Abstract
SARS-CoV-2 enters host cells through its viral spike protein binding to angiotensin-converting enzyme 2 (ACE2) receptors on the host cells. Here, we show that functionalized nanoparticles, termed “Nanotraps,” completely inhibited SARS-CoV-2 infection by blocking the interaction between the spike protein of SARS-CoV-2 and the ACE2 of host cells. The liposomal-based Nanotrap surfaces were functionalized with either recombinant ACE2 proteins or anti-SARS-CoV-2 neutralizing antibodies and phagocytosis-specific phosphatidylserines. The Nanotraps effectively captured SARS-CoV-2 and completely blocked SARS-CoV-2 infection to ACE2-expressing human cell lines and primary lung cells; the phosphatidylserine triggered subsequent phagocytosis of the virus-bound, biodegradable Nanotraps by macrophages, leading to the clearance of pseudotyped and authentic virus in vitro. Furthermore, the Nanotraps demonstrated an excellent biosafety profile in vitro and in vivo. Finally, the Nanotraps inhibited pseudotyped SARS-CoV-2 infection in live human lungs in an ex vivo lung perfusion system. In summary, Nanotraps represent a new nanomedicine for the inhibition of SARS-CoV-2 infection.
Keywords: SARS-CoV-2, inhibition, treatment, macrophage, nanomedicine, nanoparticle, COVID-19, antiviral
Graphical abstract
Progress and potential
To address the need for treatments for SARS-CoV-2 infection, we devised a nanomedicine termed “Nanotraps” that can completely capture and eliminate SARS-CoV-2. The Nanotraps integrate protein engineering, immunology, and nanotechnology, and are effective (10-times more so than their soluble counterparts), biocompatible, safe, stable, and feasible for mass production. The Nanotraps have the potential to be formulated into a nasal spray or inhaler for easy administration and direct delivery to the respiratory system, as an oral or ocular liquid, or subcutaneous, intramuscular, or intravenous injection to target different sites of SARS-CoV-2 exposure, thus offering flexibility in administration and treatment. More broadly, the highly versatile Nanotrap platform could be further developed into new vaccines and therapeutics against a broad range of diseases by incorporating different small-molecule drugs, RNA, DNA, peptides, recombinant proteins, or antibodies.
Presentation of a nanoparticle entitled “Nanotrap” that inhibits entry of SARS-CoV-2 to ACE2-expressing cells and triggers subsequent phagocytosis by macrophages to clear the virus. The Nanotraps showed a neutralizing capacity of more than 10-times that of its soluble ACE2 or neutralizing antibody counterparts. The Nanotraps showed an excellent biosafety profile in vitro and in vivo and completely inhibited pseudotyped SARS-CoV-2 infection in living human lungs in an ex vivo lung perfusion system, demonstrating high potential for clinical translation.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) caused the global pandemic of coronavirus disease 2019 (COVID-19). As of January 8, 2021, SARS-CoV-2 has spread to over 180 countries and has resulted in more than 88.2 million infections and over 1.9 million deaths globally.1 Despite tremendous efforts devoted to drug development, safe and effective medicines to treat SARS-CoV-2 infection are largely lacking. Given that the virus is within nanoscale, nanomaterial-based delivery systems are expected to play a paramount role in the success of prophylactic or therapeutic approaches.2 , 3 To combat this highly contagious virus, here we set out to devise a nanomedicine termed “Nanotraps” to inhibit SARS-CoV-2 infection.
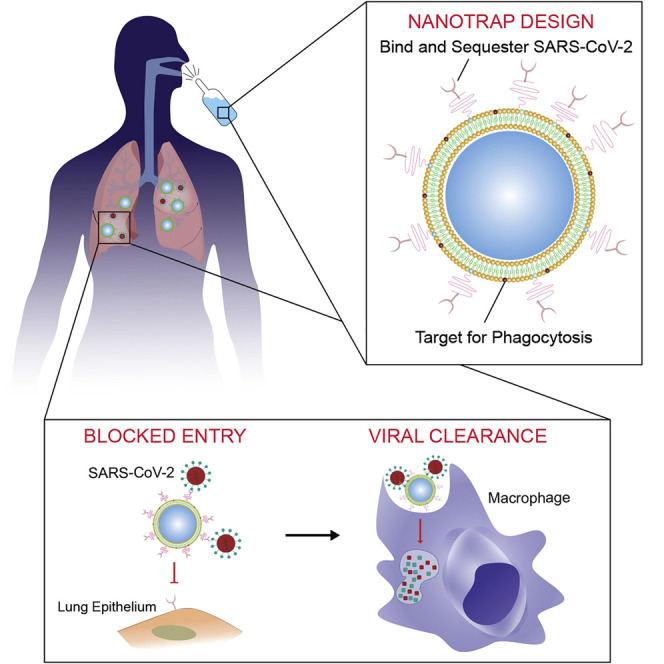
To gain entry to host cells for infection, SARS-CoV-2 surface spike protein binds to its receptor human angiotensin-converting enzyme 2 (ACE2) with high affinity.4, 5, 6, 7 Blocking entry of SARS-CoV-2 to host cells is one of the most effective ways to prevent infection. To achieve this goal, both soluble recombinant ACE2 proteins8 , 9 and anti-SARS-CoV-2 neutralizing antibodies10, 11, 12, 13, 14 have been developed to inhibit the interaction between SARS-CoV-2 spike protein and cell-surface ACE2, although they show limited potency.8 , 9 , 15 Nanomaterial-based detection methods have been developed for COVID-19 diagnosis and monitoring,16, 17, 18 but very few nanomaterial-based therapeutic strategies have been reported.19 Cellular membrane-based nanoscale vesicles have also been developed to inhibit infection,20 , 21 but limited efficacy has been achieved. Inspired by tumor cells secreting PD-L1 exosomes to attenuate T cell effector functions,22 here we designed a therapeutic nanoparticle termed “Nanotrap” to inhibit SARS-CoV-2 infection. The Nanotrap surfaces were functionalized with either ACE2 recombinant proteins or anti-SARS-CoV-2 neutralizing antibodies with high surface density. This design endowed the Nanotraps with high avidity to outperform soluble ACE2 or antibody counterparts to capture and contain SARS-CoV-2. Thus, the high avidity, small size, and high diffusivity of our newly engineered Nanotraps efficiently blocked the binding of SARS-CoV-2 to ACE2-expressing host cells including epithelial cells in the respiratory system, resulting in abrogation of SARS-CoV-2 entry.
Furthermore, we aimed to clear the virus after containment by Nanotrap-mediated macrophage phagocytosis. The role of macrophages in the control of infections has long been documented,23 and recent single-cell RNA sequencing found abundant monocyte-derived macrophages in the bronchoalveolar lavage fluid of COVID-19 patients.24 As professional phagocytes, macrophages engulf apoptotic cells by recognizing phosphatidylserine on the outer leaflet of the plasma membrane of apoptotic cells.25, 26, 27 Phosphatidylserine coatings have been previously employed to enhance the uptake of liposomal nanoparticles by macrophages.28 , 29 Thus, our Nanotraps were further designed to guide the phosphatidylserine-specific phagocytosis by macrophages, enabling not only the containment but also the clearance of SARS-CoV-2 through binding and subsequent phagocytosis.
Here, we engineered Nanotraps composed of a Food and Drug Administration (FDA)-approved polylactic acid (PLA) polymeric core, a liposome shell, surface ACE2/neutralizing antibodies, and phosphatidylserine ligands. Our Nanotraps completely blocked pseudotyped SARS-CoV-2 entry into susceptible ACE2-overexpressing HEK293T cells, lung epithelial A549 cells, and human primary lung cells, as well as authentic SARS-CoV-2 infection of Vero E6 cells. The neutralizing capacity of the Nanotraps was shown to be ~10-times more effective than its soluble counterparts. Subsequently, macrophages efficiently engulfed and neutralized virus-bound, biodegradable Nanotraps through phosphatidylserine-guided phagocytosis without causing infection to macrophages in vitro. Furthermore, in vitro cell culture and in vivo intratracheal administration of Nanotraps to immunocompetent mice demonstrated an excellent biosafety profile. Lastly, the Nanotraps completely inhibited infection of SARS-CoV-2 pseudovirus in live human lungs maintained under normothermic physiologic conditions on a clinically applicable ex vivo lung perfusion (EVLP) system,30 , 31 confirming the therapeutic efficacy. Our Nanotraps are safe, effective, biocompatible, ready for mass production, and convenient to use, and represent a new type of nanomedicine to effectively contain and clear SARS-CoV-2 for the prevention and treatment of COVID-19.
Results
Design, synthesis, and characterization of Nanotraps
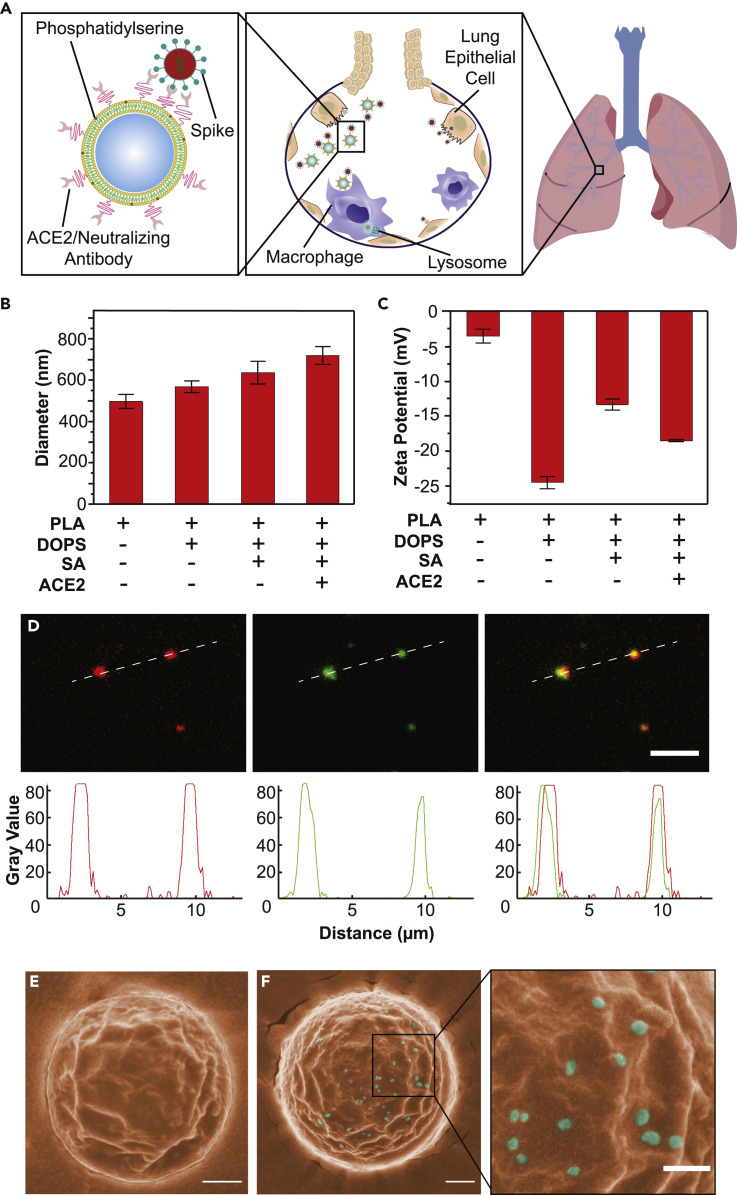
SARS-CoV-2 gains entry into host cells via surface spike proteins that bind to human ACE2 receptors on host cells with very high affinity.6 , 32, 33, 34 To inhibit SARS-CoV-2 infection, we set out to engineer a family of nano-enabled virus-trapping particles, termed “Nanotraps,” to contain and clear SARS-CoV-2 (Figure 1 A). We used an FDA-approved, biodegradable PLA polymeric core and liposome shell materials to synthesize the Nanotraps. Nanotraps with different diameters (200, 500, and 1,200 nm) were synthesized by varying polymer concentrations (see experimental procedures for details) (Figure S1A). The solid PLA core acts as a “cytoskeleton” to provide mechanical stability, controlled morphology, biodegradability, and large surface area for nanoscale membrane coating and surface modification. The lipid shell enveloping the PLA core exhibits behavior similar to that of cell membranes. The lipid shell provides a nanoscopic platform and can interact with a wide variety of molecules35, 36, 37 either within the membrane or on the surface.38 , 39 Thus, we aimed to functionalize the Nanotrap surface with a molecular bait (a recombinant ACE2 protein or an anti-SARS-CoV-2 neutralizing antibody) and a phagocytosis-inducing ligand (phosphatidylserine). We hypothesized that (1) the high-density ACE2 or neutralizing antibodies on the Nanotraps can outcompete low-expression ACE2 on host cells in capturing SARS-CoV-2, thus enabling selective virus containment by the Nanotraps, and that (2) surface phosphatidylserine ligands on suitably sized Nanotraps can trigger subsequent phosphatidylserine-mediated phagocytosis by professional phagocytes, such as macrophages, thus enabling viral clearance (Figure 1A). The resultant structures were monodispersed and significantly smaller than mammalian cells, yet still large enough to bind several SARS-CoV-2 virions (Figures 1B–1F).
