RM, Coppens J, van Veldhuijzen J, Bos J, Hoedemaker H, Negrello M, Willuhn I, De Zeeuw CI, et al. (2020). NINscope, a versatile miniscope for multi-region circuit investigations. eLife 9:e49987. doi: 10.7554/eLife.49987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehghani H, White BR, Zeff BW, and Culver JP (2008). Depth Sensitivity Analysis of high-density imaging arrays for mapping brain function with Diffuse Optical Tomography. Paper presented at: Biomedical Optics (St. Petersburg, Florida: Optical Society of America; ). [Google Scholar]
- Denk W, Strickler JH, and Webb WW (1990). Two-photon laser scanning fluorescence microscopy. Science 248, 73–76. [DOI] [PubMed] [Google Scholar]
- Ding R, Liao X, Li J, Zhang J, Wang M, Guang Y, Qin H, Li X, Zhang K, Liang S, et al. (2017). Targeted Patching and Dendritic Ca(2+) Imaging in Nonhuman Primate Brain in vivo. Sci Rep 7, 2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducros M, Goulam Houssen Y, Bradley J, de Sars V, and Charpak S. (2013). Encoded multisite two-photon microscopy. Proceedings of the National Academy of Sciences of the United States of America 110, 13138–13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara MS, Pisanello M, Sileo L, De Vittorio M, and Pisanello F. (2018). A Wireless Head- mountable Device with Tapered Optical Fiber-coupled Laser Diode for Light Delivery in Deep Brain Regions. IEEE Trans Biomed Eng. vol. 66, no. 7, pp. 1996–2009, July 2019, doi: 10.1109/TBME.2018.2882146. [DOI] [PubMed] [Google Scholar]
- Engelbrecht CJ, Voigt F, and Helmchen F. (2010). Miniaturized selective plane illumination microscopy for high-contrast in vivo fluorescence imaging. Optics Letters 35, 1413–1415. [DOI] [PubMed] [Google Scholar]
- Fang Q, Liow TY, Song JF, Tan CW, Yu MB, Lo GQ, and Kwong DL (2010). Suspended optical fiber-to-waveguide mode size converter for silicon photonics. Optics Express 18, 7763–7769. [DOI] [PubMed] [Google Scholar]
- Felder CB, Graul RC, Lee AY, Merkle HP, and Sadee W. (1999). The Venus flytrap of periplasmic binding proteins: an ancient protein module present in multiple drug receptors. AAPS PharmSci 1, 7–26. 10.1208/ps010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler TM (2019). Silicon Neural Probes for Stimulation of Neurons and the Excitation and Detection of Proteins in the Brain. Dissertation (PhD), California Institute of Technology. [Google Scholar]
- Gerstein GL, and Clark WA (1964). Simultaneous Studies of Firing Patterns in Several Neurons. Science 143, 1325–1327. [DOI] [PubMed] [Google Scholar]
- Ghosh KK, Burns LD, Cocker ED, Nimmerjahn A, Ziv Y, Gamal AE, and Schnitzer MJ (2011). Miniaturized integration of a fluorescence microscope. Nature Methods 8, 871–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girkin JM, Poland S, and Wright AJ (2009). Adaptive optics for deeper imaging of biological samples. Current Opinion in Biotechnology 20, 106–110. [DOI] [PubMed] [Google Scholar]
- Goldin MA, and Mindlin GB (2017). Temperature manipulation of neuronal dynamics in a forebrain motor control nucleus. PLoS Comput Biol 13, e1005699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Huang C, Li JZ, Grewe BF, Zhang Y, Eismann S, and Schnitzer MJ (2015). High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science 350, 1361–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorin A, Jaouad A, Grondin E, Aimez V, and Charette P. (2008). Fabrication of silicon nitride waveguides for visible-light using PECVD: a study of the effect of plasma frequency on optical properties. Optics Express 16, 13509–13516. [DOI] [PubMed] [Google Scholar]
- Gray CM, Maldonado PE, Wilson M, and McNaughton B. (1995). Tetrodes markedly improve the reliability and yield of multiple single-unit isolation from multi-unit recordings in cat striate cortex. Journal of Neuroscience Methods 63, 43–54. [DOI] [PubMed] [Google Scholar]
