Abstract
Excessive use of chemical fertilizers in agricultural practices have demonstrated a significant impact on microbial diversity and community in soil by altering soil physical and chemical properties, thereby leading to a certain degree of soil salinization and nutritional imbalances. As an organic amendment, maize straw has been widely used to improve soil quality; however, its effect on the soil bacterial community remains limited in Calcarie-Fluvie Cambisols soil in semi-humid arid plateau of North China. In the present experiment, we investigated the effects of continuous straw utilization and fertilization on bacterial communities in Shouyang, Shanxi province, China. Soil samples were collected from 5 different straw utilization and fertilization modes in the following ways: straw mulching (SM), straw crushing (SC), cattle manure (CM), in which way straw is firstly used as silage and then organic fertilizer, control with no straw return (NSR), and control without fertilizers (CK), same amount of N+P fertilizer was applied to the regimes except CK. High-throughput sequencing approaches were applied to the V3-V4 regions of the 16S ribosomal RNA for analysis of the bacterial abundance and community structures. Different long-term straw returning regimes significantly altered the physicochemical properties and bacterial communities of soil, among which CM had the most significant effects on soil fertility and bacterial diversity. Proteobacteria, Actinobacteria, Chloroflexi, Acidobacteria, and Gemmatimonadetes were consistently dominant in all soil samples, and Redundancy analysis (RDA) showed significant association of total nitrogen (TN), total phosphorus (TP) and available potassium (AK) with alternation of the bacterial community. Cattle manure had the most beneficial effects on soil fertility and bacterial diversity among different straw utilization and fertilization modes.
Introduction
As an integral part of the soil, microorganisms represent the major driving force of soil organic matter and nutrient cycling, playing a key role in soil nutrient conversion, energy transformation, and the formation of soil organic matter [1–4]. Due to the rapid response of bacteria to changes in the soil environment, bacterial communities were selected as early bio-indicators of soil quality [5]. Bacterial diversity and community composition are affected by soil pH, organic matter, soil moisture content, mineral substances etc. [6, 7], which could be influenced by fertilization management practices [8, 9].
As the largest grain producer worldwide, agriculture production has largely depended on high inputs of chemical fertilizer, while about 800 million tons of crop straw are annually produced in China [10]. Crop straw has complex components, such as high contents of lignin, cellulose, and hemicellulose, which is rich in organic carbon, nitrogen, phosphorus, potassium, silicon, and other mineral nutrients [11–13], and has been widely utilized in the field to promote soil carbon sequestration and consequently maintaining soil fertility. Previous studies have documented the positive and significant impact of straw utilization on soil microbial community structure, biomass, and activity [14–18], leading to increases in soil granular structure and the content of water-stable aggregates for better soil fertility and water conservation [19–21]. Chen et al. documented a significant increase in total phospholipid fatty acids (PLFA), bacterial biomass, and actinomycete biomass under short-term straw return in Jiangyan, China [5]. Zhao et al. reported an abundant increase of Gram-negative (Gm-) bacteria, but only after long-term maize straw returned (30 years) in a summer maize-winter wheat cropping system, one located in north-central China [13].
The Shanxi province is a semi-humid arid plateau of North China with a typical soil of Calcarie-Fluvie Cambisols. Under the local soil type and climate conditions, we hypothesized that different maize straw utilization and fertilization would affect the diversity and community structure of soil bacteria. However, the understanding of the effects of straw utilization and fertilization on bacterial communities in this region remains limited. Considering the imperfection of traditional cultivation on growing media, high-throughput sequencing approaches were applied to the V3-V4 regions of the 16S ribosomal RNA for analysis of soil bacteria to different maize straw utilization and fertilization modes. Five treatments were applied: (1) straw mulching (SM); (2) straw crushing (SC); (3) cattle manure (CM), in which way maize straw is firstly used as silage, and cattle manure is applied to field as organic fertilizer; (4) control with no straw return (NSR); and control without fertilizers (CK) same amount of N+P fertilizer was applied to the regimes except CK. Soil samples were collected for analysis of bacterial diversity. The objectives of this study were to: (1) elucidate the influences of different straw utilization and fertilization modes on bacterial diversity and community structure; (2) search for the most suitable straw utilization mode.
Materials and methods
Experimental design
The long-term straw utilization and fertilization experiment was established in Shouyang, Shanxi province, China (28°15′20″N, 116°55′30″E) in 1992. The area belongs to the continental monsoon climate in a mid-latitude, temperate-warm, and semi-humid, arid area, featuring four distinct seasons. The average annual temperature is 7.4°C with a frost-free period of about 130 days, whereas the average annual precipitation is 500 mm with an annual evaporation of 1600–1800 mm. The experimental site has a sandy loam cinnamon soil, classified as a Calcarie-Fluvie Cambisols in WRB [22, 23]. The soil layer is deep, and the groundwater level is 50 meters below the surface. Analysis of soil samples taken from the experimental area showed that the basic physical and chemical properties of the surface 15 cm of soil were as follows: pH, 8.4; organic matter (OM), 23.8 g/kg; available N (AN), 117.69 mg/kg; available P (AP), 4.84 mg/kg; and available K (AK), 100 mg/kg. The authority who issued the permission for each location is “Shanxi Shouyang Field Scientific Observation and Experimental Station”.
The long-term trial began in the spring of 1992, lasting 27 years. The five treatments were arranged in a randomized block design with three replications, with each plot being 67 m2. The five treatments were (1) SM, straw mulched on the soil in the first year and then crushed in the second year to incorporate into the soil; (2) SC, straw crushed directly to incorporate into the soil; (3) CM, cattle manure, maize straw is firstly used as silage, and cattle manure is applied to field as organic fertilizer; (4) NSR, control, no straw using, same amount of N+P fertilizer was applied to the above regimes and (5) CK, control without fertilizers. Straw in treatments SM, SC, and CM had an equivalent amount of carbon. The fertilization details are shown in Table 1. All the chemical fertilizers and straws were applied as basal fertilizers before planting.
