Abstract
Gastric cancer is one of the leading causes of cancer death worldwide. Previous studies demonstrated that activation of STAT3 is crucial for the development and progression of gastric cancer. However, the role of STAT3 in neuronal related gene methylation in gastric cancer has never been explored. In this study, by using DNA methylation microarray, we identified a potential STAT3 target, C11orf87, showing promoter hypomethylation in gastric cancer patients with lower STAT3 activation and AGS gastric cancer cell lines depleted with STAT3 activation. Although C11orf87 methylation is independent of its expression, ectopic expression of a constitutive activated STAT3 mutant upregulated its expression in gastric cancer cell line. Further bisulfite pyrosequencing demonstrated a progressive increase in DNA methylation of this target in patient tissues from gastritis, intestinal metaplasia, to gastric cancer. Intriguingly, patients with higher C11orf87 methylation was associated with better survival. Furthermore, hypermethylation of C11orf87 was also frequently observed in other GI cancers, as compared to their adjacent normal tissues. These results suggested that C11orf87 methylation may serve as a biomarker for diagnosis and prognosis of GI cancers, including gastric cancer. We further postulated that constitutive activation of STAT3 might be able to epigenetically silence C11orf87 as a possible negative feedback mechanism to protect the cells from the overactivation of STAT3. Targeted inhibition of STAT3 may not be appropriate in gastric cancer patients with promoter hypermethylation of C11orf87.
Introduction
Gastric cancer is the third leading cause of cancer death worldwide [1]. Infection with Helicobacter pylori (H. pylori), a gram-negative bacteria, is an important risk factor for gastric cancer [2]. Particularly, cytotoxin-associated gene A (CagA) positive H. pylori, which induces inflammation and activation of JAK/STAT signaling, have a higher risk of developing gastric cancer [3, 4]. Previous studies also confirmed that STAT3 activation is decisive for the initiation, and progression in gastric cancer patients [5–7]. Several studies, including ours, demonstrated that STAT3 activation may confer aberrant epigenetic modifications in gastric epithelial cells [8–12]. However, the clinical significance of these modifications is not fully explored.
Abnormal DNA methylation is being considered as a hallmark of cancers because of a crucial mechanism in regulating transcription [13]. In such, aberrant promoter DNA methylation patterns in human cancers may be linked to the specific signaling pathways that are dysregulated in response to unique carcinogens exposed to specific tumor types [14]. Additionally, DNA methylation alteration has related to cancer development [15, 16].
We have previously demonstrated methylation of multiple STAT3 targets in gastric cancer patients [8–10]. In this study, we further identified a putative STAT3 target, C11orf87, showing differential hypomethylation in gastric cancer cell lines and patient samples with lower STAT3 activation status. Differential C11orf87 methylation was also observed in gastritis, intestinal metaplasia (IM), and gastric cancer patient samples. Interestingly, our results suggested that C11orf87 methylation can be a biomarker for early detection and disease prognosis in gastric cancer.
Materials and methods
Patient samples
Patient samples were collected from Changhua Christian Hospital (CCH), Taiwan or the Medical School of Zhejiang University, Hangzhou, China from March 2013 to February 2016, including 65 samples from tumor tissues and 51 patients with matched adjacent normal tissues, 27 samples from intestinal metaplasia tissues and 11 gastritis samples. The clinical-pathological data for the tissue samples are summarized in Table 1. All human studies were approved by the Institutional Review Board of the Changhua Christian Hospital, Taiwan, and the ethics committee of Zhejiang University, Hangzhou, China. The study was carried out in strict accordance with approved guidelines. Written informed consent was obtained from all participants.
Table 1. Summary of clinic-pathological data of patient samples.
| Tumor (n = 62) | IM (n = 27) | Gastritis (n = 8) | |
|---|---|---|---|
| Age | |||
| Median | 72.6438 | 52 | 39 |
| Range | 36.7–88.7 | 33–77 | 25–73 |
| Gender | |||
| Female | ND | 18 | 5 |
| Male | ND | 9 | 3 |
| HP infected status | |||
| negative | 44 | 10 | 7 |
| positive | 2 | 17 | 1 |
| ND | 16 | ||
| Histological Grade | |||
| Low (1,2) | 23 | ||
| High (3) | 39 | ||
| Pathological Stage | |||
| Low (I, II) | 21 | ||
| High (III, IV) | 41 | ||
| Relapse | |||
| Primary | 40 | ||
| Recurrence | 22 |
ND: data not available.
Cell culture
Gastric cancer cell line (AGS, KATO III, MKN28, MKN45, SNU1, NCI-N87, purchased from ATCC, Manassas, VA) and an immortalized gastric epithelial cell line, GES (a kind gift from Dr. Jun Yu, The Chinese University of Hong Kong, Hong Kong) were maintained in RPMI 1640 (Gibco, Waltham, MA) supplemented with 10% FBS (Gibco) and 1% Penicillin-Streptomycin (Gibco). All cells were maintained at 37°C, with 5% CO2, under a humidified incubator. Cells were treated with STAT3 inhibitor, JSI-124 (Sigma, St. Louis, MO) for 2 days and harvested for RNA extraction.
Plasmid constructs and transfection
MKN28 cells were transiently transfected with empty vector (pcDNA3.1) or vector overexpressing a constitutively activated STAT3 mutant (STAT3c, a gift from James Darnell, Rockefeller University, NY) as previously described [9].
DNA extraction
DNA was extracted using a Genomic DNA Mini Kit (Geneaid, Taiwan), according to the manufacturer’s instructions. DNA was then eluted in 50μl distilled water and stored at -20°C until use.
RNA extraction and reverse-transcription PCR (RT-PCR)
Total RNA from GC cell lines or treated cell lines were extracted by Trizol (Invitrogen, Carlsbad, CA), according to the manufacturer’s protocol. 1 μg of total RNA was pre-treated with DNase I (Invitrogen), and followed with cDNA synthesis by using EpiScript™ reverse transcriptase (Lucigen, Madison, WI). Reverse-transcription PCR (qRT-PCR) was performed using Platinum™ Taq DNA Polymerase (Invitrogen). Primers used to amplify C11orf87 and ACTB cDNA can be found in Table 2.