- Grewe BF, Langer D, Kasper H, Kampa BM, and Helmchen F. (2010). High-speed in vivo calcium imaging reveals neuronal network activity with near-millisecond precision. Nature Methods 7, 399–405. [DOI] [PubMed] [Google Scholar]
- Grienberger C, and Konnerth A. (2012). Imaging calcium in neurons. Neuron 73, 862–885. [DOI] [PubMed] [Google Scholar]
- Gyongy I, Calder N, Davies A, Dutton NAW, Duncan RR, Rickman C, Dalgarno P, and Henderson RK (2018). A 256*256, 100-kfps, 61% Fill-Factor SPAD Image Sensor for Time- Resolved Microscopy Applications. EEE Transactions on Electron Devices 65, 547–554. [Google Scholar]
- Hamill OP, Marty A, Neher E, Sakmann B, and Sigworth FJ (1981). Improved patch- clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflugers Arch 391, 85–100. [DOI] [PubMed] [Google Scholar]
- Haslehurst P, Yang Z, Dholakia K, and Emptage N. (2018). Fast volume-scanning light sheet microscopy reveals transient neuronal events. Biomedical optics express 9, 2154–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herreras O. (2016). Local Field Potentials: Myths and Misunderstandings. Frontiers in Neural Circuits 10, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman EMC, Voleti V, Li W, and Yu H. (2019). Light-Sheet Microscopy in Neuroscience. Annual Review of Neuroscience 42, 295–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogerwerf AC, and Wise KD (1994). A three-dimensional microelectrode array for chronic neural recording. IEEE Trans Biomed Eng 41, 1136–1146. [DOI] [PubMed] [Google Scholar]
- Horton NG, Wang K, Kobat D, Clark CG, Wise FW, Schaffer CB, and Xu C. (2013). In vivo three-photon microscopy of subcortical structures within an intact mouse brain. Nat Photonics 7, 205–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisken J, Swoger J, Del Bene F, Wittbrodt J, and Stelzer EH (2004). Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007–1009. [DOI] [PubMed] [Google Scholar]
- Hulse BK, Moreaux LC, Lubenov EV, and Siapas AG (2016). Membrane Potential Dynamics of CA1 Pyramidal Neurons during Hippocampal Ripples in Awake Mice. Neuron 89, 800–813. Inscopix. https://www.inscopix.com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob AD, Ramsaran AI, Mocle AJ, Tran LM, Yan C, Frankland PW, and Josselyn SA (2018). A Compact Head-Mounted Endoscope for In Vivo Calcium Imaging in Freely Behaving Mice. Curr Protoc Neurosci 84, e51. [DOI] [PubMed] [Google Scholar]
- Jans H, O’Brien P, Artundo I, Porcel MAG, Hoofman R, Geuzebroek D, Dumon P, van der Vliet M, Witzens J, Bourguignon E, et al. (2018). Integrated bio-photonics to revolutionize health care enabled through PIX4life and PIXAPP. Paper presented at: Proc SPIE 10506, Nanoscale Imaging, Sensing, and Actuation for Biomedical Applications XV, 105060V. [Google Scholar]
- Jeong JW, Shin G, Park SI, Yu KJ, Xu L, and Rogers JA (2015). Soft materials in neuroengineering for hard problems in neuroscience. Neuron 86, 175–186. [DOI] [PubMed] [Google Scholar]
- Jiang X, Shen S, Cadwell CR, Berens P, Sinz F, Ecker AS, Patel S, and Tolias AS (2015). Principles of connectivity among morphologically defined cell types in adult neocortex. Science 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Wang G, Lee AJ, Stornetta RL, and Zhu JJ (2013). The organization of two new cortical interneuronal circuits. Nature Neuroscience 16, 210–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun JJ, Steinmetz NA, Siegle JH, Denman DJ, Bauza M, Barbarits B, Lee AK, Anastassiou CA, Andrei A, Aydin Q, et al. (2017). Fully integrated silicon probes for high- density recording of neural activity. Nature 551, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katona G, Szalay G, Maak P, Kaszas A, Veress M, Hillier D, Chiovini B, Vizi ES, Roska B, and Rozsa B. (2012). Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nature Methods 9, 201–208. [DOI] [PubMed] [Google Scholar]
- Keller PJ, and Ahrens MB (2015). Visualizing whole-brain activity and development at the single-cell level using light-sheet microscopy. Neuron 85, 462–483. [DOI] [PubMed] [Google Scholar]