Table 1. Experimental treatments and fertilization.
| Treatments | Inorganic fertilizer (kg/hm2) | Straw (kg/hm2) | Cattle Manure (kg/hm2) | Total nutrient (kg/hm2) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | P2O5 | K2O | N | P2O5 | K2O | N | P2O5 | K2O | N | P2O5 | K2O | |
| SM | 150 | 84 | 0 | 46 | 11 | 82 | 0 | 0 | 0 | 196 | 95 | 82 |
| SC | 150 | 84 | 0 | 46 | 11 | 82 | 0 | 0 | 0 | 196 | 95 | 82 |
| CM | 150 | 84 | 0 | 0 | 0 | 0 | 350 | 236 | 756 | 500 | 320 | 756 |
| NSR | 150 | 84 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 150 | 84 | 0 |
| CK | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
SM: straw mulched; SC, straw crushed; CM, cattle manure; NSR, fertilizer with no straw return; and CK, no fertilizer and no straw return.
Soil sampling and analysis of soil properties
Soil was randomly sampled from 5 points in each plot on October 1, 2018 and mixed to form one composite sample per plot. Each mixed soil sample was placed in sterile bag and transported to a laboratory in an icebox. After removing visible stones and plant debris, one part was stored at -80°C for DNA extraction the other part was gently broken along its natural breakpoints, sieved through a 2 mm screen, thoroughly mixed for physicochemical properties analysis.
Soil physicochemical analytical procedures
Soil total nitrogen (TN), total phosphorus (TP), and total kalium (TK) were analyzed using a Vario Max element analyzer (Elementar Vario PYRO cube and Isoprime100, Germany). Soil pH was determined with a soil (air-dried) to water (w/w) ratio of 1:2.5 with a pH meter. OM was determined by K2Cr2O7 oxidation method. Soil AN was determined by the alkaline hydrolysis diffusion method. AP was determined by NaHCO3-extracted method. AK was analyzed by CH3COONH4-extracted method.
Soil DNA extraction and high-throughput sequencing
Total DNA was extracted from 0.5 g soil of each sample using the Fast DNA SPIN Isolation Kit (MP Biomedicals, Santa Ana, CA, United States) according to the manufacturer’s instructions. The quantity and quality of DNA were accessed using a NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and on 0.8% agarose gel electrophoresis, respectively.
The bacterial V3-V4 hypervariable region of 16S rRNA was amplified using primers 338F (5’-ACTCCTACGGGAGGCAGCA-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’). Specific barcodes of 7-bp were incorporated into the primers for multiplex sequencing. PCR reactions were performed in a system of 25 μL mixture containing 5 μL of 5×Q5 Reaction Buffer, 5 μL of 5×Q5 High-Fidelity GC Buffer, 2 μL of dNTPs (5U/μL), 1 μL of each primer, 0.25 μL of Q5 High-Fidelity DNA Polymerase (Transgen, China), 2μL of temple DNA, and 8.75 μL of ddH2O.
PCR was conducted as follows: 2 min of initial denaturation at 98°C, followed by 25 cycles of denaturation at 98°C for 15 s, annealing at 55°C for 30 s and extension at 72°C for 30 s, and a final extension of 5 min at 72°C. PCR amplicons were purified with Agencourt AMPure Beads (Beckman Coulter, Indianapolis, IN) and quantified using the PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). After purification and being pooled at equal concentrations, the PCR products were pair-end sequenced with 2*300 bp on the Illumina MiSeq platform with MiSeq Reagent Kit v3 at Personal Biotechnology (Shanghai, China).
Bioinformatics analysis
Raw sequencing data was processed with Quantitative Insights Into Microbial Ecology (QIIME, v1.8.0) software [24]. Raw sequencing reads with exact matches to the barcodes were assigned to respective samples and identified as valid sequences. The low-quality sequences were filtered and paired-end reads were assembled using FLASH [25]. After chimera detection, the high-quality sequences were clustered with UCLUST [26] into operational taxonomic units (OTUs) at 97% similarity. A representative sequence was selected from each OTU using default parameters. OTU taxonomic classification was conducted by BLAST searching the representative sequences set against the Silva 132 Database [27] using the best hit [28]. An OTU table was generated to record the abundance and taxonomy of each OTU in each sample. OTUs with less than 0.001% of total sequences across all samples were discarded. To minimize the difference of sequencing depth across samples, an averaged, rounded, and rarefied OTU table was generated by averaging 100 evenly resampled OTU subsets under the 90% of the minimum sequencing depth for further analysis.
Sequence data analyses were mainly performed using QIIME and R packages. OTU-level α-diversity indices, such as Chao1 richness estimator, ACE metric (Abundance-based Coverage Estimator), Shannon diversity index, and Simpson index were calculated using the OTU table in QIIME. OTU-level ranked abundance curves were generated to compare the richness and evenness of OTUs among samples. β-diversity analysis was performed to investigate the structural variation of microbial communities across samples, and nonmetric multidimensional scaling (NMDS) based on OTU was used [29]. The significance of differentiation of microbiota structure among groups was assessed by permutational multivariate analysis of variance (PERMANOVA) [30] and analysis of similarities (ANOSIM) [31, 32]. The taxonomy compositions and abundances were visualized using MEGAN [33]. A Venn diagram was generated to visualize the shared and unique OTUs among samples or groups based on the occurrence of OTUs across groups, and regardless of their relative abundance [34]. Taxa abundances at the phylum, class, order, family and genus levels were statistically compared among samples by Metastats [35]. Linear discriminant analysis (LDA) effect size (LEfSe) was performed to detect differentially abundant taxa across groups using the default parameters [36].
Statistical analysis
A one-way analysis of variance (ANOVA) was performed to determine the effects of different straw utilization and fertilization regimes on soil physicochemical characteristics with Microsoft Excel (Microsoft Corporation, USA) and SPSS Windows version 11.0 (SPSS Inc., Chicago, USA) packages. Significant differences between soil physicochemical characteristics were determined with the least significant difference (LSD) test at the p = 0.05 level.
Results
Soil physicochemical characteristics
Compared with no straw return (NSR) and no fertilizer (CK), all nutrient indices including the contents of total nitrogen (TN), total phosphorus (TP), total kalium (TK), available N (AN), available P (AP), available K (AK), and organic matter (OM) were significantly increased (p<0.05) except soil pH, indicating that significant changes in soil chemical properties were caused by the long-term straw return. All nutrient indices were the highest in CM treatment, followed by SC treatment, SM treatment, and NSR treatment. Soil pH (H2O) values were significantly decreased by the addition of crushed straw (SC) and cattle manure (CM), as well as by no straw returning (NSR), compared with CK and mulching straw (SM). The pH in CM treatment was more neutral compared with other treatments. The contents of AN and TN in SM treatment were significantly lower than in SC treatment; however, AP, AK, OM, and TK were significantly higher in SM treatment than in SC treatment (Table 2).