Table 2. Primer sequences used in this study.
| Sequence 5’-3’ | |
|---|---|
| C11orf87 | For bisulfite pyrosequencing |
| Forward | GGTTGTTTTATTTGTTGAGTAATTTGTATT |
| Reverse | ggtcgtcagactgtcgatgaagccACCCTCTAAACACACTACCTAAACTA |
| Sequencing | AGGTTGTTGGTGGTT |
| UB04 universal primer | Ggtcgtcagactgtcgatgaagcc |
| C11orf87 | For RT-PCR |
| Forward | TCTGGTCCATTCACTCCACG |
| Reverse | TGCAGAGGCACCAGGCT |
| ACTB | For RT-PCR |
| Forward | TGCGTGACATTAAGGAGAAG |
| Reverse | GCTCGTAGCTCTTCTCCA |
Infinium methylation microarray analysis
Bisulfite-modified DNA from three-paired of gastric cancer patient samples with high (n = 3) or low (n = 3) STAT3 activation, based on STAT3 IHC score, as well as AGS gastric cancer cell lines and subline depleted of STAT3 [8] were subjected to Illumina 850K methylation microarray analysis (Health GeneTech Corp, Taiwan). The methylation level of each probe (β-value) was defined by the intensity of the methylated allele (M) / (intensity of the unmethylated allele (U) + the intensity of the methylated allele (M) + 100). The microarray data has been deposited in the Gene Expression Omnibus database (accession number: GSE109541).
Bisulfite modification and bisulfite pyrosequencing
1μg of genomic DNA was bisulfite-modified using EZ DNA Methylation Kit (ZYMO research, Orange, CA). The bisulfite modified DNA was subjected to PCR amplification using a biotin-labeled universal primer added as a tailed reverse primer. PCR and sequencing primers were designed using PyroMark Assay Design 2.0 software (Qiagen GmbH, Hilden, Germany). The specific primers were shown in Table 2 and were used with Invitrogen PlatinumTM DNA Polymerase (Invitrogen) in a 25μL reaction for PCR amplification. Before pyrosequencing, 2μL of each PCR reaction was analyzed on 1.5% agarose gel. Pyrosequencing was performed on the PyroMark Q24 (Qiagen) using Pyro Gold Reagent (Qiagen) following the manufacturer’s protocol. The methylation level of specific CpG sites was measured. The methylation percentage of each cytosine was determined by dividing the fluorescence intensity of cytokines with the sum of the fluorescence intensity for cytosines and thymines at each CpG site. In vitro methylated DNA (IVD, ZYMO research) and ultrapure distilled water (Invitrogen) was included as a positive and negative control for pyrosequencing, respectively.
Statistical analysis
A comparison of non-parametric variables was assessed by the Mann-Whitney test. The comparison of cancer and adjacent normal was assessed by paired t-test. DNA methylation levels were performed for receiver operating characteristic curve by R package (pROC) under R 3.5.1. The cut-off for survival analyses was defined by ROC curve. Recurrence-free survival (RFS) is defined as the date of surgery to the date of recurrence or last follow-up date, while overall survival (OS) is defined as the date of surgery to the date of death or the last follow-up date. Both RFS and OS were assessed by Kaplan-Meier analysis. All statistical analysis was performed by GraphPad Prism version 5.0 for windows (GraphPad Software, La Jolla, CA, USA). P < 0.05 was considered as significant.
Results
Identification of differential methylated STAT3 target
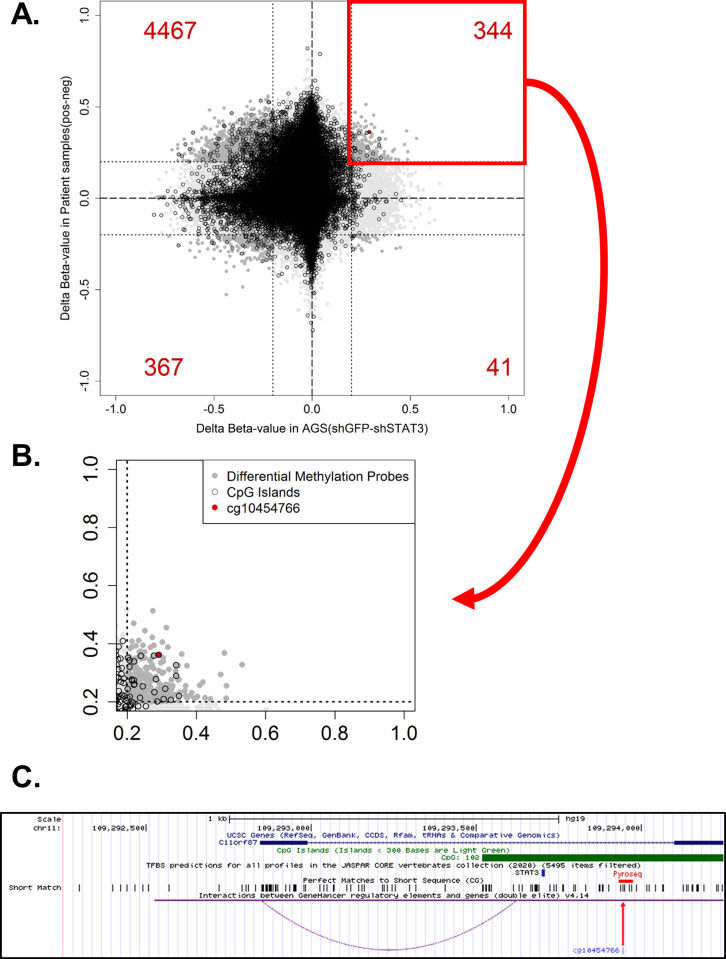
We have previously compared the methylation profile between constitutive STAT3-activated AGS gastric cancer cells, and the subline depleted of STAT3 using Illumina 850K methylation microarray [8, 9]. Additionally, in this study, we compared the methylation profile of three pairs of gastric cancer patients, based on the STAT3 activation status. Integrative computational analyses identified several putative STAT3 targets showing promoter hypomethylation in STAT3-depleted AGS cells and gastric cancer patients showing lower STAT3 activation (as determined by STAT3 nuclear translocation). We further selected probes located within promoter CpG island, showing lower methylation in normal samples but higher methylation in tumor samples in the TCGA gastric cancer dataset. Probes with more than 20% methylation changes in both cell line and patient samples would be defined as significant differential methylation (Fig 1A). One of the probes, cg10454766, showed a significant hypomethylation in both STAT3-depleted AGS cells and patients with lower STAT3 activation (Fig 1B). Further analysis identified a putative STAT3 binding which is in close proximity to cg10454766 (242bp, Fig 1C, blue arrow), and may link to the transcription start site of C11orf87, as demonstrated by GeneHancer (Fig 1C, purple dash line). This probe, cg10454766, was then selected for further analysis.