Figure 1.
Schematic design, synthesis, and characterization of Nanotraps for SARS-CoV-2
(A) Schematic illustration showing the process of the Nanotraps with polymeric core coated with lipid bilayer functionalized with ACE2 protein/neutralizing antibody. Following intratracheal administration, Nanotraps efficiently accumulated and trapped SARS-CoV-2 virions in the lung tissue forming virus-Nanotrap complexes, which can be cleared by macrophages via phagocytosis, thereby blocking viral cell entry.
(B and C) Dynamic light scattering (B) and zeta-potential measurements (C) during different stages of Nanotrap preparation. Data are presented as average ± SD.
(D) Fluorescent images of the prepared Nanotraps with PLA polymeric core (DiD, red) and ACE2 (anti-ACE2-AF488, green). Dashed lines represent displayed plot profile below. Scale bar represents 5 μm.
(E and F) Pseudocolored SEM images of Nanotraps alone (E, orange) or with SARS-CoV-2 pseudovirus (F, cyan). To better visualize the selectivity for viral binding, we imaged larger Nanotraps. Scale bars represent 300 nm.
To characterize the Nanotraps, we first used dynamic light scattering (DLS) to measure the size dispersity of the constituent nanoparticles. Controlling particle size is important for tuning the phagocytosis efficacy, reproducible mechanical characteristics, and material biocompatibility.40 , 41 The hydrolyzed diameter of the Nanotraps measured by DLS increased with the addition of each molecule (Figure 1B). The zeta potential, which reflects the surface charges of the Nanotraps,42 was found to change slightly with the addition of each molecule to the Nanotrap surface (Figure 1C). We next used fluorescent labeling and total internal reflection fluorescence microscopy (TIRFM) to confirm the presence of the ACE2 moiety on the Nanotraps. The PLA polymeric core of each Nanotrap was labeled with a lipophilic carbocyanine dye, 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine, 4-chlorobenzenesulfonate salt (DiD, red). The Nanotrap-ACE2 was further stained with an anti-ACE2 antibody labeled with Alexa Fluor 488 dye (AF488, green). The TIRFM images clearly showed excellent co-localization between the Nanotrap core and surface ACE2 at the single-particle level, confirmed by line scans of the fluorescent channels corresponding to each component (Figure 1D). These results demonstrated that we have successfully functionalized the Nanotraps with recombinant ACE2 protein. We employed scanning electron microscopy (SEM) and transmission electron microscopy (TEM) to image the Nanotraps at the subnanometer level. SEM images showed that the Nanotraps were spherical and well dispersed (Figure S1B). The Nanotraps appeared slightly crenellated in the SEM images, as the lipid layer may have shrunk due to the drying sample preparation procedure before imaging (Figure 1E). As expected, after incubation with the pseudotyped SARS-CoV-2 for 1 h at 37°C, the Nanotraps effectively captured the virus as evidenced by single virions clearly visualized on the surface of a Nanotrap; no freestanding virions were observed outside of the Nanotrap (Figure 1F). The representative TEM image clearly shows the core-shell structure of the Nanotrap (Figure S1C). Importantly, Nanotraps that were lyophilized, sealed, and stored at −20°C for 6 months demonstrated no change in surface integrity (Figure S1D) or size (Figure S1E), indicating excellent stability over time.
We further quantified the surface molecular densities of the two Nanotraps by flow cytometry: the Nanotrap-ACE2 has (3.59 ± 0.43) × 105 ACE2 molecules per Nanotrap, and the Nanotrap-Antibody has (2.47 ± 0.2) × 104 neutralizing antibodies per Nanotrap (Figures S1F and S1G). We hypothesize that the lower concentration of surface antibodies is due to (1) the lower efficiency conjugation method of N-hydroxysuccinimide (NHS) esters versus the site-specific, high-affinity (10−14 M)43 biotin-streptavidin conjugation used for ACE2 conjugation in aqueous solution, and (2) imperfect antibody orientation leading to blockage of the antibody binding site to virus spike protein (Figure S1H). However, future studies can easily manipulate this surface density as needed by simply adjusting the molar ratios of 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[biotinyl(polyethylene glycol)2000] (DSPE-PEG2000-biotin) or DSPE-PEG2000-NHS on the lipid shell for Nanotrap synthesis (see experimental procedures).
Phagocytosis of Nanotraps by macrophages
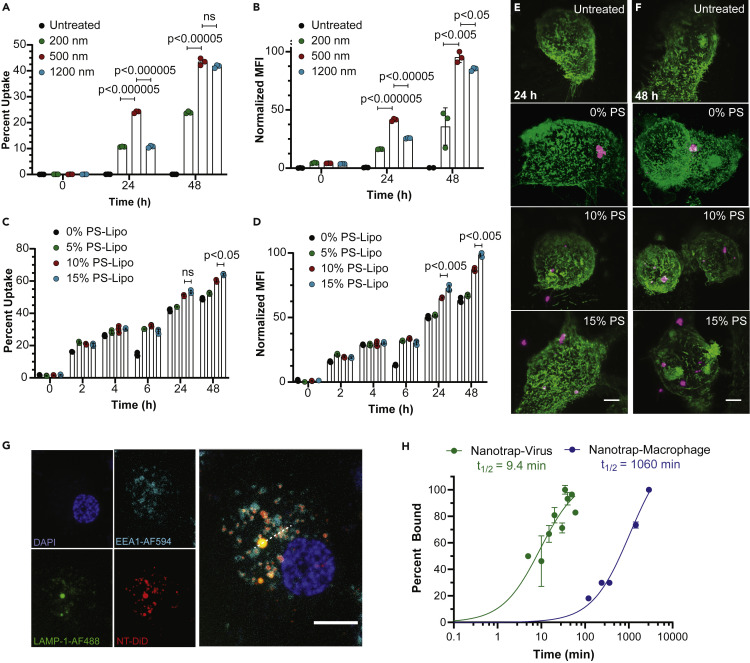
Macrophages are a class of phagocytes that engulf and clear cell debris, pathogens, microbes, cancer cells, and other foreign intruders.23 Macrophages specialize in the removal of dying or dead cells by recognizing phosphatidylserine on the outer leaflet of the plasma membrane of apoptotic cells.25, 26, 27 Phosphatidylserine coatings have been previously employed to enhance macrophage uptake of liposomal nanoparticles.44, 45, 46 Because phagocytosis by macrophages is highly dependent on size and surface phosphatidylserine, we determined the optimal size and surface phosphatidylserine density of Nanotraps. We first synthesized Nanotraps labeled with 3,3′-dioctadecyloxacarbocyanine perchlorate (DiO) fluorescent dye on the PLA polymeric core with varying diameters of 200, 500, and 1,200 nm (Figure S2A). After incubating Nanotraps with differentiated THP-1 (dTHP-1) macrophages47 for varying time durations, we examined the size-dependent phagocytosis by dTHP-1 macrophages using flow cytometry. We found that the 500-nm Nanotraps outperformed the 200-nm and 1,200-nm counterparts after 24-h and 48-h incubations, as demonstrated by the percent uptake (Figure 2 A) and the mean fluorescent intensity of DiO dye (Figures 2A and S2A–S2E) in dTHP-1 macrophages. Accordingly, 500-nm PLA-core Nanotraps were chosen for all further experiments.
Figure 2.
Phagocytosis of Nanotraps by macrophages
Nanotraps of varying sizes and phosphatidylserine (PS) densities were incubated with dTHP-1 macrophages.
(A and B) Percent uptake (A) and mean fluorescent intensity (B) quantification of flow-cytometry measurement of the internalization of Nanotraps with varying diameters. Data are shown as mean ± SD; unpaired t tests were conducted from three replicates.
(C and D) Percent uptake (C) and mean fluorescent intensity (D) quantification of flow-cytometry measurements of the internalization of Nanotraps with varying PS ratios. Data are shown as mean ± SD; unpaired t tests were conducted from three replicates.
(E and F) Lattice light-sheet microscopy images of macrophages (WGA-CF488, green) phagocytosing Nanotraps (DiD, magenta) after 24 h (E) or 48 h (F). Scale bars represent 5 μm.
(G) Confocal microscopy image of dTHP-1 cell stained for DAPI (blue), lysosomes (LAMP-1-AF488, green), and endosomes (EEA1-AF594, cyan) incubated with Nanotrap-ACE2 (DiD, red) for 6 h. Dotted line indicates line scan analyzed in Figure S2F. Scale bar represents 10 μm.
(H) Viral particles labeled with lipophilic dye DiO incubated with Nanotraps (DiD) at 37°C for various times (green) and dTHP-1 macrophages incubated with Nanotraps (DiO) at 37°C for various times (blue) were measured by flow cytometry. Double-positive events were gated, and the mean fluorescence intensity of DiO was measured over time until saturation was reached. Data were fitted with a curve, and t1/2 was extrapolated as the time at which 50% of Nanotraps were bound or engulfed.
We next functionalized the Nanotraps with varying surface densities of phosphatidylserine to induce phagocytosis by macrophages. The overall negative charge of the Nanotraps increases as the percentage of phosphatidylserine ratio increases, confirming the presence of phosphatidylserine (Figure S2B). We found that phagocytosis by macrophage was roughly correlated to the percentage of phosphatidylserine (Figures 2C and 2D). The phosphatidylserine-dependent phagocytosis of Nanotraps by dTHP-1 macrophages were further demonstrated by three-dimensional lattice light-sheet microscopy images (Figures 2E and 2F; Video S1). These experiments not only confirmed successful functionalization of the Nanotraps but also identified the optimal surface density of phosphatidylserine on the Nanotrap surfaces. Thus, we utilized the 500-nm core size and 15% surface phosphatidylserine Nanotraps for the following experiments to maximize viral capture and macrophage phagocytosis.
Three-dimensional image reconstruction of macrophages (WGA-CF488, green) phagocytosing Nanotraps (DiD, magenta). Video shows macrophages treated with 10% phosphatidylserine Nanotraps for 24 h and untreated macrophages.