Table 2. Soil pH and nutrient contents in a utilization and fertilization experiment.
| Treatments | pH | AN (mg/kg) | AP (mg/kg) | AK (mg/kg) | OM (g/kg) | TN (g/kg) | TP (g/kg) | TK (g/kg) |
|---|---|---|---|---|---|---|---|---|
| SM | 8.66±0.12a | 40.95±0.05c | 32.97±0.02b | 85.06±0.02b | 30.43±0.01b | 1.10±0.01c | 0.74±0.03b | 16.19±0.08b |
| SC | 8.52±0.02b | 45.05±0.06b | 32.25±0.04c | 78.69±0.12c | 28.28±0.02c | 1.20±0.02b | 0.74±0.12b | 15.37±0.12c |
| CM | 8.32±0.05c | 72.31±0.13a | 73.72±0.12a | 124.34±0.05a | 36.02±0.12a | 1.42±0.31a | 0.93±0.22a | 20.89±0.13a |
| NSR | 8.54±0.01b | 38.9±0.09d | 28.88±0.05d | 50.02±0.06d | 24.62±0.31d | 1.02±0.09d | 0.63±0.07c | 15.04±0.01d |
| CK | 8.65±0.10a | 32.76±0.12e | 4.56±0.03e | 21.68±0.04e | 12.90±0.07e | 0.80±0.10e | 0.50±0.29d | 14.67±0.09e |
AN: available N; AP: available P; AK: available K; OM: organic matter; TN: total N; TP: total P; TK: total K; Different letters in the same column indicate a significant difference (ANOVA followed by Turkey-Kramer post hoc test, n = 3, p<0.05, average value, SD standard deviation).
Bacterial alpha-diversity
A total of 376,833 clean sequences were generated and deposited in the NCBI Sequence Read Archive (PRJNA588667). The number of species observed in the sample was not significantly increased with the addition of new samples (S1 Fig), indicating that the sample size employed was large enough to characterize the bacterial communities.
The diversity indices Chao1 estimator, abundance-based and coverage estimator (ACE), Shannon index (Hʼ), and the Simpson index (D) were estimated as community richness indices using a randomly selected subset of about 12000 sequences per soil sample. As shown in Table 3, the diversity indices were the lowest in CK but the highest in CM treatment. However, no significant differences in bacterial richness or diversity were detected among different treatments, as indicated by Chao1, H’, ACE, and D indices.
Table 3. Richness and alpha-diversity indices of the different OTUs under the different treatments.
| Treatments | Chao1 | ACE | Shannon | Simpson | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| SM | 3144.86a | 274.72 | 3320.63a | 327.77 | 10.76a | 0.05 | 0.99883a | 6.7E-05 |
| SC | 3051.50a | 309.06 | 3233.18a | 388.58 | 10.76a | 0.06 | 0.99891a | 7.27E-05 |
| CM | 3148.42a | 442.18 | 3334.90a | 473.97 | 10.76a | 0.02 | 0.99897a | 2.35E-05 |
| NSR | 3122.67a | 354.57 | 3273.54a | 434.61 | 10.80a | 0.02 | 0.99888a | 1.47E-05 |
| CK | 2931.36b | 482.38 | 3019.21a | 594.90 | 10.71a | 0.08 | 0.99863a | 9.76E-05 |
Different letters in the same column indicate a significant difference (ANOVA followed by Turkey-Kramer post hoc test, n = 3, p<0.05, average value, SD standard deviation).
Soil bacterial community composition
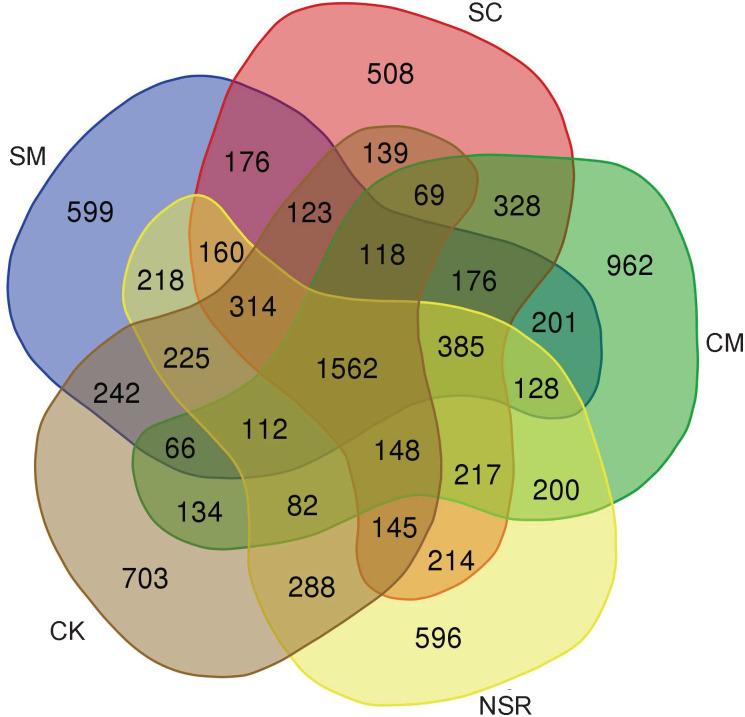
As shown in Fig 1, 1562 OTUs were shared by different treatments. The number of OTUs specific to treatment SM, SC, CM, NSR, and CK were 599, 508, 962, 596, and 703, respectively. Compared with treatment SM, SC, NSR, and CK, unique OTUs in treatment CM were increased by 60%, 89%, 61%, and 37%, respectively.
Fig 1. Venn diagram showing unique and overlapped OTUs between different straw return treatments.