- Keller PJ, Schmidt AD, Wittbrodt J, and Stelzer EH (2008). Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322, 1065–1069. [DOI] [PubMed] [Google Scholar]
- Kleinfeld D, Luan L, Mitra PP, Robinson JT, Sarpeshkar R, Shepard K, Xie C, and Harris TD (2019). Can One Concurrently Record Electrical Spikes from Every Neuron in a Mammalian Brain? Neuron 103, 1005–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodandaramaiah SB, Franzesi GT, Chow BY, Boyden ES, and Forest CR (2012). Automated whole-cell patch-clamp electrophysiology of neurons in vivo. Nature methods 9, 585–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothapalli S-R, Sakadzic S, Kim C, and Wang LV (2007). Imaging optically scattering objects with ultrasound-modulated optical tomography. Optics letters 32, 2351–2353. [DOI] [PubMed] [Google Scholar]
- Lecoq J, Orlova N, and Grewe BF (2019). Wide. Fast. Deep: Recent Advances in Multiphoton Microscopy of In Vivo Neuronal Activity. The Journal of neuroscience : the official journal of the Society for Neuroscience 39, 9042–9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Taal AJ, Choi J, Kim K, Tien K, Moreaux LM, Roukes ML, and Shepard KL (2019). A 512-Pixel 3kHz-frame-rate dual-shank lensless filterless single-photon-avalanche- diode CMOS neural imaging probe. In 2019 IEEE International Solid-State Circuits Conference- (ISSCC), 198–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leopold AV, Shcherbakova DM, and Verkhusha VV (2019). Fluorescent Biosensors for Neurotransmission and Neuromodulation: Engineering and Applications. Front Cell Neurosci 13, 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberti WA, Perkins LN, Leman DP, and Gardner TJ (2017). An open source, wireless capable miniature microscope system. J Neural Eng 14, 045001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MZ, and Schnitzer MJ (2016). Genetically encoded indicators of neuronal activity. Nature Neuroscience 19, 1142–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Fu TM, Cheng Z, Hong G, Zhou T, Jin L, Duvvuri M, Jiang Z, Kruskal P, Xie C, et al. (2015). Syringe-injectable electronics. Nat Nanotechnol 10, 629–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W-W, and Li P-C (2020). Photoacoustic imaging of cells in a three-dimensional microenvironment. Journal of Biomedical Science 27, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Liang Y, Meng G, Zhou P, Svoboda K, Paninski L, and Ji N. (2020). Rapid mesoscale volumetric imaging of neural activity with synaptic resolution. Nature Methods 17, 291–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig KA, Langhals NB, Joseph MD, Richardson-Burns SM, Hendricks JL, and Kipke DR (2011). Poly(3,4-ethylenedioxythiophene) (PEDOT) polymer coatings facilitate smaller neural recording electrodes. J Neural Eng 8, 014001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Callaway EM, and Svoboda K. (2018). Genetic Dissection of Neural Circuits: A Decade of Progress. Neuron 98, 256–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marblestone AH, Zamft BM, Maguire YG, Shapiro MG, Cybulski TR, Glaser JI, Amodei D, Stranges PB, Kalhor R, Dalrymple DA, et al. (2013). Physical principles for scalable neural recording. Front Comput Neurosci 7, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin JS, Scholl B, Wilson DE, Podgorski K, Kazemipour A, Muller JA, Schoch S, Quiroz FJU, Rebola N, Bao H, et al. (2018). Stability, affinity, and chromatic variants of the glutamate sensor iGluSnFR. Nature Methods 15, 936–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin JS, Shimoda Y, Magloire V, Leite M, Kawashima T, Jensen TP, Kolb I, Knott EL, Novak O, Podgorski K, et al. (2019). A genetically encoded fluorescent sensor for in vivo imaging of GABA. Nature Methods 16, 763–770. [DOI] [PubMed] [Google Scholar]
- Kea Mauna, Inc. http://www.maunakeatech.com.