We further tested whether the engulfed Nanotraps would fuse with endosomes or lysosomes within the macrophages. As shown in Figure 2G, Nanotraps (DiD, red) in various regions in the cell co-localize with either endosomes (cyan) or lysosomes (green), suggesting they will in fact be degraded over time. In addition, we tested the kinetics of (1) the Nanotrap binding to the virus and (2) the macrophage engulfment of Nanotraps (Figure 2H). The binding between the pseudovirus and the Nanotraps saturated within an hour (t 1/2 9.4 min), whereas the macrophage engulfment did not reach saturation with the Nanotraps until 48 h later (t 1/2 1,060 min), suggesting that the Nanotraps bind to virus before being engulfed by the macrophages. Finally, we tested whether proinflammatory cytokine interleukin-6 (IL-6) was secreted by the macrophages following the Nanotrap engulfment (Figure S2G). No significant differences were found among the following conditions: (1) macrophages alone; (2) macrophages and Nanotraps; (3) macrophages and epithelial cells; (4) macrophages, epithelial cells, and virus; (5) macrophages, epithelial cells, and Nanotraps; and (6) macrophages, epithelial cells, virus, and Nanotraps. These data indicate that the Nanotraps do not promote a proinflammatory response by the macrophages at the treatment dosage.
Nanotraps neutralize SARS-CoV-2 infection in vitro
We next generated multiple types of Nanotraps to test their efficacy. First, avi-tagged biotinylated ACE2 was conjugated to the Nanotrap surface via biotin-streptavidin interactions to make Nanotrap-ACE2. In addition, we synthesized Nanotrap-Antibody by conjugating a SARS-CoV-2 neutralizing antibody to the Nanotrap surface via NHS esters (for details see experimental procedures). Finally, to test the specificity of the Nanotraps, we made Nanotrap-Blank without a virus-binding epitope.
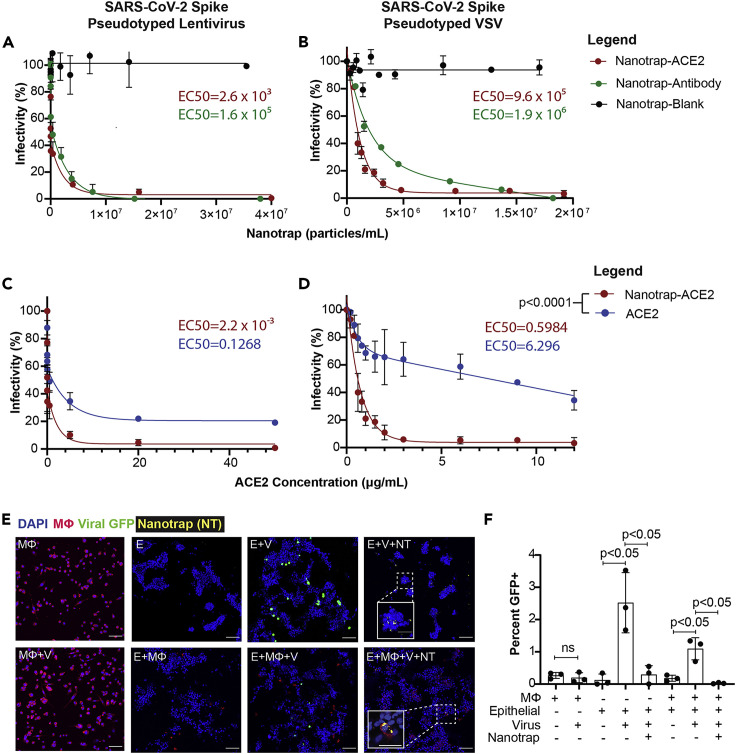
We next examined whether the Nanotraps could effectively capture and contain SARS-CoV-2 in vitro. All three Nanotraps were incubated with SARS-CoV-2 spike pseudotyped lentivirus or vesicular stomatitis virus (VSV) for 1 h before adding to HEK293T-ACE2 cells for 24 h and 72 h, respectively. Both Nanotrap-ACE2 and Nanotrap-Antibody completely blocked SARS-CoV-2 pseudovirus infection, while the Nanotrap-Blank did not, indicating both the specificity and functionality of our Nanotraps (Figures 3A, 3B, S3A, S3B, and S3E). Interestingly, despite the fact that neutralizing antibodies have a higher affinity for spike protein (0.07 nM) than that of ACE2 (22 nM),48 the Nanotrap-ACE2 blocks SARS-CoV-2 pseudovirus infection more efficiently than the Nanotrap-Antibody. This is most likely due to the lower molecular density (Figures S1F and S1G) and the random orientation (Figure S1H) of neutralizing antibody on the Nanotraps compared with that of the ACE2. Thus, the overall avidity will be lower on the Nanotrap-Antibody than on the Nanotrap-ACE2. This could be corrected in future studies by using the higher-efficiency biotin-streptavidin conjugation rather than the NHS-amine conjugation method used here. In sharp contrast to the Nanotraps, soluble recombinant ACE2 protein only partially inhibited infection to HEK293T-ACE2 cells with both SARS-CoV-2 spike pseudotyped lentivirus (Figures 3C and S3C) and VSV (Figures 3D and S3D), despite the previous use of soluble ACE2 protein.8 , 9
Figure 3.
Inhibition of SARS-CoV-2 viral infection of host cells
(A and B) HEK293T-ACE2 cells were treated with SARS-CoV-2 spike pseudotyped lentivirus (A) or VSV (B) and Nanotrap-ACE2, Nanotrap-Antibody, or Nanotrap-Blank for 72 h and 24 h, respectively. Particle densities were counted with a hemocytometer. Data are presented as mean ± SD and fitted with a two-phase decay model.
(C and D) HEK293T-ACE2 cells were treated with SARS-CoV-2 spike pseudotyped lentivirus (C) or VSV (D) and Nanotrap-ACE2 or soluble ACE2 for 72 h and 24 h, respectively. Data are presented as mean ± SD and fitted with a trend curve. For both SARS-CoV-2 spike pseudotyped lentivirus (C) and VSV (D), the Nanotrap-ACE2 and soluble ACE2 curves differ with p < 0.0001, as tested by sum-of-squares F tests.
(E) Confocal microscopy of pseudotyped VSV infection (GFP, green) in dTHP1 macrophages (WGA, red) and A549 epithelial cells (DAPI, blue); Nanotrap-ACE2 is displayed in yellow. MФ, macrophages; V, virus; E, epithelial cells; NT, Nanotraps. Scale bars represent 100 μm, with inset scale bars representing 40 μm.
(F) Quantification of (E). Data are shown as mean ± SD; unpaired t tests were conducted from three independent experiments. ns, not significant.
Macrophages play a key role in controlling SARS-CoV-2 infection.47 We thus further determined whether human macrophages could efficiently engulf and degrade the virus-bound Nanotraps-ACE2 without becoming infected (Figures 1E and 1F). Importantly, after incubating SARS-CoV-2 spike pseudotyped VSV with dTHP-1 macrophages for 24 h, no infection was found in the macrophages (Figures 3E and 3F, comparing “MФ” with “MФ + V”). This experiment demonstrated the feasibility of utilizing macrophages to clear the viral infection. We then infected a human lung epithelial cell line, A549, which expresses physiological levels of surface ACE2, with SARS-CoV-2 spike pseudotyped VSV in the absence or presence of dTHP-1 macrophages. These data suggest that macrophages significantly reduced the viral infection but could not completely eradicate it (Figures 3E and 3F, comparing “E + V” with “E + MФ + V”). Finally, we determined whether our engineered Nanotraps triggered phosphatidylserine-mediated phagocytosis by dTHP-1 macrophages for the clearance of virus. After adding Nanotrap-ACE2 into the co-culture of epithelial cells, macrophages, and SARS-CoV-2 spike pseudotyped VSV, the viral infection was completely inhibited (Figures 3E and 3F, comparing “E + V + NT” and “E + MФ + V + NT” with “E + MФ + V”). We further observed incorporation of Nanotrap-ACE2 into the macrophage cell body but not into the epithelial cells, indicating successful macrophage-specific phagocytosis (Figures 2E and 3E, “E + MФ + V + NT” inset; Video S1).
In sum, our in vitro neutralizing experiments demonstrated that our Nanotraps not only served as a sponge to capture and contain SARS-CoV-2 but also utilized the phagocytosis and sterilization machinery of macrophages to defend the host cells from infection, as we depicted in our original experimental design (Figure 1A).
In vivo local delivery to lungs and biosafety profile of Nanotraps in mice
To assess the safety of Nanotrap treatment, we first examined in vitro cytotoxicity on human cell lines. Neither HEK293T-ACE2 nor A549 cells displayed significant cytotoxicity with the addition of Nanotrap-Blank, Nanotrap-ACE2, or Nanotrap-Antibody, as evaluated by a CCK8 cytotoxicity assay (Figures S4A and S4B).
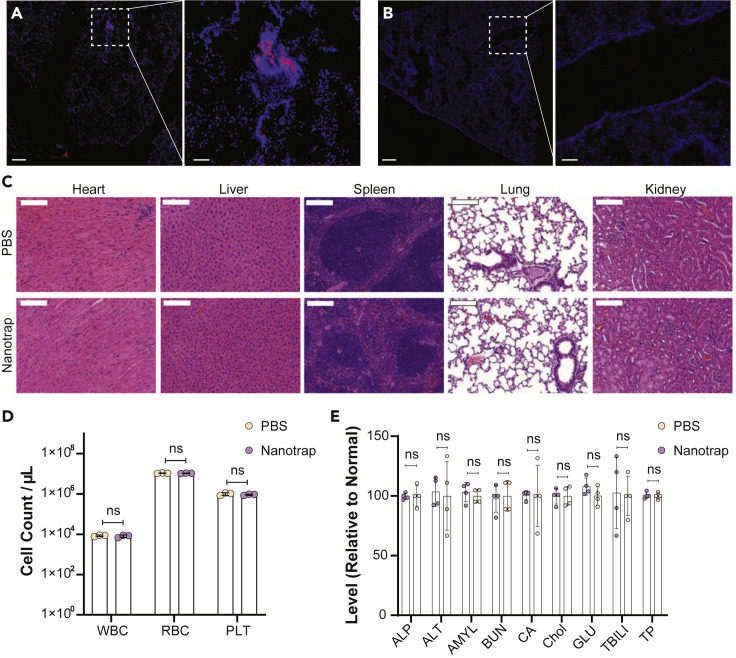
We next examined the delivery of Nanotraps to mouse lungs and evaluated the biosafety of Nanotraps in vivo. We intratracheally injected immunocompetent mice with Nanotrap-ACE2 (labeled with DiD) at a dose of 10 mg/kg. Mice were sacrificed 3 days post injection. Delivery of Nanotraps to mouse lungs was confirmed with cryosectioned mouse lung tissues: significant Nanotrap accumulation and distribution were found in the lung tissues, particularly in regions around bronchioles in the respiratory tracts (Figure 4 A). As expected, no Nanotraps were found in the lungs of PBS-treated mice (Figure 4B).
Figure 4.
Murine in vivo biosafety profile of Nanotrap treatment
WT B6 mice were treated intratracheally with 10 mg/kg Nanotrap-ACE2 or PBS for 72 h (n = 4, 2 male + 2 female).
(A and B) Representative fluorescent images of Nanotraps (DiD, red) accumulating in the lung tissues (DAPI, blue) of NT-treated mice (A) or PBS-treated mice (B) 72 h post intratracheal administration. Scale bars represent 250 μm; inset scale bars represent 50 μm.
(C) Representative H&E staining of major organ sections of heart, liver, spleen, lung, and kidney. Scale bars represent 500 μm.
(D) Blood cell counts of white blood cells (WBC), red blood cells (RBC), and platelets (PLT) 72 h after Nanotrap or PBS treatment. Data are shown as mean ± SD; unpaired t tests were conducted from four replicates. ns, not significant.
(E) Comprehensive blood chemistry panels comparing Nanotrap- and PBS-treated mice. ALP, alkaline phosphatase; ALT, alanine aminotransferase; AMYL, amylase; BUN, urea nitrogen; CA, calcium; Chol, cholesterol; GLU, glucose; TBIL, total bilirubin; TP, total protein. Data are shown as mean ± SD; unpaired t tests were conducted from four replicates. ns, not significant.