Effects of different treatments on soil bacterial communities
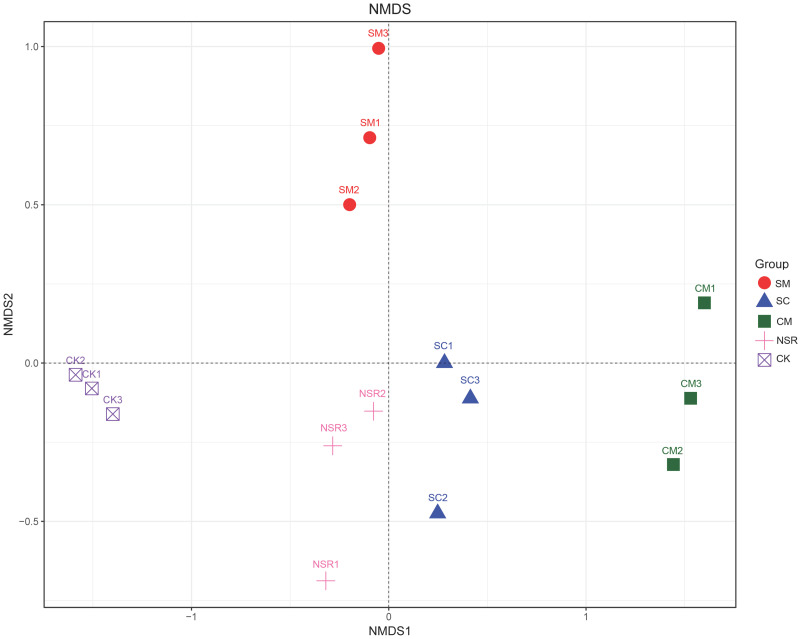
Nonmetric multidimensional scaling (NMDS) was used to compare the similarity of soil bacterial communities among different treatments at the OTU level (Fig 2). The three repeats of each of the treatments were clustered together, showing good repeatability. The samples of treatment CM were located in the right part of the graph, whereas the samples of CK were gathered at the left and samples of treatment SM were gathered in the upper part. Samples of SC and NSR distributed separately along the vertical axis, which were relatively closer among all the treatments. The results showed a significant difference in the bacterial community structures among treatment SM, CM, and CK at the OTU level.
Fig 2. Nonmetric multidimensional scaling of the bacterial community compositions in soil under different treatments based on OTUs.
Taxonomic composition analysis at the phylum and genus level
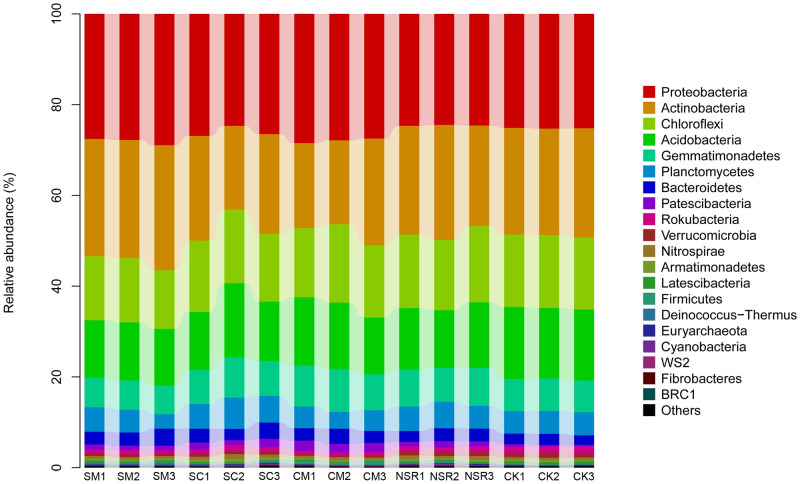
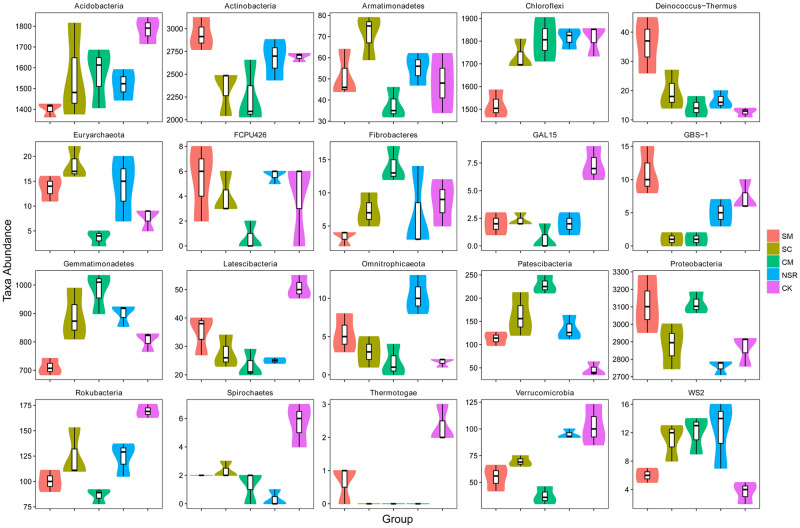
The taxonomic distributions of bacterial communities were evaluated at different levels of classification. As shown in Fig 3, Proteobacteria, Actinobacteria, Chloroflexi, Acidobacteria, Gemmatimonadetes, Planctomycetes, Bacteroidetes, Patescibateria, Rokubacteria, and Verrucomicrobia were the most abundant at the phylum level, accounting for approximately 97.5% of the microorganisms detected in all 15 soil samples. Compared with treatment NSR, the percentage of Proteobacteria was increased by 3.50% in treatment SM, 1.43% in treatment SC, and 3.33% in treatment CM. The top 20 phyla with significant differences among all the treatments are shown in Fig 4. Among the top 5 phyla, Proteobacteria, Chloroflexi, Acidobacteria, and Gemmatimonadetes increased by 3.33%, 0.02%, 0.51%, and 0.78% in treatment CM, respectively.
Fig 3. Taxonomic composition and abundance distribution of bacteria at the phylum level under different treatments.
Fig 4. The top 20 phyla with significant differences between the five treatments.