Fig 1. Identification of cg10454766 as a STAT3-mediated hypermethylation loci.
(A) Scatter plot showing differential methylation differences in gastric cancer cell lines and patient samples. AGS gastric cancer cell lines (shGFP vs shSTAT3) and gastric cancer patients (STAT3 positive vs STAT3 negative) were subjected to Illumina 850K methylation microarray. The significant differences were defined by 20% methylation changes (0.2 delta beta-value). The amount of significant differential probes showed in red numbers for each quadrat. (B) The selected quadrat denoted STAT3-related hypermethylation. Differential methylation probes showed in dark gray. The probes which reside within CpG island were showed in black circle. The selected probe, cg10454766, with most significant difference was shown in red dot. (C) Genomic landscape of cg10454766 (C11orf87) obtained from UCSC Genome browser. The predictive STAT3 binding site were performed by JASPAR2020, as shown in blue bar. The corresponding CG dinucleotides are shown in short match. The region for bisulfite pyrosequencing validation is showed in red bar, including the selected probe, cg10454766 (red arrow). Genomic interactions were performed by GeneHancer, as shown in purple curve.
The expression and methylation of C11orf87
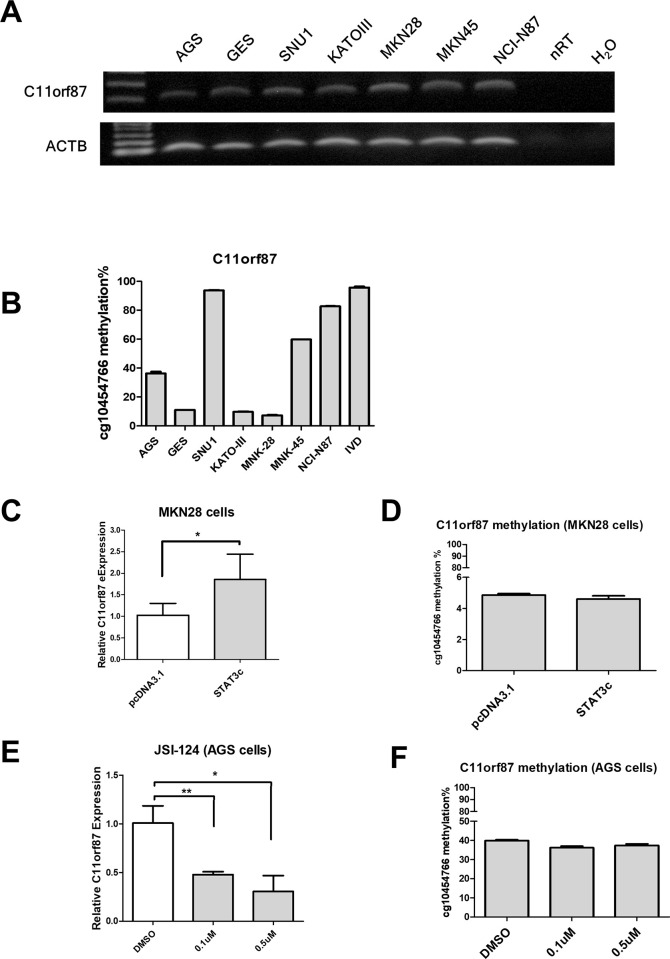
To examine the relationship between expression and methylation on C11orf87, we performed bisulfite pyrosequencing and RT-PCR in GC cell lines and an immortalized normal cell, GES. The results showed that those cell lines exhibited various C11orf87 expression (Fig 2A). Unexpectedly, further bisulfite pyrosequencing also showed various C11orf87 methylation without correlation with its expression (Fig 2B). These results suggested that C11orf87 expression is independent of its methylation.
Fig 2. The expression and methylation of C11orf87 in gastric cancer cell lines.
(A) Expression and (B) methylation of C11orf87 was determined by RT-PCR and bisulfite pyrosequencing, respectively in GC cell lines and an immortalized gastric epithelial cell lines, GES. MKN28 cells were transiently transfected with empty vector (pcDNA3.1) or vector expressing constitutive activated STAT3 mutant (STAT3c). C11orf87 expression and methylation were determined by (C) qRT-PCR and (D) bisulfite pyrosequencing, respectively. AGS gastric cancer cells were treated with DMSO or STAT3 inhibitor (JSI-124) for two days. Expression and methylation of C11orf87 were determined by (E) qRT-PCR and (F) bisulfite pyrosequencing. Significant differences between groups were indicated by *P<0.05, **P<0.01, as determined by unpaired t-test. The original gel images can be found in S1 Fig.