In vivo safety was next analyzed. Hematoxylin and eosin (H&E) staining of major organs including lung, heart, liver, spleen, and kidney showed no histological differences in the Nanotrap-treated mice when compared with the PBS-treated control group (Figure 4C). Furthermore, complete blood counts were performed to evaluate white blood cells (WBCs), red blood cells (RBCs), and platelets (PLTs). The cell counts were similar between Nanotrap- and PBS-treated groups (Figure 4D). Next, comprehensive metabolic panels of mouse blood sera were examined to provide an overall picture of the chemical balance and metabolism. No statistical differences were found between Nanotrap- and PBS-treated mice for glucose levels, electrolyte and fluid balance, kidney function, or liver function (Figure 4E). These results demonstrated the safety of Nanotraps when delivered in vivo.
The therapeutic efficacy of Nanotraps in ex vivo human lungs
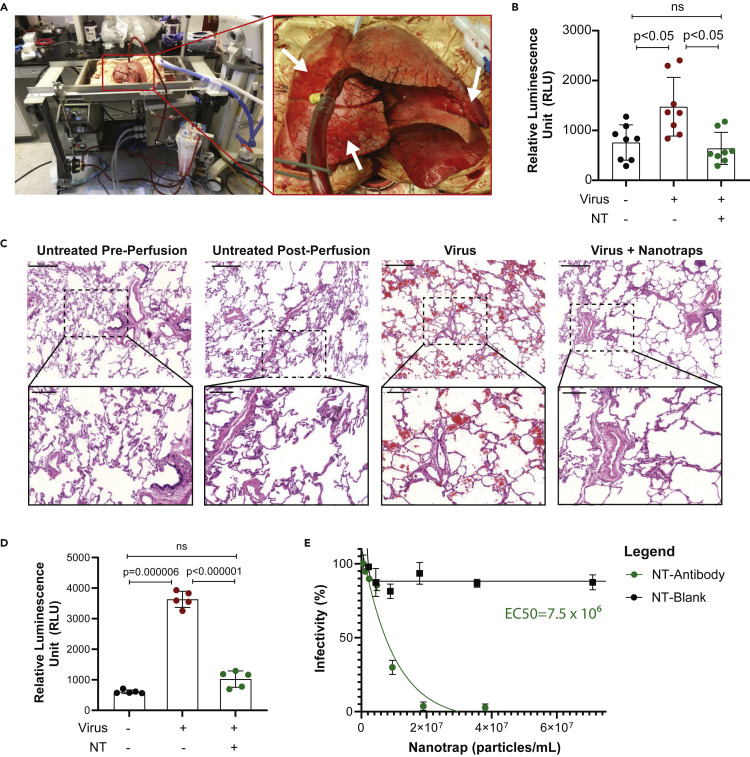
We next examined the therapeutic efficacy of Nanotraps in inhibiting pseudotyped SARS-CoV-2 infection in healthy, non-transplantable human donor lungs using an EVLP system (Figure 5 A; Video S2). EVLP allows a lung to be perfused and ventilated ex vivo after organ retrieval by maintaining lungs at normothermic physiologic conditions and is thus an excellent platform on which to model lung diseases.42
Figure 5.
Ex vivo human lung perfusion system for evaluating the neutralizing ability of the Nanotrap-Antibody
(A) Photos of human ex vivo lung perfusion (EVLP). Arrows indicate untreated (right upper lobe), virus only (right middle lobe), and virus + Nanotrap (lingula) regions.
(B) Quantification of luciferase expression in EVLP samples 8 h post infection in three regions indicated in the right panel of (A). Data are shown as mean ± SD; unpaired t tests were conducted from 8 replicates. ns, not significant.
(C) H&E staining of EVLP. Scale bars represent 500 μm; inset scale bars represent 200 μm.
(D) Quantification of luciferase expression in in vitro primary human cells 48 h post infection. Data are shown as mean ± SD; unpaired t tests were conducted from 5 replicates. ns, not significant.
(E) Vero E6 cells treated with authentic SARS-CoV-2 and Nanotrap-Blank or Nanotrap-Antibody. Particle densities were counted with a hemocytometer. Data are shown as mean ± SD fitted with a trend curve.
Video shows lungs from Figure 5 attached to the EVLP ventilator system.
The infection potential of the SARS-CoV-2 spike pseudotyped lentivirus at different doses over time in primary human lung cells was first tested in vitro, and infection was observed within 8 h (Figures S5A and S5B). After confirming infection potential, we tested our Nanotraps on an EVLP system with a pair of non-transplantable human lungs. Static lung compliance and oxygenation capacity was measured over time (Figure S5C). SARS-CoV-2 pseudovirus carrying a luciferase reporter gene was injected into the lingula of left upper lung lobe, and pseudovirus plus Nanotrap-Antibody was injected into the right middle lobe; the right upper lung lobe was used as an untreated control (Figure 5C, arrows). Human lung tissue samples were collected after perfusing for 8 h. Single-cell suspensions were generated and luciferase expressions were determined (Figure 5B). The results showed that (1) the pseudovirus infected the lung tissues and (2) the Nanotraps completely inhibited the viral infection. Furthermore, H&E staining showed significant RBC infiltration in the virus-treated sample, which was not present in the virus-plus-Nanotrap-treated region (Figures 5C, S5D, and S5E).
As our EVLP system maintains lung viability for less than 12 h, we treated single-cell suspensions of healthy, untreated lung from the right upper lobe in vitro for 48 h to confirm that the Nanotraps can function for longer-term incubations in human tissue. Again, Nanotrap-Antibody was able to fully inhibit the virus (Figure 5D). Finally, since the EVLP could not be conducted under BSL-3 conditions in order to use authentic SARS-CoV-2, we tested the ability of the Nanotraps to prevent authentic SARS-CoV-2 from infecting Vero E6 cells, which are highly susceptible to SARS-CoV-2 infection.49 Indeed, Nanotrap-Antibody was able to completely inhibit infection of authentic SARS-CoV-2, as expected (Figure 5E). Importantly, the Nanotrap-Antibody outperformed the soluble antibody in the authentic SARS-CoV-2 infection (Figure S5F; Equation S1), despite the fact that this assay cannot show the impact of the phagocytic targeting of our Nanotraps, which should further enhance the virus clearance (Figures 3E and 3F).
Taken together, our EVLP experiments demonstrated that (1) SARS-CoV-2 pseudovirus can infect human lung and (2) our newly engineered Nanotraps can completely block the viral infection, thus paving the way for future clinical trials using Nanotraps for the inhibition of SARS-CoV-2 infection.
Discussion
The highly contagious SARS-CoV-2 has caused the global COVID-19 pandemic, so effective and safe treatments are urgently needed. Remdesivir has been approved by the FDA to treat severe COVID-1950 despite its inconsistent clinical benefits and various reported adverse effects.51 , 52 Transfusion of convalescent plasma from recovered patients has shown clinical benefits in some COVID-19 patients;53 however, this approach is challenged by the limited availability of donor plasma and appropriate medical facilities.54 Simultaneously, tremendous efforts have been devoted to the development of vaccines, neutralizing antibodies, and other drugs for the prevention and treatment of COVID-19. While recently developed vaccines are being dispersed to the population, safe and effective medicines to treat SARS-CoV-2 infection are largely lacking. For example, Nanosponges have been developed,55 but they cannot completely inhibit infection. They are also not a viable treatment option: as each patient's human leukocyte antigen is different, Nanosponges would have to be personalized for each patient. Large amounts of uninfected primary cells would have to be collected from each patient to make personalized Nanosponges, similar to chimeric antigen receptor T (CAR-T) cell therapy, which has resulted in exorbitant costs and is thus unattainable by many patients.56 In addition, recombinant soluble ACE2 or ACE2-immunoglobulin G (IgG) proteins have been proposed for use as treatments,8 , 14 but soluble proteins are known to be much more liable to degradation than nanoparticles.57 Although some existing nanomedicines have been shown to inhibit viral entry,20 , 21 they do not trigger any specific clearing mechanism, leaving the risk of infection unaddressed. In contrast, our Nanotraps both inhibit cell entry and trigger the phagocytic clearance of the virus. The design of our Nanotraps was inspired by the ability of tumor cells to secrete PD-L1 exosomes, which bind to and suppress T cell immune functions and thus prevent the killing and clearance of the tumor cells.22 , 58 In a similar fashion, the synthetic Nanotraps can mimic the target cells to ensnare the virus. Meanwhile, the synthetic polymer-lipid complexes take advantage of both the stability from polymers and surface flexibility of lipids,59 thereby providing a well-controlled nanomaterial with a high capacity to trap the virus by mimicking target cells. We thus created Nanotraps to bind and inhibit SARS-CoV-2 infection to host cells.
To block the interaction between the SARS-CoV-2 spike protein and the host ACE2 receptors, we coated the Nanotrap surfaces with a high molecular density of either recombinant ACE2 proteins or anti-SARS-CoV-2 neutralizing antibodies (Figure 1A). In principle, the high binding avidity, high diffusivity, and small size of Nanotraps should enable them to easily outcompete low-ACE2-expressing host cells in capturing the SARS-CoV-2, thus effectively containing the viruses on their surfaces. Indeed, our experiments demonstrated that viral infection of both pseudotyped and authentic SARS-CoV-2 across human cell lines, lung primary cells, and lung organs can be completely inhibited by Nanotrap-ACE2 or Nanotrap-Antibody (Figures 3 and 5). Notably, the Nanotraps were 10 times more efficient than their soluble counterparts at containing SARS-CoV-2,8 , 9 attributed to their high binding avidity (Figures 3C, 3D, and S5F). We would also like to note that there are both advantages and disadvantages to bivalent/Fc-containing proteins, such as the antibody, versus monovalent/Fc-free proteins, such as ACE2. For example, bivalent proteins have increased binding capacity per molecule: while we showed that the Nanotrap-Antibody had a lower density than the Nanotrap-ACE2 (Figure S1F), the two Nanotraps still had comparable inhibitory effects (Figures 3A and 3B). However, it is possible that the Fc region could contribute to the aggravation of COVID-19 due to antibody-dependent enhancement.60 , 61 Future studies to optimize the choice of targeting molecules for human use are required.
Furthermore, our Nanotraps harness the immune system to clear the SARS-CoV-2 (Figures 2 and 3). By incorporating the phagocyte-specific phosphatidylserine ligands onto the Nanotrap surfaces, macrophages readily engulfed the virus-bound Nanotraps without becoming infected themselves (Figures 3E and 3F). While macrophages were used as a proof of principle in this study, other professional phagocytes such as neutrophils, monocytes, and dendritic cells should be able to similarly clear the virus-bound Nanotraps. In particular, macrophages and dendritic cells are professional antigen-presenting cells, which present engulfed antigens to the adaptive immune system.62 Since the Nanotraps are able to engage antigen-presenting cells, it is possible that they may also elicit virus-specific adaptive immune responses. Future studies will evaluate whether Nanotraps can prime adaptive immune responses, thereby promoting vaccine-like protection.63
In addition, we purposely designed Nanotraps to be biocompatible, biodegradable, and safe. The Nanotraps were composed of FDA-approved polymers and lipids, which provides the possibility for safe administration in a clinical setting. Indeed, our biosafety experiments have demonstrated an excellent safety profile in vitro and in vivo (Figure 4).
Lastly, we tested the efficacy of the Nanotraps in a human EVLP system. Superior to lung organoids, which cannot reproduce whole-organ response to viral infection,9 , 64 , 65 and non-human primate models, which are extremely costly,66, 67, 68 the EVLP system is a clinically relevant model. We showed that our Nanotraps can completely inhibit viral infection in living human lungs (Figure 5). As current biosafety regulations preclude the testing of authentic SARS-CoV-2 in the EVLP, we further confirmed that our Nanotraps can completely inhibit authentic virus in vitro (Figure 5E). These experiments together suggest that our Nanotraps could potentially be used to treat SARS-CoV-2 infection in the clinic.