At the genus level, the dominant genera were Sphingomonas, MND1, RB41, AKYG587, Nocardioides, Blastococcus, Micromonospora, Actinoplanes, Haliangium, and Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium (S2 Fig). The relative abundance of Sphingomonas was 1.13% in treatment SM, 0.42% in treatment SC, and 0.09% in treatment CM, as compared to treatment NSR. The percentage of MND1 increased by 0.46% in treatment SM, 0.22% in treatment SC, and 0.13% in treatment CM, as compared with treatment NSR. The top 20 genera with a significant difference among all the treatments are shown in S3 Fig. The taxonomic composition and abundance distribution of bacterias at the class, order, family level under different treatments are shown in S4–S6 Figs in the supporting information.
LEfSe revealed 7, 1, 6, 0, and 1 taxa with significant differences (LDA score>4, and p<0.05) among five treatments (S7 Fig), which might serve as biomarkers for different treatments.
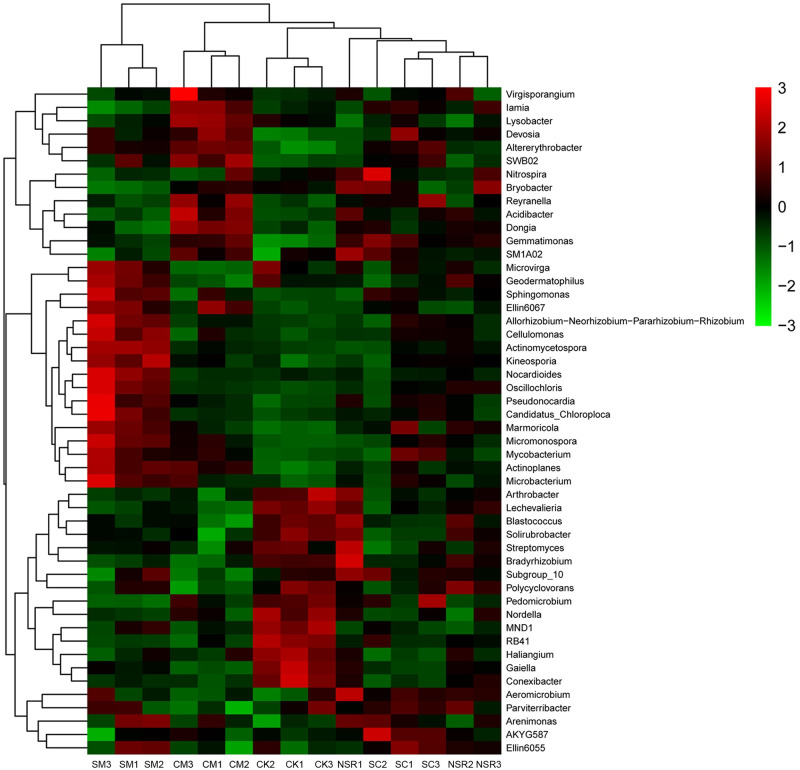
Heat map of community composition combined with cluster analysis
The genera with the top 50 abundance were clustered, showing large variation among the five treatments (Fig 5). Genera Nocardioides, Oscillochloris, Pseudonocardia, Candidatus-Chloroploca, and Microbacterium were relatively higher in abundance in treatment SM, whereas Virgisporangium, Lamia, Lysobacter, Devosia, and SWB02 were relatively higher in abundance in treatment CM.
Fig 5. Hierarchically clustered heat map analysis of community composition in the soil samples under different treatments.
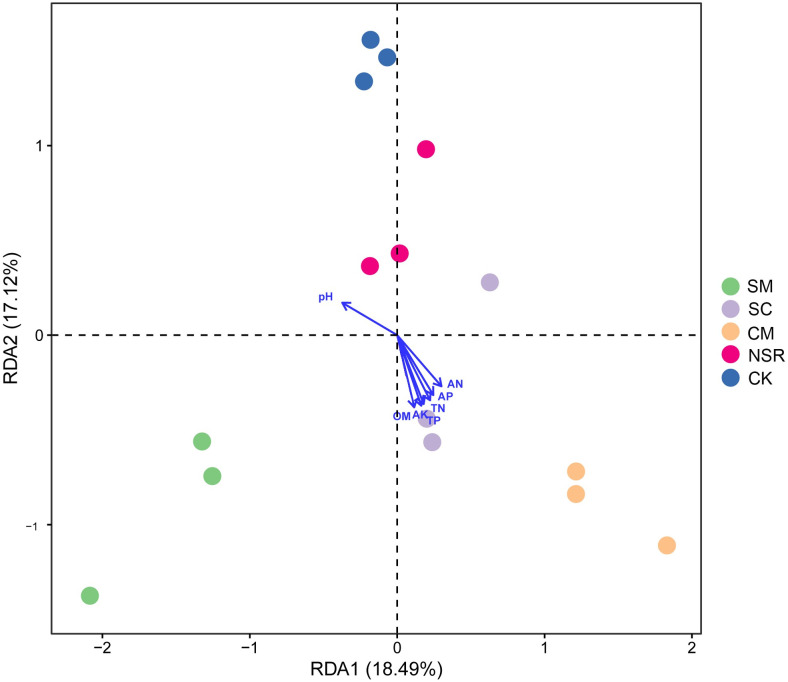
Relationship between soil bacterial communities and environmental variables
Redundancy analysis (RDA) was conducted to explore the possible physicochemical properties (pH, OM, TP, TN, TK, AP, AN, AK) differentiating soil bacterial community structures. As shown in Fig 6, eight environmental variables (pH, OM, TP, TN, TK, AP, AN, AK) together explained 35.61% of the total variance. The first two RDA axes explained most of the variation, in which the first axis accounted for 18.49% and the second axis explained 17.12% of the variation (Fig 6). The Monte Carlo permutation test (S1 Table) revealed that TN, TP and AK were the main environmental factors affecting soil bacterial distribution.
Fig 6. Redundancy analysis between soil samples based on OTUs.