- McNaughton BL, O’Keefe J, and Barnes CA (1983). The stereotrode: a new technique for simultaneous isolation of several single units in the central nervous system from multiple unit records. Journal of Neuroscience Methods 8, 391–397. [DOI] [PubMed] [Google Scholar]
- Mertz J. (2011). Optical sectioning microscopy with planar or structured illumination. Nature Methods 8, 811–819. [DOI] [PubMed] [Google Scholar]
- Miesenböck G. (2011). Optogenetic control of cells and circuits. Annu Rev Cell Dev Biol 27, 731–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittmann W, Wallace DJ, Czubayko U, Herb JT, Schaefer AT, Looger LL, Denk W, and Kerr JN (2011). Two-photon calcium imaging of evoked activity from L5 somatosensory neurons in vivo. Nature Meuroscience 14, 1089–1093. [DOI] [PubMed] [Google Scholar]
- Mohanty A, Li Q, Tadayon MA, Roberts SP, Bhatt GR, Shim E, Ji X, Cardenas J, Miller SA, Kepecs A, et al. (2020). Reconfigurable nanophotonic silicon probes for sub-millisecond deep-brain optical stimulation. Nat Biomed Eng 4, 223–231. [DOI] [PubMed] [Google Scholar]
- Moreaux L, and Laurent G. (2007). Estimating firing rates from calcium signals in locust projection neurons in vivo. Frontiers in Neural Circuits Nov 2;1:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreaux L, and Laurent G. (2008). A simple method to reconstruct firing rates from dendritic calcium signals. Front Neurosci 2, 176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto K, and Charbon E. (2020). High fill-factor miniaturized SPAD arrays with a guard- ring-sharing technique. Optics Express 28, 13068–13080. [DOI] [PubMed] [Google Scholar]
- Moser E, Mathiesen I, and Andersen P. (1993). Association between brain temperature and dentate field potentials in exploring and swimming rats. Science 259, 1324–1326. [DOI] [PubMed] [Google Scholar]
- Mukamel EA, Nimmerjahn A, and Schnitzer MJ (2009). Automated analysis of cellular signals from large-scale calcium imaging data. Neuron 63, 747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musk E. (2019). An integrated brain-machine interface platform with thousands of channels. bioRxiv, 703801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafi K, Ji J, and Wise KD (1990). Scaling limitations of silicon multichannel recording probes. IEEE Trans Biomed Eng 37, 1–11. [DOI] [PubMed] [Google Scholar]
- Navaraj WT, Gupta S, Lorenzelli L, and Dahiya R. (2018). Wafer Scale Transfer of Ultrathin Silicon Chips on Flexible Substrates for High Performance Bendable Systems. Adv Elec Materials 4, 1700277. [Google Scholar]
- O’Hara PJ, Sheppard PO, Thogersen H, Venezia D, Haldeman BA, McGrane V, Houamed KM, Thomsen C, Gilbert TL, and Mulvihill ER (1993). The ligand-binding domain in metabotropic glutamate receptors is related to bacterial periplasmic binding proteins. Neuron 11, 41–52. [DOI] [PubMed] [Google Scholar]
- Oe Y, Wang X, Patriarchi T, Konno A, Ozawa K, Yahagi K, Hirai H, Tsuboi T, Kitaguchi T, Tian Lv et al. (2020). Distinct temporal integration of noradrenaline signaling by astrocytic second messengers during vigilance. Nature communications 11, 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oheim M, Beaurepaire E, Chaigneau E, Mertz J, and Charpak S. (2001). Two-photon microscopy in brain tissue: parameters influencing the imaging depth. Journal of neuroscience methods 111, 29–37. [DOI] [PubMed] [Google Scholar]
- Ouzounov DG, Wang T, Wang M, Feng DD, Horton NG, Cruz-Hernandez JC, Cheng YT, Reimer J, Tolias AS, Nishimura Nv et al. (2017). In vivo three-photon imaging of activity of GCaMP6-labeled neurons deep in intact mouse brain. Nature methods 14, 388–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen SF, Liu MH, and Kreitzer AC (2019). Thermal constraints on in vivo optogenetic manipulations. Nature neuroscience 22, 1061–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painchaud Y, Mailloux A, Morin M, Verreault S, and Beaudry P. (1999). Time-domain optical imaging: discrimination between scattering and absorption. Appl Opt 38, 3686–3693. [DOI] [PubMed] [Google Scholar]