To investigate whether STAT3 control C11orf87 expression, we ectopically expressed a constitutive active STAT3 mutant (STAT3c) in MKN28, a STAT3 inactive cell line [8, 9]. However, ectopic expression of STAT3c promoted C11orf87 expression in MKN28 cells (Fig 2C), without affecting its methylation (Fig 2D). In this regard, we hypothesized that STAT3 may contribute to methylation maintenances rather than de novo methylation. Indeed, treatment of STAT3 inhibitor, JSI-124 [17], resulted in a downregulation of C11orf87 expression in AGS gastric cancer cells (Fig 2E). Surprisingly, treatment of JSI-124 did not affect C11orf87 methylation (Fig 2F). This results suggested that long term STAT3 depletion may be required to disrupt C11orf87 methylation, as previously observed in NR4A3, a STAT3-downregulated target [9]. Taken together, these results suggested that the C11orf87 methylation may be a passenger effect under the STAT3-mediated C11orf87 expression.
The C11orf87 methylation is related to early gastric cancer initiation
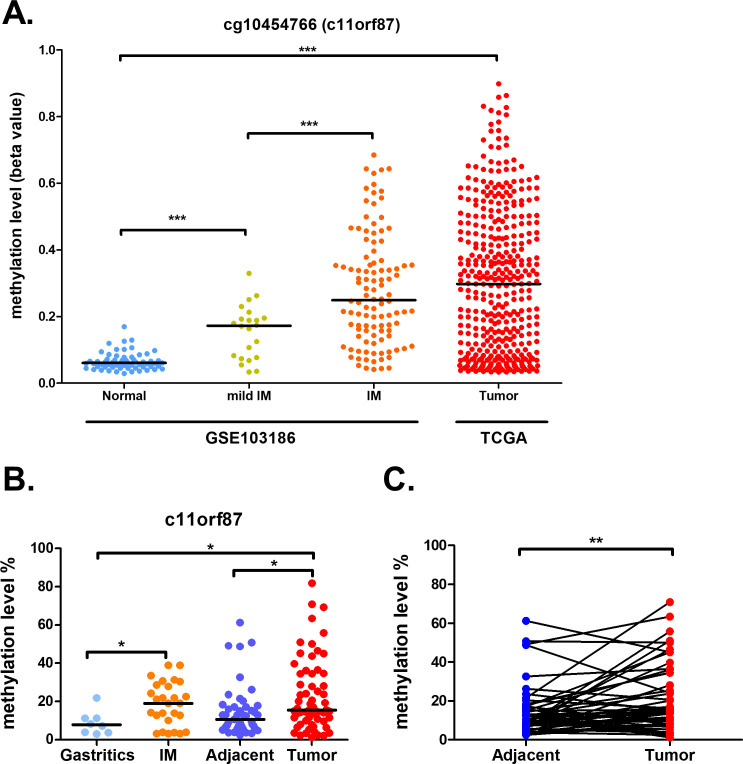
As the methylation wasn’t crucial for controlling C11orf87 expression, we then investigated whether the methylation of C11orf87 could serve as a biomarker for gastric cancer. We first analyzed the DNA methylation of cg10454766 from publicly available dataset obtained from TCGA and a previous study (GSE103186) [18], both of them were performed by Infinium HumanMethylation450 BeadChip. Overall, there was a progressive increase in DNA methylation of cg10454766 from normal gastric epithelium, mild intestinal metaplasia (IM), IM to gastric cancer (Fig 3A). Then, we performed bisulfite pyrosequencing of the CpG sites (chr11:109,293,939–109,293,966, Fig 1C, red bar) in the promoter region of C11orf87 covering cg10454766 in our in-house samples. In agreement with TCGA and the previous study, C11orf87 promoter showed a progressive increase in methylation upon disease progression (Fig 3B). Pairwise analysis of C11orf87 methylation also showed higher methylation in cancer as compared to corresponding adjacent normal (Fig 3C). These results suggested that the methylation of C11orf87 could serve as a diagnostic marker for gastric cancer.
Fig 3. Progressive hypermethylation of cg10454766 in gastric carcinogenesis.
(A) Methylation profile of cg10454766 from two datasets, GSE103186 and TCGA gastric cancer, including normal samples (n = 61), mild IM (n = 22), IM (n = 108) and cancer (n = 395). (B) Methylation of C11orf87 from our in-house samples including gastritis (n = 8), intestinal metaplasia (n = 27), tumor-adjacent normal (n = 47) to gastric tumor patients. (n = 62), as determined by bisulfite pyrosequencing. (C) Pairwise analysis of C11orf87 methylation in tumor-adjacent normal and tumor tissues (n = 47). Black lines denoted median values. Significant differences between groups and pairwise samples were indicated by *P<0.05, **P<0.01, ***P<0.005, as determined by Mann-Whitney U-test or paired t-test, wherever appropriate.
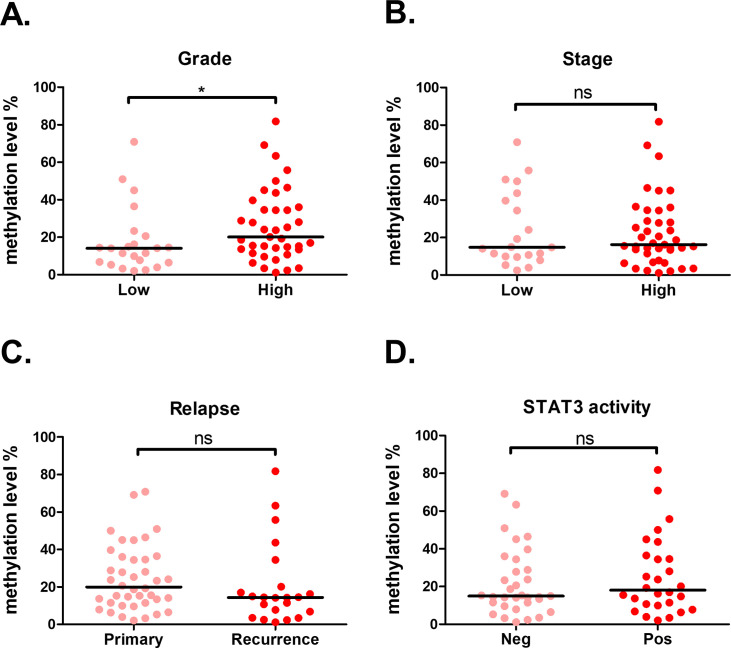
For disease progression analysis, comparison of tumor grade showed that a significant higher C11orf87 methylation was observed in patient samples with higher grade, as compared to low grade samples (Fig 4A). However, C11orf87 methylation was not associated with tumor stage, relapse or STAT3 activity (Fig 4B–4D). As activation of STAT3 was related to poor prognosis in gastric cancer [19], these results suggested that methylation of C11orf87 might independent to cancer progression.