Discussion
Effects of straw return on the soil chemical properties
The soil bacterial communities generally change when nutrients are added to the soil. In the present study, treatment CK had not added fertilizers for 27 years, leading to the lowest chemical properties. Compared with CK and NSR, different straw utilization and fertilization regimes significantly improved the soil chemical properties including OM, TP, TN, TK, AP, AN, and AK, among which treatment CM performed the best. However, treatment CM reduced soil pH, consistent with the findings of Liu et al., who reported a decline in soil pH with the application of manure, resulting in an increased risk of soil acidification [37]. The reason might be that cattle manure contains a large amount of nutrients, particularly AN. Moreover, the contents of AP, AK, OM, and TK were significantly higher in treatment SM than in treatment SC, which might be caused by straw that was decomposed in treatment SM, as opposed to the fresh straw in treatment SC. Notably, the soil pH had no significant difference between the straw mulching and CK over 27 years. Previous studies demonstrated that decomposition facilitated the release of nutrients of the straw [38, 39]. SM has been proven to improve soil water retention, thereby enhancing soil moisture conservation under arid and semiarid conditions [17, 40] to promote plant growth [41].
Effects of straw utilization and fertilization on the soil bacterial community
Consistent with the previous results reported in the wheat-rice rotation system [42], alpha diversity analysis showed no significant effects of long-term straw utilization and fertilization on bacterial community richness and diversity. NMDS showed significant differences (p<0.05) in bacterial community structure under different treatments, which is consistent with the results of Bei et al., who documented clear separation of bacterial and fungal community composition between the straw utilization and no straw utilization [43]. Therefore, straw utilization directly affects the structure of soil microorganisms by promoting or inhibiting the change of soil microorganism composition, further impacting the biological activity of microorganisms in the soil. Among different straw utilization and fertilization regimes, treatment CM showed the most significant effect on bacterial community structure.
Maize straw is composed by cellulose (35%), hemicellulose (27%), lignin (12%), and other complex substances, and is mainly decomposed by soil microorganisms in two phases. Our study demonstrated no significant effect of different long-term maize straw utilization and fertilization regimes on the bacterial community structure at the phylum level, which is consistent with the results of Xia et al., who reported no significant change of bacterial community structure with straw using [44]. Proteobacteria, Actinobacteria, Chloroflexi, Acidobacteria, and Gemmatimonadetes were the dominant phyla in our study, accounting for approximately 86.7%, which is consistent with the previous results reported in maize planting land, wheat-maize rotation system [43], forest, vineyard, copper mine, river, lake, and greenhouse [45–50]. Among these phyla, Proteobacteria and Gemmatimonadetes mainly participated in the initial stage of straw decomposition, whereas Actinobacteria is involved in almost all of the straw degradation phase. The addition of straw changes the soil pH, thereby favoring the growth of Actinobacteria. Chloroflexi is generally believed to produce energy through photosynthesis and have a certain degradation function for soil environmental pollution [51].
Under long-term straw utilization and fertilization conditions, Sphingomonas, MND1, RB41, AKYG587, Nocardioides, Blastococcus, Micromonospora, Actinoplanes, Haliangium, and Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium together occupied the dominant position. Sphingomonas was one of the most effective microorganisms for degradation of soil toxic substances, promotion of nutrient absorption and resistance to multiple pathogens. Previous studies reported several Sphingomonas strains with the characteristics of nitrogen fixation and dehydrogenation, playing an important role in maintaining the nitrogen balance of soil [52]. In addition to the dominant known bacteria, unclassified bacteria still accounted for a certain proportion in maize soil. Further study is needed to characterize unclassified bacteria and their function in long-term straw-using maize soil.
The LEfSe analysis demonstrated that Gemmatimonadetes (Gemmatimonadetes, Gemmatimonadales and Gemmatimonadetes) and Chloroflexi (Anaerolineae and SBR1031) might serve as indicators for treatment CM. Gemmatimonadetes were ubiquitous in soil, comprising ~2% of soil bacterial communities [53] and playing an important role in terrestrial ecosystems. However, the distribution of Gemmatimonadetes was more dependent on moisture availability in soil, and it was unable to resist moisture fluctuations induced by wet/dry cycling [53]. Additionally, the distribution of Gemmatimonadetes was also constrained by soil. Chloroflexi was distributed widely in terrestrial and aquatic ecosystems [54], whereas Anaerolineae was abundant in active sludge and other nutrient rich environments [55]. Among different straw utilization and fertilization regimes, long-term cattle manure was shown to be the most favorable for the growth and distribution of these bacteria.
Conclusion
In the present study, soil under continuously long-term straw utilization and fertilization were used to characterize physicochemical properties and bacterial communities. The results indicated that different straw utilization and fertilization had significant effects on soil properties and bacterial communities. Among different straw utilization regimes, cattle manure, which was used as silage first and then organic fertilizer, showed the richest nutrients and highest bacterial diversity. Therefore, cattle manure appears to be the most promising agricultural practice for the improvement of soil fertility.
Supporting information
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(DOCX)
Acknowledgments
We are grateful for Personalbio (Shanghai) Co., Ltd. for its assistance in sequencing analysis and bioinformatics analysis.
Data Availability
All the raw data files are available in the NCBI Sequence Read Archive (PRJNA588667).