- Park H, Kim S, Park J, Joo J, and Kim G. (2013). A fiber-to-chip coupler based on Si/SiON cascaded tapers for Si photonic chips. Optics Express 21, 29313–29319. [DOI] [PubMed] [Google Scholar]
- Patriarchi T, Cho JR, Merten K, Howe MW, Marley A, Xiong WH, Folk RW, Broussard GJ, Liang R, Jang MJ, et al. (2018). Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 360(6396). eaat4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perin R, Berger TK, and Markram H. (2011). A synaptic organizing principle for cortical neuronal groups. Proceedings of the National Academy of Sciences of the United States of America 108, 5419–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatkevich KD, Bensussen S, Tseng HA, Shroff SN, Lopez-Huerta VG, Park D, Jung EE, Shemesh OA, Straub C, Gritton HJ, et al. (2019). Population imaging of neural activity in awake behaving mice. Nature 574, 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisanello F, Mandelbaum G, Pisanello M, Oldenburg IA, Sileo L, Markowitz JE, Peterson RE, Della Patria A, Haynes TM, Emara MS, et al. (2017). Dynamic illumination of spatially restricted or large brain volumes via a single tapered optical fiber. Nature neuroscience 20, 1180–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisanello F, Sileo L, Oldenburg IA, Pisanello M, Martiradonna L, Assad JA, Sabatini BL, and De Vittorio M. (2014). Multipoint-emitting optical fibers for spatially addressable in vivo optogenetics. Neuron 82, 1245–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisano F, Pisanello M, Lee SJ, Lee J, Maglie E, Balena A, Sileo L, Spagnolo B, Bianco M, Hyun M, et al. (2019). Depth-resolved fiber photometry with a single tapered optical fiber implant. Nature methods 16, 1185–1192. [DOI] [PubMed] [Google Scholar]
- Pnevmatikakis EA, Soudry D, Gao Y, Machado TA, Merel J, Pfau D, Reardon T, Mu Y, Lacefield C, Yang W, et al. (2016). Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron 89, 285–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power RM, and Huisken J. (2017). A guide to light-sheet fluorescence microscopy for multiscale imaging. Nature methods 14, 360–373. [DOI] [PubMed] [Google Scholar]
- Puszka A, Di Sieno L, Mora AD, Pifferi A, Contini D, Boso G, Tosi A, Hervé L, Planat- Chrétien A, Koenig A, et al. (2013). Time-resolved diffuse optical tomography using fast-gated single-photon avalanche diodes. Biomedical optics express 4, 1351–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravotto L, Duffet L, Zhou X, Weber B, and Patriarchi T. (2020). A Bright and Colorful Future for G-Protein Coupled Receptor Sensors. Front Cell Neurosci 14, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recce M, and O’Keefe J. (1989). The tetrode: a new technique for multi-unit extracellular recording. In Soc Neurosci, S. Neurosci, ed. [Google Scholar]
- Resnik SG, Steenbergen W, and Boccara AC (2012). State-of-the-art of acoust--optic sensing and imaging of turbid media. J Biomed Optics 17, 040901. [DOI] [PubMed] [Google Scholar]
- Rios G, Lubenov EV, Chi D, Roukes ML, and Siapas AG (2016). Nanofabricated Neural Probes for Dense 3-D Recordings of Brain Activity. Nano Lett 16, 6857–6862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roukes ML (2011). Brain-machine interface based on photonic neural probe arrays. U.S.P.T.O. Patent No. 10,638,933, Issued May 5, 2020, Priority Date: December 8, 2011.
- Roukes ML, Cotton RJ, Moreaux LC, Shepard KL, Siapas A, and Tolias A. (2016). One-photon integrated neurophotonic systems. U.S.P.T.O. Patent Application 20160150963, June 2, 2016.