Fig 4. Relationship between C11orf87 methylation of cg10454766 and patients’ clinical parameters.
Comparisons of C11orf87 methylation in gastric cancer patients with different tumor (A) grade (Low: n = 23; High: n = 39), (B) stage (Low: n = 21; High: n = 41), (C) relapse (Primary: n = 40; Recurrence: n = 22), and (D) STAT3 activity (Neg: n = 32; Pos: n = 28, as determined by IHC staining). The black lines denoted median values. Significant differences between groups are indicated by *P<0.05, **P<0.01, ***P<0.005, as determined by Mann-Whitney U-test.
Biomarker evaluation in methylation of C11orf87 from gastric cancer patients and non-cancerous patients
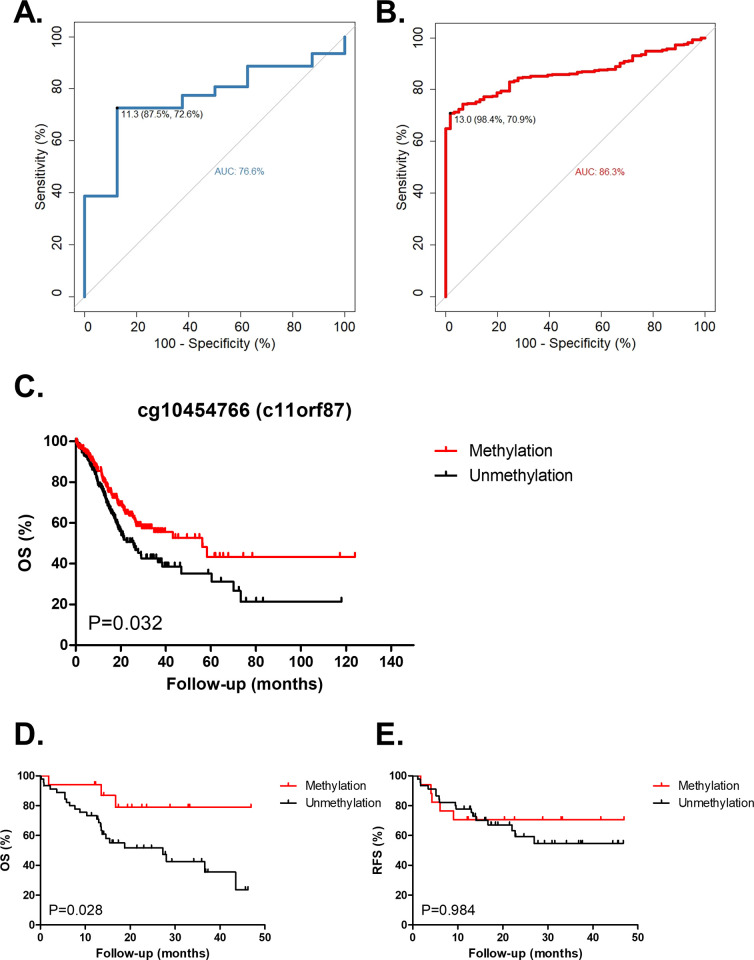
To assess if C11orf87 methylation can be a biomarker for discriminating gastric cancer vs non-cancer, receiver operating characteristics (ROC) curve was performed using the bisulfite pyrosequencing results from our in-house samples (8 gastritis and 62 gastric cancer, Fig 5A), showing an area under curve (AUC) of 76.6%. Using a cut-off of 11.3%, the sensitivity and specificity of cancer detection is 72.6% and 87.5%, respectively.
Fig 5. The methylation of C11orf87 served as a biomarker for diagnosis and diseases outcome.
(A) ROC curve of C11orf87 methylation in our in-house samples, including 8 gastritis and 62 gastric cancer samples. (B) ROC curve of cg10454766 methylation in combined datasets (GSE103186 and TCGA), including 61 normal samples from GSE103186 and 385 tumor samples from TCGA. The best cut-off methylation value for specificity and sensitivity is shown (black dot). (C) Kaplan-Meier analysis of patients with differential C11orf87 methylation status for overall survival (OS) in gastric cancer TCGA dataset (n = 385). Patients were divided into two groups according to the median value of methylation (median = 29.77%). Kaplan-Meier analysis of (D) OS and (E) recurrence-free survival (RFS) in gastric cancer patients from Taiwan. Patients were divided according to same median value from TCGA cohort. P-value is shown from the Log-rank (Mantel-Cox) test.
Besides, we also combined the results from GSE103186 and TCGA gastric cancer dataset, which utilized the same microarray platform, for ROC curve analysis (Fig 5B). ROC curve using 61 normal samples from GSE103186 and 385 cancer samples from the TCGA gastric cancer dataset, showed an area under curve of 86.3%. Using a cut-off of 13.0%, the sensitivity and specificity of cancer detection as positive, the sensitivity and specificity is 70.9% and 98.4%, respectively. These results suggested that C11orf87 methylation could be considered as a novel biomarker with well specificity for gastric cancer diagnosis.