Funding Statement
This work was financially supported by the Shanxi Key R&D Plan (201703D2110023), Shanxi Youth Fund(201901D211557)and Doctor fund of Shanxi Academy of Agricultural Sciences(YBSJJ2012). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Postma-Blaauw MB, de Goede RGM, Bloem J, Faber JH, Brussaard L. Soil biota community structure and abundance under agricultural intensification and extensification. Ecology. 2010; 91(2), 460–473. 10.1890/09-0666.1 [DOI] [PubMed] [Google Scholar]
- 2.Ollivier J, Towe S, Bannert A, Hai B, Kastl EM, Meyer A, et al. Nitrogen turnover in soil and global change. FEMS Microbiol Ecol. 2011; 78(1), 3–16. 10.1111/j.1574-6941.2011.01165.x [DOI] [PubMed] [Google Scholar]
- 3.Chaparro JM, Sheflin AM, Manter DK, Vivanco JM. Manipulating the soil microbiome to increase soil health and plant fertility. Biol Fert Soils. 2012; 48(5), 489–499. 10.1007/s00374-012-0691-4 [DOI] [Google Scholar]
- 4.Nicolás C, Hernández T, García C. Organic amendments as strategy to increase organic matter in particlesize fractions of a semi-arid soil. Appl Soil Ecol. 2012; 57, 50–58. 10.1016/j.apsoil.2012.02.018 [DOI] [Google Scholar]
- 5.Chen Z, Wang H, Liu X, Zhao X, Lu D, Zhou J. Changes in soil microbial community and organic carbon fractions under short-term straw return in a rice–wheat cropping system. Soil & Tillage Res. 2017; 165, 121–127. 10.1016/j.still.2016.07.018 [DOI] [Google Scholar]
- 6.Bünemann EK, Schwenke GD, Van Zwieten L. Impact of agricultural inputs on soil organisms—a review. Aust J Soil Res. 2006; 44: 379–406. 10.1071/SR05125 [DOI] [Google Scholar]
- 7.Strom PF. Effect of temperature on bacterial species diversity in thermophilic solid-waste composting. Applied & Environmental Microbiology, 1985, 50, 899–905. 10.1128/AEM.50.4.899-905.1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartmann M, Frey B, Mayer J, Mäder P, and Widmer F. Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 2014; 9, 1177–1194. 10.1038/ismej.2014.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luo PY, Han XR, Wang Y, Han M, Shi H, Liu N, et al. Influence of long-term fertilization on soil microbial biomass, dehydrogenase activity, and bacterial and fungal community structure in a brown soil of northeast China. Annals Microbiol. 2015; 65, 533–542. 10.1007/s13213-014-0889-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clare A, Shackley S, Joseph S, Hammond J, Pan GX, Bloom A. Competing uses for China’s straw: the economic and carbon abatement potential of biochar. Gcb Bioenergy. 2015; 7, 1272–1282. 10.1111/gcbb.12220 [DOI] [Google Scholar]
- 11.Lal R. The role of residues management in sustainable agricultural systems. J Sustain. Agric. 1995; 5, 51–78. 10.1300/J064v05n04_06 [DOI] [Google Scholar]
- 12.Duiker SW, Lal R. Crop residue and tillage effects on carbon sequestration in a Luvisol in central Ohio. Soil Till Res. 1999; 52, 73–81. 10.1016/s0167-1987(99)00059-8 [DOI] [Google Scholar]
- 13.Zhao S, Li K, Zhou W, Qiu S, Huang S, He P. Changes in soil microbial community, enzyme activities and organic matter fractions under long-term straw return in north central China. Agr Ecosyst Environ. 2016; 216, 82–88. 10.1016/j.agee.2015.09.028 [DOI] [Google Scholar]
- 14.Ma LJ, Kong FX, Wang Z, Luo Y, Lv XB, Zhou ZG, et al. Growth and yield of cotton as affected by different straw returning modes with an equivalent carbon input. Field Crops Res. 2019; 243: 1–10. 10.1016/j.fcr.2019.107616 [DOI] [Google Scholar]
- 15.Powlson DS, Riche AB, Coleman K, Glendining MJ, Whitmore AP. Carbon sequestration in European soils through straw incorporation: limitations and alternatives. Waste Manage Res. 2008; 28(4): 741–746. 10.1016/j.wasman.2007.09.024 [DOI] [PubMed] [Google Scholar]
- 16.Procházková B, Málek J, Dovrtel J. Effect of different straw management practices on yields of continuous spring barley. Rostlinná Výroba. 2002; 48(1): 27–32. [Google Scholar]
- 17.Blanco-Canqui H, Lal R. Crop residue removal impacts on soil productivity and environmental quality. Crit Rev Plant Sci. 2009; 28(3): 139–163. 10.1080/07352680902776507 [DOI] [Google Scholar]
- 18.Yu CR, Wang XJ, Hu B, Yang CQ, Sui N, Liu RX, et al. Effects of wheat straw incorporation in cotton-wheat double cropping system on nutrient status and growth in cotton. Field Crops Res. 2016; 197: 39–51. 10.1016/j.fcr.2016.08.003 [DOI] [Google Scholar]
- 19.Tian SZ, Ning TY, Wang Y, Li HJ, Zhong WL, Li ZJ. Effect of different tillage methods and straw returning on soil organic carbon content in a winter wheat field. Chin Journal Appl Ecol. 2010; 21(2): 373–378. [PubMed] [Google Scholar]
- 20.Fierer N. Embracing the unknown: disentangling the complexities of the soil microbiome. Nat Rev Microbiol. 2017; 15: 579–590. 10.1038/nrmicro.2017.87 [DOI] [PubMed] [Google Scholar]
- 21.Yin W, Yu AZ, Chai Q, Hu FL, Feng FX, Gan YT. Wheat and maize relay planting with straw covering increases water use efficiency up to 46%. Agron Sustain Dev. 2015; 35(2): 815–825. 10.1007/s13593-015-0286-1 [DOI] [Google Scholar]
- 22.ISS-CAS, Institute of Soil Science, Chinese Academy of Science (2003) Scientific data base. China soil data base, http://www.soil.csdb.cn/ Last Updated: March, 2003. Institute of Soil Science, CAS, China
- 23.IUSS Working Group WRB (2006) World reference base for soil resources 2006. A framework for international classification, correlation and communication, 2nd edn. World Soil Resources Reports No. 103. FAO, Rome
- 24.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010; 7(5): 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magoc T and Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011; 27(21): 2957–2963. 10.1093/bioinformatics/btr507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010; 26(19): 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- 27.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nuclc Acids Research, 2012; 41(1): 590–596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul SF, Madden TL, Schaffer AA, Zhang J-H, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25(17): 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramette A. Multivariate analyses in microbial ecology. FEMS Microbiology. Ecology. 2007; 62(2): 142–160. 10.1111/j.1574-6941.2007.00375.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McArdle BH and Anderson MJ. Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology. 2001; 82(1): 290–297. 10.2307/2680104 [DOI] [Google Scholar]