In summary, we developed a new type of potent, effective nanomedicine, Nanotraps, to contain and clear SARS-CoV-2 by harnessing and integrating the power of nanotechnology and immunology. The Nanotraps completely inhibited the SARS-CoV-2 infection to human cells and lung organs. The Nanotraps are effective, biocompatible, safe, stable, and feasible for mass production. It is reasonable to hypothesize that Nanotraps could be easily formulated into a nasal spray or inhaler for easy administration and direct delivery to the respiratory system, or as an oral or ocular liquid, or subcutaneous, intramuscular, or intravenous injection to target different sites of SARS-CoV-2 exposure, thus offering flexibility in administration. Furthermore, the design of our Nanotraps is highly versatile: they can be modified to incorporate small-molecule drugs or protein/mRNA vaccines to their core, and different human ACE2 recombinant proteins,8 , 9 human anti-SARS-CoV-2 neutralizing antibodies,10, 11, 12, 13, 14 or any developed therapeutic proteins or peptides can be conjugated to the surface, thus easily extending their applications beyond our current study. Overall, we thus expect continuous development of this nanomedicine for clinical use to prevent and treat SARS-CoV-2 infection.
Experimental procedures
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jun Huang (huangjun@uchicago.edu).
Materials availability
The materials generated in this study are available from the corresponding author upon request.
Data and code availability
The data used to support the findings of this study are available from the corresponding author upon request.
Synthesis of Nanotraps
To synthesize the Nanotraps, we employed a two-step method developed for polymer-lipid hybrid nanoparticles whereby the polymer and lipid components were prepared separately and combined at the end of the process. The PLA nanoparticles were prepared in accordance with existing methods69 through an oil-in-water emulsion solvent evaporation process. One hundred milligrams of PLA and 100 μL of perfluorooctyl bromide (PFOB)70 were dissolved in 3.5 mL of dichloromethane. The organic phase was mixed with 20 mL of 2.0% polyvinyl alcohol (PVA) solution. The mixture was emulsified by sonication (Fisher Sonics) on ice for 2 min. The dichloromethane in the emulsified mixture was evaporated under magnetic stirring at 300 rpm for 3–4 h at room temperature. The resulting solution was centrifuged and the pellets were washed with PBS three times (5,000 × g for 10 min), after which the pellets were lyophilized and stored at 4°C before use. For any fluorescence labeling, 0.05 mg of DiD or DiO was added into the 100-mg PLA organic phase when preparing the Nanotraps. For PLA nanoparticles with size 200 nm, 100 mg of PLA was emulsified with 2.0% PVA and the supernatant was collected after centrifugation instead of the pellet. For PLA nanoparticles with size 500 nm, 100 mg of PLA and 100 μL of PFOB were mixed and emulsified with 2.0% PVA, and the pellets were collected after centrifugation. For PLA nanoparticles with size 1,200 nm, 100 mg of PLA and 100 μL of PFOB were mixed and emulsified with 1.0% PVA and the pellets were collected after centrifugation. For further details on the synthesis of PLA nanoparticles with varying sizes, see Table S1.
Functionalized PLA nanoparticles composed of 15% phosphatidylserine and 0.5% DSPE-PEG2000-biotin were prepared by dissolving 1,2-dioleoyl-sn-glycero-3-phospho-L-serine (DOPS) (0.388 μmol), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC) (1.292 μmol), cholesterol (0.775 μmol), DSPE-mPEG2000 (0.166 μmol), and DSPE-PEG2000-biotin (0.013 μmol) at a molar ratio of 10:120:60:9:1 in dichloromethane. The dichloromethane solvent was slowly evaporated by heating the lipid solution at 55°C to remove the solvent and further dried in a vacuum drying oven to produce a dried lipid film. The lipid film was reconstituted in 2 mL of PBS (pH 7.4) containing 0.2 mg of PLA nanoparticles, and the contents were hydrated at 60°C under ultrasonication (Branson CPX5800H). The mixture was then sonicated with a sonicator probe (Fisher Sonics) for 2 min (100 W, 22.5 kHz, 30% amplitude). Formulations for Nanotraps with different phosphatidylserine surface densities are listed in Table S2.
For synthesis of Nanotrap-ACE2, 1 mg of the biotin functionalized PLA nanoparticles was then incubated with streptavidin (34.25 μL, 2 g/L) on ice for 40 min under magnetic stirring. The resulting solution was centrifuged at 5,000 × g for 10 min and washed with 1 mL of PBS three times to remove excess streptavidin. The pellet was resuspended in 100 μL of PBS and incubated with biotinylated ACE2 (Bioss Antibodies) for 30 min on ice, and any free ACE2 was removed by washing with PBS against an Ultra-centrifugal tube (100 kDa) three times (3,000 × g for 10 min). The resulting PLA@DOPS/biotin~SA~ACE2 nanoparticle was termed Nanotrap-ACE2.
Nanotrap-Antibody was synthesized by combining 15% DOPS (0.313 mg, 0.386 μmol), 50% DSPC (1.018 mg, 1.287 μmol), 30% cholesterol (0.300 mg, 0.773 μmol), and 5% DSPE-PEG2000-NHS (0.370 mg, 0.129 μmol) in dichloromethane. The lipid mixtures were vacuum dried overnight, and the resulting thin film was hydrated with PLA-NPs PBS solution in an ultrasonic water bath and further reacted with 100 μg of SARS-CoV-2 neutralizing antibody (Sino Biological) for 4 h at 4°C. Any free antibodies were removed by washing with PBS three times (5,000 × g for 10 min).
Characterization of Nanotraps
The sizes of Nanotraps were measured by a DLS particle size analyzer (Malvern Zetasizer). In brief, 1 μL of Nanotraps was dispersed in 0.1× PBS and further dispersed in an ultrasonic water bath for 10 min before testing. The size measurement was carried at 25°C with count rates within 300–500 kcps and measured three times. The zeta potentials of Nanotraps were performed by a Möbiuζ system (Wyatt Technology). The data are presented as mean ± SD.
AF488-labeled anti-ACE2 antibody (Santa Cruz Biotechnology) was added to the Nanotrap-ACE2 for 30 min on ice and centrifuged at 5,000 × g for 10 min, and the pellet was washed with PBS three times. The resulting Nanotraps were resuspended in 50% glycol and imaged by TIRFM (Nikon) with 488-nm and 647-nm excitation lasers and 200-ms exposure. Line scans were performed in Fiji.
The sizes and morphologies of the Nanotraps were studied by SEM. In brief, 10 μL of Nanotrap-ACE2 was diluted in MilliQ water and further dispersed in an ultrasonic water bath for 10 min before adding onto a silicon chip. Forty microliters of SARS-CoV-2 spike pseudotyped lentivirus was fixed by 4% paraformaldehyde (PFA) at 37°C for 30 min; the virus was then washed with PBS three times using an Amicon Ultra-15 Centrifugal Filter (pore size 100 kDa) at 3,000 × g for 10 min. The resulting fixed virus was incubated with 10 μL of Nanotrap-ACE2 at 37°C for 1 h and added onto the 1-cm2 silicon chip followed by air-drying overnight. After dehydration, the samples were coated with 8 nm of platinum/palladium by sputter coater (Cressington 208HR). The scanning electron microscope (Carl Zeiss Merlin) was used to image the morphology of the Nanotraps with an accelerating voltage of 2.0 kV. For each sample, more than ten measurements with different magnification were performed to ensure the repeatability of the results. For TEM imaging, 10 μL of the Nanotrap solution was drop-cast on a carbon film-supported TEM grid (Ted Pella, 01843) pre-treated with oxygen plasma. The grid was then gently rinsed with deionized water droplets and stained with 1% uranyl acetate aqueous solution for 1 min. The resultant solution was gently removed with filter paper. The sample was then imaged by an FEI Tecnai G2 F30 electronic microscope at 300 kV after thorough drying.
For long-term storage, the Nanotraps were freeze-dried into solid powder using a lyophilizer (Freezone 6, Labconco). In brief, the Nanotrap solution was transferred into Eppendorf tubes and frozen on dry ice. The lid of the frozen tube was removed and quickly embedded with parafilm; holes were punctured in the parafilm using pipette tips. The tubes were placed in the glass tank connected to a lyophilizer with lids up. The lyophilizer was vacuumed using an oil pump such that any water in the tube was sublimated under −53.3°C for 24 h, resulting in the Nanotrap powder. The tubes with Nanotrap powder were sealed by parafilm and immediately stored in a −20°C freezer. The Nanotrap powder was reconstructed 6 months later by adding PBS solution followed by ultrasonication.
The density of surface ACE2 or neutralizing antibody on the Nanotrap surface was measured by flow cytometry.71 DiD-loaded Nanotrap-ACE2 was stained with anti-ACE2-PE (Sino Biological) for 30 min on ice. DiD-loaded Nanotrap-Antibody was stained with anti-IgG-PE (BioLegend) for 30 min on ice. DiD+PE+ double-positive populations were gated, and PE Quantitation Beads (BD Quantibrite) were used to calculate surface densities according to the manufacturer's instructions.
Flow cytometry analysis of Nanotrap phagocytosis
Macrophage differentiation was conducted as follows.72 THP-1 cells were treated with 150 nM phorbol 12-myristate 13-acetate for 24 h and replaced by fresh culture medium for another 24 h. The differentiated THP-1 cells were harvested as dTHP-1 macrophages and maintained in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. The dTHP-1 macrophages were then released from the plate with Trypsin-EDTA (0.25%) and the cell number was counted by a hemocytometer. One million dTHP-1 macrophages were seeded into each 6-well plate overnight, and DiO-labeled Nanotraps (200, 500, and 1,200 nm) were incubated with dTHP-1 macrophages for 0, 24, or 48 h. For phosphatidylserine investigation, dTHP-1 macrophages were incubated with Nanotraps containing different phosphatidylserine molar ratios (0%, 5%, 10%, 15%) for 0, 2, 4, 6, 24, or 48 h. The cells were harvested and washed three times with PBS at 300 × g for 5 min, and stained with a Zombie NIR Fixable Viability Kit (BioLegend) on ice for 10 min. Cells were then washed with FACS buffer (PBS, 10% FBS, 0.1% NaN3) two times and resuspended in 200 μL of FACS buffer. Flow cytometry was carried out on a BD LSRFortessa Flow Cytometer. Live and single cells were gated (Figure S2C), and the DiO fluorescence channel was used to indicate the phagocytosis efficiency of different Nanotraps. The data were further analyzed by FlowJo (BD) and Prism (GraphPad) software.
Lattice light-sheet microcopy imaging analysis of Nanotrap phagocytosis
For lattice light-sheet imaging, 4 × 104 dTHP-1 macrophages were seeded onto each coverslip and DiD-labeled Nanotraps with different phosphatidylserine molar ratios (0%, 10%, and 15%) were added for 24- or 48-h incubation. The cells were washed with PBS three times, fixed with 4% PFA, stained with 5 μg/mL CF488 Wheat Germ Agglutinin (WGA) Conjugates (Biotium), and washed three times with Hank’s balanced salt solution. Coverslips were imaged by lattice light-sheet microscopy (3i) using z+ objective scanning. Imaging was conducted with 488-nm and 647-nm lasers, with dither set to 3-ms and 20-ms exposures. Z-steps (60) were collected with a 0.4-μm step size. Resulting images were deconvolved as described previously73 , 74 using LLSpy (cudaDeconv) used under license from Howard Hughes Medical Institute, Janelia Research Campus. Image reconstruction videos were made in Imaris (Bitplane).