Hypermethylation of C11orf87 is unexpectedly associated with better survival in gastric cancer
To further investigate the methylation of C11orf87 for clinical outcome, we first examined the overall survival (OS) and recurrence free survival (RFS) in our in-house samples (Fig 5C and 5D). Unexpectedly, patients with higher cg10454766 methylation showed a significant better OS, but not RFS in gastric cancer. Additionally, we also performed overall survival (OS) from TCGA gastric cancer cohort (Fig 5E). In agreement with our in-house samples, patients with higher cg10454766 methylation were also associated with better OS in TCGA cohort.
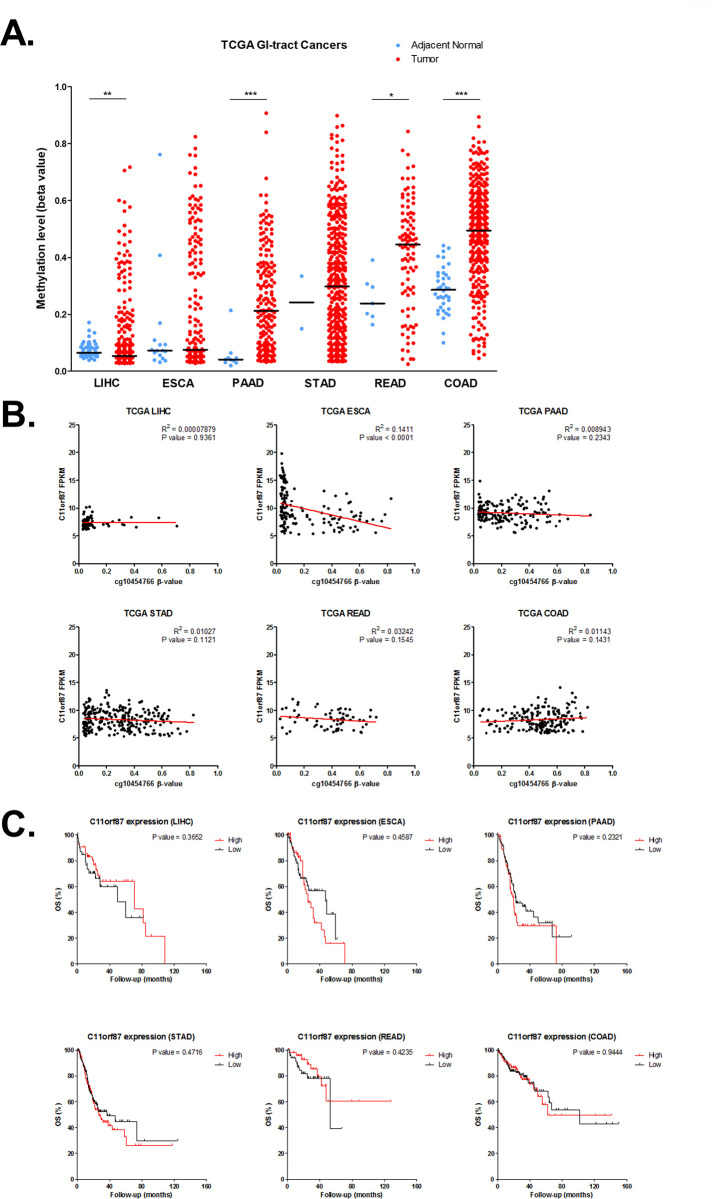
Hypermethylation of C11orf87 is frequently observed in GI-tract cancers
We also analyzed C11orf87 methylation in all of the gastrointestinal tract (GI) cancer from TCGA, including liver hepatocellular carcinoma (LIHC), esophageal carcinoma (ESCA), pancreatic adenocarcinoma (PAAD), stomach adenocarcinoma (STAD), Rectum Adenocarcinoma (READ) and colon adenocarcinoma (COAD). Surprisingly, C11orf87 methylation demonstrated a hypermethylation in most of the GI-tract cancer, as compared to adjacent normal (Fig 6A). This result suggested that C11orf87 methylation may be potentially useful for the detection of all GI cancers. In agreement with our cell lines result, there was no relationship between C11orf87 expression and methylation in most of GI-tract cancers, except for esophageal carcinoma (ESCA) (Fig 6B). Additionally, the expression of C11orf87 in OS from all of GI-tract cancers didn’t show any correlation, which further suggested the biological role of C11orf87 might not relate to clinical outcome (Fig 6C).
Fig 6. The methylation and expression profiles of C11orf87 in GI-tract cancers.
(A) Methylation of C11orf87 in all of GI-tract cancers, including liver hepatocellular carcinoma (LIHC), esophageal carcinoma (ESCA), pancreatic adenocarcinoma (PAAD), stomach adenocarcinoma (STAD), rectum adenocarcinoma (READ), and colon adenocarcinoma (COAD). Blue dots denote adjacent normal while red dots denote cancer. Black lines denote median values. Significant differences between groups are indicated by *P<0.05, **P<0.01, ***P<0.005, as determined by Mann-Whitney U-test. (B) The relationship between C11orf87 expression and methylation in all GI-tract cancers (LIHC: n = 84; ESCA: n = 146; PAAD: n = 160; STAD: n = 247; READ: n = 64; COAD: n = 189). (C) Kaplan-Meier analysis of patients with differential C11orf87 expression for overall survival (OS) in GI-tract cancers from TCGA dataset (LIHC: n = 82; ESCA: n = 145; PAAD: n = 159; STAD: n = 261; READ: n = 98; COAD: n = 248). Patients were divided into two groups according to the median value of expression, while the expression value shown as 0 or missing, were excluded from the analysis.