- 31.Clarke KR. Non-parametric multivariate analyses of changes in community structure. Aust J Ecology. 1993; 18(1): 117–143. 10.1111/j.1442-9993.1993.tb00438.x [DOI] [Google Scholar]
- 32.Warton DI, Wright ST and Wang Y. Distance-based multivariate analyses confound location and dispersion effects. Methods Ecol Evol. 2012; 3(1): 89–101. 10.1111/j.2041-210x.2011.00127.x [DOI] [Google Scholar]
- 33.Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011; 21(9): 1552–1560. 10.1101/gr.120618.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaura E, Keijser BJF, Huse SM, Crielaard W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009; 9(1): 1–12. 10.1186/1471-2180-9-259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White JR, Nagarajan N, Pop M. Statistical Methods for Detecting Differentially Abundant Features in Clinical Metagenomic Samples. PloS Comput Biol. 2009; 5(4): 1–11. 10.1371/journal.pcbi.1000352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011; 12(6), 1–18. 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu E., Yan C., Mei X., He W., Bing S. H., & Ding L., et al. (2010). Long-term effect of chemical fertilizer, straw, and manure on soil chemical and biological properties in northwest china. Geoderma, 158(3–4), 0–180. [Google Scholar]
- 38.Yadvinder S, Bijay S, Timsina J. Crop residue management for nutrient cycling and improving soil productivity in rice-based cropping systems in the tropics. Adv Agron. 2005; 85(4): 269–407. 10.1016/S0065-2113(04)85006-5 [DOI] [Google Scholar]
- 39.Powlson DS, Clendining ML, Coleman K, Whitmore AP. Implications for soil properties of removing cereal straw: results from long-term studies. Agron J. 2011; 103(1): 279–287. 10.2134/agronj2010.0146s [DOI] [Google Scholar]
- 40.Serraj R, Siddique KHM. Conversation agriculture in dry areas. Field Crop Res. 2012; 132: 1–6. Available at http://hdl.handle.net/10919/70012. [Google Scholar]
- 41.Pittelkow CM, Liang X, Linquist BA, van Groenigen KJ, Lee J, Lundy ME, et al. Productivity limits and potentials of the principles of conservation agriculture. Nature. 2015; 517(7534):365–368. 10.1038/nature13809 [DOI] [PubMed] [Google Scholar]
- 42.Yu D, Wen Z, Li X, Song X, Wu H, Yang P. Effects of straw return on bacterial communities in a wheat-maize rotation system in the North China Plain. PLoS ONE. 2018; 13(6): e0198087. 10.1371/journal.pone.0198087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bei SK, Zhang YL, Li TT, Christie P, Li XL, Zhang JL. Response of the soil microbial community to different fertilizer inputs in a wheat-maize rotation on a calcareous soil. Agr, Ecosyst & Environ. 2018; 260:58–69. 10.1016/j.agee.2018.03.014 [DOI] [Google Scholar]
- 44.Xia XY, Zhang PP, He LL, Gao XX, Li WJ, Zhou YY, et al. Effects of tillage managements and maize straw returning on soil microbiome using 16S rDNA sequencing. J Integr Plant Biol. 2019; 61(6): 765–777. 10.1111/jipb.12802 [DOI] [PubMed] [Google Scholar]
- 45.Miyashita NT, Hiroko I, Suliana C, Bilian D, John S, Lucy C. Soil bacterial community structure in five tropical forests in Malaysia and one temperate forest in Japan revealed by pyrosequencing analyses of 16S rRNA gene sequence variation. Genes Genet Syst. 2013; 88(2): 93–103. 10.1266/ggs.88.93 [DOI] [PubMed] [Google Scholar]
- 46.Burns KN, Kluefel DA, Strauss SL, Bokulich NA, Cantu D, Steenwerth KL. Vineyard soil bacteria diversity and composition revealed by 16S rRNA genes: Differentiation by geographic features. Soil Biol Biochem. 2015; 91: 232–247. 10.1016/j.soilbio.2015.09.002 [DOI] [Google Scholar]
- 47.Ligi T, Oopkaup K, Truu M, Preem JK, Nõlvak H, Mitsch WJ, et al. Characterization of bacterial communities in soil and sediment of a created riverine wetland complex using high-throughoutput 16S rRNA amplicon sequencing. Ecol Eng. 2014; 72: 56–66. 10.1016/j.ecoleng.2013.09.007 [DOI] [Google Scholar]
- 48.Rodrigues VD, Torres TT, Ottoboni LM. Bacterial diversity assessment in soil of an active Brazilian copper mine using high-throughput sequencing of 16S rDNA amplicons. Anton van Leeuw. 2014; 106(5): 879–890. 10.1007/s10482-014-0257-6 [DOI] [PubMed] [Google Scholar]
- 49.Song LY, Wang YQ. Investigation of microbial community structure of a shallow lake after one season copper sulfate algaecide treatment. Microbiol Res. 2015; 170: 105–113. 10.1016/j.micres.2014.08.008 [DOI] [PubMed] [Google Scholar]
- 50.Li HX, Cai XX, Gong JY, Xu T, Ding GC, Li J. Long-Term Organic Farming Manipulated Rhizospheric Microbiome and Bacillus Antagonism Against Pepper Blight (Phytophthoracapsici). Front Microbiol. 2019; 10: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chang AR, Li J, Zhang S, Zhan JW, Wei DH, Wang PW, et al. Analysis of bacterial community structure in rhizosphere soil of tobacco based on the metagenomics 16S rDNA sequencing technology. J Agr Sci Tech. 2017; 19(2): 43–50. [Google Scholar]
- 52.Sampaio VS, De Araujo Jean Luiz Simões, Luciana DSR, Baldani Vera Lúcia Divan, Baldani José Ivo. Occurrence and diversity of nitrogen-fixing Sphingomonas bacteria associated with rice plants grown in Brazil, FEMS Microbiol Letters, 2008; 293(1): 11–19. 10.1111/j.1574-6968.2008.01475.x [DOI] [PubMed] [Google Scholar]
- 53.DeBruyn JM, Nixon LT, Fawaz MN, Johnson AM, and Radosevich M. Global biogeography and quantitative seasonal dynamics of Gemmatimonadetes in soil. Appl Environ Microbiol. 2011; 77, 6295–6300. 10.1128/AEM.05005-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, et al. Crossbiome metagenomic analyses of soil microbial communities and their functional attributes. P Nat Acad Sci. 2012; 109(52): 21390–21395. 10.1073/pnas.1215210110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoon DN, Park SJ, Kim SJ, Jeon CO, Chae JC, Rhee SK. Isolation, characterization, and abundance of filamentous members of Caldilineaein activated sludge. J Microbiol. 2010; 48(3): 275–283. 10.1007/s12275-010-9366-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(TIF)
(DOCX)
Data Availability Statement
All the raw data files are available in the NCBI Sequence Read Archive (PRJNA588667).