Confocal microscopy of endosomes and lysosomes
For confocal imaging, 3 × 104 dTHP-1 cells were seeded into 8-well chambers and 500 nm of 15% phosphatidylserine DiD-labeled Nanotraps were added for 6 h at 37°C. The cells were washed with PBS three times, fixed with 4% PFA, and washed three times with PBS. The cells were then stained with 10 μg/mL anti-LAMP-1-AF488 (BioLegend) and 0.5 μg/mL anti-EEA1-AF594 (Abcam) for 30 min, then washed three times with PBS. 4′,6-Diamidino-2-phenylindole DAPI (300 nM) was then added for 10 min and washed three times with PBS. Confocal imaging was conducted on a Leica SP8 with a white-light laser and 100× oil-immersion objective with 1.5× zoom. Z-stacks were acquired with a z-step of 300 nm. Images were z-projected and despeckled in Fiji. Line scan plot profiling was conducted in Fiji.
Kinetics assays
For Nanotrap-virus binding, SARS-CoV-2 spike pseudotyped lentivirus was labeled with lipophilic dye DiO75 for 20 min, fixed with 4% PFA, then washed three times with PBS in an Amicon Ultra-centrifugal tube (100 kDa). DiD-loaded Nanotraps were prepared and incubated with the DiO-labeled pseudotyped lentivirus for various times. DiD+DiO+ double-positive events were gated, and the mean fluorescence intensity of DiO was recorded by flow cytometry until saturation was achieved.
For Nanotrap-macrophage binding, macrophages were prepared as described above and incubated with DiO-loaded Nanotraps for varying times until saturation was achieved. The cells were harvested and washed three times with PBS at 300 × g for 5 min, and stained with a Zombie NIR Fixable Viability Kit (BioLegend) on ice for 10 min. The cells were then washed with FACS buffer two times and resuspended in 200 μL of FACS buffer. Flow cytometry was carried on a BD LSRFortessa Flow Cytometer. Live and single cells were gated, and the DiO fluorescence channel was used to indicate the phagocytosis efficiency at various time points.
Enzyme-linked immunosorbent assay
For the IL-6 enzyme-linked immunosorbent assay (ELISA), six conditions were conducted to test macrophage activation: (1) macrophages alone; (2) macrophages and Nanotraps; (3) macrophages and epithelial cells; (4) macrophages, epithelial cells, and virus; (5) macrophages, epithelial cells, and Nanotraps; and (6) macrophages, epithelial cells, virus, and Nanotraps. For all wells, 1 × 105 dTHP-1 cells were plated into 12-well plates overnight. The next day, to wells containing epithelial cells, 2 × 105 HEK293-ACE2 cells were added. To wells that included Nanotraps, 10 μg/mL Nanotraps was added. To wells that included Nanotraps and virus, 10 μg/mL Nanotraps was added to 500 focus-forming units (FFU) of pseudotyped VSV, incubated for 1 h at 37°C, and added to the cells. Plates were incubated for 24 h and then centrifuged at 300 × g for 5 min, and supernatants were harvested. IL-6 ELISA (BioLegend) was conducted according to the manufacturer's instructions.
Production of SARS-CoV-2 spike pseudotyped VSV
Packaging cells (HEK293T) in serum-free Dulbecco’s modified Eagle’s medium (DMEM) were transfected with 9 μg of pCAGGS SARS-CoV-2 spike expression plasmid using polyethylenimine (PEI). After 24 h, 3 × 107 FFU VSVdG∗G-GFP virus was added to the HEK293T cells and incubated for another 48 h. Media were collected and spun at 500 × g for 3 min to remove cell debris, then passed through a 0.45-μm pore filter. The virus was then stored at −80°C. To check the infection rate, HEK293T-ACE2 cells (Integral, cat. #C-HA102) were incubated with the final VSVdG-GFP∗CoV2 pseudovirus and visualized by the presence of GFP-positive cells through direct microscopic imaging or flow cytometry.
SARS-CoV-2 pseudovirus neutralizing assay
For the SARS-CoV-2 spike pseudotyped VSV neutralizing assay, 4 × 104 HEK293T-ACE2 cells (maintained in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin) were seeded in a 96-well plate overnight. Different concentrations of ACE2 proteins or Nanotraps (containing 0.2, 0.4, 0.6, 0.8, 1.0, 1.5, 2.0, 3.0, 6.0, 9.0, and 12 μg/mL ACE2) were incubated with 500 FFU of SARS-CoV-2 spike pseudotyped VSV for 1 h in 37°C. The virus-Nanotrap solution was added into the HEK293T-ACE2 cells and incubated for 24 h (n = 3 for each group). The cells were imaged with a fluorescence microscope (Nikon) using 10×/0.30 numerical aperture objective. The excitation wavelengths were 470 ± 25 nm (Spectra X, Lumencor). The emissions of GFP were captured by an Andor iXon Ultra 888 back-illuminated EMCCD camera (Oxford Instruments). The number of GFP-positive cells was counted manually by objective three times. The viral infection rates were calculated as the ratio of GFP-positive cells in the group incubated to that of the group incubated with virus alone.
For the SARS-CoV-2 spike pseudotyped lentivirus neutralizing assay, 1 × 104 HEK293T-ACE2 cells were seeded onto a 384-well plate overnight. Nanotraps or ACE2 was added to SARS-CoV-2 pseudovirus (4 μL per well) and incubated for 1 h at 37°C. The virus-Nanotrap solution was added to each well (n = 3 for each group). Seventy-two hours later, the plate was centrifuged for 5 min at 500 × g to prevent cell loss. Supernatant was aspirated and 35 μL of PBS was added. PBS was carefully aspirated, leaving ~15 μL of liquid behind. Renilla-Glo Assay Substrate was added to the assay buffer at a 1:100 dilution, then 15 μL of the substrate/buffer was added to each well of a 384-well plate. Bioluminescence was recorded by a microplate reader (Fisher Scientific BioTek Cytation 5) with an exposure of 200 ms. Wells infected with pseudovirus only were normalized as 100%.
Co-culture assay
THP-1 cells were differentiated into macrophages as described above. Co-culture was carried out in a macrophage to A549 cell ratio of 1:5. A549 cells (4 × 104) were seeded in an 18-well microslide (Vivid) overnight and 8 × 103 dTHP-1 macrophages were added onto the A549 cells for another 6 h. SARS-CoV-2 spike pseudotyped VSV (500 FFU) was incubated with Nanotraps or PBS in 37°C for 1 h before adding to the co-culture cells. Twenty-four hours later, the cells were fixed with 4% PFA and stained with CF532 WGA Conjugates (Biotium) and DAPI, and imaged under a confocal microscope (Leica SP8). Percent infectivity was quantified in Fiji by dividing GFP+ cells by total cell number (DAPI-stained nuclei). Each channel was processed as follows: Image > Threshold (“Huang” preset76); Image > Binary > Fill Holes; Image > Binary > Watershed; Analyze > Count Particles.
In vitro cytotoxicity assay
A549 or HEK293T-ACE2 cells (both maintained in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin) were seeded in a 96-well plate at a density of 1 × 104 cells/well in 100 μL of culture medium overnight. Nanotraps (3.8 × 107 particles/mL) were added into cells, and the cells were cultured in a CO2 incubator at 37°C for 72 h. Ten microliters of CCK-8 (MedChem Express) solution was added to each well of the plate. The plate was incubated for 2 h in the incubator and put into a microplate reader (Fisher Scientific BioTek Cytation 5), and the plate was gently shaken for 1 min before measuring the absorbance at 450 nm. The cytotoxicity was calculated by cell viability, i.e., the relative absorbance from the control wells without Nanotraps was normalized as 100%. The Nanotrap concentrations (particles/mL) were manually counted by a hemocytometer. In brief, the freshly prepared Nanotraps were vortexed and diluted in PBS at 1:1,000 dilution. Ten microliters of the diluted solution was further mixed with 10 μL of trypan blue for enhanced contrast and added onto the hemocytometer, then the number of the Nanotraps was counted under a microscope.
In vivo biosafety assays
C57BL/6NHsd mice at the age of 6 weeks were purchased from Envigo and maintained at the Animal Facility of the University of Chicago. The animal study protocols were approved by the Institutional Animal Care and Use Committee of the University of Chicago. For evaluation of the safety of the Nanotraps, two male and two female 6- to 10-week-old C57BL/6NHsd mice were intratracheally administered with 10 mg/kg Nanotrap-ACE2 in 50 μL of PBS. Blood samples were collected by submandibular vein via cheek punch using a commercially available 4-mm point lancet after 3 days. A small aliquot of approximately 100 μL of blood was collected into EDTA-containing heparinized tubes, and RBCs, WBCs, and platelets were counted by a hematology analyzer (Beckman Coulter Act Diff 5 CP) according to the manufacturer's instructions. For the comprehensive chemistry panels, blood was allowed to coagulate at 4°C for 2 h, and serum was collected after centrifugation (1,000 × g for 15 min) for analysis. Serum alkaline phosphatase, alanine aminotransferase, amylase, urea nitrogen, calcium, cholesterol, glucose, total bilirubin, and total proteins were determined by a Vet Axcel blood chemistry analyzer (Alfa Wasserman). Lungs, heart, liver, spleen, and kidney were collected from the same mice, fixed in 10% formalin for 24 h, and embedded in paraffin. The resulting blocks were cut into 5-μm sections and further stained with H&E by the University of Chicago Human Tissue Research Center. Fluorescent imaging samples were collected in Tissue-Tek OCT Compound on dried ice and stored at −80°C before cryosectioning on a cryostat (Leica). The obtained 10-μm-thick tissue slides were then further stained with DAPI. The histology and fluorescence slides were scanned by a CRi Pannoramic MIDI 20× whole-slide scanner and analyzed using QuPath software.
Ex vivo lung perfusion assay
Non-transplantable human lungs were obtained from deceased individuals provided by the organ procurement organization Gift of Hope. All specimens and data were de-identified prior to receipt. This study was deemed exempt by the University of Chicago Institutional Review Board (IRB19-1942).
Lung harvest
Lungs unsuitable for transplantation were harvested in standard clinical fashion72 from deceased patients. Figure 5A shows the lung of a 56-year-old male patient (87.2 kg, cause of death: brain death). Lungs were transported to the laboratory at 4°C.
Lung inoculation
Tissue samples were collected from the edge of the right upper lobe before perfusion as an untreated control; 500 μL of SARS-CoV-2 spike pseudotyped lentivirus (Integral Molecular, RVP 701) was resuspended in 5 mL of PBS and injected into the lingula of the left lung; 500 μL of SARS-CoV-2 spike pseudotyped lentivirus was first incubated with Nanotrap-Antibody (7.6×106 particles/mL, 2.5 mL) for 1 h at 37°C before inoculation. This mixture was then injected into the right middle lobe. Tissue samples at all three sites were collected. Some samples were immersed in MACS buffer for tissue dissociation, and other samples were fixed in 4% PFA, sliced with thickness of 5 μm, and stained with H&E.
Lung perfusion
The lung bloc was perfused according to published techniques.31 , 77 A centrifugal pump was used to perfuse the pulmonary artery with deoxygenated cellular perfusate (1× DMEM containing 4.5 g/K D-glucose, L-glutamine, and 110 mg/L of sodium pyruvate, with addition of 5% BSA and 2 units of packed RBCs). The left atrium was left open for gravity drainage. The trachea was incubated and the lung was ventilated with volume control ventilation (tidal volume set at 6–8 mL/weight of ideal body weight [kg] of donor, respiratory rate of 8–13, and fraction of inspired oxygen set at 21%). Sweep gas composed of 8% CO2, 3% O2, and 89% N2 was connected to the hollow-fiber deoxygenator heat exchanger to remove oxygen and add CO2 into the perfusate returning back to the lung. After initiation of perfusion with gradual warming and increasing pump flow over 30 min, the lung bloc was maintained at 37°C with pump flow calibrated to pulmonary artery pressure of 10–20 mmHg for 8 h.