Discussions
Activation of JAK/STAT signaling plays an important role in gastric carcinogenesis. In particular, STAT3 can serve as a transcriptional activator for oncogene expression [20–22]. However, the role of STAT3 as a transcriptional repressor and epigenetic regulator was less explored. By integrated experimental and bioinformatic analyses, we have previously demonstrated that depletion of STAT3 resulted in hypomethylation of STAT3 targets and tumor suppressors, NR4A3 [9], and SPG20 [8], in a gastric cancer cell line with constitutive activation of JAK/STAT signaling. Higher NR4A3 methylation could be observed in gastric cancer patients with STAT3 activation. Patients with higher NR4A3 methylation were associated with poor survival. These results may be due to the recruitment of DNMT, via STAT3, to the STAT3 targets [23]. Previous studies indicated that acetylation on lysine 685 of STAT3 was crucial for the interaction with DNMT1 [24]. Targeted inhibition of STAT3 acetylation may be considered as a novel epigenetic strategy for reactivation of tumor suppressor genes in human cancer [25].
In the current study, by DNA methylation microarray, we further identified a potential STAT3 target, C11orf87, showing hypomethylation in AGS gastric cancer cells depleted with STAT3 and patients with lower STAT3 activation. Although STAT3 can promote the expression of C11orf87, there is no relationship between its expression and methylation. These results, interestingly, contradict with our previous finding that STAT3 acts as an oncogenic protein in the epigenetic silencing of tumor suppressors in gastric cancer [8, 9]. Recently, a study demonstrated that SIRT1, a histone deacetylase that participated in STAT3 deacetylation, was found to be upregulated in advanced gastric cancer [26]. The authors suggested that SIRT1 upregulation may compensate for the damaging effect induced by constitutive activation of STAT3 in gastric cancer. In this regard, SIRT1 may disrupt the interaction between STAT3 and DNMT1 by deacetylation on Lys685, which further limited methylation maintenances. Herein, we postulated that hypermethylation of C11orf87 may serve as a “vestigial marker” for constitutive activation of STAT3 in gastric cancer. This hypothesis may further explain our clinical observation that C11orf87 hypermethylation was related to better survival.
C11orf87, known as neuronal integral membrane protein 1, was found to be predominantly expressed in the brain tissue [27]. However, the involvement of C11orf87 in human cancer has not been characterized. A recent study in head and neck cancer found that p53 mutated tumors could promote differentiation of nerve fibers, which then promoted tumor growth in this tumor microenvironment [28]. As the expression of C11orf87 was controlled by STAT3, we, therefore, postulate that aberrant STAT3 activation may involve in promoting differentiation of nerve fibers via upregulation of C11orf87. Although p53 mutation is frequent in gastric cancer [29], how neuronal-related gene control gastric cancer progression still requires further investigation.
In conclusion, we are the first to demonstrate the progressive increase in C11orf87 methylation in IM and gastric cancer. Hypermethylation of C11orf87 was associated with better prognosis. Importantly, methylation of C11orf87 may act as a novel diagnostic biomarker for gastric cancer. Additionally, C11orf87 methylation is frequently observed in GI cancers, suggesting that it may also be useful for the detection of GI cancers. Besides, methylation of this potential STAT3 target may serve as a marker for the response and efficacy of targeting STAT3 in gastric cancer, as it may respect to STAT3 activity in early carcinogenesis. However, the functional and clinical role of C11orf87 in gastric cancer warrants further investigation.
Supporting information
(PDF)
Data Availability
The methylation array data has been deposited into in the Gene Expression Omnibus database (accession number: GSE109541).
Funding Statement
This study was supported by grants from the Ministry of Science and Technology, Taiwan (MOST 106-2314-B-194-001-MY3; 107-2314-B-194-001; 108-2314-B-194-001; 108-2314-B-194-003- MY2) to MWYC, Changhua Christian Hospital, Taiwan (106-CCH-IRP-064) to SHL, and the Center for Innovative Research on Aging Society (CIRAS) from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by Ministry of Education (MOE) in Taiwan.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. 10.3322/caac.21492 . [DOI] [PubMed] [Google Scholar]
- 2.Polk DB, Peek RM Jr. Helicobacter pylori: gastric cancer and beyond. Nat Rev Cancer. 2010;10(6):403–14. 10.1038/nrc2857 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piao JY, Lee HG, Kim SJ, Kim DH, Han HJ, Ngo HK, et al. Helicobacter pylori Activates IL-6-STAT3 Signaling in Human Gastric Cancer Cells: Potential Roles for Reactive Oxygen Species. Helicobacter. 2016;21(5):405–16. 10.1111/hel.12298 . [DOI] [PubMed] [Google Scholar]
- 4.Lu R, Zhang YG, Sun J. STAT3 activation in infection and infection-associated cancer. Mol Cell Endocrinol. 2017;451:80–7. 10.1016/j.mce.2017.02.023 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson CB, Judd LM, Menheniott TR, Kronborg I, Dow C, Yeomans ND, et al. Augmented gp130-mediated cytokine signalling accompanies human gastric cancer progression. J Pathol. 2007;213(2):140–51. 10.1002/path.2218 . [DOI] [PubMed] [Google Scholar]