- Rousche PJ, Pellinen DS, Pivin DP Jr., Williams JC, Vetter RJ, and Kipke DR (2001). Flexible polyimide-based intracortical electrode arrays with bioactive capability. IEEE Trans Biomed Eng 48, 361–371. [DOI] [PubMed] [Google Scholar]
- Rungta RL, Osmanski BF, Boido D, Tanter M, and Charpak S. (2017). Light controls cerebral blood flow in naive animals. Nature Communications 8, 14191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustami E, Sasagawa K, Sugie K, Ohta Y, Haruta M, Noda T, Tokuda T, and Ohta J.(2020). Needle-Type Imager Sensor With Band-Pass Composite Emission Filter and Parallel Fiber-Coupled Laser Excitation. In IEEE Transactions on Circuits and Systems I: Regular Papers (IEEE). vol. 67, no. 4, pp. 1082–1091, April 2020, doi: 10.1109/TCSI.2019.2959592. [DOI] [Google Scholar]
- Sacher WD, Liu XY, Almog IF, Fomenko A, Lordello T, Chen FD, Moradi-Chameh H, Naderian A, Chang M, Fowler TM, et al. (2019a). Nanophotonic neural probes for in vivo light sheet imaging (Conference on Lasers and Electo-Optics (CLEO)). 5 May 2019 – 10 May 2019. Paper SM4H.6. Published by Optical Society of America; (ISBN 978–1-943580–57-6). [Google Scholar]
- Sacher WD, Liu XY, Chen FD, Moradi-Chameh H, Almog IF, Lordello T, Chang M, Naderian A, Fowler TM, Segev E, et al. (2019b). Beam-steering nanophonic phased-array neural probes. Paper presented at: Conference on Lasers and Electo-Optics (CLEO). 5 May 2019 – 10 May 2019. Paper ATh4I.4. Published by Optical Society of America (ISBN 978–1-943580–57-6). [Google Scholar]
- Sacher WD, Luo X, Yang Y, Chen FD, Lordello T, Mak JCC, Liu X, Hu T, Xue T, Guo-Qiang Lo P, et al. (2019c). Visible-light silicon nitride waveguide devices and implantable neurophotonic probes on thinned 200 mm silicon wafers. Optics Express 27, 37400–37418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher WD, Mikkelsen JC, Dumais P, Jiang J, Goodwill D, Luo X, Huang Y, Yang Y, Bois A, Lo PG, et al. (2017). Tri-layer silicon nitride-on-silicon photonic platform for ultra- low-loss crossings and interlayer transitions. Optics Express 25, 30862–30875. [DOI] [PubMed] [Google Scholar]
- Sacher WD, Chen FD, Moradi-Chameh H, Luo X, Fomenko A, Shah P, Lordello T, Liu X, Felts Almog I, Straguzzi JN, Fowler TM, Jung Y, Hu T, Jeong J, Lozano AM, Guo- Qiang Lo P, Valiante TA, Moreaux LC, Poon JKS, Roukes ML (2020). Implantable photonic neural probes for light-sheet fluorescence brain imaging. (bioRxiv, uploaded, awaiting link; will provide by 29 Sep 2020.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasagawa K, Kimura A, Haruta M, Noda T, Tokuda T, and Ohta J. (2018). Highly sensitive lens-free fluorescence imaging device enabled by a complementary combination of interference and absorption filters. Biomedical Optics Express 9, 4329–4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segev E, Moreaux LM, Fowler TM, Faraon A, and Roukes ML (2015). Implantable, highly collimated light-emitters for biological applications, U.S.P.T.O., ed. (U.S.A.). Patent No. 10,471,273, Issue date: 12 November 2019; Priority date: 16 October 2015.
- Segev E, Reimer J, Moreaux LC, Fowler TM, Chi D, Sacher WD, Lo M, Deisseroth K, Tolias AS, Faraon A, et al. (2017). Patterned photostimulation via visible-wavelength photonic probes for deep brain optogenetics. Neurophotonics 4, 011002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senova S, Scisniak I, Chiang C-C, Doignon I, Palfi S, Chaillet A, Martin C, and Pain F. (2017). Experimental assessment of the safety and potential efficacy of high irradiance photostimulation of brain tissues. Scientific Reports 7, 43997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahrjerdi D, and Bedell SW (2013). Extremely flexible nanoscale ultrathin body silicon integrated circuits on plastic. Nano Lett 13, 315–320. [DOI] [PubMed] [Google Scholar]
- Shao Z, Chen Y, Chen H, Zhang Y, Zhang F, Jian J, Fan Z, Liu L, Yang C, Zhou L, et al. (2016). Ultra-low temperature silicon nitride photonic integration platform. Optics Express 24, 1865–1872. [DOI] [PubMed] [Google Scholar]