Sampling
Perfusate was sampled at serial time points from the pulmonary artery and left atrium. Differences in oxygen content between perfusate samples were used to calculate the oxygenation capacity of the lung. Airway pressure was measured periodically to calculate lung compliance based on tidal volume. Tissue samples were collected from treated and untreated lobes of the lung at time 0 and time 8 h of perfusion.
Sample processing
After 8 h of perfusion, tissues were harvested as described above. Tissue dissociation was performed by mechanical digestion in DMEM treated with 2.5 U/mL DNase; samples were passed through 70-μm cell strainers (Fisher Scientific). RBCs were lysed in 10 mL of RBC lysis buffer (Life Technologies), washed three times with DMEM (300 × g for 3 min), and resuspended in 10 mL of DMEM. Cells were counted with a hemocytometer and immediately used for luciferase assay or cryopreserved in Cell Banker medium (Amsbio) at a density of 4 × 106 cells/mL.
Samples that were not dissociated were fixed in 4% PFA and sliced with thickness of 5 μm. Some samples were stained for H&E and imaged on a CRi Pannoramic MIDI 20× whole-slide scanner. Tissue slides used for RBC quantification were stained for DAPI and imaged on a CRi Pannoramic MIDI 20× whole-slide scanner with 488-nm and 560-nm lasers. Both channels were quantified in Fiji as follows: Image > Threshold (“Otsu” preset78); Image > Binary > Fill Holes; Analyze > Measure.
For luciferase assay, the cells harvested as described above were washed with PBS (300 × g for 3 min) three times, after which 2 × 104 cells were seeded onto a 96-well plate. Renilla-Glo Assay Substrate was added to the assay buffer at a 1:100 dilution, then 30 μL of the substrate buffer was added to each well for 10 min. The bioluminescence from each well was detected by a microplate reader (Fisher Scientific BioTek Cytation 5) with an exposure of 200 ms.
Primary cells harvested from the untreated right superior lobe of the human lung were further used for infection analyses. In brief, 2 × 104 primary lung cells were seeded onto a 384-well plate. Twenty-five microliters of Nanotraps-Antibody (6.08 × 107 particles/mL) was added to 20 μL of SARS-CoV-2 spike pseudotyped lentivirus (Integral Molecular, RVP 701) per well and incubated for 1 h at 37°C before adding to the cells. For the virus-only group, 10 μL of SARS-CoV-2 spike pseudotyped lentivirus was added to the cells. Forty-eight hours later, the plate was centrifuged for 5 min at 500 × g to prevent cell loss. Supernatant was aspirated and 35 μL of PBS was added. PBS was carefully aspirated, leaving ~15 μL of liquid behind. Renilla-Glo Assay Substrate was added to the assay buffer at a 1:100 dilution, then 15 μL of the substrate/buffer was added to each well of a 384-well plate. The bioluminescence was recorded by a microplate reader (Fisher Scientific BioTek Cytation 5) with an exposure of 200 ms. The infectivity was calculated by the relative luminescence intensity: wells infected with SARS-CoV-2 pseudovirus only were normalized as 100%.
Authentic SARS-CoV-2 neutralizing assay
All SARS-CoV-2 infections were performed in biosafety level 3 conditions at the University of Chicago Howard T. Ricketts Regional Biocontainment Laboratory. African green monkey kidney (Vero E6) cells were maintained in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin. Nanotraps or neutralizing antibodies were serially diluted 2-fold and mixed with 400 plaque-forming units of SARS-CoV-2 (nCoV/Washington/1/2020, kindly provided by the National Biocontainment Laboratory, Galveston, Texas) for 1 h at 37°C, then used to infect Vero E6 cells for 3 days. Cells were fixed with 3.7% formalin and stained with 0.25% crystal violet. Crystal violet-stained cells were then quantified by absorbance at 595 nm with a Tecan m200 microplate reader. Cell survival was calculated by normalizing untreated cells to 100%.
Acknowledgments
This work was supported by NIH New Innovator award 1DP2AI144245 (J.H.), NSF Career award 1653782 (J.H.), and NIDDK RC2DK122394 (E.C.). M.C. is partially supported by the UChicago Big Ideas Generator Grant. J.R. is supported by the NSF Graduate Research Fellowships Program DGE-1746045. We would like to thank Dr. Nicholas Ankenbruck, Yifei Hu, Guoshuai Cao, Ruyi Wang, Joe Reda, and Aaron Alpar for their experimental guidance. P.P.M. is supported by DP2DA051912. We would also like to thank Dr. Aaron Esser-Kahn for kindly providing THP-1 cells, Dr. Jeffery Hubbell for the use of laboratory equipment, and Dr. Shann Yu for kindly ordering C57BL/6NHsd mice during the pandemic. We also acknowledge the support from the University of Chicago Human Tissue Research Center, University of Chicago Integrated Light Microscopy Core, and the Soft Matter Characterization Facility of the Pritzker School of Molecular Engineering for sample processing and characterization.
Author contributions
Conceptualization, M.C. with input from J.H.; methodology, M.C. with input from J.H.; investigation/experiments, M.C., J.R., X.C., A.C.H.L., J.S., M.N., T.W., V.M., A.J.E., J.F., J.S.D., K.S., Y.L., G.R., B.T., and M.L.M.; SARS-CoV-2 pseudotyped VSV generation and discussions, P.P.-M.; formal analysis, M.C. and J.R. with input from J.H.; manuscript writing, M.C., J.H., and J.R.; funding acquisition, J.H. and E.C.; supervision, J.H.; manuscript editing and review, all authors.
Declaration of interests
The authors have filed a provisional patent application around the technology described in this article.
Inclusion and diversity
We worked to ensure sex balance in the selection of non-human subjects. We worked to ensure diversity in experimental samples through the selection of the cell lines. One or more of the authors of this paper received support from a program designed to increase minority representation in science. The author list of this paper includes contributors from the location where the research was conducted who participated in the data collection, design, analysis, and/or interpretation of the work.
Published: April 22, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.matt.2021.04.005.
Supplemental information
References
- 1.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Florindo H.F., Kleiner R., Vaskovich-Koubi D., Acúrcio R.C., Carreira B., Yeini E., Tiram G., Liubomirski Y., Satchi-Fainaro R. Immune-mediated approaches against COVID-19. Nat. Nanotechnol. 2020;15:630–645. doi: 10.1038/s41565-020-0732-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shin M.D., Shukla S., Chung Y.H., Beiss V., Chan S.K., Ortega-Rivera O.A., Wirth D.M., Chen A., Sack M., Pokorski J.K., et al. COVID-19 vaccine development and a potential nanomaterial path forward. Nat. Nanotechnol. 2020;15:646–655. doi: 10.1038/s41565-020-0737-y. [DOI] [PubMed] [Google Scholar]
- 4.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vabret N., Britton G.J., Gruber C., Hegde S., Kim J., Kuksin M., Levantovsky R., Malle L., Moreira A., Park M.D., et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shang J., Ye G., Shi K., Wan Y., Luo C., Aihara H., Geng Q., Auerbach A., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U. S. A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lei C., Qian K., Li T., Zhang S., Fu W., Ding M., Hu S. Neutralization of SARS-CoV-2 spike pseudotyped virus by recombinant ACE2-Ig. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-16048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monteil V., Kwon H., Prado P., Hagelkrüys A., Wimmer R.A., Stahl M., Leopoldi A., Garreta E., Hurtado del Pozo C., Prosper F., et al. Inhibition of SARS-CoV-2 infections in engineered human tissues using clinical-grade soluble human ACE2. Cell. 2020;181:905–913. doi: 10.1016/j.cell.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J., et al. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- 13.Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X., Li R., Pan Z., Qian C., Yang Y., You R., Zhao J., Liu P., Gao L., Li Z., et al. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell. Mol. Immunol. 2020;17:647–649. doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with covid-19. N. Engl. J. Med. 2020;384:229–237. doi: 10.1056/nejmoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen W., Cai B., Geng Z., Chen F., Wang Z., Wang L., Chen X. Reducing false negatives in COVID-19 testing by using microneedle-based oropharyngeal swabs. Matter. 2020;3:1589–1600. doi: 10.1016/j.matt.2020.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torrente-Rodríguez R.M., Lukas H., Tu J., Min J., Yang Y., Xu C., Rossiter H.B., Gao W. SARS-CoV-2 rapidPlex: a graphene-based multiplexed telemedicine platform for rapid and low-cost COVID-19 diagnosis and monitoring. Matter. 2020;3:1981–1998. doi: 10.1016/j.matt.2020.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin Z., Peng R., Baravik I.K., Liu X. Fighting COVID-19: Integrated micro- and nanosystems for viral infection diagnostics. Matter. 2020;3:628–651. doi: 10.1016/j.matt.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai X., Prominski A., Lin Y., Ankenbruck N., Rosenberg J., Chen M., Shi J., Chang E.B., Penaloza-MacMaster P., Tian B., et al. A neutralizing antibody-conjugated photothermal nanoparticle captures and inactivates SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.11.30.404624. [DOI] [Google Scholar]
- 20.Rao L., Xia S., Xu W., Tian R., Yu G., Gu C., Pan P., Meng Q.F., Cai X., Qu D., et al. Decoy nanoparticles protect against COVID-19 by concurrently adsorbing viruses and inflammatory cytokines. Proc. Natl. Acad. Sci. U. S. A. 2020;117:27141–27147. doi: 10.1073/pnas.2014352117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Q., Honko A., Zhou J., Gong H., Downs S.N., Vasquez J.H., Fang R.H., Gao W., Griffiths A., Zhang L. Cellular nanosponges inhibit SARS-CoV-2 infectivity. Nano Lett. 2020;20:5570–5574. doi: 10.1021/acs.nanolett.0c02278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daassi D., Mahoney K.M., Freeman G.J. The importance of exosomal PDL1 in tumour immune evasion. Nat. Rev. Immunol. 2020;20:209–215. doi: 10.1038/s41577-019-0264-y. [DOI] [PubMed] [Google Scholar]
- 23.Gordon S. Phagocytosis: an immunobiologic process. Immunity. 2016;44:463–475. doi: 10.1016/j.immuni.2016.02.026. [DOI] [PubMed] [Google Scholar]
- 24.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 25.Fadok V.A., Bratton D.L., Frasch S.C., Warner M.L., Henson P.M. The role of phosphatidylserine in recognition of apoptotic cells by phagocytes. Cell Death Differ. 1998;5:551–562. doi: 10.1038/sj.cdd.4400404. [DOI] [PubMed] [Google Scholar]
- 26.Wu Y., Tibrewal N., Birge R.B. Phosphatidylserine recognition by phagocytes: a view to a kill. Trends Cell Biol. 2006;16:189–197. doi: 10.1016/j.tcb.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 27.Aderem A., Underhill D.M. Mechanisms of phagocytosis in macrophages. Annu. Rev. Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 28.Johnstone S.A., Masin D., Mayer L., Bally M.B. Surface-associated serum proteins inhibit the uptake of phosphatidylserine and poly(ethylene glycol) liposomes by mouse macrophages. Biochim. Biophys. Acta - Biomembr. 2001;1513:25–37. doi: 10.1016/S0005-2736(01)00292-9. [DOI] [PubMed] [Google Scholar]
- 29.Shah N.K., Gupta S.K., Wang Z., Meenach S.A. Enhancement of macrophage uptake via phosphatidylserine-coated acetylated dextran nanoparticles. J. Drug Deliv. Sci. Technol. 2019;50:57–65. doi: 10.1016/j.jddst.2019.01.013. [DOI] [Google Scholar]