- 6.Rocha GA, Rocha AM, Gomes AD, Faria CL Jr., Melo FF, Batista SA, et al. STAT3 polymorphism and Helicobacter pylori CagA strains with higher number of EPIYA-C segments independently increase the risk of gastric cancer. BMC Cancer. 2015;15:528. 10.1186/s12885-015-1533-1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bronte-Tinkew DM, Terebiznik M, Franco A, Ang M, Ahn D, Mimuro H, et al. Helicobacter pylori cytotoxin-associated gene A activates the signal transducer and activator of transcription 3 pathway in vitro and in vivo. Cancer Res. 2009;69(2):632–9. 10.1158/0008-5472.CAN-08-1191 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei KL, Chou JL, Chen YC, Jin H, Chuang YM, Wu CS, et al. Methylomics analysis identifies a putative STAT3 target, SPG20, as a noninvasive epigenetic biomarker for early detection of gastric cancer. PLoS One. 2019;14(6):e0218338. 10.1371/journal.pone.0218338 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yeh CM, Chang LY, Lin SH, Chou JL, Hsieh HY, Zeng LH, et al. Epigenetic silencing of the NR4A3 tumor suppressor, by aberrant JAK/STAT signaling, predicts prognosis in gastric cancer. Sci Rep. 2016;6:31690. 10.1038/srep31690 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu CS, Wei KL, Chou JL, Lu CK, Hsieh CC, Lin JM, et al. Aberrant JAK/STAT Signaling Suppresses TFF1 and TFF2 through Epigenetic Silencing of GATA6 in Gastric Cancer. Int J Mol Sci. 2016;17(9). 10.3390/ijms17091467 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi H, Yang Z, Dai C, Wang R, Ke X, Zhang S, et al. STAT3 activates MSK1-mediated histone H3 phosphorylation to promote NFAT signaling in gastric carcinogenesis. Oncogenesis. 2020;9(2):15. 10.1038/s41389-020-0195-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan YM, Wang CG, Zhu M, Xing R, Cui JT, Li WM, et al. STAT3 signaling drives EZH2 transcriptional activation and mediates poor prognosis in gastric cancer. Mol Cancer. 2016;15(1):79. 10.1186/s12943-016-0561-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flavahan WA, Gaskell E, Bernstein BE. Epigenetic plasticity and the hallmarks of cancer. Science. 2017;357(6348). 10.1126/science.aal2380 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biswas S, Rao CM. Epigenetics in cancer: Fundamentals and Beyond. Pharmacol Ther. 2017;173:118–34. 10.1016/j.pharmthera.2017.02.011 . [DOI] [PubMed] [Google Scholar]
- 15.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature reviews Genetics. 2012;13(7):484–92. 10.1038/nrg3230 . [DOI] [PubMed] [Google Scholar]
- 16.Luczak MW, Jagodzinski PP. The role of DNA methylation in cancer development. Folia histochemica et cytobiologica. 2006;44(3):143–54. . [PubMed] [Google Scholar]
- 17.Blaskovich MA, Sun J, Cantor A, Turkson J, Jove R, Sebti SM. Discovery of JSI-124 (cucurbitacin I), a selective Janus kinase/signal transducer and activator of transcription 3 signaling pathway inhibitor with potent antitumor activity against human and murine cancer cells in mice. Cancer Res. 2003;63(6):1270–9. . [PubMed] [Google Scholar]
- 18.Huang KK, Ramnarayanan K, Zhu F, Srivastava S, Xu C, Tan ALK, et al. Genomic and Epigenomic Profiling of High-Risk Intestinal Metaplasia Reveals Molecular Determinants of Progression to Gastric Cancer. Cancer cell. 2018;33(1):137–50 e5. 10.1016/j.ccell.2017.11.018 . [DOI] [PubMed] [Google Scholar]
- 19.Kim DY, Cha ST, Ahn DH, Kang HY, Kwon CI, Ko KH, et al. STAT3 expression in gastric cancer indicates a poor prognosis. Journal of gastroenterology and hepatology. 2009;24(4):646–51. 10.1111/j.1440-1746.2008.05671.x . [DOI] [PubMed] [Google Scholar]
- 20.Levy DE, Darnell JE Jr, Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3(9):651–62. 10.1038/nrm909 . [DOI] [PubMed] [Google Scholar]
- 21.Polak KL, Chernosky NM, Smigiel JM, Tamagno I, Jackson MW. Balancing STAT Activity as a Therapeutic Strategy. Cancers (Basel). 2019;11(11). 10.3390/cancers11111716 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim CP, Cao X. Structure, function, and regulation of STAT proteins. Mol Biosyst. 2006;2(11):536–50. 10.1039/b606246f . [DOI] [PubMed] [Google Scholar]
- 23.Zhang Q, Wang HY, Marzec M, Raghunath PN, Nagasawa T, Wasik MA. STAT3- and DNA methyltransferase 1-mediated epigenetic silencing of SHP-1 tyrosine phosphatase tumor suppressor gene in malignant T lymphocytes. Proc Natl Acad Sci U S A. 2005;102(19):6948–53. 10.1073/pnas.0501959102 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee H, Zhang P, Herrmann A, Yang C, Xin H, Wang Z, et al. Acetylated STAT3 is crucial for methylation of tumor-suppressor gene promoters and inhibition by resveratrol results in demethylation. Proc Natl Acad Sci U S A. 2012;109(20):7765–9. 10.1073/pnas.1205132109 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang HJ, Yi YW, Hou SJ, Kim HJ, Kong Y, Bae I, et al. Disruption of STAT3-DNMT1 interaction by SH-I-14 induces re-expression of tumor suppressor genes and inhibits growth of triple-negative breast tumor. Oncotarget. 2017;8(48):83457–68. 10.18632/oncotarget.4054 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang S, Yang Y, Huang S, Deng C, Zhou S, Yang J, et al. SIRT1 inhibits gastric cancer proliferation and metastasis via STAT3/MMP-13 signaling. J Cell Physiol. 2019;234(9):15395–406. 10.1002/jcp.28186 . [DOI] [PubMed] [Google Scholar]
- 27.Etemadikhah M, Niazi A, Wetterberg L, Feuk L. Transcriptome analysis of fibroblasts from schizophrenia patients reveals differential expression of schizophrenia-related genes. Sci Rep. 2020;10(1):630. 10.1038/s41598-020-57467-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amit M, Takahashi H, Dragomir MP, Lindemann A, Gleber-Netto FO, Pickering CR, et al. Loss of p53 drives neuron reprogramming in head and neck cancer. Nature. 2020;578(7795):449–54. 10.1038/s41586-020-1996-3 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhyu MG, Park WS, Jung YJ, Choi SW, Meltzer SJ. Allelic deletions of MCC/APC and p53 are frequent late events in human gastric carcinogenesis. Gastroenterology. 1994;106(6):1584–8. 10.1016/0016-5085(94)90414-6 . [DOI] [PubMed] [Google Scholar]