- Shemesh OA, Linghu C, Piatkevich KD, Goodwin D, Celiker OT, Gritton HJ, Romano MF, Gao R, Yu CJ, Tseng HA, et al. (2020). Precision Calcium Imaging of Dense Neural Populations via a Cell-Body-Targeted Calcium Indicator. Neuron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shichida Y, and Matsuyama T. (2009). Evolution of opsins and phototransduction. Philos Trans R Soc Lond B Biol Sci 364, 2881–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shobe JL, Claar LD, Parhami S, Bakhurin KI, and Masmanidis SC (2015). Brain activity mapping at multiple scales with silicon microprobes containing 1,024 electrodes. J Neurophysiol 114, 2043–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuman T, Aharoni D, Cai DJ, Lee CR, Chavlis S, Page-Harley L, Vetere LM, Feng Y, Yang CY, Mollinedo-Gajate I, et al. (2020). Breakdown of spatial coding and interneuron synchronization in epileptic mice. Nature Neuroscience 23, 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sileo L, Bitzenhofer SH, Spagnolo B, Popplau JA, Holzhammer T, Pisanello M, Pisano F, Bellistri E, Maglie E, De Vittorio M, et al. (2018). Tapered Fibers Combined With a Multi-Electrode Array for Optogenetics in Mouse Medial Prefrontal Cortex. Front Neurosci 12, 771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skocek O, Nobauer T, Weilguny L, Martinez Traub F, Xia CN, Molodtsov MI, Grama A, Yamagata M, Aharoni D, Cox DD, et al. (2018). High-speed volumetric imaging of neuronal activity in freely moving rodents. Nature methods 15, 429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroniew NJ, Flickinger D, King J, and Svoboda K. (2016). A large field of view two- photon mesoscope with subcellular resolution for in vivo imaging. eLife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram S, Partlow WD, and Liu CS (1983). Low-loss optical waveguides using plasma- deposited silicon nitride. Appl Opt 22, 3664–3665. [DOI] [PubMed] [Google Scholar]
- Steinmetz NA, Koch C, Harris KD, and Carandini M. (2018). Challenges and opportunities for large-scale electrophysiology with Neuropixels probes. Curr Opin Neurobiol 50, 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz NA, Zatka-Haas P, Carandini M, and Harris KD (2019). Distributed coding of choice, action and engagement across the mouse brain. Nature 576, 266–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson IH, and Kording KP (2011). How advances in neural recording affect data analysis. Nature Neuroscience 14, 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Zeng J, Jing M, Zhou J, Feng J, Owen SF, Luo Y, Li F, Wang H, Yamaguchi T, et al. (2018). A Genetically Encoded Fluorescent Sensor Enables Rapid and Specific Detection of Dopamine in Flies, Fish, and Mice. Cell 174, 481–496 e419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SM, Masukawa LM, and Prince DA (1985). Temperature dependence of intrinsic membrane properties and synaptic potentials in hippocampal CA1 neurons in vitro. The Journal of neuroscience : the official journal of the Society for Neuroscience 5, 817–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, Chalasani SH, Petreanu L, Akerboom J, McKinney SA, Schreiter ER, et al. (2009). Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nature methods 6, 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolias AS The iARPA MICrONS project - functional imaging achievements. [Google Scholar]
- Ulku AC, Bruschini C, Antolovic IM, Charbon E, Kuo Y, Ankri R, Weiss S, and Michalet X. (2019). A 512×512 SPAD Image Sensor with Integrated Gating for Widefield FLIM. IEEE J Sel Top Quantum Electron 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villette V, Chavarha M, Dimov IK, Bradley J, Pradhan L, Mathieu B, Evans SW, Chamberland S, Shi D, Yang R, et al. (2019). Ultrafast Two-Photon Imaging of a High-Gain Voltage Indicator in Awake Behaving Mice. Cell 179, 1590–1608 e1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voleti V, Patel KB, Li W, Perez Campos C, Bharadwaj S, Yu H, Ford C, Casper MJ, Yan RW, Liang W, et al. (2019). Real-time volumetric microscopy of in vivo dynamics and large-scale samples with SCAPE 2.0. Nature Methods 16, 1054–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker EY, Sinz FH, Cobos E, Muhammad T, Froudarakis E, Fahey PG, Ecker AS, Reimer J, Pitkow X, and Tolias AS (2019). Inception loops discover what excites neurons most using deep predictive models. Nature Neuroscience 22, 2060–2065. [DOI] [PubMed] [Google Scholar]