- 30.Divithotawela C., Cypel M., Martinu T., Singer L.G., Binnie M., Chow C.W., Chaparro C., Waddell T.K., De Perrot M., Pierre A., et al. Long-term outcomes of lung transplant with ex vivo lung perfusion. JAMA Surg. 2019;154:1143–1150. doi: 10.1001/jamasurg.2019.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cypel M., Yeung J.C., Hirayama S., Rubacha M., Fischer S., Anraku M., Sato M., Harwood S., Pierre A., Waddell T.K., et al. Technique for prolonged normothermic ex vivo lung perfusion. J. Hear. Lung Transpl. 2008;27:1319–1325. doi: 10.1016/j.healun.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Wang Q., Zhang Y., Wu L., Niu S., Song C., Zhang Z., Lu G., Qiao C., Hu Y., Yuen K.Y., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang S.S., Lim J., Yu Z., Kong P., Sefik E., Xu H., Harman C.C.D., Kim L.K., Lee G.R., Li H.B., et al. MRNA destabilization by BTG1 and BTG2 maintains T cell quiescence. Science. 2020;367:1255–1260. doi: 10.1126/science.abb2507. [DOI] [PubMed] [Google Scholar]
- 34.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 35.Torchilin V.P. Recent advances with liposomes as pharmaceutical carriers. Nat. Rev. Drug Discov. 2005;4:145–160. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- 36.Riley R.S., June C.H., Langer R., Mitchell M.J. Delivery technologies for cancer immunotherapy. Nat. Rev. Drug Discov. 2019;18:175–196. doi: 10.1038/s41573-018-0006-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Allen T.M., Cullis P.R. Liposomal drug delivery systems: from concept to clinical applications. Adv. Drug Deliv. Rev. 2013;65:36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 38.Torchilin V.P. Multifunctional, stimuli-sensitive nanoparticulate systems for drug delivery. Nat. Rev. Drug Discov. 2014;13:813–827. doi: 10.1038/nrd4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suk J.S., Xu Q., Kim N., Hanes J., Ensign L.M. PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliv. Rev. 2016;99:28–51. doi: 10.1016/j.addr.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He C., Hu Y., Yin L., Tang C., Yin C. Effects of particle size and surface charge on cellular uptake and biodistribution of polymeric nanoparticles. Biomaterials. 2010;31:3657–3666. doi: 10.1016/j.biomaterials.2010.01.065. [DOI] [PubMed] [Google Scholar]
- 41.Champion J.A., Walker A., Mitragotri S. Role of particle size in phagocytosis of polymeric microspheres. Pharm. Res. 2008;25:1815–1821. doi: 10.1007/s11095-008-9562-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doane T.L., Chuang C.H., Hill R.J., Burda C. Nanoparticle ζ-potentials. Acc. Chem. Res. 2012;45:317–326. doi: 10.1021/ar200113c. [DOI] [PubMed] [Google Scholar]
- 43.Howarth M., Chinnapen D.J.F., Gerrow K., Dorrestein P.C., Grandy M.R., Kelleher N.L., El-Husseini A., Ting A.Y. A monovalent streptavidin with a single femtomolar biotin binding site. Nat. Methods. 2006;3:267–273. doi: 10.1038/nmeth861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toita R., Kawano T., Murata M., Kang J.H. Anti-obesity and anti-inflammatory effects of macrophage-targeted interleukin-10-conjugated liposomes in obese mice. Biomaterials. 2016;110:81–88. doi: 10.1016/j.biomaterials.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 45.Harel-Adar T., Mordechai T. Ben, Amsalem Y., Feinberg M.S., Leor J., Cohen S. Modulation of cardiac macrophages by phosphatidylserine-presenting liposomes improves infarct repair. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1827–1832. doi: 10.1073/pnas.1015623108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han C.Z., Juncadella I.J., Kinchen J.M., Buckley M.W., Klibanov A.L., Dryden K., Onengut-Gumuscu S., Erdbrügger U., Turner S.D., Shim Y.M., et al. Macrophages redirect phagocytosis by non-professional phagocytes and influence inflammation. Nature. 2016;539:570–574. doi: 10.1038/nature20141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lund M.E., To J., O’Brien B.A., Donnelly S. The choice of phorbol 12-myristate 13-acetate differentiation protocol influences the response of THP-1 macrophages to a pro-inflammatory stimulus. J. Immunol. Methods. 2016;430:64–70. doi: 10.1016/j.jim.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 48.Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Herbert A.S., Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369:1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsuyama S., Nao N., Shirato K., Kawase M., Saito S., Takayama I., Nagata N., Sekizuka T., Katoh H., Kato F., et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc. Natl. Acad. Sci. U. S. A. 2020;117:7001–7003. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., et al. Remdesivir for the treatment of Covid-19—final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/nejmoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grein J., Ohmagari N., Shin D., Diaz G., Asperges E., Castagna A., Feldt T., Green G., Green M.L., Lescure F.-X., et al. Compassionate use of remdesivir for patients with severe Covid-19. N. Engl. J. Med. 2020;382:2327–2336. doi: 10.1056/nejmoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen C., Wang Z., Zhao F., Yang Y., Li J., Yuan J., Wang F., Li D., Yang M., Xing L., et al. Treatment of 5 critically Ill patients with COVID-19 with convalescent plasma. J.A.M.A. 2020;323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pérez-Cameo C., Marín-Lahoz J. Serosurveys and convalescent plasma in COVID-19. EClinicalMedicine. 2020;23 doi: 10.1016/j.eclinm.2020.100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Q., Honko A., Zhou J., Gong H., Downs S.N., Vasquez J.H., Fang R.H., Gao W., Griffiths A., Zhang L. Cellular nanosponges inhibit SARS-CoV-2 infectivity. Nano Lett. 2020;20:5570–5574. doi: 10.1021/acs.nanolett.0c02278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fiorenza S., Ritchie D.S., Ramsey S.D., Turtle C.J., Roth J.A. Value and affordability of CAR T-cell therapy in the United States. Bone Marrow Transplantat. 2020;55:1706–1715. doi: 10.1038/s41409-020-0956-8. [DOI] [PubMed] [Google Scholar]
- 57.Yu M., Wu J., Shi J., Farokhzad O.C. Nanotechnology for protein delivery: overview and perspectives. J. Control Release. 2016;240:24–37. doi: 10.1016/j.jconrel.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen G., Huang A.C., Zhang W., Zhang G., Wu M., Xu W., Yu Z., Yang J., Wang B., Sun H., et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature. 2018;560:382–386. doi: 10.1038/s41586-018-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mukherjee A., Waters A.K., Kalyan P., Achrol A.S., Kesari S., Yenugonda V.M. Lipid-polymer hybrid nanoparticles as a next-generation drug delivery platform: state of the art, emerging technologies, and perspectives. Int. J. Nanomedcine. 2019;14:1937–1952. doi: 10.2147/IJN.S198353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bournazos S., Gupta A., Ravetch J.V. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 2020;20:633–643. doi: 10.1038/s41577-020-00410-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee W.S., Wheatley A.K., Kent S.J., DeKosky B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020;5:1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PubMed] [Google Scholar]
- 62.Janeway C., Travers P., Walport M., Shlomchik M.J. Garland Science; 2008. Janeway’s Immunobiology. [Google Scholar]
- 63.Klichinsky M., Ruella M., Shestova O., Lu X.M., Best A., Zeeman M., Schmierer M., Gabrusiewicz K., Anderson N.R., Petty N.E., et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat. Biotechnol. 2020;38:947–953. doi: 10.1038/s41587-020-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suzuki T., Itoh Y., Sakai Y., Saito A., Okuzaki D., Motooka D., Minami S., Kobayashi T., Yamamoto T., Okamoto T., et al. Generation of human bronchial organoids for SARS-CoV-2 research. bioRxiv. 2020 doi: 10.1101/2020.05.25.115600. [DOI] [Google Scholar]
- 65.Han Y., Yang L., Duan X., Duan F., Nilsson-Payant B., Yaron T., Wang P., Tang X., Zhang T., Zhao Z., et al. Identification of candidate COVID-19 therapeutics using hPSC-derived Lung organoids. bioRxiv. 2020 doi: 10.1101/2020.05.05.079095. [DOI] [Google Scholar]
- 66.Chandrashekar A., Liu J., Martino A.J., McMahan K., Mercad N.B., Peter L., Tostanosk L.H., Yu J., Maliga Z., Nekorchuk M., et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science. 2020;369:812–817. doi: 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu J., Tostanosk L.H., Peter L., Mercad N.B., McMahan K., Mahrokhia S.H., Nkolol J.P., Liu J., Li Z., Chandrashekar A., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rockx B., Kuiken T., Herfst S., Bestebroer T., Lamers M.M., Munnink B.B.O., De Meulder D., Van Amerongen G., Van Den Brand J., Okba N.M.A., et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science. 2020;368:1012–1015. doi: 10.1126/science.abb7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li S., Gou T., Wang Q., Chen M., Chen Z., Xu M., Wang Y., Han D., Cao R., Liu J., et al. Ultrasound/optical dual-modality imaging for evaluation of vulnerable atherosclerotic plaques with osteopontin targeted nanoparticles. Macromol. Biosci. 2020;20:1900279. doi: 10.1002/mabi.201900279. [DOI] [PubMed] [Google Scholar]
- 70.Mattrey R.F., Brown J.J., Shelton R.E., Ogino M.T., Johnson K.K., Mitten R.M. Use of perfluorooctylbromide (PFOB) to detect liver abscesses with computed tomography: safety and efficacy. Invest. Radiol. 1991;26:792–798. doi: 10.1097/00004424-199109000-00003. [DOI] [PubMed] [Google Scholar]
- 71.Huang J., Edwards L.J., Evavold B.D., Zhu C. Kinetics of MHC-CD8 interaction at the T cell membrane. J. Immunol. 2007;179:7653–7662. doi: 10.4049/jimmunol.179.11.7653. [DOI] [PubMed] [Google Scholar]
- 72.Hsu H.Y., Nicholson A.C., Hajjar D.P. Inhibition of macrophage scavenger receptor activity by tumor necrosis factor-α is transcriptionally and post-transcriptionally regulated. J. Biol. Chem. 1996;271:7767–7773. doi: 10.1074/jbc.271.13.7767. [DOI] [PubMed] [Google Scholar]
- 73.Rosenberg J., Huang J. Visualizing surface T-cell receptor dynamics four-dimensionally using lattice light-sheet microscopy. J. Vis. Exp. 2020:e59914. doi: 10.3791/59914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rosenberg J., Cao G., Borja-Prieto F., Huang J. Lattice light-sheet microscopy multi-dimensional analyses (LaMDA) of T-cell receptor dynamics predict T-cell signaling states. Cell Syst. 2020;10:433–444. doi: 10.1016/j.cels.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu S.L., Wang Z.G., Xie H.Y., Liu A.A., Lamb D.C., Pang D.W. Single-virus tracking: from imaging methodologies to virological applications. Chem. Rev. 2020;120:1936–1979. doi: 10.1021/acs.chemrev.9b00692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang L.-K., Wang M.-J.J. Image thresholding by minimizing the measures of fuzziness. Pattern Recognit. 1995;28:41–51. doi: 10.1016/0031-3203(94)E0043-K. [DOI] [Google Scholar]
- 77.Ross J.T., Nesseler N., Lee J.W., Ware L.B., Matthay M.A. The ex vivo human lung: research value for translational science. JCI Insight. 2019;4:e128833. doi: 10.1172/jci.insight.128833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Otsu N. A Threshold Selection Method from Gray-Level Histograms. IEEE transactions on systems, man, and cybernetics. 1979;9:62–66. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Three-dimensional image reconstruction of macrophages (WGA-CF488, green) phagocytosing Nanotraps (DiD, magenta). Video shows macrophages treated with 10% phosphatidylserine Nanotraps for 24 h and untreated macrophages.
Video shows lungs from Figure 5 attached to the EVLP ventilator system.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.