- Wang A, Gill P, and Molnar A. (2009). Light field image sensors based on the Talbot effect. Appl Opt 48, 5897–5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Jacques SL, and Zhao X. (1995). Continuous-wave ultrasonic modulation of scattered laser light to image objects in turbid media. Optics Letters 20, 629–631. [DOI] [PubMed] [Google Scholar]
- Wang L, and Zhao X. (1997). Ultrasound-- - modulated optical tomography of absorbing objects buried in dense tissue-simulating turbid media. Appl Opt 36, 7277–7282. [DOI] [PubMed] [Google Scholar]
- Wang LV (2001). Mechanisms of Ultrasonic Modulation of Multiply Scattered Coherent Light: An Analytic Model. Physical Review Letters 87, 043903. [DOI] [PubMed] [Google Scholar]
- Wang M, Wu C, Sinefeld D, Li B, Xia F, and Xu C. (2018). Comparing the effective attenuation lengths for long wavelength in vivo imaging of the mouse brain. Biomedical optics Express 9, 3534–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XF, Uchida T, Coleman DM, and Minami S. (1991). A two-dimensional fluorescence lifetime imaging system using a gated image intensifier. Appl Spectrosc 45, 360–366. [Google Scholar]
- Wheelock M, Culver J, and Eggebrecht A. (2019). High-density diffuse optical tomography for imaging human brain function. Review of Scientific Instruments 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MA, and McNaughton BL (1993). Dynamics of the hippocampal ensemble code for space. Science 261, 1055–1058. [DOI] [PubMed] [Google Scholar]
- Wise KD, Angell JB, and Starr A. (1970). An integrated-circuit approach to extracellular microelectrodes. IEEE Trans Biomed Eng 17, 238–247. [DOI] [PubMed] [Google Scholar]
- Wise KD, Sodagar AM, Yao Y, Gulari MN, Gayatri EP, and Najafi K. (2008). Microelectrodes, microelectronics, and implantable neural microsystems. In Proc IEEE, pp. 1184–1202. [Google Scholar]
- Wu F, Stark E, Ku PC, Wise KD, Buzsaki G, and Yoon E. (2015). Monolithically Integrated muLEDs on Silicon Neural Probes for High-Resolution Optogenetic Studies in Behaving Animals. Neuron 88, 1136–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Liang Y, Chen S, Hsu CL, Chavarha M, Evans SW, Shi D, Lin MZ, Tsia KK, and Ji N. (2020). Kilohertz two-photon fluorescence microscopy imaging of neural activity in vivo. Nature Methods 17, 287–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, and Wang LV (2006). Photoacoustic imaging in biomedicine. Review of Scientific Instruments 77, 041101. [Google Scholar]
- Yaqoob Z, Psaltis D, Feld MS, and Yang C. (2008). Optical Phase Conjugation for Turbidity Suppression in Biological Samples. Nat Photonics 2, 110–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko D, Moreaux LC, Choi J, Tolias AS, Shepard KL, and Roukes ML (2020). Signal separability in integrated neurophotonics. (bioRxiv, uploaded, awaiting link; will provide by 29 Sep 2020.). [Google Scholar]
- Ye F, Avants BW, and Robinson JT (2016). Light sheet illumination with an integrated photonic probe. In Conference on Lasers and Electro-Optics, JW2A142 (Optical Society of America; ). [Google Scholar]
- Yuste R. (2010). Imaging: A Laboratory Manual (Cold Spring Harbor Laboratory Press; ). [Google Scholar]
- Yuste R, and Katz LC (1991). Control of postsynaptic Ca2+ influx in developing neocortex by excitatory and inhibitory neurotransmitters. Neuron 6, 333–344. [DOI] [PubMed] [Google Scholar]
- Zhang F, Aravanis AM, Adamantidis A, de Lecea L, and Deisseroth K. (2007). Circuit- breakers: optical technologies for probing neural signals and systems. Nat Rev Neurosci 8, 577–581. [DOI] [PubMed] [Google Scholar]
- Zhou P, Resendez SL, Rodriguez-Romaguera J, Jimenez JC, Neufeld SQ, Giovannucci A, Friedrich J, Pnevmatikakis EA, Stuber GD, Hen R, et al. (2018). Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. eLife 7, e28728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong W, Wu R, Li M, Hu Y, Li Y, Li J, Rong H, Wu H, Xu Y, Lu Y., et al. (2017). Fast high-resolution miniature two-photon microscopy for brain imaging in freely behaving mice. Nature Methods 14, 713–719. [DOI] [PubMed] [Google Scholar]
- Zorzos AN, Scholvin J, Boyden ES, and Fonstad CG (2012). Three-dimensional multiwaveguide probe array for light delivery to distributed brain circuits. Optics Letters 37, 4841–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]